- 1Department of Gastrointestinal Surgery, Osaka University Graduate School of Medicine, Suita, Japan
- 2Department of Medical Data Science, Center of Medical Innovation and Translational Research, Osaka University Graduate School of Medicine, Suita, Japan
Introduction
Recent single-cell level sequence analysis in pancreatic cancer, which is a representative of intractable gastrointestinal cancers, revealed that the tumor tissue comprised of not only epithelial malignant cancer cells but also stromal activated fibroblasts and infiltrated immune cells. This indicates that various changes in gene expression occur in cells, including changes in cell-to-cell communication, to form a microenvironment that is characteristic of cancer (1–6). Epithelial malignant cancer cells harbor “big 4” driver mutations, that is, substitutions or alterations of nucleotides in KRAS proto-oncogene, GTPase (KRAS), tumor protein P53 (TP53), and cyclin-dependent kinase inhibitor 2A (CDKN2A), and mothers against decapentaplegic homolog 4 (SMAD4) commonly occurs in pancreatic ductal adenocarcinoma (PDAC) (https://portal.gdc.cancer.gov), which can be useful for predicting survival in patients with resected PDAC (7). Conversely, numerous gene expression alterations, due to cancer-specific transcription, RNA processing, and translations, are demonstrated by mesenchymal components, which include activated fibroblasts, vascular endothelial cells, and immune cells (8, 9). Importantly, this cancer-specific alterations induce the production of abnormal peptides and proteins with deleterious degenerations, which led to anti-apoptotic and pro-survival signals in cell-to-cell communication among tumor-component cells, contributing to biologically malignant phenotypes such as epithelial-to-mesenchymal transition phenotypes, invasion, and metastasis (8, 9). Thus, the aberrant protein production is important for the development of cancer-specific therapeutic approaches. Eventually, studies on mutation-prone tumors indicated that the mismatch-repair status predicted the clinical benefit of immune checkpoint blockade with anti-programmed cell death 1 reagents (10–12), suggesting that genetic mutations and resultant production of aberrant peptides or proteins may sensitize the response to cancer therapy. Recent studies have emerged that the importance of the regulation of aberrant protein production (RAPP) in many secretary and membrane proteins (13–16), playing a role in epithelial cancer cell and activated fibroblast or immune cell communication, the process associated with patient survival (6). In this article, we focused on the mechanism of RAPP in gastrointestinal cancer, especially pancreatic cancer, and explored the possibility of an innovative approach against intractable cancers.
RAPP in cancer
The process of RAPP is mediated by a signal recognition particle (SRP), a ubiquitous initiator of protein translation (17), which is composed of six protein subunits arranged on a noncoding RNA, 7SL1 (16). The Alu domain is associated with SRP9 (9 kDa; encoded at the cytogenetic band 1q42.12) and SRP14 (14 kDa; encoded at the cytogenetic band 15q22) proteins, whereas the signal recognition domain is bounded by SRP19 (19 kDa; encoded at the cytogenetic band 5q22.2), SRP54 (54 kDa; encoded at the cytogenetic band 14q13.2), SRP68 (68 kDa; encoded at the cytogenetic band 17q25.1), and SRP72 (72 kDa; encoded at the cytogenetic band 4q12) proteins (16). Given that an SRP is a complex composed of multiple proteins encoded by multiple regions, it is suggested that it is responsible for a finely regulated mechanism (16, 17). This mechanism is also conserved across species, of which disruption can lead to various human diseases (16, 17). Here, we focused on the role of SRP in gastrointestinal cancers.
