- Department of Internal Medicine I, Gastroenterology, Hepatology, Endocrinology, Rheumatology, and Infectious Diseases, University Hospital Regensburg, Regensburg, Germany
Hepatocellular carcinoma (HCC) is one of the most common and deadly tumors worldwide. Management of HCC depends on reliable biomarkers for screening, diagnosis, and monitoring of the disease, as well as predicting response towards therapy and safety. To date, imaging has been the established standard technique in the diagnosis and follow-up of HCC. However, imaging techniques have their limitations, especially in the early detection of HCC. Therefore, there is an urgent need for reliable, non/minimal invasive biomarkers. To date, alpha-fetoprotein (AFP) is the only serum biomarker used in clinical practice for the management of HCC. However, AFP is of relatively rather low quality in terms of specificity and sensitivity. Liquid biopsies as a source for biomarkers have become the focus of clinical research. Our review highlights alternative biomarkers derived from liquid biopsies, including circulating tumor cells, proteins, circulating nucleic acids, and exosomes, and their potential for clinical application. Using defined combinations of different biomarkers will open new perspectives for diagnosing, treating, and monitoring HCC.
1 Hepatocellular carcinoma – a high disease burden
Hepatocellular carcinoma (HCC) is one of the most frequent tumor types worldwide. Its clinical importance has been rising tremendously in the past two decades: The incidence increased about 75% (1). Concurrently, HCC is ranked 5th most common tumor worldwide (2). In 2020 905,677 new cases of liver cancer worldwide were diagnosed. About 80% of them were HCC (2).
Regarding death caused by cancer, HCC ranks 3rd place (2). The World Health Organization expects more than one million deaths due to liver cancer by 2030 (3). Men are almost three times more likely to be affected than women.
73% of HCC cases occur in Asia (2). The reason for this geographic variance is the endemic occurrence of hepatitis viruses, the most common risk factor for HCC. Despite the decrease in the number of Hepatitis B virus (HBV) infections due to the global introduction of vaccination programs, one-third of all HCC cases worldwide are still based on chronic HBV infection (33%) (1, 4). Hepatitis C (HCV) infection is the second most important risk factor. Even after the introduction of new, highly efficient combination therapies with polymerase inhibitors, protease inhibitors, and non-structural protein 5A (NS5A) inhibitors in 2013, more than one-fifth of HCCs are due to HCV infection (1). Another relevant risk factor with a substantial global disease burden and potential to grow is the metabolic syndrome. Since 1975, overweight and obesity have tripled worldwide (5). Other risk factors are alcohol-related liver cirrhosis (30%), primary biliary cholangitis, primary sclerosing cholangitis, hemochromatosis, and alpha1-antitrypsin deficiency (1). Aflatoxin is a mycotoxin that can also trigger HCC and is mainly found in nuts, dried fruits, and spices.
Physicians should regularly monitor the risk groups depicted above for the occurrence of HCC. Given the high burden of disease, there is a tremendous need for valid and cost-effective biomarkers that can
● identify individual risks for developing HCC,
● detect HCC in an early stage,
● predict therapy response to specific therapies,
● monitor response to therapy and predict adverse side effects of cancer therapies,
● and predict cancer recurrence.
These biomarkers should be easy to obtain and easy to analyze routinely.
2 What characterizes an optimal biomarker?
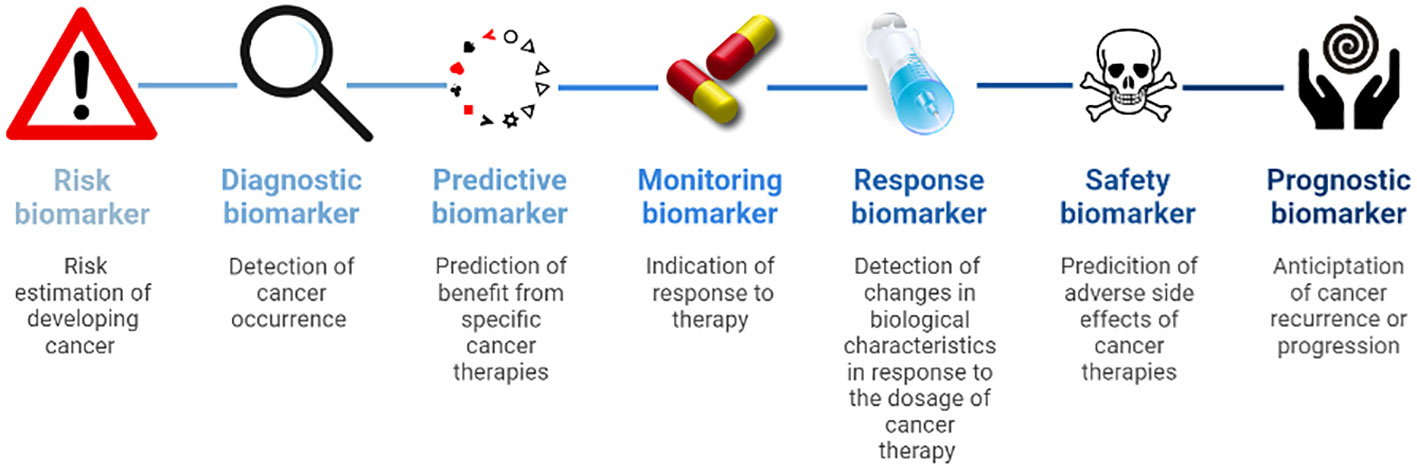
Modern medicine depends on reliable biomarkers for screening, diagnosis, disease monitoring, prognosis, predicting therapy success, response to therapy, and treatment safety. The BEST (Biomarkers, EndpointS, and other Tools) glossary of the FDA-NIH Biomarker Working Group defines biomarkers as a characteristic that is measured as an indicator of normal biological processes, pathogenic processes or biological responses to an exposure or intervention, including therapeutic interventions.” (6). They contain molecular, histologic, radiographic or physiologic characteristics.” (6, 7). The FDA-NIH Biomarker Working Group distinguishes between (6):
i. Susceptibility or risk biomarkers are associated with the chance of developing cancer and are essential to define risk populations for surveillance.
ii. Diagnostic biomarkers detect cancer occurrence.
iii. Predictive biomarkers identify patients that might benefit from specific cancer therapies.
iv. Monitoring biomarkers indicate the activity of the disease and response to therapy.
v. Pharmacodynamics or response biomarkers show changes in biological characteristics in response to the dosage of cancer therapy.
vi. Safety biomarkers predict adverse side effects of cancer therapies.
vii. Prognostic biomarkers anticipate cancer recurrence or progression (Figure 1).
For clinical applicability, biomarkers must meet the following criteria:
● The biomarker has to be valid, reliable, and objective.
● The sample collection must only involve a minimal risk for the patient.
● The biomarker has to be easy to collect.
● The sample must be stable under clinical and laboratory conditions.
● Samples should be able to be analyzed on a routine basis.
● Analysis has to be feasible and rapid.
3 State of the old – conventional biomarkers recommended by guidelines
Unfortunately, there are only a few biomarkers for HCC in daily practice so far: alpha-fetoprotein (AFP), HSP70 (HSPA7), glypican 3 (GPC3), and glutamine synthetase (GS). They are not even considered in all major guidelines due to their limited diagnostic quality. In the following, we will briefly review the use of biomarkers for HCC in daily practice:
3.1 Susceptibility biomarkers
Up to date, no susceptibility biomarkers are recommended by HCC guidelines and in daily clinical practice for stratifying the risk of HCC occurrence.
3.2 Diagnostic biomarkers
AFP is the most established clinical biomarker in clinical practice for detecting HCC. It is a transport protein for copper, nickel, fatty acids, and bilirubin expressed during the embryonic phase in human cells. Serum AFP levels diminish rapidly after birth and remain very low during adulthood (8). Of clinical relevance, in adults, AFP is mainly found in tumor cells of the liver, testes, and ovaries. It has been shown that AFP dampens the immune response and can enhance immunological tolerance toward tumors. In detail, secreted AFP interferes with the maturation and function of dendritic cells, leading to a decreased antigen presentation and induction of immune responses (9, 10). Furthermore, AFP interferes with T cell proliferation and shifts T cell response to a more regulatory phenotype. Thus, AFP promotes the immune system’s tolerance toward the tumor (11, 12).
Although AFP is commonly used as a biomarker for detection, it imposes enormous limitations: AFP has a very low-test sensitivity and specificity. Several studies showed disappointing or even contradictory results (9). AFP expression is absent in around 80% of early HCC (13). In many cases, liver damage also leads to the upregulation of AFP expression and secretion (14, 15). Especially in patients with high viral load, the determination of AFP displays low reliability. Low cut-off levels above 20 ng/ml show high sensitivity but low specificity, whereas high cut-off levels above 200 ng/ml raise specificity but lower sensitivity (16). Thus, serum AFP levels cannot reliably discriminate between chronic liver damage (e.g., fibrosis and cirrhosis) and HCC. Therefore, the Practice Guidance of the American Association for the Study of Liver Diseases (AASLD) and the Pan-Asian adapted European Society for Medical Oncology (ESMO) Clinical Practice Guidelines recommend biannual ultrasound with or even without determination of AFP levels for patients with liver cirrhosis and hepatitis-virus-infected patients (17, 18). Child Pugh-Stage and co-morbidities should allow curative or palliative therapy. The European Association for the Study of the Liver (EASL) even explicitly opposes the determination of AFP for the reasons given above (19). Nevertheless, according to new meta-analyses, the combination of ultrasound and AFP is a reliable diagnostic approach to detect HCC (7, 20). Zhang et al. recommended 400 ng/ml AFP protein in terms of sensitivity and specificity, whether AFP is used alone or combined with ultrasound (21).
As for AFP, the EASL guidelines assess other conventional diagnostic biomarkers, AFP-L3 and Des-gamma-carboxy-prothrombin (DCP), as unsuitable for cost-efficiency reasons. Nonetheless, studies of last years revealed that a combination of the clinical markers gender and age with the biomarkers AFP-L3, AFP, and DCP, is a good diagnostic marker (22). The combination is named the GALAD score after the initials of the biomarkers.
AFP-L3, an isoform of AFP, binds the lectin lens culinaris agglutinin (LCA) and is produced by malignant hepatocytes (23). A meta-analysis has shown that AFP-L3 has high specificity but low sensitivity for the diagnosis of early-stage HCC, suggesting that AFP-L3 is more valuable for ruling out HCC in conditions with elevated AFP levels than for diagnosing early HCC (24).
DCP is a defective prothrombin and results from a lack of post-translational carboxylation of the prothrombin precursor in HCC cells. Most large-scale studies have been performed in patients mainly of HCV- or HCB-related etiology (25–28).
Johnson et al. developed the score initially (22). In their study, the performance of the GALAD model was significantly better than the simple combination of AFP-3, AFP, and DCP alone. In a prospective phase 3 cohort study by Tayob et al. of 50 HCC patients and 484 controls, the GALAD score was associated with a substantial improvement in sensitivity for detecting HCC. However, in this study, a limitation of the GALAD score also shows up since the advantage of increased sensitivity was offset by a high number of false-positive results (29). In contrast, the study of Best et al. showed a high specificity of 93.3% (30) and was reliable in the subgroup of HCV and NAFLD patients. In another case-control study from Germany and a pilot cohort study in Japan, the GALAD score has been shown to detect early-stage HCC with high accuracy in patients with NASH, with and without cirrhosis (30–32) and thus could facilitate the monitoring of patients with NASH. In addition, another study shows that the GALAD score is also excellent for distinguishing HCC from chronic liver disease in an HCV subgroup of a cohort of Chinese patients (33). Therefore, despite limitations, the GALAD score seems one of the best options for HCC detection and will likely find its way into the guidelines.
If the diagnosis of HCC cannot be confirmed by typical contrast agent behavior in imaging, the EASL guidelines recommend a biopsy (19), with subsequent staining of HSP70, Glypican-3 (GPC3), and glutamine synthetase (GS).
HSP70 is a chaperone involved in protein folding, protein translocation, and regulation of transcription. In contrast to normal cells, many tumor entities, including HCC, overexpress HSP70 and secrete it into the extracellular matrix. Expression profiling identified HSP70 as a molecular marker for the detection of early HCC (34, 35). In addition, HSP70 serum levels enable discrimination between chronic hepatitis, cirrhosis, and HCC. However, this observation is limited by analyzing a relatively small cohort (86 healthy donors, 50 donors with chronic hepatitis, and 47 HCC patients) (36), and HSP70 is secreted by other tumors as well.
GPC3 belongs to a family of glycosylphosphatidylinositol-anchored cell surface heparin-sulfate proteoglycans. Sung et al. showed that GPC3 is upregulated in HCC tissue and plays an important role in the proliferation of malignant cells (37). Hippo et al. demonstrated that, compared to AFP, GCP3 is a more reliable marker to distinguish between patients with small, well-differentiated HCC and liver cirrhosis (38). However, a meta-analysis by Xu et al. revealed that GCP3 is inferior to AFP in the differential diagnosis between HCC and liver cirrhosis (39). Indeed, GCP3 expression displays no correlation to the expression of AFP.
The enzyme GS catalyzes the synthesis of glutamine, the primary energy source of tumor cells, from glutamate and ammonia in the liver. GS’s mRNA, protein, and activity were increasingly upregulated in precancerous lesions to advanced HCC (40, 41). GS displays a 50-59% sensitivity and specificity of 86-90% (42, 43).
A combination of the three markers, GCP3, the chaperone HSP70, and glutamine synthase for early detection of HCC revealed a sensitivity of 72% and a specificity of 100% (42). Thus, the combination of multiple markers results in advanced sensitivity and specificity.
3.3 Predictive biomarkers
Predictive biomarkers play almost no role in the choice of HCC therapy. There are two exceptions:
The curative treatment options for HCC are resection, local ablation procedures, and liver transplantation. Due to the pervasive organ shortage, bridging therapy to transplantation is required, and patients must be carefully selected. According to national guidelines, liver transplantation should not be considered if AFP levels are above 1000 ng/ml due to poor postoperative prognosis (18, 44–46).
Palliative treatment options for advanced-stage HCC include immunotherapies (atezolizumab/bevacizumab, tremelimumab/durvalumab, nivolumab/ipilimumab, pembrolizumab) and protein tyrosine kinase inhibitors (sorafenib, lenvatinib, donafenib, regorafenib, cabozantinib, apatinib). Ramucirumab is a recombinant human monoclonal antibody that inhibits vascular endothelial growth factor receptor (VEGFR) and is approved for second-line therapy. The REACH-2 trial demonstrated that ramucirumab improved overall survival compared to placebo in patients with hepatocellular carcinoma and AFP levels of at least 400 ng/ml who had previously been treated with sorafenib (47). Patients with lower AFP levels did not benefit.
3.4 Monitoring biomarkers
Imaging techniques primarily assess the response of patients with HCC to local or drug therapies. None of the guidelines recommend specific biomarkers for assessing response to therapies. Bruix et al. analyzed two phase 3 studies of prognostic factors and predictors of the benefit of sorafenib in patients with HCC. AFP was not a predictive biomarker of sorafenib benefit (48). However, a recent meta-analysis showed that post-treatment AFP response was significantly associated with overall and recurrence-free survival (49).
3.5 Prognostic biomarkers
Like the assessment of response to treatment, screening for relapse after curative treatment is mainly performed by imaging with MRI, CT, or ultrasound. Imamura et al. showed in a study of 249 patients undergoing hepatectomy that AFP levels above 32 ng/ml indicate relapse (50).
In summary, AFP is the only biomarker used for early detection, prediction, and monitoring of response to treatment and disease recurrence. It has significant limitations in sensitivity and specificity, especially when used alone without combination with other biomarkers. Therefore, there is a great need for other or complementary biomarkers to improve the quality of care for patients with HCC.
