
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 25 November 2022
Sec. Thoracic Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.1043862
 Lu-Shan Peng1†
Lu-Shan Peng1† Sai-Li Duan2†
Sai-Li Duan2† Run-Qi Li3
Run-Qi Li3 Dan Wang1
Dan Wang1 Ying-Ying Han1
Ying-Ying Han1 Tao Huang1
Tao Huang1 Yu-Pei Yu1
Yu-Pei Yu1 Chun-Lin Ou1,4*
Chun-Lin Ou1,4* Jun-Pu Wang1,3,5*
Jun-Pu Wang1,3,5*Background: The GSDM family includes six members, GSDMA, GSDMB, GSDMC, GSDMD, GSDME (DFNA5), and PJVK (Pejvakin, DFNB59), which can induce pyroptosis, thereby regulating the tumorigenesis of various cancers. However, the clinical characteristics and role of the GSDM family in LUAD are not well understood.
Methods: In this study, several important bioinformatics databases were used to integrate the analysis of the expression, prognostic value, and immune infiltration of GSDMs in LUAD. These databases include UALCAN, DiseaseMeth, GEPIA, THPA, cBioPortal, TIMER, WebGestalt, STRING database, and Cytoscape.
Results: The findings from the UALCAN database revealed that the expression of all six GSDMs based on the tumor stage in LUAD was increased (particularly GSDMD). Our IHC results verified it. Additionally, the DiseaseMeth database showed that the methylation levels of GSDMA, GSDMB, GSDMC, and GSDMD were decreased. The expression of six GSDMs was related to shorter overall survival in patients with LUAD, according to the GEPIA database. The cBioPortal database was further used to explore the alteration rate and correlated genes in LUAD. Subsequently, these genes were subjected to functional enrichment and protein-protein interaction network analyses. We identified that the GSDM family regulate several signaling pathways, including immune-associated signaling pathways. According to tumor-infiltrating immune cell analysis from the TIMER database, GSDM family members are associated with the infiltration of important immune cells and their signature markers.
Conclusions: GSDM family may be prognostic markers and novel strategies for the treatment of LUAD.
Lung cancer is the leading cause of cancer-related deaths (18.0%) among the top ten cancer types causing over 1.7 million deaths worldwide in 2020 (1). Lung adenocarcinoma (LUAD) recently surpassed lung squamous cell carcinoma as the most common lung cancer subtype (2), accounting for 30%–35% of the total number of lung cancer cases. Consequently, novel biomarker exploration may be necessary for diagnosing and treating LUAD (3).
In humans, there are six gasdermins (GSDMs), including GSDMA, GSDMB, GSDMC, GSDMD, GSDME (DFNA5), and PJVK (Pejvakin, DFNB59). In addition to PJVK, the other five GSDMs promote cell death mainly through pyroptosis, a lytic and pro-inflammatory form of cell death (4). Pyroptosis is also known as “gasdermin-mediated programmed cell death” (5). In the past, cell death was mainly divided into apoptosis and necrosis. In recent years, scientists have proposed that cell death also involves other forms of programmed necroptosis, including pyroptosis (6).Pyroptosis can be accompanied by immune responses and the release of pro-inflammatory factors, which play a vital role in various genetic diseases, autoinflammatory diseases, and tumors (7, 8). A recent study found that GSDMs are a family of histones that play a decisive role in pyroptosis (9). They can induce inflammatory responses, assist the transition from apoptosis to pyroptosis (10, 11), and play an important role in the occurrence and development of various tumors (12, 13). For example, Zhou et al. found that gasdermin-mediated pyroptosis may enhance antitumor immunity through pyroptosis-induced inflammation (14, 15). Similar function was suggested in GSDMB and GSDME (16) (17) (18). However, the exact mechanism of the role of GSDM family tumors is not clear. There are also reports on GSDMs family members about their cancer-promoting effect. GSDMD could be used as a prognostic marker in different cancers, including adrenocortical carcinoma, kidney renal clear cell carcinoma, brain lower grade glioma, hepatocellular carcinoma, skin cutaneous melanoma, and rectal adenocarcinoma (19).High GSDMC expression was significantly associated with the tumorigenesis of clear cell renal cell carcinoma (20). In LUAD, a study found that high GSDMD expression may promote the occurrence and development of the tumors. This indicates a poor prognosis of LUAD and may serve as an independent prognostic biomarker (21). Upregulated GSDMC expression is an independent indicator of poor first progression (FP) and poor overall survival (OS) in patients with LUAD, which is regarded as a promising predictor of poor prognosis (22). Therefore, the GSDM family might play a role in promoting tumorigenesis in LUAD. In LUAD, there are only 2 researches, both of them believe that GSDM could promote the development of LUAD. A study found that high GSDMD expression may promote the occurrence and development of the tumors. This indicates a poor prognosis of LUAD and may serve as an independent prognostic biomarker (21). Upregulated GSDMC expression is an independent indicator of poor first progression (FP) and poor overall survival (OS) in patients with LUAD, which is regarded as a promising predictor of poor prognosis (22). We think different role of GSDM family members toward tumor is related to the heterogeneity of tumors. Subsequently, we speculated that the GSDM family might play a role in promoting tumorigenesis in LUAD. However, detailed, and systematic studies on the role of the GSDM family in LUAD is lacking.
In the present study, the function of GSDMs in LUAD was analyzed using several bioinformatics databases, such as gene expression profiling interactive analysis (GEPIA), UALCAN, and cBioPortal to explore the role of GSDMs in LUAD, which might help identify the role of GSDMs in LUAD and provide new strategies for LUAD treatment.
We use Gene Expression Profiling Interactive Analysis (GEPIA), a cancer big data analysis website based on TCGA and GTEx data, to assess the expression of the GSDMs family in LUAD tissues and in normal tissues. The analysis contents include tumor/normal differential expression profiling, expression distribution, pathological staging, survival analysis, similar genes, gene expression correlation and dimensionality reduction analysis, etc. (23). Also, the roles of GSDMs family in pathological stage and prognosis are evaluated by using the GEPIA.
The Human Protein Atlas (HPA) is a useful database for studying protein localization and expression in human tissues and cells, which contains more than 10 million images (24). By using this database, we obtain the immunohistochemistry images of all six GSDMs family members in LUAD and in normal tissues.
UALCAN is an interactive web-portal which perform in-depth analyses of TCGA gene expression data. TCGA level 3 RNA-seq and 31 cancer types clinical data are used in this database (25). We use this database to assess the mRNA expression of six GSDMs family members according to the LUAD stage, and the mRNA expression of six GSDMs family members in different LUAD histology subtypes.
The cBioPortal is a prevalent open-source translational research platform, providing genomics from more than 200 cancers (26, 27). We use Cbioportal to explore the coexpression profiles and genetic alterations of the GSDMs family in LUAD.
WebGestalt is a tool which can interpretate the gene lists derived from large scale -omics studies. This database is often used for functional enrichment analysis (28). We use this database to analyze the functional enrichment of GSDMs co-expression genes.
Timer is a comprehensive database with visualization functions of tumor infiltrating immune cells (29). In this study, Timer was used to get the scatterplots that show the correlation of GSDMs with 6 types of infiltrated immune cells including dendritic cells, macrophages, CD4+ T cells, CD8+ T cells, B cells, and neutrophils, and their signature markers.