SRP
Studies on the Alu-domain associated with SRP9 and SRP14 indicated that they are involved in several human cancers and can be used as diagnostic markers. The proteomic expression analysis of human colorectal cancer showed the upregulation of SRP9 with hypoxic adaptation of the tumor microenvironment of heterogeneous primary human tumor tissues (18), suggesting the role of SRP9 in RAPP in cancer. Interestingly, in adenosine-to-inosine RNA editing, 10 recurrent nonsynonymous RNA editing candidates were identified in nine genes, including the gene for SRP9 (19), indicating that the mechanism of those genes not directly encoded in the genomic DNA is implicated in colorectal cancer. The nucleotide sequence analysis of non-Hodgkin B-cell lymphoma, a hematopoietic malignancy, allowed the identification of an aberrant fusion gene of SRP9 conjoined with epoxide hydrolase 1 (EPHX1, encoded at the cytogenetic band 1q42.12) (20), suggesting that the mechanism that converts epoxides from the degradation of aromatic compounds to trans-dihydrodiols resulting in secretion from cells may be involved in this disease. The study of dysregulated RNA binding proteins identified 11 candidates of RNA binding proteins, including SRP14, which are involved in the hepatitis B virus (HBV)-related hepatocellular carcinoma prognosis (21), indicating that RAPP may be involved in the mechanism such as the HBV lifecycle and the progression of this disease. The study of pancreatic cancer PSN-1 cells indicated that the knockdown of SRP72 resulted in a significant increase in sensitization to radiation therapy as measured by colony formation assays, indicating that SRP72 is a marker of radiotherapy resistance against cancer (22).
7SL1
Previous studies indicated that 7SL1, a noncoding RNA that binds to proteins to execute potent post-transcriptional regulation, is upregulated in cancer cells (23). It was demonstrated that 7SL1 binds to the 3’-untranslated region (UTR) of mRNA of tumor suppressor TP53 and that the interaction of 7SL1 with TP53 mRNA reduced the translation of p53 protein. On the other hand, the silencing of 7SL1 resulted in the increased binding of embryonic lethal, abnormal vision, Drosophila (ELAV)-like 1, Hu Antigen R (HuR) to TP53 mRNA, an interaction that led to the promotion of p53 translation, which reveals the competitive mechanism of 7SL1 and HuR for binding to TP53 3’-UTR (23). HuR selectively binds to adenylate-uridylate-(AU)-rich elements (AREs) found in the 3’-UTRs of various mRNAs. Given that AREs stimulate the degradation of mRNAs, HuR plays a role in stabilizing ARE-containing mRNAs by inhibiting degradation (24). The HuR is highly expressed in many cancers, including pancreatic cancer, functions as the post-transcriptional regulator of core metabolic enzymes, and is critical for survival under acute glucose deprivation in pancreatic cancer cells (25). In hepatocellular carcinoma, Wilms tumor 1-associating protein (WTAP) is significantly upregulated, and avian erythroblastosis virus E26 (V-Ets) oncogene homolog-1 (ETS1) was identified as the downstream effector of WTAP, suggesting that WTAP-guided m6A modification contributes to the progression of hepatocellular carcinoma via the HuR-ETS1-cyclin dependent kinase inhibitor 1A (p21)/cyclin dependent kinase inhibitor 1B (p27) axis (26). In pancreatic cancer, the role of mRNA modification at m6A in polo-like kinase 1 in cell cycle homeostasis was demonstrated (27), although the involvement of HuR and 7SL1 was elusive, suggesting that 7SL1 is involved, at least partially, in the process of antagonizing to p53 (Figure 1).