4 Promising new biomarkers for clinical application
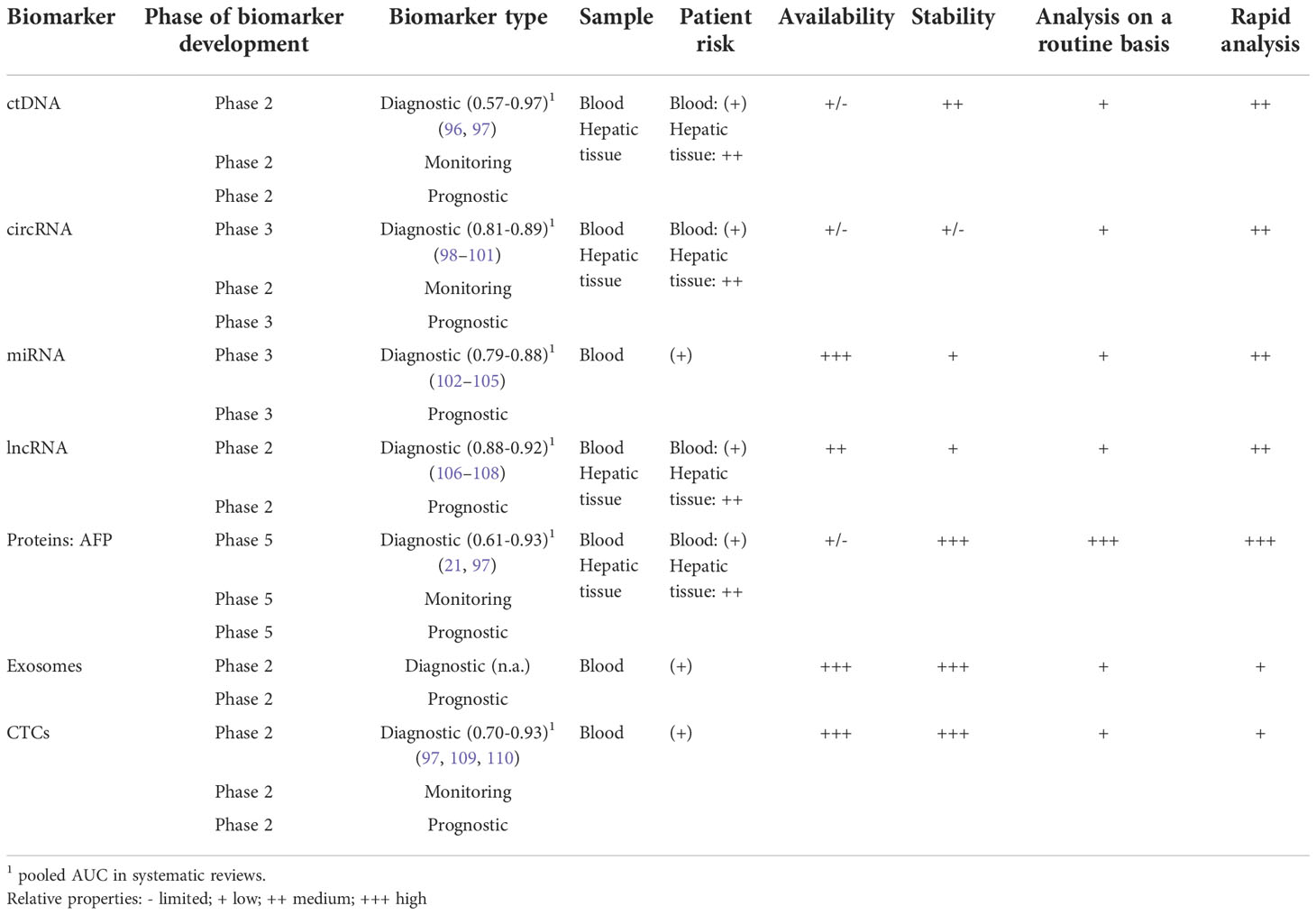
Conventional tissue biopsies are invasive and associated with risks for patients. Sometimes tissue biopsies are impossible or very difficult to perform due to the location of the tumor or the presence of multiple lesions. In contrast, biomarkers from body fluids are a promising tool for diagnosing and monitoring tumor diseases, especially because they are not or only minimally invasive and can therefore be obtained without or with low risk. The idea of liquid biopsy is based on molecular analysis of circulating cells that have been shed from the tumor and products from malignant tissue that have been released into biological fluids, such as the bloodstream. Thus, liquid biopsies provide access to tumor-derived materials, including circulating nucleic acids, proteins, exosomes, and circulating tumor cells (CTCs). However, the differences between these various groups of markers in terms of accessibility, stability, and detection must be considered.
4.1 Circulating nucleic acids
Nucleic acids are released into the bloodstream after induction of apoptotic or necrotic cell death of tumor cells. These circulating nucleic acids can be divided into two subgroups: (i) circulating tumor desoxyribonucleic acid (ctDNA) and (ii) cell-free ribonucleic acid (RNA).
4.1.1 Circulating tumor DNA
In 1948 Mandel et al. described for the first time that freely circulating DNA is released from dying cells into the peripheral blood (51). Later Leon et al. observed that circulating DNA appeared more frequently in the serum and plasma of cancer patients (52) and reflected the tumor burden (53). Furthermore, ctDNA provides direct access to molecular key information, including genomic (point mutations or copy number variations [CNV]) as well as epigenetic data (changes in DNA methylation) (54).
Apoptotic and necrotic cells release DNA into the extracellular matrix. This DNA can be detected as circulating cell-free DNA in the blood. Solid tumors often show large necrotic areas due to undersupply of oxygen and glucose. Therefore, various tumors, including HCC, can be detected by an increased level of cell-free DNA. Iizuka et al.’s study highlighted the potential diagnostic value of monitoring the amount of cell-free DNA to detect HCC (55). However, elevated cell-free DNA levels have been observed in multiple cancers. Therefore, the amount of cell-free DNA is not HCC-specific. Nevertheless, combining the determination of the amount of cell-free DNA with the detection of HCC-specific protein biomarkers like AFP results in a sensitivity of 87% and specificity of 100% to detect HCC (56). A recent study presents a novel computer-based prediction model that uses comprehensive fragmentomic profiling of cell-free DNA in plasma for early detection of liver tumors. The model showed excellent performance with a sensitivity of 98.8% and 96.8% specificity in detecting primary liver cancer (sensitivity for HCC 96.2% and intrahepatic cholangiocarcinoma 100%). In addition, early-stage primary liver cancer detection was 95.9% (stage I) and 97.9% (stage II). For tumors less than 3 cm in size, the sensitivity was 98.2% (57).
The use of modern technologies such as next-generation sequencing (NGS) allows the detection of mutations in DNA isolated from the blood (58). For example, tumor-specific point mutations of various genes have been detected in the ctDNA of HCC patients. NGS analyses revealed that seven genes and DNA regions derived from ctDNA harbor the most important mutations associated with poor survival in HCC: (i) TERT promoter, (ii) TP53, (iii) CTNNB1, (iv) AXIN1, (v) JAK1, (vi) EPS15 and (vii) CACNA2D4 (59–67). Several studies have shown that analysis of point mutations in these genes is a valuable tool with a clinically relevant impact on prognosis and early detection of HCC (59, 68, 69).
However, the use of somatic mutations previously detected in primary tumor tissue as biomarkers is limited by their variability and low concentration in plasma (70). Besides detection and analysis of point mutations, CNV can be used as early biomarkers and prognostic parameters for HCC (63–66, 71–78).
For example, a characteristic CNV was found in preresection plasma samples of patients with HCC, whereas this CNV was almost absent in post-resection plasma samples. Thus, CNV is an additional marker for detection and treatment surveillance (79, 80).
Furthermore, genomic alterations and epigenetic modifications (e.g., DNA methylation) were detected in ctDNA (81–85). DNA methylation is a mechanism to regulate gene expression and control DNA stability and DNA-protein interactions. DNA methylation is essential in cancer development, especially in HCC formation (54, 86–88). The methylation status of several tumor suppressor genes correlates with HCC occurrence and progression. These genes include p15 and p16, APC, SPINT2, SFRP1, TFP12, GSTP1, and RASSF1A (86, 89, 90). Moreover, specific hypomethylation of the long interspersed nuclear element-1 (LINE-1) (91), methylation of the insulin-like growth factor-binding protein 7 (IGFBP7) gene (92) and hypomethylation of the promotor region of the transcriptional repressor CTCFL were associated with reduced survival in HCC patients (93).
Therefore, combined determination of (i) the amount of circulating cell-free DNA, (ii) additional biomarkers, and (iii) genetic analysis of ctDNA, including detection of tumor-specific point mutations, CNV, and alteration in methylation patterns, are essential tools for detection and prognosis of HCC. Furthermore, genetic aberrations detected in ctDNA provide molecular information about tumor development and possibly response towards treatment. Recent developments allow the analysis of small amounts of ctDNA. This advance will facilitate analyses of ctDNA in the future.
Conclusion: ctDNA provides a comprehensive representation of genomic alterations of different tumor regions. Isolation procedures are well-established. Improvements in technology allow for higher sensitivity of analytical assays. The short half-life of ctDNA allows real-time monitoring of cancer development with more accurate clinical correlations. The amounts of blood required for isolation are practical from the clinical point of view. The number of patients and the control groups are also large enough in most studies, and international studies are available in addition to many Asian studies. Most studies with ctDNA are in biomarker development phase 1 or 2 (57, 84, 94, 95). The limitations are that it is difficult to distinguish between ctDNA and circulating free DNA (released from non-malignant cells) and the low concentration of ctDNA in blood. In addition, the short half-life, which on the one hand allows real-time monitoring, is on the other hand a challenge for the analysis and storage of the samples. In comparison to CTCs ctDNA is not suitable for functional assays. Furthermore, analysis is time-consuming and costly, and most emerging assays have not yet been clinically validated (Table 1; Figure 2).
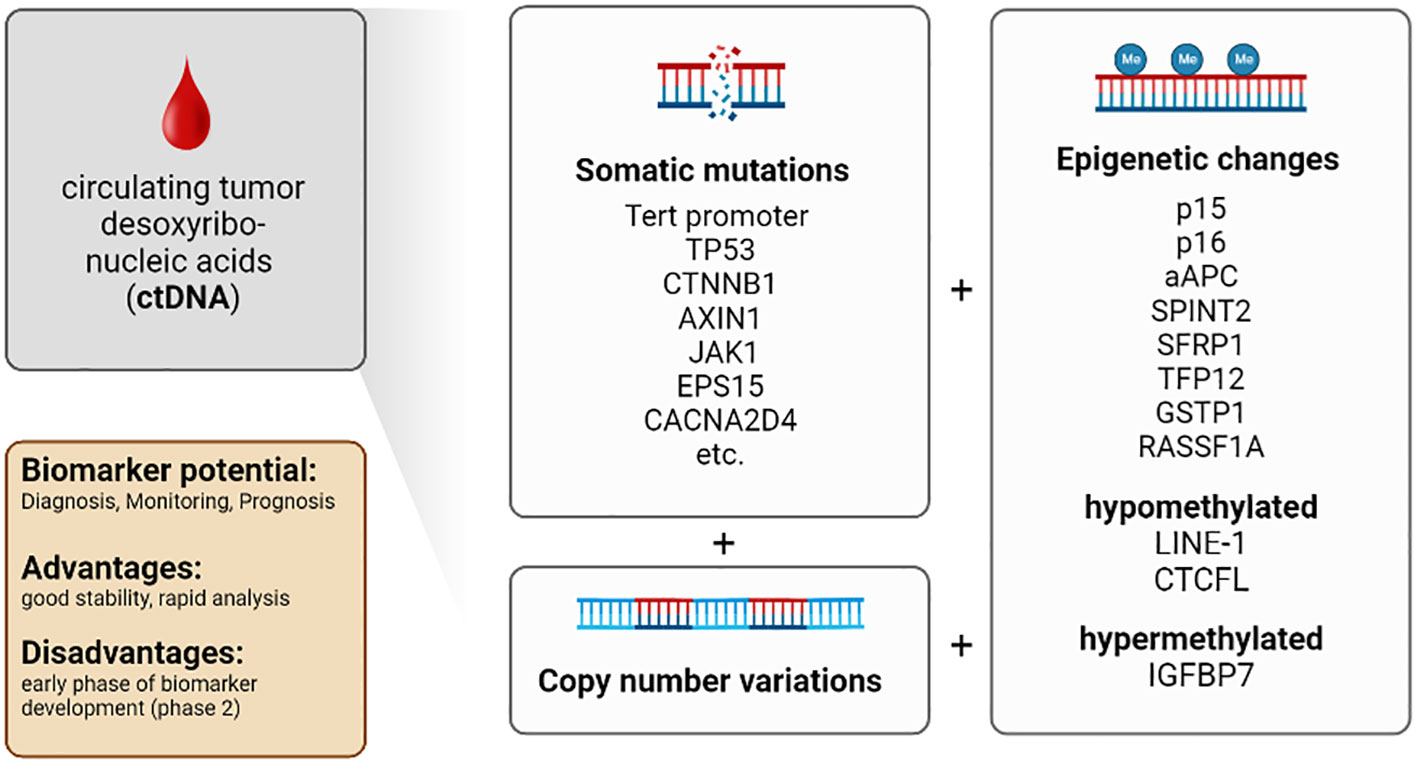
Figure 2 Characteristics of the biomarker class circulating tumor desoxyribonucleic acids (ctDNA) in patients with HCC.
4.1.2 Cell-free tumor RNA
In addition to DNA, RNA is also released into the extracellular space. It should be highlighted that RNA is less stable compared to DNA. Therefore, cell-free RNA is associated with proteins, proteolipids, or encapsulated into exosomes preventing its degradation. Three different groups of RNA can be detected in the bloodstream and used as potential biomarkers: circular RNA, micro (mi)RNA, and long-non-coding (lnc)RNA (Figure 3).

Figure 3 Characteristics of the biomarker class circulating tumor ribonucleic acids (cell-free RNA) in patients with HCC.
4.1.2.1 Circular RNA
Circular RNAs are differentially expressed in various cancer tissues (111–114), including HCC (115), and they are closely associated with the initiation and development of cancer. Circular RNAs arise from aberrant by-products or abnormally spliced transcripts (116). Most up-regulated circular RNAs are positively associated with HCC progression, whereas down-regulation usually displays suppressive effects and prevents HCC development. Most data on circular RNA are based on tissue analysis. Therefore, circular RNAs can also be used as markers in normal biopsies. The particular advantage, however, is that these RNAs can also be found in blood and thus be sampled minimally invasively (117, 118).
Several circular mRNAs showed highly interesting results in context with early detection and diagnosis of HCC (115, 119–123). For example, the circular RNAs hsa_circ_001565 (B4GALT2), hsa_circ_000224 (C17orf107), and hsa_circ_000520 (VIM) display a sensitivity of up to 97% and a specificity of up to 92% in the detection of HCC (124). Very similar results were obtained for hsa_circRNA_104075 (NUP153), hsa_circ_0005075 (EIF4G3), hsa_circ_0028502 (SLC24A6) and hsa_circ_0076251 (ZFAND3). They showed comparable sensitivities of up to 96% and specificities of up to 98% (115, 122, 125).
Besides their potential as biomarkers for the early detection of HCC, circular RNAs are of high prognostic value (124, 126–132). Exemplary is a study showing that hsa_circ_0001727 (circZK - SCAN1) expression was positively correlated with HCC prognosis (129). Moreover, the detection of hsa_circ_001565 (B4GALT2), hsa_circ_000224 (C17orf107), and hsa_circ_000520 (VIM) was associated with prolonged relapse-free survival (124). In contrast, low expression of hsa_circRNA8662-12 (TRIM33-12) was closely correlated with poor prognosis (133).
Conclusion: Circulating free RNA provides an up-to-date snapshot of the transcriptome. It can indicate cancer and trace it back to its site of origin. Limitations are sample instability and high variability of circulating free RNA expression between different individuals. Therefore, studies aiming to define panels of cell-free circulating RNAs that can be used as general biomarkers for HCC are needed. In addition to the small amount and not yet very detailed purification protocols, the high variability and heterogeneity of cell-free circulating RNA are problematic and currently do not allow for clinical application. In addition, there is always a risk that samples will be contaminated with transcellular mRNA. Unfortunately, there are only very few studies from Europe or the USA. Most of the studies originate from East Asia and are in development phase 3 (121, 123, 131, 134) (Table 1). Thus, there is still much development work to be done here.
4.1.2.2 Cell-free micro (mi)RNA
In 2008 Lawrie et al. were the first to describe microRNA (miRNA) as tumor biomarker (135). miRNAs are a member of endogenous non-protein-coding RNA with a size of approximately 20-22 nucleotides. miRNAs are relatively resistant to RNase degradation, boiling, and freeze-thaw cycles (136, 137). miRNAs can be used as markers in tissues and the blood. In terms of patient safety, miRNAs obtained in a liquid biopsy are of particular interest. Especially the stability of these RNAs and their release into the bloodstream makes them attractive as a biomarker. Over the past ten years, miRNAs have become the most intensively studied nucleic acid biomarkers in HCC and have proven valuable in the diagnosis and prognosis of the disease. Nevertheless, many studies have conceptual weaknesses, such as very different non-validated purification methods for miRNAs or unclear sequencing and identification protocols.
miRNAs bind to the corresponding 3´UTR of their target messenger RNA (mRNA) and induce mRNA degradation. Interestingly, there is a correlation between abnormal circulating miRNA levels and pathological characteristics of certain tumors (138–140), including HCC (141). So far, over 70 miRNAs have been proposed as potential biomarkers for HCC (86, 142–159). A meta-analysis evaluated miRNA-21 as a biomarker for early diagnosis of HCC with a sensitivity of up to 88% and a specificity of up to 87% (160). Interestingly, the sensitivity and specificity of the combined miRNA panel miR-29a, miR-29c, miR-133a, miR-143, miR-145, miR-192, and miR-505 were significantly higher than the sensitivity and specificity of the established biomarker AFP regarding detection of small (AUC: 0.833 vs. 0.727) and early-stage HCC (AUC: 0.824 vs. 0.754). Another panel of miRNAs (miR-192, miRNA-125b, and miR-23a) was suitable for predicting the survival time of HCC patients (161). Huang et al. developed in a phase 3 study an HCC risk score consisting of 5 miRNAs (miR-18a, miR-26a, miR-27a, miR-222, miR-223) that correlates with an increased risk of HCC development in cirrhotic patients (162). Thus, miRNA combinations represent a strategy to develop novel diagnostic tools for HCC and improve treatment surveillance.
