- 1Department of Disease Prevention, Hainan Province Hospital of Traditional Chinese Medicine, Haikou, China
- 2Hainan Clinical Research Center for Preventive Treatment of Diseases, Haikou, China
- 3Department of Food Science and Technology Centers, National University of Singapore (Suzhou) Research Institute, Suzhou, China
- 4Department of Research and Development, Zhejiang Zhongwei Medical Research Center, Hangzhou, China
Diabetes mellitus often results in several complications, such as diabetic kidney disease (DKD) and end-stage renal diseases (ESRDs). Cancer patients often have the dysregulated glucose metabolism. Abnormal glucose metabolism can enhance the tumor malignant progression. Recently, lncRNAs have been reported to regulate the key proteins and signaling pathways in DKD development and progression and in cancer patients with diabetes. In this review article, we elaborate the evidence to support the function of lncRNAs in development of DKD and diabetes-associated cancer. Moreover, we envisage that lncRNAs could be diagnosis and prognosis biomarkers for DKD and cancer patients with diabetes. Furthermore, we delineated that targeting lncRNAs might be an alternative approach for treating DKD and cancer with dysregulated glucose metabolism.
Introduction
Noncoding RNAs have been known to play an essential role in development of many diseases (1, 2). Noncoding RNAs include short non-coding RNAs, such as microRNAs (miRNAs), small interfering RNA (siRNAs), piwi-interacting RNA (piRNAs), transfer RNA (tRNAs), small nuclear RNA (snRNAs) and small nucleolar RNA (snoRNAs), and long non-coding RNAs (lncRNAs), which is based on their length (3). LncRNAs often have more than 200 nucleotides and cancer serve as signal molecules, decoy molecules, guide molecules, and scaffold molecules to perform their functions via regulation of gene expression at epigenetic, transcriptional and post-transcriptional levels (4, 5). Accumulated evidence has dissected that lncRNAs participate in cellular biological processes via regulation of protein degradation and governing gene transcription as well as controlling protein coding sections (6–8). Dysregulated lncRNAs have been reported to participate in numerous diseases, including cancer, inflammatory bowel disease, cardiovascular disease, neurological disorders and diabetes (9–14).
Diabetes mellitus (DM) has become a major health problem in the world, which often results in several complications, such as diabetic kidney disease (DKD) (15). DKD is often known as diabetic nephropathy. DM has three types: type 1 diabetes, type 2 diabetes and gestational diabetes (GDM). Type 1 diabetes is insulin-dependent and often appears during childhood and adolescence. Type 2 diabetes often appears in older adults due to that pancreas does not make enough insulin or cells respond poorly to insulin. GDM often happens during the pregnancy after insulin secretion is not enough. DKD is one of causes to develop end stage kidney disease (ESKD) and kidney failure (16). It has been known that chronic stimuli such as high glucose in the bloodstream can lead to pathological gene modulation and DKD in diabetic patients (17). EMT and endothelial-mesenchymal transition (EndMT) have been characterized to integrate into the fibrosis and DKD (18, 19). EMT is a process in which epithelial cells acquire mesenchymal characteristics after various stimulations. Similarly, EndMT is a process in which endothelial cells have the phenotype toward mesenchymal cells, which often appears in cardiovascular diseases. Cancer patients often have the dysregulated glucose metabolism. Abnormal glucose metabolism can enhance the tumor malignant progression (20).
Recently, noncoding RNAs, including lncRNAs, have been reported to regulate the key proteins and signaling pathways in DM and DKD development and progression as well as in cancer with diabetes (21–24). In this review article, we elaborate the evidence to support the function of lncRNAs in development of DKD and cancer patients with diabetes. Moreover, we envisage that lncRNAs could be diagnostic and prognosis biomarkers for DKD and diabetes-related cancers. Furthermore, we delineated that targeting lncRNAs might be an alternative approach for treating DKD and diabetes-associated cancer.
Role of lncRNAs in DKD
Emerging evidence has suggested that lncRNAs are useful for precision medicine in DKD (25–28). Zhang and colleagues used the integrate biological, computational, and statistical strategies to analyze the pathogenesis and progression of DKD through analysis of regulatory networks including miRNAs, lncRNAs and mRNAs (29). This study reported that 127 lncRNAs were changes in DKD, among which 26 were decreased and 101 were increased. In particular, this work identified that miR-223-3p might be a biomarker for prediction of DKD disease process (29).
LncRNA HOTAIR
Evidence showed that lncRNA HOTAIR is critically involved in DKD development (30). One group used several mouse models, such as podocyte-specific Hotair knockout mice, streptozotocin-induced diabetes in mice, and the db/db mouse model of type 2 diabetes. In these mouse models, glomerular HOTAIR was upregulated. Depletion of Hotair in podocytes did not affect structure, ultrastructure, function of kidneys (30). In mouse podocytes, high glucose treatment increased the expression of HOTAIR. Interestingly, silencing of HOTAIR did not affect the kidney damage in diabetic mice. Moreover, HOTAIR expression was linked to HOXC11 expression in human kidney tissues according to a bioinformatic assay (30). Notably, the serum level of HOTAIR was increased in type 2 DM patients (31). HOTAIR can be a useful biomarker in prediction of diabetic retinopathy and DKD in patients with type 2 DM. In addition, HOTAIR facilitated high glucose-mediated fibrosis and proliferation of mesangial cells via affecting miR-147a/WNT2B axis in diabetic nephropathy (32). The role of HOTAIR in DKD needs to be ascribed to validate its function in the pathogenesis of DKD.
LncRNA GAS5
Wang et al. reported that lncRNA GAS5 promoted renal tubular epithelial fibrosis via sponging miR-96-5p (33). Renal fibrosis is often observed in DKD. Higher expression of lncRNA GAS5 was reported in renal proximal tubular cells after TGF-β1 treatment. The kidneys of high-fat diet (HFD)/streptozotocin (STZ) mice had the upregulation of lncRNA GAS5 (33). Silencing of lncRNA GAS5 reduced renal fibrosis via inhibition of miR-96-5p. Consistently, DKD mice had the lower expression of miR-96-5p, leading to upregulation of fibronectin. Hence, depletion of lncRNA GAS5 could have antifibrosis via sponging miR-96-5p and regulating fibronectin. Zhang et al. found that lncRNA GAS5 attenuated TGF-β-mediated renal fibrosis by inhibition of collagen type 1 an fibronectin via targeting the Smad3/miR-142-5p axis (34). LncRNA GAS5 suppressed fibrosis and cell proliferation through attenuating miR-221 and upregulating SIRT1 expression in diabetic nephropathy (35). LncRNA GAS5 inhibited pyroptosis and oxidative stress in renal tubular cells after high glucose stimulation (36). LncRNA GAS5 alleviated fibrosis via inhibition of MMP9 by recruitment of EZH2 in diabetic nephropathy (37). Altogether, modulation of lncRNA GAS5 might be useful for preventing DKD.
LncRNA MALAT1
LncRNA MALAT1 has been identified to play key roles in DKD pathophysiology (38). One work assessed urinary albumin in 136 patients with type 2 DM and 25 normal people. This work found that urinary lncRNA MALAT1 was positively associated with urinary podocalyxin, synaptopodin, UACR (urinary albumin), NAG (N-acetyl-D-glucosaminidase), KIM-1 (kidney injury molecule 1), miR-21, miR-93, miR-29a (38). LncRNA MALAT1 was negatively correlated with eGFR, miR-29a and miR-93. In addition, urinary lncRNA MIAT was positively linked to miR-29a, miR-93 and eGFR, while lncRNA MIAT was negatively associated with miR-21, miR-124, UACR, NAG and KIM-1 (38). In line with this report, the expression of lncRNA MALAT1 in PBMC was increased in type 2 DM and DKD (39). MALAT1 was associated with ACR, HbA1c, SOD, creatinine, α1-MG and β2-MG in type 2 DM and DKD patients. MALAT1 in combination with ACR, α1-MG and creatinine could be helpful for prediction of DKD in DM patients (39). MALAT1 enhanced diabetic nephropathy via suppression of miR-15b-5p and upregulation of TLR4 signaling (40).
MALAT1 activated LIN28 and Nox4/AMPK/mTOR pathway, resulting in promotion of renal tubular injury in diabetic nephropathy (41). Huang et al. reported that MALAT1 aggravated renal fibrosis via modulation of miR-2355-3p/IL6ST axis in diabetic nephropathy (42). One study showed that podocyte injury could be due to abnormal MALAT1 expression and subsequent dysregulated let-7f and KLF5 in diabetic nephropathy (43). MALAT1 was also reported to participate in high glucose-mediated HK-2 cell EMT via activation of Wnt/β-catenin pathway and injury (44). Consistently, MALAT1 was involved in high glucose-mediated podocyte injury in diabetic nephropathy via its interaction with β-catenin (45). MALAT1 aggravated high glucose-triggered EndMT and fibrosis through regulation of miR-145/ZEB2 axis (46). Additionally, MALAT1 participated in high glucose-mediated HK-2 cell injury via interplay with Foxo1 to affect SIRT expression (47).
LncRNA MIAT
Urinary lncRNA MIAT was positively linked to miR-29a, miR-93 and eGFR, while lncRNA MIAT was negatively associated with miR-21, miR-124, UACR, NAG and KIM-1 in type 2 DM (38). Depletion of lncRNA MIAT mitigated apoptosis and inflammation in podocyte after high glycose stimulation through modulating miR-130a-3p and TLR4 pathway (48). Ablation of lncRNA MIAT ameliorated fibrosis and cell proliferation via suppression of E2F3 expression in diabetic nephropathy (49). Loss of lncRNA MIAT blocked podocyte injury and mitotic damage in diabetic nephropathy (50). However, one study showed that lncRNA MIAT blocked the high glucose-mediated cell damage and activation of NF-κB via sponging miR-182-5p and elevating the GPRC5A expression in diabetic nephropathy, leading to suppression of diabetic nephropathy progression (51).