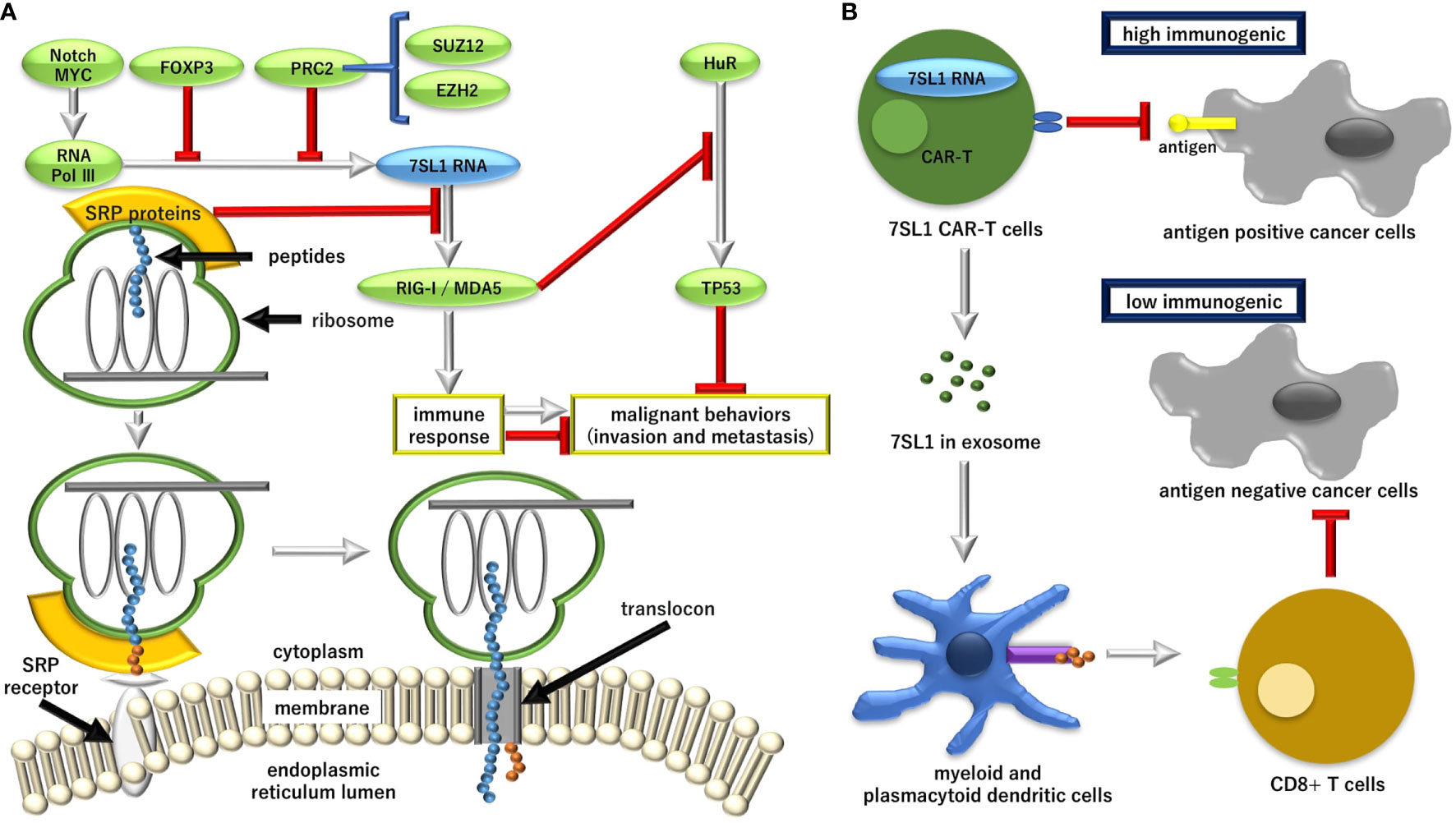
Figure 1 The role of endogenous immune stimulatory noncoding RNA 7SL1 in the pathway for the regulation of the aberrant protein production (RAPP) and its possible application in cancer therapy. (A) Transcriptional regulation of 7SL1 and the downstream mechanism for tumor suppressor tumor protein P53 (TP53). MYC, proto-oncogene, basic helix-loop-helix (BHLH) transcription factor (MYC); FOXP3, forkhead box P3; PRC2, polycomb repression complex 2; SUZ12, SUZ12 polycomb repressive complex 2 subunit; EZH2, enhancer of zeste 2 polycomb repressive complex 2 subunit; Pol II, RNA polymerase II; Pol III, RNA polymerase III; SRP, signal recognition particle; RIG-1, retinoic acid-inducible gene 1 protein; MDA5, melanoma differentiation-associated gene 5; HuR, embryonic lethal, abnormal vision, Drosophila (ELAV)-like 1, Hu antigen R. (B) Although the unshielded 7SL1-mediated stimulation of tumor tissues can result in disease progression, the exosomal delivery of 7SL1 or its combination with engineering chimeric antigen receptor (CAR)-T cells can stimulate an anti-tumor effect and lead to a favorable disease outcome. Ag, antigen.
