
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 20 January 2022
Sec. Molecular and Cellular Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.827344
Background: Renal cell carcinoma (RCC) was sex-hormone responsive, and clinical trials using progesterone significantly reduced the incidence of distal metastasis after radical nephrectomy. Recently membrane-bound progesterone receptors (mPRs) were discovered to mediate the non-genomic effect of progesterone. Aberrant expressions of these mPRs were reported in human breast, ovarian, urinary bladder, brain, uterine, and prostate cancers. However, their expression profiles in RCC are yet to be assessed.
Methods: Multiple datasets from RNA sequencing (RNA-seq), cDNA microarray, and proteomic analysis were used to compare gene expression between cancerous and normal kidney tissues. Immunohistochemistry was conducted to examine protein expression in kidney tissues. Promoter methylation levels were assessed for correlation analysis with gene expression.
Results: Of the seven membrane-bound progesterone receptor genes, the progestin and adipoQ receptor-5 (PAQR5) gene is predominantly expressed in normal kidney tissue but was significantly downregulated in RCC tissues. PAQR5 downregulation correlated with tumor stage, cancer grade, lymph node invasion, and distal metastasis only in clear cell RCC (ccRCC) tissues. PAQR5 downregulation was associated with an increased promoter DNA methylation and a poor survival outcome in ccRCC patients. In addition, PAQR5 expression inversely correlated with transforming growth factor beta-1 (TGFB1) expression, and TGFβ1 treatment significantly reduced PAQR5 gene expression.
Conclusion: PAQR5 is a novel prognostic biomarker in ccRCC and is negatively regulated by the TGFβ1 pathway.
Renal cell carcinoma (RCC) is the most common kidney cancer, and it is derived from renal tubular cells. Among the 15 histological subtypes of RCC, clear cell RCC (ccRCC) is the most common, followed by papillary RCC (pRCC) and chromophobe RCC (ChRCC). Although localized RCC cases have a relatively good prognosis with treatment with the 5-year survival rate of 74-81% for Stage 1-2 patients, metastatic diseases with Stage 3-4 at diagnosis are suffering a poor 5-year survival rate of only 53% (1). Therefore, novel biomarkers are urgently needed to understand the mechanisms leading to tumor progression and to serve as therapeutic biomarkers for better disease management.
RCC tumors have been considered as sex-hormone responsive based on clinical observation and pathological analysis (2, 3) [reviewed in ref (4)]. The sex hormone binding sites for estrogen, androgen, and progesterone were confirmed in RCC tissues (3). Expression of the receptor proteins, including the estrogen receptor (ER), progesterone receptor (PGR), and androgen receptor (AR), were observed in 30-40% of ccRCC tissues (5). However, a significantly low level of progesterone, but not estradiol-17-β (E2), was observed in RCC tissues compared to normal kidney tissues (6). Consistently, clinical trials using progesterone (Medroxyprogesterone Acetate) as a postoperative prophylactic therapy significantly reduced the incidence (10% in treatment groups vs. 35% in the control group) of distal metastasis three years after radical nephrectomy for localized patients (7). In contrast, anti-AR flutamide had no clinical benefit in disseminated RCC patients (8). These studies suggest that progesterone has a suppressive function in RCC progression.
In last two decades, a group of membrane-bound progesterone receptors (mPRs) were identified in human reproductive and neuronal tissues to mediate the non-genomic effect of progesterone (9, 10). There are two types of mPR proteins, the Class II progesterone and adipoQ receptor (PAQR) family (10) and the heme-binding protein family termed as progesterone receptor membrane component-1/2 (PGRMC1/2) (11). While most of the literature reports on these mPRs focused on their function in reproductive and neuronal biology, aberrant expressions of these mPRs were also shown in other types of human cancers, including breast, ovarian, urinary bladder, brain, uterine, and prostate (10, 12–17). In human endometrial cancers, increased PAQR5 expression was associated with a favorable patient prognosis (12), whereas increased PAQR6 expression was associated with a poor prognosis in prostate cancers (13). However, there is a lack of research on these mPR genes in human kidney cancers. In this study we conducted a comprehensive analysis of mPR gene expression profiles using multiple datasets from RNA-seq, cDNA microarray, and proteomic analysis. Our research revealed that PAQR5 expression was significantly reduced in RCC tissues but only correlated with disease progression and survival outcomes in ccRCC patients. Our study suggests that PAQR5 downregulation was associated with promoter hypermethylation and TGFB1 gene upregulation in ccRCC tissues.
