- 1Department of Pathology, West China Second Hospital of Sichuan University, Chengdu, Sichuan, China
- 2Key Laboratory of Birth Defects and Related Diseases of Women and Children (Sichuan University), Ministry of Education, Chengdu, Sichuan, China
- 3NHC Key Laboratory of Chronobiology (Sichuan University), Chengdu, Sichuan, China
CIC–DUX4 fusion-positive sarcoma is a subtype of undifferentiated small round cell sarcoma that is rarely reported. As far as we know, less than 200 cases have been reported worldwide to date. The clinicopathologic characteristics of this kind of tumor are non-specific, which makes it difficult to be diagnosed. Therefore, more cases are required to enrich the diagnosis and treatment experience. Here, we present a 17-year-old Asian girl diagnosed with CIC–DUX4 fusion-positive sarcoma after targeted next-generation sequencing. Her clinical manifestation was abdominal pain. Furthermore, a mass in the pelvic cavity and massive ascites were found after an imaging examination. After resection, the mass was sent to the pathology department for a definite diagnosis, and the micromorphology showed an undifferentiated sarcoma with massive necrosis. The tumor cells were round to spindle with clear to eosinophilic cytoplasm and vesicular nuclei. Rhabdoid cells and myxoid mesenchyme were focally shown. Immunohistochemical staining showed diffusely positive for vimentin, cyclin D1, Fli-1, and WT-1 and very focally positive for CD99. Moreover, the targeted next-generation sequencing also revealed other genetic changes in this tumor including LongInDel of POLE, copy number variation of CD79, low tumor mutational burden, and microsatellite stability. With a follow-up time of 6 months, the patient survived the disease and received chemotherapy routinely. This report presented a rare primary site CIC–DUX4 fusion-positive sarcoma (CDS) and revealed novel genetic changes that enrich the manifestation, histology, and cytogenetic scales of this rare sarcoma. In addition, we have summarized the clinicopathologic characteristics of this tumor by reviewing the literature to have a better understanding of CIC–DUX4 fusion-positive sarcomas, which may be helpful for diagnosis and treatment.
Introduction
Undifferentiated small round cell sarcomas (USRCSs) are a group of tumors with similar morphology, which makes it difficult to identify and diagnose. However, with the rapidly developing molecular pathology, it is possible to clarify them by genetic changes. CIC-rearranged sarcoma is one of the new tumor types of the USRCS, which used to be considered Ewing-like sarcoma.
Somers et al. (1) presented a case of an unusual and more aggressive cutaneous and subcutaneous primitive neuroectodermal tumor/Ewing sarcoma (PNET/ES), which showed a complex karyotype with t(4;19)(q33~q35;q13.1). Two years later, Kawamura-Saito et al. (2) found a recurrent chromosomal translocation t(4;19)(q35;q13) in two cases and recognized the fusion of CIC–DUX4. Afterward, Italiano et al. implicated that CIC was fused to copies of DUX4 gene not only on 4q35 but also on 10q26.3 (3). Moreover, more fusion partners of CIC were found, such as FOXO4 (4, 5) and NUTM1 (6, 7). More cases were included to summarize the characteristics of this kind of tumor and showed a distinct immunoprofile, karyotype, and worse outcome as compared to ES (8, 9), suggesting that it was a new pathologic entity.
To date, more than 200 cases of CIC-rearranged sarcoma have been reported, among which the CIC–DUX4 was the most common fusion (1–3, 7–32). Here, we report a case of CIC–DUX4 fusion-positive sarcoma (CDS) arising in the pelvic cavity of a 17-year-old girl with targeted next-generation sequencing results, which enriches the manifestation, histology, and cytogenetic scales of this rare sarcoma.
Case report
A 17-year-old Asian girl came to the hospital with complaints of abdominal pain. A computed tomography (CT) scan showed a cystic and solid mass measured 12.1 cm × 8.2 cm × 13.6 cm in the pelvic cavity with heterogeneous enhancement in an enhanced scan (Figure 1). Ultrasound detected fluid dark areas in multiple abdominal spaces. The serum level of carbohydrate antigen 125 was 447.6 U/ml. After adequate evaluation and preparation, the patient received surgery. During the surgery, ascites with a volume of 3,000 ml were drained from the abdominal cavity. Both the mass and ascites were sent for pathologic examination.