DiseaseMeth is a database including DNA methylation data in 162 diseases and 4,9 949 High-throughput profiles samples (30). We use this database to explore the methylation levels of the GSDMs family members.
STRING is a database which can be used to explore all publicly available sources of protein-protein interaction information, and to complement these with computational prediction (31). Cytoscape is an open-source platform for network analysis and visualization. We use STRING and Cytoscape together to construct the PPIs network.
The analysis of NK cell infiltration in LUAD was performed by single sample GSEA (ssGSEA) method from R package GSVA (version 3.6.3) (https://portal.gdc.cancer.gov/). P values were determined by the Spearman and Wilcoxon rank sum test. We think P values < 0.05 were statistically significant.
10 LUAD specimens were achieved from the department of pathology, Xiangya Hospital, Central South University. All specimens were fixed in 10% neutral-buffered formalin, embedded in paraffin, and stained with hematoxylin and eosin for histological examination. During initial case diagnosis, immunohistochemical staining was performed by using formalin-fixed paraffin-embedded tissue sections and the Ventana Benchmark automated immunostainer (Ventana Medical System Inc, Roche, Tucson, AZ, USA). We used GSDMD (Proteintech, #20770–1-AP, 1:200) as primary antibodies, and incubated with it overnight at 4 °C. The sections were then incubated with the secondary antibodies and DAB regents for staining (Zhong Shan Golden Bridge Biotechnology, Beijing, China).
First, the TIMER database was used to compare the expression levels of six GSDMs in cancerous and normal tissues. It was found that GSDMs were overexpressed in several tumors, including cholangiocarcinoma, bladder urothelial, kidney renal clear cell carcinoma, and LUAD (Figure 1). Among them, we found that in LUAD, all six GSDM family members are upregulated.
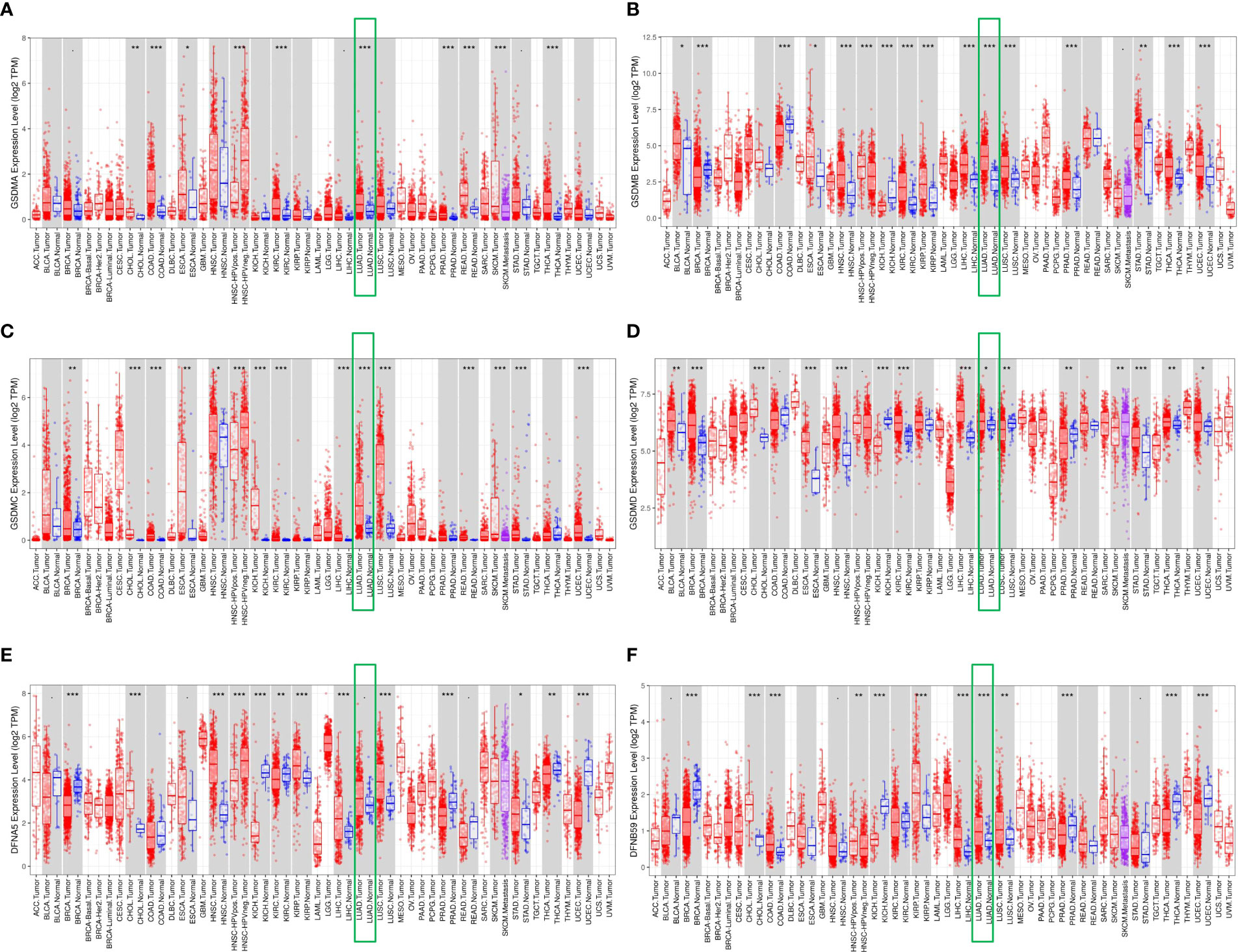
Figure 1 Differential expression of GSDMs between tumor and adjacent normal tissues. The expression of (A) GSDMA, (B) GSDMB, (C) GSDMC, (D) GSDMD, (E) GSDME, and (F) PJVK in different tumors and normal tissues based on the Timer database. * means p<0.05, ** means p<0.005, and *** means p<0.001.
For targeted research on LUAD, the UALCAN database was used to evaluate mRNA expression levels of the GSDM family in normal and LUAD tissues. The mRNA expression levels of all six GSDM family members were higher in LUAD tissues than in normal tissues. Among the six members, the expression level of GSDMD was the highest, whereas that of GSDMC was the lowest (Figure 2A). The Human Protein Atlas database (THPA) was used to analyze the protein expression of GSDMs in LUAD and normal tissues through the immunohistochemistry (IHC) staining. The results showed that the expression levels of GSDMA, GSDMB, GSDMC, GSDMD, and GSDME were increased in LUAD tissues (Figure 2B), especially GSDMD, which was significantly increased. The IHC data related to PJVK were not retrieved from the THPA database. Collectively, the expression of GSDMs was significantly high (P < 0.005) in LUAD tissues. To verify the expression of GSDMs in LUAD. We made IHC of GSDMD in 10 specimens (due to the result of the expression of GSDMD was the highest. The result supported the result retrieved from database (Figure 2C).