Conclusion: The advantage of using miRNA as a biomarker is its wide range of applications, as miRNAs are involved in many pathogenic processes and have high specificity and reproducibility. miRNAs display inherent stability, and the serum concentration is relatively high. The main challenge in establishing miRNAs as biomarkers is that although the available studies have identified a large number of miRNAs as potential markers, the miRNAs identified vary depending on the specific study. Therefore, a “universal marker” is still lacking. This is due to the lack of standardized protocols for the purification of miRNAs and the fact that the expression of these miRNAs is directly linked to the developmental stage and characteristics of the individual tumor. In addition, comorbidities can lead to an increase of unspecific miRNAs, which interfere with detecting cancer-specific miRNAs. Nevertheless, it should be possible to define miRNA panels that could act as universal markers. These panels can then be used for both prognosis and diagnostics purposes. In studies containing information about the required blood volumes, the required volumes were in a range that could be used in the clinic (≤10ml). Unfortunately, some studies lack this information. Most studies include a relatively large number of participants (>100). The control groups are also sufficiently big in most cases. Interestingly, most recent studies were conducted in East Asia (China) or North Africa (Egypt). However, the number of patients with HCC is increasing in Western countries due to lifestyle and the resulting metabolic syndrome. There is a high heterogeneity of the miRNAs found, and each study finds different miRNAs that can be used as biomarkers. Most studies are in phase 2 (155, 157), and few are in phase 3 (162). We need more studies with clearly defined protocols for the purification of miRNAs and also very precisely characterized cohorts of patients. Therefore, very little can be said about miRNAs as biomarkers for HCC. Generally, they are a potential option that fulfills all the prerequisites for a good marker (Table 1).
4.1.2.3 Cell-free long non-coding RNA (lncRNA)
Circular RNA and miRNA are not the only RNAs that can serve as biomarkers for HCC. Furthermore, long non-coding RNA (lncRNA) shifted into the focus as novel potential biomarkers in HCC. Comparable to miRNAs, lncRNAs belong to the group of non-protein-coding RNA transcripts. They exceed a length of 200 nucleotides. Like other non-protein-coding RNAs, lncRNAs can be detected in the blood. Upon isolation, lncRNAs are stable in the plasma (163, 164). They can interact with proteins, DNA, and other types of RNA. They act as modulators of gene expression by regulating transcriptional and post-transcriptional processes and controlling various cellular processes, e.g., genomic imprinting, cell cycle, cell proliferation, differentiation, and apoptosis (165, 166). Various lncRNAs are involved in cancer pathogenesis, controlling invasion, migration, and metastasis of tumor cells (167–169), and several studies showed that dysregulation of lncRNAs promoted the development of HCC (170).
Therefore, lncRNAs might contribute to the panel of beneficial biomarkers in hepatic tumors. One of the first and most studied lncRNAs in HCC is Highly Upregulated in Liver Cancer (HULC). Several studies demonstrated that circulating HULC could be used as a diagnostic marker, being up-regulated in the blood of patients with HCC (171, 172). Another lncRNA of clinical relevance is LINC00152. The amount of LINC00152 in the blood increases from healthy donors to patients with liver cirrhosis and displays the strongest up-regulation in patients with HCC. This close association with progress from liver cirrhosis to HCC underlines LINC00152 abilities as a potential diagnostic biomarker (172, 173). A further significant increase in sensitivity and specificity was achieved by combining HULC with LINC00152 or combining HULC, LINC00152, and AFP (174). However, a more detailed analysis is needed to prove their value as biomarkers for HCC (175).
Conclusion: The advantage of using lncRNA as a biomarker is its relatively high stability. lncRNAs are involved in multiple pathological processes and can obtain new insights into the progression of the disease. The disadvantages are similar to those of miRNAs. Since lncRNAs are involved in many physiological and pathophysiological processes, detecting tumor-specific lncRNA is difficult. Interference by co-morbidities cannot be excluded. Some studies directly compare tissue samples and blood samples. However, many studies involve small patient and control cohorts. In addition, most studies have been conducted in East Asia. Care should be taken to include European and American cohorts in the future. Most studies are in biomarker development phase 2 (172, 176–179). Therefore, further detailed studies are needed to improve data on individual lncRNAs and establish defined lncRNA panels that can be used as general markers for detecting HCC. The large heterogeneity of lncRNAs identified in the previous studies, which unfortunately overlap only partially, is the major obstacle to clinical application.
Overall, circulating nucleic acids occur in different forms, e.g., circulating tumor DNA (ctDNA) or cell-free RNA. Although circulating nucleic acids are relatively inert towards degradation, it is still more elaborate to purify, store and analyze them compared to proteins. The advantage of nucleic acids as markers is that they contain essential biological information and may be of prognostic value. But these properties provide a large heterogeneity in terms of expression in tumors of different patients, making clinical application difficult. Interestingly, a combination of nucleic acids and marker proteins displays the highest specificity and sensitivity in HCC diagnosis and could thus help develop new identification panels that can be used universally for detecting HCC. Therefore, combining different biomarkers, regardless of nucleic acid, protein or CTC, might be the most promising approach for the future (Table 1).
4.2 Proteins
Tumor cells secret various proteins into the extracellular matrix, e.g., proteases required for invasion and metastasis, proteins dampening the immune system, cytokines, growth factors, and angiogenic factors for proliferation. In addition, tumor cells undergo apoptotic or necrotic cell death due to a lack of oxygen and energy. These dying cells release proteins into the bloodstream.
Proteins are very stable in the blood, and there are several methods for identification by immune-linked and biochemical methods. Detecting tumor-specific proteins is less complex and expensive than identifying and purifying cell-free DNAs and RNAs. As described above, proteins are the most commonly used biomarkers in clinical routine (Figure 4).
4.2.1 Cytokeratin 19
Cytokeratin 19 belongs to the keratin family. It is a filament protein essential for the structural integrity of cells. Cytokeratin 19 has been associated with poor clinical prognosis in HCC patients in several studies. The co-expression of Cytokeratin 19, AFP, and Glypican-3 is an excellent predictive factor for metastasis and adverse treatment outcomes (180). In addition, concurrent expression of these three proteins was also associated with poor survival (181–183).
4.2.2 Golgi protein 73
Golgi protein 73 is a transmembrane protein localized in the Golgi apparatus. Its function is incompletely understood. Golgi protein 73 expression is linked to patients with liver disease, particularly HCC (184, 185). In the serum of HCC patients, Golgi protein 73 concentration is significantly elevated compared to patients with liver cirrhosis (186). Whether Golgi protein 73 alone is superior to AFP in detecting HCC and discrimination from liver cirrhosis is controversial (186, 187). However, combining both markers has improved the detection of early HCC and discrimination from liver cirrhosis (187).
4.2.3 Annexin 2
Annexin 2 is a calcium-dependent, phospholipid-binding protein linked to cell mobility and protein interaction with the actin cytoskeleton. Recent studies reported that Annexins, including Annexin 2, can interfere with immune functions and induce tolerance (188). Interestingly, Annexin 2 is upregulated in HCC and can indicate tumor malignancy (189). Annexin 2 also showed better sensitivity and specificity than AFP to detect early HCC (190). Thus, Annexin 2 might be a helpful marker for early tumor detection (189), although more detailed studies are needed to estimate the potential of Annexin 2 for clinical application.
4.2.4 Osteopontin
Osteopontin (OPN) is an extracellular matrix (ECM) protein whose elevation is associated with tumor invasion, proliferation, and metastasis in several cancers (191). Using tissue microarrays, Desert et al. analyzed 366 samples from patients with normal liver, cirrhosis, dysplastic nodules, or HCC. They show that OPN increases in expression during hepatocarcinogenesis (192). Wu et al. showed in a recent study that OPN induces JAK2/STAT3/NOX1-mediated ROS production, leading to hepatocellular carcinoma progression (193). Several phase 2 studies show osteopontin has diagnostic (194, 195) and prognostic potential (196, 197) as a biomarker.
4.2.5 Midkine
Midkine, also known as neurite growth-promoting factor 2 (NEGF2), is a secreted protein that functions as a cytokine and growth factor and mediates its signal through proteoglycan and non-proteoglycan receptors on the cell surface (198–201). Midkine enhances the angiogenic and proliferative activities of cancer cells. Expression of midkine (mRNA and protein expression) is increased in several cancers, including HCC. Thus, Midkine can serve as a biomarker in HCC, as shown in phase 3 trials (202–205). Midkine is mainly overexpressed in AFP-negative patients, so it increases detection rates of HCC (205).
4.2.6 Dickkopf-1
The glycoprotein Dickkopf-1 (DKK-1), expressed mainly in the placenta and embryonic tissues, is an antagonist of the Wnt/β-catenin signaling pathway and is elevated in several cancer types. DKK-1 shows higher efficacy for detecting HCC than AFP in phase 4 trials (206–208), but midkine is more precise than DKK-1 in cirrhotic HCV patients (209). A combination of Golgi protein 73, AFP, and Dickkopf-1 increases the sensitivity and specificity of HCC detection (210).
4.2.7 Squamous cell carcinoma antigen 1 and 2
Squamous Cell Carcinoma Antigen (SCCA) consists of two proteins, SCCA-1 and SCCA-2, which are serine protease inhibitors. Several studies investigated the diagnostic value of SCCA and its immune complex SCCA-IgM in HCC. A recent phase 4 trial with 203 cirrhotic patients revealed that patients with HCC have higher levels of SCCA-IgM than those without it during a five-year follow-up (211). Another phase 4 trial with 91 patients showed that SCCA-IgM also offers predictive value (212). But gender differences have to be considered, as low levels of SCCA-IgM after transarterial embolization indicate more prolonged survival in males and shorter in females (213). SCCA and SCCA-IgM show moderate diagnostic accuracy in several meta-analyses (214–216). Combination with AFP increases prognostic value significantly (214).
4.2.8 Alpha-l-fucosidase
Alpha-l-fucosidase (AFU) is a lysosomal enzyme that is present in low concentrations in human cells, blood, and body fluid and hydrolyzes fucose-containing sugars. Its activity is increased in the serum and tissue of HCC patients. Still, it is not specific to HCC, as high levels are also found in patients with diabetes, pancreatitis, and hypothyroidism (217). A retrospective phase 3 study with 280 HCC patients due to HBV B AFU shows good early detection prosperities of HCC (218), but midkine is a more sensitive predictor than AFU in HCC due to HCV (209). The combination of AFU with AFP raises sensitivity and specificity, especially in hepatitis-negative patients (219), but is worse than AFP alone in patients with HCC due to HBV (220).
Conclusion: The major advantage of protein biomarkers is that the detection is easy to perform, less error-prone, and inexpensive. Results can be obtained quickly and without complex equipment. Therefore, protein biomarkers are optimal for clinical use. The major disadvantage of protein biomarkers is that tumors can escape from detection due to individual differences in the protein expression pattern. However, this can be overcome by using defined combinations of different biomarkers. In the future, defined combinations of protein biomarkers with other types of markers, e.g., circulating nuclear acid and CTCs, may improve detection specificity and sensitivity as well as the prognosis of HCC.
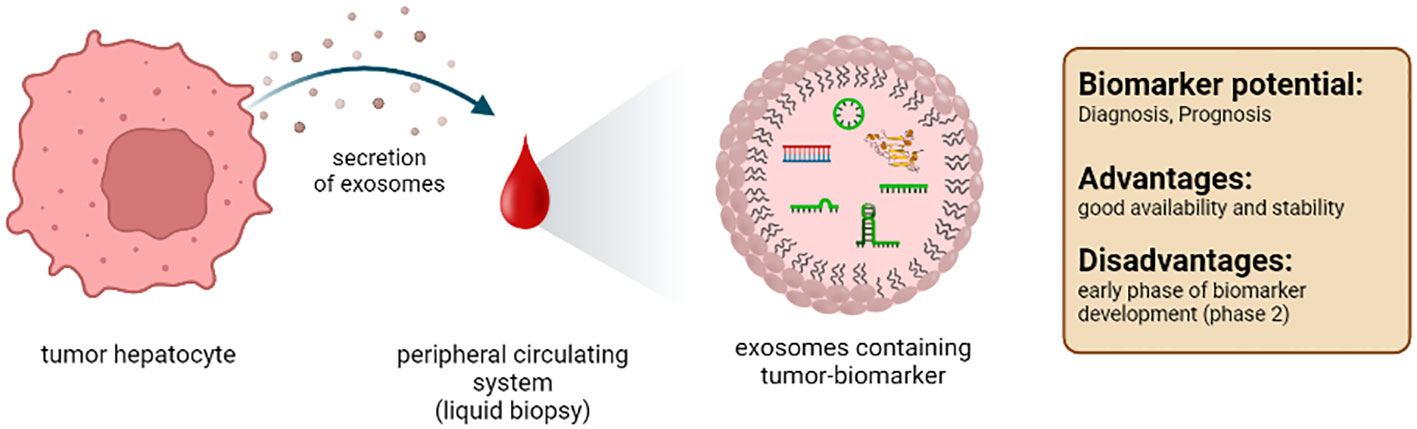
4.3 Exosomes
Exosomes are extracellular vesicles with a diameter of 30-200nm. They are formed in the endosomal compartment of eukaryotic cells. Exosomes are specifically released, facilitating intracellular transport processes, and enabling communication between cells. Their content consists of various components, such as proteins and nucleic acids (Figure 5). Therefore, exosomes give detailed information about the secreting cell or tissue (221). In the liver, mainly hepatocytes, immune cells, and non-parenchymal liver cells release exosomes (222). It should be emphasized that the administration of antibiotics from the subgroup of fluocinolones, especially ciprofloxacin, can increase the secretion of exosomes (223). Furthermore, it must be admitted that studies on exosomes as markers are still in the early stages.
4.3.1 Exosomal lipids and proteins
Exosomal membranes are composed of lipids characteristic for different tissue, including tumors. To date, mainly in vitro data are available on the potential role of exosomal lipids as biomarkers for HCC (224, 225). Thus, further analysis must show whether exosomal lipids are suitable biomarkers in vivo.
Exosomes contain macromolecules, e.g., proteins protected from extracellular degradation processes. Therefore, the exosomal content is potentially interesting for tumor detection and prognosis. One study identified an HCC-specific exosomal protein profile that included CD44, cell division cycle 42 (CDC42), RAS related protein (RRAS), MET, G protein subunit alpha 13 (GNA13), metalloproteinase domain 1 (ADAM1), GNAS complex locus (GNAS), eukaryotic translation initiation factor 4A3 (EIF4A3) and S100 family proteins (226). An additional study detected HSP70, Hsp90, glypican 3, and the well-established marker AFP specific to HCC (227). However, whether exosome-derived protein panels are more valid biomarkers than freely circulating proteins needs to be analyzed in clinical studies, comparing both parameters according to the purification method, the stability, and thus the robustness of the analysis.
Probably the closest study to a clinical application is the HCC EV ECG score, an extracellular vesicle-based protein assay for detecting early-stage hepatocellular carcinoma (228). Here, exosomes isolated from plasma are analyzed for surface expression of EpCAM, CD63, CD147, CD63, and GPC3. The analysis of this panel of surface markers results in a sensitivity of 91% and a specificity of 90% in the early detection of HCC. Thus, this score could complement current monitoring methods and improve patient outcomes.
4.3.2 Exosomal nucleic acids
In addition to proteins, exosomes also contain nucleic acids, e.g., DNA and RNA. Our knowledge of exosomal DNA as biomarkers for HCC is limited to date. Exosome-derived DNA is protected against degradation; it is of high molecular weight and, therefore, suitable as an HCC biomarker (56, 229). More intensively than DNA, exosome-derived RNA has been explored as a potential biomarker for HCC (230, 231). Several types of RNA can be found in exosomes that are protected from degradation by the exosomal membrane and thus have a significantly longer half-life than free RNAs: mRNA, circular RNA, miRNA, and lncRNA. These RNA species, alone or in combination with other exosomal components, represent interesting biomarkers for HCC. For example, RAB11A mRNA was present in exosomes purified from the serum of patients with HCC. Combined with exosomal lncRNA-RP11-513115.6 and miR-1262, it turned out to be an effective biomarker with high sensitivity and specificity in distinguishing patients with HCC from patients with chronic hepatitis C virus infection (232). In addition, exosomal circular RNAs are particularly interesting for predicting the prognosis of HCC. Many circular RNAs are abundant and stable in exosomes derived from patients with HCC, such as hsa_circ_0088030 (circPTGR1), Cerebellar degeneration-related protein 1 antisense RNA (Cdr1), and circDB. These circular RNAs promote cancer cell proliferation and metastasis and are indicators of aggressive tumors with poor prognosis (233–236). Another RNA species detected in exosomes are miRNAs. Detection of exosomal miRNA-210 and miRNA-224 is specific for HCC. Both miRNAs promote angiogenesis and enhance the proliferation and invasion of the tumor (237, 238). Exosomal lncRNAs are also potentially significant for the HCC diagnosis. For example, exosomal-derived lnc-FAM72D-3 and lnc-EPC1-4 levels are significantly increased in the serum of patients with HCC (239, 240).
Overall, exosomes contain similar biomarkers compared to those detected in the blood. The vesicle protects the marker molecules from damage and degradation. This is particularly important for nucleic acids as they are targets for degradation in the blood. Further studies must confirm that exosomes and their contents are suitable biomarkers in HCC.