LncRNA NEAT1
Evidence has suggested that lncRNA NEAT1 governed renal tubular EMT via regulation of the ERK1/2 signaling pathway in DKD (52). LncRNA NEAT1 was increased in BSA-treated HK2 cells and HFD/STZ-induced DKD mice. Depletion of NEAT1 suppressed the expression of the EMT-related markers, such as vimentin and a-SMA, and the renal fibrosis-associated markers, including TGF-β1 and CTGF (52). LncRNA NEAT1 regulated DKD progression via modulation of the ERK1/2 signaling pathway. Li et al. discovered that NEAT1 interacted with miR-129 to promote renal fibrosis via upregulation of collagen type 1 and promotion of EMT process (53). Additionally, urinary lncRNA NEAT1 was positively correlated with miR-21, miR-124, KIM-1, synaptopodin, and NAG in type 2 DM. Urinary lncRNA NEAT1 had a negative association with miR-29a, miR-93 and eGFR (38).
LncRNA NEAT1 activated Akt/mTOR pathway and accelerated cell fibrosis and proliferation in diabetic nephropathy (54). LncRNA NEAT1 enhanced EMT and accumulation of extracellular matrix in diabetic nephropathy via sponging miR-27b-3p and ZEB1 (55). Ablation of lncRNA NEAT1 attenuated proliferation, fibrosis and inflammation of mouse mesangial cells in diabetic nephropathy (56). In addition, lncRNA NEAT1 accelerated diabetic nephropathy occurrence and progression via suppression of miR-23c (57). LncRNA NEAT1 affected pyroptosis via targeting the miR-34c and NLRP3 in diabetic nephropathy (58). One group showed that lncRNA NEAT1 accelerated high glucose-triggered hypertrophy in mesangial cells through modulating miR-222-3p and CDKN1B (59). Yang et al. found that lncRNA NEAT1 enhanced tubular epithelial cell damage in kidney through regulation of mitophagy by targeting miR-150-5p and DRP1 in diabetic nephropathy (60). Moreover, lncRNA NEAT1 promoted fibrosis, inflammation, proliferation and oxidative stress by modification of the miR-423/5p and GLIPR2 pathway in diabetic nephropathy (61). Hence, lncRNA NEAT1 might be a promising therapeutic target for the treatment of DKD.
LncRNA TUG1
LncRNA TUG1 has been identified to play a crucial role in DKD progression (62). One study revealed that ChREBP controlled lncRNA TUG1 transcription when glucose levels were increased in podocytes (62). Besides ChREBP, other coregulates, such as MXD1, MLX and HDAC1, were increased at the TUG1 promoter in response to high glucose exposures. This work suggested that ChREBP coordinated glucose homeostasis via regulation of lncRNA TUG1 (62). In addition, lncRNA TUG1 was discovered to regulate mitochondrial bioenergetics via regulation of PGC-1α in podocytes in diabetic nephropathy (63, 64). Overexpression of TUG1 in podocytes ameliorated diabetes-mediated chronic kidney disease in mice (63). Zhang et al. reported that knockdown of lncRNA TUG1 retarded the EMT of renal tubular epithelial cells via targeting miR-141-3p/β-catenin (65). Another work also demonstrated that lncRNA TUG1 reduced accumulation of extracellular matrix by sponging miR-377 and targeting PPARγ in diabetic nephropathy (66). Moreover, lncRNA TUG1 interacted with miR-9 and upregulated SIRT1, resulting in protection of podocytes from high glucose-triggered apoptosis and mitochondrial dysfunction (67). Urinary lncRNA TUG1 was positively associated with miR-29a, miR-93 and eGFR in type 2 DM, while lncRNA TUG1 had a negative association with miR-21, miR-124, podocalyxin, NAG and synaptopodin (38).
LncRNA TUG1 participated in regulation of podocyte apoptosis via modulation of TRAF5 pathway in diabetic nephropathy rats (68). LncRNA TUG1 influenced podocyte apoptosis via promotion of endoplasmic reticulum stress in diabetic nephropathy progression (69). Additionally, lncRNA TUG1 repressed the PI3K/AKT pathway and suppressed the fibrosis and proliferation in mesangial cells in diabetic nephropathy (70). LncRNA TUG1 inhibited the expression of miR-21 and enhanced the TIMP3 expression, leading to ameliorating diabetic nephropathy (71). LncRNA TUG1 repressed the PU.1/RTN1 pathway and improved diabetic nephropathy (72). Notably, lncRNA TUG1 affected high glucose-stimulated renal epithelial cell injury via regulation of endoplasmic reticulum stress by targeting miR-29c-3p and SIRT1 in diabetic nephropathy (73).
LncRNA MEG3
LncRNA MEG3 has been revealed to regulate glucose metabolisms in diabetic mice (74). STZ-mediated diabetic mice had an increased expression of lncRNA MEG3, which was associated with the podocyte numbers. Mice with knockdown of MEG3 in podocyte had improved renal physiological and histopathological features (74). These mice also had a reduced mitochondrial translocation of Drp1 and a decreased podocyte damage (74). Overexpression of lncRNA MEG3 in podocyte led to podocyte injury and enhanced mitochondria damage and upregulated expression and phosphorylation of Drp1 (74). LncRNA MEG3 increased fibrosis and inflammation through regulating miR-181a, Egr-1 and TLR4 in diabetic nephropathy (75). Moreover, lncRNA MEG3 sponged miR-145 and impacted the development of diabetic nephropathy (76). Strikingly, lncRNA MEG3 inactivated the Wnt/β-catenin pathway and reduced podocyte injury in diabetic nephropathy (77). Therefore, MEG3 plays an essential role in diabetic mice and DKD.
LncRNA KCNQ1OT1
Downregulation of KCNQ1OT1 attenuated oxidative stress and inflammation and reduced pyroptosis in renal tubular epithelial cells after high glucose stimulations through regulation of miR-506-3p (78). One study showed that KCNQ1OT1 participated in governing fibrosis, apoptosis and proliferation via regulation of miR-18b-5p and SORBS2 and NF-κB in diabetic nephropathy (79). Another study revealed that KCNQ1OT1 sponged miR-18b and increased the expression of HMGA2 and led to controlling high glucose-triggered oxidative stress, proliferation and extracellular matrix promotion in mesangial cells (80). In addition, KCNQ1OT1 was reported to accelerate diabetic nephropathy development via modulating miR-93-5p/ROCK2 axis (81). Xu et al. dissected that KCNQ1OT1 governed cell oxidative stress, proliferation, inflammation and extracellular matrix enhancement through miR-147a/SOX6 pathway in diabetic nephropathy (82). Recently, KCNQ1OT1 expression in diabetic nephropathy was increased and associated with activation of MEK/ERK pathway in diabetic nephropathy (83). LncRNA KCNQ1OT1 participates in DKD development and progression.
LINC00472
Wang et al. used the data from Gene Expression Omnibus (GEO) database to explore the differentially expressed profiles between DKD patients and the normal patients. This study found that among 252 lncRNAs, 14 lncRNAs were differentially expressed. LINC00472 was identified to be differentially expressed in DKD patients, suggesting that LINC00472 could act as the diagnostic biomarkers for DKD patients (84). It is required to explore the detailed role of LINC00472 in DKD.
LncRNA NONMMUG023520.2 and NONMMUG032975.2
Smad3 has been reported to enhance the development of type 2 DM and involve in DKD pathogenesis (85–87). One group discovered the Smad3-associated genes via analysis of whole transcriptome profile in three types of transgenic mouse models, including Smad3 WT-db/db, Smad3 KO-db/db, Smad3+/- db/db mice (88). Smad3 KO-db/db mice displayed dysregulated genes involved in metabolism and RNA splicing, Smad3+/- db/db mice exhibited dysregulated genes that were associated with cell cycle and cell division (88). Two lincRNAs, NONMMUG023520.2 and NONMMUG032975.2, were further validated to be linked to the pathogenesis of diabetic nephropathy. Moreover, Upk1b, Psca and Gdf15 were identified to be correlated with diabetic nephropathy development [26. Without a doubt, further investigation is pivotal to determine the function of lncRNA NONMMUG023520.2 and NONMMUG032975.2 in DKD development and pathogenesis.
LncRNA 254693
Increased evidence has revealed that lncRNA ENSG00000254693 participated in DKD development (89). One research used RNA sequencing data and observed numerous differentially expressed lncRNAs in renal specimens of DKD. Among these dysregulated lncRNAs, lncRNA ENSG00000254693 was drastically changed. Moreover, DKD patients had higher expression of lncRNA ENSG00000254693 (89). Consistently, lncRNA ENSG00000254693 was upregulated in human podocytes after high glucose exposures. Depletion of lncRNA 254693 attenuated apoptosis, inflammation, and podocyte injury that were induced by high glucose (89). Furthermore, lncRNA 254693 was found to combine with HuR, and depletion of lncRNA 254693 reduced HuR levels. Interestingly, silencing of HuR reduced the expression and stability of lncRNA 254693 and alleviate podocyte injury, apoptosis and inflammation (89). Therefore, lncRNA 254693 might be a predicted factor for DKD treatment.