The expression profiles of PAQR5/6/7/8/9, PGR, and PGRMC1/2 genes in normal organs or tissues were assessed using the cDNA microarray datasets (NCBI GSE3526) on the Oncomine platform (18, 19). These datasets were generated with 353 normal tissues on Affymetrix U133 Plus 2.0 microarrays that measured 19,574 genes with 54,675 reporters. A total of 65 primary human organs were utilized in comparison.
The RNA-seq datasets derived from The Cancer Genome Atlas (TCGA) project (20) were used as the primary approach to compare gene expression between normal and cancerous kidney tissues. These TCGA datasets for kidney cancers contained three subtypes of RCC tissues, chRCC (KICH), ccRCC (KIRC), and pRCC (KIRP).
Tissue microarray slides containing 42 ccRCC and 10 normal kidney sections were purchased from Novus Biologicals (Centennial, CO). After deparaffinization and hydration, tissue slides were treated with 3% H2O2 for 15 minutes and then blocked with 5% bovine serum albumin (BSA) in tris-buffered saline (TBS) and tween-20 (TBS-T) for 60 minutes. The primary antibody for PAQR5 protein was derived from LSBio (Catalog LSC413057/413055) and used at 1:200 dilution in 5% BSA/TBS-T overnight at 4C with agitation. The immune signals were visualized using the DAKO LSAB2 kit obtained from Agilent (Santa Clara, CA). The immunosignal index was calculated by multiplying the immune density (weak = 1, moderate = 2, strong = 3) with the percentage positivity, as described in our previous report (21).
Protein expression in ccRCC and normal kidney tissues was assessed using the proteomic datasets derived for the Clinical Proteomic Tumor Analysis Consortium (CPTAC) (22). Proteomic profiles were available for selective proteins from 110 ccRCC tissues and 84 normal kidney tissues on the UALCAN platform (23). A z-score value was used to present the relative level of protein expression.
Gene expression analyses at the mRNA level in RCC patients were conducted using the TCGA datasets. Correlation analyses between gene expression levels and PAQR5 promoter DNA methylation were performed using the HM450 methylation data derived from the TCGA project.
The microarray dataset (NCBI GDS2975) was used for assessing the effect of TGFβ1 on PAQR gene expression. This dataset was generated on human immortalized ovarian surface epithelial (IOSE) cells as described (24). Briefly, immortalized ovarian surface epithelial (IOSE) cells were treated with TGFβ1 (Millipore-Sigma, St Louis, MO) at 10 ng/ml concentration for 0, 3, 6, 12 hours. Total RNAs from IOSE cells were isolated using the TRIzol reagent (Invitrogen, Waltham, MA) and were subjected to cDNA microarray analysis using the Human Genome U133A plus 2.0 GeneChip Oligonucleotide Array (Affymetrix, Santa Clara, CA).
Patient survival outcomes, including overall survival, disease-specific survival, and progression-free interval, were assessed using the Kaplan-Meier curve approach (25). Patients were stratified into high or low expression groups using the minimum p-value approach (26). The significance of the hazard ratio was statistically analyzed using the Log-rank test. A nomogram was constructed using the R-rms package (version 6.2-0) and the survival package (version 3.2-10) based on PAQR5 expression and patient clinicopathological parameters, as described (27).
Quantitative data for gene expression at the mRNA and protein levels were presented as the MEAN with the SEM (standard error of the mean). Differences among multiple groups were analyzed using the statistical methods described in the figure legend. Microscopic images from immunohistochemistry were representative of normal and cancerous kidney tissues. The semi-quantitative intensity of the immunosignals was analyzed using the Student t-test as described (21).