A study on forkhead box P3 (FOXP3), a transcription factor that is crucial for the development and inhibitory function of regulatory T-cells (Treg), indicated that overexpression of FOXP3 resulted in the repression of the transcription of 7SL1, whereas the knockdown of FOXP3 showed the upregulation of 7SL1 RNA transcription (28). The mechanism was confirmed by chromatin immuno-precipitation analysis and reporter assay (28). The study showed that FOXP3 promoted the expression of TP53 at the translational levels through repressing 7SL1 RNA (28). However, considering the recent study that indicated that Treg is chemoattracted to the tumor microenvironment by chemokine gradients such as C-C motif chemokine receptor 4 (CCR4)-C-C motif chemokine ligand 17 (CCL17)/CCL22, CCR8-CCL1, CCR10-CCL28, and CXCR3-CCL9/10/11, it is demonstrated that Treg cells are activated and inhibit antitumor immune responses, suggesting that strategies to deplete Treg cells and the control of Treg cell functions to increase antitumor immune responses are required in cancer immunotherapy (29), which demonstrates the multifaceted role of Treg in cancer. Given that FOXP3 unlikely represents the effect on 7SL1 exclusively in Treg cells, whether 7SL1 is an appropriate target to control Treg remains to be investigated.
Although a role of the polycomb repressive complex 2 (PRC2) component, working on the enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2)-mediated epigenetic control of RNA polymerase II (Pol II) transcribed coding gene transcription, has been well-established, a recent study on EZH2-mediated epigenetic regulation of RNA polymerase III (Pol III) transcription indicated that EZH2 is involved in the repression of Pol III transcription of tRNA(Tyr), 5S rRNA, and 7SL1 RNA genes via the interaction with transcriptional factor complex IIIC (TFIIIC) as well as SUZ12 polycomb repressive complex 2 subunit (SUZ12) (30), suggesting that basic mechanisms common to cells, which are specific mechanisms to cancer, are involved in 7SL1 regulation. Thus, it is necessary to study the upstream regulation of 7SL1 transcription in cells, which responds to various stimuli such as hypoxia and hyponutrition that are characterized by the extracellular tumor microenvironment.
Damage-associated molecular patterns in cancer-associated fibroblasts-to-cancer cell interactions
In a single-cell sequence analysis (1–6), it was well-recognized that interactions between cancer cells and CAFs in stroma generate signals for cancer progression, inflammatory responses, and therapeutic outcomes (31–33). Previous reports have indicated that the arrangement of SRPs in 7SL1 is necessary to execute a physiological function of PARP in DAMPs (34). Reportedly, SRP dysfunction resulted in the induction of invasion and metastasis of cancer cells, which is associated with the evacuation from host’s immune response. A previous study elucidated that SRPs and 7SL1 are involved in the process of such signal, which revealed the importance of endogenous RNA response acting as damage-associated molecular patterns (35). In this study, it was demonstrated that triggering of notch receptor 1-MYC proto-oncogene and basic helix-loop-helix (BHLH) transcription factor (MYC) signaling in the stroma of breast cancer results in the activation of Pol III-driven increase in 7SL1, which is normally shielded by SRP9 and SRP 14 (35). The induced 7SL1 transcription resulted in an alteration of its stoichiometry with SRP9 and SRP 14, which led to the generation of unshielded 7SL1 in the stromal exosomes (35). Moreover, the unshielded 7SL1 can drive an inflammatory response. The unshielded 7SL1 activates the retinoic acid-inducible gene 1 protein (RIG-I), a pattern recognition receptor usually reserved for viral infections, to enhance tumor growth, metastasis, and therapy resistance, suggesting that the regulation of RNA unshielding can couple stromal activation with deployment of DAMPs of RNA, which is closely associated with the aggressive features of cancer (35). This also indicates the potential of 7SL1 as a target of cancer therapy, considering the immunogenic property of this endogenous non-coding RNA. The significant roles of exosome-mediated RNA transfer was demonstrated in a breast cancer study, indicating that CAFs orchestrate an intricate crosstalk with cancer cells by utilizing exosomes (36). The significance in gastrointestinal cancer warrants further investigation.