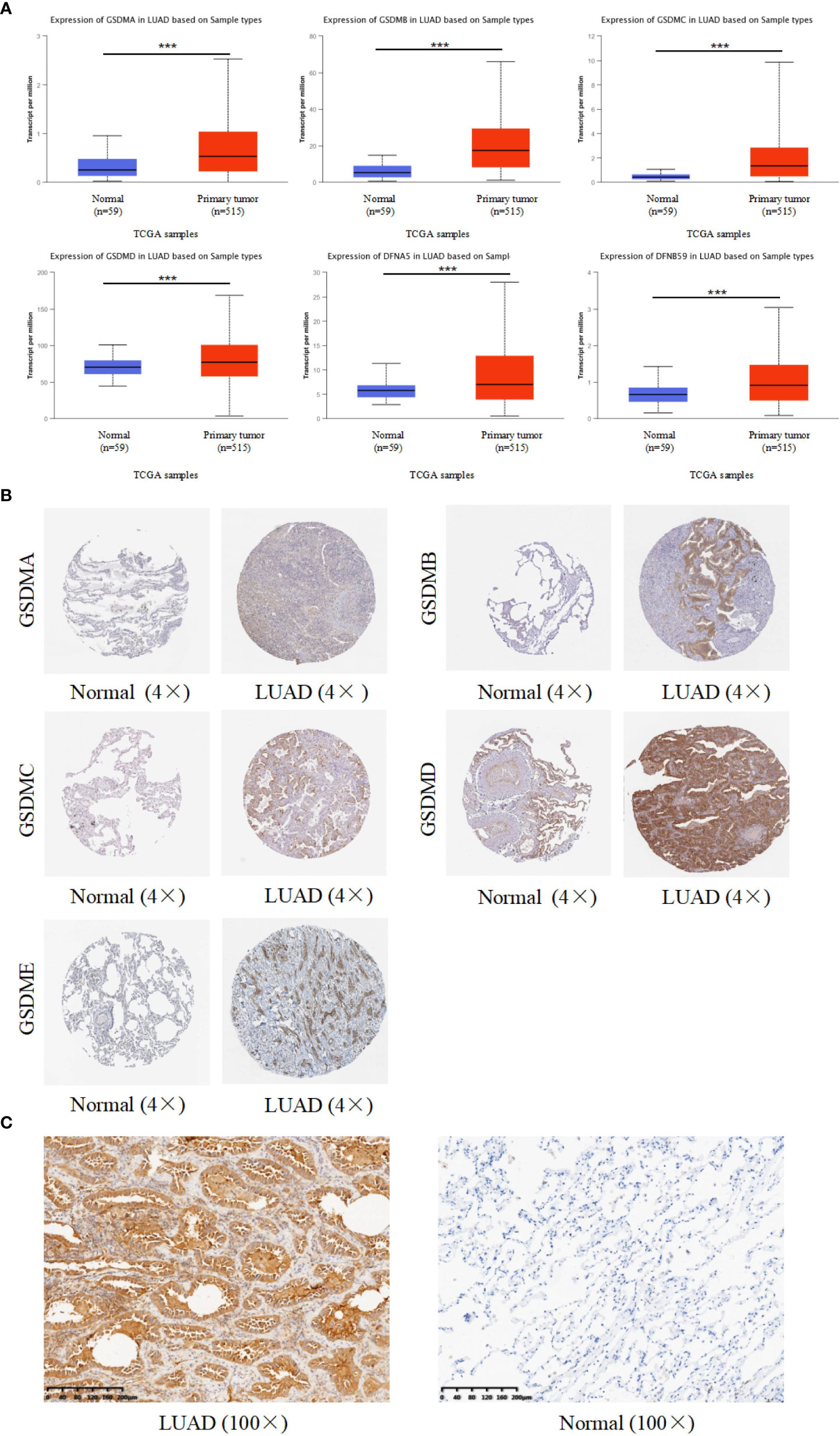
Figure 2 Expression of six GSDM family members in LUAD. (A) The UALCAN database was used to retrieve the GSDMs’ mRNA expression profiles. (B) The THPA database was used to collect IHC images of GSDM family members from LUAD tissues and normal lung tissues, representing the GSDM protein expression levels. (C) The IHC result of GSDMD expression in LAUD and adjacent normal lung tissue. *** means p<0.001.
Furthermore, the influence of GSDMs on clinical-pathological characterizations in patients with LUAD was elucidated. The tumor stage describes the severity of malignant tumors according to the primary tumor and its degree of dissemination inside the body, which helps understand the prognosis and outcome of the disease. Based on the analysis of UALCAN, the expression of all GSDM family members was increased in tumor-stage 1–3 subgroups, and the expression of GSDMA, GSDMB, GSDMC, and PJVK was higher in all four tumor-stage subgroups than in normal subgroups (Figure 3A). This indicate that the abnormal expression of GSDMs may be involved in the progression of LUAD through certain mechanisms. Furthermore, the expression of six GSDMs based on LUAD histological subtypes was analyzed (Figure 3B). The results showed that the expression of GSDMs was elevated in most of the LUAD histological subtypes. This further suggests that the abnormal expression of GSDMs has an important influence on the clinicopathological features of different types of LUAD.
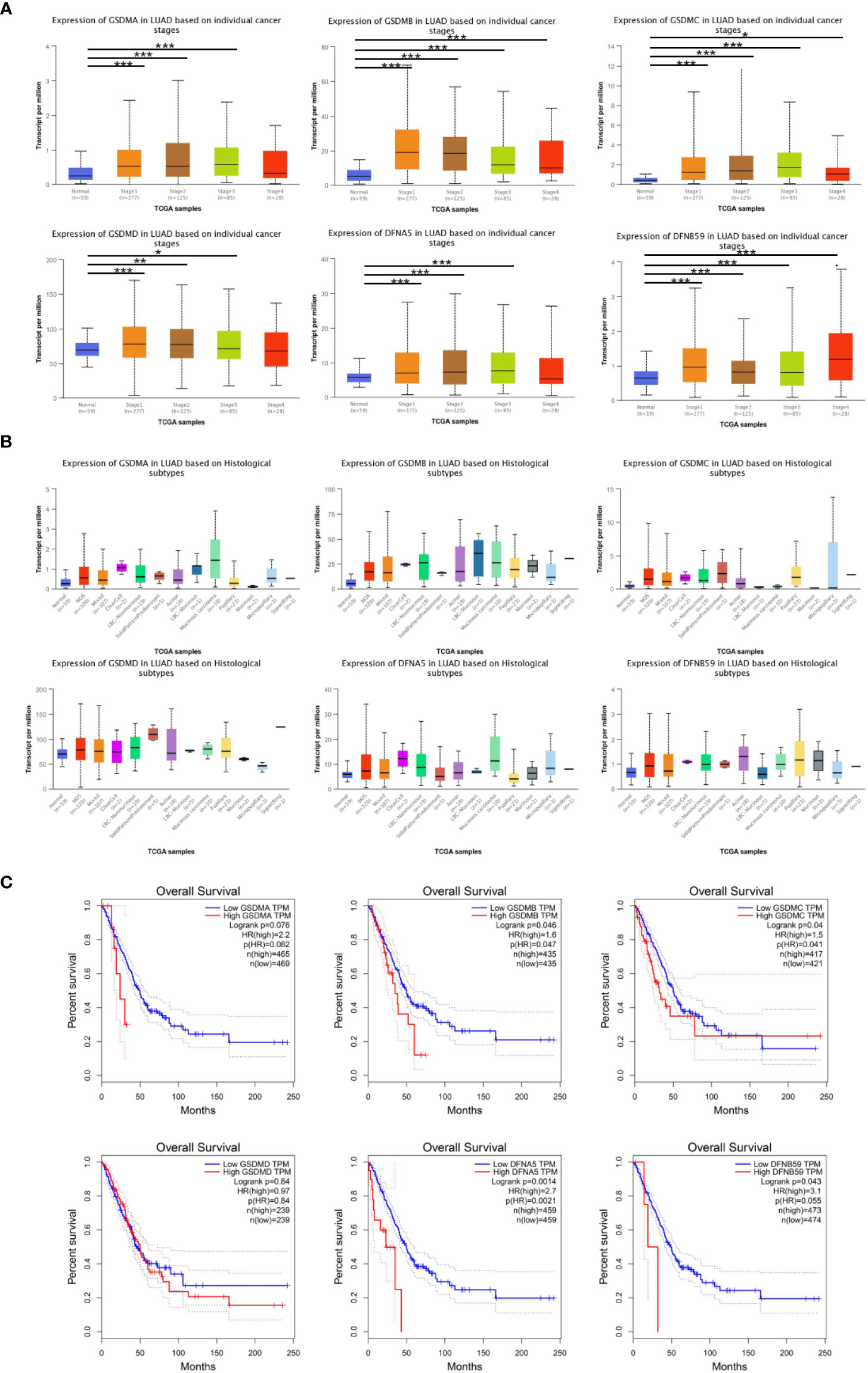
Figure 3 GSDMs family member-associated clinical pathology characteristics of patients with LUAD. (A) The associations between GSDM transcript levels and individual cancer stages of LUAD. (B) The expression of GSDM family members in different LUAD histological subtypes. (C) The GEPIA database was used to analyze the relationships between GSDM transcript levels and OS of LUAD patients. * means p<0.05, ** means p<0.005, and *** means p<0.001.