We first surveyed the expression patterns of PAQR, PGR, and PGRMC genes in normal tissues using a cDNA microarray dataset generated from 65 types of normal tissues (19). As shown in Supplementary Figure S1, the PAQR5 gene was expressed at a relatively higher level in renal tissues (Supplementary Figure S1A). In contrast, PAQR6/7/8 genes were ubiquitously expressed in all tested tissue types with variable levels (Supplementary Figures S1B–D). PAQR9 was highly expressed in the liver, heart, and testis (Supplementary Figure S1E), and PGR expression was mainly in the gonads and female reproductive tissues (Supplementary Figure S1F), as expected. Similar to PAQR6/7/8 genes, PGRMC1/2 were ubiquitously expressed in all types of tissues (Supplementary Figures S1G, H). These data indicate that the PAQR5 gene might have a critical role in the kidney than in other organs.
Since the PAQR5 gene is highly expressed in normal kidney tissue, we then focused our assessment on PAQR5 expression in kidney cancers using the RNA-seq dataset from the TCGA project. There were case-matched normal and cancerous tissue pairs from renal cell carcinoma patients, and a pair-wise comparison was conducted to analyze PAQR expression. Our results showed a significant reduction of PAQR5 expression in all three types of RCC tissues compared to the normal counterparts (Figure 1A), except few cases showed increased expression in RCC tissues. In addition to kidney cancers, other human cancers also showed significant alterations in PAQR5 gene expression, including upregulation in cancers from the breast, bile duct, liver, and uterine corpus, and downregulation in cancers from the colon, brain, lung, adrenal gland, prostate, rectal, and thyroid organs (Supplementary Figure S2).
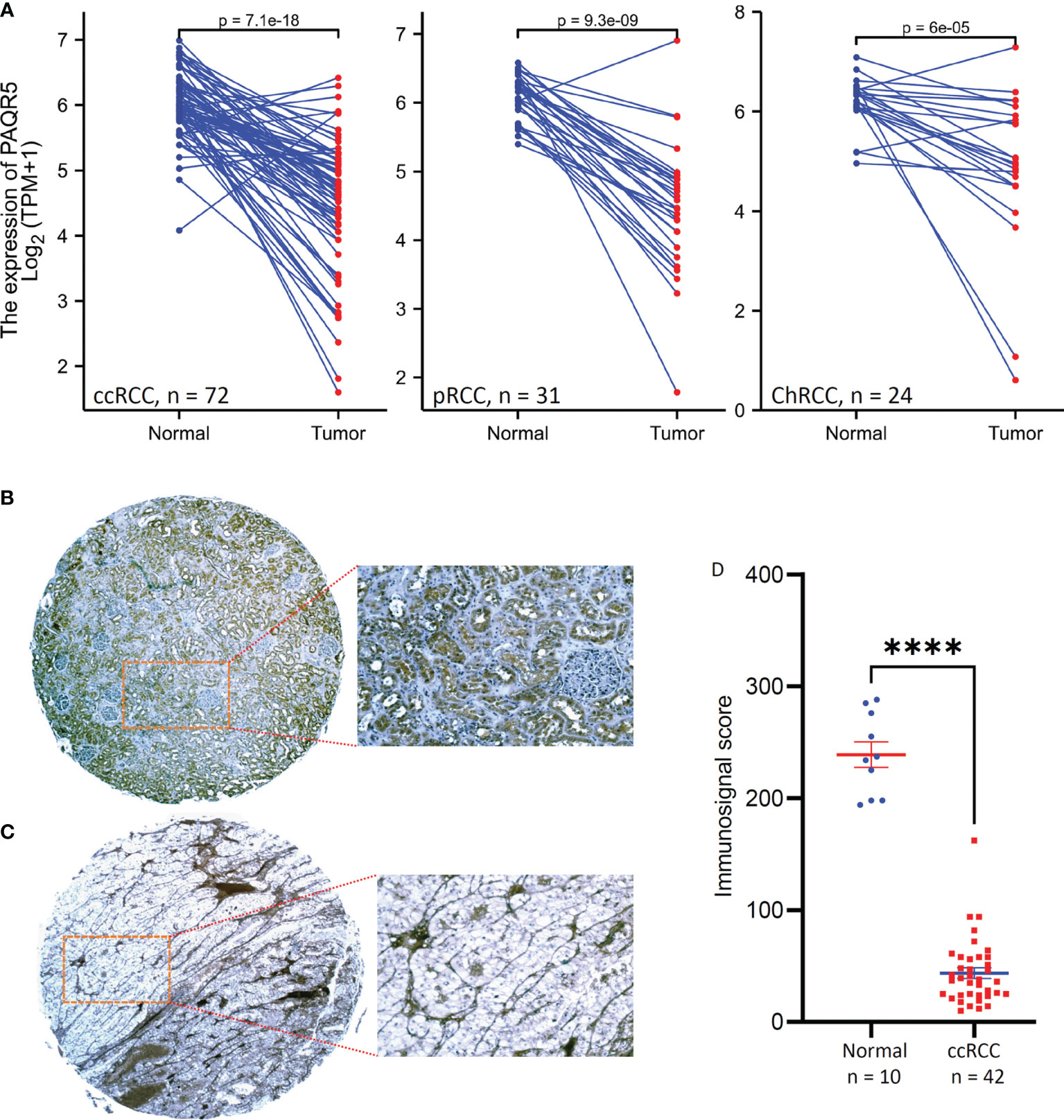
Figure 1 PAQR5 expression in RCC tissues. (A) Pair-wise comparison was conducted using the RNA-seq datasets from the TCGA project. The p-values were derived from Wilcoxon signed-rank test. (B, C) Representative microscopic images were taken from anti-PAQR5 immunohistochemistry on the normal kidney (B) and ccRCC tissue (C) sections. Magnification x 200. (D) Semi-quantitative data from immunohistochemistry staining were compared between normal kidney and ccRCC tissues. The error bars indicate the MEAN and SEM. The Quadro asterisk indicates a significant difference (Student t-test, p < 0.0001).