Application to chimeric antigen receptor-T therapy
Recently, it was demonstrated that the immune stimulatory property can be utilized for the development of CAR-T cell therapy (37). The vector-mediated gene transfer of 7SL1 activated the RIG-I/melanoma differentiation-associated gene 5 (MDA5) signaling and promoted the expansion and effector-memory differentiation of CAR-T cells (37). MDA5 binds to RNAs with a modified DExD/H-box helicase core and a C-terminal domain, thus leading to a proinflammatory response that includes interferons (38, 39). RN7SL1 restricts myeloid-derived suppressor cell development, decreases transforming growth factor beta 1 (TGFB) in myeloid cells, and fosters dendritic cell subsets, which led to the endogenous expansion of effector-memory and tumor-specific T cells (37, 40). It is suggested that the unshielded form of 7SL1 can be used in peptide antigens to enhance anti-tumor efficacy (Figure 1).
Discussion
Despite advances in surgery, standard radiation therapy, and chemotherapy, immunotherapy was added as a fourth treatment to cancer, as the discovery of immune modulation through immune check points and exploration of new combinations of cancer multimodality therapies led to overcoming long-term resistance and tumor recurrence (41). Cancer is a genetic disease involving numerous mutations (42–44), and the evidence that the mismatch-repair status predicted the clinical benefit of immune checkpoint blockade with pembrolizumab is important (10–12), as this suggests that the quantity or quality of gene mutations may influence susceptibility to immune checkpoint inhibitor therapy. Furthermore, the effect of immunotherapy can be maximized by regulating gene mutations and the consequent amino acid mutation neoantigen to eradicate cancer (45–47). If inducing a hot state in which cell-to-cell interactions are activated immunologically is possible, immune checkpoint inhibitors can not only be applied, but cancer-specific antigens can also be identified and used as vectors or employed as CAR-T therapy (48, 49). Nevertheless, methods to induce immunologically cold tumors into hot ones remain to be developed, and further investigations are necessary for application in a clinical setting.
The RAPP pathway study revealed the immune stimulatory property of endogenous non-coding RNA, 7SL1, which is a component of SRPs. A previous study suggests several important implications. First, given that the co-deploy peptide antigen with 7SL1 can exert an efficient anti-tumor effect (36), this approach may be useful to treat intractable solid tumors such as mutation-prone gastrointestinal cancer. Second, considering that unshielded 7SL1 in exosomes can be transferred in the tumor microenvironment (34), it is possible that the cell-to-cell communication is assessed by an RNA study in liquid biopsy as a companion diagnostic tool. Third, as radiotherapy can induce inflammatory microenvironment remodeling (50), the combination of RAPP pathway-mediated stimulatory cancer therapy with radiation may be useful to induce the immune response to convert an immune cold into a hot tumor, and multiple protocols have been developed (51). Fourth, nucleotide-mediated immune stimulation may be expanded as an anti-cancer therapy. Eventually, the intratumoral injection of the seasonal flu shot can generate systemic CD8+ T cell-mediated antitumor immunity and sensitizes resistant tumors to the checkpoint blockade, suggesting that it converts immunologically cold tumors to hot tumors and serves as an immunotherapy for cancer (52). To optimize the effect of treatment for intractable cancer, further studies to accumulate evidence are warranted.
Author contributions
HS, KS, and HI conceptualized the study objectives and obtained the funding, wrote the manuscript. TH, SK, YD, HE, and TS outlined content of the manuscript. TS suggested which disease information should be addressed in the manuscript. All authors have read and approved the final manuscript version for publication.
Funding
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (19K22658; 20H00541; 22H03146; 22K19559), and Japan Agency for Medical Research and Development (AMED) (17cm0106414h0002; JP19lm0203007; JP20lm0203007; JP21lm0203007). Partial support was received from Princess Takamatsu Cancer Research Fund and Mitsubishi Foundation to HI.
Acknowledgments
Authors are thankful to every lab member.
