- 1Department of Internal Medicine I for Hematology With Stem Cell Transplantation, Hemostaseology and Medical Oncology, Ordensklinikum Linz, Linz, Austria
- 2Gastrointestinal Cancer Center, Ordensklinikum Linz, Linz, Austria
- 3Johannes Kepler University Linz, Medical Faculty, Linz, Austria
Anti-EGFR targeting is one of the key strategies in the treatment of metastatic colorectal cancer (mCRC). For almost two decades oncologists have struggled to implement EGFR antibodies in the mCRC continuum of care. Both sidedness and RAS mutational status rank high among the predictive factors for the clinical efficacy of EGFR inhibitors. A prospective phase III trial has recently confirmed that anti-EGFR targeting confers an overall survival benefit only in left sided RAS-wildtype tumors when given in first line. It is a matter of discussion if more clinical benefit can be reached by considering putative primary resistance mechanisms (e.g., HER2, BRAF, PIK3CA, etc.) at this early stage of treatment. The value of this procedure in daily routine clinical utility has not yet been clearly delineated. Re-exposure to EGFR antibodies becomes increasingly crucial in the disease journey of mCRC. Yet re- induction or re-challenge strategies have been problematic as they relied on mathematical models that described the timely decay of EGFR antibody resistant clones. The advent of liquid biopsy and the implementation of more accurate next-generation sequencing (NGS) based high throughput methods allows for tracing of EGFR resistant clones in real time. These displays the spatiotemporal heterogeneity of metastatic disease compared to the former standard radiographic assessment and re-biopsy. These techniques may move EGFR inhibition in mCRC into the area of precision medicine in order to apply EGFR antibodies with the increase or decrease of EGFR resistant clones. This review critically discusses established concepts of tackling the EGFR pathway in mCRC and provides insight into the growing field of liquid biopsy guided personalized approaches of EGFR inhibition in mCRC.
1 Introduction
Colorectal cancer (CRC) remains a significant issue in global health. According to a recent analysis comparing the global cancer burden in the last decade, CRC is one of the 5 main causes of cancer-related disability-adjusted life years and is ranked 2nd after lung cancer, overtaking stomach cancer in 2019 (1). While rising cases are being reported, especially among the younger population, CRC is still a disease of the elderly. About 70% of cases occur between 50 and 80 years of age, with a mean onset of disease at the age of 72 for men and 75 for women (2). According to the Surveillance, Epidemiology, and End Results (SEER) database, 20% of patients present with primary metastatic CRC (mCRC) and 40% with relapse after previously curative intended treatment. The long-term outcome of mCRC is still poor, with a 5-year survival rate below 20% (1, 3). The treatment repertoire is stratified according to predictive biological markers of the respective tumors to leverage individualized treatment concepts. This treatment armamentarium has recently become more diverse and increased in number. These include: Monoclonal antibodies targeting the epithelial growth factor receptor (EGFR), such as Cetuximab or Panitumumab; HER2-directed agents such as Trastuzumab deruxtecan; antiangiogenic agents targeting vascular endothelial growth factor (VEGF) signalling, such as Bevacizumab, Aflibercept and Ramucirumab; as well as the broad spectrum kinase inhibitor Regorafenib. For microsatellite instable (MSI) cases, checkpoint inhibitors have evolved as a valid choice. These new treatment options have improved overall survival (OS) of patients with mCRC from approximately 1 year in the era of single agent 5-fluorouracil (5-FU) to more than 3 years with currently available options (2). This remarkable increase in survival appears to be largely based on the outcome data of left-sided mCRC (4). Left-sided mCRC has a more favourable predictive biological profile with a lower incidence of RAS mutations (RAS-MT) (5). Both sidedness and RAS mutational status rank high among the predictive factors for the clinical efficacy of EGFR inhibitors. In-depth knowledge of the appropriate integration of EGFR inhibitors in the continuum of care is required to gain maximum survival time for mCRC patients. This review will critically assess established concepts of EGFR targeting in mCRC in light of new diagnostic tools in order to shape the application of EGFR inhibitors in future clinical practice.
1.1 The EGF-Receptor
The discovery of EGFR as an oncogene is closely related to the history of our modern understanding of cancer pathogenesis (6–8). Almost half a century ago an EGFR variant that lacked sequences in the N-terminal ectodomain was found to promote aberrant cellular signalling in the absence of a binding ligand, thus transforming cells into a malignant phenotype. Some years before the findings of Cohen et al. showed that EGF ligand-dependent signalling via EGFR stimulated cellular processes like growth, proliferation and deemed worthy of Nobel prize honours (9). In the era when cancer was seen merely as a misguided signalling network, tackling aberrant EGFR signalling became one of the first goals of targeted cancer research (6, 7, 10, 11).
Decades of research later, EGFR is known to be embedded in a family of cell membrane-tagged receptor tyrosine kinases, including HER2/c-neu (ERBB2), HER3 (ERBB3) and HER4 (ERBB4). They share a common structure: a single amino acid chain protein forms an extracellular ligand binding domain, a transmembrane domain for homodimerization or heterodimerization and a tyrosine kinase intracellular portion. Each domain can initiate and drive malignant signalling; the ectodomain through binding ligands, the transmembrane domain allows ligand-independent signalling through dimerization, and amino acid modifications of the intracellular domain enables signalling regardless of ligand binding (10–12). Immunohistochemical (IHC) studies confirmed the expression of members of the ERBB family in various types of tumors. The mode of action depended on the tumor type and the isotype of the ERBB receptor (7). In breast cancer homo/-heterodimerization of ERBB2 and ERBB3 is the main route, while lung cancer is linked to tyrosine receptor mutations in the intracellular domain of ERBB1. CRC lacks activating mutations in ERBB1 and it was initially assumed that only overexpression of the physiological normal wild-type ERBB1 conferred tumorigenic activity (13). Blocking EGFR-mediated signalling in CRC through the development of antibodies competing for the binding site of physiological activators seemed a promising approach (8). Gill and Goldstein generated a mouse chimeric monoclonal antibody (IgG1) known as Cetuximab, which binds to the domain III of the extracellular domain with high affinity (14, 15). It renders the EGFR receptor in an inactive state mitigating downstream signalling pathways. Furthermore, it promotes receptor internalization, subsequent degradation and finally receptor down-regulation (15). Cetuximab is immunogenic in about 5% of patients. Therefore, a full human antibody (IgG2) against EGFR has been developed by immunization of transgenic mice (XenoMouse) known as Panitumumab. The mode of action is similar to Cetuximab. Differences in the IgG subclass that favour antibody-dependent cellular cytotoxicity and complement mediated cytotoxicity for IgG1 subtype antibodies appear negligible (15, 16).
Clinical evidence for the efficacy of EGFR inhibitors in the treatment of mCRC arose initially from phase II trials in later lines. Notably, mCRC included in these trials had to be EGFR IHC positive. Saltz et al. reported a clinical benefit rate in approximately half of the patients treated with Cetuximab as monotherapy (17). Together with Irinotecan, the overall response rate (ORR) doubled (23% vs. 11%) compared to monotherapy, as shown in the famous BOND trial by Cunningham et al. that finally prompted clinical approval of the drug by the FDA (18). Similar results for Panitumumab were obtained in 2006 by Giusti et al. that also led to FDA approval of Panitumumab in EGFR-expressing advanced mCRC after failure of first line therapy (19).
Interestingly, the BOND trial failed to confirm EGFR expression as a predictive marker for Cetuximab. Moreover, Cetuximab seemed to function even when EGFR was absent as measured by IHC and having sensitivity of IHC assays and probable tumor heterogeneity in mind. Though EGFR expression/amplification is not considered as a prerequisite for mCRC to be suitable for EGFR blockade, EGFR amplification (only 1% of mCRC) can booster EGFR inhibitors to exceptional outcomes in patients with RAS/BRAF-wildtype (WT) mCRC (20). EGFR amplification, albeit of no practical clinical significance, is the only positive predictive marker indicating exaggerated response to EGFR blockade thus far. The overall prognostic relevance of EGFR overexpression as an independent variable remains contradictory (21, 22).
2 Relevant clinical resistance mechanisms
2.1 EGFR ectodomain mutations
EGFR ectodomain (EGFR-ECD) mutations (exon 12) that prevent antibody binding rarely occur as a primary resistance mechanism. They typically evolve as a secondary resistance mechanism under the evolutionary pressure of sustained EGFR blockade (23). EGFR-ECD mutations are responsible for up to 25% of resistance mutations after failure of EGFR inhibitors (24). Usually, these patients experience deeper and longer responses to EGFR inhibitors in contrast to RAS related resistance mechanisms (23). Mutations in the extracellular domain S492, G465, S464, V441 are most prevalent, responsible for approximately 14% of all secondary resistance causes (23). Special attention should be paid to the variant S492 that has been found in 16% of patients after Cetuximab exposure and only 1% after Panitumumab treatment (25). A re-challenge approach with Panitumumab to overcome Cetuximab resistance in these cases would be rational from a molecular perspective, but prospective studies are still lacking to evaluate the efficacy of Panitumumab in the EGFR p.S492R mutant population (26).
Due to a seminal paper in 2006 the focus of main resistance mechanisms shifted to EGFR downstream pathway components (27).
2.2 RAS mutation
The MAPK pathway is one of the major downstream effectors of EGFR-based signalling in addition to the Pi3K/AKT/mTOR and PLCγ. The MAPK pathway consists of consecutive activated molecules named RAS/RAF/MEK/ERK that drive cell proliferation and malignant transformation (7, 10). In the 1980s RAS was one of the first cloned (proto)oncogenes characterized by Weinberg (28). During normal physiological states it is tethered to the plasma membrane through posttranslational modification mediated by farnesyltransferase (FTase) and acts a GTPase shifting from off to on states and back again (binary switch). This process is mainly regulated by extrinsic guanine nucleotide exchange factors (GEF) such as son of sevenless homologue 1 (SOS1) for GDP-to-GTP transition, and GTPase activating proteins (GAP) such as neurofibromin for GTP hydrolysis. A prominent RAS intrinsic GTPase activity is most prominently described for KRAS G12C and gets therapeutically exploited.
Activating mutations in the RAS family - consisting mostly of the isoforms KRAS, NRAS, and HRAS - are found in more than 20% of human cancers. In mCRC about 40% of the cases harbor activating mutations in KRAS, predominantly in exon 2 codon 12 (70-80%). Mutations in NRAS account for about 5%, particularly in exon 3 codon 61 (60%). HRAS mutations are rare in mCRC, but more dominant in head and neck cancer and urinary tract cancer. Secondary acquired RAS mutations after EGFR blockade are more often present as atypical mutations, e.g., KRAS codon 61 or codon 146 (29). The prognostic value of the RAS-MT in mCRC remains controversial, while it evolved as the crucial predictive biomarker in the treatment of mCRC (30).
In 2006 Lievre et al. published data from only 30 patients with mCRC treated with Cetuximab; they screened for mutations in KRAS, BRAF, and PIK3CA and their correlation with the response to EGFR inhibition and interestingly, none of the tumors harboring a KRAS mutation responded (27). These results were confirmed by other smaller trials, a retrospective analysis of phase III trials a metanalysis according to RAS status, and a smaller last line trial with panitumumab (19). Altogether, these results changed the treatment landscape of mCRC profoundly (31). In 2009 the FDA changed the label for Cetuximab and Panitumumab and then mandated KRAS exon 2 testing as a prerequisite. In 2009 the PRIME trial (Panitumumab randomized trial in combination with chemotherapy for metastatic CRC to determine efficacy) was first to test KRAS exon 2 mutational status prospectively as a predictive biomarker for mCRC (30). Panitumumab prolonged PFS by 1,4 months and an OS benefit of 4,4 months was only observed in KRAS exon 2-WT patients. PRIME further evaluated the possible negative predictive value of additional isoforms and exons of RAS and clearly showed that any activating mutation in KRAS or NRAS predicted resistance to Panitumumab (32). Similar data were acquired from a retrospective analysis of the CHRYSTAL or OPUS trial with Cetuximab as an EGFR antibody (33). According to recent ASCO and ESMO guidelines, expanded RAS biomarker testing (all-RAS) is standard of care. RAS analysis should comprise at least KRAS exons 2,3,4 and NRAS exons 2,3,4. All-RAS testing enhanced ORR response rates to EGFR antibodies from 20% to over 40%.