Conclusion: Exosomes contain a multitude of important information about the cells from which they originate. Their lipid composition, protein, and nucleic acid content are like a fingerprint of the (tumor) cell from which they originate. However, the purification of these small extracellular vesicles and their analysis are complex and require sophisticated equipment. The quality of the isolation of the inter-exosomal proteins or nucleic acids is highly dependent on the methodology, especially since standardized protocols are not yet available.
Several phase 2 trials show promising results for diagnostic and prognostic biomarker usage (82, 241–243). We need more international studies to prove the importance of exosomes as tumor markers, as most studies are from Asia. From a scientific and clinical point of view, exosomes seem to fulfill all the conditions for an ideal marker (Table 1).
4.4 Circulating tumor cells
In 1869, Ashworth detected “cells similar to those in the tumours” in the blood of a patient with a metastatic tumor (244). This was the first description of CTCs and the first detection of tumors by blood analysis. However, at this time, reliable detection and identification of these cells were a nearly unsolvable challenge. Furthermore, in 1895, x-ray imaging by Roentgen was developed as a novel diagnostic tool. Thus, scientists primarily focused on this new imaging method, and the idea of using CTCs to detect tumors was put on hold. Today we know that imaging tools have their limitations, and additional strategies for early detection and prognosis are urgently needed. Therefore, CTCs again became a focus as biomarkers, especially since detection, isolation, and analysis to investigate CTCs have made tremendous progress (Figure 6).
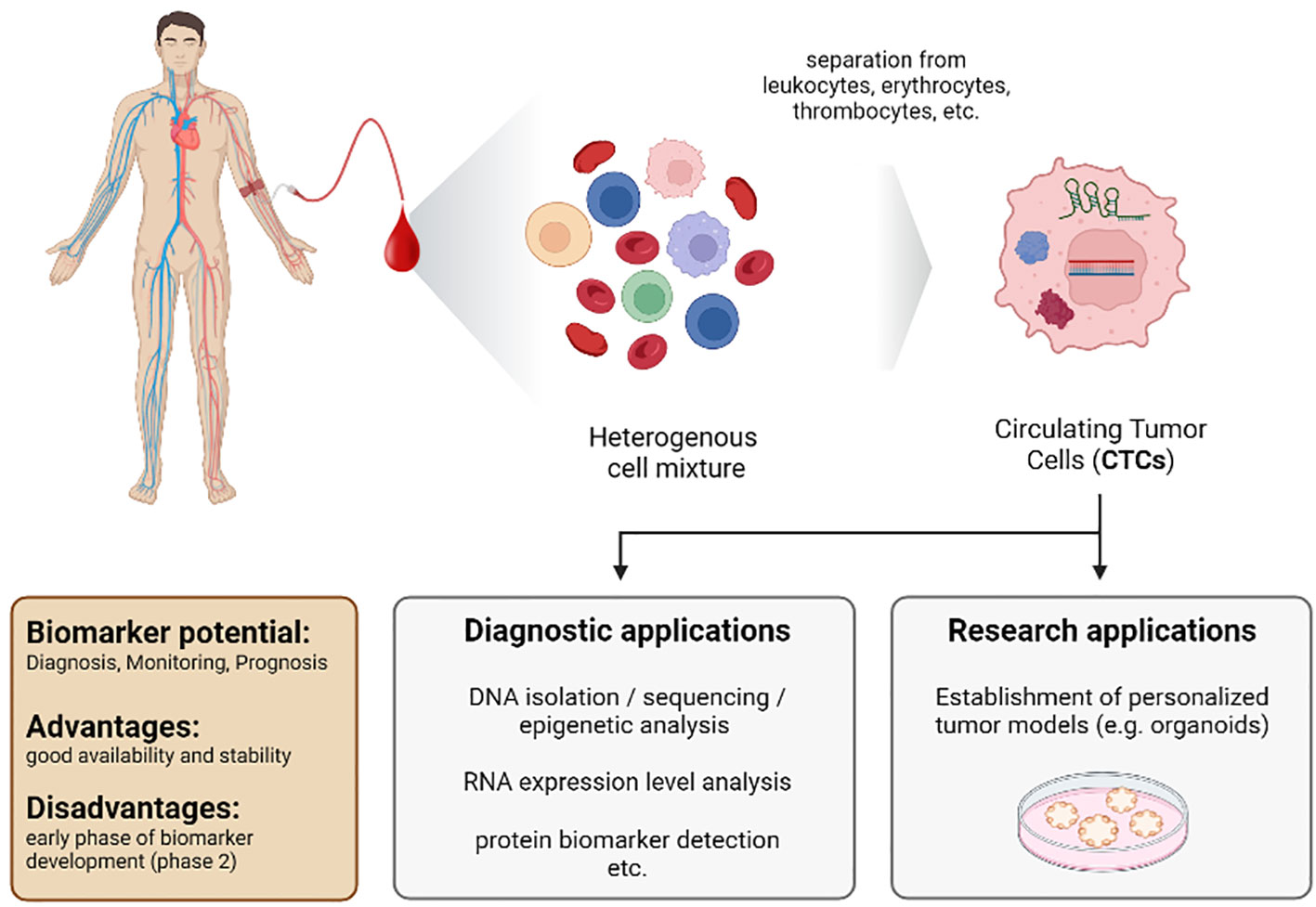
Figure 6 Characteristics of the biomarker class circulating tumor cells (CTCs) from a liquid biopsy in patients with HCC.
CTCs are cancer cells that circulate in the blood upon being shed off from the tumor. The genetic information of these cells indicates mutations and, therefore, contains hints on sensitivity and resistance towards therapy. In addition, CTCs can be used to form organoids, which serve as personalized tumor models to analyze cell signaling and mimic therapeutic approaches in vitro (245–249).
Nevertheless, the isolation and analysis of these cells are still challenging despite novel techniques. Once cells detach from the extracellular matrix, apoptotic cell death, anoikis, is induced in most cells (250). Thus, most tumor cells die within a few hours upon shedding, resulting in a low frequency of CTCs (251, 252). In the blood of patients with metastatic tumors, there is approximately one CTC per 1x109 cells (253). This low frequency makes identification and isolation extraordinarily challenging and asks for strict definitions regarding identification.
The CellSearch™ definition is considered to be the current state-of-the-art standard. This definition states that a CTC is a circulating nucleated cell larger than 4µm, expressing the epithelial cell adhesion molecule (EpCAM) and cytokeratins 8, 18, and 19. To separate CTCs from immune cells, the CTC has to be negative for the leukocyte-specific antigen CD45 (254). However, only about one-third of CTCs derived from HCC patients are positive for EpCAM and cytokeratins. Thus, the application of CellSearch™ criteria is unsuitable for all subgroups of HCC (255, 256). Another concern is that the level of CTCs correlates with tumor burden. Therefore, sensitivity in the early stage of the disease might be low (252).
Nevertheless, several studies using CellSearch™ showed interesting results for CTCs as diagnostic and prognostic markers. One study analyzed patients with HCC before and one month after liver resection. The number of CTCs was a reliable diagnostic and prognostic marker, indicating the reoccurrence of the tumor (257). Another study using CellSearch™ criteria demonstrated the frequent presence of CTCs in patients with intermediate and advanced HCC (258). Further CellSearch™-based studies showed that the appearance of EpCAM-positive CTCs in HCC could be used to predict recurrence and is associated with poor prognosis (259, 260).
In one study, the nanofiltration technique CanPatrol™ was used to identify clusters of CTCs and white blood cells. Patients exhibiting those clusters show significantly shorter disease-free survival and overall survival (261). Another study using CanPatrol™ shows that a high percentage of mesenchymal CTCs are closely related to the expression of CK19, which is associated with a poor prognosis in HCC patients (262). Qi et al. use CanPatrol™ combined with an RNA-ISH assay to enrich and classify CTCs from patients with HCC. In this study, a slightly increased percentage (≥2%) of mesenchymal CTCs before resection was shown to be a predictive factor for early tumor recurrence (263).
Interestingly, other identification criteria based on less strict definitions also revealed good results for detecting HCC. One study reported that CTCs identified only based on their morphology were associated with shorter survival in HCC (264). In another study, erythrocytes and CD45+ immune cells were depleted from the pool of circulating cells. Afterward, the expression pattern of the leftover cells was monitored by polymerase chain reaction (PCR) using differential expression of EpCAM, CD90, CD133, and CK19 as identifiers for CTCs. Of note, this analysis revealed a sensitivity of 72.5% and a very high specificity of 95% to detect HCC in healthy donors and discriminate HCC from chronic HBV infection and benign hepatic lesions. Using AFP as a biomarker displayed a sensitivity of 57% and a specificity of 90%. Of clinical relevance, this approach also performed well in patients with early-stage HCC and showed a sensitivity of 71.8% and a specificity of 95%. Thus, this method could be a risk prediction and treatment surveillance tool enabling early decision-making to adjust effective antitumor strategies (265). Results of an interesting recent study showed that the detection of CTCs in patients who have undergone LTx allows early prediction of recurrence. Therefore, serial CTC detection may be helpful in the postoperative monitoring of HCC recurrence. However, it has to be considered that the study design is limited due to its small patient cohort, relatively short follow-up of the course, and its single-center design. Therefore, further clinical studies have to follow (243).
Conclusion: The major advantage of CTCs as biomarkers is that these cells contain all the information about the tumor. Furthermore, looking for markers of epithelial-mesenchymal transition determines metastasis formation and aggressiveness of a tumor. Therefore, the analysis of CTCs will advance our understanding of the biology of metastatic diseases and the development of treatment strategies. The required blood volumes are also practical and do not exceed 10ml. The disadvantage of CTCs as biomarkers is that purification and detection are complicated, time-consuming, and costly. The necessary technical equipment may not be available in all clinical settings. Thus, further improvement in the detection and isolation of CTCs is required to use them routinely as biomarkers in the clinic. On the one hand, PCR-based methods might address the challenges in detecting and isolating CTCs and lead to less time-consuming and cost-intensive investigations. On the other hand, such strategies entirely depend on the quality and definition of target transcripts for the detection and staging of the tumor. It is important to note that the control groups in most studies of CTCs were small. In addition, most studies were conducted in East Asia (China, Japan, and Taiwan). Thus, ethnic differences cannot be excluded. Most studies are in phase 2 of biomarker development (257, 261, 263, 265–268) (Table 1). Prospective studies are needed.
5 Final conclusion
HCC is one of the most common tumor diseases with rising incidence and high mortality. Management of HCC especially requires early diagnosis and therapy. Therefore, we need reliable, valid, and objective biomarkers for screening, diagnosis, disease monitoring, prognosis, predicting response to therapy, and treatment safety. Identification of novel non-invasive biomarkers for HCC has become the focus of research. There is an urgent need to define circulating markers that can replace invasive methods like liver biopsies and provide additional information about the tumor. These markers would enable more personalized medicine, including the prediction of therapeutic response. CTCs, ctDNA, circulating RNA, and exosomes are attractive candidates for liquid biopsy since they fulfill many essential characteristics of an ideal biomarker.
A big step towards ideal biomarkers is certainly the analysis of ctDNAs. ctDNA is stable and provides epigenetic and genetic data on the tumor. Detection of mutated DNA and methylation profiling is suitable for early detection of HCC and estimation of prognosis. However, NGS and methylation profiling is complicated and time-consuming. In addition, the different studies often identify completely different markers. It is difficult to understand why there are often no similarities between the markers identified in the studies. One reason is certainly a high heterogeneity between individual tumors. However, this cannot explain the lack of common markers in the studies. The differences must also be due to different study conditions. Here we need standardized and validated protocols for ctDNA purification and analysis. The quality of the samples is the key to a valid statement on the quality of ctDNA as a potential tumor marker.
miRNA and lncRNAs have shifted into the focus of cancer research. So far, more than 70 miRNAs have been proposed as potential biomarkers for HCC, and more and more lncRNA markers have been identified. Most remarkable results for the detection and prognosis of HCC were obtained using well-defined marker panels. These combinations of different RNAs showed very good results in sensitivity and specificity. Other combinations, e.g., with protein markers, are also possible. However, there are often entirely different marker panels, and the studies are not able to confirm the panels from other studies. Again, the quality of the samples plays a crucial role. We need standardized protocols to be used in studies worldwide. This is the only way to identify unique markers that can be used universally. This is true for HCC, but also all other tumors.
Proteins, including AFP, are the best-characterized biomarkers. They do not need elaborate purification and detection methods. Thus, they are optimal to be used in clinical routine. However, they often lack specificity. This disadvantage can be overcome using a combination of different markers or a panel of protein markers and other parameters. A very good example for the combination of several markers is the GALAD score. The score shows that combining three protein markers that can be easily determined in routine clinical practice (AFP, AFP-L3, and DCP) with patient metadata can significantly improve the predictive value. The combination of the three protein markers significantly increases prediction sensitivity but decreases specificity. However, adding simple patient metadata such as age and gender to the protein markers significantly improves both sensitivity and specificity. However, other combinations of tumor markers are also conceivable. The combination of protein biomarkers and nucleic acid markers also shows initial success and leads to improved sensitivity and specificity in prediction. Again, it must be said that more studies from Europe and North America are important to support the findings and reconcile ethnical differences.
Even though fewer data are currently available, analysis of exosomes could provide novel options to detect and understand the development of HCC. Exosomes contain unique functional information, e.g., about interactions between cancer cells and distant cells or the tumor microenvironment. Thus, they are important to gain more insights into tumor physiology. There is certainly a long way to go for exosomes as biomarkers until clinical application. However, they represent a tool with many advantages. Many macromolecules are already degraded in the body or during the purification process. In exosomes, they are protected. Thus, errors can be avoided, and differences due to purification can be prevented. This could help to solve the challenges described above for nucleic acid biomarkers.
CTCs are the carbon copy of the tumor itself. On the one hand, they can provide genetic information about mutations and epigenetic alterations of the tumor. Moreover, CTCs exhibit the transcriptome and proteome of the cancer of origin. Thus, they fulfill all criteria of an ultimate biomarker. On the other hand, they occur sparsely in blood, and there are no optimal/general surface markers for HCC-derived CTCs. Thus, purification and identification are very challenging and costly. Nevertheless, the blood volumes required for purification are practical (not exceeding 10ml). Regardless, new appropriate purification and analysis methods must be developed to routinely use these biomarkers in the clinic. Additionally, it is unfortunate to note that most studies on CTCs and other tumor biomarkers were conducted in East Asia. We need more studies in the Western industrialized nations since it is precisely here that an increase in HCC is apparent. This is primarily due to lifestyle and associated malnutrition.
We are on the right track in identifying new biomarkers, but we need more and better studies. Since HCC is a worldwide challenge, we need international studies to consider ethnical differences. To establish reliable universal markers, we need standardized and validated purification, storage, and processing protocols for the corresponding macromolecules or CTCs. This is the only way to identify and develop new good tumor markers.
After developing and validating novel biomarkers, the final step has to be their integration into the clinical routine. The novel liquid biopsy-based tools will not replace the established methods but will supplement them to optimize patient care. These markers are non-invasive or minimally invasive and, therefore, easier to implement, as only small volumes of biological material (e.g., blood) are required. As mentioned before, the challenge is certainly the purification and analysis of the samples. However, there are great technological advances that help us to overcome these obstacles. Therefore, we can look optimistically into the future and assume that we will have significantly more and better biomarkers for HCC in the near future.
Author contributions
Conceptualization: MM, SoS and KG. Investigation: SoS and KG. Original draft preparation: MM, KG and SoS. Review and editing: MM, KG, CK, SoS, AK, StS, DT, HT. Table preparation: SoS, KG, BV, KN, NE. Visualization: KG, SoS and DT. Supervision: MM. All authors contributed to the article and approved the submitted version.
Acknowledgments
Pictures have been created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wild CP, Weiderpass E, Stewart BW eds. World cancer report: Cancer research for cancer prevention. Lyon, France: International Agency for Research on Cance (2020). Available at: http://publications.iarc.fr/586.
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
3. Howell J, Pedrana A, Schroeder SE, Scott N, Aufegger L, Atun R, et al. A global investment framework for the elimination of hepatitis b. J Hepatol (2021) 74(3):535–49. doi: 10.1016/j.jhep.2020.09.013
4. Debes JD, Romagnoli PA, Prieto J, Arrese M, Mattos AZ, Boonstra A, et al. Serum biomarkers for the prediction of hepatocellular carcinoma. Cancers (Basel) (2021) 13(7):1681. doi: 10.3390/cancers13071681
5. Lobstein T, Brinsden H. Obesity: missing the 2025 global targets: Trends, costs and country reports. London: World Obesity Federation (2020). Available at: https://data.worldobesity.org/publications/WOF-Missing-the-2025-Global-Targets-Report-FINAL-WEB.pdf.
6. Food and Drug Administration (US). BEST (Biomarkers, EndpointS, and other tools) resource. Silver Spring (MD): Food and Drug Administration (US); Bethesda (MD): National Institutes of Health (US) (2016).
