- Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang, China
Long non-coding RNAs (lncRNAs) are non-protein coding RNAs with more than 200 nucleic acids in length. When lncRNAs are located in the nucleus, they regulate chromosome structure, participate in chromatin remodeling, and act as transcription regulators. When lncRNAs are exported to the cytoplasm, they regulate mRNA stability, regulate translation, and interfere with post-translational modification. In recent years, more and more evidences have shown that lncRNA can regulate the biological processes of tumor proliferation, apoptosis, invasion and metastasis, and can participate in a variety of tumor signaling pathways. Long-gene non-protein coding RNA641 (LINC00641), located on human chromosome 14q11.2, is differentially expressed in a variety of tumors and is related to overall survival and prognosis, etc. Interfering the expression of LINC00641 can lead to changes in tumor cell proliferation, invasion, metastasis, apoptosis and other biological behaviors. Therefore, LINC00641 is a promising new biomarker and potential clinical therapeutic target. In this review, the biological functions, related mechanisms and clinical significance of LINC00641 in many human cancers are described in detail.
Background and Introduction
Cancer is a global and growing disease, which still has high incidence rate and mortality rate. For the exploration of cancer, from genomics (1), epigenetics (2), to tumor microenvironment (3), immunotherapy (4), to cancer cell metabolism (5) and tumor microbiota (6), people have never stopped moving forward.
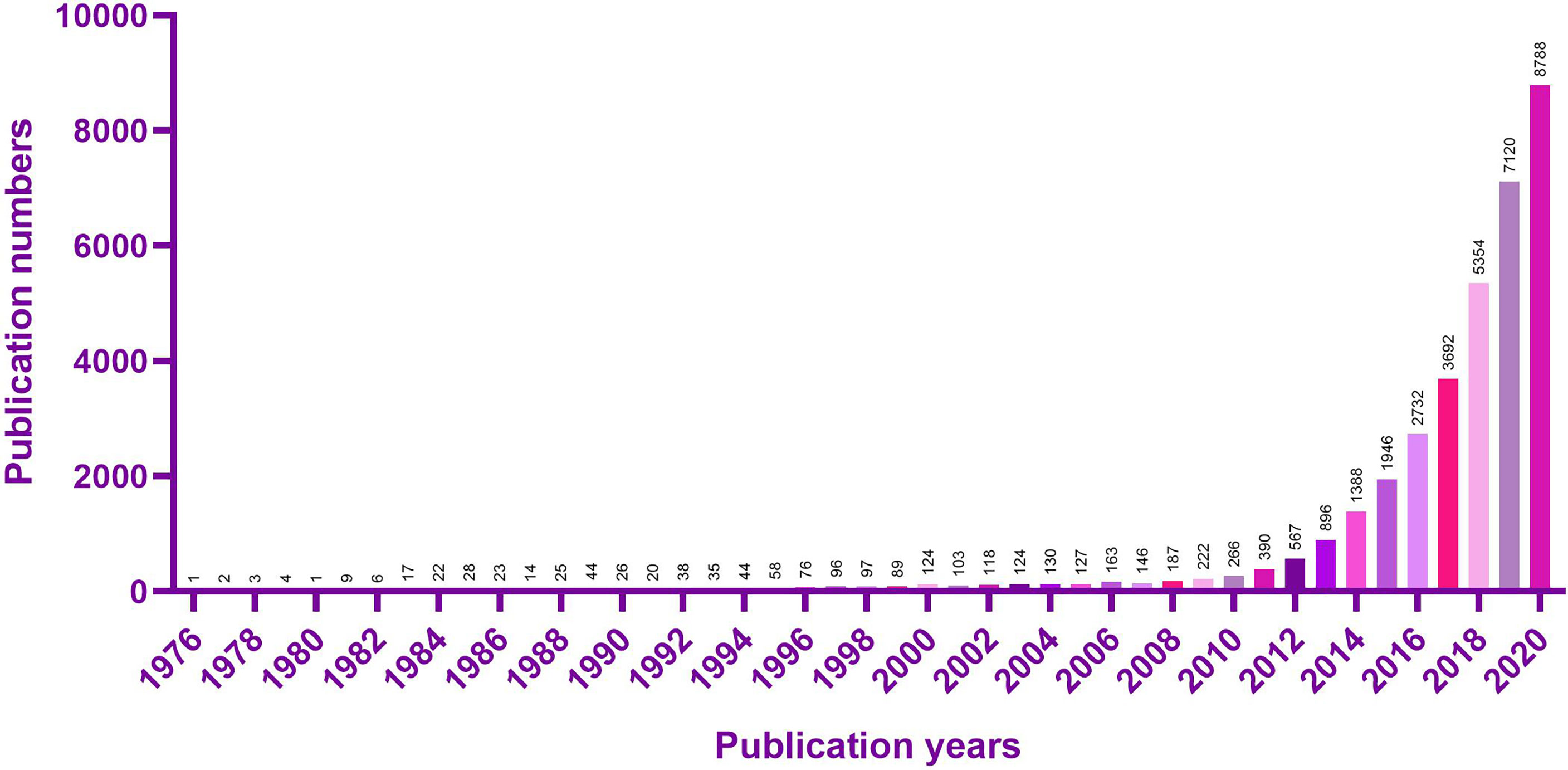
Non protein coding genes, which account for about 98% of the total genome sequence (7), have long been removed from the transcriptional “noise”. Long non-coding RNA (LncRNA) with a length of more than 200 nucleotides (8) can participate in the regulation of gene expression at epigenetic (9), transcriptional and post transcriptional levels (10), adopting the action mode of signal, bait, guide or scaffold (11). The researches on LncRNA have sprung up in recent ten years (Figure 1).