In addition, the GEPIA database was used to explore the prognostic role of the GSDM family in patients with LUAD. The overall survival (OS) curves and interactive correlations are presented in Figure 3C. This analysis suggested that, in patients with LUAD, high expression of GSDMs was significantly related to shorter OS, except for GSDMC (P < 0.05). These results suggest that GSDMs could provide prognostic information in patients with LUAD.
To gain insight into the influence of GSDMs on LUAD. genetic alterations in GSDMs were analyzed (Figure 4A). Among the 566 enrolled patients with LUAD, 257 (45%) had altered GSDM gene expression. The highest and lowest genetic alteration rates were 21% (GSDMC) and 6% (GSDMA), respectively. Amplification and high mRNA expression are the two main types of alterations in the GSDM family members.
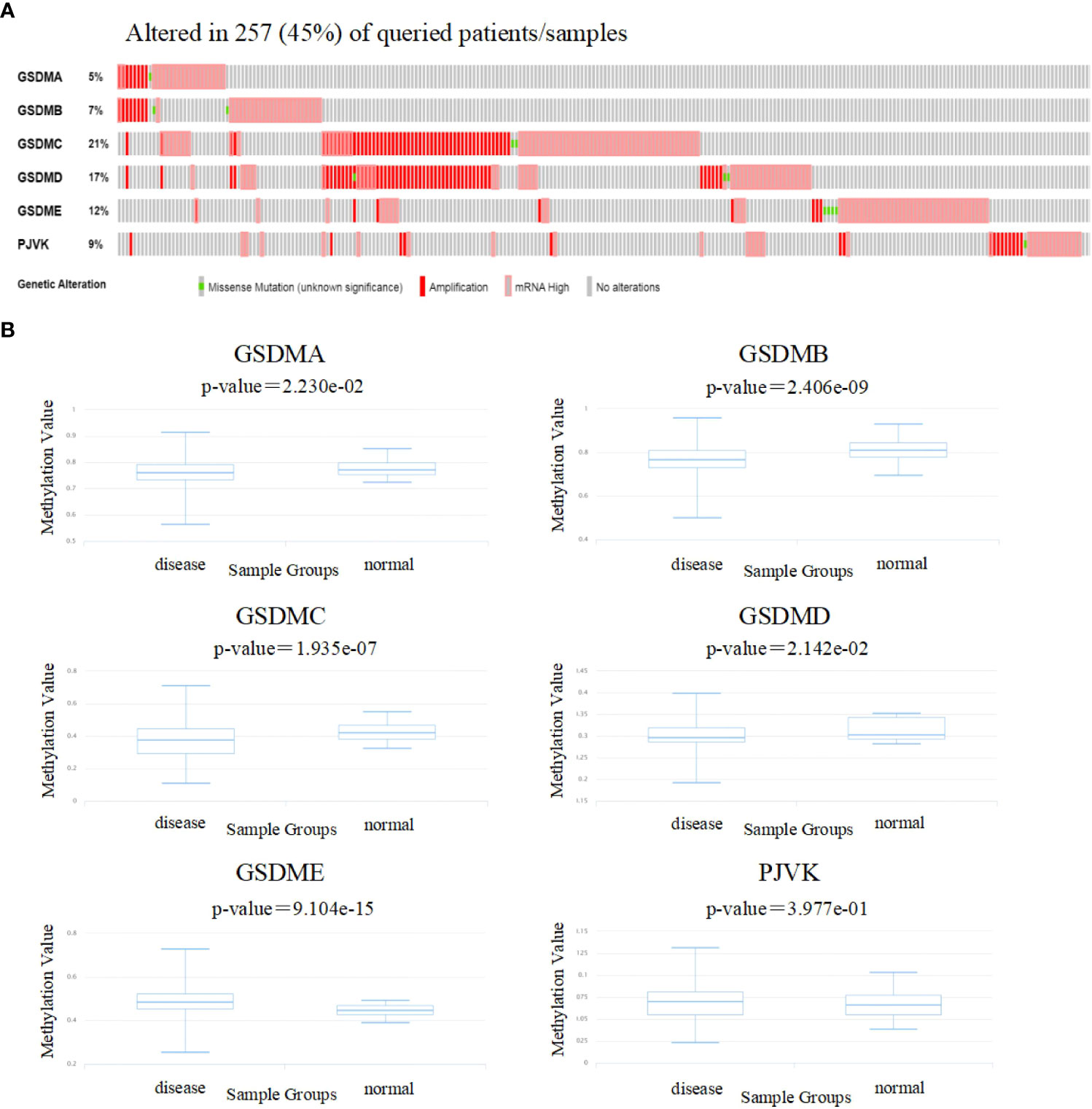
Figure 4 Genetic alterations and methylation levels of GSDM family members in LUAD. (A) The cBioPortal database was used to achieve the genetic alteration profiles of GSDMs. (B) The DiseaseMeth database was used to evaluate the DNA methylation levels of GSDM family members in LUAD.
Next, the DiseaseMeth database was used to evaluate the methylation levels of the GSDMs (Figure 4B). Several studies have shown that abnormal DNA methylation is closely associated with carcinogenesis and tumorigenesis (32, 33). Thus, oncogenes may be activated by reduced methylation levels. In this study, we found that the methylation levels of GSDMA, GSDMB, GSDMC, and GSDMD were lower in LUAD tissues compared to that in normal tissues, which may be associated with their abnormal expression in LUAD.
Based on this, the genes correlated with GSDMs were further explored using the cBioPortal database. The results showed that there were 19316 genes correlated with GSDMs.