7. Colli A, Nadarevic T, Miletic D, Giljaca V, Fraquelli M, Štimac D, et al. Abdominal ultrasound and alpha-foetoprotein for the diagnosis of hepatocellular carcinoma in adults with chronic liver disease. Cochrane Database Syst Rev (2021) 4(4):CD013346. doi: 10.1002/14651858.CD013346.pub2
8. Spangenberg HC, Thimme R, Blum HE. Serum markers of hepatocellular carcinoma. Semin Liver Dis (2006) 26(4):385–90. doi: 10.1055/s-2006-951606
9. Suryatenggara J, Wibowo H, Atmodjo WL, Mathew G. Characterization of alpha-fetoprotein effects on dendritic cell and its function as effector immune response activator. JHC (2017) 4:139–51. doi: 10.2147/JHC.S139070
10. Janikashvili N, Bonnotte B, Katsanis E, Larmonier N. The dendritic cell-regulatory T lymphocyte crosstalk contributes to tumor-induced tolerance. Clin Dev Immunol (2011) 2011:430394. doi: 10.1155/2011/430394
11. Pardee AD, Shi J, Butterfield LH. Tumor-derived α-fetoprotein impairs the differentiation and T cell stimulatory activity of human dendritic cells. J.I (2014) 193(11):5723–32. doi: 10.4049/jimmunol.1400725
12. Alisa A, Boswell S, Pathan AA, Ayaru L, Williams R, Behboudi S. Human CD4(+) T cells recognize an epitope within alpha-fetoprotein sequence and develop into TGF-beta-producing CD4(+) T cells. J.I (2008) 180(7):5109–17. doi: 10.4049/jimmunol.180.7.5109
13. Saffroy R, Pham P, Reffas M, Takka M, Lemoine A, Debuire B. New perspectives and strategy research biomarkers for hepatocellular carcinoma. Clin Chem Lab Med (2007) 45(9):1169–79. doi: 10.1515/CCLM.2007.262
14. Lok AS, Lai CL. Alpha-fetoprotein monitoring in Chinese patients with chronic hepatitis b virus infection: role in the early detection of hepatocellular carcinoma. Hepatology (1989) 9(1):110–5. doi: 10.1002/hep.1840090119
15. Tao L-Y, Cai L, He X-D, Liu W, Qu Q. Comparison of serum tumor markers for intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Am Surg (2010) 76(11):1210–3. doi: 10.1177/000313481007601119
16. Trevisani F, D'Intino PE, Morselli-Labate AM, Mazzella G, Accogli E, Caraceni P, et al. Serum α-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: influence of HBsAg and anti-HCV status. J Hepatol (2001) 34(4):570–5. doi: 10.1016/S0168-8278(00)00053-2
17. Chen L-T, Martinelli E, Cheng A-L, Pentheroudakis G, Qin S, Bhattacharyya GS, et al. Pan-Asian adapted ESMO clinical practice guidelines for the management of patients with intermediate and advanced/relapsed hepatocellular carcinoma: a TOS-ESMO initiative endorsed by CSCO, ISMPO, JSMO, KSMO, MOS and SSO. Ann Oncol (2020) 31(3):334–51. doi: 10.1016/j.annonc.2019.12.001
18. Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology (2018) 67(1):358–80. doi: 10.1002/hep.29086
19. Galle PR, Forner A, Llovet JM, Mazzaferro V, Piscaglia F, Raoul J-L, et al. EASL clinical practice guidelines: Management of hepatocellular carcinoma. J Hepatol (2018) 69(1):182–236. doi: 10.1016/j.jhep.2018.03.019
20. Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: A meta-analysis. Gastroenterology (2018) 154(6):1706–1718.e1. doi: 10.1053/j.gastro.2018.01.064
21. Zhang J, Chen G, Zhang P, Zhang J, Li X, Gan D, et al. The threshold of alpha-fetoprotein (AFP) for the diagnosis of hepatocellular carcinoma: A systematic review and meta-analysis. PloS One (2020) 15(2):e0228857. doi: 10.1371/journal.pone.0228857
22. Johnson PJ, Pirrie SJ, Cox TF, Berhane S, Teng M, Palmer D, et al. The detection of hepatocellular carcinoma using a prospectively developed and validated model based on serological biomarkers. Cancer Epidemiol Biomarkers Prev (2014) 23(1):144–53. doi: 10.1158/1055-9965.EPI-13-0870
23. Li D, Mallory T, Satomura S. AFP-L3: A new generation of tumor marker for hepatocellular carcinoma. Clinica Chimica Acta (2001) 313(1-2):15–9. doi: 10.1016/S0009-8981(01)00644-1
24. Zhou J-M, Wang T, Zhang K-H. AFP-L3 for the diagnosis of early hepatocellular carcinoma: A meta-analysis. Med (Baltimore) (2021) 100(43):e27673. doi: 10.1097/MD.0000000000027673
25. Nakamura S, Nouso K, Sakaguchi K, Ito YM, Ohashi Y, Kobayashi Y, et al. Sensitivity and specificity of des-gamma-carboxy prothrombin for diagnosis of patients with hepatocellular carcinomas varies according to tumor size. Am J Gastroenterol (2006) 101(9):2038–43. doi: 10.1111/j.1572-0241.2006.00681.x
26. Nagaoka S, Yatsuhashi H, Hamada H, Yano K, Matsumoto T, Daikoku M, et al. The des-gamma-carboxy prothrombin index is a new prognostic indicator for hepatocellular carcinoma. Cancer (2003) 98(12):2671–7. doi: 10.1002/cncr.11839
27. Song P, Feng X, Zhang K, Song T, Ma K, Kokudo N, et al. Screening for and surveillance of high-risk patients with HBV-related chronic liver disease: promoting the early detection of hepatocellular carcinoma in China. Biosci Trends (2013) 7(1):1–6. doi: 10.5582/bst.2013.v7.1.1
28. DiMarino A J. Usefulness of highly sensitive AFP-L3 and DCP in surveillance for hepatocellular carcinoma in patients with a normal alpha-fetoprotein. J Med Microb Diagn (2014) 03(01):1–6. doi: 10.4172/2161-0703.1000130
29. Tayob N, Kanwal F, Alsarraj A, Hernaez R, El-Serag HB. The performance of AFP, AFP-3, DCP as biomarkers for detection of hepatocellular carcinoma (HCC): A phase 3 biomarker study in the united states. Clin Gastroenterol Hepatol (2022) 3:S1542-3565(22)00106-9. doi: 10.1016/j.cgh.2022.01.047
30. Best J, Bilgi H, Heider D, Schotten C, Manka P, Bedreli S, et al. Der GALAD-score, ein AFP-, AFP-L3- und DCP-basierter diagnosealgorithmus verbessert die detektionsrate des hepatozellulären karzinoms im BCLC-frühstadium signifikant. Z Gastroenterol (2016) 54(12):1296–305. doi: 10.1055/s-0042-119529
31. Liu M, Wu R, Liu X, Xu H, Chi X, Wang X, et al. Validation of the GALAD model and establishment of GAAP model for diagnosis of hepatocellular carcinoma in Chinese patients. J Hepatocell Carcinoma (2020) 7:219–32. doi: 10.2147/JHC.S271790
32. Best J, Bechmann LP, Sowa J-P, Sydor S, Dechêne A, Pflanz K, et al. GALAD score detects early hepatocellular carcinoma in an international cohort of patients with nonalcoholic steatohepatitis. Clin Gastroenterol Hepatol (2020) 18(3):728–735.e4. doi: 10.1016/j.cgh.2019.11.012
33. Huang C, Fang M, Xiao X, Wang H, Gao Z, Ji J, et al. Validation of the GALAD model for early diagnosis and monitoring of hepatocellular carcinoma in Chinese multicenter study. Liver Int (2022) 42(1):210–23. doi: 10.1111/liv.15082
34. Chuma M, Sakamoto M, Yamazaki K, Ohta T, Ohki M, Asaka M, et al. Expression profiling in multistage hepatocarcinogenesis: identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology (2003) 37(1):198–207. doi: 10.1053/jhep.2003.50022
35. Wang B, Lan T, Xiao H, Chen Z-H, Wei C, Chen L-F, et al. The expression profiles and prognostic values of HSP70s in hepatocellular carcinoma. Cancer Cell Int (2021) 21(1):286. doi: 10.1186/s12935-021-01987-9
36. Gehrmann M, Cervello M, Montalto G, Cappello F, Gulino A, Knape C, et al. Heat shock protein 70 serum levels differ significantly in patients with chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Front Immunol (2014) 5:307. doi: 10.3389/fimmu.2014.00307
37. Sung YK, Hwang SY, Park MK, Farooq M, Han IS, Bae HI, et al. Glypican-3 is overexpressed in human hepatocellular carcinoma. Cancer Sci (2003) 94(3):259–62. doi: 10.1111/j.1349-7006.2003.tb01430.x
38. Hippo Y, Watanabe K, Watanabe A, Midorikawa Y, Yamamoto S, Ihara S, et al. Identification of soluble NH2-terminal fragment of glypican-3 as a serological marker for early-stage hepatocellular carcinoma. Cancer Res (2004) 64(7):2418–23. doi: 10.1158/0008-5472.CAN-03-2191
39. Xu D, Su C, Sun L, Gao Y, Li Y. Performance of serum glypican 3 in diagnosis of hepatocellular carcinoma: A meta-analysis. Ann Hepatol (2019) 18(1):58–67. doi: 10.5604/01.3001.0012.7863
40. Christa L, Simon M-T, Flinois J-P, Gebhardt R, Brechot C, Lasserre C. Overexpression of glutamine synthetase in human primary liver cancer. Gastroenterology (1994) 106(5):1312–20. doi: 10.1016/0016-5085(94)90024-8
41. Osada T, Sakamoto M, Nagawa H, Yamamoto J, Matsuno Y, Iwamatsu A, et al. Acquisition of glutamine synthetase expression in human hepatocarcinogenesis. Cancer (1999) 85(4):819–31. doi: 10.1002/(SICI)1097-0142(19990215)85:4<819::AID-CNCR9>3.0.CO;2-E
42. Di Tommaso L, Franchi G, Park YN, Fiamengo B, Destro A, Morenghi E, et al. Diagnostic value of HSP70, glypican 3, and glutamine synthetase in hepatocellular nodules in cirrhosis. Hepatology (2007) 45(3):725–34. doi: 10.1002/hep.21531
43. Tremosini S, Forner A, Boix L, Vilana R, Bianchi L, Reig M, et al. Prospective validation of an immunohistochemical panel (glypican 3, heat shock protein 70 and glutamine synthetase) in liver biopsies for diagnosis of very early hepatocellular carcinoma. Gut (2012) 61(10):1481–7. doi: 10.1136/gutjnl-2011-301862
44. Voesch S, Bitzer M, Blödt S, Follmann M, Freudenberger P, Langer T, et al. S3-leitlinie: Diagnostik und therapie des hepatozellulären karzinoms und biliärer karzinome – version 2.0 – juni 2021, AWMF-registernummer: 032-053OL. Z Gastroenterol (2022) 60(1):e131–85. doi: 10.1055/a-1589-7585
45. Hameed B, Mehta N, Sapisochin G, Roberts JP, Yao FY. Alpha-fetoprotein level 1000 ng/mL as an exclusion criterion for liver transplantation in patients with hepatocellular carcinoma meeting the Milan criteria. Liver Transpl (2014) 20(8):945–51. doi: 10.1002/lt.23904
46. Berry K, Ioannou GN. Serum alpha-fetoprotein level independently predicts posttransplant survival in patients with hepatocellular carcinoma. Liver Transpl (2013) 19(6):634–45. doi: 10.1002/lt.23652
47. Zhu AX, Kang Y-K, Yen C-J, Finn RS, Galle PR, Llovet JM, et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol (2019) 20(2):282–96. doi: 10.1016/S1470-2045(18)30937-9
48. Bruix J, Cheng A-L, Meinhardt G, Nakajima K, de SY, Llovet J. Prognostic factors and predictors of sorafenib benefit in patients with hepatocellular carcinoma: Analysis of two phase III studies. J Hepatol (2017) 67(5):999–1008. doi: 10.1016/j.jhep.2017.06.026
49. He C, Peng W, Liu X, Li C, Li X, Wen T-F. Post-treatment alpha-fetoprotein response predicts prognosis of patients with hepatocellular carcinoma: A meta-analysis. Med (Baltimore) (2019) 98(31):e16557. doi: 10.1097/MD.0000000000016557
50. Imamura H, Matsuyama Y, Tanaka E, Ohkubo T, Hasegawa K, Miyagawa S, et al. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol (2003) 38(2):200–7. doi: 10.1016/S0168-8278(02)00360-4
51. MANDEL P, METAIS P. Les Acides nucléiques du plasma sanguin chez l'homme. C R Seances Soc Biol Fil (1948) 142(3-4):241–3.
52. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res (1977) 37(3):646–50.
53. Thierry AR, Mouliere F, Gongora C, Ollier J, Robert B, Ychou M, et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res (2010) 38(18):6159–75. doi: 10.1093/nar/gkq421
54. Labgaa I, Villanueva A, Dormond O, Demartines N, Melloul E. The role of liquid biopsy in hepatocellular carcinoma prognostication. Cancers (Basel) (2021) 13(4):1–17. doi: 10.3390/cancers13040659
55. Iizuka N, Sakaida I, Moribe T, Fujita N, Miura T, Stark M, et al. Elevated levels of circulating cell-free DNA in the blood of patients with hepatitis c virus-associated hepatocellular carcinoma. Anticancer Res (2006) 26(6C):4713–9.