The advent of deep sequencing techniques allowed the detection of small RAS-MT subclones in biopsied tumor samples. The size of these subclones is commonly expressed in percentage as the number of sequence reads of a specific DNA variant divided by the overall coverage at that locus (variant allele frequency; VAF%). It can be explained as a surrogate parameter of the proportion of DNA molecules in the tumor specimen harboring this specific variant (e.g., RAS mutation). It was retrospectively observed that tumors carrying small RAS-MT subclones still benefit from EGFR blockade. The optimal cut-off value for RAS-MT tumors responding to EGFR inhibitors was determined by the ULTRA study. Tumors carrying RAS mutations below 5% VAF detected by deep sequencing (ddPCR) are still sensitive to EGFR blockade (34).
2.2.1 KRAS G12C
The prevalence of KRAS p.G12C mutation in mCRC is about 3-4% according to different cohorts.
Among KRAS mutated mCRC, KRAS G12C is associated with shorter OS compared to KRAS non-p.G12C tumors. Reasons are unclear, but might be attributed to differences in metabolism and resistance mechanisms (35).In addition to its prognostic impact, KRAS G12C is characterized by its unique biophysical properties, which render KRAS G12C as the first targetable member of the so far undruggable RAS family. First, an outstanding intrinsic GTPase activity allows the covalent binding of small molecule inhibitors (e.g., Sotorasib or Adagrasib) during the inactive GDP state of KRAS (G12C), thus arresting the KRAS GDP and abrogating downstream signaling. Second, during the GDP state a recently discovered SII pocket is transiently formed, thus leveraging covalent binding of small molecule inhibitors to the cysteine residue of KRAS G12C (36–38).
In contrast to the clinical efficacy of KRAS G12C inhibition in lung cancer, the benefit in mCRC is less pronounced. CodeBreaK100 revealed an ORR of only 9,7% and a mPFS of 4 months when Sotorasib was used as monotherapy. Adagrasib in the KRYSTAL-1 trial showed slightly better but similar results. Distinct signaling networks in lung and CRC might partly explain differences in clinical efficacy. Similar to BRAF V600E inhibition, targeting KRAS G12C and consecutive downregulation of MPAK pathway stimulates EGFR signaling via a negative feedback loop mechanism (39–41). This attenuates the efficacy of Sotorasib or Adagrasib and activates bypass mechanisms. This phenomenon is almost only observed in mCRC, reflecting its dominant dependency on EGFR signaling. To overcome these limitations a strategy resembling targeting BRAF V600E in mCRC has been tested. In the KRYSTAL-1 trial Adagrasib combined with Cetuximab more than doubled response rates, whereas mPFS was only slightly improved by one month. CodeBreaK101 investigating Sotorasib+Panitumumab presented with similar clinical efficacy. The role of these treatment combinations in daily routine practice has to be finetuned in further trials (42, 43).
2.3 BRAF mutation
BRAF lies downstream of RAS in the MAPK pathway. Usually, BRAF alterations are classified according to their mode of activation. Whereas Class III is like the WT counterpart dependent on upstream RAS activation, class II and III work on RAS independently. Class I acts as active monomers; Class II is still dimer dependent. BRAF-V600E is the most important representative of Class I and occurs in 8–10% of CRC - ranked 5th across all cancer subtypes after hairy cell leukemia, papillary thyroid cancer, melanoma and Langerhans cell histiocytosis. It confers an extremely hostile phenotype with poor prognosis in mCRC, resulting in a nearly two-fold increase in mortality compared to wild-type BRAF mCRC (median PFS in first-line about 6 months and median OS about 13 months) (44).
BRAF-V600E is associated with right-sided tumors (up to 20%), high grade histology, higher patient age, female sex, MLH1 hypermethylation, serrated adenoma pathway and predominantly peritoneal metastatic spread; notably, BRAF-V600E is mutually exclusive to RAS mutations (44).
Class II/III alterations occur in about 2% of mCRC and are often associated, contrary to BRAF-V600E, with younger age of onset, male sex, left-sided primaries and a better prognosis.
Concurrent BRAF-V600E and MLH1 hypermethylation confer a MSI phenotype that profits from checkpoint inhibition (45, 46). BRAF-V600E mutations in MSS mCRC are one of the most negative prognostic markers in mCRC. This is well established from phase III trials like CRYSTAL, OPUS, COIN and TRIBE. An assumed negative predictive value for the efficacy of EGFR antibody-based therapy still remains controversial. Two meta-analyses of RCTs conducted by Rowland et al. and Pietrantonio et al. attempted to address this topic. Rowland et al. found no statistically significant difference in OS and PFS between RAS-WT/BRAF-MT and RAS-WT/BRAF-WT tumors, abrogating the negative predictive role of BRAF-V600E mutation for the use of anti-EGFR monoclonal antibodies in RAS-WT mCRC (47). However, Pietrantonio et al. demonstrated a lack of benefit with anti-EGFR treatment in patients with BRAF-V600E mutated CRCs (48). This finding was further strengthened by Stinzing et al. in the prospective FIRE4.5 trial (49). Based on its RAS-independent mode of action class III, BRAF mutations seem to retain sensitivity to EGFR-targeting (23, 50).
In the current European daily clinical practice these controversial results are discussed by stating that only 7,5% of patients with the BRAF-V600E mutation receive an anti-EGFR-based chemo-doublette in the first-line (51). EGFR blockade remains of significant importance in second-line treatment. According to the BEACON trial, additional EGFR inhibition is paramount, as BRAF-V600E altered MAPK pathway signalling is not sufficiently abrogated by BRAF and MEK inhibitors. This pathophysiological variation is unique to mCRC compared to other entities, e.g., melanoma. In mCRC inhibition of MAPK pathway leads via adaptive feedback loops to reactivation of the EGFR and exaggerated EGFR based signaling, thereby overcoming previous inhibitory attempts (52).
2.4 Human epidermal growth factor 2
The search for optimized biomarkers predicting EGFR efficacy was first uncovered in 2011 in preclinical data and later in a retrospective analysis of HER2 amplification/overexpression as a potential negative predictive marker for Cetuximab (53–55). It was shown that Homo- or heterodimerization of HER2 leads to activation of downstream signalling networks largely shared by EGFR signalling, thereby bypassing EGFR-mediated growth inhibition.
HER2 was first established in gastric cancer in the ToGA trial in 2010, which overexpresses HER2 in up to 20% of cases as a clinically valuable target (56). In mCRC, alteration in HER2 accounts for only up to 5% of cases and appears to be enriched in RAS-WT CRC, accounting for up to 40% in cases showing resistance to EGFR base antibody therapies (57, 58).
According to results from Sartor-Bianchi et al. and Raghav et al., HER2 amplification implicates worse clinical outcomes in EGFR-based first line trials in terms of ORR, PFS or OS (59, 60). However, upfront testing of KRAS-WT CRC for HER2 amplification or mutation is currently not recommended in the ESMO or ASCO guidelines. Targeting HER2 in mCRC currently finds its place in second or later lines (58, 61). First line trials are currently recruiting. The therapeutic landscape will presumably be revolutionized with the advent of novel tyrosine kinase inhibitors and HER2- targeting antibody drug conjugates (62).
2.5 Sidedness
The impact of primary tumor location on OS in mCRC dates back to the 1990s and was observed in several subsequent trials afterward (63). However, no clinical consequence was drawn from this observation. In 2016 a retrospective analysis of the CALGB/SWOG 80405 trial at ASCO 2016 and a later metanalysis including CRYSTAL, PRIME, CALGB/SWOG 80405, PEAK, and FIRE-3 changed the way we treat mCRC comparable to the impact of RAS mutation (64, 65).
CALGB/SWOG 80405 showed an OS of 33,3 months for left vs. 19,4 months for right-sided tumors. Furthermore, although the primary endpoint of the trial comparing Bevacizumab versus Cetuximab based doublettes in RAS-WT tumors was negative, dividing the trial population according to right vs. left disease deciphered the impact of EGFR inhibition on mCRC. In left-sided tumor RAS-WT Cetuximab almost tripled OS (39,3 vs 13,6 months) compared to right-sided RAS-WT tumors. Additionally, left-sided tumor benefited significantly from the addition of Cetuximab compared to Bevacizumab in terms of OS (39,3 vs 32,6 month) and PFS (64).
Two metanalyses enforced findings from CALGB/SWOG 80405 and EGFR inhibition was found to be superior to chemo-doublette alone (pooled analysis of CRYSTAL and PRIME) as well as compared to chemo-doublette+Bevacizumab pooled analysis of PEAK, FIRE-3 and CALGB/SWOG 80405) in terms of PFS and OS, but only in left-sided tumors (65–67). Therefore, left-sided primary tumor localization evolved as a positive predictive biomarker for efficacy of EGFR inhibitor therapy in RAS-WT mCRC.
At the ASCO meeting in 2022 the PARADIGM trial was the first trial to prospectively test the superiority of Panitumumab vs. Bevacizumab in combination with FOLFOLX6 in all-RAS-WT and left-sided primary tumors. OS increased from 34,3 months in the Bevacizumab arm to an unpreceded 37,9 months in the Panitumumab arm, whereas PFS remained comparable. Furthermore, ORR and R0 resection rates were increased by Panitumumab (68).The superior OS despite similar PFS rate of EGFR blockade compared to VEGFR inhibition in left-sided RAS WT tumors might be explained by deeper and earlier responses, expressed as early tumor shrinkage (ETS) and depth of response (DpR), according to a retrospective analysis and additional mathematical modeling (69–71). The DEEPER trial (JACCRO CC-13) with DpR as the primary endpoint confirmed the superiority of Cetuximab over Bevacizumab in terms of early tumor dynamics (72).
The differential sensitivity of right vs. left sided RAS-WT mCRC to EGFR inhibition is based on a diverse biological background. Right vs. left colon can be considered almost as two organs differing in numerous ways, including embryological development, bacterial colonization, gene expression levels during cancer development. Right sided mCRC display more often a MSI phenotype and resistance mutations of RAS, BRAF, and PIK3CA are more prevalent. Left-sided mCRC is characterized by chromosomal aberrations, HER2 and EGFR overexpression and a consensus molecular subtype (CMS) 2 and CMS 4 gene-expression profile (5). These differences may only be partly linked to the altered sensitivity to EGFR inhibition. Sidedness remains a stand-alone predictive factor. This was most recently confirmed in ultraselected mCRC cases, where left sided mCRC still conferred greater clinical benefit to EGFR blockade despite excluding mCRC with rare or ultra-rare resistance alterations in the EGFR pathway (73).
2.6 Emerging resistance mechanisms
Quadruple-WT CRC, hyperselection, and ultraselection are terms that describe molecular enrichment strategies to improve the efficacy outcome of EGFR inhibition. In 2011 a large retrospective analysis by De Roock and a later prospective CAPRi-GOIM trial coined the term quadruple WT CRC, indicating wild-type KRAS/NRAS/BRAF/PIK3CA tumors. In these trials, the ORR (64.4%) and median progression free survival (mPFS; 11.3 months) compared with patients exhibiting a mutation in one of these genes (ORR 47.4% and mPFS 7.7 months) was markedly improved (74, 75). Hyperselection means further refinement provided by the PRESSING (PRimary rESiStance IN RAS and BRAF wild-type mCRC patients treated with anti-EGFR monoclonal antibodies) panel including HER2 amplification/activating mutations, MET amplification, NTRK, ROS1, ALK, RET rearrangements, PIK3CA exon 20 mutations, PTEN inactivating mutations, AKT1 mutations. PRESSING alterations were more prevalent in right-sided tumors and more often associated with MSI status. Efficacy was initially demonstrated in a small case control trial and was further investigated in an exploratory analysis of the VALENTINO trial (76, 77). PRESSING positive tumors had significantly lower ORR (59% vs. 75%), PFS (7.7 vs. 12.1 months) and OS (68.1 vs. 48.1% 2 year OS rate). The PRESSING panel served as a predictive marker only in left-sided tumors, while right-sided tumors could not be further differentiated.
A further level of granularity was introduced by the PRESSING2 panel, which in addition to the PRESSING panel included rare and ultrarare potential resistance alterations (i.e., NF1 mutations/loss, ARAF/KRAS amplification, MAP2K1/MAP2K2 and MAP2K4 mutations, IGF1R amplification, ERBB3 amplification/mutations, FGFR2 amplification, AKT1/2 amplification, MSI status and POLE exonuclease domain). This enrichment strategy is commonly referred as ultraselection. About 50% of the RAS/BRAF-WT population harbor PRESSING2 mutations. The ORR (79%) and PFS rates (13 months) were comparable to the results of the PRESSING panel in the VALENTINO trial. The mOS rate of 51.2 months in left-sided PRESSSING2 negative tumors is unprecedented so far, extending mOS compared to the recently reported PARADIGM trials by 13.3 months (73). The PRESSING2 panel predicted outcome predominantly in left-sided tumors, but in contrast to the earlier PRESSING panel also right-sided PRESSING2 negative tumors gained benefit from EGFR blockade. It is therefore tempting to speculate that a defined ultra-selected subset of right-sided tumor might benefit from EGFR blockade - a finding with potential practical clinical implications (73).