To further explore the role of GSDMs in LUAD, functional analysis was conducted. A total of 19316 correlated genes were downloaded, 177 of which were screened according to |log ratio| >0.8 and p<0.05, using the cBioPortal database (Supplementary Table 1). Based on this, the protein-protein interaction (PPI) network was constructed using the STRING database and Cytoscape (Figure 5A). Furthermore, the WebGestalt database was used to conduct Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. It was found that there were ten potential signaling pathways, and folate biosynthesis, bile secretion, and IL-17 signaling pathways were ranked in the top three, indicating that these pathways may be strongly associated with the progression of LUAD (Figure 5B). Next, Gene Ontology (GO) annotation from the same database (Figure 5C) suggested that GSDMs were mainly enriched in the membrane. Biological regulation was the most enriched biological process category. In addition, metabolic processes, responses to stimuli, multicellular organismal processes, localization, cell communication, cellular component organization, and developmental processes were also highly enriched.
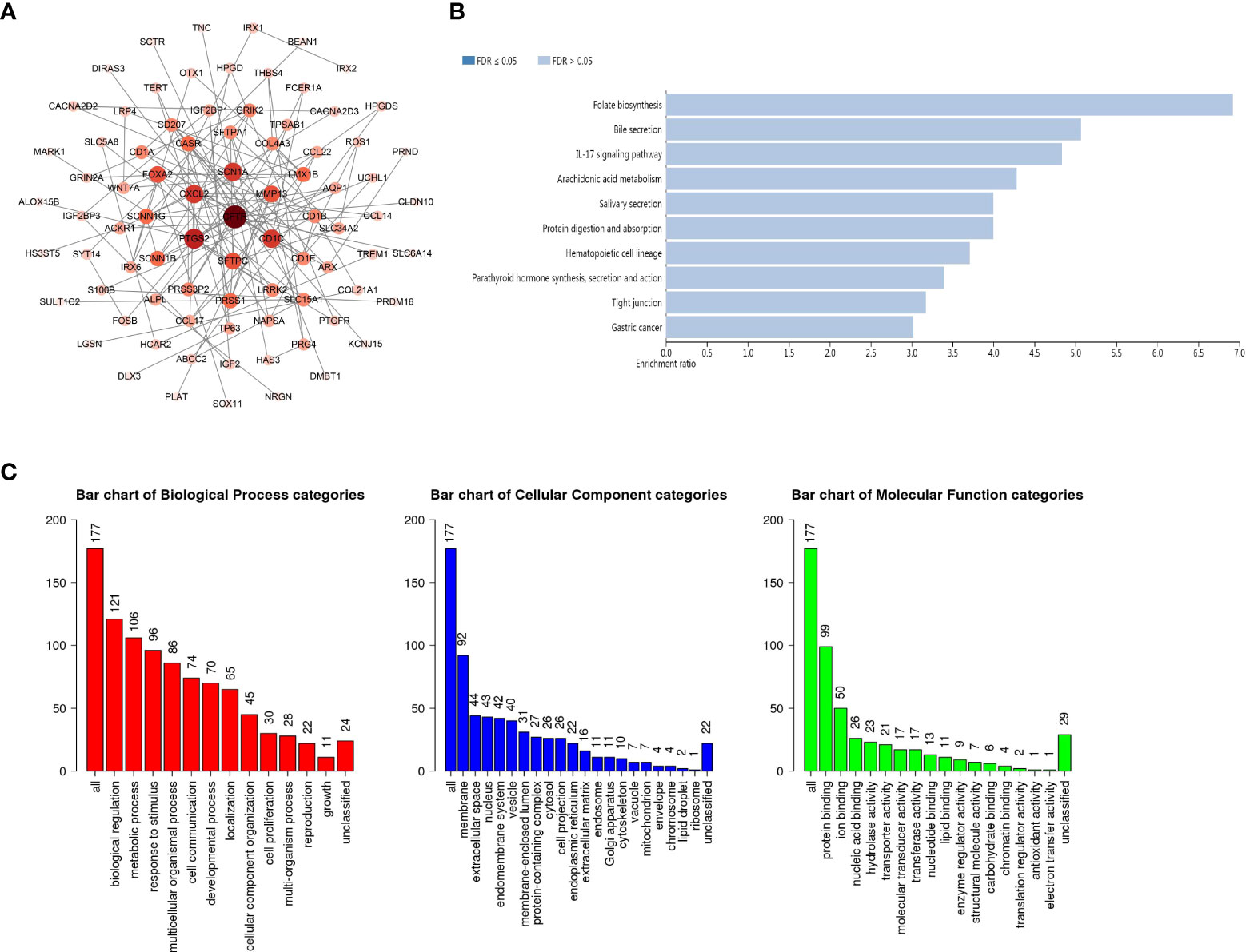
Figure 5 PPI network and the biological pathways of GSDM family members. (A) STRING and Cytoscape were used to build the PPI network. The WebGestalt database was used for the GSDM-related co-expressed molecules to analyze the GO functional enrichment analysis (B) and the KEGG pathway analysis (C).
In recent years, immune cells have been considered a crucial part of the tumor microenvironment, which is the interval environment for tumor initiation and survival (34, 35). The functional enrichment analysis suggested that some pathways and biological processes of GSDMs are related to immunity. Therefore, the Timer database was used to explore the correlation between tumor-infiltrating immune cells and the GSDM family. This study revealed that the expression of GSDMA and GSDME was positively correlated with the infiltration of all six immune cells, including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells (Figures 6A, E). GSDMB was positively correlated with the infiltration of B cells (Cor = 0.232, p < 0.05), CD4+ T cells (Cor = 0.318, p < 0.05), and dendritic cells (Cor = 0.108, p < 0.05) (Figure 6B). However, GSDMC and the infiltration of different immune cells were not closely correlated (Figure 6C). GSDMD was found to be negatively correlated with the infiltration of CD8+ T cells (Cor = -0.109, p < 0.05) but positively correlated with the infiltration of B cells (Cor = 0.223, p < 0.05) and CD4+ T cells (Cor = 0.276, p < 0.05) (Figure 6D). Figure 6F shows that PJVK was positively correlated with CD4+ T cells (Cor = 0.122, p < 0.05), whereas it was negatively correlated with other immune cells, except B cells. Natural killer cell (NK) is an important immune cell and closely related with anti-tumor response. Unlike T and B cells, NK cells are a class of lymphocytes that can non-specifically kill tumor cells without pre-sensitization. We analyzed the infiltration of NK cells in LUAD by using ssGSEA (Figure 6G). We found that GSDMC was negatively correlated with NK cells (p=0.007). While GSDMA (p < 0.001) and GSDME (p < 0.001) were found positively correlated with NK cells. These results indicated a potential relationship between the role of GSDMs in LUAD and the infiltration of different immune cells.