Other lncRNAs in DM and DKD
LncRNA CASC2 expression in renal samples and serum was identified to be downregulated in type 2 DM patients with chronic renal failure (90). Low serum level of CASC2 was associated with higher incidence of kidney failure, indicating that serum lncRNA CASC2 could be a biomarker for prediction of the occurrence of kidney failure in type 2 DM patients (90). By RT-PCR analysis in 77 type 2 DM patients, 60 diabetic nephropathy and 60 healthy people, one group found that lncRNA PANDAR in the serum was upregulated compared with healthy people (91). PANDAR expression was linked to the level of proteinuria and glomerular filtration rate. PANDAR might serve as a biomarker for judgement of DKD prognosis (91). Yang et al. reported the differential expression profiles of circulating lncRNAs in DM and DKD patients. Compared with healthy persons, 245 lncRNAs were increased, while 680 lncRNAs were decreased in the serum of DM patients. Compared with diabetes patients, 45 and 813 lncRNAs were increased and decreased in the serum of DKD patients, respectively (92). LncRNA ARAP1-AS1 expression was elevated during DM and DKD progression, while lncRNA ARAP1-AS2 was decreased in DM and DKD progression (92). Hence, circulating lncRNA ARAP1-AS1 and ARAP1-AS2 might predict the progression of DM and DKD.
Another group identified that lncRNA KCNQ1OT1 was abnormally elevated in PBMCs of diabetic nephropathy, which was correlated with the activation of MEK/ERK pathway (83). LncRNA CASC2 modulated cell proliferation, oxidative stress and extracellular matrix promotion in human mesangial cells upon high glucose treatment through regulation of miR-133b and FOXP1 expressions (93). LncRNA CASC2 mitigated diabetic nephropathy development via sponging miR-144 and regulating SOCS2 expression (94). LncRNA CASC2 ablated cell inflammation, proliferation and fibrosis in glomerular mesangial cells upon high glucose exposures via targeting miR-135a-5p/TIMP3 pathway and JNK pathway (95).
LncRNAs regulate glucose metabolism in cancer
Competing endogenous RNAs (ceRNA) can compete for shared miRNAs to modulate the expression of other RNA transcripts. A ceRNA network profile has identified the several lncRNAs for classifying diabetic pancreatic cancer form non-diabetic pancreatic cancer, including HOTAIR, CECR7, UCA1, suggesting that lncRNAs are important predictors for diabetic pancreatic cancer (96). In the following paragraphs, we will discuss the association between lncRNAs and glucose metabolisms in human cancer (Figure 1).
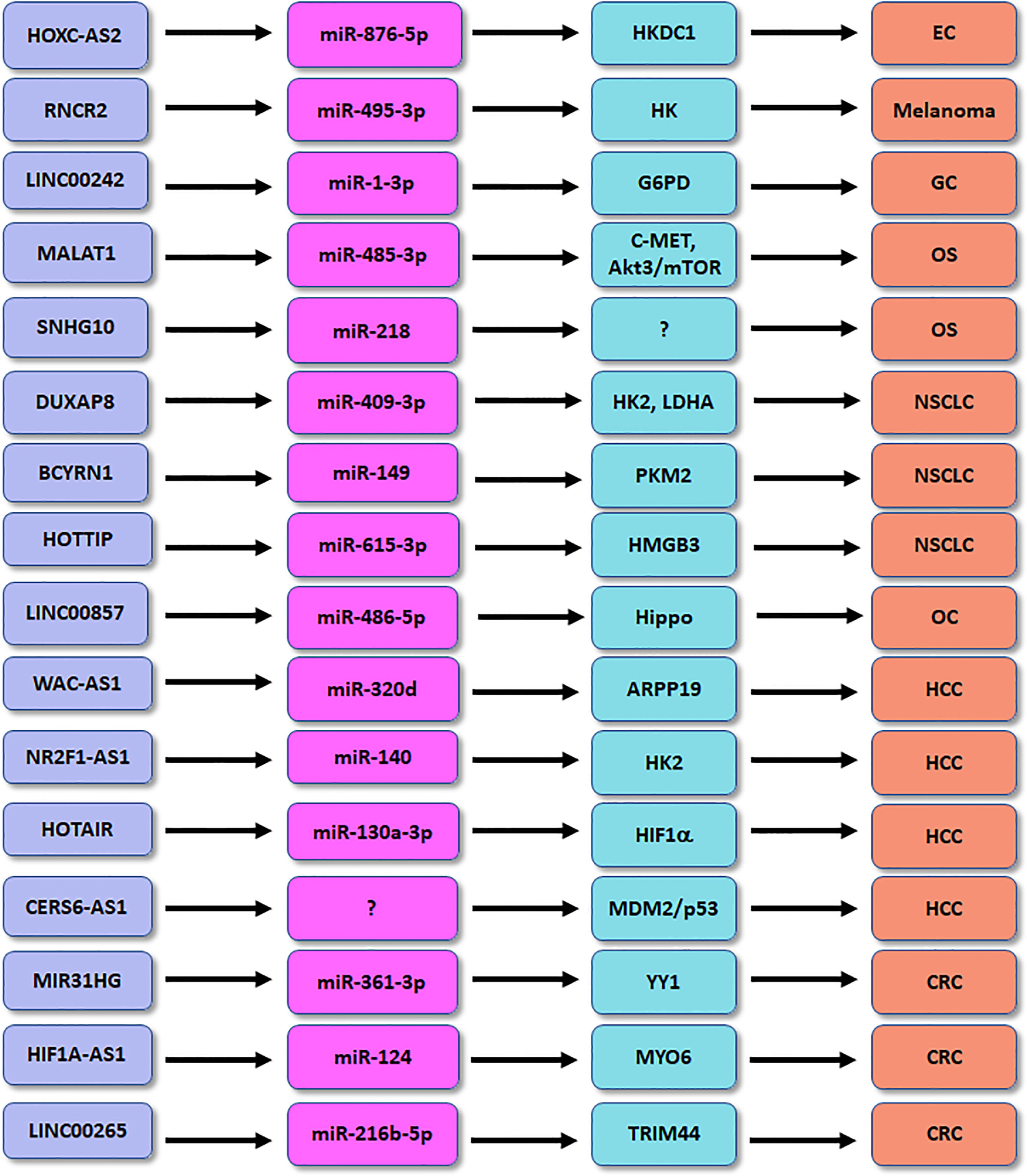
Figure 1 The role of lncRNAs in regulation of glucose metabolism in human cancers. EC, endometrial cancer; OS, osteosarcoma; HCC, hepatocellular carcinoma; CRC, colorectal cancer; GC, gastric cancer; NSCLC, non-small cell lung cancer.
LncRNAs regulate glucose metabolism in cancer
Evidence has dissected that lncRNA-associated genetic variants are shared between cancers and type 2 DM in human (97). LncRNA DRAIR has been known to involve in the development of type 2 DM (98). One study showed that the expression of lncRNA DRAIR was remarkably elevated in triple-negative breast cancer (TNBC) samples and plasma (99). High expression of DRAIR in plasma was associated with chemoresistance after therapy and tumor recurrence in TNBC patients. In vitro experiments showed that overexpression of DRAIR enhanced proliferation and viability of TNBC cells after doxorubicin treatment (99).
Accumulated evidence dissected that lncRNA HOXC-AS2 participated in the progression in high glucose-related endometrial cancer (EC) (100). EC patients with diabetes had the increased expression of HKDC1 compared with EC patients with normal glucose. HKDC1 governed pyroptosis, a highly inflammatory response of regulated cell death, via regulation of ROS and cytokine release in EC cells after high glucose stimulation (100). Moreover, miR-876-5p can inhibit the expression of HKDC1 in high glucose-related EC. LncRNA HOXC-AS2 was dissected to suppress the miR-876-5p/HKDC1 axis in high glucose-associated EC (100). HKDC1 affected the formation of TME via promotion of glycolysis, leading to accelerating EC progression. This work provided the new therapeutic strategy for EC patients with diabetes by targeting lncRNA HOXC-AS2 (100). LncRNA SNHG10 enhanced glucose uptake and increased proliferation of osteosarcoma cells via promotion of miR-218 methylation (101). LncRNA MALAT1 facilitated glycolysis and tumor metastasis via blocking miR-485-3p and upregulating c-MET and Akt3/mTOR pathways in osteosarcoma (102). LncRNA CERS6-AS1 regulated the MDM2/p53 axis and modulated glucose metabolism and progression of HCC (103). LncRNA WAC-AS1 sponged miR-320d and regulated the expression of ARPP19, which promoted glucose uptake and lactate production in HCC (104). LncRNA NR2F1-AS1 affected hypoxia-mediated glycolysis and migratory ability of HCC cells via targeting miR-140 and HK2 (105). Depletion of lncRNA HOTAIR reduced glycolysis via inhibition of miR-130a-3p and upregulation of HIF1α in HCC cells under hypoxia (106).
LncRNA MALAT1 modulated MYBL2/mTOR pathway and caused glucose metabolism changes in prostate cancer (107). LncRNA MIR31HG heightened glycolysis and tumor malignant progression via regulating miR-361-3p and YY1 transcription factor in colorectal cancer (108). LncRNA KCNQ1OT1 accelerated colorectal oncogenesis via promoting aerobic glycolysis by upregulation of HK2 (109). HNF1A-AS1 governed glycolysis, invasion and migration through targeting miR-124 and MYO6 in colorectal cancer (110). Similarly, LINC00265 enhanced glycolysis and lactate release via binding with miR-216b-5p and elevating the expression of TRIM44 in colorectal cancer (111). LncRNA RNCR2 promoted glycolysis and EMT and proliferation of melanoma cells via interacting with miR-495-3p and upregulating HK2 in melanoma (112). LINC00242 combined miR-1-3p and elevated the expression of G6PD, leading to enhancement of aerobic glycolysis and oncogenesis of gastric cancer (113). LncRNA MSC-AS1 increased glycolysis and cell growth via targeting PFKFB3 expression in gastric cancer cells (114). OIP5-AS1 heightened aerobic glycolysis and proliferation via miR-186 sponge in gastric cancer (115).