To verify PAQR5 downregulation, we examined PAQR5 protein expression in ccRCC tissues using an immunohistochemistry approach. As shown in Figure 1B, PAQR5 protein was highly expressed in normal kidney tissues, exclusively in the renal tubular cells but not in the glomerulus cells. In contrast, PAQR5 protein was only expressed in stromal cells of ccRCC tissues and were almost lost in cancer cells (Figure 1C). Summarized immunosignal data showed that PAQR5 protein expression levels were significantly reduced in ccRCC tissues compared to normal kidney tissues (Figure 1D). These results demonstrate that PAQR5 expression was significantly downregulated in RCC tissues.
To evaluate the clinical significance of PAQR5 gene downregulation, we analyzed the association between PAQR5 expression and clinicopathological parameters using the TCGA RNA-seq dataset. RCC patients were divided into PAQR5low and PAQR5high subgroups at the median levels of PAQR5 expression. The clinicopathological parameters included TNM category, pathological and clinical stages, patient gender, overall and disease-specific survival (DSS) status, and progression-free interval (PFI). Our analysis revealed that PAQR5 expression was significantly associated with all the clinicopathological parameters in ccRCC cases (Table 1) but not in pRCC patients (Supplementary Table S1). In ChRCC patients, PAQR5 expression was only significantly associated with tumor stage but not with other parameters (Supplementary Table S2). These results indicate that PAQR5 downregulation is only related to ccRCC disease progression.
To verify the significance of PAQR5 association with disease progression in ccRCC patients, we compared PAQR5 expression levels with different clinicopathological parameters. As shown in Figures 2A–E, PAQR5 expression levels were significantly lower in patients with late-stage tumors (T3-T4 vs. T1-2), lymph node invasive tumors (N1 vs. N0), distal metastasis (M1 vs. M0), late clinical stage tumors (pT3-4 vs. pT1-2), and high-grade tumors (G3-4 vs. G1-2). Interestingly, male patients also showed a significantly lower PAQR5 expression than female patients (Figure 2F). In addition, patients who were deceased or relapsed also displayed a substantially lower level of PAQR5 expression (Figures 2G–I). These data demonstrate that PAQR5 downregulation was associated with ccRCC disease progression and metastasis, as well as poor survival status.

Figure 2 PAQR5 expression is inversely associated with disease progression in ccRCC patients. (A–I) The RNAseq dataset from the TCGA project was used to compare different clinicopathological parameters. Multiple group comparison (panel A, D, E) was conducted using the Kruskal-Wallis test followed by Bonferroni modification and Dunn’s test. Two group comparison (panel B, C, F–I) was performed using the Wilcoxon rank-sum test. Case numbers in each sub-group were listed at the bottom of each panel.