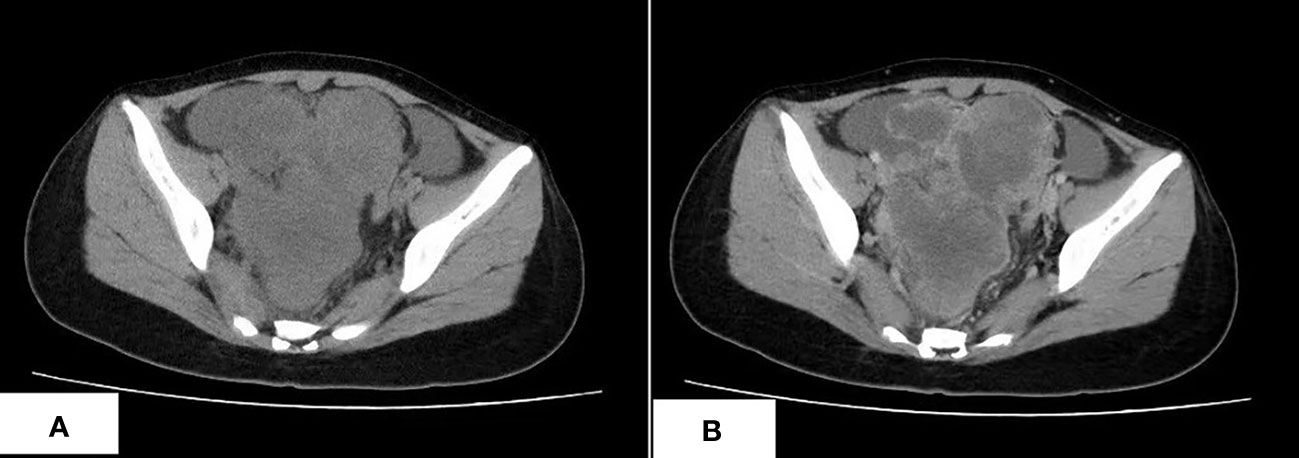
Figure 1 The CT scan of CDS. (A) An irregular lobulated mass in pelvic cavity, which has unclear margin with peripheral structure, is shown. (B) The tumor shows heterogeneous enhancement in the contrast-enhanced CT scan.
Grossly, the tumor showed a fused-nodular mass with an appearance of color and a touch of tenderness when being cut. The histological examination demonstrated the tumor consisted of round to spindle malignant cells with clear to eosinophilic cytoplasm and vesicular nuclei. Rhabdoid cells and myxoid mesenchyme were focally shown. Moreover, necrosis and pathologic mitoses were apparent (Figures 2A–F). Tumor cells were also detected in the peritoneal lavage. Immunohistochemical (IHC) staining showed diffusely positive for vimentin, Fli-1, cyclin D1, and WT-1 and very focally positive for CD99 (Figures 2G–I). Pan-CK, ER, PR, CD117, DOG-1, NSE, Syn, CD34, S100, desmin, caldesmon, CD10, and Sall-4 were all negative. The index of Ki-67 was >90%.
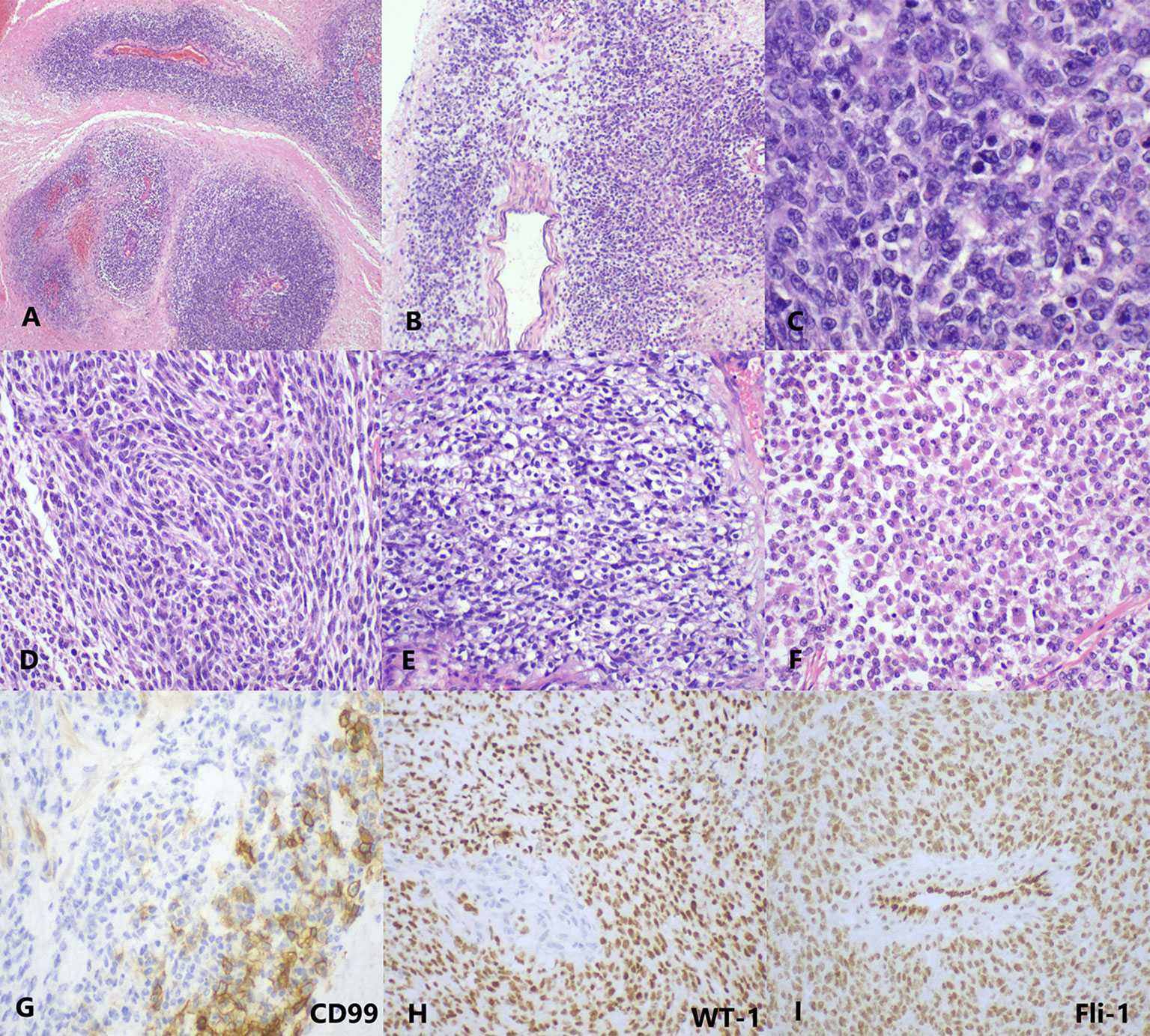
