- 1Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Key Laboratory of Cancer Prevention and Therapy, Tianjin, China
- 2Department of Pathology, Tianjin Medical University, Tianjin, China
Introduction: Pancreatic ductal adenocarcinoma (PDAC) is characterized by high aggressiveness and a hypoxic tumour microenvironment. Macrophage migration inhibitory factor (MIF) is a hypoxia-related pleiotropic cytokine that plays important roles in cancer. However, its role in PDAC progression has not been fully elucidated.
Methods: The clinical significance of MIF and hypoxia inducible factor 1 subunit alpha (HIF1A) in PDAC was analysed using immunohistochemical staining on PDAC tissues and data from KM-Plotter database. Spatial distribution of MIF and HIF1A gene expression was visualized by spatial transcriptomics in PDAC cell xenografts. To monitor the role of MIF in PDAC cell malignancy, immunostaining, lentivirus shRNA, migration assays, flow cytometry, transcriptomics and in vivo tumorigenicity were performed.
Results: The spatial distribution of MIF and HIF1A was highly correlated and that high MIF expression was associated with poor prognosis of PDAC patients. MIF knockdown impaired cell invasion, with a decrease in the expression of urokinase-type plasminogen activator receptor (uPAR). Although PLAUR transcript was not reduced, a uPAR endocytic receptor, low-density lipoprotein receptor–related protein 1 (LRP1), was upregulated at both the mRNA and protein levels after MIF knockdown. The LRP1 antagonist RAP restored uPAR expression and invasiveness. MIF attenuated the nuclear translocation of p53, a transcriptional regulator of LRP1. Furthermore, MIF downregulation blunted the growth of PDAC cell xenografts and inhibited cell proliferation under normoxia and hypoxia. Transcriptome analysis also provided evidence for the role of MIF in cancer-associated pathways.
Discussion: We demonstrate a novel link between the two pro-invasive agents MIF and uPAR and explain how MIF increases PDAC cell invasion capability. This finding provides a basis for therapeutic intervention of MIF in PDAC progression.
1 Introduction
Pancreatic ductal adenocarcinoma (PDAC), the most common type of pancreatic cancer, contributes to a five-year survival rate of only 10% (1). Due to the characteristics of invasive growth, early metastasis and general resistance to chemotherapy or radiotherapy, a potentially curative treatment is surgical resection for PDAC diagnosed early enough (2). The hypoxic tumour microenvironment of PDAC contributes to aggressive tumour biology. Hypoxia is involved in tumour cell genomic instability, immune responses, metabolic reprogramming, cancer stem cells, etc., through effects on levels of mRNA, protein expression and DNA methylation (3). Understanding the mechanisms responsible for the highly aggressive and metastatic characteristics of PDAC is required to improve treatment and disease outcomes. Under hypoxia, significant alterations in the mRNA expression of genes involved in invasion and metastasis were observed across different cancer types, including macrophage migration inhibitory factor (MIF).
MIF is a pluripotent and pleiotropic cytokine and is broadly expressed in various cell types and tissues. MIF plays roles in a multitude of disorders, such as immune abnormalities, heart disease, neurodegeneration, and the development of malignancies (4, 5). MIF is localized extracellularly and intracellularly, interacting with various cell surface and intracellular proteins, so it functions in an autocrine and paracrine way (6–9). Extracellular MIF binds to receptor complexes, and intracellular MIF modifies effector functions, leading to specific phenotypes (10, 11). MIF overexpression is frequently observed in most cancers and is a poor prognostic indicator (12). Under hypoxic conditions, the stabilization of hypoxia-inducible factor 1alpha (HIF-1a) promotes MIF expression (13), and MIF prevents the degradation of HIF-1a (14, 15), showing a collaboration between HIF-1a and MIF in MIF-associated pro/antitumour effects. Because of the multifaceted and critical functions of MIF, it has emerged as a therapeutic target (16–18).
Tumor cell invasion is a complex and multistep process, involving remodeling of the extracellular matrix (ECM) by tumor cell-associated proteases, which lead to proteolysis and dispersion of the obstacles and barriers. Of note is that MIF has emerged as an inducer of cancer cell migration and invasion by up-regulating and activating matrix metalloproteases (MMPs), such as MMP2, MMP9 and MMP13, then contributed to degradation of ECM (19–24). ECM restructure induced by the plasmin, serine protease urokinase plasminogen activator (uPA), uPA receptor (uPAR) and multiple MMPs has been well documented (25–28). The uPAR-uPA system is important for MMP2 and MMP9 activation (27, 29). The uPA has a wide range of targets, and its activation needs the binding of pro-uPA to uPAR (30). Overexpression of uPAR is significantly associated with poor prognosis in PDAC patients (31). uPAR is a membrane-anchored protein and binds to its high-affinity ligand uPA to facilitate the activation of proteases and cell signalling pathways (32, 33). Furthermore, uPA decreases the uPAR protein half-life by interacting with a multiligand endocytic receptor, low-density lipoprotein receptor–related protein 1 (LRP1). As a member of the low-density lipoprotein receptor family, which functions in receptor-mediated uptake of extracellular molecules, LRP1 is reported to regulate the abundance of proteins in the plasma membrane, including uPAR (34). By regulating cell signalling and gene expression, LRP1 is also reported to be associated with tumorigenesis and tumour progression (35).
In this study, we investigated the relationship of MIF expression with hypoxia status, cell invasiveness and MIF-related transcriptional changes in PDAC cells. Interestingly, we observed that the MIF-mediated reduction in cell invasiveness is regulated by the LRP1-uPAR interaction through the mechanism of MIF-induced inhibition of p53 and p53-induced LRP1 expression. We described a newly recognized regulatory mechanism of MIF in cell invasion, indicating potential implications for MIF-targeting therapeutic strategies.
2 Materials and methods
2.1 Clinical patient samples
Eighty-four pancreatic cancer specimens were obtained with informed consent from patients according to protocols approved by the Tianjin University Cancer Hospital Ethics Committee. Patients were diagnosed with pancreatic duct adenocarcinoma by postoperative pathology, and axillary node metastases were present in 13 patients. The median age of the patients was 59 years (range 41–72 years). The follow-up period began at the time of surgery and ended in June 2016. The collected sample tissues were fixed with neutral buffered formalin and embedded in paraffin for subsequent experiments.
2.2 Histopathology, immunohistochemistry and immunofluorescence
Haematoxylin and eosin staining (H&E) and immunohistochemical analysis were performed on pancreatic cancer and xenograft tissues following standard protocols to analyse the tumour histology and the following proteins: MIF (Abcam, ab65869) and Ki-67 (Abcam, ab279653). Immunohistochemical staining was scored as previously published (36). For immunofluorescence, adherent cells were grown on coverslips, fixed in 4% paraformaldehyde for 15 min, and stained with the following antibodies: uPAR (Abcam, ab103791) and p53 (Abcam, ab16665). Alexa Fluor 568 (Invitrogen, A10437) was used as the secondary antibody. Cell nuclei were counterstained with 4’, 6-Diamidino -2-phenylindole dihydrochloride (DAPI), and images were recorded on a Nikon A1R-A1 confocal microscope (Nikon Corporation, Tokyo, Japan).
2.3 Clinical significance of MIF and gene expression analysis
The prognostic values of MIF and HIF1A mRNA expression were evaluated using the KM-Plotter database (http://kmplot.com/analysis/). Progression-free survival (PFS) and overall survival (OS) analyses were performed in 177 PDAC patient samples according to the median mRNA expression. We also explored the prognostic values of MIF in TCGA-pancreatic cancer (PAAD) and the relationship between the expression levels of MIF and LRP1 using UCSC Xena (xena.ucsc.edu/) (37).
2.4 Spatial transcriptomics
Pancreatic cancer cell line PANC1-derived xenografts were collected as our previous study (38). Briefly, pancreatic cancer cells were subcutaneously transplanted into the groins of nude mice after left hind limb ischaemia by femoral artery ligation or sham operation (n=4 for each group). Frozen sections from the xenografts were analysed by 10x Genomics and spatial transcriptomics sequencing following the manufacturer’s procedure (Supplementary materials: CG000238_Visium Spatial Tissue Optimization User Guide, CG000239_Visium Spatial Gene Expression_User Guide_Rev_A, CG000240_Visium Spatial Protocols_Tissue Preparation Guide_Rev A and SAM000092_Planner_Visium Spatial Gene Expression Workflow Planner_Rev A).
2.5 Cell culture
The human pancreatic cancer cell lines PANC-1, SW1990, AsPC-1, BxPC-3 and CFPAC-1 were purchased from the Type Culture Collection Committee of the Chinese Academy of Sciences. Cells were maintained in Roswell Park Memorial Institute 1640 Medium (RPMI 1640), Dulbecco’s Modified Eagle Medium (DMEM), or Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C with 5% CO2 in a humidified incubator. Cobalt chloride (CoCl2)-treated cells were established as an in vitro hypoxia-mimetic condition. SW1990, BxPC-3, AsPC-1 and PANC-1cells were treated with 150 μM, 200 μM, 150 μM and 300 μM CoCl2 respectively for 24 h. To inhibit LRP1 function, pancreatic cancer cells were treated with the LRP1 antagonist receptor-associated protein (RAP) (0.5 μM) (human recombinant RAP, 553506 Sigma−Aldrich) for 3 days.
2.6 Lentivirus production and transduction
The lentivirus expressing shRNA targeting MIF (NM_002415) (targeting 5’-GACAGGGTCTACATCAACTAT-3’) and lentivirus vector for full-length MIF overexpression (NM_002415) were synthesized by GeneChem (Shanghai, China). AsPC-1 and PANC-1 cells were transduced with shRNA targeting MIF (shMIF) or a nontargeting control (shNTC: 5’-TTCTCCGAACGTGTCACGT-3’); SW1990 and BxPC-3 cells were transduced with MIF overexpression lentivirus (exMIF) or a control vector (vec). Stably transduced cell lines were generated by lentivirus infection for 12 h, followed by selection with puromycin (0.5 mg/ml) for 2 weeks.
2.7 Western blot analysis
For immunoblot analysis, cells were washed twice with PBS, harvested and lysed with RIPA buffer. Whole-cell proteins were separated by SDS−PAGE, transferred onto polyvinylidene fluoride (PVDF) membranes and immunoblotted with antibodies against MIF (Abcam, ab65869), uPAR (Abcam, ab103791), LRP1 (Abcam, ab92544) and GAPDH (Santa Cruz Biotechnology, sc25778). Blots were detected with WesternBright ECL HRP substrate (R-03031-D2, advansta) and visualized with a C-DiGit Blot Scanner (LI-COR Biosciences).
2.8 Cell migration
Cell invasion and migration were examined using transwell assays and scratch wound healing assays as previously described (39). For transwell invasion/migration assays, 2x104 cells were plated in the upper chamber (Transwell chambers, 8 mm pore size, BD Biosciences) in serum-free culture medium with or without Matrigel matrix (BD Biosciences) on the inserts. In the lower chamber, 10% FBS was used as the chemoattractant. After 24 hours, migratory cells on the lower membrane surface were stained with crystal violet and counted for three random 100x fields per well. For wound healing assays, cells were seeded in 12-well plates. Confluent cell monolayers were scraped with 200 µl pipette tips and washed twice with PBS. Wound healing images were captured at 0, 24 and 48 hours after scratching. Cell motility was assessed by measuring the distance from each side of the cell wound. Images were documented under a phase contrast microscope.