3 Treatment algorithms in first-line treatment
Choice of first-line therapy is key in optimizing long-term outcome in mCRC. Achieving deep responses and long-term remissions in first-line is the prerequisite for maintenance concepts, treatment breaks and oligometastatic concepts; in short, it is the basis for the continuum of care concept (78).
For optimal induction therapy, four parameters are crucial to know in every routine clinical practice: RAS mutation status, MSI status, BRAF-V600E status and primary tumor localization (79, 80).
The largest benefit from EGFR inhibition is derived for left-sided RAS/BRAF-WT/MSS tumors. A retrospective analysis of three first-line trials observed an OS benefit of Cetuximab or Panitumumab over Bevacizumab, which was recently prospectively confirmed by the PARADIGM trial. Notably, HR for OS in the major trials (CALGB, PEAK, FIRE-3 and PARADIGM) was consistently comparable. PFS of second line was also beneficial after EGFR-based first-line as shown in FIRE-3. The STRATEGIC trial prospectively compared EGFR followed by Bevacizumab vs. Bevacizumab post progression and a numerical benefit in OS was observed (81).. The fact that the PFS of first-line remained the same between EGFR inhibition with either Panitumumab or Cetuximab vs. Bevacizumab, but OS showed a huge difference, may be largely explained by ETS in EGFR-based therapies and lack of EGFR response after VEGFR (69, 71, 82, 83).
The value of chemo-intensification in left-sided RAS-WT/BRAF-WT tumors was answered by the TRIPLETE trial. No benefit was reported when the chemo backbone was complemented by a third agent. Therefore, the mainstay for treatment of left-sided RAS-WT/BRAF-WT tumors l is still a chemo-doublette+EGFR inhibitor (84).
The value of EGFR inhibitors in right-sided RAS/BRAF-WT tumors is still controversial. Both the FIRE-3 and PEAK trials showed detrimental effects on OS compared to Bevacizumab, while a meta-analysis accounted for favourable ORR for EGFR. Therefore, for right-sided tumors where an oligometastatic concept seems feasible, EGFR inhibitors might still be of value. Optimal treatment of BRAF-MT/RAS-WT mCRC remains challenging. According to the FIRE 4.5 trial Cetuximab has a negative impact on clinical outcome data compared to Bevacizumab (85).
In the elderly frail population de-escalation strategies sparing a chemotherapeutic component are highly recommended. The PANDA trial clearly demonstrated that Oxaliplatin can be left out without losing efficacy (86). Furthermore, it compared favourably to the long-existing standard Capecitabin/Bevacizumab in terms of ORR, while preserving PFS (87).
In second and later line settings EGFR antibodies have failed to demonstrate any OS benefit (88–90). In particular, the sequence VEGF first-line→EGFR second-line in RAS-WT patients should be avoided (91, 92).
4 Maintenance
Typically, chemo-doublette+EGFR inhibition serves as an induction treatment for 4-6 months, as ETS and DpR in first-line treatment are key to overall OS benefit. Post-induction approaches should aim to consolidate the efficacy of induction treatment, while minimizing side-effects and preserving quality of life (QoL).
Three different treatment options exist. First, induction therapy can be continued until progression. This is only recommended for Irinotecan-based chemo-backbone by ESMO, as prolonged Oxaliplatin has debilitating effects on QoL parameters. In part, the same may hold for Cetuximab or Panitumumab. Second is a combination of drug holiday and re-exposure to chemo-doublette+antibody after progression. Third is de-escalation including withdrawal of one or two components of the induction regime and escalation upon progression; this is optimal for active maintenance approaches.
Evidence that de-intensifying after successful induction is feasible is derived from several trials. MACCRO-2 trial (Cetuximab vs. FOLFOX+Cetuximab), NORDIC VII (Cetuximab vs. FLOX+Cetuximab), SAPPHIRE (5-FU+Panitumumab vs. FOLFOX Panitumumab) and ERMES (Cetuximab vs. FOLFIRI+Cetuximab) preserved efficacy, while reducing incidence of peripheral neuropathy or acneiform rash (93–96). Maintenance of monotherapy versus drug holidays was compared in COIN-B, PRODIGE 28-time UNICANCER (Cetuximab vs. observation) and FOCUS4-N (Capecitabin vs. observation) (97–99). PFS was improved in the active maintenance arm, whereas OS remained unaffected. Prospective phase II trials (VALENTINO, PANAMA) favoured the combination of Capecitabin/5-FU+EGFR inhibition (Panitumumab) over the respective single agent (Panitumumab or Capecitabin/5-FU) in terms of prolongation of PFS without compromising QoL parameters (100). Maintenance with Bevacizumab +/- 5-FU in RAS-WT tumor was investigated in MACBETH (Cetuximab vs. Bevacizumab) and in a retrospective analysis of the PEAK trial (101, 102). In PEAK the median PFS and median OS from the discontinuation of Oxaliplatin were 9,7 vs. 7 months and 33,5 vs 23,3 months in the 5-FU- Panitumumab arm compared to the 5-FU Bevacizumab arm (102).
To summarize these different approaches and often conflicting results, a recent metanalysis was performed and revealed a PFS and even OS benefit for continuation of an EGFR-based doublette or active maintenance with EGFR + 5+FU over 5-FU or EGFR inhibitor monotherapy or observation (103). These findings were confirmed by a further meta-analysis of a larger real-world cohort and individual patient data pooled observations from the PANAMA and VALENTINO trial (104–107).
Predictive markers for the benefit of maintenance concepts are scarce. It is tempting to speculate that patients with SD disease compared to responding tumors derive the greatest benefit from post-induction treatments concepts according to FOUS4-N and PANAMA data. Biologically, tumors with PR and CR after 4-6 months of induction almost always experienced a maximal tumor response; presumably by eradicating the EGFR-sensible clonal population (82, 108, 109). Tumors with SD might already confer partial resistance mechanism requiring prolonged treatment or new concepts in the future (83). It is likely that only liquid biopsy will uncover forthcoming resistance mechanism as dynamic biomarkers for maintenance treatment stratification. Novel maintenance approaches in the MODUL or FOCUS-4 trials already incorporated more sophisticated stratified concepts recognizing the molecular portrait of the tumor at the end of induction phase (e.g., HER2+EGFR blockade or BRAF inhibitor+Capecitabin+EGFR inhibitor) (110, 111).
The idea of maintenance mandates re-exposure to a full first line induction scheme after progression. Only two trials prospectively evaluated this strategy. First, the phase 2 COIN-B study randomized patients with KRAS exon 2- wild-type mCRC with to receive FOLFOX–Cetuximab for 12 weeks, followed by Cetuximab maintenance vs. observation and reintroduction of FOLFOX–Cetuximab at progressive disease. There was no difference observed among the maintenance and intermittent strategies in 10-month failure-free survival (52% vs. 50%, respectively), even if a trend towards a better post-induction PFS (5.8 vs. 3.1 months, respectively) and OS (22.2 vs. 16.8 months, respectively) was noted in favor of the maintenance treatment (88).
Second, the prospective randomized phase 2 PANAMA trial compared 5-FU/LV+Panitumumab vs. 5-FU/LV alone as maintenance strategies in RAS-WT mCRC. The PFS of maintenance therapy was significantly improved with 5-FU/LV+Panitumumab (8.8 vs. 5.7 months), with a trend towards better OS (28.7 vs. 25.7 months). It is remarkable that time to failure of strategy (TFS) was only prolonged by 18 days (112).
The value of maintenance therapy after EGFR first line was recently further questioned by the IMPROVE trial at the annual ASCO meeting in 2022. It compared a continuous vs an intermittent treatment strategy with a chemo-doublette combined with EGFR inhibition by Cetuximab and suggest a similar OS in both arms (113).
These results argue for a strategy that includes drug holidays. However, there are several issues regarding trial end points that have to be re-evaluated. Nevertheless, the concept of intermittent or continuous treatment in a biologically favourable population is highly interesting.
The weighting of potentially improving OS and/or other clinical outcome markers against the accumulation of side-effects which reduce QoL has led to intensive discussion of the applicability of maintenance strategies in clinical practice. These issues should be included in new trials.
5 Liquid biopsy
Evaluation of tumor dynamics in metastatic disease still commonly relies on regular CT scans every 8-12 weeks. In routine clinical practice a differential outcome of the observed lesions can often be documented, drawing a heterogeneous portrait concerning sensitivity to a certain systemic therapy. Therefore, more accurate modes of response assessment have been eagerly awaited that take into account the spatial and temporal heterogeneity of metastatic tumor burden. Liquid biopsy has the potential to overcome these aforementioned limitations (114).
Liquid biopsy summarizes different non-invasive techniques for detection or monitoring of cancer. Circulating tumor cells (CTCs), tumor derived exosomes and cell free DNA (cfDNA) fragments can be measured in various body excretions and in depth biological information that impacts prognosis and further therapeutic choices can be drawn from e.g., a simple blood sample. CTCs and exosomes may provide a more extended image of the tumor- besides DNA based genomic information, including exosomal microRNA’s as biomarkers for prognosis and drug sensitivity prediction or CTCs for xenografting and in vivo drug testing (115, 116). Unfortunately, they miss reasonable sensitivity for e.g., tumor genomic alterations in comparison to cfDNA and are therefore not ready for broader clinical application (116–118).
In blood plasma cfDNA consists typically of 140-170bp in length originating mostly from leukocytes. As early as 1949 it was shown that patients with tumors derive a higher plasma cfDNA concentration (119, 120). The portion of cfDNA derived from cancer is called ctDNA and is depicted as variant allele fractions (VAF) typically ranging from <0,1 to 10% or higher.
In mCRC, evidence was first drawn by the detection of cfDNA in blood or stool probes more than 30 years ago (121). Today, ctDNA is on the edge of emerging as a viable biomarker in daily routine practice. New innovations in molecular biology techniques pushed the way forward. In 1999 the group of Bert Vogelstein set a milestone with the invention of digital-PCR (122). It allowed accurate qualitative and quantitative measuring of mutations against background noise aiming at 0,01% VAF or even lower. Years later Vogelstein et al. also developed the first high throughput digital-PCR method called BEAMing (beads, emulsions, amplification and magnetics), which allowed the routine application in research questions (123). Nevertheless, despite providing a sufficient technical limit of detection (LOD), digital PCR platforms are hampered by the restricted number of mutations per assay that can be analyzed. Next generation sequencing (NGS) of ctDNA can overcome these limitations by allowing detection of an infinite number of alterations including ones unknown so far. By establishing molecular barcoding (MB) techniques, LODs that may equal or even surpass that of digital PCR seem feasible (124–127). MB technologies combined with hybridize-capture-based methods (SureSelect XT HS and HaloPlex (Agilent) or amplicon-based methods (QIAseq Targeted Panel (Qiagen), IonAmpliSeq HD (Thermo Fisher Scientific), Signatera (Natera) are already commercially available. In CRC detection of minimal residual disease (MRD) after surgery might become one the first broad clinical applications of MB-NGS techniques (Signatera).
In metastatic disease, monitoring the course of the disease by following the tides of various mutations harboring subclones has gained increasing attention. In 2008 the Vogelstein group was again the first to assess tumor dynamics in mCRC by measuring serial ctDNA (APC, TP53, KRAS mutations) compared to plasma biomarkers and radiographic evaluation (123). An early drop of ctDNA was later prospectively validated as an indicator of tumor response overtaking conventional staging modalities (128).
The ESMO precision medicine working group recently recommended for the use of ctDNA in daily routine practice in chemotherapy-naive mCRC KRAS/NRAS/BRAFV600E/MSI testing by ctDNA if tissue testing is not feasible or urgent therapeutic decision making is necessary (129). During a metastatic disease course the ESMO precision medicine working group suggests ctDNA testing for KRAS/NRAS/BRAF/EGFR-ECD/HER2 amplification. In contrast to tissue-based biopsy sampling of single lesions, ctDNA- based assays enables real-time portraits of tumor heterogeneity.
Concordance between RAS status in matched ctDNA and tumor tissue biopsy samples reaches over 90% according to various retrospective analyse and three larger prospectively conducted trials (130–137). For other mutations, concordance is quite similarly predictable, e.g., BRAF-V600E up to 100%, EGFR-ECD 99% (138). Reasons for impaired sensitivity of ctDNA plasma testing might depend on the specific ctDNA assay used, whereby OncoBEAM yielded the best results. On the other hand, ctDNA shedding seems to associate with specific tumor features. Low tumor burden, peritoneal and lung metastases and also mucinous histology hamper ctDNA release in contrast to high tumor burden and liver metastases (137).