LINC00551 inhibited glycolysis and blocked tumor progression via modulation of c-Myc-induced PKM2 expression in lung cancer (116). LncRNA CRYBG3 potentiated glycolysis via interaction with lactate dehydrogenase A (LDHA) in lung cancer (117). LncRNA DUXAP8 accelerated glycolysis, viability and migratory capacities via suppression of miR-409-3p and upregulation of HK2 and LDHA in NSCLC cells (118). LncRNA BCYRN1 accelerated glycolysis via controlling the miR-149 expression and elevating PKM2 expression in NSCLC (119). HOTTIP enhanced hypoxia-mediated glycolysis via modulation of miR-615-3p and HMGB3 in NSCLC cells (120). LINC00857 was found to regulate glycolysis and tumor progression via governing the Hippo signaling pathway by binding to miR-486-5p in ovarian cancer (121). Downregulation of lncRNA UCA1 attenuated glycolysis pathway and led to suppression of growth of pituitary cancer cells (122). Overexpression of lncRNA PCED1B-AS1 resulted in upregulation of glucose uptake, proliferation and lactate production in glioblastoma by activation of HIF-1α pathway (123). LncRNA HNF4A-AS1 elevated aerobic glycolysis and tumor progression via modulating hnRNPU/CTCF axis in neuroblastoma (124).
High/low glucose regulates lncRNAs in cancer
Some studies have demonstrated that high glucose or glucose deprivation affected the expression of lncRNAs in cancer cells. For example, U87 and LN18 glioma cells after glucose deprivation had upregulation of lncRNA TP53TG1 and glucose metabolism-associated genes, including LDHA, IDH1 and GRP79 (125). Downregulation of TP53TG1 suppressed proliferation and migration of U87 cells after glucose deprivation, while overexpression of TP53TG1 displayed the opposite functions (125). Low glucose condition promoted the efficacy of TP53TG1 compared with high glucose condition. This study suggested that glucose metabolism dysregulation can affect the expression of TP53TG1 and tumor proliferation and migration in glioma (125).
High glucose increased the expression of miR-483-3p in hepatocellular carcinoma (HCC) cells. Moreover, upregulation of miR-483-3p inhibited the expression of ER protein 29 (ERp29), resulting in promotion of proliferation and migration of HCC cells (126). Furthermore, lncRNA MEG3 can bind with miR-483-3p in HCC cells. High glucose also reduced the expression of lncRNA MEG3 in HCC cells. Consistently, silencing of lncRNA MEG3 suppressed the expression of ERp29 in HCC cells (126). This study showed that high glucose could affect the expression of lncRNA MEG3 and govern the miR-483-3p/ERp29 proteins in HCC patients, suggesting that management of lncRNA MEG3 could be promising for the treatment of HCC patients with diabetes (126). Low glucose elevated the expression of lncRNA HOXC-AS3, leading to promotion of metabolic reprogramming of breast cancer via binding to SIRT6 and inactivating HIF1α (127).
Targeting lncRNAs for treating DKD and cancer
Klotho is often known as an antiaging protein to prevent of aging. Klotho has been identified to protect renal tubular EMT during the DKD development (52). Overexpression of Klotho reduced the lncRNA NEAT1 expression in HFD/STZ-mediated DKD mice. Moreover, overexpression of Klotho attenuated the expression levels of NEAT1 in BSA-treated HK2 cells (52). On the contrary, knockdown of Klotho increased the expression of lncRNA NEAT1 in HK2 cells. Thereby, knockdown of Klotho caused upregulation of NEAT1 and activation of EMT and fibrosis in a ERK1/2-dependent manner (52). Another study showed that Klotho blocked EMT via downregulation of early growth response factor 1 (Egr-1) by suppression of the ERK1/2 pathway in DKD mice (128). Similarly, Klotho decreased Egr-1 expression via repressing TGF-β1/Smad3 pathway in human mesangial cells after high glucose exposures (129). Triptolide, a diterpenoid epoxide that is obtained from the thunder god vine, blocked renal tubular EMT via modulation of miR-188-5p-involved PI3K/Akt pathway in DKD (130). Several studies have showed that triptolide regulated the expression of multiple lncRNAs, including lncRNAs WAKMAR2, PACER, ENST00000619282, RP11-83J16.1 (131–135). Therefore, whether triptolide regulates the lncRNA expression in DKD needs to be further explored. Berberine, an isoquinoline alkaloid, has been reported to upregulate the expression of lncRNA GAS5 to reduce the mitochondrial ROS generation in HK-2 cells under high glucose environment through regulation of miR-18a-5p and C/EBPβ expression (136). The antisense oligonucleotide treatment by targeting specific lncRNAs could provide targeted medicine to cure DKD and cancer in the future.
Conclusion
In summary, burgeoning data demonstrate that lncRNAs play an essential role in the development of DKD and diabetes-associated cancer. LncRNAs could be diagnosis and prognosis biomarkers for DKD and diabetes-related cancer. Modulation of lncRNAs might be a promising strategy for treating DKD and diabetes-associated cancer. It is important to note that it is far from being fully clarified, although some studies have explored the role of lncRNAs in DKD and cancer patients with DM. A small number of lncRNAs are identified in regulation of DKD and cancer patients with abnormal glucose metabolism. Whether other lncRNAs also participate in DKD and diabetes-associated cancer need to be explored. Compared with other factors such as m6A and signaling pathways, it remains questionable whether lncRNAs are more important in modulation of DKD and diabetes-related cancers. Addressing these questions will help us understand the mechanism of lncRNAs-regulated DKD and cancers, which could provide the clues for discovering new therapeutic strategy for DKD and cancer patients with diabetes.
Author contributions
YC and XW wrote the manuscript. YX made the figures. YC and PW edited the manuscript and supervised this study. All authors read and approved the final manuscript.
Funding
This work was supported by National Natural Science Foundation of China (82260876) and Hainan Province Science and Technology Special Fund (ZDKJ2021034).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
α-SMA, alpha smooth muscle actin; α1-MG, α1-microglobulin; APEX2, ascorbate peroxidase; β2-MG, β2-microglobulin; BSA, bovine serum albumin; CASC2, cancer susceptibility candidate 2; ChREBP, carbohydrate response element binding protein; CTGF, connective tissue growth factor; DRAIR, diabetes regulated anti-inflammatory lncRNA; DKD, diabetic kidney disease; HbA1c, glycosylated hemoglobin; HDAC1, histone deacetylase 1; HKDC1, hexokinase domain-containing 1; HOTAIR, HOX antisense intergenic RNA; LncRNAs, long noncoding RNAs; MEG3, maternally expressed gene 3; MLX, MAX dimerization protein; MXD1, MAX dimerization protein 1; NEAT, nuclear-enriched abundant transcript; PBMC, peripheral blood mononuclear cells; PGC-1α, peroxisome proliferator-activated receptor γ coactivator α; SOD, superoxide dismutase; TGF-β1, transforming growth factor-β1; TME, tumor microenvironment; TUG1, taurine upregulated gene 1.