2.9 Real-time reverse transcription PCR
Total RNA isolation from pancreatic cancer cells was performed using the SPARKeasy Cell RNA Kit (Sparkjade, AC1601) followed by reverse transcription using the All-in-one First-Strand cDNA Synthesis Kit (GeneCopoeia, AORT-0020). Synthesized cDNA was subjected to SYBR Green real-time PCR (All-in-one qPCR Mix, GeneCopoeia, AORT-0600) using the following primers: LRP1 forward 5′-CTGGCGAACAAACACACTGGa-3″, LRP1 reverse 5′-CACGGTCCGGTTGTAGTTGA-3′; PLAUR forward 5′-GAGAGAAGACGTGCAGGGAC-3′. PLAUR reverse 5′-ACTCTTCCACACGGCAATCC-3′. The relative LRP1 or PLAUR mRNA expression levels were normalized to GAPDH expression. Each synthesis and amplification were run in triplicate for the target and internal control genes.
2.10 Flow cytometry
Apoptotic cells were quantified using an Annexin V-APC/propidium iodide (PI) apoptosis double staining kit (US EVERBRIGT, A6030) according to the manufacturer’s instructions. Briefly, cells were harvested using trypsin, centrifuged at 2,000 rpm for 5 min, and stained with 5 μl Annexin V-APC and 5 μl PI for 15 min in the dark. Untreated cells were used as the negative controls. Data analysis was performed using a BD AccuriR C6. Annexin V-positive and PI-negative cells were identified as early apoptotic cells, and Annexin V-positive and PI-positive cells were identified as late apoptotic cells. The extent of apoptosis was calculated as the percentage of both cell populations.
2.11 Mouse xenograft assays
Female BALB/c-nu mice (4-6 weeks of age) were obtained from Beijing HFK Biosciences (Beijing, China). Animal studies were performed according to protocols approved by the Institutional Animal Care and Use Committee at Tianjin Medical University. Pancreatic cancer cell suspensions (5x106) were prepared for subcutaneous injections into the groin after left hind limb ischaemia by femoral artery ligation or sham operation (40). Tumours were measured using a calliper every second day until harvest at 3-4 weeks, and tumour volumes (TV) were determined using the formula (TV (mm3) = 0.5 × length × (width)2).
2.12 Transcriptomics
Total RNAs of PANC-1 cells transduced with shMIF or shNTC were sent to LC-Bio Technology (Hangzhou, China) for sequencing and analysis. After RNA purification, the 2×150bp paired-end sequencing (PE150) on an Illumina Novaseq™ 6000 was performed. Reads of all samples were aligned to the homo sapiens reference genome using HISAT2 (https://daehwankimlab.github.io/hisat2/, version: hisat2-2.2.1) package. Genes differential expression analysis was performed by DESeq2 software between two different groups. The genes with the parameter of false discovery rate (FDR) below 0.005 and absolute fold change ≥ 2 were considered differentially expressed genes (DEGs). DEGs were then subjected to Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis performed by Lc-bio (https://www.lc-bio.cn/). Details are provided in the Supplementary materials.
2.13 Statistical analysis
GraphPad Prism 8.0 (GraphPad Software) was adopted for statistical analysis. All experiments were performed with at least three replicates per condition. Measurement data are reported as the mean ± standard deviation (SD). For comparisons between more than two groups, analysis of variance (ANOVA) followed by post hoc Tukey’s (for multiple comparisons) test was used. For comparisons between two groups, Student t test was performed. For data of abnormal distribution, the nonparametric Kruskal-Wallis or Mann Whitney-U test was applied. Survival time was analysed using the Kaplan–Meier method and compared by the log-rank test. For all statistical tests, P ≤0.05 was considered significant.
3 Results
3.1 HIF1A and MIF expression is associated with worse outcomes in PDAC patients
High MIF levels are associated with a poor prognosis in multiple cancer types through various mechanisms (41–45). To test the above point, we investigated the involvement of MIF in the outcomes of PDAC patients. Eighty-four patients with PDAC who underwent surgical resection were included in the study. The pathological characteristics of these PDAC tissues were detected by H&E staining. Levels of MIF expression were examined using immunohistochemical techniques, and staining scores were graded as weakly positive (5-49%) and strongly positive (>50%) based on the percentage of positively stained tumour cells (Figure 1A). Fifty-seven PDAC patients showing negative or weak tumour MIF expression had a better OS (P = 0.007) and a trend for increased PFS (P = 0.08) compared with the 27 patients whose tumour showed strong MIF expression using Kaplan−Meier analyses (Figure 1B). The clinical significance of MIF and HIF1A mRNA was also analysed in 177 PDAC samples from the KM-Plotter database. Although MIF mRNA expression and OS did not show a correlation (P = 0.25), it was associated with poor PFS (P = 0.037). The mRNA expression of HIF1A was significantly related to both the OS and PFS of PDAC patients (P = 0.048; P = 0.004). (Figure 1C). Although high MIF expression may predict a worse prognosis, no difference of MIF expression was observed in TNM (Tumor, Nodal Involvement, Metastasis) stages of 196 TCGA pancreatic cancers using UCSC Xena database (xena.ucsc.edu/) (P = 0.7862, Figure S1). These results suggest that high levels of the hypoxia-responsive factor MIF might be a predictor of worse survival of PDAC patients, which is consistent with the study by Ahmed A et al., highlighting the role of MIF as a biomarker candidate for the diagnosis and prognosis of PDAC (44).
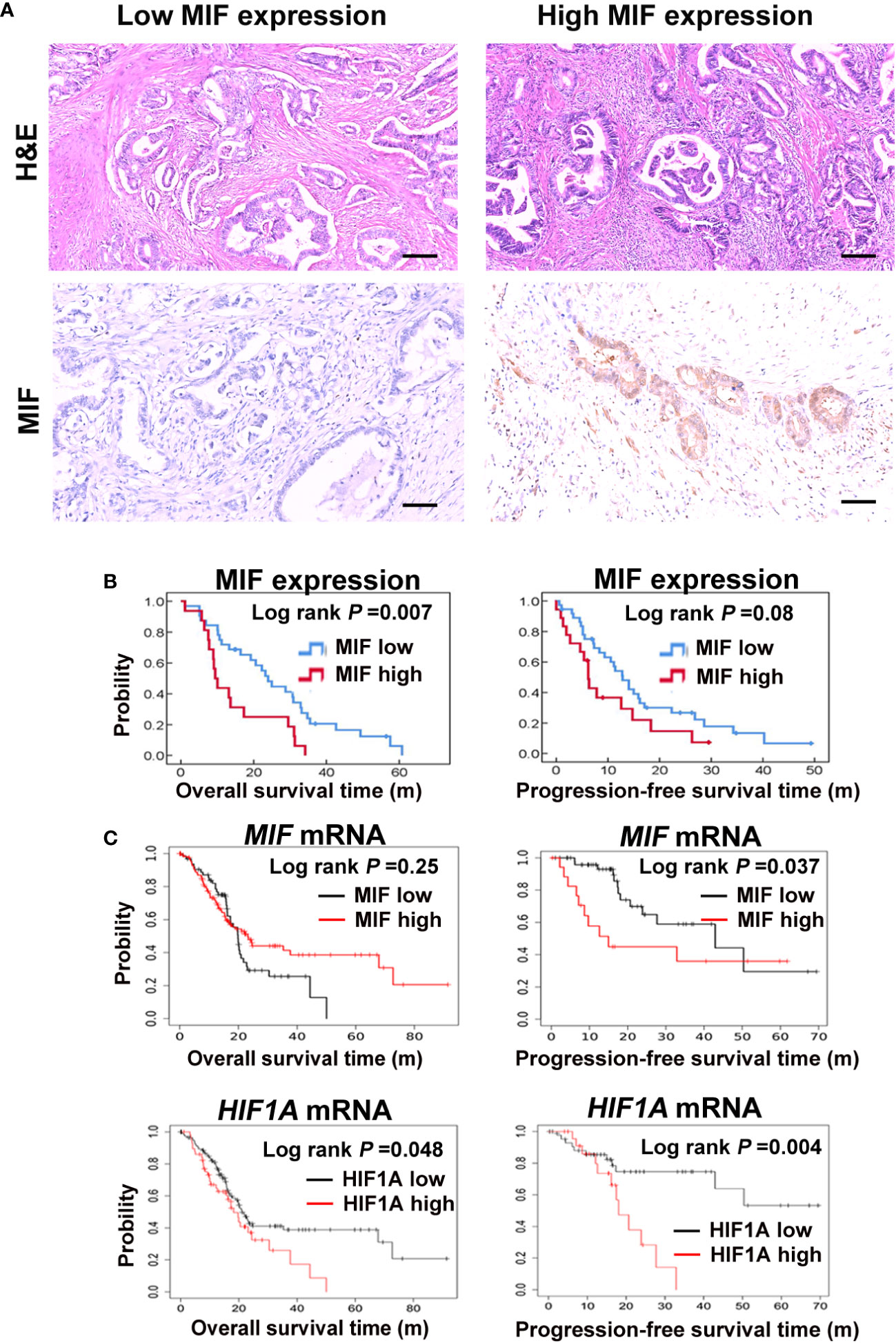
Figure 1 Elevated MIF expression correlates with a poor prognosis in PDAC patients (A) Pathological and histological assessment of 84 human PDAC sections, including haematoxylin eosin staining (H&E) and expression of MIF by immunohistochemistry staining. Representative images of MIF immunostaining with low and high intensity. Scale bars, 100 μm. (B) Kaplan−Meier plots show the correlations between MIF mRNA expression and the OS/PFS of 84 PDAC patients. (C) Survival curves show the relationships between MIF or HIF1A mRNA expression and OS/PFS of 177 PDAC patients from the KM-Plotter database.
3.2 Visualization of MIF and HIF1A gene expression in PDAC xenografts by spatial transcriptomics
Since hypoxia-induced expression and secretion of MIF has been observed before (13, 46), we next investigated the expression and spatial distributions of the hypoxia marker genes HIF1A and MIF in PDAC xenografts by spatial transcriptomics as we reported previously (38). For this, PANC-1 cells were subcutaneously injected into the mouse groin after femoral artery ligation of the left hind limb or sham operation. By 10x Genomics and spatial transcriptomics sequencing, we investigated the gene expression pattern of HIF1A and MIF in 8 sections from PDAC cell-derived xenografts. The spatially resolved expression profile and position of the HIF1A and MIF genes showed approximately the same levels and overlapping positions (Figure 2). These data display positional information on the MIF gene related to the hypoxia marker HIF1A, validating that hypoxia is a potent inducer of MIF expression in PDAC.