Figure 2 The histopathology and immunohistology of CDS. (A) At low magnification, the tumor cells are separated by massive necrosis, arranged in nest-like and perivascular (H&E, ×40). (B) Focal myxoid stroma (H&E, ×40). (C) At high magnification, the tumor cells show round-to-oval shape with lightly eosinophilic cytoplasm and vesicular nuclei; several mitotic figures are visible (H&E, ×400). Moreover, (D) spindle cell-like (H&E, ×200), (E) clear cell-like (H&E, ×200), and (F) rhabdoid plasmacytoid tumor cells (H&E, ×200) are also exhibited in some areas. (G) CD99 is focally membranous positive (IHC, ×400). (H) WT-1 (IHC, ×200) and (I) Fli-1 (IHC, ×200) are diffusely nuclear positive. IHC, immunohistochemistry.
For diagnosis and further treatment strategy, the paraffin section of the tumor was sent for targeted next-generation sequencing (NGS) on the Illumina platform. The exons of 672 and 633 genes were detected by DNA sequencing and RNA sequencing, respectively. The results were as follows: 1) fusion between CIC exon 20 and DUX4 exon 1 (Figure 3A), 2) LongInDel of POLE (exon22_exon31del) (Figure 3B), 3) copy number variation (CNV) of CD79A (5.8 copies), 4) low tumor mutational burden (TMB) at 0.5 Muts/Mb, and 5) microsatellite stability (MSS). Finally, this tumor was diagnosed as CDS.