Misale in 2012 pioneered the detection of acquired resistance to anti-EGFR therapy in mCRC. ctDNA of KRAS mutation or amplification were traced by BEAMing as early as 10 months before radiographic progression (139).
Resistance to EGFR inhibitor therapy is commonly associated with an alteration in the MAPK pathway. Emerging RAS mutated clones and EGFR-ECD mutations such as S492, G456, S464, V441 rank top among others like MET, RAS, and HER2 amplification. Other genetic alterations that develop selectively under EGFR blockade involve LRP1B, ZNF217, MAP2K1, PIK3CG, ATM, ATR, and BRCA1 (140). Furthermore, resistance mutations are by far not mutually exclusive. Rather, tumors with KRAS or EGFR mutations harbor >1 additional mutation in over 50%. Most data describing EGFR resistance mechanism are collected from EGFR application in later lines. Parseghian at ASCO 2021 demonstrated that results might be different in a strictly first line population conferring a rather low prevalence of so far established resistance mutations (141).
Resistance conferring clones are traced in plasma probes with a lead time of several months before clinical progress is visible on CT scans. Monitoring evolving clones occurring under EGFR inhibitor pressure allows earlier termination of ineffective therapies, enables strategies of continuation of EGFR blockade beyond progression and informs about potential targeted approaches, e.g., MET amplification- Crizotinib, HER2 amplification- T-DXd, KRAS-G12C mutation- Sotorasib, Adagrasib (142, 143).
Treatment with EGFR antibodies beyond progression commonly lacks valuable clinical implementation. Strategies to enrich All-RAS WT/BRAF- WT before second line EGFR blockade improved results formally but were not clinically meaningful. Resistance alteration besides RAS status, for example due to MET amplification might be relevant (144–148).
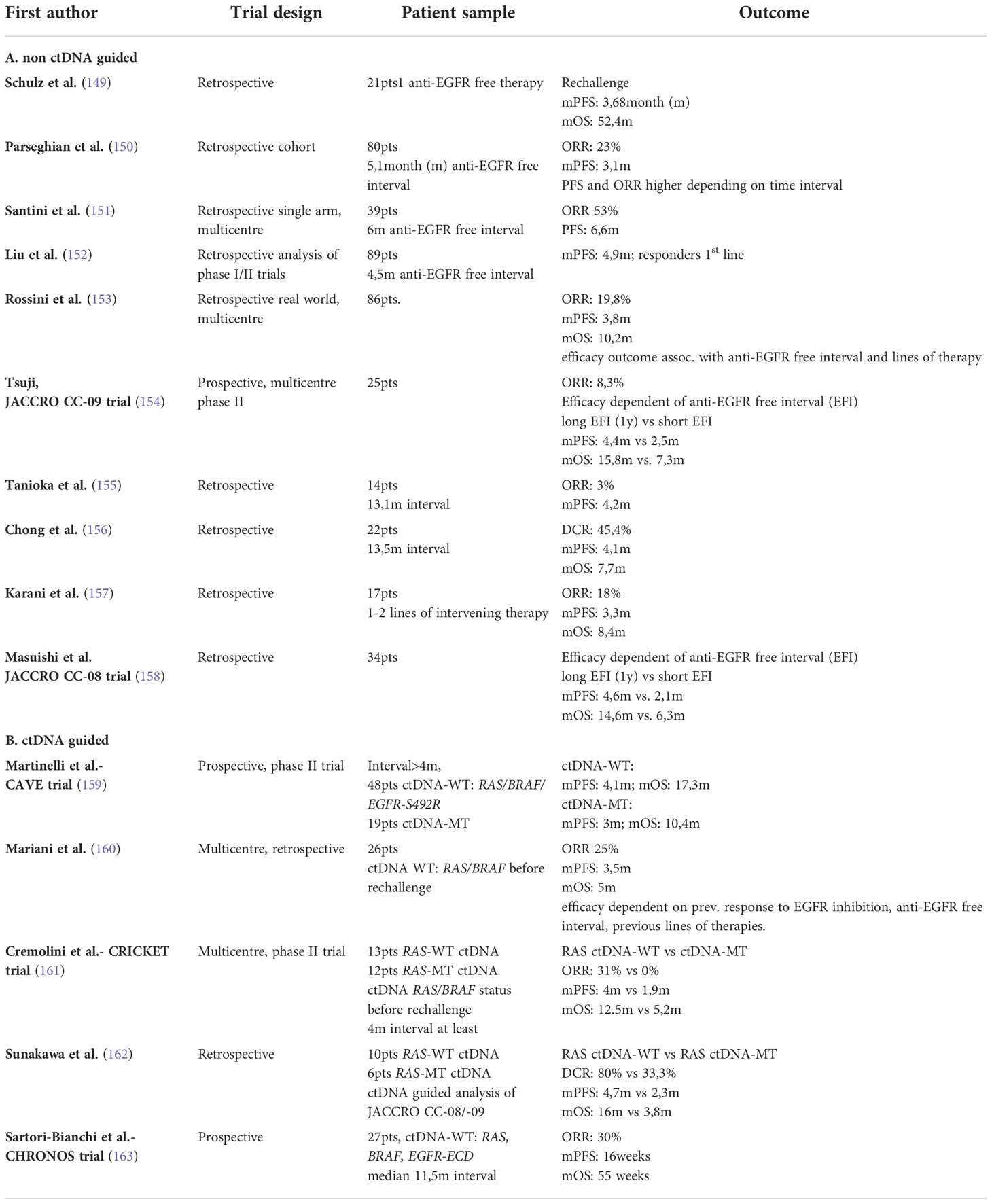
In contrast to continued EGFR blockade, rechallenging initial RAS-WT mCRC tumors with EGFR inhibitors in 3rd line or later after proper EGFR inhibitor free intervals, seems a valuable further direction, which gained broader clinical applicability through the use of liquid biopsy (Table 1). Santini et al. in 2012 were one of the first to prove retrospectively that re-exposure of initial RAS-WT tumors with Irinotecan+Cetuximab reached PFS and OS benefit only in patients who were RAS-WT on ctDNA before re-challenge (151). However, the optimal intervening time interval between two EGFR inhibitor-based therapies remained a matter of debate. Aided by serial ctDNA measurements, Parseghian et al. calculated an exponential decay of RAS mutant or EGFR-ECD with a half-life of about 4 months. They also showed that, although not significant, the use of EGFR inhibitors after a treatment-free interval of at least 2 half-life cycles yielded the greatest ORR. The biological rationale for EGFR re-challenge is based on emerging and dwindling of pre-existing or acquired resistance conferring subclones. RAS-MT subclones pre-exist from the beginning and selective EGFR inhibitor pressure on RAS-WT clones leverages outgrowth of the RAS- MT clones to become the dominant one. Mutations in EGFR-ECD clonal populations are believed to occur as secondary events. The time course of other secondary resistance mutations is less well characterized.
Until now it remains unknown if the main reason for the decay of the resistant subclones is due to the effectiveness of EGFR free treatments or if predominantly the RAS WT clone gains growth advantage in relation to the RAS-MT clone, expressed in VAF% of total ctDNA.
Recently, several phase II trials such as REMARRY and PURSUIT, CRICKET (re-challenge Irinotecan+Cetuximab) or CAVE (re-challenge Avelumab+Cetuximab) confirmed these findings in larger, more defined cohorts. Inclusion criteria for CRICKET and CAVE required at least partial remission for 6 months during first-line EGFR inhibitor therapy, an intervening second line therapy and an EGFR treatment-free interval of at least 4 months. CRICKET reported an ORR of 21% and DCR of 54% in the ITT population (28 mCRC patients) (161). Baseline ctDNA testing was performed before re-challenge in 25 patients. 13 had RAS-WT and 12 RAS-MT ctDNA. In these 13 patients mOS and mPFS were 12.5 and 4 months, respectively whereas mPFS and mOS in the 12 patients with RAS-MT ctDNA were 1.9 and 5.2 months, respectively. ORR was extended to more than 30% of WT-ctDNA versus ITT.
In CAVE the primary end point was accomplished with mOS of 11.6 months (159). Disease control was 65% with a mPFS of 3.6 months. A significant difference in mOS was observed in patients with RAS-WT/BRAF-WT ctDNA at baseline compared to patients with mutated ctDNA (17.3 vs 10.4 months). Patients with mutated ctDNA reached a mPFS of 3.0 months whereas patients with RAS-WT/BRAF-WT ctDNA reached am PFS longer than 6 months in 41% of cases. ORR was only slightly enhanced from 8% (ITT) to 9% (RAS-WT/BRAF-WT ctDNA), but almost doubled compared to RAS or BRAF -MT ctDNA. The striking differences in ORR and mOS are most probably due to the EGFR partner: Chemotherapy vs. checkpoint inhibitor.
The recently published phase II CHRONOS trial pursued an anti-EGFR re-challenge strategy based on an interventional assessment of RAS, BRAF and EGFR-ECD status in ctDNA (163). It acknowledged the fact that the decay of resistance conferring subclones follows an individual timeframe, challenging a previous recommendation to wait at least 4- 8 months - the equivalent of one to two mutant clone half-lives. In CHRONOS a “zero mutation ctDNA triage” of RAS, BRAF and EGFR-ECD had to be evident at the time of re-challenge when compared with the time of progression. Out of 52 patients, 16 (31%) harbored at least one mutation that conferred resistance to anti-EGFR therapy and were excluded. A total of 27 patients were finally enrolled. A 30% response rate compared favorable with the response rates of 8% (CAVE) and 21% (CRICKET). The median PFS was comparable with about 4 months. Remarkably, although the median time between the last dose of EGFR directed therapy and CHRONOS screening was 11.5 months, 17 patients received screening within 8 months after the last EGFR-based therapy. Ten of these 17 patients were ctDNA negative again and could be included. Four patients responded (ORR 40%), 5 had stable disease and 3 progressive disease. Despite these small numbers, CHRONOS clearly favors individualizing re-challenge intervals for the single patient, which can only be reliably performed by liquid biopsy. Further refinement was derived from the REMARRY and PURSUIT trial at ASCO 2022 (164). These trials were designed to determine the efficacy of re-challenge strategies based not only on the negative ctDNA RAS mutation status just before preexposure to EGFR, but also, in particular, on the specific resistance mutations after EGFR blockade during first line treatment. Only tumors that did not develop RAS mutations as resistance mechanism during first line therapy responded to EGFR blockade, though RAS-MT ctDNA had to be cleared before re-challenge. These findings were recently reaffirmed by Topham et al. showing that the decay times of various acquired resistance mutations follow a different time scale (140). RAS mutations present quite tenaciously in contrast to other resistance mutations conferring the MAPK pathway, e.g., EGFR-ECD, BRAF or MAP2K1 (140). Although it is too early to draw any routine clinical reasoning from these observations, one may conclude that it is highly likely that patients benefit in terms of PFS and OS from re-challenge strategies. Decisions for maintenance/re-challenge treatment strategies including EGFR inhibition can be refined by ctDNA measurements, as patients must show a negative ctDNA status as a prerequisite. Additionally, clonal resistance history and the time interval between EGFR treatments are important and can be monitored by ctDNA measurements (165).
Application of extended NGS based panels that cover multiple variants of secondary resistance such as MET/HER2 amplification and PIK3CA mutation could further enrich responses of re-challenge strategies (hyper- or ultraselection), although a minimal clone size value in % VAF determining resistance is still missing. For RAS mutation a cut-off of 5% was reported as a gross orientation; interestingly also small RA-MT subclones might affect response parameters depicting a significant linear correlation clone size and response to EGFR blockade (108).
For a broad clinical application, the implementation of re-challenge strategies is still hampered by the fact that prospective trials comparing EGFR re-exposure to phase III proven treatments like TAS-102 or Regorafenib are still lacking. It is a promising strategy for the future as, for example, a cross-trial comparison of the CHRONOS trial compared to aforementioned third line options revealed a doubling of PFS and OS. Various meta-analyses already regard re-challenging strategies as mandatory to exploit tumor vulnerabilities serving optimized continuum of care concepts (149, 166).
Conclusion
One of the main concepts that has been validated over the past 20 years to extend survival in mCRC is blocking the EGF receptor and inhibition of pathogenic MAPK signalling. Despite long-term routine use of Cetuximab or Panitumumab in everyday clinical care, the dawning of sophisticated molecular techniques including digital PCR, NGS and molecular barcoding among othersshed new light on the putative clinically relevant resistance mechanism. Use of liquid biopsy in clinical practice will optimize incorporation of EGFR inhibitors in the continuum of care of mCRC.
Author contributions
BD wrote the first draft of the manuscript and all subsequent drafts. HR provided extensive revisions. AP extensively proofread the manuscript and contributed valuable points for discussion. All authors contributed to the article and approved the final version of the manuscript.