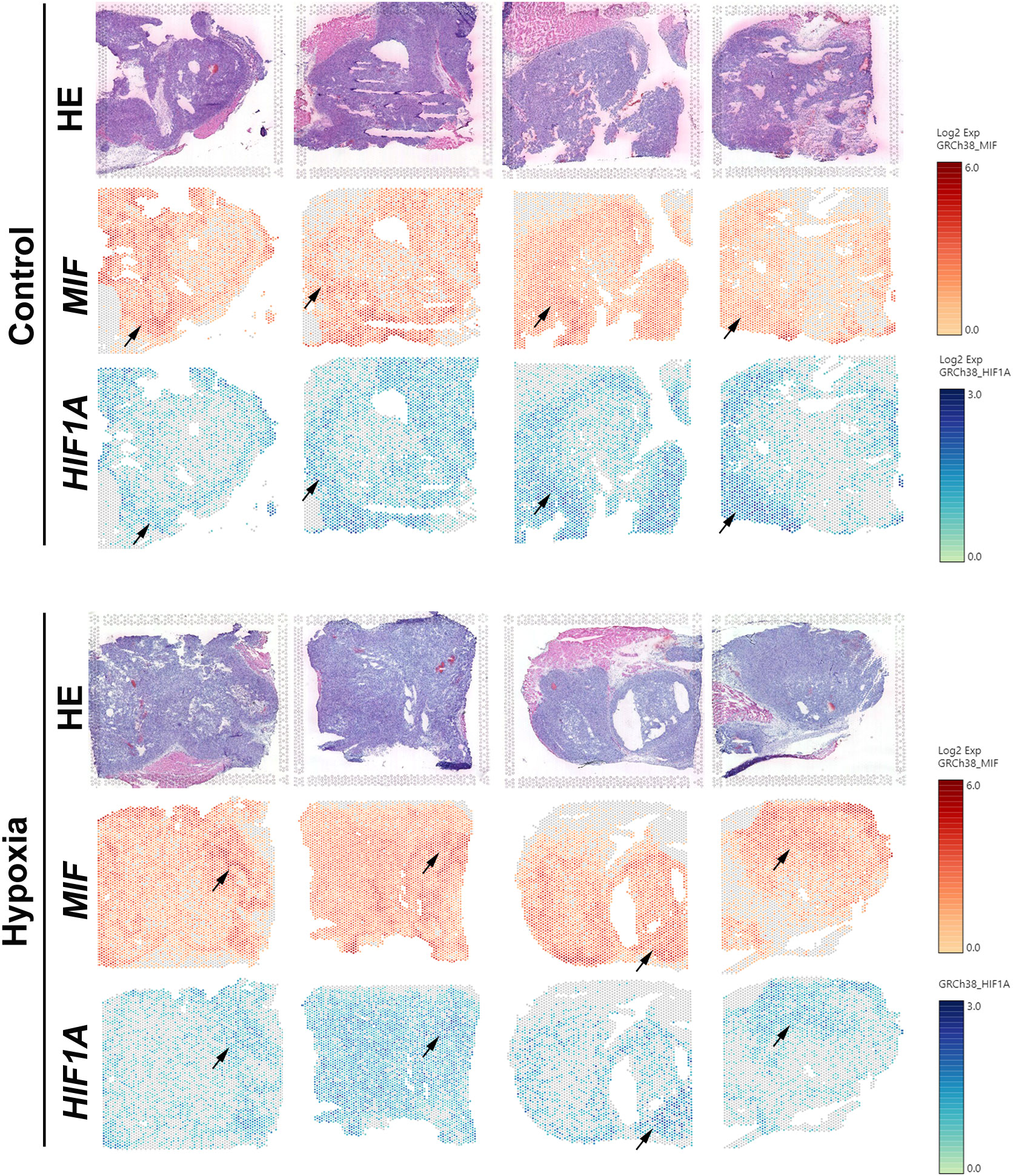
Figure 2 Visualization of the expression patterns of HIF1A and MIF genes in PDAC xenografts by spatial transcriptomics Spatially resolved expression of HIF1A and MIF in tumour sections taken from 8 PDAC xenografts subcutaneously in the mouse groin after femoral artery ligation of the left hind limb (hypoxia) or sham operation (control).
3.3 MIF is generally expressed in pancreatic cancer cell lines
To detect the expression of MIF in most used pancreatic cancer cell lines and its relationship with hypoxia in vitro, then we examined MIF protein expression in a panel of pancreatic cancer cell lines using western blotting. The results showed that MIF was generally and differentially expressed in these five pancreatic cancer cell lines (Figure 3A). Since the MIF promoter contains responsive elements for HIF-1a (47), we then determined whether there was a correlation between hypoxic conditions and MIF expression in vitro. For hypoxia induction, cells were cultured with a hypoxia-mimetic agent, CoCl2 at the indicated concentration for 24 h. The changes in MIF protein levels examined by western blotting showed that CoCl2 treatment markedly elevated MIF protein levels. (Figure 3B).
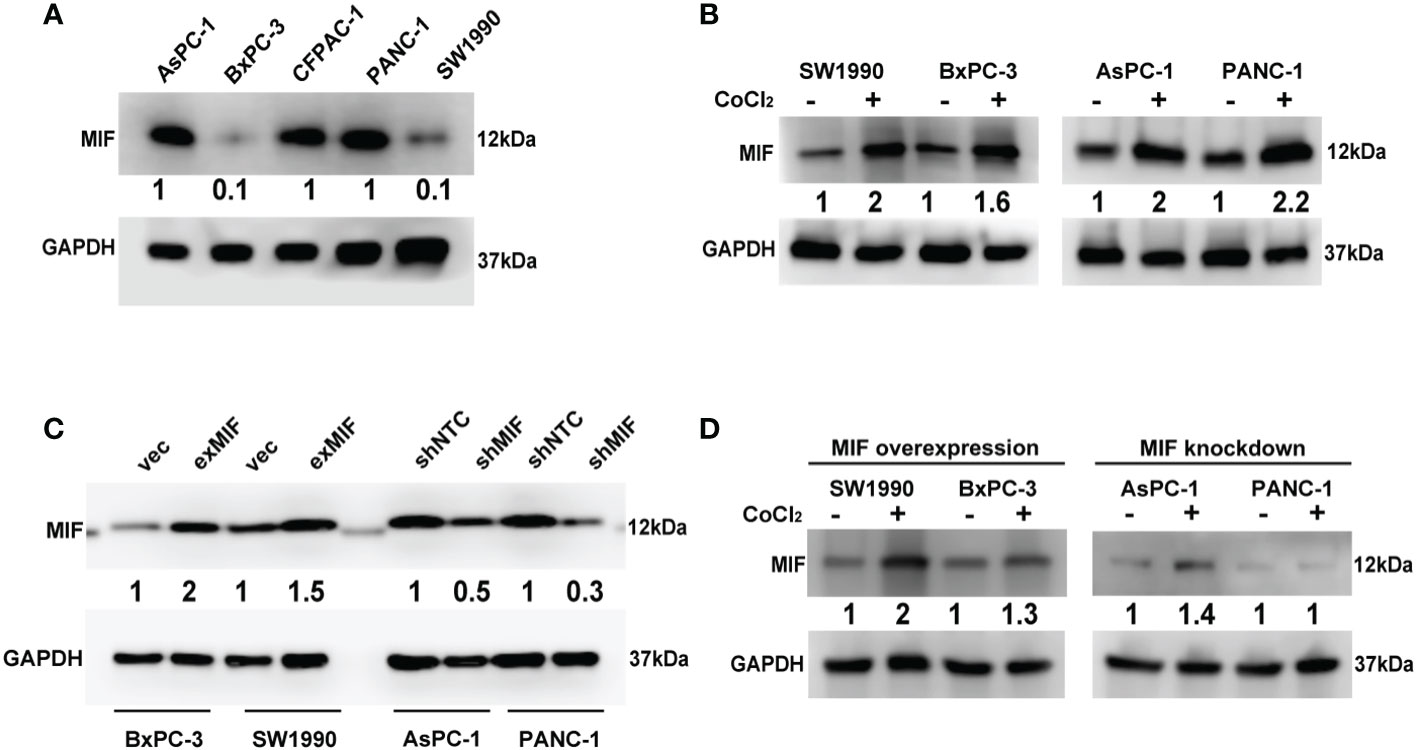
Figure 3 Detection of MIF expression in PDAC cell lines Western blot analysis of MIF protein levels in five human PDAC cell lines (A), MIF expression after CoCl2 treatment for 24 h compared with vehicle (B), transduction efficiency of lentivirus-cDNA-MIF (exMIF) or vector (vec) for MIF overexpression and lentivirus-shRNA (shMIF) or nontarget control (shNTC) for MIF knockdown (C), and MIF expression after CoCl2 treatment in stably transduced cells (D). The loading control was assessed by GAPDH blotting. The numbers below the blots are quantitative ratios of the MIF/GAPDH band densities. CoCl2 concentration used: SW1990, 150 μM; BxPC-3, 200 μM; AsPC-1, 150 μM; PANC-1, 300 μM.
To determine the effect of MIF on the malignant phenotype of pancreatic cancer cells, we started to construct MIF-overexpressing and MIF-knockdown cells according to the basal expression levels of MIF, as shown in Figure 3A. SW1990 and BxPC-3 cells were selected for stable transduction with lentivirus-MIF-cDNA (exMIF) or control vector (vec); PANC-1 and AsPC-1 cells were selected for stable transduction with MIF-shRNA (shMIF) or nontarget control shRNA (shNTC). The up/down regulation efficiency of MIF expression was quantified by western blot. As shown in Figure 3C, MIF protein was elevated in both SW1990 and BxPC-3 cells compared with the control vector; MIF-shRNA dramatically reduced MIF protein in both PANC-1 and AsPC-1 cells compared with the nontarget control. In addition, CoCl2 treatment markedly elevated the MIF protein levels in MIF-overexpressing cells, whereas in MIF knockdown cells, no significant increase in MIF expression was observed (Figure 3D). These data show that MIF is generally expressed in pancreatic cancer cell lines and can be enhanced by the hypoxia-mimicking agent CoCl2 in vitro.
3.4 MIF promotes the invasion of pancreatic cancer cells
Increasing data indicated that MIF promoted migration and invasion of cancer cells, so to investigate the effects of MIF on the invasive phenotype of pancreatic cancer cells, we sought to determine the impact of altered expression of MIF on cellular mobility. We observed that overexpression of MIF promoted the migration and invasion of SW1990 and BxPC-3 cells, whereas knockdown of MIF attenuated the migration and invasion of PANC-1 and AsPC-1 cells (Figure 4A, B). These findings provide evidence that MIF has a significant role as a positive regulator of PDAC cell mobility.
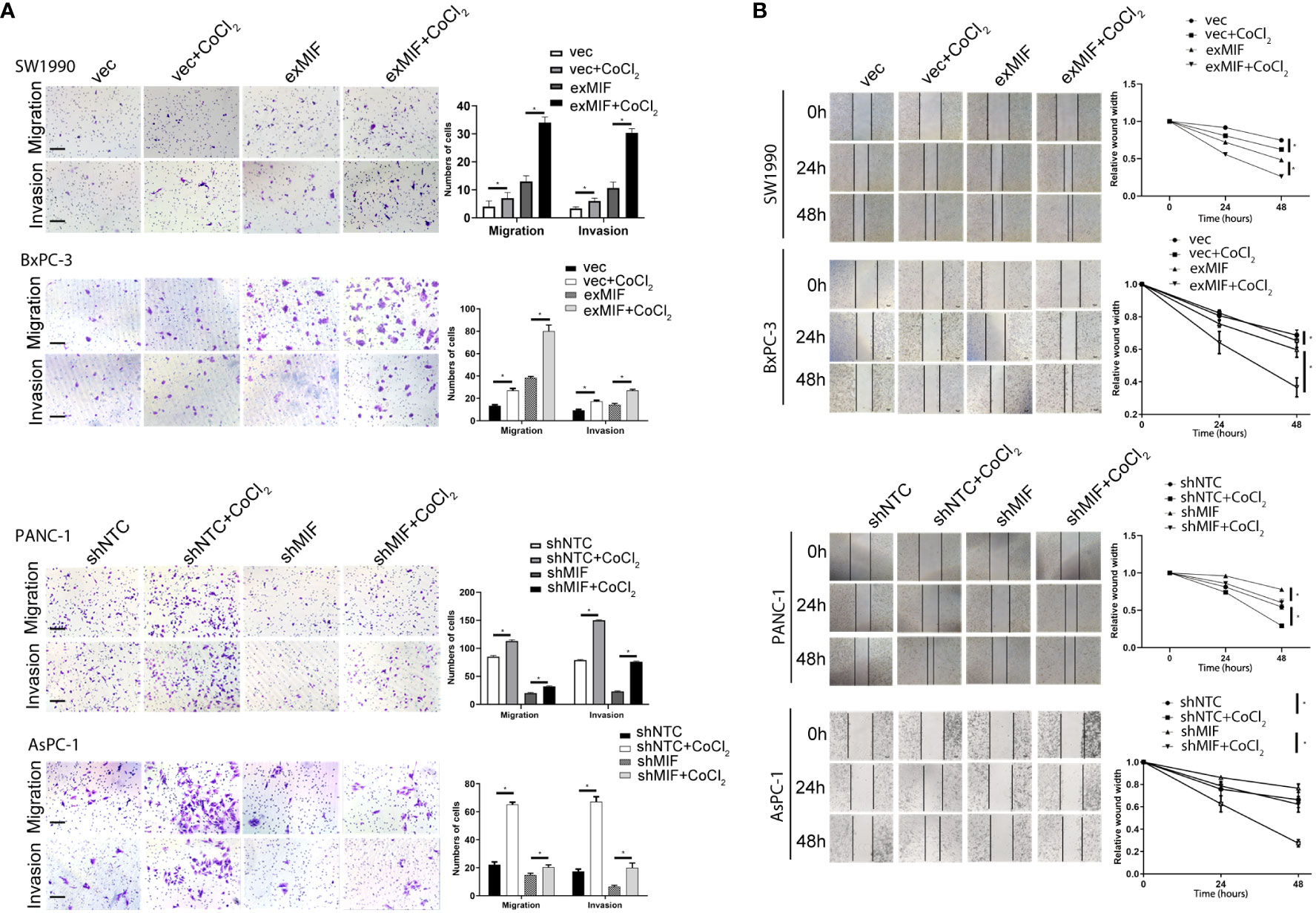
Figure 4 MIF promotes the invasion/migration of PDAC cells in vitro (A) Transwell migration and invasion assay. Images of migration and invasion of each cell group were presented. The migrating cells were stained with violet color. The bar charts showing the average migration and invasion cell number per field among different experimental groups. Data are the mean ± SD from three independent experiments, *P<0.05. Scale bars, 100 µm. (B) Wound healing assay. Representative images showing the scratch at time 0 h, 24 h and 48 h of PDAC cells in indicated conditions. The line charts showing the quantitative analysis of the relative wound width. The values were normalized by the wound width at the same area of the scratch at time 0 h. *P < 0.05.