Conflict of interest
Partial institutional endowments were received from Taiho Pharmaceutical Co., Ltd. (Tokyo, Japan), Hirotsu Bio Science Inc. (Tokyo, Japan); Kinshu-kai Medical Corporation (Osaka, Japan); Kyowa-kai Medical Corporation (Osaka, Japan); IDEA Consultants Inc. (Tokyo, Japan); Unitech Co. Ltd. (Chiba, Japan).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov (2019) 9(8):1102–23. doi: 10.1158/2159-8290.CD-19-0094
2. Lin W, Noel P, Borazanci EH, Lee J, Amini A, Han IW, et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med (2020) 12(1):80. doi: 10.1186/s13073-020-00776-9
3. Chen K, Wang Q, Li M, Guo H, Liu W, Wang F, et al. Single-cell RNA-seq reveals dynamic change in tumor microenvironment during pancreatic ductal adenocarcinoma malignant progression. EBioMedicine (2021) 66:103315. doi: 10.1016/j.ebiom.2021.103315
4. Peng J, Sun BF, Chen CY, Zhou JY, Chen YS, Chen H, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res (2019) 29(9):725–38. doi: 10.1038/s41422-019-0195-y
5. Schlesinger Y, Yosefov-Levi O, Kolodkin-Gal D, Granit RZ, Peters L, Kalifa R, et al. Single-cell transcriptomes of pancreatic preinvasive lesions and cancer reveal acinar metaplastic cells’ heterogeneity. Nat Commun (2020) 11(1):4516. doi: 10.1038/s41467-020-18207-z
6. Chijimatsu R, Kobayashi S, Takeda Y, Kitakaze M, Tatekawa S, Arao Y, et al. Establishment of a reference single-cell RNA sequencing dataset for human pancreatic adenocarcinoma. iScience (2022) 25(8):104659. doi: 10.1016/j.isci.2022.104659
7. McIntyre CA, Lawrence SA, Richards AL, Chou JF, Wong W, Capanu M, et al. Alterations in driver genes are predictive of survival in patients with resected pancreatic ductal adenocarcinoma. Cancer (2020) 126(17):3939–49. doi: 10.1002/cncr.33038
8. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell (2000) 100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9
9. Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
10. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med (2015) 372(26):2509–20. doi: 10.1056/NEJMoa1500596
11. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science (2017) 357(6349):409–13. doi: 10.1126/science.aan6733
12. Ciombor KK, Goldberg RM. Hypermutated tumors and immune checkpoint inhibition. Drugs (2018) 78(2):155–62. doi: 10.1007/s40265-018-0863-0
13. Popp MW-L, Maquat LE. Defective secretory-protein mrnas take the rapp. Trends Biochem Sci (2014) 39(4):154–6. doi: 10.1016/j.tibs.2014.02.001
14. Karamyshev AL, Patrick AE, Karamysheva ZN, Griesemer DS, Hudson H, Tjon-Kon-Sang S, et al. Inefficient SRP interaction with a nascent chain triggers a mrna quality control pathway. Cell (2014) 156(1-2):146–57. doi: 10.1016/j.cell.2013.12.017
15. Karamyshev AL, Karamysheva ZN. Lost in translation: Ribosome-associated mRNA and protein quality controls. Front Genet (2018) 9:431. doi: 10.3389/fgene.2018.00431
16. Kellogg MK, Tikhonova EB, Karamyshev AJ. Signal recognition particle in human diseases. Front Genet (2022) 13:898083. doi: 10.3389/fgene.2022.898083
17. Lütcke H. Signal recognition particle (SRP), a ubiquitous initiator of protein translocation. Eur J Biochem (1995) 228(3):531–50. doi: 10.1111/j.1432-1033.1995.tb20293.x
18. Rho JH, Qin S, Wang JY, Roehrl MH. Proteomic expression analysis of surgical human colorectal cancer tissues: Up-regulation of PSB7, PRDX1, and SRP9 and hypoxic adaptation in cancer. J Proteome Res (2008) 7(7):2959–72. doi: 10.1021/pr8000892