Funding
This publication was supported by the Johannes Kepler Open Access Publishing Fund to cover the costs of the publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
mCRC, Metastatic colorectal cancer; VEGF, Vascular endothelial growth factor; MSI , Microsatellite instable; EGFR, Epithelial growth factor receptor; OS, Overall survival; IHC, Immunohistochemical; PR, Partial remission; SD, Stable disease; PFS, Progression free survival; ORR, Overall response rate; VAF, Variant allele frequency; QoL, Quality of life; LOD, Level of detection; ETS, Early tumor shrinkage; DpR, Depthness of response; WT, Wilde-type; MT, Mutated- type; MAPK, Mitogen-activated protein kinase; ddPCR, Digital droplet PCR; EGFR-ECD, EGFR extracellular domain; CMS, Consensus molecular subtypes; MB, Molecular barcoding; CTC, Circulating tumor cells; cfDNA, Circulating free DNA; ctDNA, Circulating tumor DNA; NGS, Next generation sequencing
References
1. Kocarnik JM, Compton K, Dean FE, Fu W, Gaw BL, Harvey JD, et al. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019. JAMA Oncol (2022) 8:420–44. doi: 10.1001/jamaoncol.2021.6987
2. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet (2019) 394:1467–80. doi: 10.1016/s0140-6736(19)32319-0
3. Kocarnik JM, Compton K, Dean F, Fu W, Gaw B, Harvey J, et al. The global burden of 29 cancer groups from 2010 to 2019: A systematic analysis for the global burden of disease study 2019. J Clin Oncol (2021) 39:10577–7. doi: 10.1200/jco.2021.39.15_suppl.10577
4. Rumpold H, Hackl M, Petzer A, Wolf D. Improvement in colorectal cancer outcomes over time is limited to patients with left-sided. Disease (2021) 148:3007–14. doi: 10.1007/s00432-021-03868-0
5. Stintzing S, Tejpar S, Gibbs P, Thiebach L, Lenz H-J. Understanding the role of primary tumour localisation in colorectal cancer treatment and outcomes. Eur J Cancer (2017) 84:69–80. doi: 10.1016/j.ejca.2017.07.016
6. Citri A, Yarden Y. EGF–ERBB signalling: towards the systems level. Nat Rev Mol Cell Bio (2006) 7:505–16. doi: 10.1038/nrm1962
7. Arteaga CL, Engelman JA. ERBB receptors: From oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell (2014) 25:282–303. doi: 10.1016/j.ccr.2014.02.025
8. Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. New Engl J Med (2008) 358:1160–74. doi: 10.1056/nejmra0707704
9. Cohen S. Origins of Growth Factors: NGF and EGF. J Biol Chem (2008) 283:33793–7. doi: 10.1074/jbc.x800008200
10. Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol (2009) 21:177–84. doi: 10.1016/j.ceb.2008.12.010
11. Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer (2005) 5:341–54. doi: 10.1038/nrc1609
12. Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer (2004) 4:361–70. doi: 10.1038/nrc1360
13. Roskoski R. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res (2014) 79:34–74. doi: 10.1016/j.phrs.2013.11.002
14. Gill GN, Kawamoto T, Cochet C, Le A, Sato JD, Masui H, et al. Monoclonal anti-epidermal growth factor receptor antibodies which are inhibitors of epidermal growth factor binding and antagonists of epidermal growth factor binding and antagonists of epidermal growth factor-stimulated tyrosine protein kinase activity. J Biol Chem (1984) 259:7755–60. doi: 10.1016/s0021-9258(17)42857-2
15. Goldstein NI, Prewett M, Zuklys K, Rockwell P, Mendelsohn J. Biological efficacy of a chimeric antibody to the epidermal growth factor receptor in a human tumor xenograft model. Clin Cancer Res Off J Am Assoc Cancer Res (1995) 1:1311–8.
16. García-Foncillas J, Sunakawa Y, Aderka D, Wainberg Z, Ronga P, Witzler P, et al. Distinguishing features of cetuximab and panitumumab in colorectal cancer and other solid tumors. Front Oncol (2019) 9:849. doi: 10.3389/fonc.2019.00849
17. Saltz LB, Meropol NJ, Loehrer PJ, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol (2004) 22:1201–8. doi: 10.1200/jco.2004.10.182
18. Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. New Engl J Med (2004) 351:337–45. doi: 10.1056/nejmoa033025
19. Giusti RM, Shastri K, Pilaro AM, Fuchs C, Cordoba-Rodriguez R, Koti K, et al. U.S. food and drug administration approval: Panitumumab for epidermal growth factor receptor–expressing metastatic colorectal carcinoma with progression following fluoropyrimidine-, oxaliplatin-, and irinotecan-containing chemotherapy regimens. Clin Cancer Res (2008) 14:1296–302. doi: 10.1158/1078-0432.ccr-07-1354
20. Randon G, Yaeger R, Hechtman JF, Manca P, Fucà G, Walch H, et al. EGFR amplification in metastatic colorectal cancer. Jnci J Natl Cancer Inst (2021) 113:djab069. doi: 10.1093/jnci/djab069
21. Spano J-P, Lagorce C, Atlan D, Milano G, Domont J, Benamouzig R, et al. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol (2005) 16:102–8. doi: 10.1093/annonc/mdi006
22. Yang Z-Y, Shen W-X, Hu X-F, Zheng D-Y, Wu X-Y, Huang Y-F, et al. EGFR gene copy number as a predictive biomarker for the treatment of metastatic colorectal cancer with anti-EGFR monoclonal antibodies: A meta-analysis. J Hematol Oncol (2012) 5:52. doi: 10.1186/1756-8722-5-52
23. Emburgh BOV, Arena S, Siravegna G, Lazzari L, Crisafulli G, Corti G, et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun (2016) 7:13665. doi: 10.1038/ncomms13665
24. Montagut C, Argilés G, Ciardiello F, Poulsen TT, Dienstmann R, Kragh M, et al. Efficacy of Sym004 in patients with metastatic colorectal cancer with acquired resistance to anti-EGFR therapy and molecularly selected by circulating tumor DNA analyses: A phase 2 randomized clinical trial. JAMA Oncol (2018) 4:e175245–e175245. doi: 10.1001/jamaoncol.2017.5245
25. Price T, Ang A, Boedigheimer M, Kim TW, Li J, Cascinu S, et al. Frequency of S492R mutations in the epidermal growth factor receptor: Analysis of plasma DNA from patients with metastatic colorectal cancer treated with panitumumab or cetuximab monotherapy. Cancer Biol Ther (2020) 21:891–8. doi: 10.1080/15384047.2020.1798695
26. Pietrantonio F, Perrone F, Biondani P, Maggi C, Lampis A, Bertan C, et al. Single agent panitumumab in KRAS wild-type metastatic colorectal cancer patients following cetuximab-based regimens. Cancer Biol Ther (2013) 14:1098–103. doi: 10.4161/cbt.26343
27. Lièvre A, Bachet J-B, Corre DL, Boige V, Landi B, Emile J-F, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res (2006) 66:3992–5. doi: 10.1158/0008-5472.can-06-0191
28. Shih C, Weinberg RA. Isolation of a transforming sequence from a human bladder carcinoma cell line. Cell (1982) 29:161–9. doi: 10.1016/0092-8674(82)90100-3
29. Morelli MP, Overman MJ, Dasari A, Kazmi SMA, Mazard T, Vilar E, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol (2015) 26:731–6. doi: 10.1093/annonc/mdv005
30. Douillard J-Y, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: The PRIME study. J Clin Oncol (2010) 28:4697–705. doi: 10.1200/jco.2009.27.4860
31. Bokemeyer C, Cutsem EV, Rougier P, Ciardiello F, Heeger S, Schlichting M, et al. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: Pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer (2012) 48:1466–75. doi: 10.1016/j.ejca.2012.02.057
32. Douillard J-Y, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab–FOLFOX4 treatment and RAS mutations in colorectal cancer. New Engl J Med (2013) 369:1023–34. doi: 10.1056/nejmoa1305275
33. Cutsem EV, Lenz H-J, Köhne C-H, Heinemann V, Tejpar S, Melezínek I, et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol (2015) 33:692–700. doi: 10.1200/jco.2014.59.4812
34. Santos C, Azuara D, Viéitez JM, Páez D, Falcó E, Élez E, et al. Phase II study of high-sensitivity genotyping of KRAS, NRAS, BRAF and PIK3CA to ultra-select metastatic colorectal cancer patients for panitumumab plus FOLFIRI: the ULTRA trial. Ann Oncol (2019) 30:796–803. doi: 10.1093/annonc/mdz082
35. Fakih M, Tu H, Hsu H, Aggarwal S, Chan E, Rehn M, et al. Real-world study of characteristics and treatment outcomes among patients with KRAS p.G12C-mutated or other KRAS mutated metastatic colorectal cancer. Oncol (2022) 27:663–74. doi: 10.1093/oncolo/oyac077
36. Vasta JD, Peacock DM, Zheng Q, Walker JA, Zhang Z, Zimprich CA, et al. KRAS is vulnerable to reversible switch-II pocket engagement in cells. Nat Chem Biol (2022) 18:596–604. doi: 10.1038/s41589-022-00985-w
37. Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature (2013) 503:548–51. doi: 10.1038/nature12796
38. Ostrem JML, Shokat KM. Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat Rev Drug Discovery (2016) 15:771–85. doi: 10.1038/nrd.2016.139
39. Ryan MB, Coker O, Sorokin A, Fella K, Barnes H, Wong E, et al. KRASG12C-independent feedback activation of wild-type RAS constrains KRASG12C inhibitor efficacy. Cell Rep (2022) 39:110993. doi: 10.1016/j.celrep.2022.110993
40. Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, et al. EGFR blockade reverts resistance to KRASG12C inhibition in colorectal cancer. Cancer Discovery (2020) 10:1129–39. doi: 10.1158/2159-8290.cd-20-0187
41. Lou K, Steri V, Ge AY, Hwang YC, Yogodzinski CH, Shkedi AR, et al. KRASG12C inhibition produces a driver-limited state revealing collateral dependencies. Sci Signal (2019) 12:eaaw9450. doi: 10.1126/scisignal.aaw9450
42. Kuboki Y, Yaeger R, Fakih MG, Strickler JH, Masuishi T, Kim EJ, et al. 315O sotorasib in combination with panitumumab in refractory KRAS G12C-mutated colorectal cancer: Safety and efficacy for phase ib full expansion cohort. Ann Oncol (2022) 33:S680–1. doi: 10.1016/j.annonc.2022.07.453
43. Klempner SJ, Weiss J, Pelster M, Spira A, Barve M, Ou S-HI, et al. LBA24 KRYSTAL-1: Updated efficacy and safety of adagrasib (MRTX849) with or without cetuximab in patients with advanced colorectal cancer (CRC) harboring a KRASG12C mutation. Ann Oncol (2022) 33:S1391. doi: 10.1016/j.annonc.2022.08.020
44. Ciombor KK, Strickler JH, Bekaii-Saab TS, Yaeger R. BRAF-mutated advanced colorectal cancer: A rapidly changing therapeutic landscape. J Clin Oncol (2022) 40:2706–15. doi: 10.1200/jco.21.02541
45. Diaz LA, Shiu K-K, Kim T-W, Jensen BV, Jensen LH, Punt C, et al. Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): final analysis of a randomised, open-label, phase 3 study. Lancet Oncol (2022) 23:659–70. doi: 10.1016/s1470-2045(22)00197-8
46. Overman MJ, Lenz H-J, Andre T, Aglietta M, Wong MK, Luppi G, et al. Nivolumab (NIVO) ± ipilimumab (IPI) in patients (pts) with microsatellite instability-high/mismatch repair-deficient (MSI-H/dMMR) metastatic colorectal cancer (mCRC): Five-year follow-up from CheckMate 142. J Clin Oncol (2022) 40:3510–0. doi: 10.1200/jco.2022.40.16_suppl.3510