56. Yan L, Chen Y, Zhou J, Zhao H, Zhang H, Wang G. Diagnostic value of circulating cell-free DNA levels for hepatocellular carcinoma. Int J Infect Dis (2018) 67:92–7. doi: 10.1016/j.ijid.2017.12.002
57. Zhang X, Wang Z, Tang W, Wang X, Liu R, Bao H, et al. Ultrasensitive and affordable assay for early detection of primary liver cancer using plasma cell-free DNA fragmentomics. Hepatology (2021) 76(2):317–29. doi: 10.1002/hep.32308
58. Benesova L, Belsanova B, Suchanek S, Kopeckova M, Minarikova P, Lipska L, et al. Mutation-based detection and monitoring of cell-free tumor DNA in peripheral blood of cancer patients. Anal Biochem (2013) 433(2):227–34. doi: 10.1016/j.ab.2012.06.018
59. Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet (2012) 44(6):694–8. doi: 10.1038/ng.2256
60. Yu L, Liu X, Han C, Lu S, Zhu G, Su H, et al. XRCC1 rs25487 genetic variant and TP53 mutation at codon 249 predict clinical outcomes of hepatitis b virus-related hepatocellular carcinoma after hepatectomy: A cohort study for 10 years' follow up. Hepatol Res (2016) 46(8):765–74. doi: 10.1111/hepr.12611
61. Mathai RA, Vidya RVS, Reddy BS, Thomas L, Udupa K, Kolesar J, et al. Potential utility of liquid biopsy as a diagnostic and prognostic tool for the assessment of solid tumors: Implications in the precision oncology. J Clin Med (2019) 8(3):1–17. doi: 10.3390/jcm8030373
62. Howell JA, Sharma R. The clinical role of 'liquid biopsy' in hepatocellular carcinoma. Hepat Oncol (2016) 3(1):45–55. doi: 10.2217/hep.15.38
63. Jiao J, Watt GP, Stevenson HL, Calderone TL, Fisher-Hoch SP, Ye Y, et al. Telomerase reverse transcriptase mutations in plasma DNA in patients with hepatocellular carcinoma or cirrhosis: Prevalence and risk factors. Hepatol Commun (2018) 2(6):718–31. doi: 10.1002/hep4.1187
64. Oversoe SK, Clement MS, Pedersen MH, Weber B, Aagaard NK, Villadsen GE, et al. TERT promoter mutated circulating tumor DNA as a biomarker for prognosis in hepatocellular carcinoma. Scand J Gastroenterol (2020) 55(12):1433–40. doi: 10.1080/00365521.2020.1837928
65. Hirai M, Kinugasa H, Nouso K, Yamamoto S, Terasawa H, Onishi Y, et al. Prediction of the prognosis of advanced hepatocellular carcinoma by TERT promoter mutations in circulating tumor DNA. J Gastroenterol Hepatol (2021) 36(4):1118–25. doi: 10.1111/jgh.15227
66. Shen T, Li S-F, Wang J-L, Zhang T, Zhang S, Chen H-T, et al. TP53 R249S mutation detected in circulating tumour DNA is associated with prognosis of hepatocellular carcinoma patients with or without hepatectomy. Liver Int (2020) 40(11):2834–47. doi: 10.1111/liv.14581
67. Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res (2013) 23(9):1422–33. doi: 10.1101/gr.154492.113
68. Xiong Y, Xie C-R, Zhang S, Chen J, Yin Z-Y. Detection of a novel panel of somatic mutations in plasma cell-free DNA and its diagnostic value in hepatocellular carcinoma. Cancer Manag Res (2019) 11:5745–56. doi: 10.2147/CMAR.S197455
69. Howell J, Atkinson SR, Pinato DJ, Knapp S, Ward C, Minisini R, et al. Identification of mutations in circulating cell-free tumour DNA as a biomarker in hepatocellular carcinoma. Eur J Cancer (2019) 116:56–66. doi: 10.1016/j.ejca.2019.04.014
70. Jiang P, Sun K, Tong YK, Cheng SH, Cheng THT, Heung MMS, et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc Natl Acad Sci (2018) 115(46):E10925–33. doi: 10.1073/pnas.1814616115
71. Lin SY, Dhillon V, Jain S, Chang T-T, Hu C-T, Lin Y-J, et al. A locked nucleic acid clamp-mediated PCR assay for detection of a p53 codon 249 hotspot mutation in urine. J Mol Diagn (2011) 13(5):474–84. doi: 10.1016/j.jmoldx.2011.05.005
72. Ono A, Fujimoto A, Yamamoto Y, Akamatsu S, Hiraga N, Imamura M, et al. Circulating tumor DNA analysis for liver cancers and its usefulness as a liquid biopsy. Cell Mol Gastroenterol Hepatol (2015) 1(5):516–34. doi: 10.1016/j.jcmgh.2015.06.009
73. Liao W, Yang H, Xu H, Wang Y, Ge P, Ren J, et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget (2016) 7(26):40481–90. doi: 10.18632/oncotarget.9629
74. Hann H-W, Jain S, Park G, Steffen JD, Song W, Su Y-H. Detection of urine DNA markers for monitoring recurrent hepatocellular carcinoma. Hepatoma Res (2017) 3:105–11. doi: 10.20517/2394-5079.2017.15
75. Kim SS, Eun JW, Choi J-H, Woo HG, Cho HJ, Ahn HR, et al. MLH1 single-nucleotide variant in circulating tumor DNA predicts overall survival of patients with hepatocellular carcinoma. Sci Rep (2020) 10(1):17862. doi: 10.1038/s41598-020-74494-y
76. Felden Jv, Craig AJ, Garcia-Lezana T, Labgaa I, Haber PK, D'Avola D, et al. Mutations in circulating tumor DNA predict primary resistance to systemic therapies in advanced hepatocellular carcinoma. Oncogene (2021) 40(1):140–51. doi: 10.1038/s41388-020-01519-1
77. Cai Z, Chen G, Zeng Y, Dong X, Li Z, Huang Y, et al. Comprehensive liquid profiling of circulating tumor DNA and protein biomarkers in long-term follow-up patients with hepatocellular carcinoma. Clin Cancer Res (2019) 25(17):5284–94. doi: 10.1158/1078-0432.CCR-18-3477
78. Oh CR, Kong S-Y, Im H-S, Kim HJ, Kim MK, Yoon K-A, et al. Genome-wide copy number alteration and VEGFA amplification of circulating cell-free DNA as a biomarker in advanced hepatocellular carcinoma patients treated with sorafenib. BMC Cancer (2019) 19(1):292. doi: 10.1186/s12885-019-5483-x
79. Chan KCA, Jiang P, Zheng YWL, Liao GJW, Sun H, Wong J, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem (2013) 59(1):211–24. doi: 10.1373/clinchem.2012.196014
80. Jiang P, Chan CWM, Chan KCA, Cheng SH, Wong J, Wong VW-S, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci USA (2015) 112(11):E1317–25. doi: 10.1073/pnas.1500076112
81. Xu R-H, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater (2017) 16(11):1155–61. doi: 10.1038/nmat4997
82. Cai J, Chen L, Zhang Z, Zhang X, Lu X, Liu W, et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut (2019) 68(12):2195–205. doi: 10.1136/gutjnl-2019-318882
83. Chan KCA, Jiang P, Chan CWM, Sun K, Wong J, Hui EP, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci (2013) 110(47):18761–8. doi: 10.1073/pnas.1313995110
84. Kisiel JB, Dukek BA VSR, Kanipakam R, Ghoz HM, Yab TC, Berger CK, et al. Hepatocellular carcinoma detection by plasma methylated DNA: Discovery, phase I pilot, and phase II clinical validation. Hepatology (2019) 69(3):1180–92. doi: 10.1002/hep.30244
85. Lu C-Y, Chen S-Y, Peng H-L, Kan P-Y, Chang W-C, Yen C-J. Cell-free methylation markers with diagnostic and prognostic potential in hepatocellular carcinoma. Oncotarget (2017) 8(4):6406–18. doi: 10.18632/oncotarget.14115
86. Song T, Li L, Wu S, Liu Y, Guo C, Wang W, et al. Peripheral blood genetic biomarkers for the early diagnosis of hepatocellular carcinoma. Front Oncol (2021) 11:583714. doi: 10.3389/fonc.2021.583714
87. Hernandez-Meza G, Felden Jv, Gonzalez-Kozlova EE, Garcia-Lezana T, Peix J, Portela A, et al. DNA Methylation profiling of human hepatocarcinogenesis. Hepatology (2021) 74(1):183–99. doi: 10.1002/hep.31659
88. Villanueva A, Portela A, Sayols S, Battiston C, Hoshida Y, Méndez-González J, et al. DNA Methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology (2015) 61(6):1945–56. doi: 10.1002/hep.27732
89. Zhang C, Huang C, Sui X, Zhong X, Yang W, Hu X, et al. Association between gene methylation and HBV infection in hepatocellular carcinoma: A meta-analysis. J Cancer (2019) 10(25):6457–65. doi: 10.7150/jca.33005
90. Banini BA, Sanyal AJ. The use of cell free DNA in the diagnosis of HCC. Hepatoma Res (2019) 5. doi: 10.20517/2394-5079.2019.30
91. Yeh C-C, Goyal A, Shen J, Wu H-C, Strauss JA, Wang Q, et al. Global level of plasma DNA methylation is associated with overall survival in patients with hepatocellular carcinoma. Ann Surg Oncol (2017) 24(12):3788–95. doi: 10.1245/s10434-017-5913-4
92. Li F, Qiao C-Y, Gao S, Fan Y-C, Chen L-Y, Wang K. Circulating cell-free DNA of methylated insulin-like growth factor-binding protein 7 predicts a poor prognosis in hepatitis b virus-associated hepatocellular carcinoma after hepatectomy. Free Radic Res (2018) 52(4):455–64. doi: 10.1080/10715762.2018.1443448
93. Chen MM, Zhao RC, Chen KF, Huang Y, Liu ZJ, Wei YG, et al. Hypomethylation of CTCFL promoters as a noninvasive biomarker in plasma from patients with hepatocellular carcinoma. Neoplasma (2020) 67(4):909–15. doi: 10.4149/neo_2020_190819N789
94. Lee HW, Kim E, Cho KJ, Park HJ, Seo J, Lee H, et al. Applications of molecular barcode sequencing for the detection of low-frequency variants in circulating tumour DNA from hepatocellular carcinoma. Liver Int (2022) 42(10):2317–26. doi: 10.1111/liv.15356
95. Oussalah A, Rischer S, Bensenane M, Conroy G, Filhine-Tresarrieu P, Debard R, et al. Plasma mSEPT9: A novel circulating cell-free DNA-based epigenetic biomarker to diagnose hepatocellular carcinoma. EBioMedicine (2018) 30:138–47. doi: 10.1016/j.ebiom.2018.03.029
96. Zhang Z, Chen P, Xie H, Cao P. Using circulating tumor DNA as a novel biomarker to screen and diagnose hepatocellular carcinoma: A systematic review and meta-analysis. Cancer Med (2020) 9(4):1349–64. doi: 10.1002/cam4.2799
97. Chen VL, Xu D, Wicha MS, Lok AS, Parikh ND. Utility of liquid biopsy analysis in detection of hepatocellular carcinoma, determination of prognosis, and disease monitoring: A systematic review. Clin Gastroenterol Hepatol (2020) 18(13):2879–2902.e9. doi: 10.1016/j.cgh.2020.04.019
98. Nie G, Peng D, Li B, Lu J, Cai Y, Xiong X, et al. Diagnostic accuracy of Serum/Plasma circular RNAs and the combination of circular RNAs and α-fetoprotein for detecting hepatocellular carcinoma: A meta-analysis. Front Genet (2021) 12:722208. doi: 10.3389/fgene.2021.722208
99. Wang M, Yang Y, Xu J, Bai W, Ren X, Wu H. CircRNAs as biomarkers of cancer: a meta-analysis. BMC Cancer (2018) 18(1):303. doi: 10.1186/s12885-018-4213-0
100. Huang X, Zhang W, Shao Z. Prognostic and diagnostic significance of circRNAs expression in hepatocellular carcinoma patients: A meta-analysis. Cancer Med (2019) 8(3):1148–56. doi: 10.1002/cam4.1939
101. Hao Q, Han Y, Xia W, Wang Q, Qian H. Systematic review and meta-analysis of the utility of circular RNAs as biomarkers of hepatocellular carcinoma. Can J Gastroenterol Hepatol (2019) 2019:1684039. doi: 10.1155/2019/1684039
102. Lu F, Zhao X, Zhang Z, Xiong M, Wang Y, Sun Y, et al. The diagnostic and prognostic value of the miR-17-92 cluster in hepatocellular carcinoma: A meta-analysis. Front Genet (2022) 13:927079. doi: 10.3389/fgene.2022.927079
103. Huang Y, Chen Y, Tu S, Zhang J, Qiu Y, Yu W. Diagnostic accuracy of circulating microRNAs for hepatitis c virus-associated hepatocellular carcinoma: a systematic review and meta-analysis. BMC Infect Dis (2022) 22(1):323. doi: 10.1186/s12879-022-07292-8
104. Zhang W-T, Gil-Gómez A, Liu C-H, Gao S-S, Romero-Gómez M. Diagnostic accuracy of circulating microRNA in hepatitis b virus-related hepatocellular carcinoma: a meta-analysis based on Asian data. Rev Esp Enferm Dig (2022) 114(5):280–8. doi: 10.17235/reed.2021.8139/2021
105. Zhang H, Ding R, Chen D. Value of miR-21 levels as potential biomarkers in the early diagnosis of hepatocellular carcinoma:a meta-analysis. Biomarkers (2021) 26(7):586–97. doi: 10.1080/1354750X.2021.1955976
106. Zeng C, Tang Y, Jiang Y, Zuo Z, Tao H. Long noncoding RNAs as biomarkers for the diagnosis of hepatocellular carcinoma: A meta-analysis. Pathol Res Pract (2021) 224:153546. doi: 10.1016/j.prp.2021.153546
107. Chen S, Zhang Y, Wu X, Zhang C, Li G. Diagnostic value of lncRNAs as biomarker in hepatocellular carcinoma: An updated meta-analysis. Can J Gastroenterol Hepatol (2018) 2018:8410195. doi: 10.1155/2018/8410195
108. Hao Q-Q, Chen G-Y, Zhang J-H, Sheng J-H, Gao Y. Diagnostic value of long noncoding RNAs for hepatocellular carcinoma: A PRISMA-compliant meta-analysis. Med (Baltimore) (2017) 96(28):e7496. doi: 10.1097/MD.0000000000007496
109. Cui K, Ou Y, Shen Y, Li S, Sun Z. Clinical value of circulating tumor cells for the diagnosis and prognosis of hepatocellular carcinoma (HCC): A systematic review and meta-analysis. Med (Baltimore) (2020) 99(40):e22242. doi: 10.1097/MD.0000000000022242
110. Sun C, Liao W, Deng Z, Li E, Feng Q, Lei J, et al. The diagnostic value of assays for circulating tumor cells in hepatocellular carcinoma: A meta-analysis. Med (Baltimore) (2017) 96(29):e7513. doi: 10.1097/MD.0000000000007513
111. Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep (2016) 6(1):37982. doi: 10.1038/srep37982
112. Wan L, Zhang L, Fan K, Cheng Z-X, Sun Q-C, Wang J-J. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the wnt/β-catenin pathway. BioMed Res Int (2016) 2016:1579490. doi: 10.1155/2016/1579490
113. Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta (2017) 466:167–71. doi: 10.1016/j.cca.2017.01.025
114. Zhao J, Li L, Wang Q, Han H, Zhan Q, Xu M. CircRNA expression profile in early-stage lung adenocarcinoma patients. Cell Physiol Biochem (2017) 44(6):2138–46. doi: 10.1159/000485953
115. Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Med (Baltimore) (2016) 95(22):e3811. doi: 10.1097/MD.0000000000003811
116. Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform (2017) 18(6):984–92. doi: 10.1093/bib/bbw081
117. Wang W, Li Y, Li X, Liu B, Han S, Li X, et al. Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. BioMed Pharmacother (2020) 121:109517. doi: 10.1016/j.biopha.2019.109517
118. Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett (2020) 475:119–28. doi: 10.1016/j.canlet.2020.01.022
119. Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X, et al. A noncoding regulatory RNAs network driven by circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology (2020) 71(1):130–47. doi: 10.1002/hep.30795
120. Guan Z, Tan J, Gao W, Li X, Yang Y, Li X, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J Cell Physiol (2018) 234(1):500–8. doi: 10.1002/jcp.26612
121. Yu J, Ding W-B, Wang M-C, Guo X-G, Xu J, Xu Q-G, et al. Plasma circular RNA panel to diagnose hepatitis b virus-related hepatocellular carcinoma: A large-scale, multicenter study. Int J Cancer (2020) 146(6):1754–63. doi: 10.1002/ijc.32647
122. Jiang Z, Shen L, Wang S, Wu S, Hu Y, Guo J, et al. Hsa_circ_0028502 and hsa_circ_0076251 are potential novel biomarkers for hepatocellular carcinoma. Cancer Med (2019) 8(17):7278–87. doi: 10.1002/cam4.2584
123. Zhu K, Zhan H, Peng Y, Yang L, Gao Q, Jia H, et al. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis b virus-related hepatocellular carcinoma. Carcinogenesis (2020) 41(3):296–302. doi: 10.1093/carcin/bgz154
124. Matboli M, Shafei AE, Ali MA, Ashry AM, Kamal KM, Agag MA, et al. circRNAs (hsa_circ_00156, hsa_circ _000224, and hsa_circ _000520) are novel potential biomarkers in hepatocellular carcinoma. J Cell Biochem (2018) 120(5):7711–24. doi: 10.1002/jcb.28045
125. Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen Y, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis (2018) 9(11):1091. doi: 10.1038/s41419-018-1132-6
126. Gong Y, Mao J, Wu D, Wang X, Li L, Zhu L, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int (2018) 18:116. doi: 10.1186/s12935-018-0602-3
127. Pan H, Tang L, Jiang H, Li X, Wang R, Gao J, et al. Enhanced expression of circ_0000267 in hepatocellular carcinoma indicates poor prognosis and facilitates cell progression by sponging miR-646. J Cell Biochem (2019) 120(7):11350–7. doi: 10.1002/jcb.28411
128. Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol (2017) 11(4):422–37. doi: 10.1002/1878-0261.12045