LncRNAs perform different functions according to their localization in subcells (12). When LncRNA located in the nucleus, it regulates chromosome structure (13), participates in chromatin remodeling (14), and acts as a transcription regulator (15). When lncRNA is exported to the cytoplasm, it regulates mRNA stability (16), regulates translation (17), and interferes with post-translational modifications (18).
Mutation and misregulation of long non-coding RNA (LncRNA) play an important role in cancer (19), and participate in multiple biological processes such as tumor proliferation, apoptosis, invasion and metastasis (20–23). LncRNA can participate in a variety of tumor signaling pathways, including p53 (24) and NF-κB (25)、PI3K/AKT (26)、 Wnt/β- Catenin (27) and notch (28), etc. The most important feature of LncRNA mediated cell signal regulation is that LncRNA can act as a scaffold (29). A large number of papers have identified many LncRNAs, which can regulate gene expression in various cancers through ceRNA regulation. LncRNA can be used as a “sponge” to adsorb microRNA, blocking the inhibition of microRNA on its downstream target mRNA, and indirectly regulating the expression of genes, which functions as a microRNA decoy (30–33).
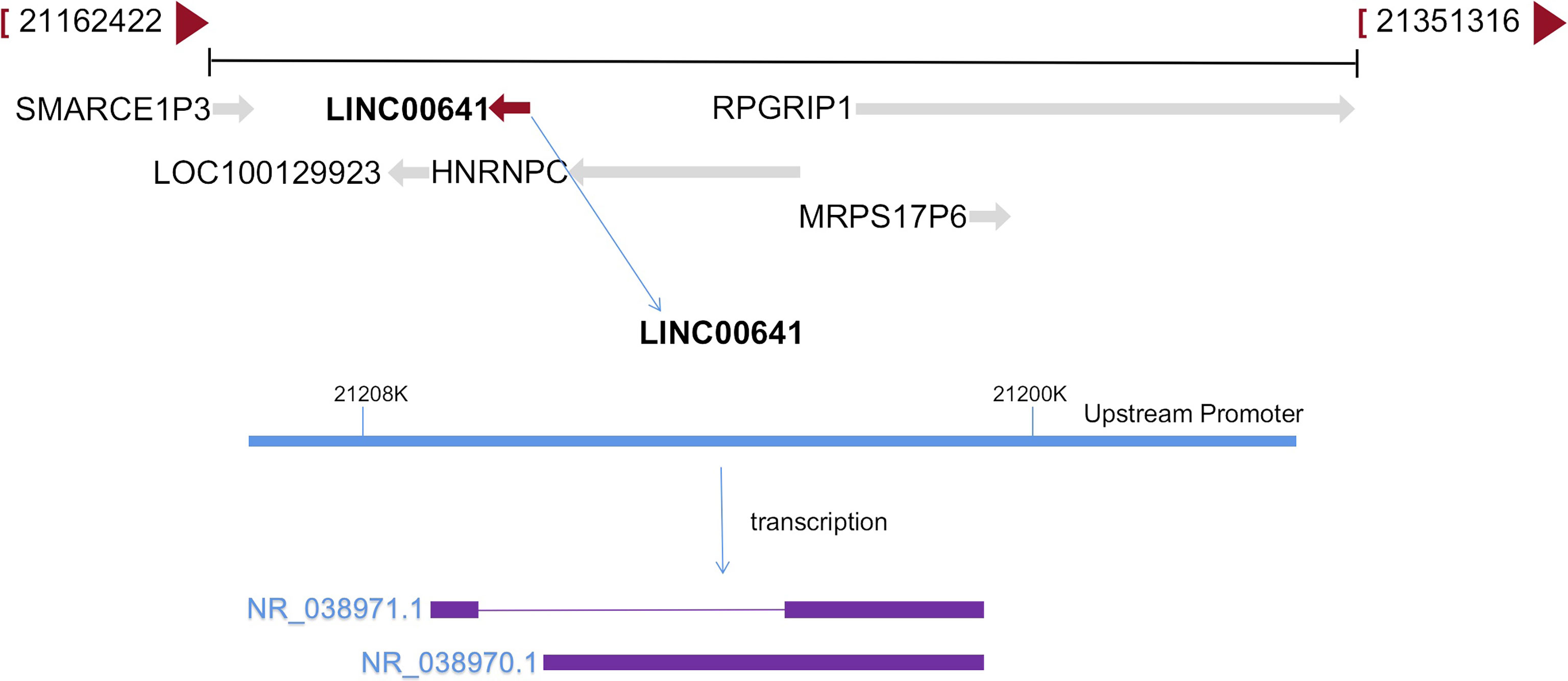
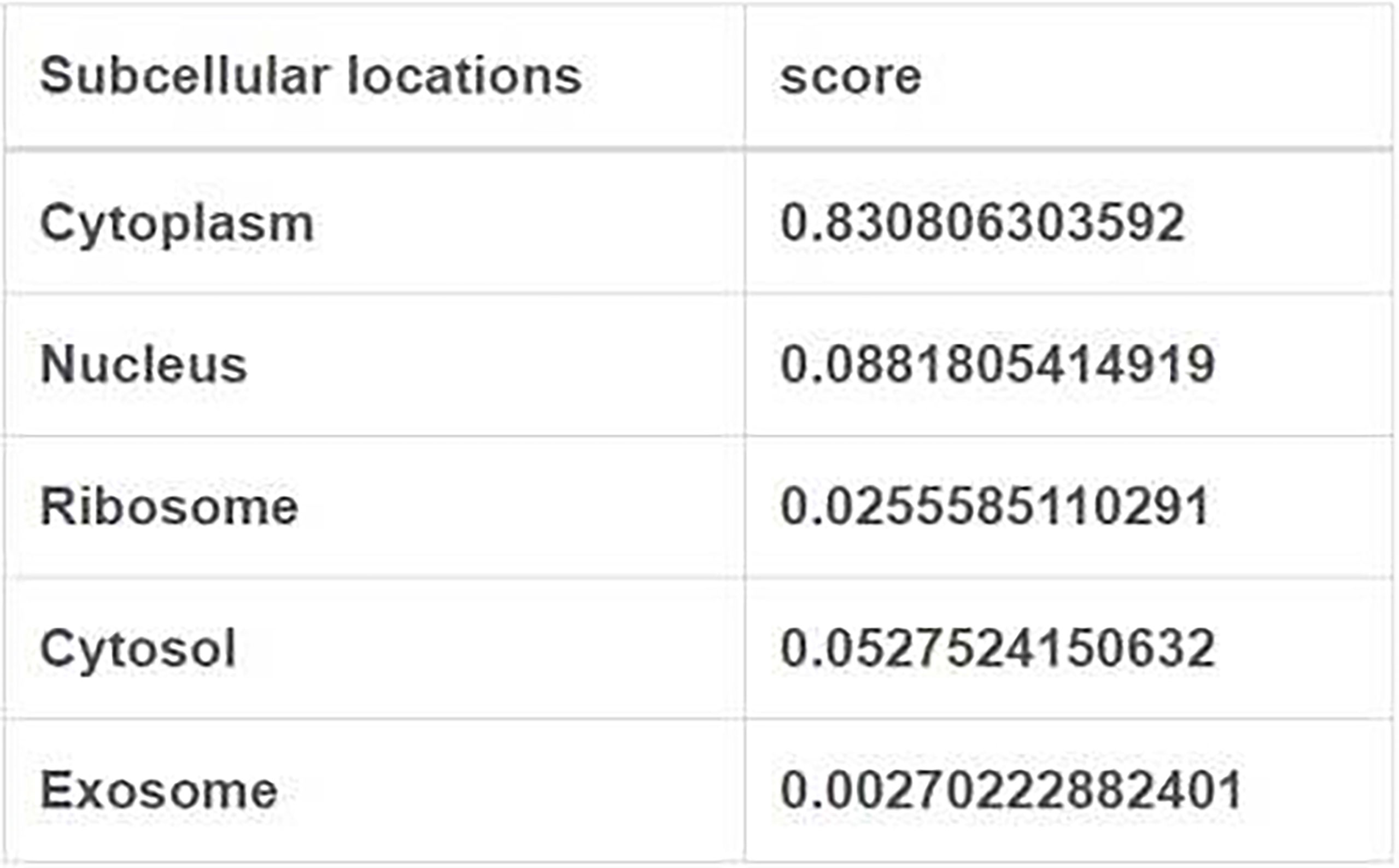
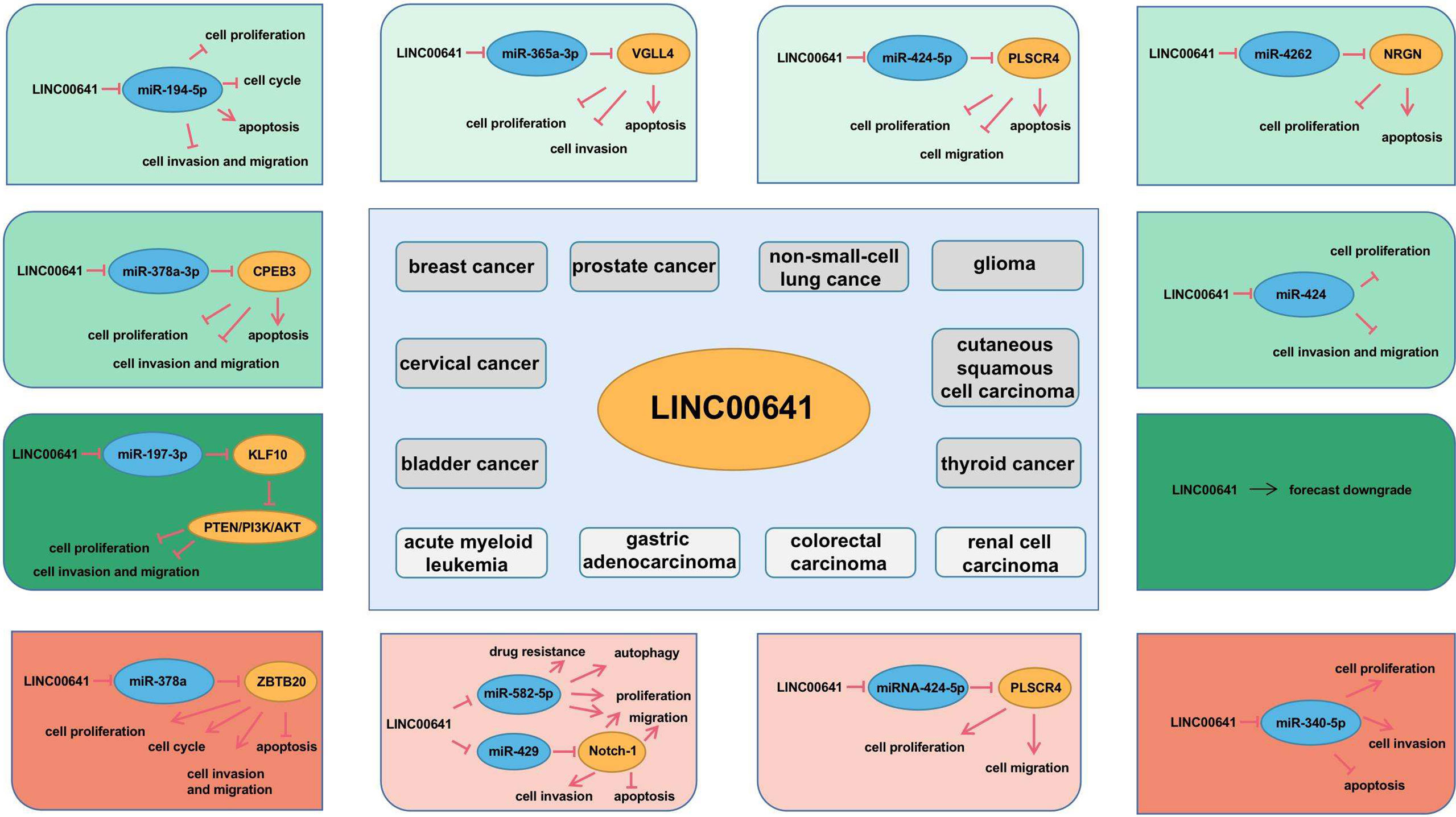
The long intergenic non protein coding RNA641 (LINC00641) is located on human chromosome 14q11.2. It is widely expressed in 22 tissues such as brain tissue and bone marrow, among which the expression in brain tissue is the highest (34). More and more evidence show that LINC00641 is differentially expressed in a variety of tumors and is associated with overall survival and prognosis (35–37). LINC00641 can participate in the regulation of proliferation, invasion, metastasis and apoptosis of a variety of tumors (38–40). A search of NCBI database revealed that LINC00641 gene sequence could be transcribed into two transcripts: NR_038970.1 and NR_038971.1 (Figure 2). According to the lnclocator database, LINC00641 is mainly located in the cytoplasm (Figure 3).