47. Rowland A, Dias MM, Wiese MD, Kichenadasse G, McKinnon RA, Karapetis CS, et al. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Brit J Cancer (2015) 112:1888–94. doi: 10.1038/bjc.2015.173
48. Pietrantonio F, Petrelli F, Coinu A, Bartolomeo MD, Borgonovo K, Maggi C, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: A meta-analysis. Eur J Cancer (2015) 51:587–94. doi: 10.1016/j.ejca.2015.01.054
49. Stintzing S, von Weikersthal L, Fuchs M, Kaiser F, Heinrich K, Modest DP, et al. Randomized study to investigate a switch maintenance concept with 5-FU plus bevacizumab after FOLFIRI plus cetuximab induction treatment versus continued treatment with FOLFIRI plus cetuximab: Report of a secondary endpoint of the phase-III FIRE-4 study (AIO KRK-0114). J Clin Oncol (2022) 40:3519–9. doi: 10.1200/jco.2022.40.16_suppl.3519
50. Schirripa M, Biason P, Lonardi S, Pella N, Pino MS, Urbano F, et al. Class 1, 2, and 3 BRAF-mutated metastatic colorectal cancer: A detailed clinical, pathologic, and molecular characterization. Clin Cancer Res (2019) 25:3954–61. doi: 10.1158/1078-0432.ccr-19-0311
51. Asselain B, Martinelli E, Cremolini C, Mazard T, Vidal J, Virchow I, et al. 438P first line treatment patterns in BRAFV600E-mutant metastatic colorectal cancer patients (mCRC): The CAPSTAN European retrospective study. Ann Oncol (2021) 32:S553. doi: 10.1016/j.annonc.2021.08.959
52. Ros J, Baraibar I, Sardo E, Mulet N, Salvà F, Argilés G, et al. BRAF, MEK and EGFR inhibition as treatment strategies in BRAF V600E metastatic colorectal cancer. Ther Adv Med Oncol (2021) 13:1758835921992974. doi: 10.1177/1758835921992974
53. Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, et al. A molecularly annotated platform of patient-derived xenografts (“Xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discovery (2011) 1:508–23. doi: 10.1158/2159-8290.cd-11-0109
54. Martin V, Landi L, Molinari F, Fountzilas G, Geva R, Riva A, et al. HER2 gene copy number status may influence clinical efficacy to anti-EGFR monoclonal antibodies in metastatic colorectal cancer patients. Brit J Cancer (2013) 108:668–75. doi: 10.1038/bjc.2013.4
55. Yonesaka K, Zejnullahu K, Okamoto I, Satoh T, Cappuzzo F, Souglakos J, et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med (2011) 3:99ra86. doi: 10.1126/scitranslmed.3002442
56. Bang Y-J, Cutsem EV, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet (2010) 376:687–97. doi: 10.1016/s0140-6736(10)61121-x
57. Bregni G, Sciallero S, Sobrero A. HER2 amplification and anti-EGFR sensitivity in advanced colorectal cancer. JAMA Oncol (2019) 5:605. doi: 10.1001/jamaoncol.2018.7229
58. Djaballah SA, Daniel F, Milani A, Ricagno G, Lonardi S. HER2 in colorectal cancer: The long and winding road from negative predictive factor to positive actionable target. Am Soc Clin Oncol Educ Book Am Soc Clin Oncol Annu Meet (2022) 42:1–14. doi: 10.1200/edbk_351354
59. Sartore-Bianchi A, Amatu A, Porcu L, Ghezzi S, Lonardi S, Leone F, et al. HER2 positivity predicts unresponsiveness to EGFR-targeted treatment in metastatic colorectal cancer. Oncol (2019) 24:1395–402. doi: 10.1634/theoncologist.2018-0785
60. Raghav K, Loree JM, Morris JS, Overman MJ, Yu R, Meric-Bernstam F, et al. Validation of HER2 amplification as a predictive biomarker for anti–epidermal growth factor receptor antibody therapy in metastatic colorectal cancer. Jco Precis Oncol (2019) 3:1–13. doi: 10.1200/po.18.00226
61. Strickler JH, Ng K, Cercek A, Fountzilas C, Sanchez FA, Hubbard JM, et al. MOUNTAINEER:open-label, phase II study of tucatinib combined with trastuzumab for HER2-positive metastatic colorectal cancer (SGNTUC-017, trial in progress). J Clin Oncol (2021) 39:TPS153–3. doi: 10.1200/jco.2021.39.3_suppl.tps153
62. Raghav KPS, Yoshino T, Guimbaud R, Chau I, Eynde MVD, Maurel J, et al. Trastuzumab deruxtecan in patients with HER2-overexpressing locally advanced, unresectable, or metastatic colorectal cancer (mCRC): A randomized, multicenter, phase 2 study (DESTINY-CRC02). J Clin Oncol (2022) 40:TPS224–4. doi: 10.1200/jco.2022.40.4_suppl.tps224
63. Bufill JA. Colorectal cancer: Evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med (1990) 113:779. doi: 10.7326/0003-4819-113-10-779
64. Venook AP, Ou F-S, Lenz H-J, Kabbarah O, Qu X, Niedzwiecki D, et al. Primary (1°) tumor location as an independent prognostic marker from molecular features for overall survival (OS) in patients (pts) with metastatic colorectal cancer (mCRC): Analysis of CALGB / SWOG 80405 (Alliance). J Clin Oncol (2017) 35:3503–3. doi: 10.1200/jco.2017.35.15_suppl.3503
65. Arnold D, Lueza B, Douillard J-Y, Peeters M, Lenz H-J, Venook A, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol (2017) 28:1713–29. doi: 10.1093/annonc/mdx175
66. Tejpar S, Stintzing S, Ciardiello F, Tabernero J, Cutsem EV, Beier F, et al. Prognostic and predictive relevance of primary tumor location in patients with RAS wild-type metastatic colorectal cancer: Retrospective analyses of the CRYSTAL and FIRE-3 trials. JAMA Oncol (2016) 3:194–201. doi: 10.1001/jamaoncol.2016.3797
67. Holch JW, Ricard I, Stintzing S, Modest DP, Heinemann V. The relevance of primary tumour location in patients with metastatic colorectal cancer: A meta-analysis of first-line clinical trials. Eur J Cancer Oxf Engl 1990 (2016) 70:87–98. doi: 10.1016/j.ejca.2016.10.007
68. Yoshino T, Watanabe J, Shitara K, Yasui H, Ohori H, Shiozawa M, et al. Panitumumab (PAN) plus mFOLFOX6 versus bevacizumab (BEV) plus mFOLFOX6 as first-line treatment in patients with RAS wild-type (WT) metastatic colorectal cancer (mCRC): Results from the phase 3 PARADIGM trial. J Clin Oncol (2022) 40:LBA1–1. doi: 10.1200/jco.2022.40.17_suppl.lba1
69. Modest DP, Laubender RP, Stintzing S, Giessen C, Schulz C, Haas M, et al. Early tumor shrinkage in patients with metastatic colorectal cancer receiving first-line treatment with cetuximab combined with either CAPIRI or CAPOX: An analysis of the German AIO KRK 0104 trial. Acta Oncol (2012) 52:956–62. doi: 10.3109/0284186x.2012.752580
70. Heinemann V, Stintzing S, Modest DP, Giessen-Jung C, Michl M, Mansmann UR. Early tumour shrinkage (ETS) and depth of response (DpR) in the treatment of patients with metastatic colorectal cancer (mCRC). Eur J Cancer (2015) 51:1927–36. doi: 10.1016/j.ejca.2015.06.116
71. Modest DP, Stintzing S, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, et al. Relation of early tumor shrinkage (ETS) observed in first-line treatment to efficacy parameters of subsequent treatment in FIRE-3 (AIOKRK0306). Int J Cancer (2017) 140:1918–25. doi: 10.1002/ijc.30592
72. Tsuji A, Ohori H, Yamaguchi T, Matsuura M, Nishioka A, Makiyama A, et al. The randomized phase II study of FOLFOXIRI plus cetuximab versus FOLFOXIRI plus bevacizumab as the first-line treatment in metastatic colorectal cancer with RAS wild-type tumors: The DEEPER trial (JACCRO CC-13). J Clin Oncol (2021) 39:3501–1. doi: 10.1200/jco.2021.39.15_suppl.3501
73. Randon G, Maddalena G, Germani MM, Pircher CC, Manca P, Bergamo F, et al. Negative ultraselection of patients with RAS/BRAF wild-type, microsatellite-stable metastatic colorectal cancer receiving anti–EGFR-Based therapy. Jco Precis Oncol (2022) 6:e2200037. doi: 10.1200/po.22.00037
74. Roock WD, Claes B, Bernasconi D, Schutter JD, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol (2010) 11:753–62. doi: 10.1016/s1470-2045(10)70130-3
75. Ciardiello F, Normanno N, Maiello E, Martinelli E, Troiani T, Pisconti S, et al. Clinical activity of FOLFIRI plus cetuximab according to extended gene mutation status by next-generation sequencing: findings from the CAPRI-GOIM trial. Ann Oncol Off J Eur Soc Med Oncol (2014) 25:1756–61. doi: 10.1093/annonc/mdu230
76. Morano F, Corallo S, Lonardi S, Raimondi A, Cremolini C, Rimassa L, et al. Negative hyperselection of patients with RAS and BRAF wild-type metastatic colorectal cancer who received panitumumab-based maintenance therapy. J Clin Oncol Off J Am Soc Clin Oncol (2019) 37:3099–110. doi: 10.1200/jco.19.01254
77. Cremolini C, Morano F, Moretto R, Berenato R, Tamborini E, Perrone F, et al. Negative hyper-selection of metastatic colorectal cancer patients for anti-EGFR monoclonal antibodies: the PRESSING case–control study. Ann Oncol (2017) 28:3009–14. doi: 10.1093/annonc/mdx546
78. Goldberg RM, Rothenberg ML, Cutsem EV, Benson AB, Blanke CD, Diasio RB, et al. The continuum of care: A paradigm for the management of metastatic colorectal cancer. Oncol (2007) 12:38–50. doi: 10.1634/theoncologist.12-1-38
79. Yoshino T, Arnold D, Taniguchi H, Pentheroudakis G, Yamazaki K, Xu R-H, et al. Pan-Asian adapted ESMO consensus guidelines for the management of patients with metastatic colorectal cancer: A JSMO–ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Ann Oncol (2018) 29:44–70. doi: 10.1093/annonc/mdx738
80. Yu IS, Aubin F, Goodwin R, Loree JM, Mather C, Sheffield BS, et al. Tumor biomarker testing for metastatic colorectal cancer: A Canadian consensus practice guideline. Ther Adv Med Oncol (2022) 14:17588359221111704. doi: 10.1177/17588359221111705
81. Chibaudel B, Dourthe L-M, Andre T, Henriques J, Bourgeois V, Etienne P-L, et al. STRATEGIC-1: Multi-line therapy trial in unresectable wild-type KRAS/NRAS/BRAF metastatic colorectal cancer–a GERCOR-PRODIGE randomized open-label phase III study. J Clin Oncol (2022) 40:3504–4. doi: 10.1200/jco.2022.40.16_suppl.3504
82. Manca P, Corallo S, Randon G, Lonardi S, Cremolini C, Rimassa L, et al. Impact of early tumor shrinkage and depth of response on the outcomes of panitumumab-based maintenance in patients with RAS wild-type metastatic colorectal cancer. Eur J Cancer (2021) 144:31–40. doi: 10.1016/j.ejca.2020.11.017
83. Claret L, Gupta M, Han K, Joshi A, Sarapa N, He J, et al. Evaluation of tumor-size response metrics to predict overall survival in Western and Chinese patients with first-line metastatic colorectal cancer. J Clin Oncol (2013) 31:2110–4. doi: 10.1200/jco.2012.45.0973
84. Cremolini C, Rossini D, Lonardi S, Antoniotti C, Pietrantonio F, Marmorino F, et al. Modified FOLFOXIRI plus panitumumab (mFOLFOXIRI/PAN) versus mFOLFOX6/PAN as initial treatment of patients with unresectable RAS and BRAF wild-type metastatic colorectal cancer (mCRC): Results of the phase III randomized TRIPLETE study by GONO. J Clin Oncol (2022) 40:LBA3505–LBA3505. doi: 10.1200/jco.2022.40.17_suppl.lba3505