3.5 MIF knockdown inhibits uPAR expression via LRP1-mediated internalization
Cellular invasion/migration is important during tumour progression, and it requires the coordination of various biological events. To move, cells have to use proteinases to break down barriers of the surrounding matrix. Numerous studies demonstrated that MIF has been involved in the migration and invasion of cancer cells by up-regulating the pro-metastatic mediators MMPs, among which MMP2 and MMP9 degrades the ECM and facilitates the tumor cell invasion and metastasis (19, 23, 48, 49). In view of these results, we investigated the influence of MIF silencing and overexpressing on the levels of MMP2 and MMP9 using western blotting assay. Different from other researches, however, no changes of MMP2 or MMP9 expression were detected (data not shown). In consideration of the role of uPA/uPAR/plasmin system-mediated MMPs activation in tumour migration and invasion, we attempted to investigate the influence of MIF silencing and overexpressing on the levels of uPAR, a plasma membrane GPI-anchored protein, that binds with high-affinity and activates the serine protease uPA, thus regulating MMPs activity. Among them, uPAR has been implicated in the invasion and metastasis of tumours and correlated with a poor prognosis in several types of tumours. As shown in Figure 5A, the levels of uPAR were reduced in shMIF cells and elevated in exMIF cells. The downregulation of uPAR protein was more obvious in shMIF cells, and subsequent experiments in vitro were performed only in shMIF cells.
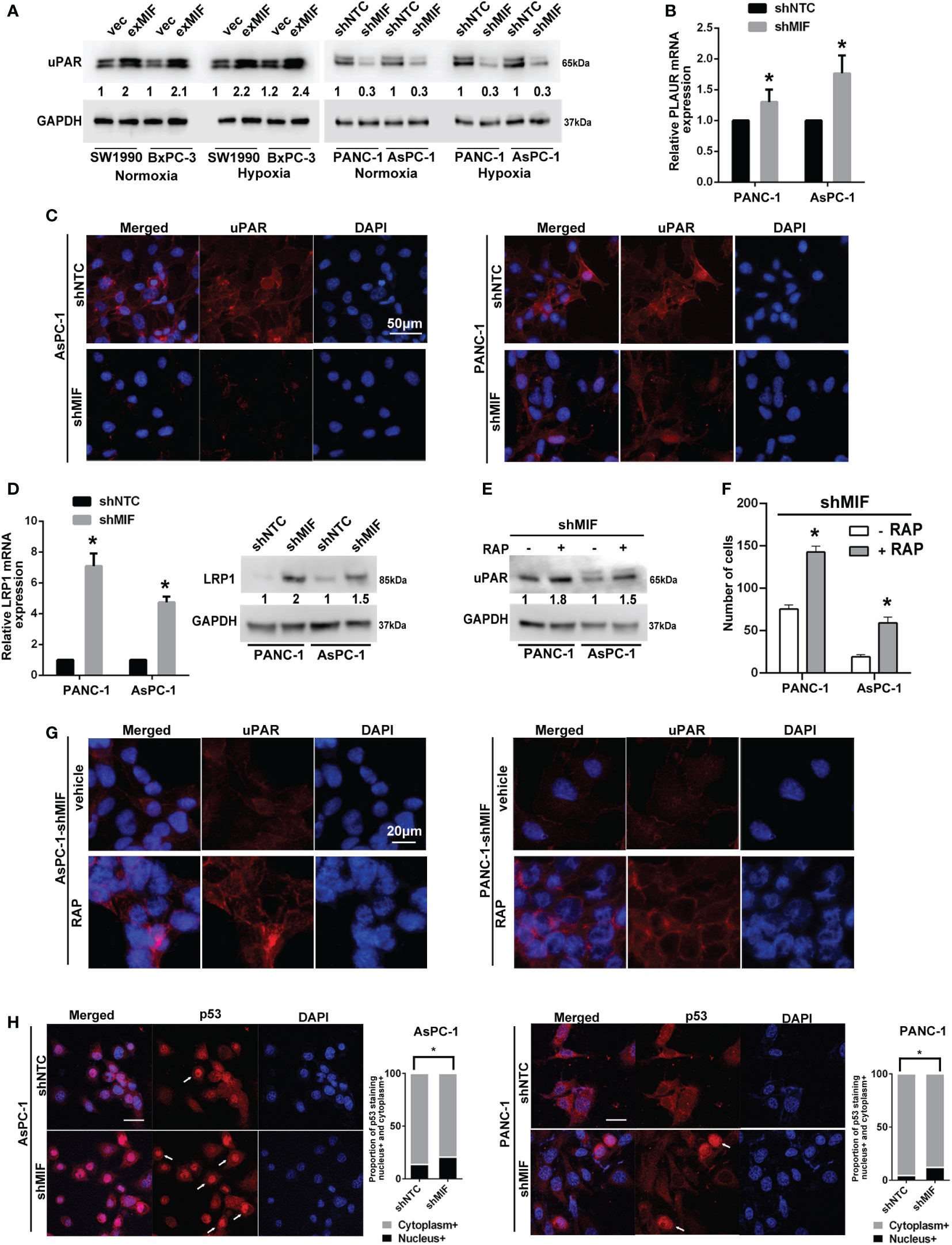
Figure 5 MIF enhances the expression of uPAR via LRP1 (A) Western blot analysis of uPAR in stably transduced PDAC cell lines after treatment with CoCl2 or vehicle for 24 h. The numbers below the blots are quantitative ratios of the uPAR/GAPDH band densities. (B) Real-time PCR analysis using specific primers for PLAUR with pancreatic cancer cell lines. (C) Confocal microscopy images of shMIF cells for uPAR expression on the cell surface and in the cytoplasm (red). Scale bars, 50 μm. (D) Real-time PCR and western blot analysis of the mRNA and protein levels of LRP1 in shMIF cells. (E) uPAR levels in shMIF cells treated with the LRP1 antagonist RAP or vehicle. (F) Quantification of invasion assays of shMIF cells treated with LRP1 antagonist, RAP or vehicle using Matrigel matrix-coated transwell chambers. (G) Immunofluorescence images of uPAR on the cell surface and in the cytoplasm with the addition of RAP or vehicle in shMIF cells. (H) Immunofluorescence images and quantification of nuclear/cytoplasmic p53 staining in AsPC-1 and PANC-1 cells. The graphs show the mean of the proportions of p53-positive staining in the nucleus versus the cytoplasm. Scale bars, 20 μm. *P < 0.05.
To determine the molecular basis for the effect of downregulated MIF expression on the levels of uPAR, we sought to detect the mRNA expression of uPAR in shMIF cells using real-time PCR. We found that the levels of uPAR mRNA were slightly elevated in shMIF cells (Figure 5B). As a multifunctional receptor, it is known that uPAR is internalized with uPA/PAI-1 (plasminogen activator inhibitor type-1) complexes via a mechanism involving LRP1 (34). Unligated uPAR is recycled back to the cell surface; however, uPAR recycling is not 100% efficient. As a result, the cell-surface abundance of uPAR can be reduced by LRP1-mediated uPAR endocytosis (50). To test whether the decreased expression of uPAR in shMIF cells is attributed to uPAR internalization, we detected the location of uPAR by immunofluorescence staining. Cell membrane and cytoplasmic uPAR were visualized by confocal microscopy imaging, which showed a moderate reduction in surface uPAR fluorescence intensity in shMIF cells compared with shNTC cells (Figure 5C).
To assess the possibility of the regulation of uPAR expression by LRP1, we detected the mRNA and protein levels of LRP1, which were elevated in shMIF cells (Figure 5D). Similarly, expression levels of MIF and LRP1 showed significantly negative correlation in 196 pancreatic cancers from TCGA using UCSC Xena database (Pearson’s rho r = -0.3718, P = 2.204e-7, Figure S2). This analysis suggested a significant negative correlation between LRP1 and MIF. To determine whether LRP1 participates in uPAR internalization, we measured the expression of uPAR with the LRP1 antagonist RAP (51, 52). We observed increased expression of uPAR in response to RAP in shMIF cells (Figure 5E). Importantly, as shown in Figures 5F, G, RAP treatment was sufficient to increase the number of invasive cells, and the signal intensity of cell surface uPAR was restored with decreased cytoplasmic uPAR in shMIF cells, suggesting that uPAR expression restoration by LRP1 inhibition leads to rescue of cell mobility. Finally, owing to previous studies suggesting p53-induced LRP1 upregulation and MIF-mediated inhibition of p53 activity (53–55), we reasoned that the increased LRP1 expression in shMIF cells may contribute to the release of p53 suppression by MIF knockdown. As expected, we observed a moderate increase in nuclear p53 fluorescence in MIF knockdown cells (Figure 5H) compared to shNTC cells, which corresponds with the effect of MIF on p53 nuclear import (56, 57). These results suggest one possible mechanism by which MIF promotes cell invasion by suppressing p53-regulated LRP1 expression and LRP1-uPAR endocytosis.
3.6 MIF and tumour development in vivo
MIF has been reported to be involved in the development and progression of a variety of tumours (58), and we next addressed the role of MIF in PDAC progression in vivo. We established subcutaneous xenografts in nude mice using MIF knockdown, MIF-overexpressing and control cells.
The volume of tumours developed in mice injected with shNTC cells was significantly higher than that in mice receiving shMIF PANC-1 cells. However, there were no significant differences in tumour volumes between the vector control and exMIF SW1990 cell groups (Figure 6). In addition, there were differences in tumour growth between the normoxic (sham operation) and hypoxic (femoral artery ligation of hind limb) groups either in shMIF vs. shNTC or in exMIF vs. vector groups. The protumorigenic ability of MIF may partially result from its prosurvival and antiapoptotic effects. As shown in Figure 7, the expression of Ki-67, a proliferating cell marker, was quantitated on the basis of the percentage. Consistently, while we did not identify significant differences in Ki-67-positive staining in the exMIF and vector control SW1990 cell groups, a low percentage of Ki-67-positive staining was observed in shMIF PANC-1 cells under normoxia and hypoxia. Meanwhile, apoptosis was determined by Annexin V-APC/PI staining analysis using flow cytometry, which showed that the percentages of apoptotic cells were higher in the MIF knockdown groups than in the shNTC groups and fewer apoptotic cells in the MIF-overexpressing groups than in the vector groups (Figure S3). With regard to discrepant results between in vivo and in vitro, we speculate that this may be due to the complex society of the tumour microenvironment in vivo, disturbing the prosurvival role of MIF. Collectively, these results indicated that the MIF-involved suppression of tumour growth may be due to its anti-apoptosis role.