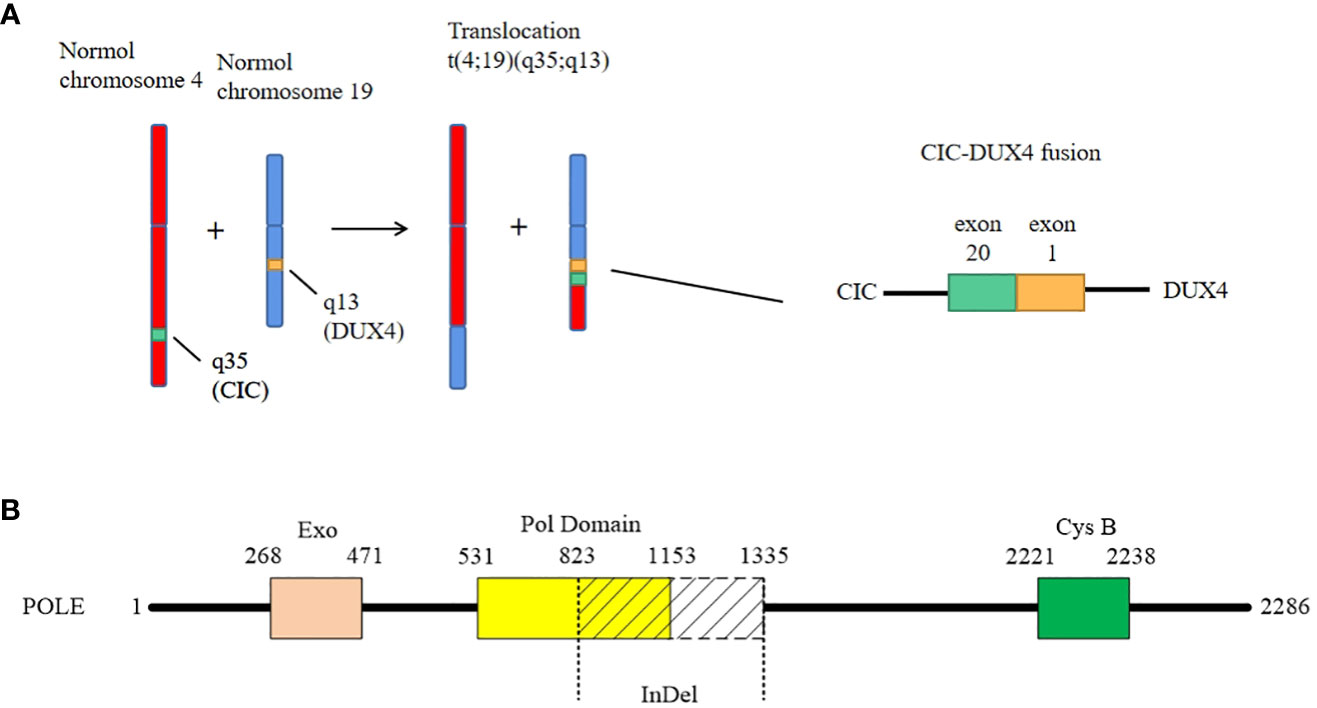
Figure 3 The genetic changes of the case. (A) The case shows a classical fusion of CIC–DUX4, which is generated by translocation of chromosome t(4;19)(q35;q13). The breakpoints are located in exon 20 of CIC and exon 1 of DUX4, respectively. (B) The LongInDel of POLE (exon22_exon31del) causes deletion of amino acid residues 823–1,335 of POLE protein, which may lead to frameshift and loss of polymerase domain 25 and CysB motif (amino acid residues 2,221–2,238).
After the surgery, the patient went to another medical institution for further treatment. At a follow-up of 6 months, the patient was still alive.
Discussion
In the latest (2020) World Health Organization (WHO) classification of tumors of soft tissue and bone, CIC-rearranged sarcomas were categorized as a new entity of USRCS (33). So far, less than 200 cases have been reported, including 157 cases of CDS, among which 88 cases are available for clinicopathologic information. The age ranged from 8 to 69 years (average at 30.7 and median at 29.5). Gender, 41 male (46.6%) and 47 female (53.4%), did not yield much difference. The most common primary sites are the soft tissue of the limbs (43/88, 48.9%), followed by the soft tissue of the trunk (35/88, 39.8%) and parenchymal organs (7/88, 7.9%). Only three cases originated from bones (3/88, 3.4%) (18, 26), which was distinguished from ES. Only two cases arose from the pelvic cavity: one is a 45-year-old man, and the other is the current case report. The maximum diameter of tumors was mentioned in 35 cases, which ranged from 1.5 to 20 cm (mean at 8.2 cm and median at 8 cm). The manifestations are usually not special, mostly occasionally discovery of mass. However, some unique manifestations are also reported, such as mimicking a phlegmon (27), cardiac tamponade (31), vaginal bleeding (28), and massive ascites in this report, which are caused by the tumor invasion of particular organs. The most common metastasis organ of CDS is the lung, followed by the brain and liver.
For morphology, the typical CDS usually consists of undifferentiated small round-to-oval tumor cells with clear to eosinophilic cytoplasm and vesicular nucleus, arranged in sheets and nests. Pathologic mitoses and geographic necrosis are easy to see. Focal myxoid matrix (1, 3, 7–9, 11, 13, 14, 18, 25, 26, 30) and spindle tumor cells (3, 8–11, 14, 18, 19, 21, 27) are also reported in some studies. Other less common morphologies include rhabdoid appearance or plasmacytoid appearance (3, 9, 12, 18, 19, 25, 32), epithelioid (18, 32), formation of microcystic spaces or pseudoacinar arrangements (8, 9, 15, 21, 23, 32), or single cell cords (13). There is one case in that focal hyaline cartilage formations appeared after chemotherapy (19).