References
1. Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov (2021) 20(8):629–51. doi: 10.1038/s41573-021-00219-z
2. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol (2021) 22(2):96–118. doi: 10.1038/s41580-020-00315-9
3. Zhang P, Wu W, Chen Q, Chen M. Non-coding RNAs and their integrated networks. J Integr Bioinform (2019) 16(3):20190027. doi: 10.1515/jib-2019-0027
4. Andergassen D, Rinn JL. From genotype to phenotype: Genetics of mammalian long non-coding RNAs in vivo. Nat Rev Genet (2022) 23(4):229–43. doi: 10.1038/s41576-021-00427-8
5. Gao N, Li Y, Li J, Gao Z, Yang Z, Li Y, et al. Long non-coding RNAs: The regulatory mechanisms, research strategies, and future directions in cancers. Front Oncol (2020) 10:598817. doi: 10.3389/fonc.2020.598817
6. Xie W, Chu M, Song G, Zuo Z, Han Z, Chen C, et al. Emerging roles of long noncoding RNAs in chemoresistance of pancreatic cancer. Semin Cancer Biol (2022) 83:303–18. doi: 10.1016/j.semcancer.2020.11.004
7. Jiang W, Xia J, Xie S, Zou R, Pan S, Wang ZW, et al. Long non-coding RNAs as a determinant of cancer drug resistance: Towards the overcoming of chemoresistance via modulation of lncRNAs. Drug Resist Update (2020) 50:100683. doi: 10.1016/j.drup.2020.100683
8. Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer (2021) 21(7):446–60. doi: 10.1038/s41568-021-00353-1
9. Liu J, Shang G. The roles of noncoding RNAs in the development of osteosarcoma stem cells and potential therapeutic targets. Front Cell Dev Biol (2022) 10:773038. doi: 10.3389/fcell.2022.773038
10. Chen T, Liu J, Zhang H, Li J, Shang G. Long intergenic noncoding RNA00265 enhances cell viability and metastasis via targeting miR-485-5p/USP22 axis in osteosarcoma. Front Oncol (2022) 12:907472. doi: 10.3389/fonc.2022.907472
11. Chen X, Liu Y, Liu H, Wang ZW, Zhu X. Unraveling diverse roles of noncoding RNAs in various human papillomavirus negative cancers. Pharmacol Ther (2022) 238:108188. doi: 10.1016/j.pharmthera.2022.108188
12. Jiang W, Pan S, Chen X, Wang ZW, Zhu X. The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in cancer immunotherapy. Mol Cancer (2021) 20(1):116. doi: 10.1186/s12943-021-01406-7
13. Jiang H, Zhang Y, Yue J, Shi Y, Xiao B, Xiao W, et al. Non-coding RNAs: The neuroinflammatory regulators in neurodegenerative diseases. Front Neurol (2022) 13:929290. doi: 10.3389/fneur.2022.929290
14. Nadhan R, Isidoro C, Song YS, Dhanasekaran DN. Signaling by LncRNAs: Structure, cellular homeostasis, and disease pathology. Cells (2022) 11(16):2517. doi: 10.3390/cells11162517
15. Tomic D, Shaw JE, Magliano DJ. The burden and risks of emerging complications of diabetes mellitus. Nat Rev Endocrinol (2022) 18(9):525–39. doi: 10.1038/s41574-022-00690-7
16. Thomas MC, Brownlee M, Susztak K, Sharma K, Jandeleit-Dahm KA, Zoungas S, et al. Diabetic kidney disease. Nat Rev Dis Primers (2015) 1:15018. doi: 10.1038/nrdp.2015.18
17. Antonetti DA, Silva PS, Stitt AW. Current understanding of the molecular and cellular pathology of diabetic retinopathy. Nat Rev Endocrinol (2021) 17(4):195–206. doi: 10.1038/s41574-020-00451-4
18. Chen Y, Zou H, Lu H, Xiang H, Chen S. Research progress of endothelial-mesenchymal transition in diabetic kidney disease. J Cell Mol Med (2022) 26(12):3313–22. doi: 10.1111/jcmm.17356
19. Hills CE, Squires PE. The role of TGF-beta and epithelial-to mesenchymal transition in diabetic nephropathy. Cytokine Growth Factor Rev (2011) 22(3):131–9. doi: 10.1016/j.cytogfr.2011.06.002
20. Hay N. Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat Rev Cancer (2016) 16(10):635–49. doi: 10.1038/nrc.2016.77
21. Leti F, DiStefano JK. Long noncoding RNAs as diagnostic and therapeutic targets in type 2 diabetes and related complications. Genes (Basel) (2017) 8(8):207. doi: 10.3390/genes8080207
22. Leti F, Morrison E, DiStefano JK. Long noncoding RNAs in the pathogenesis of diabetic kidney disease: Implications for novel therapeutic strategies. Per Med (2017) 14(3):271–8. doi: 10.2217/pme-2016-0107
23. Kato M. Noncoding RNAs as therapeutic targets in early stage diabetic kidney disease. Kidney Res Clin Pract (2018) 37(3):197–209. doi: 10.23876/j.krcp.2018.37.3.197
24. Hu M, Ma Q, Liu B, Wang Q, Zhang T, Huang T, et al. Long non-coding RNAs in the pathogenesis of diabetic kidney disease. Front Cell Dev Biol (2022) 10:845371. doi: 10.3389/fcell.2022.845371
25. Panchapakesan U, Pollock C. Long non-coding RNAs-towards precision medicine in diabetic kidney disease? Clin Sci (Lond) (2016) 130(18):1599–602. doi: 10.1042/CS20160261
26. Guo J, Liu Z, Gong R. Long noncoding RNA: An emerging player in diabetes and diabetic kidney disease. Clin Sci (Lond) (2019) 133(12):1321–39. doi: 10.1042/CS20190372
27. Gu YY, Lu FH, Huang XR, Zhang L, Mao W, Yu XQ, et al. Non-coding RNAs as biomarkers and therapeutic targets for diabetic kidney disease. Front Pharmacol (2020) 11:583528. doi: 10.3389/fphar.2020.583528
28. Zhao Y, Yan G, Mi J, Wang G, Yu M, Jin D, et al. The impact of lncRNA on diabetic kidney disease: Systematic review and in silico analyses. Comput Intell Neurosci (2022) 2022:8400106. doi: 10.1155/2022/8400106
29. Zhang L, Li R, He J, Yang Q, Wu Y, Huang J, et al. Co-Expression analysis among microRNAs, long non-coding RNAs, and messenger RNAs to understand the pathogenesis and progression of diabetic kidney disease at the genetic level. Methods (2017) 124:46–56. doi: 10.1016/j.ymeth.2017.05.023
30. Majumder S, Hadden MJ, Thieme K, Batchu SN, Niveditha D, Chowdhury S, et al. Dysregulated expression but redundant function of the long non-coding RNA HOTAIR in diabetic kidney disease. Diabetologia (2019) 62(11):2129–42. doi: 10.1007/s00125-019-4967-1
31. Wang H, Xia Y, Zhang Y. Diagnostic significance of serum lncRNA HOTAIR and its predictive value for the development of chronic complications in patients with type 2 diabetes mellitus. Diabetol Metab Syndr (2021) 13(1):97. doi: 10.1186/s13098-021-00719-3
32. Wang X, Wang W, HuangFu W, Liu Z, Zhao F. LncRNA HOTAIR facilitates high glucose-induced mesangial cell proliferation, fibrosis and oxidative stress in diabetic nephropathy via regulating miR-147a/WNT2B axis. Diabetol Metab Syndr (2022) 14(1):33. doi: 10.1186/s13098-022-00802-3
33. Wang W, Jia YJ, Yang YL, Xue M, Zheng ZJ, Wang L, et al. LncRNA GAS5 exacerbates renal tubular epithelial fibrosis by acting as a competing endogenous RNA of miR-96-5p. BioMed Pharmacother (2020) 121:109411. doi: 10.1016/j.biopha.2019.109411
34. Zhang YY, Tan RZ, Yu Y, Niu YY, Yu C. LncRNA GAS5 protects against TGF-beta-induced renal fibrosis via the Smad3/miRNA-142-5p axis. Am J Physiol Renal Physiol (2021) 321(4):F517–26. doi: 10.1152/ajprenal.00085.2021
35. Ge X, Xu B, Xu W, Xia L, Xu Z, Shen L, et al. Long noncoding RNA GAS5 inhibits cell proliferation and fibrosis in diabetic nephropathy by sponging miR-221 and modulating SIRT1 expression. Aging (Albany NY) (2019) 11(20):8745–59. doi: 10.18632/aging.102249
36. Xie C, Wu W, Tang A, Luo N, Tan Y. lncRNA GAS5/miR-452-5p reduces oxidative stress and pyroptosis of high-Glucose-Stimulated renal tubular cells. Diabetes Metab Syndr Obes (2019) 12:2609–17. doi: 10.2147/DMSO.S228654
37. Zhang L, Zhao S, Zhu Y. Long noncoding RNA growth arrest-specific transcript 5 alleviates renal fibrosis in diabetic nephropathy by downregulating matrix metalloproteinase 9 through recruitment of enhancer of zeste homolog 2. FASEB J (2020) 34(2):2703–14. doi: 10.1096/fj.201901380RR
38. Petrica L, Hogea E, Gadalean F, Vlad A, Vlad M, Dumitrascu V, et al. Long noncoding RNAs may impact podocytes and proximal tubule function through modulating miRNAs expression in early diabetic kidney disease of type 2 diabetes mellitus patients. Int J Med Sci (2021) 18(10):2093–101. doi: 10.7150/ijms.56551
39. Zhou LJ, Yang DW, Ou LN, Guo XR, Wu BL. Circulating expression level of LncRNA Malat1 in diabetic kidney disease patients and its clinical significance. J Diabetes Res (2020) 2020:4729019. doi: 10.1155/2020/4729019
40. Yang Z, Song D, Wang Y, Tang L. lncRNA MALAT1 promotes diabetic nephropathy progression via miR-15b-5p/TLR4 signaling axis. J Immunol Res (2022) 2022:8098001. doi: 10.1155/2022/8098001
41. Song P, Chen Y, Liu Z, Liu H, Xiao L, Sun L, et al. LncRNA MALAT1 aggravates renal tubular injury via activating LIN28A and the Nox4/AMPK/mTOR signaling axis in diabetic nephropathy. Front Endocrinol (Lausanne) (2022) 13:895360. doi: 10.3389/fendo.2022.895360