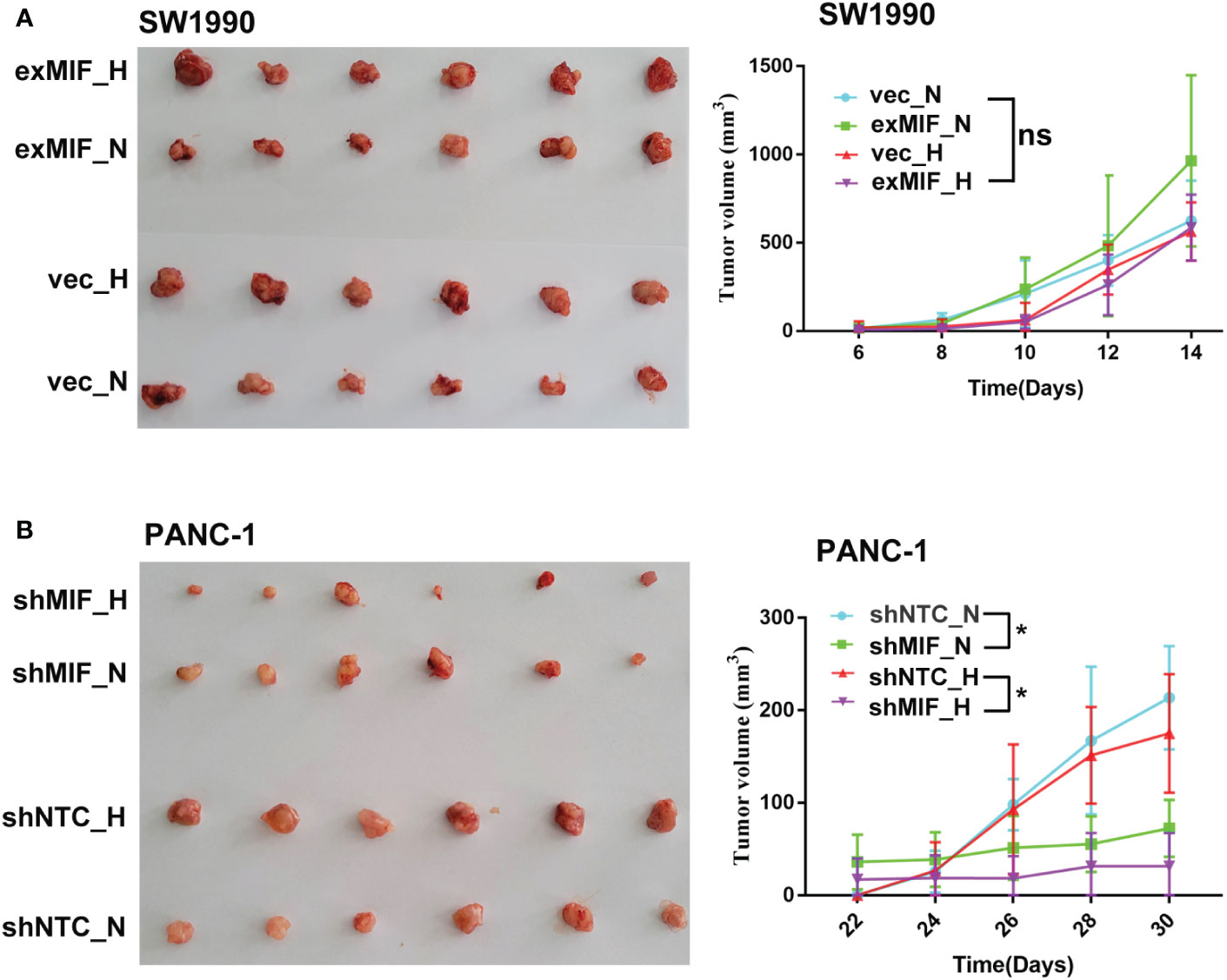
Figure 6 MIF knockdown attenuates the tumorigenesis of PDAC cells in vivo Representative images and quantification of subcutaneous xenograft tumours (n = 6 in each group) from stably transduced cell lines SW1990 (A) and PANC-1 (B) in nude mice after sham operation (N, normoxia) or femoral artery ligation (H, hypoxia). ns, no significance; *P < 0.05.
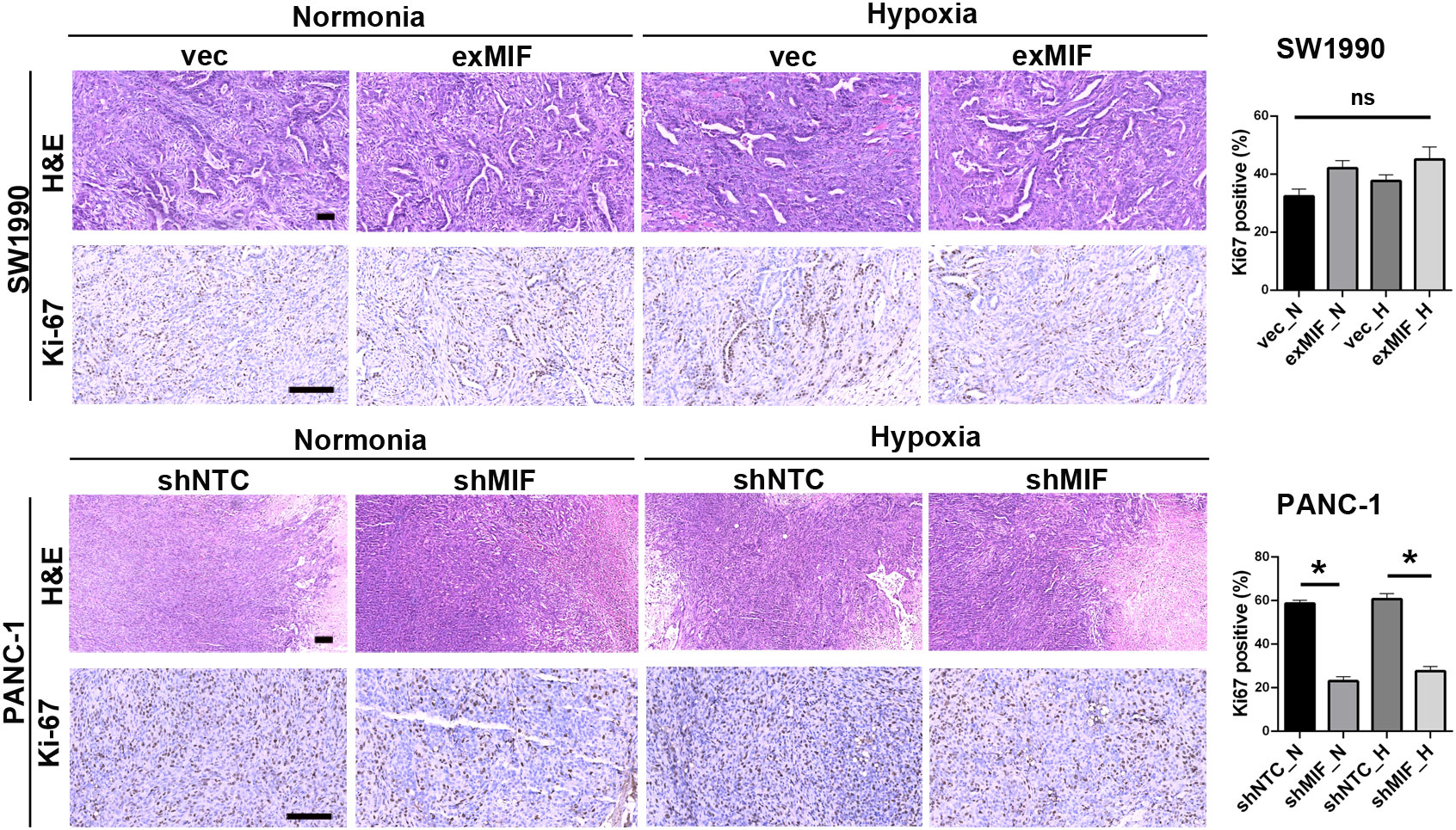
Figure 7 MIF knockdown inhibits proliferation of PDAC cells Representative images of xenograft tumours stained for H&E and Ki-67. The histograms show the quantification of Ki-67 staining; n = 6 per group. *P < 0.05.
3.7 MIF knockdown triggers multiple tumour-associated processes in PDAC cells
To further explore the mechanisms underlying the MIF knockdown-induced cell invasive phenotype attenuation, we performed transcriptome RNA-seq analysis in MIF knockdown PANC-1 cells. After screening, 3438 and 2802 significantly DEGs of shMIF vs. shNTC PANC-1 cells in normoxia (2259 up, 1179 down) and hypoxia (985 up, 1817 down) were shown in the volcano figures (Figures 8A, B). Then, the DEGs were analysed for their functions and enrichment pathways by GO and KEGG pathway analysis. Several biological processes associated with cancer progression were enriched in shMIF cells, including pathways in cancer, transcriptional misregulation in cancer, innate immune response, regulation of apoptosis and cell adhesion (Figure 8C-F). These findings correspond with our in vivo analysis and provide more comprehensive explanations for the role of MIF in the malignant behaviour of PDAC cells.
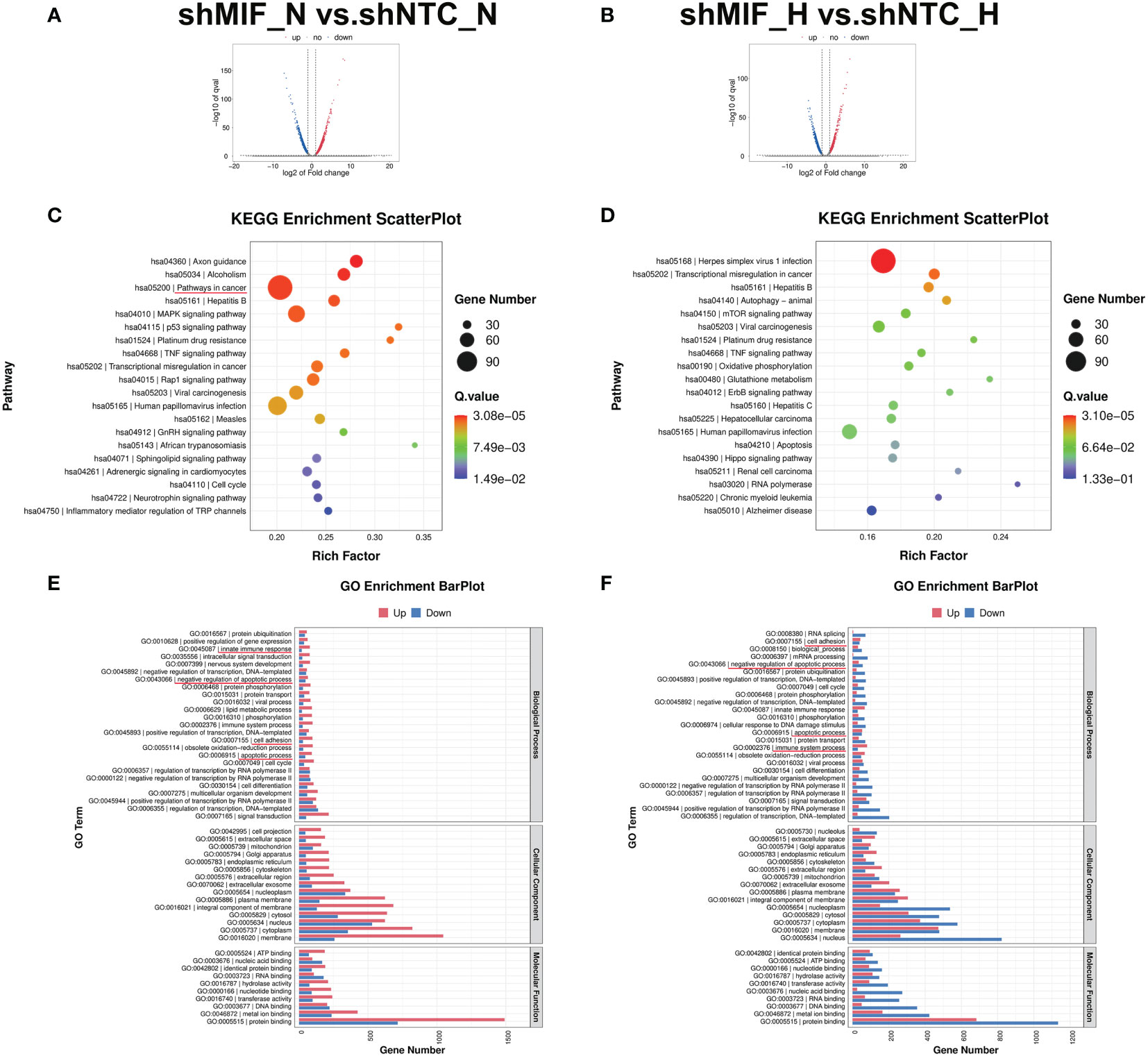
Figure 8 RNA-seq analysis in MIF knockdown PANC-1 cells (A, B) Volcano maps demonstrate the DEGs (fold-change >2 and FDR <0.005) regulated by MIF knockdown in PANC-1 cells. (C, D) KEGG pathway enrichment of the DEGs. (E, F) GO enrichment analysis of DEGs divided into three functional groups: biological process, cellular component, and molecular function.