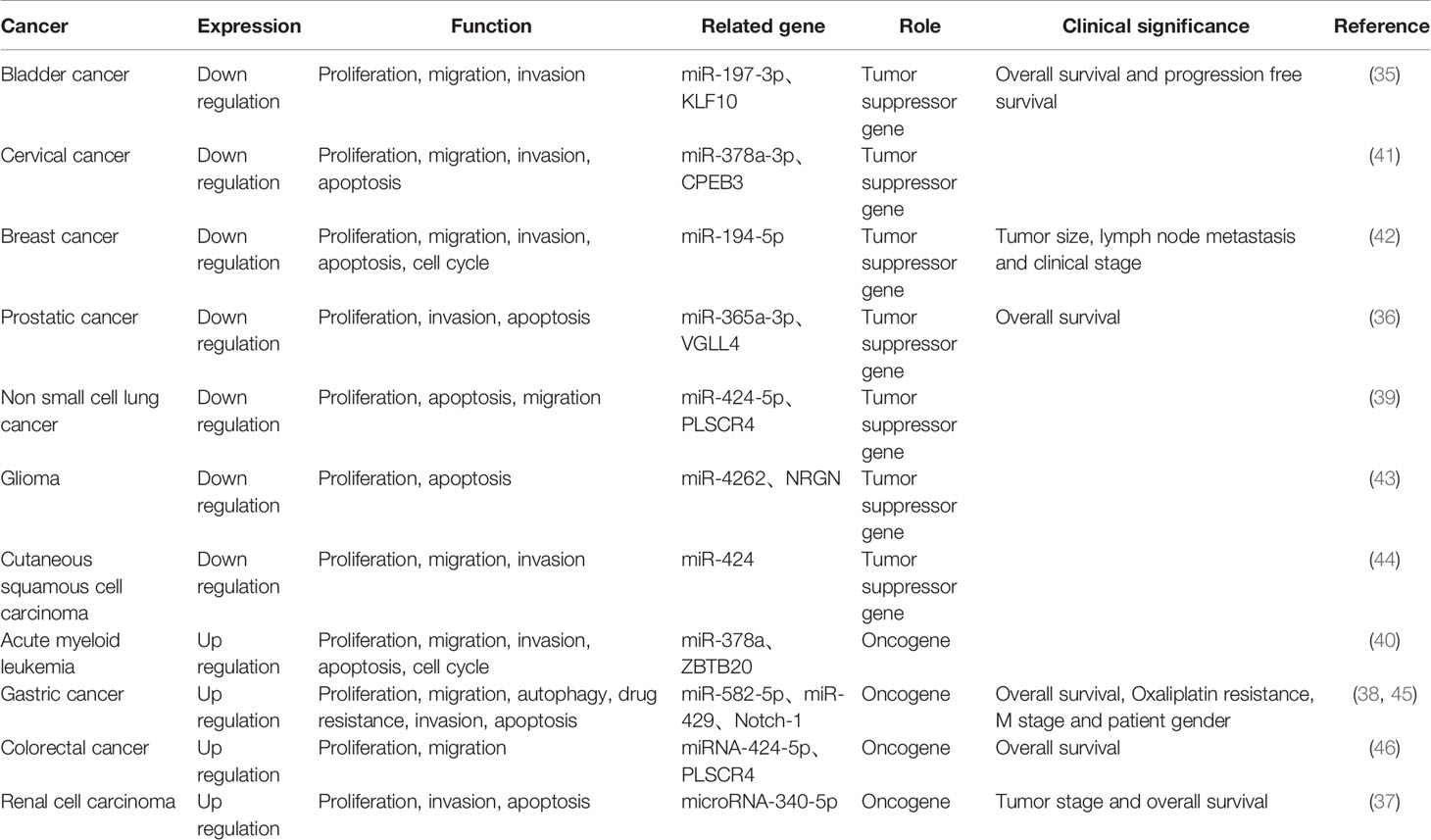
LINC00641 is downregulated in bladder cancer, cervical cancer, breast cancer, prostate cancer, non-small cell lung cancer, glioma and cutaneous squamous cell carcinoma, and can be used as a tumor suppressor gene, while it is up-regulated in acute myeloid leukemia, gastric cancer, rectal cancer and renal cell carcinoma, and can be used as oncogene. Considering the different expression of LINC00641 in different tumors, it may be related to tissue specificity. In this paper, we elaborate the biological function, related mechanism and clinical significance of LINC00641 in human cancer (Table 1).
Role of LINC00641 in Cancer
Bladder Cancer
Bladder cancer is a common malignant tumor in women and the fourth most common malignancy tumor in men (47). Worldwide, there are an estimated 500,000 new cases and 200,000 deaths each year (48). Research into cancer genomics, risk factors and immunotherapy is the key to combat this malignant disease (49). It is very important to explore the molecular mechanism of bladder cancer development. Li et al. (35) analyzed the expression of LINC00641 in 39 pairs of bladder cancer tissues and normal tissues and found that LINC00641 was significantly decreased in bladder cancer tissues, and the low expression of LINC00641 was associated with overall survival and progression-free survival of patients. In vitro, the overexpression of LINC00641 reduced the proliferation, migration and invasion of bladder cancer cell lines. In vivo, LINC00641 overexpression significantly inhibited tumor volume. Starbase bioinformatics analysis showed that LINC00641 may interact with miR-197-3p and using Targetscan 7 tool, it was determined that miR-197-3p may target KLF10. The results showed that increasing the expression of LINC00641 inhibited miR-197-3p, while miR-197-3p inhibited the expression of KLF10 in cells. In a myeloma study, KLF10 inactivated the PTEN/PI3K/AKT pathway. The experiment also demonstrated that LINC00641 promoted KLF10 expression by targeting miR-197-3p, thereby inhibiting PTEN/PI3K/AKT pathway. In conclusion, LINC00641 can competitively bind to miR-197-3p through the ceRNA mechanism to increase the expression of KLF10, further inhibit the activation of PTEN/PI3K/AKT pathway, and inhibit the proliferation, migration and invasion of bladder cancer cells. In general, LINC00641 is a tumor suppressor factor in bladder cancer. LINC00641/miR-197-3p/KLF10/PTEN/PI3K/AKT cascade may be a promising target for bladder cancer treatment.
Cervical Cancer
Cervical cancer is the fourth most common cancer among women worldwide, with more than 85% of cervical cancer deaths occurring in less developed regions of the world (50). Although HPV vaccine is one of the major breakthroughs in modern medicine, and much is known about HPV induced carcinogenesis, the clinical outcomes have been stagnated for decades (51). More and more cervical cancer genome analyses have revealed new potential therapeutic targets for cervical cancer (52). By studying the molecular mechanism of cervical cancer, we aim to provide some methods for targeted therapy. Zhang et al. (41) found that the expression of LINC00641 was significantly down regulated in cervical cancer cell lines compared with normal cervical cells. Upregulation of LINC00641 inhibited cell proliferation, increased apoptosis, decreased invasion and migration, and inhibited EMT. Fish showed that LINC00641 was abundant in the cytoplasm of cervical cancer cells. After further exploring the mechanism of LINC00641, combined with Starbase bioinformatic analysis and experiments, it was proved that miR-378a-3p was the target of LINC00641 and LINC00641 inhibited the growth of cervical cancer cells by reducing the expression of miR-378a-3p. Starbase database was used to predict the opposite changes of CPEB3 and miR-378a-3p. The experiment proved that LINC00641 competed with CPEB3 and bound to miR-378a-3p, confirming that LINC00641 regulated the progression of cervical cancer by up regulating CPEB3. In conclusion, LINC00641 can competitively bind miR-378a-3p through the ceRNA mechanism, so as to increase the expression of CPEB3 mRNA and inhibit the proliferation, migration and invasion of cervical cancer cells. In general, LINC00641 is a tumor suppressor of cervical cancer and plays an important role in its occurrence and development. LINC00641 deserves to be regarded as a new biomarker in the treatment of cervical cancer.
Breast Cancer
Breast cancer is the most frequently diagnosed cancer among women, ranking the second among the causes of cancer-related deaths in women (53). Although the use of adjuvant chemotherapy, the widespread use of hormonal drugs and the application of targeted drugs have reduced the mortality of breast cancer, biomarkers are still an important method for assisting diagnosis and monitoring prognosis. The American Society of Clinical Oncology once recommended CA 15-3, carcinoembryonic antigen, estrogen and progesterone receptor for clinical diagnosis (54). For the treatment of breast cancer, molecular mechanism and potential therapeutic target have become a research hotspot (55). Mao et al. (42) found that LINC00641 expression in breast cancer tissues was significantly lower than that in normal adjacent tissues, and was significantly correlated with tumor size, lymph node metastasis and clinical stage. Over expression of LINC00641 inhibits cell proliferation, migration, invasion and G1/S phase transition and promotes cell apoptosis in breast cancer cell lines. According to starbase 3 and miRcode analysis, miR-194-5p may bind to LINC00641. Previous studies (56) have found that miR-194-5p can promote the growth and metastasis of breast cancer cells and act as an oncogene. It is confirmed that miR-194-5p is a direct target of LINC00641 and the overexpression of miR-194-5p can reverse the inhibition of LINC00641 up-regulation on the biological behavior of breast cancer cells. Therefore, the molecular mechanism of LINC00641 overexpression inhibiting proliferation, migration and invasion of breast cancer cells can be elucidated through the sponge action of miR-194-5p, and LINC00641/miR-194-5p may contribute to the treatment of breast cancer as a favorable target.