During immunohistochemistry, almost all CDS showed vimentin positivity, and most showed weak and focal membranous CD99 positivity. Specht et al. (8) found that WT-1 was strongly positive in all CDS cases but negative in all ES cases. Therefore, they proposed that consistent WT-1 expression might provide a useful clue in the diagnosis of CDS. However, it may be a pitfall in diagnosis when CDS is primarily in the kidney. There is a report that a renal CDS was originally diagnosed as a stromal Wilms tumor (WT) because of the expression of WT-1 and rare recognition of CDS (21). In addition, CDS also frequently expresses Fli-1 and ERG, the ETS transcription factor family members (14). Cytokeratin is usually very focal positive. Bcl-2 positivity (3, 20, 21), ETV4 positivity (24), and C-MYC protein expression (15) were also reported. Moreover, Yoshimoto and his colleagues found that cyclin D2 and MUC5AC were novel biomarkers and were useful for distinguishing CDS from ES (34).
For diagnosis, although clinical, histological, and immunochemical characteristics of CDS are well recognized, none of them are entirely sensitive or specific. Therefore, a molecular testing result of CIC–DUX4 fusion is necessary to reach the final diagnosis. Fluorescence in situ hybridization (FISH) and targeted RNA sequencing are the most popular methods to detect CIC rearrangement. However, Yoshida et al. described four cases that showed negative results by CIC break-apart FISH assays but identified CIC–DUX4 and its fusion breakpoint through high-throughput RNA sequencing (24), suggesting there may be a false-negative rate for FISH to detect CDS. Moreover, ETV transcriptional upregulation (22), DNA methylation profiling, and next-generation sequencing (29) were suggested to be more sensitive than FISH and RNA sequencing. Therefore, when histology and immunoprofiles highly indicate the diagnosis of CDS but the result of FISH or RNA sequencing is negative, conclusions must not be drawn yet; it is necessary to consider another detection technique.
For pathogenesis, Kawamura-Saito and his colleagues found that CIC–DUX4 protein transforms NIH 3T3 cells and is a strong transcriptional activator that upregulates PEA3 family genes (2). Other researchers revealed that CIC–DUX4 sarcomas demonstrate frequent MYC amplification and ETS-family transcription factor expression (14). These studies uncovered the downstream targets of CIC–DUX4 protein. Subsequently, the upstream factors were found by Lin et al. (35), who suggested that negative MAPK-ERK regulation sustained CIC–DUX4 oncoprotein expression. Moreover, targeted NGS was also performed to characterize potential somatic driver alterations of CDS. Vega et al. analyzed 11 CDS cases by targeted NGS. Although no recurrent somatic mutations were identified, they detected low mutational burden and recurrent chromosome 1p loss (36). Other researchers also found a low mutational burden by targeted NGS, as well as stable microsatellite status (28), which is consistent with this report. However, we found mutations of POLE and CD79A, which have not been reported before. Moreover, Ricker and her colleagues noticed the overexpression of the translation factor eEF1A1 when whole-genome sequencing and RNA sequencing were performed in a case of CDS (30) (Table 1). Furthermore, cell lines were established for drug sensitivity tests, and bortezomib, crizotinib (37), and inhibition of the CCNE–CDK2 complex (38) seemed to be able to suppressed the growth of CDS cells. However, more studies are needed to better understand this rare sarcoma.
What is worthy to be mentioned is that we found a novel mutation of POLE. It is well known that exonuclease domain mutations of POLE were mostly reported in some epithelial tumors such as colorectal cancer and endometrial carcinoma, which lead to a deficiency of proofreading activity and usually high TMB (39). However, in this case, the mutation of POLE is LongInDel from exon 22 to exon 31 (823–1,335 amino acid residues), which causes the loss of part of the polymerase domain and CysB motif (2,221–2,238 amino acid residues) (Figure 3B). The site of mutation is beyond the exonuclease domain, which means it is a non-proofreading mutation. A study suggested that tumors with POLE proofreading mutation showed a significantly higher TMB than tumors with non-proofreading mutations (40), which may explain why this sarcoma has a POLE mutation but low TMB. Moreover, we revealed the CNV of CD79A, which may lead to the increase of its mRNA and protein expression levels and influence the B lymphocyte antigen. However, the significance of these mutations needs more exploration in the future.
Conclusions
We report a case of CDS arising from the pelvic cavity, which is a rare primary site for the tumor. Moreover, the results of the targeted NGS, which is LongInDel of POLE and CNV of CD79A, were the first to be reported. Therefore, our case enriches the clinical and genetic scales of CDS.