42. Huang H, Zhang G, Ge Z. lncRNA MALAT1 promotes renal fibrosis in diabetic nephropathy by targeting the miR-2355-3p/IL6ST axis. Front Pharmacol (2021) 12:647650. doi: 10.3389/fphar.2021.647650
43. Zhang H, Yan Y, Hu Q, Zhang X. LncRNA MALAT1/microRNA let-7f/KLF5 axis regulates podocyte injury in diabetic nephropathy. Life Sci (2021) 266:118794. doi: 10.1016/j.lfs.2020.118794
44. Zhang J, Jiang T, Liang X, Shu S, Xiang X, Zhang W, et al. lncRNA MALAT1 mediated high glucose-induced HK-2 cell epithelial-to-mesenchymal transition and injury. J Physiol Biochem (2019) 75(4):443–52. doi: 10.1007/s13105-019-00688-2
45. Hu M, Wang R, Li X, Fan M, Lin J, Zhen J, et al. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with beta-catenin. J Cell Mol Med (2017) 21(11):2732–47. doi: 10.1111/jcmm.13189
46. Liu B, Qiang L, Wang GD, Duan Q, Liu J. LncRNA MALAT1 facilities high glucose induced endothelial to mesenchymal transition and fibrosis via targeting miR-145/ZEB2 axis. Eur Rev Med Pharmacol Sci (2019) 23(8):3478–86. doi: 10.26355/eurrev_201904_17713
47. Zhou L, Xu DY, Sha WG, Shen L, Lu GY. Long non-coding RNA MALAT1 interacts with transcription factor Foxo1 to regulate SIRT1 transcription in high glucose-induced HK-2cells injury. Biochem Biophys Res Commun (2018) 503(2):849–55. doi: 10.1016/j.bbrc.2018.06.086
48. Zhang M, Zhao S, Xu C, Shen Y, Huang J, Shen S, et al. Ablation of lncRNA MIAT mitigates high glucose-stimulated inflammation and apoptosis of podocyte via miR-130a-3p/TLR4 signaling axis. Biochem Biophys Res Commun (2020) 533(3):429–36. doi: 10.1016/j.bbrc.2020.09.034
49. Ji TT, Qi YH, Li XY, Tang B, Wang YK, Zheng PX, et al. Loss of lncRNA MIAT ameliorates proliferation and fibrosis of diabetic nephropathy through reducing E2F3 expression. J Cell Mol Med (2020) 24(22):13314–23. doi: 10.1111/jcmm.15949
50. Wang Z, Chang Y, Liu Y, Liu B, Zhen J, Li X, et al. Inhibition of the lncRNA MIAT prevents podocyte injury and mitotic catastrophe in diabetic nephropathy. Mol Ther Nucleic Acids (2022) 28:136–53. doi: 10.1016/j.omtn.2022.03.001
51. Dong Q, Wang Q, Yan X, Wang X, Li Z, Zhang L. Long noncoding RNA MIAT inhibits the progression of diabetic nephropathy and the activation of NF-kappaB pathway in high glucose-treated renal tubular epithelial cells by the miR-182-5p/GPRC5A axis. Open Med (Wars) (2021) 16(1):1336–49. doi: 10.1515/med-2021-0328
52. Yang YL, Xue M, Jia YJ, Hu F, Zheng ZJ, Wang L, et al. Long noncoding RNA NEAT1 is involved in the protective effect of klotho on renal tubular epithelial cells in diabetic kidney disease through the ERK1/2 signaling pathway. Exp Mol Med (2020) 52(2):266–80. doi: 10.1038/s12276-020-0381-5
53. Li C, Liu YF, Huang C, Chen YX, Xu CY, Chen Y. Long noncoding RNA NEAT1 sponges miR-129 to modulate renal fibrosis by regulation of collagen type I. Am J Physiol Renal Physiol (2020) 319(1):F93–F105. doi: 10.1152/ajprenal.00552.2019
54. Huang S, Xu Y, Ge X, Xu B, Peng W, Jiang X, et al. Long noncoding RNA NEAT1 accelerates the proliferation and fibrosis in diabetic nephropathy through activating Akt/mTOR signaling pathway. J Cell Physiol (2019) 234(7):11200–7. doi: 10.1002/jcp.27770
55. Wang X, Xu Y, Zhu YC, Wang YK, Li J, Li XY, et al. LncRNA NEAT1 promotes extracellular matrix accumulation and epithelial-to-mesenchymal transition by targeting miR-27b-3p and ZEB1 in diabetic nephropathy. J Cell Physiol (2019) 234(8):12926–33. doi: 10.1002/jcp.27959
56. Ma J, Zhao N, Du L, Wang Y. Downregulation of lncRNA NEAT1 inhibits mouse mesangial cell proliferation, fibrosis, and inflammation but promotes apoptosis in diabetic nephropathy. Int J Clin Exp Pathol (2019) 12(4):1174–83.
57. Li N, Jia T, Li YR. LncRNA NEAT1 accelerates the occurrence and development of diabetic nephropathy by sponging miR-23c. Eur Rev Med Pharmacol Sci (2020) 24(3):1325–37. doi: 10.26355/eurrev_202002_20190
58. Zhan JF, Huang HW, Huang C, Hu LL, Xu WW. Long non-coding RNA NEAT1 regulates pyroptosis in diabetic nephropathy via mediating the miR-34c/NLRP3 axis. Kidney Blood Press Res (2020) 45(4):589–602. doi: 10.1159/000508372
59. Liao L, Chen J, Zhang C, Guo Y, Liu W, Liu W, et al. LncRNA NEAT1 promotes high glucose-induced mesangial cell hypertrophy by targeting miR-222-3p/CDKN1B axis. Front Mol Biosci (2020) 7:627827. doi: 10.3389/fmolb.2020.627827
60. Yang DY, Zhou X, Liu ZW, Xu XQ, Liu C. LncRNA NEAT1 accelerates renal tubular epithelial cell damage by modulating mitophagy via miR-150-5p-DRP1 axis in diabetic nephropathy. Exp Physiol (2021) 106(7):1631–42. doi: 10.1113/EP089547
61. Wu X, Fan D, Chen B. LncRNA NEAT1 accelerates the proliferation, oxidative stress, inflammation, and fibrosis and suppresses the apoptosis through the miR-423-5p/GLIPR2 axis in diabetic nephropathy. J Cardiovasc Pharmacol (2022) 79(3):342–54. doi: 10.1097/FJC.0000000000001177
62. Long J, Galvan DL, Mise K, Kanwar YS, Li L, Poungavrin N, et al. Role for carbohydrate response element-binding protein (ChREBP) in high glucose-mediated repression of long noncoding RNA Tug1. J Biol Chem (2020) 295(47):15840–52. doi: 10.1074/jbc.RA120.013228
63. Long J, Badal SS, Ye Z, Wang Y, Ayanga BA, Galvan DL, et al. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J Clin Invest (2016) 126(11):4205–18. doi: 10.1172/JCI87927
64. Li SY, Susztak K. The long noncoding RNA Tug1 connects metabolic changes with kidney disease in podocytes. J Clin Invest (2016) 126(11):4072–5. doi: 10.1172/JCI90828
65. Zhang B, Zhao C, Hou L, Wu Y. Silencing of the lncRNA TUG1 attenuates the epithelial-mesenchymal transition of renal tubular epithelial cells by sponging miR-141-3p via regulating beta-catenin. Am J Physiol Renal Physiol (2020) 319(6):F1125–34. doi: 10.1152/ajprenal.00321.2020
66. Duan LJ, Ding M, Hou LJ, Cui YT, Li CJ, Yu DM. Long noncoding RNA TUG1 alleviates extracellular matrix accumulation via mediating microRNA-377 targeting of PPARgamma in diabetic nephropathy. Biochem Biophys Res Commun (2017) 484(3):598–604. doi: 10.1016/j.bbrc.2017.01.145
67. Lei M, Ke G, Wang Y, Luo D, Hu Y. Long non-coding RNA TUG1 sponges microRNA-9 to protect podocytes from high glucose-induced apoptosis and mitochondrial dysfunction via SIRT1 upregulation. Exp Ther Med (2022) 23(3):236. doi: 10.3892/etm.2022.11161
68. Lei X, Zhang L, Li Z, Ren J. Astragaloside IV/lncRNA-TUG1/TRAF5 signaling pathway participates in podocyte apoptosis of diabetic nephropathy rats. Drug Des Devel Ther (2018) 12:2785–93. doi: 10.2147/DDDT.S166525
69. Shen H, Ming Y, Xu C, Xu Y, Zhao S, Zhang Q. Deregulation of long noncoding RNA (TUG1) contributes to excessive podocytes apoptosis by activating endoplasmic reticulum stress in the development of diabetic nephropathy. J Cell Physiol (2019) 234(9):15123–33. doi: 10.1002/jcp.28153
70. Zang XJ, Li L, Du X, Yang B, Mei CL. LncRNA TUG1 inhibits the proliferation and fibrosis of mesangial cells in diabetic nephropathy via inhibiting the PI3K/AKT pathway. Eur Rev Med Pharmacol Sci (2019) 23(17):7519–25. doi: 10.26355/eurrev_201909_18867
71. Wang F, Gao X, Zhang R, Zhao P, Sun Y, Li C. LncRNA TUG1 ameliorates diabetic nephropathy by inhibiting miR-21 to promote TIMP3-expression. Int J Clin Exp Pathol (2019) 12(3):717–29.
72. Meng D, Wu L, Li Z, Ma X, Zhao S, Zhao D, et al. LncRNA TUG1 ameliorates diabetic nephropathy via inhibition of PU.1/RTN1 signaling pathway. J Leukoc Biol (2022) 111(3):553–62. doi: 10.1002/JLB.6A1020-699RRR
73. Wang S, Yi P, Wang N, Song M, Li W, Zheng Y. LncRNA TUG1/miR-29c-3p/SIRT1 axis regulates endoplasmic reticulum stress-mediated renal epithelial cells injury in diabetic nephropathy model in vitro. PloS One (2021) 16(6):e0252761. doi: 10.1371/journal.pone.0252761
74. Deng Q, Wen R, Liu S, Chen X, Song S, Li X, et al. Increased long noncoding RNA maternally expressed gene 3 contributes to podocyte injury induced by high glucose through regulation of mitochondrial fission. Cell Death Dis (2020) 11(9):814. doi: 10.1038/s41419-020-03022-7
75. Zha F, Qu X, Tang B, Li J, Wang Y, Zheng P, et al. Long non-coding RNA MEG3 promotes fibrosis and inflammatory response in diabetic nephropathy via miR-181a/Egr-1/TLR4 axis. Aging (Albany NY) (2019) 11(11):3716–30. doi: 10.18632/aging.102011
76. Li J, Jiang X, Duan L, Wang W. Long non-coding RNA MEG3 impacts diabetic nephropathy progression through sponging miR-145. Am J Transl Res (2019) 11(10):6691–8.