Prostatic Cancer
Prostate cancer is the major disease affecting men’s health all over the world and is the second most common form of cancer in men, second only to non-melanoma skin cancer (57). Since Wang et al. first purified the protein in 1979, the diagnostic strategy of population screening for prostate cancer using prostate specific antigen (PSA) has been controversial and hotly debated (58). At present, the treatment of prostate cancer patients increases the demand for reliable biomarkers and pursues individual patient-centered oncology approach (59). Liu et al. (36) found that the expression of LINC00641 was down regulated in prostate cancer tissues and correlated with the prognosis of patients after comparing prostate cancer tissues with normal tissues. Overexpression of LINC00641 inhibited cell proliferation, invasion and promotes apoptosis in prostate cancer cell lines. When further exploring the mechanism of LINC00641 in prostate cancer, it was found that miR-365a-3p was a downstream target of LINC00641 and miR-365a-3p was up-regulated in prostate cancer and negatively correlated with LINC00641. Then it was found that VGLL4, as the downstream target gene of miR-365a-3p, was down regulated in prostate cancer and negatively correlated with miR-365a-3p. In vitro experiments demonstrated that LINC00641 regulates cell proliferation and invasion through miR-365a-3p/VGLL4 axis. In conclusion, LINC00641 can be used as a tumor suppressor of prostate cancer, which can increase the expression of VGLL4 and inhibit the proliferation and invasion of prostate cancer cells by competitively binding miR-365a-3p through the ceRNA mechanism. Huang et al. (60) retrieved prostate cancer tissues and normal tissues from the Cancer Genome Atlas (TCGA) database, screened 14083 lncRNAs, and finally identified 6 lncRNAs with independent prognostic factors, including LINC00641. A risk scoring model was established to effectively evaluate the survival and prognosis of patients. These results indicated that lncRNA has certain predictive effect on the occurrence and prognosis of prostate cancer and can be used as a new biomarker for prostate cancer survival and potential therapeutic targets.
Non Small Cell Lung Cancer
Lung cancer is the most common cause of cancer death worldwide, with an estimated 1.6 million deaths per year (61), and non-small cell lung cancer(NSCLC) accounts for about 85%. Over the past two decades, important advances have been made in the treatment of NSCLC, increasing our understanding of disease biology and tumor progression mechanism, and promoted early detection (62). As a biomarker, lncRNA can be combined with traditional tumor markers such as CEA, SCCA and CYFRA21-1 in the diagnosis of NSCLC with great potential (63). Li et al. (39) found that LINC00641 expression was significantly reduced in non-small cell lung cancer cell lines compared with human normal bronchial epithelial cells. Overexpression of LINC00641 in the cell line decreased the ability of proliferation and migration and increased apoptosis. Starbase database predicted that miR-424-5p was a downstream target of LINC00641, further demonstrating that miR-424-5p could be directly bind to LINC00641 and was negatively regulated by LINC00641. The TargetScan database predicted PLSCR4 as a candidate target of miR-424-5p. Experiments proved that LINC00641, as the ceRNA of PLSCR4, increased the expression of PLSCR4 by adsorbing miR-424-5p. Therefore, LINC00641 as a tumor suppressor may play a role in the treatment of non-small cell lung cancer. But there is also the opposite voice. Dong et al. (64) comprehensively analyzed the expression of mRNAs, lncRNAs and miRNAs by using the data of 509 lung adenocarcinoma, 473 lung squamous cell carcinoma tissues and 49 adjacent non-cancerous lung tissues in TCGA database and constructed the ceRNA network mechanism of lncRNA-miRNA-mRNA. It is predicted that LINC00641, as an oncogene, can competitively combine miR-1285-3p to regulate the expression of downstream target genes PIGA, AHCYL1 and ATP1B2, or combine miR-6860 to regulate the expression of MAP3K3, SHRK004M and FGD3, so as to affect the progression of lung cancer. Comparing the two literatures, the former expounds the antitumor effect of LINC00641 in non-small cell lung cancer at the cellular level but lacks tissue validation and in vivo experiments. While the latter predicts LINC00641 as an oncogene in lung adenocarcinoma and squamous cell carcinoma, but it is only a biological information prediction and lacks experimental evidence. In view of the opposite results in the two literatures, only by further expanding the sample size and perfecting the histological verification can we obtain a more accurate mechanism of LINC00641 in non-small cell lung cancer.
Glioma
Glioma is the most common primary intracranial tumor, accounting for 81% of malignant brain tumors, which can lead to significant mortality and morbidity (65). Progress and reports have been made in the treatment of glioma from aspects of molecular genetics (66), epigenetics (67) and cell metabolism (68). As for silent messenger lncRNAs, based on the increasing number of functional lncRNAs abnormally expressed in glioma tissues and cell lines, lncRNAs may be crucial for the occurrence, progression and other malignant phenotypes of gliomas (69). Yang et al. (43) found that the expression of LINC00641 decreased significantly in glioma and cell lines and overexpression of LINC00641 in glioma cells inhibited cell proliferation and promoted apoptosis. When further exploring the mechanism of LINC00641, it was found that the expression of NRGN was down-regulated in glioma and positively correlated with LINC00641. According to the lnclocator database, LINC00641 is mainly located in the cytoplasm, and the regulation mechanism of lncRNA in the cytoplasm is mostly ceRNA. Combined with Starbase v3.0 prediction, miR-4262 was found to bind to both LINC00641 and NRGN. Further experiments demonstrated that LINC00641 enhanced the expression of NRGN in glioma cells through the absorption of miR-4262. In conclusion, LINC00641 competitively binds miR-4262 through the ceRNA mechanism to release NRGN, which plays an anti-tumor role in the progression of glioma, providing a new therapeutic target for glioma patients. Zhang et al. (70) obtained clinical information from the Chinese glioma Genome Atlas (CGGA) and Cancer Genome Atlas (TCGA) databases, identified five lncRNAs through weighted gene coexpression network analysis (WGCNA) to construct Cox regression model, which showed high accuracy in predicting the survival rate of glioma patients.
Cutaneous Squamous Cell Carcinoma
Cutaneous squamous cell carcinoma is the second most common non melanoma skin cancer, accounting for 20% of skin cancers (71). A variety of LncRNAs have been reported to be abnormally expressed in skin squamous cell carcinoma (72), participating in the complex cancer signal network in skin malignant tumors (73). Studying the signal pathway of tumor development can generate targeted therapy molecules. Liu et al. (44) found that the expression of LINC00641 was significantly reduced in cutaneous squamous cell carcinoma cell lines compared with normal human immortalized keratinocytes. Overexpression of LINC00641 reduced cell proliferation, migration and invasion. In vivo experiments also proved that overexpression of LINC00641 inhibited tumor formation in nude mice. Starbase predicted that LINC00641 could target miR-424 in the mechanism research. LINC00641 can be paired with miR-424. Both in vivo and in vitro experiments showed that the overexpression of miR-424 reversed the inhibitory effect of LINC00641 overexpression on the proliferation, invasion and migration of cutaneous squamous cell carcinoma. Therefore, LINC00641 can inhibit the development of cutaneous squamous cell carcinoma by down regulating miR-424 in cellular and animal studies, which is of great significance for the pathogenesis and therapeutic targets of squamous cell carcinoma.
Acute Myeloid Leukemia
Acute myeloid leukemia (AML) is the most common acute leukemia in adults, accounting for approximately 80% of the cases in this group, with incidence increasing with age (74). The detection of cytogenetic markers and other molecular markers such as point mutation, epigenetic and proteomic profiles are playing an emerging role in disease risk prediction, diagnosis and prognosis (75). Of course, there are many lncRNAs, which play an important role in hematopoietic cell transformation, disease progression and drug resistance (76). Wang et al. (40) found that the expression of LINC00641 increased in acute myeloid leukemia tissues and cell lines compared with normal tissues and cells. In leukemia cell lines, silencing LINC00641 inhibited the proliferation, migration and invasion of cancer cells, caused cell cycle arrest and induced apoptosis. When exploring the mechanism of LINC00641 in acute myeloid leukemia, LINC00641 was first located in the cytoplasm, and most LncRNAs in the cytoplasm were ceRNA mechanisms. Then, Starbase predicted that LINC00641 had a binding site with miR-378a. It was demonstrated that LINC00641 regulates the malignant biological behavior of leukemia cells through sponge miR-378a. Finally, ZBTB20 was predicted to be the target gene of miR-378a through miRDB, TargetScan and Starbase databases. Cell experiments verified that LINC00641 competitively combined with miR-378a to increase the expression of ZBTB20, thereby affecting the biological behavior of leukemia cells. In conclusion, LINC00641 acts through miR-378a/ZBTB20 signaling pathway and is expected to become a therapeutic target for acute myeloid leukemia.
Gastric Cancer
Gastric cancer is the fifth most common cancer and the third leading cause of cancer death in the world. The risk factors of the disease include Helicobacter pylori infection, age, high salt intake, etc. (77). The most common tumor markers clinically used for early detection of gastric cancer include CEA, CA19-9 and CA72-4, but the positive rate of these three is about 20-30% (78). Due to low specificity and sensitivity, more and more studies have been conducted on the molecular mechanism of gastric cancer. Among them, lncRNA shows clinical potential in biomarkers, diagnosis and prognosis of gastric cancer (79). Hu et al. (45) found that LINC00641 was highly expressed in 173 gastric cancer tissues compared with adjacent tissues and could be used as a biomarker to predict the overall survival rate of gastric cancer patients. In vitro experiments demonstrated that LINC00641 promoted cell proliferation and migration by inhibiting miR-582-5p. Meanwhile, compared with normal gastric cancer cell lines, the expression of LINC00641 in oxaliplatin resistant cell lines was increased, while the expression of miR-582-5p was decreased. When further exploring the mechanism of drug resistance, it was found that after down regulating LINC00641, the expression of LC3II in oxaliplatin resistant cells was inhibited, the expression of LC3 I and p62 was enhanced, and autophagy was inhibited, making gastric cancer cells more sensitive to oxaliplatin. Combined with the down-regulation of miR-582-5p can reverse the above expression, so it is concluded that LINC00641 can regulate oxaliplatin resistance by inhibiting the expression of miR-582-5p and inducing autophagy enhancement. LINC00641 and miR-582-5p participate in the regulation of oxaliplatin resistance by altering autophagy in gastric adenocarcinoma, providing a direction for the treatment of gastric cancer. Hang et al. (38) also found that silencing LINC00641 in gastric cancer cell line reduced the proliferation, migration and invasion of cancer cells and induced apoptosis. After down regulating LINC00641 in cells, the expression of miR-429 increased. Combined with bioinformatics prediction, Notch-1 may be a potential target of miR-429. Finally, the experiment proved that LINC00641 increased the expression of Notch-1 by competitive binding to miR-429 and increased the biological behaviors of gastric cancer cell proliferation, migration and invasion. In conclusion, LINC00641 can function in gastric cancer by targeting miR-429/Notch-1 axis. Due to its carcinogenic function, LINC00641 can be used for future clinical application in the treatment of gastric cancer.