Data availability statement
The original contributions presented in the study are included in the article/supplementary materials. Further inquiries can be directed to the corresponding author.
Ethics statement
Ethical review and approval were not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the minor(s)’ legal guardian/next of kin for the publication of any potentially identifiable images or data included in this article.
Author contributions
QW wrote the manuscript. YH diagnosed the case and revised the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This study was supported by Natural Science Foundation of Sichuan Province(2022NSFSC0708).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Somers GR, Shago M, Zielenska M, Chan H, Bo YN. Primary subcutaneous primitive neuroectodermal tumor with aggressive behavior and an unusual karyotype: Case report. Pediatr Dev Pathol (2004) 7:538–45. doi: 10.1007/s10024-004-2024-6
2. Kawamura-Saito M, Yamazaki Y, Kaneko K, Kawaguchi N, Kanda H, Mukai H, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t (4,19)(q35;q13) translocation. Hum Mol Genet (2006) 15:2125–37. doi: 10.1093/hmg/ddl136
3. Italiano A, Yun SS, Lei Z, Singer S, Maki RG, Coindre JI, et al. High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer (2012) 51:207–18. doi: 10.1002/gcc.20945
4. Sugita S, Arai Y, Tonooka A, Hama N, Totoki Y, Fujii T, et al. A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma: a genetically distinct variant of Ewing-like sarcoma. Am J Surg Pathol (2014) 38:1571–6. doi: 10.1097/PAS.0000000000000286
5. Solomon DA, Brohl AS, Khan J, Miettinen M. Clinicopathologic features of a second patient with Ewing-like sarcoma harboring CIC-FOXO4 gene fusion. Am J Surg Pathol (2014) 38:1724–5. doi: 10.1097/PAS.0000000000000335
6. Le Loarer F, Pissaloux D, Watson S, Godfraind C, Galmiche-Rolland L, Silva K, et al. Clinicopathologic features of CIC-NUTM1 sarcomas, a new molecular variant of the family of CIC-fused sarcomas. Am J Surg Pathol (2019) 43:268–76. doi: 10.1097/PAS.0000000000001187
7. Yan Y, Liu L, Kong F, Yan T, Shen D. Clinicopathological and molecular genetic features of 72 cases of small round cell sarcoma of bone and soft tissue. CH J Pathol (2021) 50:919–23. doi: 10.3760/cma.j.cn112151-20201108-00833
8. Specht K, Sung YS, Zhang L, Richter G, Fletcher CD, Antonescu CR. Distinct transcriptional signature and immunoprofile of CIC-DUX4 fusion–positive round cell tumors compared to EWSR1-rearranged ewing sarcomas: Further evidence toward distinct pathologic entities. Genes Chromosomes Cancer (2014) 53:622–33. doi: 10.1002/gcc.22172
9. Antonescu CR, Owosho AA, Zhang L, Chen S, Deniz K, Huryn JM, et al. Sarcomas with CIC-rearrangements are a distinct pathologic entity with aggressive outcome: A clinicopathologic and molecular study of 115 cases. Am J Surg Pathol (2017) 41:941–9. doi: 10.1097/PAS.0000000000000846
10. Rakheja D, Goldman S, Wilson KS, Lenarsky C, Schultz RA. Translocation (4;19)(q35;q13.1)–associated primitive round cell sarcoma: Report of a case and review of the literature. Pediatr Dev Pathol (2008) 11:239–44. doi: 10.2350/07-06-0296.1
11. Graham C, Chilton-Macneill S, Zielenska M, Somers GR. The CIC-DUX4 fusion transcript is present in a subgroup of pediatric primitive round cell sarcomas. Hum Pathol (2012) 43:180–9. doi: 10.1016/j.humpath.2011.04.023
12. Machado I, Cruz J, Lavernia J, Rubio L, Campos J, Barrios M, et al. Superficial EWSR1-negative undifferentiated small round cell sarcoma with CIC/DUX4 gene fusion: a new variant of Ewing-like tumors with locoregional lymph node metastasis. Virchows Archiv (2013), 463; 837–42. doi: 10.1007/s00428-013-1499-9