Since promoter hypermethylation is one of the central mechanisms for gene silencing in human cancers (28), we analyzed the methylation levels of PAQR5 gene promoters based on the TCGA data (29, 30). We first compared PAQR5 promoter methylation levels in ccRCC tissue versus normal kidney tissues. As shown in Figure 3A, PAQR5 promoter methylation levels were significantly higher in ccRCC tissues than in normal kidney tissue. The increased promoter methylation was also observed in late-stage (Figure 3B) and high-grade tumors (Figure 3C). A strong inverse correlation (Person r = -0.66) was observed between PAQR5 expression and promoter methylation in ccRCC tissues (Figure 3D). Detailed analysis on different promoter regions revealed that the methylation levels within the -997/-315 region upstream of the transcription starting site (TSS) exhibited the strongest correlation with PAQR5 expression (Figures 3F–H) over other regions (Figures 3E, I, J), which peaked at the TSS-997 & TSS-400 regions (Figures 3F, G). These data indicate that promoter methylation represents a significant mechanism for PAQR5 downregulation in ccRCC tissues, although further mechanistic study is warranted for verification.

Figure 3 PAQR5 promoter methylation is increased in ccRCC tissues. (A–C) Promoter DNA methylation on the PAQR5 gene was analyzed using the TCGA dataset (HM450 BeadChip) derived from ccRCC tissues on the UALCAN platform (29, 30). Tumor stage and cancer grade were set as the variables for multiple group comparison with ANOVA analysis followed by Student t-test. Case numbers were listed in each sub-group at the bottom. (D) Pearson coefficient analysis was conducted using the TCGA Firehose Legacy RNAseq dataset (HM27 BeadChip) downloaded from the cBioportal platform (case number = 217). (E–J) Pearson coefficient analysis was conducted between PAQR5 expression and the DNA methylation levels in a different region upstream of the TSS site using the TCGA RNAseq dataset (HM450 BeadChip).
We next evaluated the impact of PAQR5 expression on patient survival outcomes. Kaplan-Meier survival analysis using the TCGA dataset showed that PAQR5 downregulation had a significant negative impact on patient overall survival outcomes (HR = 0.36), disease-specific survival (HR = 0.18), and progression-free interval (HR = 0.25) (Figures 4A–C). Although a univariant regression analysis showed that PAQR5 and traditional clinicopathological parameters were significant prognostic factors for overall survival of patients, multivariate regression analysis showed that only PAQR5 