85. Stintzing S, Heinrich K, Tougeron D, Modest DP, Schwaner I, Euker J, et al. Randomized study to investigate FOLFOXIRI plus either bevacizumab or cetuximab as first-line treatment of BRAF V600E-mutant mCRC: The phase-II FIRE-4.5 study (AIO KRK-0116). J Clin Oncol (2021) 39:3502–2. doi: 10.1200/jco.2021.39.15_suppl.3502
86. Battaglin F, Schirripa M, Buggin F, Pietrantonio F, Morano F, Boscolo G, et al. The PANDA study: A randomized phase II study of first-line FOLFOX plus panitumumab versus 5FU plus panitumumab in RAS and BRAF wild-type elderly metastatic colorectal cancer patients. BMC Cancer (2018) 18:98. doi: 10.1186/s12885-018-4001-x
87. Cunningham D, Lang I, Marcuello E, Lorusso V, Ocvirk J, Shin DB, et al. Bevacizumab plus capecitabine versus capecitabine alone in elderly patients with previously untreated metastatic colorectal cancer (AVEX): An open-label, randomised phase 3 trial. Lancet Oncol (2013) 14:1077–85. doi: 10.1016/s1470-2045(13)70154-2
88. Sobrero AF, Maurel J, Fehrenbacher L, Scheithauer W, Abubakr YA, Lutz MP, et al. EPIC: Phase III trial of cetuximab plus irinotecan after fluoropyrimidine and oxaliplatin failure in patients with metastatic colorectal cancer. J Clin Oncol (2008) 26:2311–9. doi: 10.1200/jco.2007.13.1193
89. Peeters M, Price TJ, Cervantes A, Sobrero AF, Ducreux M, Hotko Y, et al. Final results from a randomized phase 3 study of FOLFIRI ± panitumumab for second-line treatment of metastatic colorectal cancer. Ann Oncol (2014) 25:107–16. doi: 10.1093/annonc/mdt523
90. Seymour MT, Brown SR, Middleton G, Maughan T, Richman S, Gwyther S, et al. Panitumumab and irinotecan versus irinotecan alone for patients with KRAS wild-type, fluorouracil-resistant advanced colorectal cancer (PICCOLO): A prospectively stratified randomised trial. Lancet Oncol (2013) 14:749–59. doi: 10.1016/s1470-2045(13)70163-3
91. Bennouna J, Hiret S, Borg C, Bertaut A, Bouche O, Deplanque G, et al. Bevacizumab (Bev) or cetuximab (Cet) plus chemotherapy after progression with bevacizumab plus chemotherapy in patients with wild-type (WT) KRAS metastatic colorectal cancer (mCRC): Final analysis of a French randomized, multicenter, phase II study (PRODIGE 18). Ann Oncol (2017) 28:v159–60. doi: 10.1093/annonc/mdx393.004
92. Hecht JR, Cohn A, Dakhil S, Saleh M, Piperdi B, Cline-Burkhardt M, et al. SPIRITT: A randomized, multicenter, phase II study of panitumumab with FOLFIRI and bevacizumab with FOLFIRI as second-line treatment in patients with unresectable wild type KRAS metastatic colorectal cancer. Clin Colorectal Canc (2014) 14:72–80. doi: 10.1016/j.clcc.2014.12.009
93. Aranda E, García-Alfonso P, Benavides M, Ruiz AS, Guillén-Ponce C, Safont MJ, et al. First-line mFOLFOX plus cetuximab followed by mFOLFOX plus cetuximab or single-agent cetuximab as maintenance therapy in patients with metastatic colorectal cancer: Phase II randomised MACRO2 TTD study. Eur J Cancer (2018) 101:263–72. doi: 10.1016/j.ejca.2018.06.024
94. Guren TK, Thomsen M, Kure EH, Sorbye H, Glimelius B, Pfeiffer P, et al. Cetuximab in treatment of metastatic colorectal cancer: final survival analyses and extended RAS data from the NORDIC-VII study. Brit J Cancer (2017) 116:1271–8. doi: 10.1038/bjc.2017.93
95. Munemoto Y, Nakamura M, Takahashi M, Kotaka M, Kuroda H, Kato T, et al. SAPPHIRE: A randomised phase II study of planned discontinuation or continuous treatment of oxaliplatin after six cycles of modified FOLFOX6 plus panitumumab in patients with colorectal cancer. Eur J Cancer (2019) 119:158–67. doi: 10.1016/j.ejca.2019.07.006
96. Pinto C, Orlandi A, Normanno N, Maiello E, Calegari MA, Antonuzzo L, et al. LBA22 phase III study with FOLFIRI/cetuximab versus FOLFIRI/cetuximab followed by cetuximab (Cet) alone in first-line therapy of RAS and BRAF wild-type (wt) metastatic colorectal cancer (mCRC) patients: The ERMES study. Ann Oncol (2022) 33:S1390. doi: 10.1016/j.annonc.2022.08.018
97. Adams RA, Fisher DJ, Graham J, Seligmann JF, Seymour M, Kaplan R, et al. Capecitabine versus active monitoring in stable or responding metastatic colorectal cancer after 16 weeks of first-line therapy: Results of the randomized FOCUS4-n trial. J Clin Oncol (2021) 39:3693–704. doi: 10.1200/jco.21.01436
98. Wasan H, Meade AM, Adams R, Wilson R, Pugh C, Fisher D, et al. Intermittent chemotherapy plus either intermittent or continuous cetuximab for first-line treatment of patients with KRAS wild-type advanced colorectal cancer (COIN-b): A randomised phase 2 trial. Lancet Oncol (2014) 15:631–9. doi: 10.1016/s1470-2045(14)70106-8
99. Boige V, FRANCOIS E, Abdelghani MB, Phelip JM, Brun-Ly VL, Mineur L, et al. Maintenance treatment with cetuximab versus observation in RAS wild-type metastatic colorectal cancer: Results of the randomized phase II PRODIGE 28-time UNICANCER study. J Clin Oncol (2021) 39:15–5. doi: 10.1200/jco.2021.39.3_suppl.15
100. Pietrantonio F, Morano F, Corallo S, Miceli R, Lonardi S, Raimondi A, et al. Maintenance therapy with panitumumab alone vs panitumumab plus fluorouracil-leucovorin in patients with RAS wild-type metastatic colorectal cancer. JAMA Oncol (2019) 5:1268–75. doi: 10.1001/jamaoncol.2019.1467
101. Cremolini C, Antoniotti C, Lonardi S, Aprile G, Bergamo F, Masi G, et al. Activity and safety of cetuximab plus modified FOLFOXIRI followed by maintenance with cetuximab or bevacizumab for RAS and BRAF wild-type metastatic colorectal cancer: A randomized phase 2 clinical trial. JAMA Oncol (2018) 4:529–36. doi: 10.1001/jamaoncol.2017.5314
102. Modest DP, Rivera F, Bachet J, de Braud F, Pietrantonio F, Koukakis R, et al. Panitumumab-based maintenance after oxaliplatin discontinuation in metastatic colorectal cancer: A retrospective analysis of two randomised trials. Int J Cancer (2019) 145:576–85. doi: 10.1002/ijc.32110
103. Parisi A, Ghidini M, Giampieri R, Tomasello G, Luciani A, Ferri C, et al. Post-induction strategies in metastatic colorectal cancer patients treated with first-line anti-EGFR-based treatment: A systematic review and meta-analysis. Clin Colorectal Canc (2022) 21:e162–70. doi: 10.1016/j.clcc.2021.12.005
104. da Silva LL, Aguiar PN, D’Alpino R, Dienstmann R. Maintenance therapy following an anti-EGFR-based induction regimen in metastatic colorectal cancer (mCRC): A network meta-analysis of clinical trials. J Clin Oncol (2022) 40:128–8. doi: 10.1200/jco.2022.40.4_suppl.128
105. Raimondi A, Morano F, Trarbach T, Karthaus M, Lonardi S, Fruehauf S, et al. SO-21 optimal maintenance treatment strategy following an anti-EGFR-based first-line induction therapy in patients with RAS wild type metastatic colorectal cancer: An individual patient data pooled analysis of clinical trials. Ann Oncol (2022) 33:S366. doi: 10.1016/j.annonc.2022.04.420
106. Artac M, Cubukcu E, Bozkurt O, Bilici A, Celik S, Ozcelik M, et al. P-92 real-life experience with maintenance chemotherapy plus biologics after the first-line treatment of RAS wild-type metastatic colon cancer (mCRC): A multicenter onco-colon Turkey study. Ann Oncol (2022) 33:S281–2. doi: 10.1016/j.annonc.2022.04.182
107. Hu H, Liu X, Cai W, Wu D, Xu J, Yuan Y. A retrospective exploration of targeted maintenance therapy in advanced colorectal cancer: Based on the background of Chinese patient assistance program. Front Oncol (2020) 10:522. doi: 10.3389/fonc.2020.00522
108. Manca P, Corallo S, Busico A, Lonardi S, Corti F, Antoniotti C, et al. The added value of baseline circulating tumor DNA profiling in patients with molecularly hyperselected, left-sided metastatic colorectal cancer. Clin Cancer Res Off J Am Assoc Cancer Res (2021) 27:2505–14. doi: 10.1158/1078-0432.ccr-20-4699
109. Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med (2015) 21:795–801. doi: 10.1038/nm.3870
110. Tabernero J, Grothey A, Arnold D, Ducreux M, O’Dwyer PJ, Perdicchio M, et al. Exploratory biomarker findings from cohort 2 of MODUL: An adaptable, phase 2, signal-seeking trial of fluoropyrimidine + bevacizumab ± atezolizumab maintenance therapy for BRAF wt metastatic colorectal cancer. J Clin Oncol (2021) 39:3570–0. doi: 10.1200/jco.2021.39.15_suppl.3570
111. Seligmann JF, Fisher DJ, Brown LC, Adams RA, Graham J, Quirke P, et al. Inhibition of WEE1 is effective in TP53- and RAS-mutant metastatic colorectal cancer: A randomized trial (FOCUS4-c) comparing adavosertib (AZD1775) with active monitoring. J Clin Oncol (2021) 39:3705–15. doi: 10.1200/jco.21.01435
112. Modest DP, Karthaus M, Fruehauf S, Graeven U, Müller L, König AO, et al. Panitumumab plus fluorouracil and folinic acid versus fluorouracil and folinic acid alone as maintenance therapy in RAS wild-type metastatic colorectal cancer: The randomized PANAMA trial (AIO KRK 0212). J Clin Oncol (2022) 40:72–82. doi: 10.1200/jco.21.01332
113. Avallone A, Giuliani F, Nasti G, Montesarchio V, Santabarbara G, Leo S, et al. Randomized intermittent or continuous panitumumab plus FOLFIRI (FOLFIRI/PANI) for first-line treatment of patients (pts) with RAS/BRAF wild-type (wt) metastatic colorectal cancer (mCRC): The IMPROVE study. J Clin Oncol (2022) 40:3503–3. doi: 10.1200/jco.2022.40.16_suppl.3503
114. Internò V, Tucci M, Pezzicoli G, Silvestris F, Porta C, Mannavola F. Liquid biopsy as a tool exploring in real-time both genomic perturbation and resistance to EGFR antagonists in colorectal cancer. Front Oncol (2020) 10:581130. doi: 10.3389/fonc.2020.581130
115. Li X, Wang Q, Wang R. Roles of exosome genomic DNA in colorectal cancer. Front Pharmacol (2022) 13:923232. doi: 10.3389/fphar.2022.923232
116. Patelli G, Vaghi C, Tosi F, Mauri G, Amatu A, Massihnia D, et al. Liquid biopsy for prognosis and treatment in metastatic colorectal cancer: Circulating tumor cells vs circulating tumor DNA. Target Oncol (2021) 16:309–24. doi: 10.1007/s11523-021-00795-5
117. Lucchetti D, Zurlo IV, Colella F, Ricciardi-Tenore C, Salvatore MD, Tortora G, et al. Mutational status of plasma exosomal KRAS predicts outcome in patients with metastatic colorectal cancer. Sci Rep-uk (2021) 11:22686. doi: 10.1038/s41598-021-01668-7
118. Hao Y-X, Li Y-M, Ye M, Guo Y-Y, Li Q-W, Peng X-M, et al. KRAS and BRAF mutations in serum exosomes from patients with colorectal cancer in a Chinese population. Oncol Lett (2017) 13:3608–16. doi: 10.3892/ol.2017.5889
120. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res (1977) 37:646–50.