129. Zhu Y-J, Zheng B, Luo G-J, Ma X-K, Lu X-Y, Lin X-M, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics (2019) 9(12):3526–40. doi: 10.7150/thno.32796
130. Yu J, Xu Q-G, Wang Z-G, Yang Y, Zhang L, Ma J-Z, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol (2018) 68(6):1214–27. doi: 10.1016/j.jhep.2018.01.012
131. Qiao G-L, Chen L, Jiang W-H, Yang C, Yang C-M, Song L-N, et al. Hsa_circ_0003998 may be used as a new biomarker for the diagnosis and prognosis of hepatocellular carcinoma. OTT (2019) 12:5849–60. doi: 10.2147/OTT.S210363
132. Chen D, Zhang C, Lin J, Song X, Wang H. Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag Res (2018) 10:1275–83. doi: 10.2147/CMAR.S166740
133. Zhang P-F, Wei C-Y, Huang X-Y, Peng R, Yang X, Lu J-C, et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer (2019) 18(1):105. doi: 10.1158/1535-7163.TARG-19-LB-C10
134. Li Z, Zhou Y, Yang G, He S, Qiu X, Zhang L, et al. Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin Chim Acta (2019) 492:37–44. doi: 10.1016/j.cca.2019.02.001
135. Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large b-cell lymphoma. Br J Haematol (2008) 141(5):672–5. doi: 10.1111/j.1365-2141.2008.07077.x
136. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res (2008) 18(10):997–1006. doi: 10.1038/cr.2008.282
137. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci (2008) 105(30):10513–8. doi: 10.1073/pnas.0804549105
138. Bertoli G, Cava C, Castiglioni I. MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics (2015) 5(10):1122–43. doi: 10.7150/thno.11543
139. Casanova-Salas I, Rubio-Briones J, Calatrava A, Mancarella C, Masiá E, Casanova J, et al. Identification of miR-187 and miR-182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomy. J Urol (2014) 192(1):252–9. doi: 10.1016/j.juro.2014.01.107
140. Li J, Yang H, Li Y, Liu Y, Chen S, Qi C, et al. microRNA-146 up-regulation predicts the prognosis of non-small cell lung cancer by miRNA in situ hybridization. Exp Mol Pathol (2014) 96(2):195–9. doi: 10.1016/j.yexmp.2013.11.004
141. Xu J, An P, Winkler CA, Yu Y. Dysregulated microRNAs in hepatitis b virus-related hepatocellular carcinoma: Potential as biomarkers and therapeutic targets. Front Oncol (2020) 10:1271. doi: 10.3389/fonc.2020.01271
142. Mocan T, Simão AL, Castro RE, Rodrigues CMP, Słomka A, Wang B, et al. Liquid biopsies in hepatocellular carcinoma: Are we winning? J Clin Med (2020) 9(5):1541. doi: 10.3390/jcm9051541
143. Cho HJ, Eun JW, Baek GO, Seo CW, Ahn HR, Kim SS, et al. Serum exosomal MicroRNA, miR-10b-5p, as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. J Clin Med (2020) 9(1):1–16. doi: 10.3390/jcm9010281
144. Liu AM, Yao T-J, Wang W, Wong K-F, Lee NP, Fan ST, et al. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open (2012) 2(2):e000825. doi: 10.1136/bmjopen-2012-000825
145. Li L, Guo Z, Wang J, Mao Y, Gao Q. Serum miR-18a: a potential marker for hepatitis b virus-related hepatocellular carcinoma screening. Dig Dis Sci (2012) 57(11):2910–6. doi: 10.1007/s10620-012-2317-y
146. Rashad NM, El-Shal AS, Shalaby SM, Mohamed SY. Serum miRNA-27a and miRNA-18b as potential predictive biomarkers of hepatitis c virus-associated hepatocellular carcinoma. Mol Cell Biochem (2018) 447(1-2):125–36. doi: 10.1007/s11010-018-3298-8
147. Qu J, Yang J, Chen M, Cui L, Wang T, Gao W, et al. MicroRNA-21 as a diagnostic marker for hepatocellular carcinoma: A systematic review and meta-analysis. Pak J Med Sci (2019) 35(5):1466–71. doi: 10.12669/pjms.35.5.685
148. Zhuang C, Jiang W, Huang D, Xu L, Yang Q, Zheng L, et al. Serum miR-21, miR-26a and miR-101 as potential biomarkers of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol (2016) 40(4):386–96. doi: 10.1016/j.clinre.2015.11.002
149. El Mahdy HA, Abdelhamid IA, Amen AI, Abdelsameea E, Hassouna MM. MicroRNA-215 as a diagnostic marker in Egyptian patients with hepatocellular carcinoma. Asian Pac J Cancer Prev (2019) 20(9):2723–31. doi: 10.31557/APJCP.2019.20.9.2723
150. Amr KS, Elmawgoud Atia HA, Elazeem Elbnhawy RA, Ezzat WM. Early diagnostic evaluation of miR-122 and miR-224 as biomarkers for hepatocellular carcinoma. Genes Dis (2017) 4(4):215–21. doi: 10.1016/j.gendis.2017.10.003
151. Liu M, Wang L, Zhu H, Rong W, Wu F, Liang S, et al. A preoperative measurement of serum MicroRNA-125b may predict the presence of microvascular invasion in hepatocellular carcinomas patients. Trans Oncol (2016) 9(3):167–72. doi: 10.1016/j.tranon.2016.03.002
152. Chen S, Chen H, Gao S, Qiu S, Zhou H, Yu M, et al. Differential expression of plasma microRNA-125b in hepatitis b virus-related liver diseases and diagnostic potential for hepatitis b virus-induced hepatocellular carcinoma. Hepatol Res (2017) 47(4):312–20. doi: 10.1111/hepr.12739
153. Shaker OG, Abdelwahed MY, Ahmed NA, Hassan EA, Ahmed TI, Abousarie MA, et al. Evaluation of serum long noncoding RNA NEAT and MiR-129-5p in hepatocellular carcinoma. IUBMB Life (2019) 71(10):1571–8. doi: 10.1002/iub.2096
154. Zhang W, Fu T, Guo Z, Zhang Y, Zhang L, Su H, et al. Serum miR-375 levels are closely related to disease progression from HBV infection to HBV-related hepatocellular carcinoma. BioMed Res Int (2020) 2020:5819385. doi: 10.1155/2020/5819385
155. Wen Y, Han J, Chen J, Dong J, Xia Y, Liu J, et al. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer (2015) 137(7):1679–90. doi: 10.1002/ijc.29544
156. Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis b virus-related hepatocellular carcinoma. JCO (2011) 29(36):4781–8. doi: 10.1200/JCO.2011.38.2697
157. Lin X-J, Chong Y, Guo Z-W, Xie C, Yang X-J, Zhang Q, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol (2015) 16(7):804–15. doi: 10.1016/S1470-2045(15)00048-0
158. Cho HJ, Kim SS, Nam JS, Kim JK, Lee JH, Kim B, et al. Low levels of circulating microRNA-26a/29a as poor prognostic markers in patients with hepatocellular carcinoma who underwent curative treatment. Clin Res Hepatol Gastroenterol (2017) 41(2):181–9. doi: 10.1016/j.clinre.2016.09.011
159. Yerukala Sathipati S, Ho S-Y. Novel miRNA signature for predicting the stage of hepatocellular carcinoma. Sci Rep (2020) 10(1):14452. doi: 10.1038/s41598-020-71324-z
160. Li J, Jin B, Wang T, Li W, Wang Z, Zhang H, et al. Serum microRNA expression profiling identifies serum biomarkers for HCV-related hepatocellular carcinoma. CBM (2019) 26(4):501–12. doi: 10.3233/CBM-181970
161. Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. a comprehensive review. EMBO Mol Med (2012) 4(3):143–59. doi: 10.1002/emmm.201100209
162. Huang Y-H, Liang K-H, Chien R-N, Hu T-H, Lin K-H, Hsu C-W, et al. A circulating MicroRNA signature capable of assessing the risk of hepatocellular carcinoma in cirrhotic patients. Sci Rep (2017) 7(1):523. doi: 10.1038/s41598-017-00631-9
163. Shi T, Gao G, Cao Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers (2016) 2016:9085195. doi: 10.1155/2016/9085195
164. Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res (2013) 33(8):3185–93.
165. Kung JTY, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics (2013) 193(3):651–69. doi: 10.1534/genetics.112.146704
166. Beermann J, Piccoli M-T, Viereck J, Thum T. Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol Rev (2016) 96(4):1297–325. doi: 10.1152/physrev.00041.2015
167. Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol (2011) 21(6):354–61. doi: 10.1016/j.tcb.2011.04.001
168. Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: a new player in cancer. J Hematol Oncol (2013) 6(1):37. doi: 10.1186/1756-8722-6-37
169. Qiu M-T, Hu J-W, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumor Biol (2013) 34(2):613–20. doi: 10.1007/s13277-013-0658-6
170. Chen Z, Huang L, Wu Y, Zhai W, Zhu P, Gao Y. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat Commun (2016) 7(1):12598. doi: 10.1038/ncomms12598
171. Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. BioMed Res Int (2013) 2013:136106. doi: 10.1155/2013/136106
172. Huang J, Zheng Y, Xiao X, Liu C, Lin J, Zheng S, et al. A circulating long noncoding RNA panel serves as a diagnostic marker for hepatocellular carcinoma. Dis Markers (2020) 2020:5417598. doi: 10.1155/2020/5417598
173. Yuan W, Sun Y, Liu L, Zhou B, Wang S, Gu D. Circulating LncRNAs serve as diagnostic markers for hepatocellular carcinoma. Cell Physiol Biochem (2017) 44(1):125–32. doi: 10.1159/000484589
174. Li J, Wang X, Tang J, Jiang R, Zhang W, Ji J, et al. HULC and Linc00152 act as novel biomarkers in predicting diagnosis of hepatocellular carcinoma. Cell Physiol Biochem (2015) 37(2):687–96. doi: 10.1159/000430387
175. Sukowati CHC, Cabral LKD, Tiribelli C, Pascut D. Circulating long and circular noncoding RNA as non-invasive diagnostic tools of hepatocellular carcinoma. Biomedicines (2021) 9(1):1–21. doi: 10.3390/biomedicines9010090
176. Lu J, Xie F, Geng L, Shen W, Sui C, Yang J. Investigation of serum lncRNA-uc003wbd and lncRNA-AF085935 expression profile in patients with hepatocellular carcinoma and HBV. Tumor Biol (2015) 36(5):3231–6. doi: 10.1007/s13277-014-2951-4
177. Motawi TMK, El-Maraghy SA, Sabry D, Mehana NA. The expression of long non coding RNA genes is associated with expression with polymorphisms of HULC rs7763881 and MALAT1 rs619586 in hepatocellular carcinoma and HBV Egyptian patients. J Cell Biochem (2019) 120(9):14645–56. doi: 10.1002/jcb.28726
178. Xie Z, Zhou F, Yang Y, Li L, Lei Y, Lin X, et al. Lnc-PCDH9-13:1 is a hypersensitive and specific biomarker for early hepatocellular carcinoma. EBioMedicine (2018) 33:57–67. doi: 10.1016/j.ebiom.2018.06.026
179. Wang Z-F, Hu R, Pang J-M, Zhang G-Z, Yan W, Li Z-N. Serum long noncoding RNA LRB1 as a potential biomarker for predicting the diagnosis and prognosis of human hepatocellular carcinoma. Oncol Lett (2018) 16(2):1593–601. doi: 10.3892/ol.2018.8825
180. Feng J, Zhu R, Chang C, Yu L, Cao F, Zhu G, et al. CK19 and glypican 3 expression profiling in the prognostic indication for patients with HCC after surgical resection. PloS One (2016) 11(3):e0151501. doi: 10.1371/journal.pone.0151501
181. Sun D, Zhang Y, Sun X, Chen Y, Qiu W, Ji M, et al. Prognostic value of cytokeratin 19 in hepatocellular carcinoma: A meta-analysis. Clin Chim Acta (2015) 448:161–9. doi: 10.1016/j.cca.2015.06.027
182. Yu J-P, Xu X-G, Ma R-J, Qin S-N, Wang C-R, Wang X-B, et al. Development of a clinical chemiluminescent immunoassay for serum GPC3 and simultaneous measurements alone with AFP and CK19 in diagnosis of hepatocellular carcinoma. J Clin Lab Anal (2015) 29(2):85–93. doi: 10.1002/jcla.21733
183. Lee JI, Lee J-W, Kim JM, Kim JK, Chung HJ, Kim YS. Prognosis of hepatocellular carcinoma expressing cytokeratin 19: comparison with other liver cancers. WJG (2012) 18(34):4751–7. doi: 10.3748/wjg.v18.i34.4751
184. Kladney RD, Cui X, Bulla GA, Brunt EM, Fimmel CJ. Expression of GP73, a resident golgi membrane protein, in viral and nonviral liver disease. Hepatology (2002) 35(6):1431–40. doi: 10.1053/jhep.2002.32525
185. Wang Y, Wan Y-JY. Golgi protein 73, hepatocellular carcinoma and other types of cancers. Liver Res (2020) 4(4):161–7. doi: 10.1016/j.livres.2020.09.003
186. Marrero JA, Romano PR, Nikolaeva O, Steel L, Mehta A, Fimmel CJ, et al. GP73, a resident golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. J Hepatol (2005) 43(6):1007–12. doi: 10.1016/j.jhep.2005.05.028
187. Tian L, Wang Y, Xu D, Gui J, Jia X, Tong H, et al. Serological AFP/Golgi protein 73 could be a new diagnostic parameter of hepatic diseases. Int J Cancer (2011) 129(8):1923–31. doi: 10.1002/ijc.25838
188. Bode K, Bujupi F, Link C, Hein T, Zimmermann S, Peiris D, et al. Dectin-1 binding to annexins on apoptotic cells induces peripheral immune tolerance via NADPH oxidase-2. Cell Rep (2019) 29(13):4435–46.e9. doi: 10.1016/j.celrep.2019.11.086
189. Zhang H, Yao M, Wu W, Qiu L, Sai W, Yang J, et al. Up-regulation of annexin A2 expression predicates advanced clinicopathological features and poor prognosis in hepatocellular carcinoma. Tumour Biol (2015) 36(12):9373–83. doi: 10.1007/s13277-015-3678-6
190. Sun Y, Gao G, Cai J, Wang Y, Qu X, He L, et al. Annexin A2 is a discriminative serological candidate in early hepatocellular carcinoma. Carcinogenesis (2013) 34(3):595–604. doi: 10.1093/carcin/bgs372
191. Moorman HR, Poschel D, Klement JD, Lu C, Redd PS, Liu K. Osteopontin: A key regulator of tumor progression and immunomodulation. Cancers (Basel) (2020) 12(11):1–31. doi: 10.3390/cancers12113379
192. Desert R, Ge X, Song Z, Han H, Lantvit D, Chen W, et al. Role of hepatocyte-derived osteopontin in liver carcinogenesis. Hepatol Commun (2022) 6(4):692–709. doi: 10.1002/hep4.1845
193. Wu Q, Li L, Miao C, Hasnat M, Sun L, Jiang Z, et al. Osteopontin promotes hepatocellular carcinoma progression through inducing JAK2/STAT3/NOX1-mediated ROS production. Cell Death Dis (2022) 13(4):341. doi: 10.1038/s41419-022-04806-9
194. Zhu M, Zheng J, Wu F, Kang B, Liang J, Heskia F, et al. OPN is a promising serological biomarker for hepatocellular carcinoma diagnosis. J Med Virol (2020) 92(12):3596–603. doi: 10.1002/jmv.25704
195. Abdel-Hafiz SM, Hamdy HEM, Khorshed FM, Aboushousha TS, Safwat G, Saber MA, et al. Evaluation of osteopontin as a biomarker in hepatocellular carcinomas in Egyptian patients with chronic HCV cirrhosis. Asian Pac J Cancer Prev (2018) 19(4):1021–7. doi: 10.22034/APJCP.2018.19.4.1021
196. Rong W, Zhang Y, Yang L, Feng L, Wei B, Wu F, et al. Post-surgical resection prognostic value of combined OPN, MMP7, and PSG9 plasma biomarkers in hepatocellular carcinoma. Front Med (2019) 13(2):250–8. doi: 10.1007/s11684-018-0632-1
197. Byeon H, Lee SD, Hong E-K, Lee DE, Kim BH, Seo Y, et al. Long-term prognostic impact of osteopontin and dickkopf-related protein 1 in patients with hepatocellular carcinoma after hepatectomy. Pathol Res Pract (2018) 214(6):814–20. doi: 10.1016/j.prp.2018.05.002
198. Huang Y, Hoque MO, Wu F, Trink B, Sidransky D, Ratovitski EA. Midkine induces epithelial-mesenchymal transition through Notch2/Jak2-Stat3 signaling in human keratinocytes. Cell Cycle (2008) 7(11):1613–22. doi: 10.4161/cc.7.11.5952
199. Sakaguchi N, Muramatsu H, Ichihara-Tanaka K, Maeda N, Noda M, Yamamoto T, et al. Receptor-type protein tyrosine phosphatase ζ as a component of the signaling receptor complex for midkine-dependent survival of embryonic neurons. Neurosci Res (2003) 45(2):219–24. doi: 10.1016/S0168-0102(02)00226-2
200. Weckbach LT, Gola A, Winkelmann M, Jakob SM, Groesser L, Borgolte J, et al. The cytokine midkine supports neutrophil trafficking during acute inflammation by promoting adhesion via β2 integrins (CD11/CD18). Blood (2014) 123(12):1887–96. doi: 10.1182/blood-2013-06-510875
201. Muramatsu H, Zou K, Sakaguchi N, Ikematsu S, Sakuma S, Muramatsu T. LDL receptor-related protein as a component of the midkine receptor. Biochem Biophys Res Commun (2000) 270(3):936–41. doi: 10.1006/bbrc.2000.2549
202. Hodeib H, ELshora O, Selim A, Sabry NM, El-Ashry HM. Serum midkine and osteopontin levels as diagnostic biomarkers of hepatocellular carcinoma. Electron Physician (2017) 9(1):3492–8. doi: 10.19082/3492
203. Zheng L, Li H, Huang J, Shin J, Luo S, Guo C, et al. Serum midkine levels for the diagnosis and assessment of response to interventional therapy in patients with hepatocellular carcinoma. J Interv Med (2021) 4(1):39–45. doi: 10.1016/j.jimed.2020.10.009