13. Choi E, Thomas DG, Mchugh JB, Patel RM, Lucas DR. Undifferentiated small round cell sarcoma with t(4;19)(q35;q13.1) CIC-DUX4 fusion: a novel highly aggressive soft tissue tumor with distinctive histopathology. Am J Surg Pathol (2013) 37:1379–86. doi: 10.1097/PAS.0b013e318297a57d
14. Smith SC, Buehler D, Choi E-YK, McHugh JB, Rubin BP, Billings SD, et al. CIC-DUX sarcomas demonstrate frequent MYC amplification and ETS-family transcription factor expression. Modern Pathol (2015), 28; 57–68. doi: 10.1038/modpathol.2014.83
15. Tardío JC, Machado I, Navarro L, Idrovo F, Sanz-Ortega J, Pellín A, et al. Ewing-Like sarcoma with CIC-DUX4 gene fusion in a patient with neurofibromatosis type 1. a hitherto unreported association. Pathol - Res Pract (2015) 211:1–6. doi: 10.1016/j.prp.2015.08.003
16. Haidar A, Arekapudi S, DeMattia F, Abu-Isa E, Kraut M. High-grade undifferentiated small round cell sarcoma with t(4;19)(q35;q13.1) CIC-DUX4 fusion: Emerging entities of soft tissue tumors with unique histopathologic features – a case report and literature review. Am J Case Rep (2015) 16:87–94. doi: 10.12659/AJCR.892551
17. Chebib I, Vickie Y. Round cell sarcoma with CIC-DUX4 gene fusion: Discussion of the distinctive cytomorphologic, immunohistochemical, and molecular features in the differential diagnosis of round cell tumors. Cancer Cytopathol (2016) 124:350–61. doi: 10.1002/cncy.21685
18. Gambarotti M, Benini S, Gamberi G, Cocchi S, Palmerini E, Sbaraglia M, et al. CIC-DUX4 fusion-positive round-cell sarcomas of soft tissue and bone: a single-institution morphological and molecular analysis of seven cases. Histopathol (2016) 69:624–34. doi: 10.1111/his.12985
19. Yoshida A, Goto K, Kodaira M, Kobayashi E, Kawai A. CIC-rearranged sarcomas: A study of 20 cases and comparisons with Ewing sarcomas. Am J Surg Pathol (2015) 40:313–23. doi: 10.1097/PAS.0000000000000570
20. Bergerat S, Barthelemy P, Mouracade P, Lang H, Saussine C, Lindner V, et al. Primary CIC-DUX4 round cell sarcoma of the kidney: A treatment-refractory tumor with poor outcome. Pathol Res Pract (2017) 213:154–60. doi: 10.1016/j.prp.2016.11.015
21. Mangray S, Somers GR, He J, Zhong S, Shago M, Treaba DO, et al. Primary undifferentiated sarcoma of the kidney harboring a novel variant of CIC-DUX4 gene fusion. Am J Surg Pathol (2016) 40:1298. doi: 10.1097/PAS.0000000000000688
22. Kao Y-C, Sung Y-S, Chen C-L, Zhang L, Dickson BC, Swanson D, et al. ETV transcriptional upregulation is more reliable than RNA sequencing algorithms and FISH in diagnosing round cell sarcomas with CIC gene rearrangements. Gene Chromo Canc (2017) 56:501–10. doi: 10.1002/gcc.22454
23. Owosho AA, Estilo CL, Huryn JM, Zhang L, Fletcher CDM, Antonescu CR. Head and neck round cell sarcomas: A comparative clinicopathologic analysis of 2 molecular subsets: Ewing and CIC-rearranged sarcomas. Head Neck Pathol (2017) 11:450–9. doi: 10.1007/s12105-017-0808-z
24. Yoshida A, Arai Y, Kobayashi E, Yonemori K, Ogura K, Hama N, et al. CIC break-apart fluorescence in-situ hybridization misses a subset of CIC-DUX4 sarcomas: a clinicopathological and molecular study. Histopathol Off J Br Division Int Acad Pathol (2017) 71: 461–469. doi: 10.1111/his.13252
25. Donahue JE, Yakirevich E, Zhong S, Treaba DO, Lakis NS, Ali SM, et al. Primary spinal epidural CIC-DUX4 undifferentiated sarcoma in a child. Pediatr Devel Pathol (2018) 4:411–7. doi: 10.1177/1093526617707856
26. Brcic I, Brodowicz T, Cerroni L, Kashofer K, Serbanescu GL, Kasseroler MT, et al. Undifferentiated round cell sarcomas with CIC-DUX4 gene fusion: expanding the clinical spectrum. Pathol (2020) 52:236–42. doi: 10.1016/j.pathol.2019.09.015
27. Donthi D, Malik P, Prenshaw KL, Hong H. A rare case of round cell sarcoma with CIC-DUX4 mutation mimicking a phlegmon: Review of literature. Am J Case Rep (2020) 21:1–4. doi: 10.12659/AJCR.925683