4 Discussion
Tumour aggressiveness and treatment resistance are partially due to the complex milieu characterized by hypoxia, which is an important microenvironment feature in PDAC (38). The highly invasive and metastatic nature of pancreatic cancer requires the development of effective therapeutic interventions. Under hypoxia, multiple adaptive changes occur through hypoxic effects on the levels of mRNA, protein expression and DNA methylation. Buffa, F. M. et al. validated a common hypoxia signature in multiple cancers (46). By investigating hypoxia-induced spatial transcriptome distribution in human PDAC cells engrafted into mouse ischaemic hind limbs, we also identified multiple hypoxia-associated genes. Among them, a proinflammatory cytokine, MIF, attracted our attention.
It has been clear for the regulatory role for HIF-1α in the expression of MIF, and the role for MIF in HIF-1α stabilization (13–15). The reprogramming of cancer cells during hypoxic adaptation, in the respects of gene expression alterations, metabolism regulation, ECM deposition, remodeling and degradation, etc., has been extensively reviewed (59–62). In the research field of regulation of cancer cell invasion and metastasis, MIF showed similar effect or intersection with hypoxia, such as association with an increased expression of MMPs and PLAUR (19, 63–65) and macrophage recruitment (66, 67). Given the importance of MIF in malignant disease progression at hypoxia, in-depth studies of MIF directed therapeutics in cancer development are critical. MIF is ubiquitously and constitutively expressed in almost all cells, including numerous human cancer types, and it is involved in numerous biological processes (18). Emerging evidence has demonstrated the biological effects of MIF on the malignant characteristics of PDAC (68–72). Consistently, in the present study, we also observed the role of MIF in pro-invasiveness, anti-apoptosis and prognostic prediction. Interestingly, the main finding and focus of the current study was to investigate the involvement of LRP1-mediated uPAR endocytosis in the MIF-related invasive phenotype of PDAC.
uPAR has been shown to be upregulated in cancer cells, to play a critical role in driving plasminogen-dependent matrix degradation and to affect a variety of physiological and pathological processes, including cell migration, invasion, adhesion, survival and proliferation (73). In this report, we found a decreased protein abundance but not mRNA of uPAR in MIF-knockdown PDAC cells, suggesting that MIF may not affect PLAUR transcription. It is well known that internalization of cell uPAR, followed by either recycling back to the cell surface or degradation, is crucial for its homeostasis and functional signalling. Hence, to test the possibility that MIF altered the stability of uPAR, we examined its cellular location. Our data suggest that MIF knockdown regulated the cellular distribution of uPAR from the cytomembrane to the cytoplasm. The endocytosis of uPAR has been identified through LRP1, which is an endocytic receptor for multiple ligands. Next, we demonstrated the overexpression of LRP1 at both the mRNA and protein levels in MIF knockdown PDAC cells. Based on these results, we hypothesize that LRP1 is involved in the MIF-related decrease in the cell-surface abundance of uPAR. To investigate this possibility, we used the LRP1 antagonist RAP, which blocks the binding of soluble ligands to LRP1, subsequently blocking uPAR internalization. Our data showed that both the total and cell-surface abundance of uPAR protein were restored in the presence of RAP. Collectively, this evidence supports the role of LRP1 as a ubiquitous endocytic cell surface receptor that recognizes a wide range of ligands (34, 74).
Studies have indicated the role of LRP-1 in cancer progression. Although several studies have reported that a low level of LRP1 was associated with the aggressive phenotype of certain types of cancer (75–77), other studies have demonstrated that upregulation of LRP1 expression was observed in the progression of certain types of malignancy (78, 79). These results suggested that LRP1 expression may be dependent on the cancer subtypes and stages. Understanding how LRP1 is regulated is important for unscrambling its pleiotropic role. Data about the basal molecular mechanisms of LRP1 regulation have not been clearly characterized. A review by H. Emonard et al. described the regulation of LRP1 expression at the gene and transcript levels (80). In addition, Leslie PL et al. demonstrated that LRP1 was upregulated at the transcriptional level and protein level through direct promoter binding by p53 and p53-activating stresses (53). Based on previous work confirming the role of MIF in stabilizing the binding of p53-Mdm2, leading to suppression of p53 activity in apoptosis and cell cycle arrest, it is reasonable to presume that the elevated mRNA expression of LRP1 in MIF knockdown cells in the present study might result from MIF-induced inhibition of p53 activity (54, 81–83). More rigorous studies are needed to address this regulatory mechanism.
It is noteworthy that the present study is limited in the following aspects, which should be paid attention to during our further exploration in the effects of MIF on tumour progression: 1) Given MIF as a cytokine that secretes into the environment, considering and exploring its effect on the surrounding cells and cell-cell interactions should be a major concern and await the results of further studies; 2) The influence of MIF-p53-LRP1-uPAR signaling pathway on the ECM remodeling are need to be confirmed; 3) Although CoCl2-induced chemical hypoxia is one of the most commonly used models mimicking decreased oxygen concentration, there are limitations to this model. Firstly, the mechanisms by which CoCl2 stabilizes HIF1a/2a is not completely understood; Secondly, even HIF1a/2a is stabilized and similar transcriptional activities occurs, the transcription of distinct sets of genes not affected by low oxygen has been observed by CoCl2 treatment. We should make sure that the differences between effects of low oxygen concentration and CoCl2 in cell metabolism, transcriptional regulation and signaling pathways are adapted to our study purpose. Thirdly, in the CoCl2-induced chemical hypoxia model, oxygen is still present in cell culture. We must keep in mind the complex relationship between oxygen- and CoCl2-induced cellular responses (84–86).
In conclusion, our current study provides the first evidence regarding the association of LRP1-mediated uPAR endocytosis with the MIF-related invasive phenotype. It will be important to extend the invasion mechanism identified in this study to other types of malignancies. Our study also provides the potential for targeting the novel MIF-p53-LRP1-uPAR pathway (Figure 9) to improve the therapeutic outcome for PDAC patients.
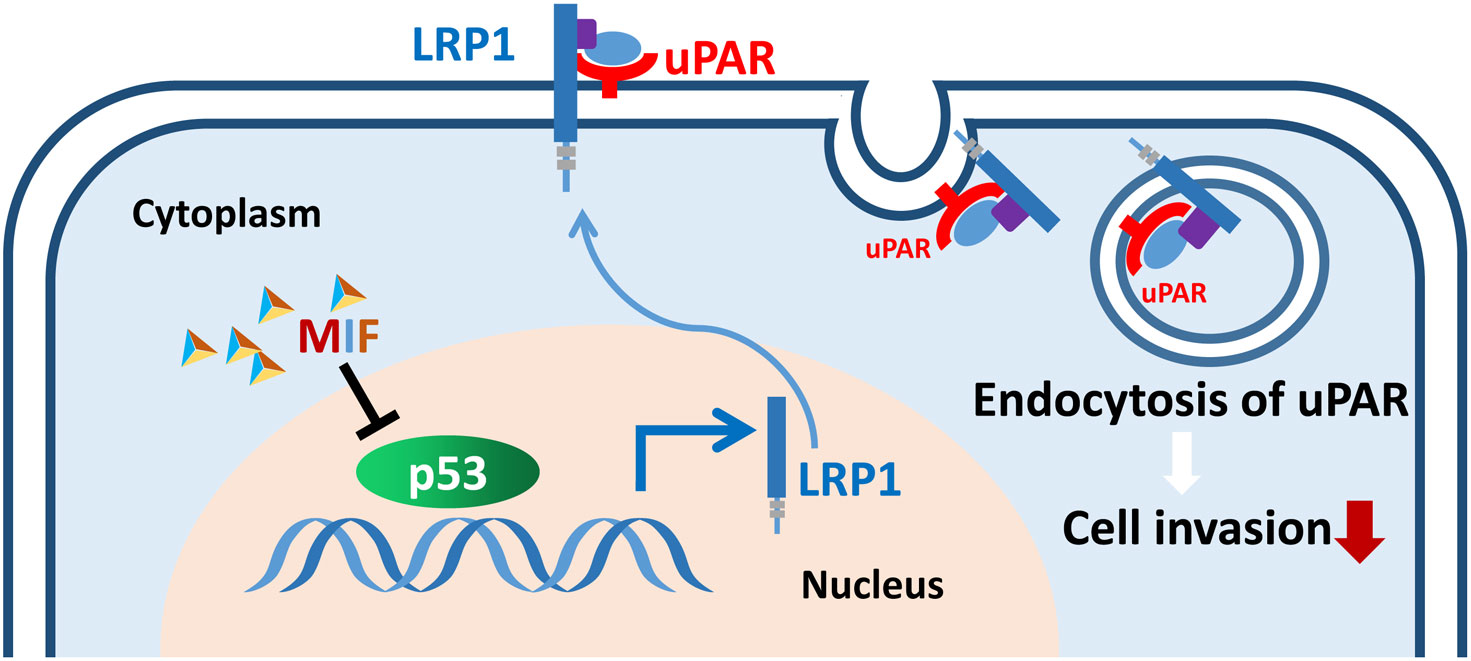
Figure 9 Schematic of the proposed mechanism by which the MIF-p53-LRP1-uPAR interaction promotes the invasion of PDAC cells. The interaction of MIF-p53 inhibits the degradation of p53, which upregulates the transcription of LRP1, and LRP1 then induces the internalization of uPAR and consequently weakens cell invasion.
Data availability statement
The original contributions presented in the study are included in the article/supplementary materials. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee of Tianjin University Cancer Hospital. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Institutional Animal Care and Use Committee at the Tianjin Medical University.