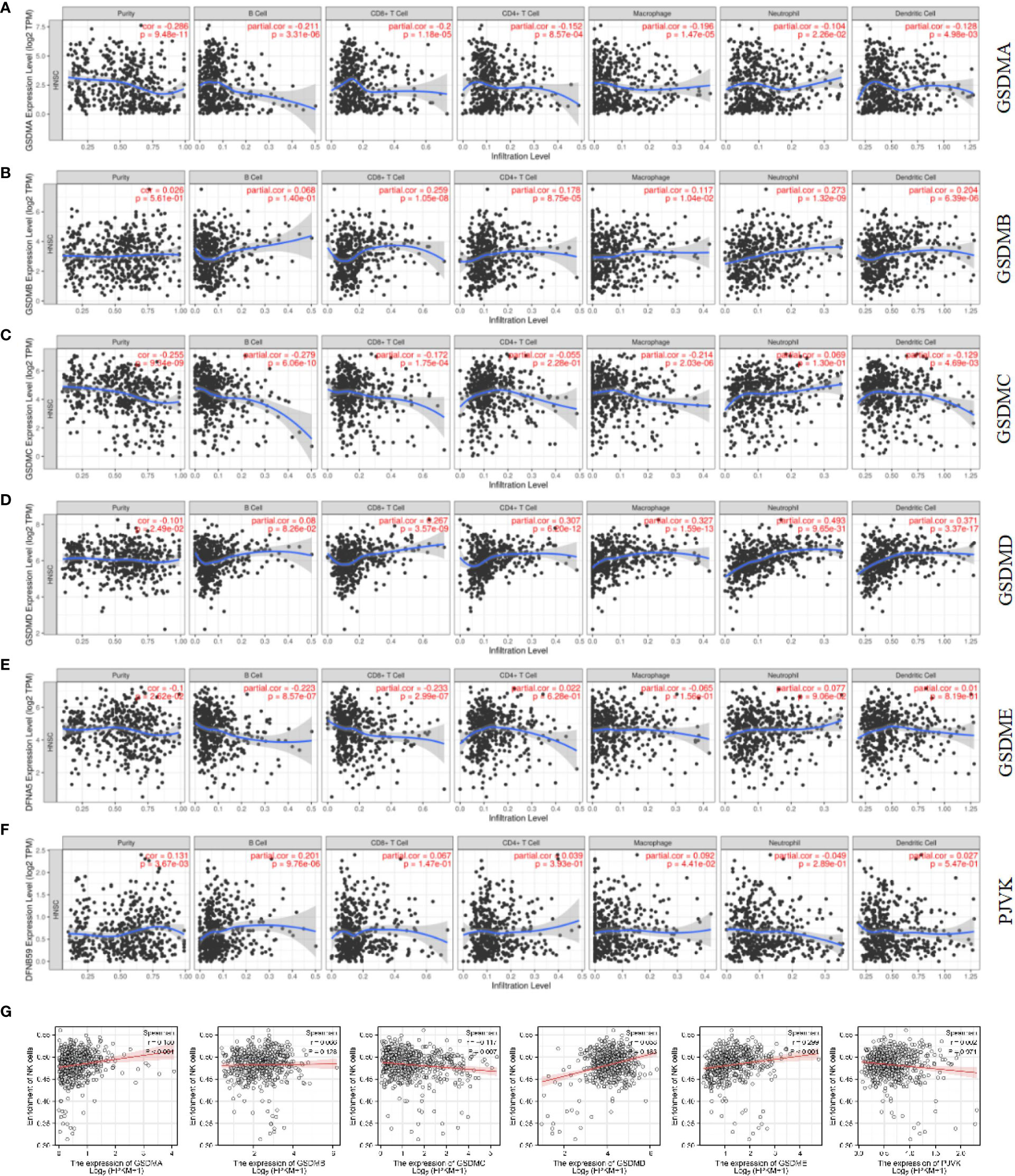
Figure 6 Correlation between immune cell infiltration and GSDM mRNA expression levels. The TIMER database was used to evaluate the association of GSDMA (A), GSDMB (B), GSDMC (C), GSDMD (D), GSDME (E), and PJVK (F) expression and immune cell infiltration. The ssGSEA method from R package GSVA was used to evaluate the association of six GSDM family members and NK cell (G).
Additionally, the relationship between GSDMs and signature markers of various immune infiltrating cells in LUAD was explored using the TIMER database (Table 1). The results showed that, among the six members, the expression of GSDMA, GSDMB, GSDMD, and GSDME was significantly correlated with most signature marker genes of various immune cells in LUAD (GSDME expression was negatively correlated).
On the basis, we used STRING database to explore the relationship between gasdermin family and immune check-points related genes in LUAD to clarify whether this family could adjust tumorigenesis through immune related mechanism. We found that GSDMA (Figure 7A), GSDMB (Figure 7B), and GSDMD (Figure 7C) are correlated with immune related gene, including IKZF3, IL18R1, IL1RL1, IL18, NLRP9, and CASP5. Among them, NLRP9 and CASP5 could work as mediators of programmed cell death. So, we further speculated that GSDM family members play roles in immune related mechanism in LUAD.
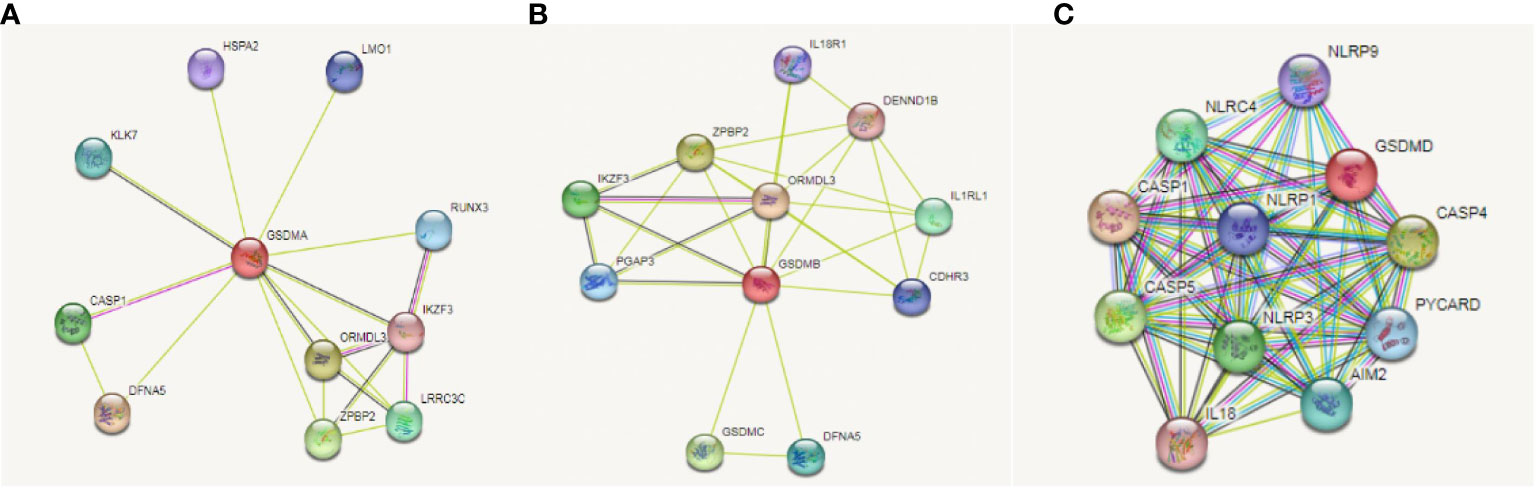
Figure 7 Correlation between GSDMs and immune check-points related genes in LUAD. We used the STRING database to analyze the correlation between GSDMA (A), GSDMB (B), and GSDMD (C) and immune related genes.