77. Che X, Deng X, Xie K, Wang Q, Yan J, Shao X, et al. Long noncoding RNA MEG3 suppresses podocyte injury in diabetic nephropathy by inactivating wnt/beta-catenin signaling. PeerJ (2019) 7:e8016. doi: 10.7717/peerj.8016
78. Zhu B, Cheng X, Jiang Y, Cheng M, Chen L, Bao J, et al. Silencing of KCNQ1OT1 decreases oxidative stress and pyroptosis of renal tubular epithelial cells. Diabetes Metab Syndr Obes (2020) 13:365–75. doi: 10.2147/DMSO.S225791
79. Jie R, Zhu P, Zhong J, Zhang Y, Wu H. LncRNA KCNQ1OT1 affects cell proliferation, apoptosis and fibrosis through regulating miR-18b-5p/SORBS2 axis and NF-kB pathway in diabetic nephropathy. Diabetol Metab Syndr (2020) 12:77. doi: 10.1186/s13098-020-00585-5
80. Li J, Li M, Bai L. KCNQ1OT1/miR-18b/HMGA2 axis regulates high glucose-induced proliferation, oxidative stress, and extracellular matrix accumulation in mesangial cells. Mol Cell Biochem (2021) 476(1):321–31. doi: 10.1007/s11010-020-03909-1
81. Zhao L, Chen H, Wu L, Li Z, Zhang R, Zeng Y, et al. LncRNA KCNQ1OT1 promotes the development of diabetic nephropathy by regulating miR-93-5p/ROCK2 axis. Diabetol Metab Syndr (2021) 13(1):108. doi: 10.1186/s13098-021-00726-4
82. Xu Y, Zhan X. lncRNA KCNQ1OT1 regulated high glucose-induced proliferation, oxidative stress, extracellular matrix accumulation, and inflammation by miR-147a/SOX6 in diabetic nephropathy (DN). Endocr J (2022) 69(5):511–22. doi: 10.1507/endocrj.EJ21-0514
83. Huang X, Tan J, Li Y, Su H, Huang M, Huang F, et al. Expression of LncRNA KCNQ1Ot1 in diabetic nephropathy and its correlation with MEK/ERK signaling pathway. Am J Transl Res (2022) 14(3):1796–806.
84. Wang YZ, Zhu DY, Xie XM, Ding M, Wang YL, Sun LL, et al. EA15, MIR22, LINC00472 as diagnostic markers for diabetic kidney disease. J Cell Physiol (2019) 234(6):8797–803. doi: 10.1002/jcp.27539
85. Zhang H, Wei X, Lu S, Lin X, Huang J, Chen L, et al. Protective effect of DMDD, isolated from the root of averrhoa carambola l., on high glucose induced EMT in HK-2 cells by inhibiting the TLR4-BAMBI-Smad2/3 signaling pathway. BioMed Pharmacother (2019) 113:108705. doi: 10.1016/j.biopha.2019.108705
86. Ren H, Shao Y, Wu C, Lv C, Zhou Y, Wang Q. VASH-1 regulates oxidative stress and fibrosis in diabetic kidney disease via SIRT1/HIF1alpha and TGFbeta1/Smad3 signaling pathways. Front Mol Biosci (2020) 7:137. doi: 10.3389/fmolb.2020.00137
87. Xu BH, Sheng J, You YK, Huang XR, Ma RCW, Wang Q, et al. Deletion of Smad3 prevents renal fibrosis and inflammation in type 2 diabetic nephropathy. Metabolism (2020) 103:154013. doi: 10.1016/j.metabol.2019.154013
88. Zhou Q, Guo H, Yu C, Huang XR, Liang L, Zhang P, et al. Identification of Smad3-related transcriptomes in type-2 diabetic nephropathy by whole transcriptome RNA sequencing. J Cell Mol Med (2021) 25(4):2052–68. doi: 10.1111/jcmm.16133
89. Yu Q, Lin J, Ma Q, Li Y, Wang Q, Chen H, et al. Long noncoding RNA ENSG00000254693 promotes diabetic kidney disease via interacting with HuR. J Diabetes Res (2022) 2022:8679548. doi: 10.1155/2022/8679548
90. Wang L, Su N, Zhang Y, Wang G. Clinical significance of serum lncRNA cancer susceptibility candidate 2 (CASC2) for chronic renal failure in patients with type 2 diabetes. Med Sci Monit (2018) 24:6079–84. doi: 10.12659/MSM.909510
91. Zhao C, Hu J, Wang Z, Cao ZY, Wang L. Serum LncRNA PANDAR may act as a novel serum biomarker of diabetic nephropathy in patients with type 2 diabetes. Clin Lab (2020) 66(6). doi: 10.7754/Clin.Lab.2019.191032
92. Yang Y, Lv X, Fan Q, Wang X, Xu L, Lu X, et al. Analysis of circulating lncRNA expression profiles in patients with diabetes mellitus and diabetic nephropathy: Differential expression profile of circulating lncRNA. Clin Nephrol (2019) 92(1):25–35. doi: 10.5414/CN109525
93. Zhang XL, Zhu HQ, Zhang Y, Zhang CY, Jiao JS, Xing XY. LncRNA CASC2 regulates high glucose-induced proliferation, extracellular matrix accumulation and oxidative stress of human mesangial cells via miR-133b/FOXP1 axis. Eur Rev Med Pharmacol Sci (2020) 24(2):802–12. doi: 10.26355/eurrev_202001_20063
94. Min XQ, Xie Y. LncRNA CASC2 alleviates the progression of diabetic nephropathy by regulating the miR-144/SOCS2 signalling axis. Kidney Blood Press Res (2020) 45(6):837–49. doi: 10.1159/000508078
95. Zhu D, Wu X, Xue Q. Long non-coding RNA CASC2 restrains high glucose-induced proliferation, inflammation and fibrosis in human glomerular mesangial cells through mediating miR-135a-5p/TIMP3 axis and JNK signaling. Diabetol Metab Syndr (2021) 13(1):89. doi: 10.1186/s13098-021-00709-5
96. Yao K, Wang Q, Jia J, Zhao H. A competing endogenous RNA network identifies novel mRNA, miRNA and lncRNA markers for the prognosis of diabetic pancreatic cancer. Tumour Biol (2017) 39(6):1010428317707882. doi: 10.1177/1010428317707882
97. Giordo R, Gulsha R, Kalla S, Calin GA, Lipovich L. LncRNA-associated genetic etiologies are shared between type 2 diabetes and cancers in the UAE population. Cancers (Basel) (2022) 14(14):3313. doi: 10.3390/cancers14143313
98. Reddy MA, Amaram V, Das S, Tanwar VS, Ganguly R, Wang M, et al. lncRNA DRAIR is downregulated in diabetic monocytes and modulates the inflammatory phenotype via epigenetic mechanisms. JCI Insight (2021) 6(11):e143289. doi: 10.1172/jci.insight.143289
99. Wang P, Peng W, Peng J. Diabetes regulated anti-inflammatory lncRNA is overexpressed in triple-negative breast cancer and predicts chemoresistance and tumor recurrence. Bioengineered (2022) 13(5):12718–25. doi: 10.1080/21655979.2022.2068821
100. Guo J, Ye F, Xie W, Zhang X, Zeng R, Sheng W, et al. The HOXC-AS2/miR-876-5p/HKDC1 axis regulates endometrial cancer progression in a high glucose-related tumor microenvironment. Cancer Sci (2022) 113(7):2297–310. doi: 10.1111/cas.15384
101. He P, Xu Y, Wang Z. LncRNA SNHG10 increases the methylation of miR-218 gene to promote glucose uptake and cell proliferation in osteosarcoma. J Orthop Surg Res (2020) 15(1):353. doi: 10.1186/s13018-020-01865-6
102. Wang Q, Liu MJ, Bu J, Deng JL, Jiang BY, Jiang LD, et al. miR-485-3p regulated by MALAT1 inhibits osteosarcoma glycolysis and metastasis by directly suppressing c-MET and AKT3/mTOR signalling. Life Sci (2021) 268:118925. doi: 10.1016/j.lfs.2020.118925
103. Xu B, Wei Y, Liu F, Li L, Zhou S, Peng Y, et al. Long noncoding RNA CERS6-AS1 modulates glucose metabolism and tumor progression in hepatocellular carcinoma by promoting the MDM2/p53 signaling pathway. Cell Death Discov (2022) 8(1):348. doi: 10.1038/s41420-022-01150-x
104. Xia X, Zhang H, Xia P, Zhu Y, Liu J, Xu K, et al. Identification of glycolysis-related lncRNAs and the novel lncRNA WAC-AS1 promotes glycolysis and tumor progression in hepatocellular carcinoma. Front Oncol (2021) 11:733595. doi: 10.3389/fonc.2021.733595
105. Li X, Li Y, Bai S, Zhang J, Liu Z, Yang J. NR2F1-AS1/miR-140/HK2 axis regulates hypoxia-induced glycolysis and migration in hepatocellular carcinoma. Cancer Manag Res (2021) 13:427–37. doi: 10.2147/CMAR.S266797
106. Hu M, Fu Q, Jing C, Zhang X, Qin T, Pan Y. LncRNA HOTAIR knockdown inhibits glycolysis by regulating miR-130a-3p/HIF1A in hepatocellular carcinoma under hypoxia. BioMed Pharmacother (2020) 125:109703. doi: 10.1016/j.biopha.2019.109703
107. Mu X, Shen Z, Lin Y, Xiao J, Xia K, Xu C, et al. LncRNA-MALAT1 regulates cancer glucose metabolism in prostate cancer via MYBL2/mTOR axis. Oxid Med Cell Longev (2022) 2022:8693259. doi: 10.1155/2022/8693259