19. Lee S-H, Kim H-P, Kang J-K, Song S-H, Han S-W, Kim T-Y. Identification of diverse adenosine-to-Inosine RNA editing subtypes in colorectal cancer. Cancer Res Treat (2017) 49(4):1077–87. doi: 10.4143/crt.2016.301
20. Matsumoto Y, Tsukamoto T, Chinen Y, Shimura Y, Sasaki N, Nagoshi H, et al. Detection of novel and recurrent conjoined genes in non-Hodgkin b-cell lymphoma. J Clin Exp Hematop (2021) 61(2):71–7. doi: 10.3960/jslrt.20033
21. Li M, Liu Z, Wang J, Liu H, Gong H, Li S, et al. Systematic analysis identifies a specific RNA-binding protein-related gene model for prognostication and risk-adjustment in HBV-related hepatocellular carcinoma. Front Genet (2021) 12:707305. doi: 10.3389/fgene.2021.707305
22. Prevo R, Tiwana GS, Maughan TS, Buffa FM, McKenna WG, Higgins GS. Depletion of signal recognition particle 72kDa increases radiosensitivity. Cancer Biol Ther (2017) 18(6):425–32. doi: 10.1080/15384047.2017.1323587
23. Abdelmohsen K, Panda AC, Kang M-J, Guo R, Kim J, Grammatikakis I, et al. 7SL RNA represses P53 translation by competing with hur. Nucleic Acids Res (2014) 42(15):10099–111. doi: 10.1093/nar/gku686
24. Brody JR, Dixon DA. Complex hur function in pancreatic cancer cells. Wiley Interdiscip Rev RNA (2018) 9(3):e1469. doi: 10.1002/wrna.1469
25. Burkhart RA, Pineda DM, Chand SN, Romeo C, Londin ER, Karoly ED, et al. HuR is a post-transcriptional regulator of core metabolic enzymes in pancreatic cancer. RNA Biol (2013) 10(8):1312–23. doi: 10.4161/rna.25274
26. Chen Y, Peng C, Chen J, Chen D, Yang B, He B, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-Dependent epigenetic silencing of ETS1. Mol Cancer (2019) 18(1):127. doi: 10.1186/s12943-019-1053-8
27. Tatekawa S, Tamari K, Chijimatsu R, Konno M, Motooka D, Mitsufuji S, et al. N(6)-methyladenosine methylation-regulated polo-like kinase 1 cell cycle homeostasis as a potential target of radiotherapy in pancreatic adenocarcinoma. Sci Rep (2022) 12(1):11074. doi: 10.1038/s41598-022-15196-5
28. Yang Y, Cheng J, Ren H, Zhao H, Gong W, Shan C. Tumor FOXP3 represses the expression of long noncoding RNA 7SL. Biochem Biophys Res Commun (2016) 472(3):432–6. doi: 10.1016/j.bbrc.2015.12.082
29. Ohue Y, Nishikawa H, Regulatory T. (Treg) cells in cancer: Can treg cells be a new therapeutic target? Cancer Sci (2019) 110(7):2080–9. doi: 10.1111/cas.14069
30. Liu C, Li S, Dai X, Ma J, Wan J, Jiang H, et al. PRC2 regulates RNA polymerase III transcribed non-translated RNA gene transcription through EZH2 and SUZ12 interaction with tfiiic complex. Nucleic Acids Res (2015) 43(13):6270–84. doi: 10.1093/nar/gkv574
31. Biffi G, Tuveson DA. Diversity and biology of cancer-associated fibroblasts. Physiol Rev (2021) 101(1):147–76. doi: 10.1152/physrev.00048.2019
32. Chen Y, McAndrews KM, Kalluri R. Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat Rev Clin Oncol (2021) 18(12):792–804. doi: 10.1038/s41571-021-00546-5
33. Chen X, Song E. Turning foes to friends: Targeting cancer-associated fibroblasts. Nat Rev Drug Discovery (2019) 18(2):99–115. doi: 10.1038/s41573-018-0004-1
34. Krysko DV, Garg AD, Kaczmarek A, Krysko O, Agostinis P, Vandenabeele P. Immunogenic cell death and damps in cancer therapy. Nat Rev Cancer (2012) 12(12):860–75. doi: 10.1038/nrc3380
35. Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon T, Kim BC, et al. Exosome rna unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell (2017) 170(2):352–66.e13. doi: 10.1016/j.cell.2017.06.031
36. Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y, Yoon T, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell (2014) 159(3):499–513. doi: 10.1016/j.cell.2014.09.051