The GSDM family regulates various physiological and pathological processes such as cell differentiation, coagulation, inflammation, and tumorigenesis, which is thought to be related to pyroptosis (36). Further studies have confirmed the irreplaceable role of GSDMs in tumor progression and development (37, 38). In recent years, LUAD gradually replaced lung squamous cell carcinoma as the most common subtype of malignant lung tumors (39).It is also the most common type of malignant tumor in the worldwide population (40).Currently, there are limited studies on GSDMs in LUAD. A study found that GSDMD deficiency reduces the cytolytic capacity of CD8+ T cells, and GSDMD is required for an optimal cytotoxic T lymphocytes response in cancer cells (41). The high expression of GSDMD may promote the occurrence and development of LUAD. This indicates a poor prognosis of LUAD and can be seen as an independent prognostic biomarker (21). Upregulated GSDMC expression is an independent indicator of poor FP and OS in patients with LUAD and is regarded as a promising predictor of prognosis (22). These studies confirmed the conclusions drawn from a series of databases used in the current study.
After comparing the expression levels of six GSDM in cancerous and normal tissues, this study revealed that the mRNA expression of GSDMs was significantly increased in LUAD, indicating that GSDMs may be involved in the tumorigenesis of LUAD. Our results showed that among the six members, GSDMD has the highest expression level, while GSDMC has the lowest expression level. Additionally, in terms of protein expression, IHC results showed that the expression of GSDMB, GSDMC, GSDMD, and GSDME in LUAD tissues was higher than that in normal tissues, particularly GSDMD. To validate this result, we performed IHC of GSDMD on 10 specimens. The experimental results support the search results from the database that GSDMD is indeed highly expressed in LUAD tissues. Next, a study on the prognosis of patients with LUAD, based on the expression levels of GSDMs, was conducted. The results showed that expression of GSDMs was elevated in most histological subtypes of LUAD. This further suggests that the abnormal expression of GSDMs has an important impact on the clinicopathological characteristics of different types of LUAD. The OS curves showed that the GSDMB, GSDMC, GSDMD, and GSDME were significantly associated with the poor prognosis of patients with LUAD. Their expression levels were almost all significantly elevated at different stages of LUAD. It was further found that the methylation levels of GSDMC and GSDMD in these four genes were decreased in LUAD. The mutation rates of GSDMC and GSDMD in LUAD were as high as 16% and 19%, respectively. Through KEGG enrichment analysis, we found that 10 potential signaling pathways were associated with GSDM, among which folic acid biosynthesis, bile secretion and IL-17 signaling pathways ranked the top three, suggesting that these pathways may be closely related to the progression of LUAD. Combined with previous studies, it was speculated that GSDMC and GSDMD have the potential to become new biomarkers and therapeutic targets for LUAD, and these two genes may deserve better and deeper exploration.
Based on the studies discussed, the cBioPortal database was used to explore the correlated genes of the GSDM family, and, thereafter, the WebGestalt database was used for functional enrichment analysis. This study demonstrated that some GSDM-associated pathways and biological processes are related to cellular immunity, which is crucial for tumorigenesis. Subsequently, the TIMER database was used to study several important immune cells and the relationship between the signature markers of these immune cells and the expression of GSDMs in LUAD. The results revealed that GSDM expression is correlated with various immune cells and most of their marker genes in LUAD. Later, we explored the relationship between the gasdermin family and immune checkpoint-related genes in LUAD to elucidate whether this family can regulate tumorigenesis through immune-related mechanisms. We found that GSDMA, GSDMB, and GSDMD were associated with immune-related genes. Among these genes, NLRP9 and CASP5 can act as mediators of programmed cell death. Therefore, we further speculated that GSDM family members play a role in the immune-related mechanisms of LUAD. We plan to explore the critical role of immune cell infiltration in antitumor immunity in the future.
In conclusion, the present study assessed the molecular profiles of the GSDM family using bioinformatic databases. The results suggested that the expression of GSDMs (especially GSDMD) may be involved in the development and progression of LUAD. These findings may help enhance our understanding of LUAD and provide further details on prognosis prediction and treatment strategies for patients with LUAD.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
C-LO and J-PW contributed to the conception and design of the study. L-SP and S-LD performed resource analysis, and wrote the first draft of the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by the National Natural Science Foundation of China (No. 81602167 and 81903032), the China Postdoctoral Science Foundation (2020M672520), the Hunan Provincial Natural Science Foundation of China (No. 2021JJ31100 and 2021JJ4101), and the Science and Technology Program Foundation of Changsha City (No. kq2004085).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1043862/full#supplementary-material
GSDM, Gasdermin; LUAD, lung adenocarcinoma; GEPIA, gene expression profiling interactive analysis; THPA, the Human Protein Atlas; TIMER, tumor immune estimation resource; FP, first progression; OS, overall survival; KEGG, Encyclopedia of Genes and Genomes; GO, Gene Ontology.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Bray, global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
2. Tomášková H, Horáček J, Šlachtová H, Šplíchalová A, Riedlová P, Dalecká A, et al. Analysis of histopathological findings of lung carcinoma in Czech black coal miners in association with coal workers' pneumoconiosis. Int J Environ Res Public Health (2022) 19(2):710. doi: 10.3390/ijerph19020710
3. Wang Y, Nie H, He X, Liao Z, Zhou Y, Zhou J, et al. The emerging role of super enhancer-derived noncoding RNAs in human cancer. Theranostics (2020) 10:11049–62. doi: 10.7150/thno.49168
4. Sarrió D, Martínez-Val J, Molina-Crespo Á., Sánchez L, Moreno-Bueno G. The multifaceted roles of gasdermins in cancer biology and oncologic therapies. Biochim Biophys Acta Rev Cancer (2021) 1876:188635. doi: 10.1016/j.bbcan.2021.188635
5. Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ (2018) 25:486–541. doi: 10.1038/s41418-017-0012-4
6. Tsuchiya K. Switching from apoptosis to pyroptosis: Gasdermin-elicited inflammation and antitumor immunity. Int J Mol Sci (2021) 22(1):426. doi: 10.3390/ijms22010426
7. Jorgensen I, Rayamajhi M, Miao EA. Programmed cell death as a defence against infection. Nat Rev Immunol (2017) 17:151–64. doi: 10.1038/nri.2016.147