28. Sedighim S, Burke J, Schneider D, Kamdjou T, Diaz-Perez JA, Trent J, et al. CIC-rearranged round cell (Ewing-like) sarcoma of the uterus: Review of the literature. Gyneco Oncol Rep (2020) 33:100592. doi: 10.1016/j.gore.2020.100592
29. Miele E, Vito RD, Ciolfi A, Pedace L, Russo I, Pasquale MDD, et al. DNA Methylation profiling for diagnosing undifferentiated sarcoma with capicua transcriptional receptor (CIC) alterations. Int J Mol Sci (2020) 21:1818. doi: 10.3390/ijms21051818
30. Ricker CA, Berlow NE, Crawford KA, Georgopapadakos T, Huelskamp AN, Woods AD, et al. Undifferentiated small round cell sarcoma in a young male: a case report. Mol Case Stud (2020) 6:1–22. doi: 10.1101/mcs.a004812
31. Maekawa A, Matsunobu T, Nabeshima A, Fukushima S, Makihara K, Hisaoka M, et al. Cardiac tamponade as an unusual initial clinical manifestation of CIC-DUX4 sarcoma. Am J Case Rep (2021) 22:e929349. doi: 10.12659/AJCR.929349
32. Vieira AC, Xavier CB, Vieira TD, Carvalho FM, Scaranti M, Munhoz RR, et al. CIC-DUX4 rearranged uterine cervix round-cell sarcoma exhibiting near-complete pathologic response following radiation and neoadjuvant chemotherapy: A case report. Gyneco Oncol Rep (2021) 36:100745–5. doi: 10.1016/j.gore.2021.100745
33. Antonescu CR, Yoshida A. Undifferentiated small round cell sarcomas of bone and soft tissue/CIC-rearranged sarcoma. In: WHO classification of tumours of soft tissue and bone 5th edition, Lyon(France): International Agency for Research on Cancer 2020. vol. 3. (2020). p. 330.
34. Yoshimoto T, Tanaka M, Homme M, Yamazaki Y, Takazawa Y, Antonescu CR, et al. CIC-DUX4 induces small round cell sarcomas distinct from Ewing sarcoma. Cancer Res (2017) 77:2927–37. doi: 10.1158/0008-5472.CAN-16-3351
35. Kawe Lin Y, Wu W, Ponce RK, Kim JW, Okimoto RA. Negative MAPK-ERK regulation sustains CIC-DUX4 oncoprotein expression in undifferentiated sarcoma. P Natl A Sci India B (2020) 117:20776–84. doi: 10.1073/pnas.2009137117
36. Lazo de la Vega L, Hovelson DH, Cani AK, Liu C-J, McHugh JB, Lucas DR, et al. Targeted next-generation sequencing of CIC-DUX4 soft tissue sarcomas demonstrates low mutational burden and recurrent chromosome 1p loss. Hum Pathol (2016) 58:161–70. doi: 10.1016/j.humpath.2016.09.004
37. Oyama R, Takahashi M, Yoshida A, Sakumoto M, Takai Y, Kito F, et al. Generation of novel patient-derived CIC- DUX4 sarcoma xenografts and cell lines. Sci Rep (2017) 7:4712–2. doi: 10.1038/s41598-017-04967-0
38. Okimoto RA, Wu W, Nanjo S, Olivas V, Lin YK, Ponce RK, et al. CIC-DUX4 oncoprotein drives sarcoma metastasis and tumorigenesis via distinct regulatory programs. J Clin Invest (2019) 129:3401–6. doi: 10.1172/JCI126366
39. Kane DP, Shcherbakova PV. A common cancer-associated DNA polymerase ϵ mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res (2014) 74:1895–901. doi: 10.1158/0008-5472.Can-13-2892
Keywords: CIC–DUX4, undifferentiated small round cell sarcoma, target next generation sequencing, low tumor mutational burden, POLE
Citation: Wu Q and He Y (2022) A case report of CIC–DUX4 fusion-positive sarcoma in the pelvic cavity with targeted next-generation sequencing results. Front. Oncol. 12:1018992. doi: 10.3389/fonc.2022.1018992
Received: 24 August 2022; Accepted: 24 November 2022;
Published: 15 December 2022.
Edited by:
Athina Markou, National and Kapodistrian University of Athens, GreeceReviewed by:
Congwang Zhang, Shenzhen Longhua District Central Hospital, ChinaJagadheshwar Balan, Mayo Clinic, United States
David Robert Shorthouse, MRC Cancer Unit, University of Cambridge, United Kingdom
Copyright © 2022 Wu and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying He, aGV5aW5nNjI2QDE2My5jb20=
 Qian Wu
Qian Wu Ying He1,2,3*
Ying He1,2,3*