Colorectal Cancer
Colorectal cancer is the third most common cancer, which causes nearly 700000 deaths annually, with the highest incidence rate in developed countries, and is the fourth most common cause of cancer-related death (80, 81). LncRNAs play an important role in the growth and metastasis of colorectal cancer, especially as competitive endogenous RNAs (ceRNAs), targeting miRNAs to regulate downstream target genes of colorectal cancer (82). Xue et al. (46) found that the expression of LINC00641 in rectal cancer tissues and cells was higher than that in normal rectal tissues and cells. When LINC00641 was knocked down, the proliferation and migration of rectal cancer cells decreased. Bioinformation predicted that miRNA-424-5p was the downstream target of LINC00641, and cell assay proved that the expression of miRNA-424-5p was increased after LINC00641 knockdown. PLSCR4 was predicted to be the downstream target gene of miRNA-424-5p. After knocking down miRNA-424-5p in rectal cancer cells, PLSCR4 expression was up-regulated, and PLSCR4 was positively correlated with LINC00641. In conclusion, LINC00641 competitively binds miRNA-424-5p to relieve the inhibition of PLSCR4. LINC00641 plays a carcinogenic role in rectal cancer through miRNA-424-5p/PLSCR4 axis, which can provide a new target for the treatment of rectal cancer.
Renal Cell Carcinoma
Globally, renal cell carcinoma is the ninth most common cancer, with rates varying geographically, and the incidence rate of developed countries is the highest (83). To date, there are no screening procedures or reliable biomarkers for the early diagnosis of renal cell carcinoma (84). Datas about lncRNA as a diagnostic biomarker of renal cell carcinoma are limited (85). However, some serum and urine biomarkers have been proposed as potential screening tools. Circulating lncRNA, as an easily accessible blood based biomarker, has been recommended for urogenital malignancies, such as prostate, bladder and kidney cancer (86). Zhang et al. (37) detected 48 cases of renal cell carcinoma and adjacent normal tissues and found that the expression of LINC00641 was increased in renal cell carcinoma, which was related to stage and prognosis. Knockdown of LINC00641 in renal cell carcinoma cell line decreased cell proliferation, invasion and increased apoptosis. It is predicted by Starbase that miR-340-5p can be used as a potential target of LINC00641. Cell experiments demonstrated the interaction between LINC00641 and miR-340-5p, and the expression of miR-340-5p was up-regulated after silencing LINC00641. Inhibition of miR-340-5p could eliminate the pro-apoptotic and anti-invasive effects of LINC00641 silencing on renal cell carcinoma cell line but had no significant effect on cell proliferation. In vivo experiments showed that inhibition of LINC00641 reduced tumor volume and increased miR-340-5p expression in nude mice. In conclusion, LINC00641, as a carcinogen of renal cell carcinoma, plays a role by targeting mir-340-5p through ceRNA mechanism, and can be used as a potential target for the treatment of renal cell carcinoma.
Thyroid Cancer
Thyroid cancer is a common endocrine malignancy tumor. Its incidence rate continues to rise worldwide, and remains one of the lowest mortality rates of human cancers (87). LncRNA is now considered to be an important regulatory molecule involved in the progression of thyroid cancer (88). Rao et al. (89) obtained samples from Co-LncRNA database, including 12 normal samples and 83 thyroid cancer samples, and identified differentially expressed LncRNAs by linear model of microarray analysis (limma), and finally identified 6 up-regulated and 85 down-regulated lncrnas. It is predicted that LINC00641 is down-regulated in thyroid cancer and is associated with longer disease-free survival, but it is still lack of experimental verification.
Clinical Significance of LINC00641
Serum cancer biomarkers are limited and nonspecific, with limitations (90). Therefore, exploring more targets will contribute to the early diagnosis, treatment and prognosis of cancer. The mechanism of LINC00641 in cancer has been concentrated in recent 5 years, which can be used as an emerging biomarker of cancer prognosis.
As a Biomarker of Cancer Prognosis
Li (35) divided 39 bladder cancer tissues into LINC00641 high group and LINC00641 low group according to the median LINC00641 level. Kaplan-Meier curve analysis showed that low expression of LINC00641 was associated with reduced overall survival and progression free survival. Mao (42) analyzed the expression of LINC00641 in 166 breast cancer tissues, and statistical analysis showed that LINC00641 was significantly correlated with tumor size, lymph node metastasis and clinical stage (P < 0.01). Liu (36) analyzed 23 prostate cancer patients and found that the low LINC00641 expression group had a lower survival rate than the high LINC00641 expression group (P < 0.05). Hu (38) detected the expression of LINC00641 in 173 cases of gastric cancer, which was related to M stage (P < 0.01). Cox regression model including LINC00641, miR-582-5p, CEA and age could predict the 1-year and 3-year survival rates of gastric cancer patients (C index was 0.876 ± 0.06). Xue’s study (46) included 50 cases of rectal cancer and made a survival curve, and found that the high LINC00641 expression group had worse survival (P < 0.05). Zhang (37) analyzed 48 cases of renal cell carcinoma and found that the expression of LINC00641 was related to clinical T stage (P < 0.01) and metastasis (P < 0.05), and Kaplan-Meier survival analysis showed that the high LINC00641 expression group had poor prognosis (P < 0.05). In conclusion, the differential expression of LINC00641 is related to survival time and can be used as a biomarker of cancer prognosis, such as combining with known molecular biomarkers of cancer, to increase the reliability of predictive value.
As a Target for Cancer Treatment
As a tumor suppressor gene: the over expression of LINC00641 in bladder cancer cell lines reduces cell proliferation, migration and invasion ability (35). In vivo, LINC00641 overexpression significantly inhibited tumor volume. In cervical cancer cell lines, up-regulation of LINC00641 inhibits cell proliferation, increases apoptosis, reduces invasion and migration ability, and inhibits EMT (41). In breast cancer cell lines, overexpression of LINC00641 inhibits cell proliferation, migration and invasion, stagnates cells in G1 phase and promotes apoptosis (42). In prostate cancer cell lines, over expression of LINC00641 inhibits cell proliferation, invasion and promotes apoptosis (36). In non-small cell lung cancer cell lines, overexpression of LINC00641 reduces the ability of proliferation and migration and increases apoptosis (39). Overexpression of LINC00641 in glioma cells inhibits cell proliferation and promotes apoptosis (43). In cutaneous squamous cell carcinoma, LINC00641 overexpression decreased cell proliferation, migration and invasion ability in vitro, and the tumor volume of LINC00641 overexpression group was smaller in nude mouse tumorigenesis experiment (44).
As an oncogene: in leukemia cell lines, silencing LINC00641 inhibits the proliferation, migration and invasion of cancer cells, causes cell cycle arrest and induces apoptosis (40). Silencing LINC00641 reduces the proliferation, migration and invasion of cancer cells and induces apoptosis in gastric cancer cell lines (38), and LINC00641 participates in regulating oxaliplatin resistance by changing autophagy in gastric adenocarcinoma, providing a direction for the treatment of gastric cancer (45). When LINC00641 was knocked down, the proliferation and migration of rectal cancer cells decreased (46). Knockdown LINC00641 in renal cell carcinoma cell line decreased cell proliferation, invasion and increased apoptosis (37). Therefore, LINC00641 can change the sensitivity of tumor to drugs by regulating the biological behavior of tumor cell proliferation and invasion, and it is a promising target for cancer treatment.
Many LncRNAs have been shown to be potential biomarkers and targets for cancer diagnosis and treatment (91). The expression characteristics and clinical significance of LncRNA in tumors have been proved by in vivo and in vitro experiments, but it still needs some time to be applied in clinical practice. One of the difficulties is that it is currently impossible to determine the sequences and structural elements that allow long-chain non-coding RNA molecules to perform their cellular functions (92). Secondly, new effective and stable genome editing strategies, as well as more effective and less toxic gene therapy delivery systems, need to be developed before LncRNAs can become potential therapies (80). Moreover, the expression of LncRNA is usually low, which increases the difficulty of detection. Although a variety of methods for detecting cancer biomarkers have been created, most of them are only conceptual demonstrations, which benefit from highly optimized laboratory conditions (93). However, with the rapid improvement of experimental technology, exploring LncRNA as a biomarker of cancer is of great significance to clarify the diagnosis of cancer and improve the prognosis of cancer.