Author contributions
Conceptualization, HS, DZ and CH. Methodology, HS and DZ. Software, RC and YLL. Validation, HS, YG, FL and JM. Formal analysis, YLL and RC. Investigation, HS, YL and XB. Resources, HS. Writing—original draft preparation, HS and JM. Writing—review and editing, JM and CH. Visualization, DZ, FL and JM. Supervision, CH. Project administration, CH and JM. Funding acquisition, CH. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by National Nature Science Foundation of China, grant number 82072659.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1028070/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Kardosh A, Lichtensztajn DY, Gubens MA, Kunz PL, Fisher GA, Clarke CA. Long-term survivors of pancreatic cancer: A California population-based study. Pancreas (2018) 47(8):958–66. doi: 10.1097/Mpa.0000000000001133
3. Shah VM, Sheppard BC, Sears RC, Alani AW. Hypoxia: Friend or foe for drug delivery in pancreatic cancer. Cancer Lett (2020) 492:63–70. doi: 10.1016/j.canlet.2020.07.041
4. Bach JP, Rinn B, Meyer B, Dodel R, Bacher M. Role of mif in inflammation and tumorigenesis. Oncology-Basel (2008) 75(3-4):127–33. doi: 10.1159/000155223
5. Nobre CC, de Araujo JM, Fernandes TA, Cobucci RN, Lanza DC, Andrade VS, et al. Macrophage migration inhibitory factor (Mif): Biological activities and relation with cancer. Pathol Oncol Res (2017) 23(2):235–44. doi: 10.1007/s12253-016-0138-6
6. Wadgaonkar R, Somnay K, Garcia JG. Thrombin induced secretion of macrophage migration inhibitory factor (Mif) and its effect on nuclear signaling in endothelium. J Cell Biochem (2008) 105(5):1279–88. doi: 10.1002/jcb.21928
7. Lue H, Thiele M, Franz J, Dahl E, Speckgens S, Leng L, et al. Macrophage migration inhibitory factor (Mif) promotes cell survival by activation of the akt pathway and role for Csn5/Jab1 in the control of autocrine mif activity. Oncogene (2007) 26(35):5046–59. doi: 10.1038/sj.onc.1210318
8. Xie J, Yang L, Tian L, Li W, Yang L, Li L. Macrophage migration inhibitor factor upregulates mcp-1 expression in an autocrine manner in hepatocytes during acute mouse liver injury. Sci Rep (2016) 6:27665. doi: 10.1038/srep27665
9. Ma H, Wang J, Thomas DP, Tong C, Leng L, Wang W, et al. Impaired macrophage migration inhibitory factor-Amp-Activated protein kinase activation and ischemic recovery in the senescent heart. Circulation (2010) 122(3):282–92. doi: 10.1161/CIRCULATIONAHA.110.953208
10. Jankauskas SS, Wong DWL, Bucala R, Djudjaj S, Boor P. Evolving complexity of mif signaling. Cell Signal (2019) 57:76–88. doi: 10.1016/j.cellsig.2019.01.006
11. Noe JT, Mitchell RA. Mif-dependent control of tumor immunity. Front Immunol (2020) 11:609948. doi: 10.3389/fimmu.2020.609948
12. Lippitz BE. Cytokine patterns in patients with cancer: A systematic review. Lancet Oncol (2013) 14(6):e218–28. doi: 10.1016/S1470-2045(12)70582-X
13. Baugh JA, Gantier M, Li L, Byrne A, Buckley A, Donnelly SC. Dual regulation of macrophage migration inhibitory factor (Mif) expression in hypoxia by creb and hif-1. Biochem Biophys Res Commun (2006) 347(4):895–903. doi: 10.1016/j.bbrc.2006.06.148
14. Winner M, Koong AC, Rendon BE, Zundel W, Mitchell RA. Amplification of tumor hypoxic responses by macrophage migration inhibitory factor-dependent hypoxia-inducible factor stabilization. Cancer Res (2007) 67(1):186–93. doi: 10.1158/0008-5472.CAN-06-3292
15. Bemis L, Chan DA, Finkielstein CV, Qi L, Sutphin PD, Chen X, et al. Distinct aerobic and hypoxic mechanisms of hif-alpha regulation by Csn5. Genes Dev (2004) 18(7):739–44. doi: 10.1101/gad.1180104
16. O'Reilly C, Doroudian M, Mawhinney L, Donnelly SC. Targeting mif in cancer: Therapeutic strategies, current developments, and future opportunities. Med Res Rev (2016) 36(3):440–60. doi: 10.1002/med.21385
17. Kindt N, Journe F, Laurent G, Saussez S. Involvement of macrophage migration inhibitory factor in cancer and novel therapeutic targets. Oncol Lett (2016) 12(4):2247–53. doi: 10.3892/ol.2016.4929
18. Sumaiya K, Langford D, Natarajaseenivasan K, Shanmughapriya S. Macrophage migration inhibitory factor (Mif): A multifaceted cytokine regulated by genetic and physiological strategies. Pharmacol Ther (2022) 233:108024. doi: 10.1016/j.pharmthera.2021.108024
19. Wang SS, Zheng M, Pang X, Zhang M, Yu XH, Wu JB, et al. Macrophage migration inhibitory factor promotes the invasion and metastasis of oral squamous cell carcinoma through matrix metalloprotein-2/9. Mol carcinogenesis (2019) 58(10):1809–21. doi: 10.1002/mc.23067
20. Luo Y, Yi H, Huang X, Lin G, Kuang Y, Guo Y, et al. Inhibition of macrophage migration inhibitory factor (Mif) as a therapeutic target in bleomycin-induced pulmonary fibrosis rats. Am J Physiol Lung Cell Mol Physiol (2021) 321(1):L6–L16. doi: 10.1152/ajplung.00288.2020
21. Lu H, Bai Y, Wu L, Hong W, Liang Y, Chen B, et al. Inhibition of macrophage migration inhibitory factor protects against inflammation and matrix deposition in kidney tissues after injury. Mediators Inflammation (2016) 2016:2174682. doi: 10.1155/2016/2174682
22. Kong YZ, Yu X, Tang JJ, Ouyang X, Huang XR, Fingerle-Rowson G, et al. Macrophage migration inhibitory factor induces mmp-9 expression: Implications for destabilization of human atherosclerotic plaques. Atherosclerosis (2005) 178(1):207–15. doi: 10.1016/j.atherosclerosis.2004.08.030
23. Meyer-Siegler K. Macrophage migration inhibitory factor increases mmp-2 activity in Du-145 prostate cells. Cytokine (2000) 12(7):914–21. doi: 10.1006/cyto.2000.0682
24. Onodera S, Kaneda K, Mizue Y, Koyama Y, Fujinaga M, Nishihira J. Macrophage migration inhibitory factor up-regulates expression of matrix metalloproteinases in synovial fibroblasts of rheumatoid arthritis. J Biol Chem (2000) 275(1):444–50. doi: 10.1074/jbc.275.1.444
25. Welch DR, Hurst DR. Defining the hallmarks of metastasis. Cancer Res (2019) 79(12):3011–27. doi: 10.1158/0008-5472.CAN-19-0458
26. Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol (2011) 21(4):228–37. doi: 10.1016/j.tcb.2010.12.002
27. He Y, Liu XD, Chen ZY, Zhu J, Xiong Y, Li K, et al. Interaction between cancer cells and stromal fibroblasts is required for activation of the upar-Upa-Mmp-2 cascade in pancreatic cancer metastasis. Clin Cancer Res (2007) 13(11):3115–24. doi: 10.1158/1078-0432.CCR-06-2088
28. Baricos WH, Cortez SL, el-Dahr SS, Schnaper HW. Ecm degradation by cultured human mesangial cells is mediated by a Pa/Plasmin/Mmp-2 cascade. Kidney Int (1995) 47(4):1039–47. doi: 10.1038/ki.1995.150
29. Zhou HM, Nichols A, Meda P, Vassalli JD. Urokinase-type plasminogen activator and its receptor synergize to promote pathogenic proteolysis. EMBO J (2000) 19(17):4817–26. doi: 10.1093/emboj/19.17.4817
30. Behrendt N, Ronne E, Dano K. The structure and function of the urokinase receptor, a membrane protein governing plasminogen activation on the cell surface. Biol Chem Hoppe Seyler (1995) 376(5):269–79.
31. Kumar AA, Buckley BJ, Ranson M. The urokinase plasminogen activation system in pancreatic cancer: Prospective diagnostic and therapeutic targets. Biomolecules (2022) 12(2):152. doi: 10.3390/Biom12020152
32. Smith HW, Marshall CJ. Regulation of cell signalling by upar. Nat Rev Mol Cell Biol (2010) 11(1):23–36. doi: 10.1038/nrm2821
33. Blasi F, Carmeliet P. Upar: A versatile signalling orchestrator. Nat Rev Mol Cell Biol (2002) 3(12):932–43. doi: 10.1038/nrm977
34. Gonias SL, Gaultier A, Jo M. Regulation of the urokinase receptor (Upar) by ldl receptor-related protein-1 (Lrp1). Curr Pharm Des (2011) 17(19):1962–9. doi: 10.2174/138161211796718224
35. Montel V, Gaultier A, Lester RD, Campana WM, Gonias SL. The low-density lipoprotein receptor-related protein regulates cancer cell survival and metastasis development. Cancer Res (2007) 67(20):9817–24. doi: 10.1158/0008-5472.CAN-07-0683
36. Nakagawa S, Schielzeth H. Repeatability for Gaussian and non-Gaussian data: A practical guide for biologists. Biol Rev Camb Philos Soc (2010) 85(4):935–56. doi: 10.1111/j.1469-185X.2010.00141.x
37. Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data Via the xena platform. Nat Biotechnol (2020) 38(6):675–8. doi: 10.1038/s41587-020-0546-8
38. Sun H, Zhang D, Huang C, Guo Y, Yang Z, Yao N, et al. Hypoxic microenvironment induced spatial transcriptome changes in pancreatic cancer. Cancer Biol Med (2021) 18(2):616–30. doi: 10.20892/j.issn.2095-3941.2021.0158
39. Mo J, Zhao X, Wang W, Zhao N, Dong X, Zhang Y, et al. Tfpi2 promotes perivascular migration in an angiotropism model of melanoma. Front Oncol (2021) 11:662434. doi: 10.3389/fonc.2021.662434
40. Couffinhal T, Silver M, Zheng LP, Kearney M, Witzenbichler B, Isner JM. Mouse model of angiogenesis. Am J Pathol (1998) 152(6):1667–79.