204. Omran MM, Farid K, Omar MA, Emran TM, El-Taweel FM, Tabll AA. A combination of α-fetoprotein, midkine, thioredoxin and a metabolite for predicting hepatocellular carcinoma. Ann Hepatol (2020) 19(2):179–85. doi: 10.1016/j.aohep.2019.09.002
205. Vongsuvanh R, van der Poorten D, Iseli T, Strasser SI, McCaughan GW, George J. Midkine increases diagnostic yield in AFP negative and NASH-related hepatocellular carcinoma. PloS One (2016) 11(5):e0155800. doi: 10.1371/journal.pone.0155800
206. Jiang X, Hui F, Qin X, Wu Y, Liu H, Gao J, et al. Diagnosis accuracy and prognostic significance of the dickkopf-1 protein in gastrointestinal carcinomas: Systematic review and network meta-analysis. J Cancer (2020) 11(24):7091–100. doi: 10.7150/jca.49970
207. Li Z, Mou L, Gao H, Zeng Y, Tang X, Deng X, et al. Diagnostic accuracy of serum dickkopf-1 protein in diagnosis hepatocellular carcinoma: An updated meta-analysis. Med (Baltimore) (2019) 98(32):e16725. doi: 10.1097/MD.0000000000016725
208. Younis YS, Alegaily HS, Elagawy W, Semeya AA, Abo-Amer YE-E, El-Abgeegy M, et al. Serum dickopff 1 as a novel biomarker in hepatocellular carcinoma diagnosis and follow up after ablative therapy. Cancer Manag Res (2019) 11:10555–62. doi: 10.2147/CMAR.S218532
209. El-Shayeb AF, El-Habachi NM, Mansour AR, Zaghloul MS. Serum midkine is a more sensitive predictor for hepatocellular carcinoma than dickkopf-1 and alpha-l-fucosidase in cirrhotic HCV patients. Med (Baltimore) (2021) 100(17):e25112. doi: 10.1097/MD.0000000000025112
210. ElZefzafy WM, Hussien M, Mohmmed ZAZ, Abd Elbaky NM. The diagnostic value of golgi protien-73 and DICKKOPF-1 in hepatocellular carcinoma. J Immunoassay Immunochem (2021) 42(2):174–87. doi: 10.1080/15321819.2020.1844750
211. Gil-Gómez A, Rojas Á, Liu C-H, Gallego-Duran R, Muñoz-Hernandez R, Fassina G, et al. Combination of squamous cell carcinoma antigen immunocomplex and alpha-fetoprotein in mid- and long-term prediction of hepatocellular carcinoma among cirrhotic patients. World J Gastroenterol (2021) 27(48):8343–56. doi: 10.3748/wjg.v27.i48.8343
212. Cagnin M, Biasiolo A, Martini A, Ruvoletto M, Quarta S, Fasolato S, et al. Serum squamous cell carcinoma antigen-immunoglobulin m complex levels predict survival in patients with cirrhosis. Sci Rep (2019) 9(1):20126. doi: 10.1038/s41598-019-56633-2
213. Pelizzaro F, Soldà F, Cardin R, Imondi A, Sartori A, Penzo B, et al. SCCA-IgM in hepatocellular carcinoma patients treated with transarterial chemoembolization: gender-related differences. Biomarkers Med (2020) 14(10):855–67. doi: 10.2217/bmm-2019-0564
214. Liu C-H, Gil-Gómez A, Ampuero J, Romero-Gómez M. Diagnostic accuracy of SCCA and SCCA-IgM for hepatocellular carcinoma: A meta-analysis. Liver Int (2018) 38(10):1820–31. doi: 10.1111/liv.13867
215. Yu J, Wang Z-J, Chen L-H, Dong W-Z. Diagnostic value of serum squamous cell carcinoma antigen for hepatocellular carcinoma: a systematic review and meta-analysis. Scand J Clin Lab Invest (2017) 77(1):8–14. doi: 10.1080/00365513.2016.1238504
216. Zhu H. Squamous cell carcinoma antigen: Clinical application and research status. Diagn (Basel) (2022) 12(5):1–16. doi: 10.3390/diagnostics12051065
217. Conti F, Dall'Agata M, Gramenzi A, Biselli M. Biomarkers for the early diagnosis of bacterial infection and the surveillance of hepatocellular carcinoma in cirrhosis. Biomarkers Med (2015) 9(12):1343–51. doi: 10.2217/bmm.15.100
218. Zhang W, Chen Z, Xue C, Zhang Y, Wu L, Zhu J, et al. The applicability of ADA, AFU, and LAC in the early diagnosis and disease risk assessment of hepatitis b-associated liver cirrhosis and hepatocellular carcinoma. Front Med (Lausanne) (2021) 8:740029. doi: 10.3389/fmed.2021.740029
219. Liu D, Luo Y, Chen L, Chen L, Zuo D, Li Y, et al. Diagnostic value of 5 serum biomarkers for hepatocellular carcinoma with different epidemiological backgrounds: A large-scale, retrospective study. Cancer Biol Med (2021) 18(1):256–70. doi: 10.20892/j.issn.2095-3941.2020.0207
220. Xing H, Qiu H, Ding X, Han J, Li Z, Wu H, et al. Clinical performance of α-l-fucosidase for early detection of hepatocellular carcinoma. Biomarkers Med (2019) 13(7):545–55. doi: 10.2217/bmm-2018-0414
221. Aheget H, Mazini L, Martin F, Belqat B, Marchal JA, Benabdellah K. Exosomes: Their role in pathogenesis, diagnosis and treatment of diseases. Cancers (Basel) (2020) 13(1):1–45. doi: 10.3390/cancers13010084
222. Wu Q, Zhou L, Lv D, Zhu X, Tang H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J Hematol Oncol (2019) 12(1):53. doi: 10.1186/s13045-019-0739-0
223. Németh A, Orgovan N, Sódar BW, Osteikoetxea X, Pálóczi K, Szabó-Taylor KÉ, et al. Antibiotic-induced release of small extracellular vesicles (exosomes) with surface-associated DNA. Sci Rep (2017) 7(1):8202. doi: 10.1038/s41598-017-08392-1
224. Chapuy-Regaud S, Dubois M, Plisson-Chastang C, Bonnefois T, Lhomme S, Bertrand-Michel J, et al. Characterization of the lipid envelope of exosome encapsulated HEV particles protected from the immune response. Biochimie (2017) 141:70–9. doi: 10.1016/j.biochi.2017.05.003
225. Haraszti RA, Didiot M-C, Sapp E, Leszyk J, Shaffer SA, Rockwell HE, et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J Extracellular Vesicles (2016) 5(1):32570. doi: 10.3402/jev.v5.32570
226. Wang S, Xu M, Li X, Su X, Xiao X, Keating A, et al. Exosomes released by hepatocarcinoma cells endow adipocytes with tumor-promoting properties. J Hematol Oncol (2018) 11(1):82. doi: 10.1186/s13045-018-0625-1
227. Rao Q, Zuo B, Lu Z, Gao X, You A, Wu C, et al. Tumor-derived exosomes elicit tumor suppression in murine hepatocellular carcinoma models and humans in vitro. Hepatology (2016) 64(2):456–72. doi: 10.1002/hep.28549
228. Sun N, Zhang C, Lee Y-T, Tran BV, Wang J, Kim H, et al. HCC EV ECG score: An extracellular vesicle-based protein assay for detection of early-stage hepatocellular carcinoma. Hepatology (2022). doi: 10.1002/hep.32692
229. Bernard V, Kim DU, San Lucas FA, Castillo J, Allenson K, Mulu FC, et al. Circulating nucleic acids are associated with outcomes of patients with pancreatic cancer. Gastroenterology (2019) 156(1):108–118.e4. doi: 10.1053/j.gastro.2018.09.022
230. Li L-M, Liu Z-X, Cheng Q-Y. Exosome plays an important role in the development of hepatocellular carcinoma. Pathol Res Pract (2019) 215(8):152468. doi: 10.1016/j.prp.2019.152468
231. Fang T, Lv H, Lv G, Li T, Wang C, Han Q, et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun (2018) 9(1):191. doi: 10.1038/s41467-017-02583-0
232. Abd El Gwad A, Matboli M, El-Tawdi A, Habib EK, Shehata H, Ibrahim D, et al. Role of exosomal competing endogenous RNA in patients with hepatocellular carcinoma. J Cell Biochem (2018) 119(10):8600–10. doi: 10.1002/jcb.27109
233. Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine (2019) 40:432–45. doi: 10.1016/j.ebiom.2018.12.062
234. Wang H, Lu Z, Zhao X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol (2019) 12(1):133. doi: 10.1186/s13045-019-0806-6
235. Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY) (2019) 11(19):8183–203. doi: 10.18632/aging.102312
236. Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene (2019) 38(15):2844–59. doi: 10.1038/s41388-018-0619-z
237. Lin X-J, Fang J-H, Yang X-J, Zhang C, Yuan Y, Zheng L, et al. Hepatocellular carcinoma cell-secreted exosomal MicroRNA-210 promotes angiogenesis In vitro and in vivo. Mol Ther - Nucleic Acids (2018) 11:243–52. doi: 10.1016/j.omtn.2018.02.014
238. Cui Y, Xu H-F, Liu M-Y, Xu Y-J, He J-C, Zhou Y, et al. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. WJG (2019) 25(15):1890–8. doi: 10.3748/wjg.v25.i15.1890
239. Yao Z, Jia C, Tai Y, Liang H, Zhong Z, Xiong Z, et al. Serum exosomal long noncoding RNAs lnc-FAM72D-3 and lnc-EPC1-4 as diagnostic biomarkers for hepatocellular carcinoma. Aging (Albany NY) (2020) 12(12):11843–63. doi: 10.18632/aging.103355
240. Chen W, Mao Y, Liu C, Wu H, Chen S. Exosome in hepatocellular carcinoma: an update. J Cancer (2021) 12(9):2526–36. doi: 10.7150/jca.54566
241. von Felden J, Garcia-Lezana T, Dogra N, Gonzalez-Kozlova E, Ahsen ME, Craig A, et al. Unannotated small RNA clusters associated with circulating extracellular vesicles detect early stage liver cancer. Gut (2022) 71:2069–80. doi: 10.1136/gutjnl-2021-325036
242. Lu Y, Duan Y, Xu Q, Zhang L, Chen W, Qu Z, et al. Circulating exosome-derived bona fide long non-coding RNAs predicting the occurrence and metastasis of hepatocellular carcinoma. J Cell Mol Med (2020) 24(2):1311–8. doi: 10.1111/jcmm.14783
243. Wang T, Zhu H, Xiao M, Zhou S. Serum exosomal long noncoding RNA CRNDE as a prognostic biomarker for hepatocellular carcinoma. J Clin Lab Anal (2021) 35(11):e23959. doi: 10.1002/jcla.23959
244. Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Med J Aust (1869) 14):146–7.
245. Yu JH, Ma S. Organoids as research models for hepatocellular carcinoma. Exp Cell Res (2022) 411(1):112987. doi: 10.1016/j.yexcr.2021.112987
246. Geurts MH, de Poel E, Pleguezuelos-Manzano C, Oka R, Carrillo L, Andersson-Rolf A, et al. Evaluating CRISPR-based prime editing for cancer modeling and CFTR repair in organoids. Life Sci Alliance (2021) 4(10):1–12. doi: 10.26508/lsa.202000940
247. Chan LH, Zhou L, Ng KY, Wong TL, Lee TK, Sharma R, et al. PRMT6 regulates RAS/RAF binding and MEK/ERK-mediated cancer stemness activities in hepatocellular carcinoma through CRAF methylation. Cell Rep (2018) 25(3):690–701.e8. doi: 10.1016/j.celrep.2018.09.053
248. Zhang Q, Rong Y, Yi K, Huang L, Chen M, Wang F. Circulating tumor cells in hepatocellular carcinoma: single-cell based analysis, preclinical models, and clinical applications. Theranostics (2020) 10(26):12060–71. doi: 10.7150/thno.48918
249. Crignis E, Hossain T, Romal S, Carofiglio F, Moulos P, Khalid MM, et al. Application of human liver organoids as a patient-derived primary model for HBV infection and related hepatocellular carcinoma. Elife (2021) 10:1–32. doi: 10.7554/eLife.60747.sa2
250. Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ (2018) 25(3):486–541. doi: 10.1038/s41418-017-0012-4
251. Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer (2019) 18(1):114. doi: 10.1186/s12943-019-1043-x
252. Okajima W, Komatsu S, Ichikawa D, Miyamae M, Ohashi T, Imamura T, et al. Liquid biopsy in patients with hepatocellular carcinoma: Circulating tumor cells and cell-free nucleic acids. WJG (2017) 23(31):5650–68. doi: 10.3748/wjg.v23.i31.5650
253. Miller MC, Doyle GV, Terstappen LWMM. Significance of circulating tumor cells detected by the CellSearch system in patients with metastatic breast colorectal and prostate cancer. J Oncol (2010) 2010:617421. doi: 10.1155/2010/617421
254. van de Stolpe A, Pantel K, Sleijfer S, Terstappen LW, den Toonder JMJ. Circulating tumor cell isolation and diagnostics: toward routine clinical use. Cancer Res (2011) 71(18):5955–60. doi: 10.1158/0008-5472.CAN-11-1254
255. Kelley RK, Magbanua MJM, Butler TM, Collisson EA, Hwang J, Sidiropoulos N, et al. Circulating tumor cells in hepatocellular carcinoma: a pilot study of detection, enumeration, and next-generation sequencing in cases and controls. BMC Cancer (2015) 15(1):206. doi: 10.1186/s12885-015-1195-z
256. Ogle LF, Orr JG, Willoughby CE, Hutton C, McPherson S, Plummer R, et al. Imagestream detection and characterisation of circulating tumour cells - a liquid biopsy for hepatocellular carcinoma? J Hepatol (2016) 65(2):305–13. doi: 10.1016/j.jhep.2016.04.014
257. Sun Y-F, Xu Y, Yang X-R, Guo W, Zhang X, Qiu S-J, et al. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology (2013) 57(4):1458–68. doi: 10.1002/hep.26151
258. Schulze K, Gasch C, Staufer K, Nashan B, Lohse AW, Pantel K, et al. Presence of EpCAM-positive circulating tumor cells as biomarker for systemic disease strongly correlates to survival in patients with hepatocellular carcinoma. Int J Cancer (2013) 133(9):2165–71. doi: 10.1002/ijc.28230
259. von Felden J, Schulze K, Krech T, Ewald F, Nashan B, Pantel K, et al. Circulating tumor cells as liquid biomarker for high HCC recurrence risk after curative liver resection. Oncotarget (2017) 8(52):89978–87. doi: 10.18632/oncotarget.21208
260. Wang L, Li Y, Xu J, Zhang A, Wang X, Tang R, et al. Quantified postsurgical small cell size CTCs and EpCAM+ circulating tumor stem cells with cytogenetic abnormalities in hepatocellular carcinoma patients determine cancer relapse. Cancer Lett (2018) 412:99–107. doi: 10.1016/j.canlet.2017.10.004
261. Luo Q, Wang C, Peng B, Pu X, Cai L, Liao H, et al. Circulating tumor-Cell-Associated white blood cell clusters in peripheral blood indicate poor prognosis in patients with hepatocellular carcinoma. Front Oncol (2020) 10:1758. doi: 10.3389/fonc.2020.01758
262. Zhang Q, Xing W, Zhang J, Hu J, Qi L, Xiang B. Circulating tumor cells undergoing the epithelial-mesenchymal transition: Influence on prognosis in cytokeratin 19-positive hepatocellular carcinoma. OTT (2021) 14:1543–52. doi: 10.2147/OTT.S298576
263. Qi L-N, Xiang B-D, Wu F-X, Ye J-Z, Zhong J-H, Wang Y-Y, et al. Circulating tumor cells undergoing EMT provide a metric for diagnosis and prognosis of patients with hepatocellular carcinoma. Cancer Res (2018) 78(16):4731–44. doi: 10.1158/0008-5472.CAN-17-2459
264. Vona G, Estepa L, Béroud C, Damotte D, Capron F, Nalpas B, et al. Impact of cytomorphological detection of circulating tumor cells in patients with liver cancer. Hepatology (2004) 39(3):792–7. doi: 10.1002/hep.20091
265. Guo W, Sun Y-F, Shen M-N, Ma X-L, Wu J, Zhang C-Y, et al. Circulating tumor cells with stem-like phenotypes for diagnosis, prognosis, and therapeutic response evaluation in hepatocellular carcinoma. Clin Cancer Res (2018) 24(9):2203–13. doi: 10.1158/1078-0432.CCR-17-1753
266. Wang P-X, Xu Y, Sun Y-F, Cheng J-W, Zhou K-Q, Wu S-Y, et al. Detection of circulating tumour cells enables early recurrence prediction in hepatocellular carcinoma patients undergoing liver transplantation. Liver Int (2021) 41(3):562–73. doi: 10.1111/liv.14734
267. Zhou J, Zhang Z, Zhou H, Leng C, Hou B, Zhou C, et al. Preoperative circulating tumor cells to predict microvascular invasion and dynamical detection indicate the prognosis of hepatocellular carcinoma. BMC Cancer (2020) 20(1):1047. doi: 10.1186/s12885-020-07488-8
Keywords: hepatocellular carcinoma, biomarker, personalized medicine, alpha-fetoprotein, liquid biopsy, circulating tumor cells, circulating nucleic acids, exosomes
Citation: Schlosser S, Tümen D, Volz B, Neumeyer K, Egler N, Kunst C, Tews HC, Schmid S, Kandulski A, Müller M and Gülow K (2022) HCC biomarkers – state of the old and outlook to future promising biomarkers and their potential in everyday clinical practice. Front. Oncol. 12:1016952. doi: 10.3389/fonc.2022.1016952
Received: 11 August 2022; Accepted: 04 November 2022;
Published: 28 November 2022.
Edited by:
Emilio Francesco Giunta, Università degli Studi della Campania Luigi Vanvitelli, ItalyReviewed by:
Mamdouh M. El-Shishtawy, Mansoura University, EgyptNurbubu Moldogazieva, I.M. Sechenov First Moscow State Medical University, Russia
Copyright © 2022 Schlosser, Tümen, Volz, Neumeyer, Egler, Kunst, Tews, Schmid, Kandulski, Müller and Gülow. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karsten Gülow, a2Fyc3Rlbi5ndWVsb3dAdWtyLmRl
 Sophie Schlosser
Sophie Schlosser Deniz Tümen
Deniz Tümen Claudia Kunst
Claudia Kunst Hauke Christian Tews
Hauke Christian Tews Stephan Schmid
Stephan Schmid Martina Müller
Martina Müller