8. Peng L, Wang D, Han Y, Huang T, He X, Wang J, et al. Emerging role of cancer-associated fibroblasts-derived exosomes in tumorigenesis. Front Immunol (2021) 12:795372. doi: 10.3389/fimmu.2021.795372
9. Du T, Gao J, Li P, Wang Y, Qi Q, Liu X, et al. Pyroptosis, metabolism, and tumor immune microenvironment. Clin Transl Med (2021) 11:e492. doi: 10.1002/ctm2.492
10. He H, Yi L, Zhang B, Yan B, Xiao M, Ren J, et al. USP24-GSDMB complex promotes bladder cancer proliferation via activation of the STAT3 pathway. Int J Biol Sci (2021) 17:2417–29. doi: 10.7150/ijbs.54442
11. Wang CJ, Tang L, Shen DW, Wang C, Yuan QY, Gao W, et al. The expression and regulation of DFNA5 in human hepatocellular carcinoma DFNA5 in hepatocellular carcinoma. Mol Biol Rep (2013) 40:6525–31. doi: 10.1007/s11033-013-2581-8
12. Privitera G, Rana N, Scaldaferri F, Armuzzi A, Pizarro TT. Novel insights into the interactions between the gut microbiome, inflammasomes, and gasdermins during colorectal cancer. Front Cell Infect Microbiol (2021) 11:806680. doi: 10.3389/fcimb.2021.806680
13. Bourdonnay E, Henry T. Transcriptional and epigenetic regulation of gasdermins. J Mol Biol (2022) 434:167253. doi: 10.1016/j.jmb.2021.167253
14. Liu X, Xia S, Zhang Z, Wu H, Lieberman J. Channelling inflammation: gasdermins in physiology and disease. Nat Rev Drug Discov (2021) 20:384–405. doi: 10.1038/s41573-021-00154-z
15. Wang Q, Wang Y, Ding J, Wang C, Zhou X, Gao W, et al. A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature (2020) 579:421–6. doi: 10.1038/s41586-020-2079-1
16. Zhang Z, Zhang Y, Xia S, Kong Q, Li S, Liu X, et al. Gasdermin e suppresses tumour growth by activating anti-tumour immunity. Nature (2020) 579:415–20. doi: 10.1038/s41586-020-2071-9
17. Zhou Z, He H, Wang K, Shi X, Wang Y, Su Y, et al. Granzyme a from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science (2020) 368(6494):eaaz7548. doi: 10.1126/science.aaz7548
18. Liu Y, Fang Y, Chen X, Wang Z, Liang X, Zhang T, et al. Gasdermin e-mediated target cell pyroptosis by CAR T cells triggers cytokine release syndrome. Sci Immunol 5 (2020) 5(43):eaax7969. doi: 10.1126/sciimmunol.aax7969
19. Qiu S, Hu Y, Dong S. Pan-cancer analysis reveals the expression, genetic alteration and prognosis of pyroptosis key gene GSDMD. Int Immunopharmacol (2021) 101:108270. doi: 10.1016/j.intimp.2021.108270
20. Cui YQ, Meng F, Zhan WL, Dai ZT, Liao X. High expression of GSDMC is associated with poor survival in kidney clear cell cancer. BioMed Res Int (2021) 2021:5282894. doi: 10.1155/2021/5282894
21. Gao J, Qiu X, Xi G, Liu H, Zhang F, Lv T, et al. Downregulation of GSDMD attenuates tumor proliferation via the intrinsic mitochondrial apoptotic pathway and inhibition of EGFR/Akt signaling and predicts a good prognosis in non−small cell lung cancer. Oncol Rep (2018) 40:1971–84. doi: 10.3892/or.2018.6634
22. Wei J, Xu Z, Chen X, Wang X, Zeng S, Qian L, et al. Overexpression of GSDMC is a prognostic factor for predicting a poor outcome in lung adenocarcinoma. Mol Med Rep (2020) 21:360–70. doi: 10.3892/mmr.2019.10837
23. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45 (2017) 45(W1):W98–102. doi: 10.1093/nar/gkx247
24. Thul PJ, Lindskog C. The human protein atlas: A spatial map of the human proteome. Protein Sci (2018) 27:233–44. doi: 10.1002/pro.3307
25. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (2017) 19:649–58. doi: 10.1016/j.neo.2017.05.002
26. Unberath P, Knell C, Prokosch HU, Christoph J. Developing new analysis functions for a translational research platform: Extending the cBioPortal for cancer genomics. Stud Health Technol Inform (2019) 258:46–50.
27. Nie H, Wang Y, Yang X, Liao Z, He X, Zhou J, et al. Clinical significance and integrative analysis of the SMC family in hepatocellular carcinoma. Front Med (Lausanne) (2021) 8:727965. doi: 10.3389/fmed.2021.727965
28. Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47 (2019) 47(W1):W199–205. doi: 10.1093/nar/gkz401
29. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res (2020) 48:W509–w514. doi: 10.1093/nar/gkaa407
30. Xing J, Zhai R, Wang C, Liu H, Zeng J, Zhou D, et al. DiseaseMeth version 3.0: a major expansion and update of the human disease methylation database. Nucleic Acids Res (2022) 50:D1208–15. doi: 10.1093/nar/gkab1088
31. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res (2019) 47:D607–13. doi: 10.1093/nar/gky1131
32. Huang Y, Yu Z, Zheng M, Yang X, Huang H, Zhao L. Methylation−associated inactivation of JPH3 and its effect on prognosis and cell biological function in HCC. Mol Med Rep (2022) 25(4):124. doi: 10.3892/mmr.2022.12640
33. Fan X, Tao S, Li Q, Deng B, Tan QY, Jin H. The miR-23a/27a/24-2 cluster promotes postoperative progression of early-stage non-small cell lung cancer. Mol Ther Oncolytics (2022) 24:205–17. doi: 10.1016/j.omto.2021.12.014
34. Shen L, Xiao Y, Tian J, Lu Z. Remodeling metabolic fitness: Strategies for improving the efficacy of chimeric antigen receptor T cell therapy. Cancer Lett (2022) 529:139–52. doi: 10.1016/j.canlet.2022.01.006
35. Qu Y, Wang X, Bai S, Niu L, Zhao G, Yao Y, et al. The effects of TNF-α/TNFR2 in regulatory T cells on the microenvironment and progression of gastric cancer. Int J Cancer (2022) 150:1373–91. doi: 10.1002/ijc.33873
36. Zou J, Zheng Y, Huang Y, Tang D, Kang R, Chen R. The versatile gasdermin family: Their function and roles in diseases. Front Immunol (2021) 12:751533. doi: 10.3389/fimmu.2021.751533
37. Sun Q, Yang J, Xing G, Sun Q, Zhang L, He F. Expression of GSDML associates with tumor progression in uterine cervix cancer. Transl Oncol (2008) 1:73–83. doi: 10.1593/tlo.08112
38. Miguchi M, Hinoi T, Shimomura M, Adachi T, Saito Y, Niitsu H, et al. Gasdermin c is upregulated by inactivation of transforming growth factor β receptor type II in the presence of mutated apc, promoting colorectal cancer proliferation. PloS One (2016) 11:e0166422. doi: 10.1371/journal.pone.0166422
39. Zhou H, He X, He Y, Ou C, Cao P. Exosomal circRNAs: Emerging players in tumor metastasis. Front Cell Dev Biol (2021) 9:786224. doi: 10.3389/fcell.2021.786224
40. Shi Q, Shi QN, Xu JW, Wang HY, Li YJ, Zhang XX, et al. rs9390123 and rs9399451 influence the DNA repair capacity of lung cancer by regulating PEX3 and PHACTR2−AS1 expression instead of PHACTR2. Oncol Rep (2022) 47(3):59. doi: 10.3892/or.2022.8270
Keywords: GSDM family, lung adenocarcinoma, methylation, immune infiltration, biomarker, prognosis
Citation: Peng L-S, Duan S-L, Li R-Q, Wang D, Han Y-Y, Huang T, Yu Y-P, Ou C-L and Wang J-P (2022) Prognostic value and immune infiltration of the gasdermin family in lung adenocarcinoma. Front. Oncol. 12:1043862. doi: 10.3389/fonc.2022.1043862
Received: 14 September 2022; Accepted: 09 November 2022;
Published: 25 November 2022.
Edited by:
Nguyen Quoc Khanh Le, Taipei Medical University, TaiwanReviewed by:
Shiyu Xia, California Institute of Technology, United StatesCopyright © 2022 Peng, Duan, Li, Wang, Han, Huang, Yu, Ou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun-Lin Ou, b3VjaHVubGluQGNzdS5lZHUuY24=; Jun-Pu Wang, d2FuZy1qcDIwMTNAY3N1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.