41. Richard V, Kindt N, Saussez S. Macrophage migration inhibitory factor involvement in breast cancer (Review). Int J Oncol (2015) 47(5):1627–33. doi: 10.3892/ijo.2015.3185
42. Hamatake M, Yoshino I, Tomiyasu M, Miura N, Okazaki H, Ohba T, et al. Intratumoral expression of macrophage migration inhibitory factor is correlated with serum c-reactive protein and interleukin-6 in patients with non-small cell lung cancer. Surg Today (2008) 38(10):921–5. doi: 10.1007/s00595-008-3765-z
43. Hagemann T, Robinson SC, Thompson RG, Charles K, Kulbe H, Balkwill FR. Ovarian cancer cell-derived migration inhibitory factor enhances tumor growth, progression, and angiogenesis. Mol Cancer Ther (2007) 6(7):1993–2002. doi: 10.1158/1535-7163.MCT-07-0118
44. Ahmed A, Kohler S, Klotz R, Giese N, Lasitschka F, Hackert T, et al. Peripheral blood and tissue assessment highlights differential tumor-circulatory gradients of Il2 and mif with prognostic significance in resectable pancreatic ductal adenocarcinoma. Oncoimmunology (2021) 10(1):1962135. doi: 10.1080/2162402X.2021.1962135
45. Jager B, Klatt D, Plappert L, Golpon H, Lienenklaus S, Barbosa PD, et al. Cxcr4/Mif axis amplifies tumor growth and epithelial-mesenchymal interaction in non-small cell lung cancer. Cell Signal (2020) 73:109672. doi: 10.1016/j.cellsig.2020.109672
46. Buffa FM, Harris AL, West CM, Miller CJ. Large Meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br J Cancer (2010) 102(2):428–35. doi: 10.1038/sj.bjc.6605450
47. Elsby LM, Donn R, Alourfi Z, Green LM, Beaulieu E, Ray DW. Hypoxia and glucocorticoid signaling converge to regulate macrophage migration inhibitory factor gene expression. Arthritis Rheum (2009) 60(8):2220–31. doi: 10.1002/art.24659
48. Xu X, Wang B, Ye C, Yao C, Lin Y, Huang X, et al. Overexpression of macrophage migration inhibitory factor induces angiogenesis in human breast cancer. Cancer Lett (2008) 261(2):147–57. doi: 10.1016/j.canlet.2007.11.028
49. Yu X, Lin SG, Huang XR, Bacher M, Leng L, Bucala R, et al. Macrophage migration inhibitory factor induces mmp-9 expression in macrophages Via the mek-erk map kinase pathway. J Interferon Cytokine Res (2007) 27(2):103–9. doi: 10.1089/jir.2006.0054
50. Nykjaer A, Conese M, Christensen EI, Olson D, Cremona O, Gliemann J, et al. Recycling of the urokinase receptor upon internalization of the Upa:Serpin complexes. EMBO J (1997) 16(10):2610–20. doi: 10.1093/emboj/16.10.2610
51. Iadonato SP, Bu G, Maksymovitch EA, Schwartz AL. Interaction of a 39 kda protein with the low-Density-Lipoprotein-Receptor-Related protein (Lrp) on rat hepatoma cells. Biochem J (1993) 296(Pt 3):867–75. doi: 10.1042/bj2960867
52. Mantuano E, Lam MS, Gonias SL. Lrp1 assembles unique Co-receptor systems to initiate cell signaling in response to tissue-type plasminogen activator and myelin-associated glycoprotein. J Biol Chem (2013) 288(47):34009–18. doi: 10.1074/jbc.M113.509133
53. Leslie PL, Franklin DA, Liu Y, Zhang Y. P53 regulates the expression of Lrp1 and apoptosis through a stress intensity-dependent microrna feedback loop. Cell Rep (2018) 24(6):1484–95. doi: 10.1016/j.celrep.2018.07.010
54. Jung H, Seong HA, Ha H. Critical role of cysteine residue 81 of macrophage migration inhibitory factor (Mif) in mif-induced inhibition of P53 activity. J Biol Chem (2008) 283(29):20383–96. doi: 10.1074/jbc.M800050200
55. Kong F, Deng X, Kong X, Du Y, Li L, Zhu H, et al. Zfpm2-As1, a novel lncrna, attenuates the P53 pathway and promotes gastric carcinogenesis by stabilizing mif. Oncogene (2018) 37(45):5982–96. doi: 10.1038/s41388-018-0387-9
56. Fukaya R, Ohta S, Yaguchi T, Matsuzaki Y, Sugihara E, Okano H, et al. Mif maintains the tumorigenic capacity of brain tumor-initiating cells by directly inhibiting P53. Cancer Res (2016) 76(9):2813–23. doi: 10.1158/0008-5472.CAN-15-1011
57. Mitchell RA, Liao H, Chesney J, Fingerle-Rowson G, Baugh J, David J, et al. Macrophage migration inhibitory factor (Mif) sustains macrophage proinflammatory function by inhibiting P53: Regulatory role in the innate immune response. Proc Natl Acad Sci United States America (2002) 99(1):345–50. doi: 10.1073/pnas.012511599
58. Bifulco C, McDaniel K, Leng L, Bucala R. Tumor growth-promoting properties of macrophage migration inhibitory factor. Curr Pharm Des (2008) 14(36):3790–801. doi: 10.2174/138161208786898608
59. Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer (2008) 8(12):967–75. doi: 10.1038/nrc2540
60. Finger EC, Giaccia AJ. Hypoxia, inflammation, and the tumor microenvironment in metastatic disease. Cancer Metastasis Rev (2010) 29(2):285–93. doi: 10.1007/s10555-010-9224-5
61. Semenza GL. Molecular mechanisms mediating metastasis of hypoxic breast cancer cells. Trends Mol Med (2012) 18(9):534–43. doi: 10.1016/j.molmed.2012.08.001
62. Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: Drivers of tumour metastasis. Nat Rev Cancer (2014) 14(6):430–9. doi: 10.1038/nrc3726
63. Munoz-Najar UM, Neurath KM, Vumbaca F, Claffey KP. Hypoxia stimulates breast carcinoma cell invasion through Mt1-mmp and mmp-2 activation. Oncogene (2006) 25(16):2379–92. doi: 10.1038/sj.onc.1209273
64. Choi JY, Jang YS, Min SY, Song JY. Overexpression of mmp-9 and hif-1alpha in breast cancer cells under hypoxic conditions. J Breast Cancer (2011) 14(2):88–95. doi: 10.4048/jbc.2011.14.2.88
65. Buchler P, Reber HA, Tomlinson JS, Hankinson O, Kallifatidis G, Friess H, et al. Transcriptional regulation of urokinase-type plasminogen activator receptor by hypoxia-inducible factor 1 is crucial for invasion of pancreatic and liver cancer. Neoplasia (2009) 11(2):196–206. doi: 10.1593/neo.08734
66. Casazza A, Laoui D, Wenes M, Rizzolio S, Bassani N, Mambretti M, et al. Impeding macrophage entry into hypoxic tumor areas by Sema3a/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell (2013) 24(6):695–709. doi: 10.1016/j.ccr.2013.11.007
67. Barnes MA, McMullen MR, Roychowdhury S, Madhun NZ, Niese K, Olman MA, et al. Macrophage migration inhibitory factor is required for recruitment of scar-associated macrophages during liver fibrosis. J Leukoc Biol (2015) 97(1):161–9. doi: 10.1189/jlb.3A0614-280R
68. Denz A, Pilarsky C, Muth D, Ruckert F, Saeger HD, Grutzmann R. Inhibition of mif leads to cell cycle arrest and apoptosis in pancreatic cancer cells. J Surg Res (2010) 160(1):29–34. doi: 10.1016/j.jss.2009.03.048
69. Yang S, He P, Wang J, Schetter A, Tang W, Funamizu N, et al. A novel mif signaling pathway drives the malignant character of pancreatic cancer by targeting Nr3c2. Cancer Res (2016) 76(13):3838–50. doi: 10.1158/0008-5472.CAN-15-2841
70. Funamizu N, Hu C, Lacy C, Schetter A, Zhang G, He P, et al. Macrophage migration inhibitory factor induces epithelial to mesenchymal transition, enhances tumor aggressiveness and predicts clinical outcome in resected pancreatic ductal adenocarcinoma. Int J Cancer (2013) 132(4):785–94. doi: 10.1002/ijc.27736
71. Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol (2015) 17(6):816–26. doi: 10.1038/ncb3169
72. Wang D, Wang R, Huang A, Fang Z, Wang K, He M, et al. Upregulation of macrophage migration inhibitory factor promotes tumor metastasis and correlates with poor prognosis of pancreatic ductal adenocarcinoma. Oncol Rep (2018) 40(5):2628–36. doi: 10.3892/or.2018.6703
73. Alfano D, Franco P, Stoppelli MP. Modulation of cellular function by the urokinase receptor signalling: A mechanistic view. Front Cell Dev Biol (2022) 10:818616. doi: 10.3389/fcell.2022.818616
74. Herz J, Strickland DK. Lrp: A multifunctional scavenger and signaling receptor. J Clin Invest (2001) 108(6):779–84. doi: 10.1172/JCI13992
75. Wang B, Shen C, Li Y, Zhang T, Huang H, Ren J, et al. Oridonin overcomes the gemcitabine resistant panc-1/Gem cells by regulating gst pi and Lrp/1 Erk/Jnk signalling. Onco Targets Ther (2019) 12:5751–65. doi: 10.2147/OTT.S208924
76. Huang XY, Shi GM, Devbhandari RP, Ke AW, Wang Y, Wang XY, et al. Low level of low-density lipoprotein receptor-related protein 1 predicts an unfavorable prognosis of hepatocellular carcinoma after curative resection. PLoS One (2012) 7(3):e32775. doi: 10.1371/journal.pone.0032775
77. Meng H, Chen G, Zhang X, Wang Z, Thomas DG, Giordano TJ, et al. Stromal Lrp1 in lung adenocarcinoma predicts clinical outcome. Clin Cancer Res (2011) 17(8):2426–33. doi: 10.1158/1078-0432.CCR-10-2385
78. Yamamoto M, Ikeda K, Ohshima K, Tsugu H, Kimura H, Tomonaga M. Increased expression of low density lipoprotein receptor-related Protein/Alpha2-macroglobulin receptor in human malignant astrocytomas. Cancer Res (1997) 57(13):2799–805.
79. Catasus L, Llorente-Cortes V, Cuatrecasas M, Pons C, Espinosa I, Prat J. Low-density lipoprotein receptor-related protein 1 (Lrp-1) is associated with highgrade, advanced stage and P53 and P16 alterations in endometrial carcinomas. Histopathology (2011) 59(3):567–71. doi: 10.1111/j.1365-2559.2011.03942.x
80. Emonard H, Theret L, Bennasroune AH, Dedieu S. Regulation of lrp-1 expression: Make the point. Pathol Biol (Paris) (2014) 62(2):84–90. doi: 10.1016/j.patbio.2014.02.002
81. Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the P53 pathway by small-molecule antagonists of Mdm2. Science (2004) 303(5659):844–8. doi: 10.1126/science.1092472
82. Costa AF, Gomes SZ, Lorenzon-Ojea AR, Martucci M, Faria MR, Pinto Ddos S Jr., et al. Macrophage migration inhibitory factor induces phosphorylation of Mdm2 mediated by phosphatidylinositol 3-Kinase/Akt kinase: Role of this pathway in decidual cell survival. Placenta (2016) 41:27–38. doi: 10.1016/j.placenta.2016.03.001
83. Chen T, Wang F, Wei S, Nie Y, Zheng X, Deng Y, et al. Fgfr/Rack1 interacts with Mdm2, promotes P53 degradation, and inhibits cell senescence in lung squamous cell carcinoma. Cancer Biol Med (2021) 18(3):665–74. doi: 10.20892/j.issn.2095-3941.2020.0389
84. Munoz-Sanchez J, Chanez-Cardenas ME. The use of cobalt chloride as a chemical hypoxia model. J Appl Toxicol (2019) 39(4):556–70. doi: 10.1002/jat.3749
85. Befani C, Mylonis I, Gkotinakou IM, Georgoulias P, Hu CJ, Simos G, et al. Cobalt stimulates hif-1-Dependent but inhibits hif-2-Dependent gene expression in liver cancer cells. Int J Biochem Cell Biol (2013) 45(11):2359–68. doi: 10.1016/j.biocel.2013.07.025
Keywords: MIF, uPAR, LRP1, hypoxia, spatial transcriptomics
Citation: Sun H, Cheng R, Zhang D, Guo Y, Li F, Li Y, Li Y, Bai X, Mo J and Huang C (2023) MIF promotes cell invasion by the LRP1-uPAR interaction in pancreatic cancer cells. Front. Oncol. 12:1028070. doi: 10.3389/fonc.2022.1028070
Received: 25 August 2022; Accepted: 20 December 2022;
Published: 10 January 2023.
Edited by:
Robert Cheng, National Cancer Institute at Frederick (NIH), United StatesReviewed by:
Yunlong Yang, Fudan University, ChinaGerard Bannenberg, Global Organization for EPA and DHA Omega-3s (GOED), United States
Wei Huang, Sichuan University, China
Copyright © 2023 Sun, Cheng, Zhang, Guo, Li, Li, Li, Bai, Mo and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chongbiao Huang, Y2hodWFuZ0B0bXUuZWR1LmNu; Jing Mo, bW9qaW5nQHRtdS5lZHUuY24=
 Huizhi Sun1
Huizhi Sun1 Runfen Cheng
Runfen Cheng Danfang Zhang
Danfang Zhang Fan Li
Fan Li Yanlei Li
Yanlei Li Yue Li
Yue Li Xiaoyu Bai
Xiaoyu Bai Jing Mo
Jing Mo