Conclusions, Limitations and Future Prospects
The mechanism of LINC00641 in different tumors has been summarized above. Current studies focus on the ceRNA mechanism. LINC00641 was predicted in the database to be 83% localized in the cytoplasm, conforming to the localization of ceRNA mechanism. Through the database prediction and verification in cell experiments, LINC00641 can be used as miR-197-3p、miR-378a-3p、miR-194-5p、miR-365a-3p、miR-424-5p、miR-4262、miR-424、miR-378a、miR-582-5p、miR-429、miRNA-424-5p、microRNA-340-5p sponges (Figure 4). LINC00641 is involved in tumorigenesis in more than one way, so it may be difficult to use as a therapeutic target for different tissues. In a review of the mechanism of lncRNA in the cytoplasm, ceRNA has been studied the most. LncRNA mediated ceRNA network plays a key role in the proliferation, invasion, apoptosis and autophagy of varieties of tumors, as well as distant metastasis, epithelial mesenchymal transformation and chemoresistance. The inclusion of abnormally expressed lncRNA, miRNA and mRNA in tumors and the construction of ceRNA network help to clarify the regulatory mechanism of lncRNA as ceRNA to promote tumorigenesis, and further evaluate the potential therapeutic targets and prognostic biomarkers of cancer (82, 94–96). At present, there is still much room for exploring the research mechanism of LINC00641. For example, LncRNA containing Alu element can mediate mRNA decay in the cytoplasm by recruiting STAU1 protein (97), and LncRNA can also act as RBP molecular bait involved in mRNA decay. For example, lncRNA OCC-1 can regulate the level of a large number of mRNA at the post transcriptional level by regulating the stability of RBP HuR (98). Secondly, LncRNA can also regulate translation or post-translational modification in the cytoplasm. For example, lincRNA-p21 plays a post transcriptional function as a translation regulator (99). Post translational modification mediated by LncRNA is the hub of many cellular signaling pathways. LncRNA can act on post-translational modification of metabolic enzymes, transcription factors or other proteins involved in energy metabolism pathways, including ubiquitination and acetylation, which provides a new idea for cancer energy metabolism (100). In addition to the above mechanism of action in the cytoplasm, although LINC00641 does not account for a high proportion in the nucleus, the role of LINC00641 in the nucleus can be further explored. Such a large number of long chain non-coding RNAs can interact with genes, non-coding genes and proteins to build a crisscross network in the abnormal regulation of cancer.
The above study of LINC00641 was performed in tissue cells. It is an invasive operation to detect the gene expression level in solid tumors, which needs to be obtained from surgical or biopsy specimens. Liquid biopsy, which detects cancer genes from blood or body fluids, has attracted more and more attention (101). LncRNA can be secreted out of cells by exosomes and regulate the occurrence and progression of cancer by changing the tumor microenvironment (102). Exosome mediated LncRNA can regulate malignant biological behaviors such as tumor apoptosis, invasion and migration, promote lymph node metastasis of cancer and affect the drug resistance of tumor drugs (103–106). In conclusion, exosomal LncRNA can be regarded as a novel biomarker of cancer progression and a potential therapeutic target for clinical application (107).
At present, the research data on LINC00641 in tumors are limited. In view of the numerous mechanisms of LncRNA, we look forward to more research on LINC00641 in tumors, enrich the significance of LINC00641 and contribute to its early clinical application.
Author Contributions
Conceptualization: XH and SZ. Methodology: SZ. Data curation: XH and SZ. Writing—original draft preparation: XH. Writing—review and editing: XH and SZ. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Stratton MR, Campbell PJ, Futreal PA. The Cancer Genome. Nature (2009) 458(7239):719–24. doi: 10.1038/nature07943
2. Dawson MA, Kouzarides T. Cancer Epigenetics: From Mechanism to Therapy. Cell (2012) 150(1):12–27. doi: 10.1016/j.cell.2012.06.013
3. Casey SC, Amedei A, Aquilano K, Azmi AS, Benencia F, Bhakta D, et al. Cancer Prevention and Therapy Through the Modulation of the Tumor Microenvironment. Semin Cancer Biol (2015) 35 Suppl:S199–223. doi: 10.1016/j.semcancer.2015.02.007
4. Miller JF, Sadelain M. The Journey From Discoveries in Fundamental Immunology to Cancer Immunotherapy. Cancer Cell (2015) 27(4):439–49. doi: 10.1016/j.ccell.2015.03.007
5. Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab (2016) 23(1):27–47. doi: 10.1016/j.cmet.2015.12.006
6. Roy S, Trinchieri G. Microbiota: A Key Orchestrator of Cancer Therapy. Nat Rev Cancer (2017) 17(5):271–85. doi: 10.1038/nrc.2017.13
7. International Human Genome Sequencing, C. Finishing the Euchromatic Sequence of the Human Genome. Nature (2004) 431(7011):931–45. doi: 10.1038/nature03001
8. Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell (2009) 136(4):629–41. doi: 10.1016/j.cell.2009.02.006
9. Wei JW, Huang K, Yang C, Kang CS. Non-Coding RNAs as Regulators in Epigenetics (Review). Oncol Rep (2017) 37(1):3–9. doi: 10.3892/or.2016.5236
10. Dykes IM, Emanueli C. Transcriptional and Post-Transcriptional Gene Regulation by Long Non-Coding RNA. Genomics Proteomics Bioinf (2017) 15(3):177–86. doi: 10.1016/j.gpb.2016.12.005
11. Wang KC, Chang HY. Molecular Mechanisms of Long Noncoding RNAs. Mol Cell (2011) 43(6):904–14. doi: 10.1016/j.molcel.2011.08.018
12. Yao RW, Wang Y, Chen LL. Cellular Functions of Long Noncoding RNAs. Nat Cell Biol (2019) 21(5):542–51. doi: 10.1038/s41556-019-0311-8
13. Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, et al. LncRNA Khps1 Regulates Expression of the Proto-Oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol Cell (2015) 60(4):626–36. doi: 10.1016/j.molcel.2015.10.001
14. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long Non-Coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature (2010) 464(7291):1071–6. doi: 10.1038/nature08975
15. Li XL, Subramanian M, Jones MF, Chaudhary R, Singh DK, Zong X, et al. Long Noncoding RNA PURPL Suppresses Basal P53 Levels and Promotes Tumorigenicity in Colorectal Cancer. Cell Rep (2017) 20(10):2408–23. doi: 10.1016/j.celrep.2017.08.041
16. Jing F, Ruan X, Liu X, Yang C, Wang D, Zheng J, et al. The PABPC5/HCG15/ZNF331 Feedback Loop Regulates Vasculogenic Mimicry of Glioma via STAU1-Mediated mRNA Decay. Mol Ther Oncolytics (2020) 17:216–31. doi: 10.1016/j.omto.2020.03.017
17. Chen R, Liu Y, Zhuang H, Yang B, Hei K, Xiao M, et al. Quantitative Proteomics Reveals That Long Non-Coding RNA MALAT1 Interacts With DBC1 to Regulate P53 Acetylation. Nucleic Acids Res (2017) 45(17):9947–59. doi: 10.1093/nar/gkx600
18. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al. A Cytoplasmic NF-kappaB Interacting Long Noncoding RNA Blocks IkappaB Phosphorylation and Suppresses Breast Cancer Metastasis. Cancer Cell (2015) 27(3):370–81. doi: 10.1016/j.ccell.2015.02.004
19. Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res (2017) 77(15):3965–81. doi: 10.1158/0008-5472.CAN-16-2634
20. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell (2016) 29(4):452–63. doi: 10.1016/j.ccell.2016.03.010
21. Cao HL, Liu ZJ, Huang PL, Yue YL, Xi JN. lncRNA-RMRP Promotes Proliferation, Migration and Invasion of Bladder Cancer via miR-206. Eur Rev Med Pharmacol Sci (2019) 23(3):1012–21. doi: 10.26355/eurrev_201902_16988
22. Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR Influences Cell Growth, Migration, Invasion, and Apoptosis via the miR-20a-5p/HMGA2 Axis in Breast Cancer. Cancer Med (2018) 7(3):842–55. doi: 10.1002/cam4.1353
23. Fu C, Li D, Zhang X, Liu N, Chi G, Jin X. LncRNA PVT1 Facilitates Tumorigenesis and Progression of Glioma via Regulation of MiR-128-3p/GREM1 Axis and BMP Signaling Pathway. Neurotherapeutics (2018) 15(4):1139–57. doi: 10.1007/s13311-018-0649-9
24. Zhang A, Xu M, Mo YY. Role of the lncRNA-P53 Regulatory Network in Cancer. J Mol Cell Biol (2014) 6(3):181–91. doi: 10.1093/jmcb/mju013