121. Sidransky D, Tokino T, Hamilton SR, Kinzler KW, Levin B, Frost P, et al. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science (1992) 256:102–5. doi: 10.1126/science.1566048
122. Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci (1999) 96:9236–41. doi: 10.1073/pnas.96.16.9236
123. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med (2008) 14:985–90. doi: 10.1038/nm.1789
124. Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci (2003) 100:8817–22. doi: 10.1073/pnas.1133470100
125. Newman AM, Bratman SV, To J, Wynne JF, Eclov NCW, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med (2014) 20:548–54. doi: 10.1038/nm.3519
126. Cohen JD, Douville C, Dudley JC, Mog BJ, Popoli M, Ptak J, et al. Detection of low-frequency DNA variants by targeted sequencing of the Watson and crick strands. Nat Biotechnol (2021) 39:1220–7. doi: 10.1038/s41587-021-00900-z
127. Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci (2011) 108:9530–5. doi: 10.1073/pnas.1105422108
128. Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol (2015) 26:1715–22. doi: 10.1093/annonc/mdv177
129. Pascual J, Attard G, Bidard F-C, Curigliano G, Mattos-Arruda LD, Diehn M, et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO precision medicine working group. Ann Oncol (2022) 33:750–68. doi: 10.1016/j.annonc.2022.05.520
130. Salem ME, Puccini A, Tie J. Redefining colorectal cancer by tumor biology. Am Soc Clin Oncol Educ Book (2020) 40:147–59. doi: 10.1200/edbk_279867
131. Bachet JB, Bouché O, Taieb J, Dubreuil O, Garcia ML, Meurisse A, et al. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: the AGEO RASANC prospective multicenter study. Ann Oncol Off J Eur Soc Med Oncol (2018) 29:1211–9. doi: 10.1093/annonc/mdy061
132. Grasselli J, Elez E, Caratù G, Matito J, Santos C, Macarulla T, et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol (2017) 28:1294–301. doi: 10.1093/annonc/mdx112
133. Vidal J, Muinelo L, Dalmases A, Jones F, Edelstein D, Iglesias M, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol (2017) 28:1325–32. doi: 10.1093/annonc/mdx125
134. García-Foncillas J, Tabernero J, Élez E, Aranda E, Benavides M, Camps C, et al. Prospective multicenter real-world RAS mutation comparison between OncoBEAM-based liquid biopsy and tissue analysis in metastatic colorectal cancer. Brit J Cancer (2018) 119:1464–70. doi: 10.1038/s41416-018-0293-5
135. Schmiegel W, Scott RJ, Dooley S, Lewis W, Meldrum CJ, Pockney P, et al. Blood-based detection of RAS mutations to guide anti-EGFR therapy in colorectal cancer patients: concordance of results from circulating tumor DNA and tissue-based RAS testing. Mol Oncol (2017) 11:208–19. doi: 10.1002/1878-0261.12023
136. Normanno N, Abate RE, Lambiase M, Forgione L, Cardone C, Iannaccone A, et al. RAS testing of liquid biopsy correlates with the outcome of metastatic colorectal cancer patients treated with first-line FOLFIRI plus cetuximab in the CAPRI-GOIM trial. Ann Oncol (2018) 29:112–8. doi: 10.1093/annonc/mdx417
137. Benavides M, Alcaide-Garcia J, Torres E, Gil-Calle S, Sevilla I, Wolman R, et al. Clinical utility of comprehensive circulating tumor DNA genotyping compared with standard of care tissue testing in patients with newly diagnosed metastatic colorectal cancer. Esmo Open (2022) 7:100481. doi: 10.1016/j.esmoop.2022.100481
138. Janku F, Angenendt P, Tsimberidou AM, Fu S, Naing A, Falchook GS, et al. Actionable mutations in plasma cell-free DNA in patients with advanced cancers referred for experimental targeted therapies. Oncotarget (2015) 6:12809–21. doi: 10.18632/oncotarget.3373
139. Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature (2012) 486:532–6. doi: 10.1038/nature11156
140. Topham JT, O’Callaghan CJ, Feilotter H, Kennecke HF, Lee YS, Li W, et al. Circulating tumor DNA identifies diverse landscape of acquired resistance to anti-epidermal growth factor receptor therapy in metastatic colorectal cancer. J Clin Oncol Off J Am Soc Clin Oncol (2022) JCO2200364 - Online ahead of print. doi: 10.1200/jco.22.00364
141. Parseghian CM, Sun R, Napolitano S, Morris VK, Henry J, Willis J, et al. Rarity of acquired mutations (MTs) after first-line therapy with anti-EGFR therapy (EGFRi). J Clin Oncol (2021) 39:3514–4. doi: 10.1200/jco.2021.39.15_suppl.3514
142. Dasari A, Morris VK, Allegra CJ, Atreya C, Benson AB, Boland P, et al. ctDNA applications and integration in colorectal cancer: An NCI colon and rectal–anal task forces whitepaper. Nat Rev Clin Oncol (2020) 17:757–70. doi: 10.1038/s41571-020-0392-0
143. Lieu CH, Corcoran RB, Overman MJ. Integrating biomarkers and targeted therapy into colorectal cancer management. Am Soc Clin Oncol Educ Book Am Soc Clin Oncol Annu Meet (2019) 39:207–15. doi: 10.1200/edbk_240839
144. Ciardiello F, Normanno N, Martinelli E, Troiani T, Pisconti S, Cardone C, et al. Cetuximab continuation after first progression in metastatic colorectal cancer (CAPRI-GOIM): A randomized phase II trial of FOLFOX plus cetuximab versus FOLFOX. Ann Oncol (2016) 27:1055–61. doi: 10.1093/annonc/mdw136
145. Aparicio J, Manrique ACV, Capdevila J, Boza FM, Galván P, Richart P, et al. Randomized phase II trial of FOLFIRI-panitumumab compared with FOLFIRI alone in patients with RAS wild-type circulating tumor DNA metastatic colorectal cancer beyond progression to first-line FOLFOX-panitumumab: the BEYOND study (GEMCAD 17-01). Clin Transl Oncol (2022) 24:2155–65. doi: 10.1007/s12094-022-02868-x
146. Watanabe J, Maeda H, Nagasaka T, Yokota M, Hirata K, Akazawa N, et al. Multicenter, single-arm, phase II study of the continuous use of panitumumab in combination with FOLFIRI after FOLFOX for RAS wild-type metastatic colorectal cancer: Exploratory sequential examination of acquired mutations in circulating cell-free DNA. Int J Cancer (2022) 151:2172–81. doi: 10.1002/ijc.34184
147. Li Q-H, Wang Y-Z, Tu J, Liu C-W, Yuan Y-J, Lin R, et al. Anti-EGFR therapy in metastatic colorectal cancer: mechanisms and potential regimens of drug resistance. Gastroenterol Rep (2020) 8:179–91. doi: 10.1093/gastro/goaa026
148. Li D, Wang F, Xu S, Li K, Meng X, Huang Y, et al. Continuing cetuximab vs bevacizumab plus chemotherapy after first progression in wild-type KRAS, NRAS and BRAF V600E metastatic colorectal cancer: A randomized phase II trial. J Cancer (2021) 12:5268–74. doi: 10.7150/jca.60014
149. Schulz MS, Wolf S, Struck V, Thomas N, Husman G, Zeuzem S, et al. Anti-EGFR reintroduction and rechallenge in metastatic colorectal cancer (mCRC): A real-world analysis. Cancers (2022) 14:1641. doi: 10.3390/cancers14071641
150. Parseghian CM, Loree JM, Morris VK, Liu X, Clifton KK, Napolitano S, et al. Anti-EGFR-resistant clones decay exponentially after progression: implications for anti-EGFR re-challenge. Ann Oncol (2019) 30:243–9. doi: 10.1093/annonc/mdy509
151. Santini D, Vincenzi B, Addeo R, Garufi C, Masi G, Scartozzi M, et al. Cetuximab rechallenge in metastatic colorectal cancer patients: how to come away from acquired resistance? Ann Oncol (2012) 23:2313–8. doi: 10.1093/annonc/mdr623
152. Liu X, George GC, Tsimberidou AM, Naing A, Wheler JJ, Kopetz S, et al. Retreatment with anti-EGFR based therapies in metastatic colorectal cancer: impact of intervening time interval and prior anti-EGFR response. BMC Cancer (2015) 15:713. doi: 10.1186/s12885-015-1701-3
153. Rossini D, Germani MM, Pagani F, Pellino A, Dell’Aquila E, Bensi M, et al. Retreatment with anti-EGFR antibodies in metastatic colorectal cancer patients: A multi-institutional analysis. Clin Colorectal Canc (2020) 19:191–199.e6. doi: 10.1016/j.clcc.2020.03.009
154. Tsuji A, Nakamura M, Watanabe T, Manaka D, Matsuoka H, Kataoka M, et al. Phase II study of third-line panitumumab rechallenge in patients with metastatic wild-type KRAS colorectal cancer who obtained clinical benefit from first-line panitumumab-based chemotherapy: JACCRO CC-09. Target Oncol (2021) 16:753–60. doi: 10.1007/s11523-021-00845-y
155. Tanioka H, Asano M, Yoshida R, Waki N, Uno F, Ishizaki M, et al. Cetuximab retreatment in patients with metastatic colorectal cancer who exhibited a clinical benefit in response to prior cetuximab: A retrospective study. Oncol Lett (2018) 16:3674–80. doi: 10.3892/ol.2018.9127
156. Chong LC, Hardingham JE, Townsend AR, Piantadosi C, Rico GT, Karapetis C, et al. Rechallenge with anti-EGFR therapy in metastatic colorectal cancer (mCRC): Results from south Australia mCRC registry. Target Oncol (2020) 15:751–7. doi: 10.1007/s11523-020-00760-8
157. Karani A, Felismino TC, Diniz L, Macedo MP, Silva V, Mello CA. Is there a role for rechallenge and reintroduction of anti-EGFR plus chemotherapy in later lines of therapy for metastatic colorectal carcinoma? a retrospective analysis. Ecancermedicalscience (2020) 14:1069. doi: 10.3332/ecancer.2020.1069
158. Masuishi T, Tsuji A, Kotaka M, Nakamura M, Kochi M, Takagane A, et al. Phase 2 study of irinotecan plus cetuximab rechallenge as third-line treatment in KRAS wild-type metastatic colorectal cancer: JACCRO CC-08. Brit J Cancer (2020) 123:1490–5. doi: 10.1038/s41416-020-01042-w
159. Martinelli E, Martini G, Famiglietti V, Troiani T, Napolitano S, Pietrantonio F, et al. Cetuximab rechallenge plus avelumab in pretreated patients with RAS wild-type metastatic colorectal cancer. JAMA Oncol (2021) 7:1529. doi: 10.1001/jamaoncol.2021.2915
160. Mariani S, Puzzoni M, Giampieri R, Ziranu P, Pusceddu V, Donisi C, et al. Liquid biopsy-driven cetuximab rechallenge strategy in molecularly selected metastatic colorectal cancer patients. Front Oncol (2022) 12:852583. doi: 10.3389/fonc.2022.852583
161. Cremolini C, Rossini D, Dell’Aquila E, Lonardi S, Conca E, Re MD, et al. Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan. JAMA Oncol (2019) 5:343–50. doi: 10.1001/jamaoncol.2018.5080
162. Sunakawa Y, Nakamura M, Ishizaki M, Kataoka M, Satake H, Kitazono M, et al. RAS mutations in circulating tumor DNA and clinical outcomes of rechallenge treatment with anti-EGFR antibodies in patients with metastatic colorectal cancer. Jco Precis Oncol (2020) 4:898–911. doi: 10.1200/po.20.00109
163. Sartore-Bianchi A, Pietrantonio F, Lonardi S, Mussolin B, Rua F, Crisafulli G, et al. Circulating tumor DNA to guide rechallenge with panitumumab in metastatic colorectal cancer: the phase 2 CHRONOS trial. Nat Med (2022) 28:1612–8. doi: 10.1038/s41591-022-01886-0
164. Kagawa Y, Kotani D, Bando H, Takahashi N, Hamaguchi T, Kanazawa A, et al. Plasma RAS dynamics and anti-EGFR rechallenge efficacy in patients with RAS/BRAF wild-type metastatic colorectal cancer: REMARRY and PURSUIT trials. J Clin Oncol (2022) 40:3518–8. doi: 10.1200/jco.2022.40.16_suppl.3518
165. Vlachou MS, Mauri D, Zarkavelis G, Ntellas P, Tagkas C, Gkoura S, et al. Plasma ctDNA RAS status selects patients for anti-EGFR treatment rechallenge in metastatic colorectal cancer: A meta-analysis. Exp Oncol (2021) 43:252–6. doi: 10.32471/exp-oncology.2312-8852.vol-43-no-3.16592
Keywords: Metastatic colorectal cancer, epidermal growth factor receptor, clonal evolution, liquid biopsy, systemic treatment, molecular oncology, maintenance
Citation: Doleschal B, Petzer A and Rumpold H (2022) Current concepts of anti-EGFR targeting in metastatic colorectal cancer. Front. Oncol. 12:1048166. doi: 10.3389/fonc.2022.1048166
Received: 19 September 2022; Accepted: 26 October 2022;
Published: 17 November 2022.
Edited by:
Andreas Seeber, Innsbruck Medical University, AustriaReviewed by:
Davide Ciardiello, University of Campania Luigi Vanvitelli, ItalyKeith R. Laderoute, Consultant, Redwood City, CA, United States
Maria Ouzounova, Institut Curie, France
Copyright © 2022 Doleschal, Petzer and Rumpold. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Holger Rumpold, aG9sZ2VyLnJ1bXBvbGRAb3JkZW5za2xpbmlrdW0uYXQ=
†ORCID: Holger Rumpold, orcid.org/0000-0002-4118-0111
 Bernhard Doleschal
Bernhard Doleschal Andreas Petzer1
Andreas Petzer1 Holger Rumpold
Holger Rumpold