37. Johnson LR, Lee DY, Eacret JS, Ye D, June CH, Minn AJ. The immunostimulatory RNA RN7SL1 enables car-T cells to enhance autonomous and endogenous immune function. Cell (2021) 184(19):4981–95.e14. doi: 10.1016/j.cell.2021.08.004
38. Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature (2006) 441(7089):101–5. doi: 10.1038/nature04734
39. Dias Junior AG, Sampaio NG, Rehwinkel JA. Balancing act: MDA5 in antiviral immunity and autoinflammation. Trends Microbiol (2019) 27(1):75–85. doi: 10.1016/j.tim.2018.08.007
40. Nutt WS, Srivastava S. Special delivery! car-T cells transport RN7SL1 to the tumor microenvironment. Trends Mol Med (2021) 27(11):1019–21. doi: 10.1016/j.molmed.2021.09.002
41. Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity (2020) 52(1):17–35. doi: 10.1016/j.immuni.2019.12.011
42. Devarakonda S, Rotolo F, Tsao M-S, Lanc I, Brambilla E, Masood A, et al. Tumor mutation burden as a biomarker in resected non–Small-Cell lung cancer. J Clin Oncol (2018) 36(30):2995–3006. doi: 10.1200/JCO.2018.78.1963
43. Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann Oncol (2019) 30(1):44–56. doi: 10.1093/annonc/mdy495
44. Lindeboom RGH, Vermeulen M, Lehner B, Supek F. The impact of nonsense-mediated mrna decay on genetic disease, gene editing and cancer immunotherapy. Nat Genet (2019) 51(11):1645–51. doi: 10.1038/s41588-019-0517-5
45. Bonaventura P, Shekarian T, Alcazer V, Valladeau-Guilemond J, Valsesia-Wittmann S, Amigorena S, et al. Cold tumors: A therapeutic challenge for immunotherapy. Front Immunol (2019) 10:168. doi: 10.3389/fimmu.2019.00168
46. Kakoti S, Sato H, Laskar S, Yasuhara T, Shibata A. DNA Repair and signaling in immune-related cancer therapy. Front Mol Biosci (2020) 7:205. doi: 10.3389/fmolb.2020.00205
47. Liu Y-T, Sun Z-J. Turning cold tumors into hot tumors by improving T-cell infiltration. Theranostics (2021) 11(11):5365–86. doi: 10.7150/thno.58390
48. Martinez M, Moon EK. CAR T cells for solid tumors: New strategies for finding, infiltrating, and surviving in the tumor microenvironment. Front Immunol (2019) 10:128. doi: 10.3389/fimmu.2019.00128
49. Ma S, Li X, Wang X, Cheng L, Li Z, Zhang C, et al. Current progress in CAR-T cell therapy for solid tumors. Int J Biol Sci (2019) 15(12):2548–60. doi: 10.7150/ijbs.34213
50. McLaughlin M, Patin EC, Pedersen M, Wilkins A, Dillon MT, Melcher AA, et al. Inflammatory microenvironment remodelling by tumour cells after radiotherapy. Nat Rev Cancer (2020) 20(4):203–17. doi: 10.1038/s41568-020-0246-1
51. Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discovery (2019) 18(3):197–218. doi: 10.1038/s41573-018-0007-y
Keywords: SRP, RAPP, 7SL1, therapy, gastrointestinal cancer
Citation: Sato H, Sasaki K, Hara T, Kobayashi S, Doki Y, Eguchi H, Satoh T and Ishii H (2022) Targeting the regulation of aberrant protein production pathway in gastrointestinal cancer treatment. Front. Oncol. 12:1018333. doi: 10.3389/fonc.2022.1018333
Received: 13 August 2022; Accepted: 15 September 2022;
Published: 21 October 2022.
Edited by:
Imayavaramban Lakshmanan, University of Nebraska Medical Center, United StatesReviewed by:
Ya Cui, University of California, Irvine, United StatesCopyright © 2022 Sato, Sasaki, Hara, Kobayashi, Doki, Eguchi, Satoh and Ishii. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hideshi Ishii, aGlzaGlpQGdlc3VyZy5tZWQub3Nha2EtdS5hYy5qcA==
 Hiromichi Sato1,2
Hiromichi Sato1,2 Kazuki Sasaki
Kazuki Sasaki Tomoaki Hara
Tomoaki Hara Taroh Satoh
Taroh Satoh Hideshi Ishii
Hideshi Ishii