108. Guo T, Liu D, Peng S, Wang M, Li Y. A positive feedback loop of lncRNA MIR31HG-miR-361-3p -YY1 accelerates colorectal cancer progression through modulating proliferation, angiogenesis, and glycolysis. Front Oncol (2021) 11:684984. doi: 10.3389/fonc.2021.684984
109. Chen C, Wei M, Wang C, Sun D, Liu P, Zhong X, et al. Long noncoding RNA KCNQ1OT1 promotes colorectal carcinogenesis by enhancing aerobic glycolysis via hexokinase-2. Aging (Albany NY) (2020) 12(12):11685–97. doi: 10.18632/aging.103334
110. Guo X, Zhang Y, Liu L, Yang W, Zhang Q. HNF1A-AS1 regulates cell migration, invasion and glycolysis via modulating miR-124/MYO6 in colorectal cancer cells. Onco Targets Ther (2020) 13:1507–18. doi: 10.2147/OTT.S231249
111. Sun S, Li W, Ma X, Luan H. Long noncoding RNA LINC00265 promotes glycolysis and lactate production of colorectal cancer through regulating of miR-216b-5p/TRIM44 axis. Digestion (2020) 101(4):391–400. doi: 10.1159/000500195
112. Duan BX, Geng XR, Wu YQ. lncRNA RNCR2 facilitates cell proliferation and epithelial-mesenchymal transition in melanoma through HK2-mediated warburg effect via targeting miR-495-3p. Neoplasma (2021) 68(4):692–701. doi: 10.4149/neo_2021_201120N1255
113. Deng P, Li K, Gu F, Zhang T, Zhao W, Sun M, et al. LINC00242/miR-1-3p/G6PD axis regulates warburg effect and affects gastric cancer proliferation and apoptosis. Mol Med (2021) 27(1):9. doi: 10.1186/s10020-020-00259-y
114. Jin X, Qiao L, Fan H, Liao C, Zheng J, Wang W, et al. Long non-coding RNA MSC-AS1 facilitates the proliferation and glycolysis of gastric cancer cells by regulating PFKFB3 expression. Int J Med Sci (2021) 18(2):546–54. doi: 10.7150/ijms.51947
115. Huang J, Hou S, Xu J, Wu J, Yin J. Long non-coding RNA OIP5-AS1 promotes cell proliferation and aerobic glycolysis in gastric cancer through sponging miR-186. Arch Med Sci (2021) 17(6):1742–51. doi: 10.5114/aoms.2019.87213
116. Wang L, Wang H, Wu B, Zhang C, Yu H, Li X, et al. Long noncoding RNA LINC00551 suppresses glycolysis and tumor progression by regulating c-Myc-Mediated PKM2 expression in lung adenocarcinoma. Onco Targets Ther (2020) 13:11459–70. doi: 10.2147/OTT.S273797
117. Chen H, Pei H, Hu W, Ma J, Zhang J, Mao W, et al. Long non-coding RNA CRYBG3 regulates glycolysis of lung cancer cells by interacting with lactate dehydrogenase a. J Cancer (2018) 9(14):2580–8. doi: 10.7150/jca.24896
118. Yin D, Hua L, Wang J, Liu Y, Li X. Long non-coding RNA DUXAP8 facilitates cell viability, migration, and glycolysis in non-Small-Cell lung cancer via regulating HK2 and LDHA by inhibition of miR-409-3p. Onco Targets Ther (2020) 13:7111–23. doi: 10.2147/OTT.S243542
119. Lang N, Wang C, Zhao J, Shi F, Wu T, Cao H. Long noncoding RNA BCYRN1 promotes glycolysis and tumor progression by regulating the miR149/PKM2 axis in nonsmallcell lung cancer. Mol Med Rep (2020) 21(3):1509–16. doi: 10.3892/mmr.2020.10944
120. Shi J, Wang H, Feng W, Huang S, An J, Qiu Y, et al. Long non-coding RNA HOTTIP promotes hypoxia-induced glycolysis through targeting miR-615-3p/HMGB3 axis in non-small cell lung cancer cells. Eur J Pharmacol (2019) 862:172615. doi: 10.1016/j.ejphar.2019.172615
121. Lin X, Feng D, Li P, Lv Y. LncRNA LINC00857 regulates the progression and glycolysis in ovarian cancer by modulating the hippo signaling pathway. Cancer Med (2020) 9(21):8122–32. doi: 10.1002/cam4.3322
122. Liu G, Wang L and Li Y. Inhibition of lncRNA-UCA1 suppresses pituitary cancer cell growth and prolactin (PRL) secretion via attenuating glycolysis pathway. Vitro Cell Dev Biol Anim (2020) 56(8):642–9. doi: 10.1007/s11626-020-00494-x
123. Yao Z, Zhang Q, Guo F, Guo S, Yang B, Liu B, et al. Long noncoding RNA PCED1B-AS1 promotes the warburg effect and tumorigenesis by upregulating HIF-1alpha in glioblastoma. Cell Transplant (2020) 29:963689720906777. doi: 10.1177/0963689720906777
124. Song H, Li D, Wang X, Fang E, Yang F, Hu A, et al. HNF4A-AS1/hnRNPU/CTCF axis as a therapeutic target for aerobic glycolysis and neuroblastoma progression. J Hematol Oncol (2020) 13(1):24. doi: 10.1186/s13045-020-00857-7
125. Chen X, Gao Y, Li D, Cao Y, Hao B. LncRNA-TP53TG1 participated in the stress response under glucose deprivation in glioma. J Cell Biochem (2017) 118(12):4897–904. doi: 10.1002/jcb.26175
126. Li X, Cheng T, He Y, Zhou S, Wang Y, Zhang K, et al. High glucose regulates ERp29 in hepatocellular carcinoma by LncRNA MEG3-miRNA 483-3p pathway. Life Sci (2019) 232:116602. doi: 10.1016/j.lfs.2019.116602
127. Zhu W, Chen X, Guo X, Liu H, Ma R, Wang Y, et al. Low glucose-induced overexpression of HOXC-AS3 promotes metabolic reprogramming of breast cancer. Cancer Res (2022) 82(5):805–18. doi: 10.1158/0008-5472.CAN-21-1179
128. Li Y, Xue M, Hu F, Jia Y, Zheng Z, Yang Y, et al. Klotho prevents epithelial-mesenchymal transition through egr-1 downregulation in diabetic kidney disease. BMJ Open Diabetes Res Care (2021) 9(1):e002038. doi: 10.1136/bmjdrc-2020-002038
129. Li Y, Hu F, Xue M, Jia YJ, Zheng ZJ, Wang L, et al. Klotho down-regulates egr-1 by inhibiting TGF-beta1/Smad3 signaling in high glucose treated human mesangial cells. Biochem Biophys Res Commun (2017) 487(2):216–22. doi: 10.1016/j.bbrc.2017.04.036
130. Xue M, Cheng Y, Han F, Chang Y, Yang Y, Li X, et al. Triptolide attenuates renal tubular epithelial-mesenchymal transition via the MiR-188-5p-mediated PI3K/AKT pathway in diabetic kidney disease. Int J Biol Sci (2018) 14(11):1545–57. doi: 10.7150/ijbs.24032
131. Piao X, Zhou J, Xue L. Triptolide decreases rheumatoid arthritis fibroblast-like synoviocyte proliferation, invasion, inflammation and presents a therapeutic effect in collagen-induced arthritis rats via inactivating lncRNA RP11-83J16.1 mediated URI1 and beta-catenin signaling. Int Immunopharmacol (2021) 99:108010. doi: 10.1016/j.intimp.2021.108010
132. Wen J, Liu J, Wang X, Wang J. Triptolide promotes the apoptosis and attenuates the inflammation of fibroblast-like synoviocytes in rheumatoid arthritis by down-regulating lncRNA ENST00000619282. Phytother Res (2021) 35(8):4334–46. doi: 10.1002/ptr.7129
133. Tamgue O, Chia JE, Brombacher F. Triptolide modulates the expression of inflammation-associated lncRNA-PACER and lincRNA-p21 in mycobacterium tuberculosis-infected monocyte-derived macrophages. Front Pharmacol (2021) 12:618462. doi: 10.3389/fphar.2021.618462
134. Zhou X, Xie D, Huang J, Lu A, Wang R, Jin Y, et al. Therapeutic effects of (5R)-5-Hydroxytriptolide on fibroblast-like synoviocytes in rheumatoid arthritis via lncRNA WAKMAR2/miR-4478/E2F1/p53 axis. Front Immunol (2021) 12:605616. doi: 10.3389/fimmu.2021.605616
135. Xiong S, Li Y, Xiang Y, Peng N, Shen C, Cai Y, et al. Dysregulation of lncRNA and circRNA expression in mouse testes after exposure to triptolide. Curr Drug Metab (2019) 20(8):665–73. doi: 10.2174/1389200220666190729130020
Keywords: lncRNAs, cancer, diabetes, DKD, miRNAs, treatment
Citation: Cheng Y, Wu X, Xia Y, Liu W and Wang P (2022) The role of lncRNAs in regulation of DKD and diabetes-related cancer. Front. Oncol. 12:1035487. doi: 10.3389/fonc.2022.1035487
Received: 02 September 2022; Accepted: 19 September 2022;
Published: 13 October 2022.
Edited by:
Aamir Ahmad, University of Alabama at Birmingham, United StatesCopyright © 2022 Cheng, Wu, Xia, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yawei Cheng, eWF3ZWljaGVuZzE5ODJAMTYzLmNvbQ==; Peter Wang, d2FuZ3BldGVyMkBob3RtYWlsLmNvbQ==
 Yawei Cheng
Yawei Cheng Xiaowen Wu1
Xiaowen Wu1 Wenjun Liu
Wenjun Liu Peter Wang
Peter Wang