25. Gupta SC, Awasthee N, Rai V, Chava S, Gunda V, Challagundla KB. Long Non-Coding RNAs and Nuclear factor-kappaB Crosstalk in Cancer and Other Human Diseases. Biochim Biophys Acta Rev Cancer (2020) 1873(1):188316. doi: 10.1016/j.bbcan.2019.188316
26. Wu Y, Zhang Y, Qin X, Geng H, Zuo D, Zhao Q. PI3K/AKT/mTOR Pathway-Related Long Non-Coding RNAs: Roles and Mechanisms in Hepatocellular Carcinoma. Pharmacol Res (2020) 160:105195. doi: 10.1016/j.phrs.2020.105195
27. Yang G, Shen T, Yi X, Zhang Z, Tang C, Wang L, et al. Crosstalk Between Long non-Coding RNAs and Wnt/beta-Catenin Signalling in Cancer. J Cell Mol Med (2018) 22(4):2062–70. doi: 10.1111/jcmm.13522
28. Reicher A, Foßelteder J, Kwong LN, Pichler M. Crosstalk Between the Notch Signaling Pathway and Long non-Coding RNAs. Cancer Lett (2018) 420:91–6. doi: 10.1016/j.canlet.2018.01.070
29. Peng WX, Koirala P, Mo YY. LncRNA-Mediated Regulation of Cell Signaling in Cancer. Oncogene (2017) 36(41):5661–7. doi: 10.1038/onc.2017.184
30. Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci (2018) 19(5):1310. doi: 10.3390/ijms19051310
31. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in Cancer: Possible Functions and Clinical Implications. J Med Genet (2015) 52(10):710–8. doi: 10.1136/jmedgenet-2015-103334
32. Jiang J, Bi Y, Liu XP, Yu D, Yan X, Yao J, et al. To Construct a ceRNA Regulatory Network as Prognostic Biomarkers for Bladder Cancer. J Cell Mol Med (2020) 24(9):5375–86. doi: 10.1111/jcmm.15193
33. Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, et al. STAT3-Mediated Upregulation of lncRNA HOXD-AS1 as a ceRNA Facilitates Liver Cancer Metastasis by Regulating SOX4. Mol Cancer (2017) 16(1):136. doi: 10.1186/s12943-017-0680-1
34. Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the Human Tissue-Specific Expression by Genome-Wide Integration of Transcriptomics and Antibody-Based Proteomics. Mol Cell Proteomics (2014) 13(2):397–406. doi: 10.1074/mcp.M113.035600
35. Li Z, Hong S, Liu Z. LncRNA LINC00641 Predicts Prognosis and Inhibits Bladder Cancer Progression Through miR-197-3p/KLF10/PTEN/PI3K/AKT Cascade. Biochem Biophys Res Commun (2018) 503(3):1825–9. doi: 10.1016/j.bbrc.2018.07.120
36. Liu WH, Lu JJ, Yu RK, Zhou L, Yu Q, Li DF, et al. LINC00641 Regulates Prostate Cancer Cell Growth and Apoptosis via the miR-365a-3p/VGLL4 Axis. Eur Rev Med Pharmacol Sci (2021) 25(1):108–15. doi: 10.26355/eurrev_202101_24354
37. Zhang J, Jin S, Xiao W, Zhu X, Jia C, Lin Z. Long Noncoding RNA LINC00641 Promotes Renal Cell Carcinoma Progression via Sponging microRNA-340-5p. Cancer Cell Int (2021) 21(1):210. doi: 10.1186/s12935-021-01895-y
38. Hang Q, Lu J, Zuo L, Liu M. Linc00641 Promotes the Progression of Gastric Carcinoma by Modulating the miR-429/Notch-1 Axis. Aging (Albany NY) (2021) 13(6):8497–509. doi: 10.18632/aging.202661
39. Li Y, Zhao L, Zhao P, Liu Z. Long non-Coding RNA LINC00641 Suppresses non-Small-Cell Lung Cancer by Sponging miR-424-5p to Upregulate PLSCR4. Cancer Biomark (2019) 26(1):79–91. doi: 10.3233/CBM-190142
40. Wang J, Liu ZH, Yu LJ. Long non-Coding RNA LINC00641 Promotes Cell Growth and Migration Through Modulating miR-378a/ZBTB20 Axis in Acute Myeloid Leukemia. Eur Rev Med Pharmacol Sci (2019) 23(17):7498–509. doi: 10.26355/eurrev_201909_18864
41. Zhang Y, Yu R, Li L. LINC00641 Hinders the Progression of Cervical Cancer by Targeting miR-378a-3p/CPEB3. J Gene Med (2020) 22(9):e3212. doi: 10.1002/jgm.3212
42. Mao Q, Lv M, Li L, Sun Y, Liu S, Shen Y, et al. Long Intergenic Noncoding RNA 00641 Inhibits Breast Cancer Cell Proliferation, Migration, and Invasion by Sponging miR-194-5p. J Cell Physiol (2020) 235(3):2668–75. doi: 10.1002/jcp.29170
43. Yang J, Yu D, Liu X, Changyong E, Yu S. LINC00641/miR-4262/NRGN Axis Confines Cell Proliferation in Glioma. Cancer Biol Ther (2020) 21(8):758–66. doi: 10.1080/15384047.2020.1776581
44. Liu W, Liu X. LINC00641 Inhibits the Development of Cutaneous Squamous Cell Carcinoma By Downregulating miR-424 in A431 Cells. Cancer Biother Radiopharm (2021). doi: 10.1089/cbr.2020.4325
45. Hu Y, Su Y, Lei X, Zhao H, Wang L, Xu T, et al. LINC00641/miR-582-5p Mediate Oxaliplatin Resistance by Activating Autophagy in Gastric Adenocarcinoma. Sci Rep (2020) 10(1):14981. doi: 10.1038/s41598-020-70913-2
46. Xue D, Xue YF, Zhang LJ, Cui LZ, Guo KQ, Lian J. LINC00641 Induces the Malignant Progression of Colorectal Carcinoma Through the miRNA-424-5p/PLSCR4 Feedback Loop. Eur Rev Med Pharmacol Sci (2021) 25(2):749–57. doi: 10.26355/eurrev_202101_24636
47. Lenis AT, Lec PM, Chamie K, Mshs MD. Bladder Cancer: A Review. JAMA (2020) 324(19):1980–91. doi: 10.1001/jama.2020.17598
48. Richters A, Aben KKH, Kiemeney L. The Global Burden of Urinary Bladder Cancer: An Update. World J Urol (2020) 38(8):1895–904. doi: 10.1007/s00345-019-02984-4
50. Atun R, Jaffray DA, Barton MB, Bray F, Baumann M, Vikram B, et al. Expanding Global Access to Radiotherapy. Lancet Oncol (2015) 16(10):1153–86. doi: 10.1016/S1470-2045(15)00222-3
51. Gaffney DK, Hashibe M, Kepka D, Maurer KA, Werner TL. Too Many Women are Dying From Cervix Cancer: Problems and Solutions. Gynecol Oncol (2018) 151(3):547–54. doi: 10.1016/j.ygyno.2018.10.004
52. Cancer Genome Atlas Research Network, Albert Einstein College of Medicine, Analytical Biological Services, Barretos Cancer Hospital, Baylor College of Medicine, Beckman Research Institute of City of Hope, et al. Integrated Genomic and Molecular Characterization of Cervical Cancer. Nature (2017) 543(7645):378–84. doi: 10.1038/nature21386
53. Fahad Ullah M. Breast Cancer: Current Perspectives on the Disease Status. Adv Exp Med Biol (2019) 1152:51–64. doi: 10.1007/978-3-030-20301-6_4
54. Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, et al. American Society of Clinical Oncology 2007 Update of Recommendations for the Use of Tumor Markers in Breast Cancer. J Clin Oncol (2007) 25(33):5287–312. doi: 10.1200/JCO.2007.14.2364
55. Liang Y, Zhang H, Song X, Yang Q. Metastatic Heterogeneity of Breast Cancer: Molecular Mechanism and Potential Therapeutic Targets. Semin Cancer Biol (2020) 60:14–27. doi: 10.1016/j.semcancer.2019.08.012
56. Yang F, Xiao Z, Zhang S. Knockdown of miR-194-5p Inhibits Cell Proliferation, Migration and Invasion in Breast Cancer by Regulating the Wnt/beta-Catenin Signaling Pathway. Int J Mol Med (2018) 42(6):3355–63. doi: 10.3892/ijmm.2018.3897
57. Nguyen-Nielsen M, Borre M. Diagnostic and Therapeutic Strategies for Prostate Cancer. Semin Nucl Med (2016) 46(6):484–90. doi: 10.1053/j.semnuclmed.2016.07.002
58. Rao AR, Motiwala HG, Karim OM. The Discovery of Prostate-Specific Antigen. BJU Int (2008) 101(1):5–10. doi: 10.1111/j.1464-410X.2007.07138.x
59. Kretschmer A, Tilki D. Biomarkers in Prostate Cancer - Current Clinical Utility and Future Perspectives. Crit Rev Oncol Hematol (2017) 120:180–93. doi: 10.1016/j.critrevonc.2017.11.007
60. Huang H, Tang Y, Ye X, Chen W, Xie H, Chen S. The Influence of lncRNAs on the Prognosis of Prostate Cancer Based on TCGA Database. Transl Androl Urol (2021) 10(3):1302–13. doi: 10.21037/tau-21-154
61. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global Cancer Statistics, 2012. CA Cancer J Clin (2015) 65(2):87–108. doi: 10.3322/caac.21262
62. Herbst RS, Morgensztern D, Boshoff C. The Biology and Management of non-Small Cell Lung Cancer. Nature (2018) 553(7689):446–54. doi: 10.1038/nature25183
63. Xie Y, Zhang Y, Du L, Jiang X, Yan S, Duan W, et al. Circulating Long Noncoding RNA Act as Potential Novel Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer. Mol Oncol (2018) 12(5):648–58. doi: 10.1002/1878-0261.12188
64. Dong R, Liu J, Sun W, Ping W. Comprehensive Analysis of Aberrantly Expressed Profiles of lncRNAs and miRNAs With Associated ceRNA Network in Lung Adenocarcinoma and Lung Squamous Cell Carcinoma. Pathol Oncol Res (2020) 26(3):1935–45. doi: 10.1007/s12253-019-00780-4
65. Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, et al. The Epidemiology of Glioma in Adults: A "State of the Science" Review. Neuro Oncol (2014) 16(7):896–913. doi: 10.1093/neuonc/nou087
66. Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the Molecular Genetics of Gliomas - Implications for Classification and Therapy. Nat Rev Clin Oncol (2017) 14(7):434–52. doi: 10.1038/nrclinonc.2016.204
67. Gusyatiner O, Hegi ME. Glioma Epigenetics: From Subclassification to Novel Treatment Options. Semin Cancer Biol (2018) 51:50–8. doi: 10.1016/j.semcancer.2017.11.010
68. Bi J, Chowdhry S, Wu S, Zhang W, Masui K, Mischel PS. Altered Cellular Metabolism in Gliomas - an Emerging Landscape of Actionable Co-Dependency Targets. Nat Rev Cancer (2020) 20(1):57–70. doi: 10.1038/s41568-019-0226-5
69. Peng Z, Liu C, Wu M. New Insights Into Long Noncoding RNAs and Their Roles in Glioma. Mol Cancer (2018) 17(1):61. doi: 10.1186/s12943-018-0812-2
70. Zhang C, Liu H, Xu P, Tan Y, Xu Y, Wang L, et al. Identification and Validation of a five-lncRNA Prognostic Signature Related to Glioma Using Bioinformatics Analysis. BMC Cancer (2021) 21(1):251. doi: 10.1186/s12885-021-07972-9
71. Waldman A, Schmults C. Cutaneous Squamous Cell Carcinoma. Hematol Oncol Clin North Am (2019) 33(1):1–12. doi: 10.1016/j.hoc.2018.08.001
72. Wang Y, Sun B, Wen X, Hao D, Du D, He G, et al. The Roles of lncRNA in Cutaneous Squamous Cell Carcinoma. Front Oncol (2020) 10:158. doi: 10.3389/fonc.2020.00158
73. Piipponen M, Nissinen L, Kahari VM. Long non-Coding RNAs in Cutaneous Biology and Keratinocyte Carcinomas. Cell Mol Life Sci (2020) 77(22):4601–14. doi: 10.1007/s00018-020-03554-3
74. De Kouchkovsky I, Abdul-Hay M. 'Acute Myeloid Leukemia: A Comprehensive Review and 2016 Update'. Blood Cancer J (2016) 6(7):e441. doi: 10.1038/bcj.2016.50
75. Prada-Arismendy J, Arroyave JC, Rothlisberger S. Molecular Biomarkers in Acute Myeloid Leukemia. Blood Rev (2017) 31(1):63–76. doi: 10.1016/j.blre.2016.08.005
76. Cruz-Miranda GM, Hidalgo-Miranda A, Bárcenas-López DA, Núñez-Enríquez JC, Ramírez-Bello J, Mejía-Aranguré JM, et al. Long Non-Coding RNA and Acute Leukemia. Int J Mol Sci (2019) 20(3):735. doi: 10.3390/ijms20030735
77. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric Cancer. Lancet (2020) 396(10251):635–48. doi: 10.1016/S0140-6736(20)31288-5
78. Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical Significance of Serum Tumor Markers for Gastric Cancer: A Systematic Review of Literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer (2014) 17(1):26–33. doi: 10.1007/s10120-013-0259-5
79. Yuan L, Xu ZY, Ruan SM, Mo S, Qin JJ, Cheng XD. Long Non-Coding RNAs Towards Precision Medicine in Gastric Cancer: Early Diagnosis, Treatment, and Drug Resistance. Mol Cancer (2020) 19(1):96. doi: 10.1186/s12943-020-01219-0
80. Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int J Mol Sci (2017) 18(1):197. doi: 10.3390/ijms18010197
82. Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long Noncoding RNA (lncRNA)-Mediated Competing Endogenous RNA Networks Provide Novel Potential Biomarkers and Therapeutic Targets for Colorectal Cancer. Int J Mol Sci (2019) 20(22):5758. doi: 10.3390/ijms20225758
83. Jonasch E, Gao J, Rathmell WK. Renal Cell Carcinoma. BMJ (2014) 349:g4797. doi: 10.1136/bmj.g4797
84. Rossi SH, Klatte T, Usher-Smith J, Stewart GD. Epidemiology and Screening for Renal Cancer. World J Urol (2018) 36(9):1341–53. doi: 10.1007/s00345-018-2286-7
85. Outeiro-Pinho G, Barros-Silva D, Correia MP, Henrique R, Jerónimo C. Renal Cell Tumors: Uncovering the Biomarker Potential of ncRNAs. Cancers (Basel) (2020) 12(8):2214. doi: 10.3390/cancers12082214
86. Barth DA, Drula R, Ott L, Fabris L, Slaby O, Calin GA, et al. Circulating Non-Coding RNAs in Renal Cell Carcinoma-Pathogenesis and Potential Implications as Clinical Biomarkers. Front Cell Dev Biol (2020) 8:828. doi: 10.3389/fcell.2020.00828
87. Roman BR, Morris LG, Davies L. The Thyroid Cancer Epidemic, 2017 Perspective. Curr Opin Endocrinol Diabetes Obes (2017) 24(5):332–6. doi: 10.1097/MED.0000000000000359
88. Cao J, Zhang M, Zhang L, Lou J, Zhou F, Fang M. Non-Coding RNA in Thyroid Cancer - Functions and Mechanisms. Cancer Lett (2021) 496:117–26. doi: 10.1016/j.canlet.2020.08.021
89. Rao Y, Liu H, Yan X, Wang J. In Silico Analysis Identifies Differently Expressed lncRNAs as Novel Biomarkers for the Prognosis of Thyroid Cancer. Comput Math Methods Med (2020) 2020:3651051. doi: 10.1155/2020/3651051
90. Yotsukura S, Mamitsuka H. Evaluation of Serum-Based Cancer Biomarkers: A Brief Review From a Clinical and Computational Viewpoint. Crit Rev Oncol Hematol (2015) 93(2):103–15. doi: 10.1016/j.critrevonc.2014.10.002
91. Bai Y, Long J, Liu Z, Lin J, Huang H, Wang D, et al. Comprehensive Analysis of a ceRNA Network Reveals Potential Prognostic Cytoplasmic lncRNAs Involved in HCC Progression. J Cell Physiol (2019) 234(10):18837–48. doi: 10.1002/jcp.28522
92. Huarte M. The Emerging Role of lncRNAs in Cancer. Nat Med (2015) 21(11):1253–61. doi: 10.1038/nm.3981
93. Wu L, Qu X. Cancer Biomarker Detection: Recent Achievements and Challenges. Chem Soc Rev (2015) 44(10):2963–97. doi: 10.1039/C4CS00370E
94. Long J, Bai Y, Yang X, Lin J, Yang X, Wang D, et al. Construction and Comprehensive Analysis of a ceRNA Network to Reveal Potential Prognostic Biomarkers for Hepatocellular Carcinoma. Cancer Cell Int (2019) 19:90. doi: 10.1186/s12935-019-0817-y
95. Zhou RS, Zhang EX, Sun QF, Ye ZJ, Liu JW, Zhou DH, et al. Integrated Analysis of lncRNA-miRNA-mRNA ceRNA Network in Squamous Cell Carcinoma of Tongue. BMC Cancer (2019) 19(1):779. doi: 10.1186/s12885-019-5983-8
96. Wang JD, Zhou HS, Tu XX, He Y, Liu QF, Liu Q, et al. Prediction of Competing Endogenous RNA Coexpression Network as Prognostic Markers in AML. Aging (Albany NY) (2019) 11(10):3333–47. doi: 10.18632/aging.101985
97. Park E, Maquat LE. Staufen-Mediated mRNA Decay. Wiley Interdiscip Rev RNA (2013) 4(4):423–35. doi: 10.1002/wrna.1168
98. Lan Y, Xiao X, He Z, Luo Y, Wu C, Li L, et al. Long Noncoding RNA OCC-1 Suppresses Cell Growth Through Destabilizing HuR Protein in Colorectal Cancer. Nucleic Acids Res (2018) 46(11):5809–21. doi: 10.1093/nar/gky214
99. Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-P21 Suppresses Target mRNA Translation. Mol Cell (2012) 47(4):648–55. doi: 10.1016/j.molcel.2012.06.027
100. Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA-Mediated Posttranslational Modifications and Reprogramming of Energy Metabolism in Cancer. Cancer Commun (Lond) (2021) 41(2):109–20. doi: 10.1002/cac2.12108
101. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid Biopsy: Monitoring Cancer-Genetics in the Blood. Nat Rev Clin Oncol (2013) 10(8):472–84. doi: 10.1038/nrclinonc.2013.110
102. Sun Z, Yang S, Zhou Q, Wang G, Song J, Li Z, et al. Emerging Role of Exosome-Derived Long non-Coding RNAs in Tumor Microenvironment. Mol Cancer (2018) 17(1):82. doi: 10.1186/s12943-018-0831-z
103. Zheng R, Du M, Wang X, Xu W, Liang J, Wang W, et al. Exosome-Transmitted Long non-Coding RNA PTENP1 Suppresses Bladder Cancer Progression. Mol Cancer (2018) 17(1):143. doi: 10.1186/s12943-018-0880-3
104. Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu H, et al. Exosomal Long Noncoding RNA LNMAT2 Promotes Lymphatic Metastasis in Bladder Cancer. J Clin Invest (2020) 130(1):404–21. doi: 10.1172/JCI130892
105. Dong H, Wang W, Chen R, Zhang Y, Zou K, Ye M, et al. Exosome-Mediated Transfer of Lncrnasnhg14 Promotes Trastuzumab Chemoresistance in Breast Cancer. Int J Oncol (2018) 53(3):1013–26. doi: 10.3892/ijo.2018.4467
106. Cheng J, Meng J, Zhu L, Peng Y. Exosomal Noncoding RNAs in Glioma: Biological Functions and Potential Clinical Applications. Mol Cancer (2020) 19(1):66. doi: 10.1186/s12943-020-01189-3
Keywords: lncRNA - long noncoding RNA, LINC00641, cancer, biomarker, therapeutic target
Citation: Han X and Zhang S (2022) Role of Long Non-Coding RNA LINC00641 in Cancer. Front. Oncol. 11:829137. doi: 10.3389/fonc.2021.829137
Received: 05 December 2021; Accepted: 29 December 2021;
Published: 27 January 2022.
Edited by:
Jiayi Wang, Shanghai Jiaotong University, ChinaCopyright © 2022 Han and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shitai Zhang, zhangshitai@126.com
†ORCID: Xue Han, orcid.org/0000-0002-1754-4173
Shitai Zhang, orcid.org/0000-0003-2170-4958
 Xue Han
Xue Han Shitai Zhang
Shitai Zhang