and distal metastasis were significantly associated with overall survival of patients (Table 2). A receiver operator characteristic (ROC) curve analysis indicated that PAQR5 expression is a robust prognostic factor with very high specificity and sensitivity (Figure 4D). A nomograph was constructed based on the PAQR5 expression data and clinicopathological parameters, which can be used to predict a 3-, 5- or 10-year survival probability for ccRCC patients (Figure 4E).
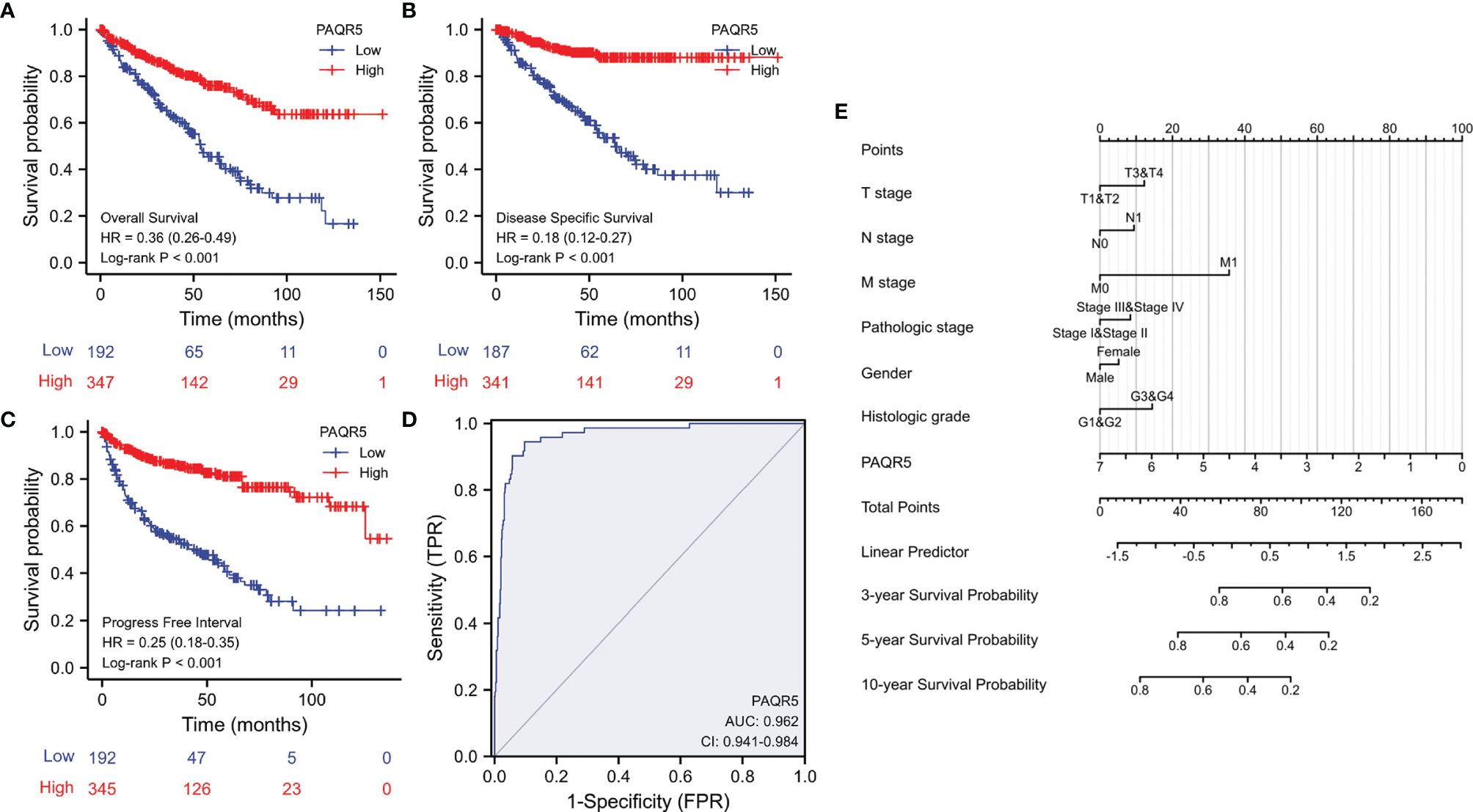
Figure 4 PAQR5 downregulation is associated with poor survival outcomes in ccRCC patients. (A–C) Kaplan-Meier survival analysis was conducted in ccRCC patients using the RNAseq data from the TCGA project. Patients were stratified using the minimum p-value approach (26). (D) A ROC curve analysis was conducted for overall survival prediction using PAQR5 expression levels in ccRCC patients. (E) A nomograph for survival prediction was constructed using the PAQR5 expression levels and the clinicopathological parameters. The nomograph is a calculator tool to predict patient survival rate using clinicopathological data plus the expression levels of biomarker gene. For instance, a male patient had a Grade-1 RCC of T1/N0/M0 at Stage-1, and his PAQR5 expression level was at 3.5. All these parameters will get their nomograph’s points (at the top line). The sum of these points will used to find a linear predictor value, which is applied to find the survival rate. In this case, his total point is 90, then his linear predictor is about 0.75, which indicates his survival rate for 3-year at 60%, 5-year at 40%, and 10-year at 20%.
In exploring the potential signaling pathways involved in PAQR5 downregulation, we analyzed the Spearman correlation coefficients between PAQR5 and the entire transcriptome (20020 genes). PAQR5 was negatively correlated with 2563 genes (Spearman r < -0.3) and was positively correlated 1646 genes (Spearman r > 0.3) (Supplementary Table S3). Gene enrichment analysis revealed that KEGG pathways of progesterone-mediated oocyte maturation and oocyte meiosis were significantly enriched among those negatively correlated genes (Supplementary Table S4), which was in line with PAQR5 function (10, 31). Additional enriched KEGG pathways included ribosome, cell cycle, cancer-related pathways, focal adhesion, and intracellular signal transduction pathways of PI3K-Akt, MAPK, and mTOR. Most interestingly, TGFβ1 signaling pathway was also enriched, and TGFB1 expression was negatively correlated with PAQR5 expression levels (Figure 5A).
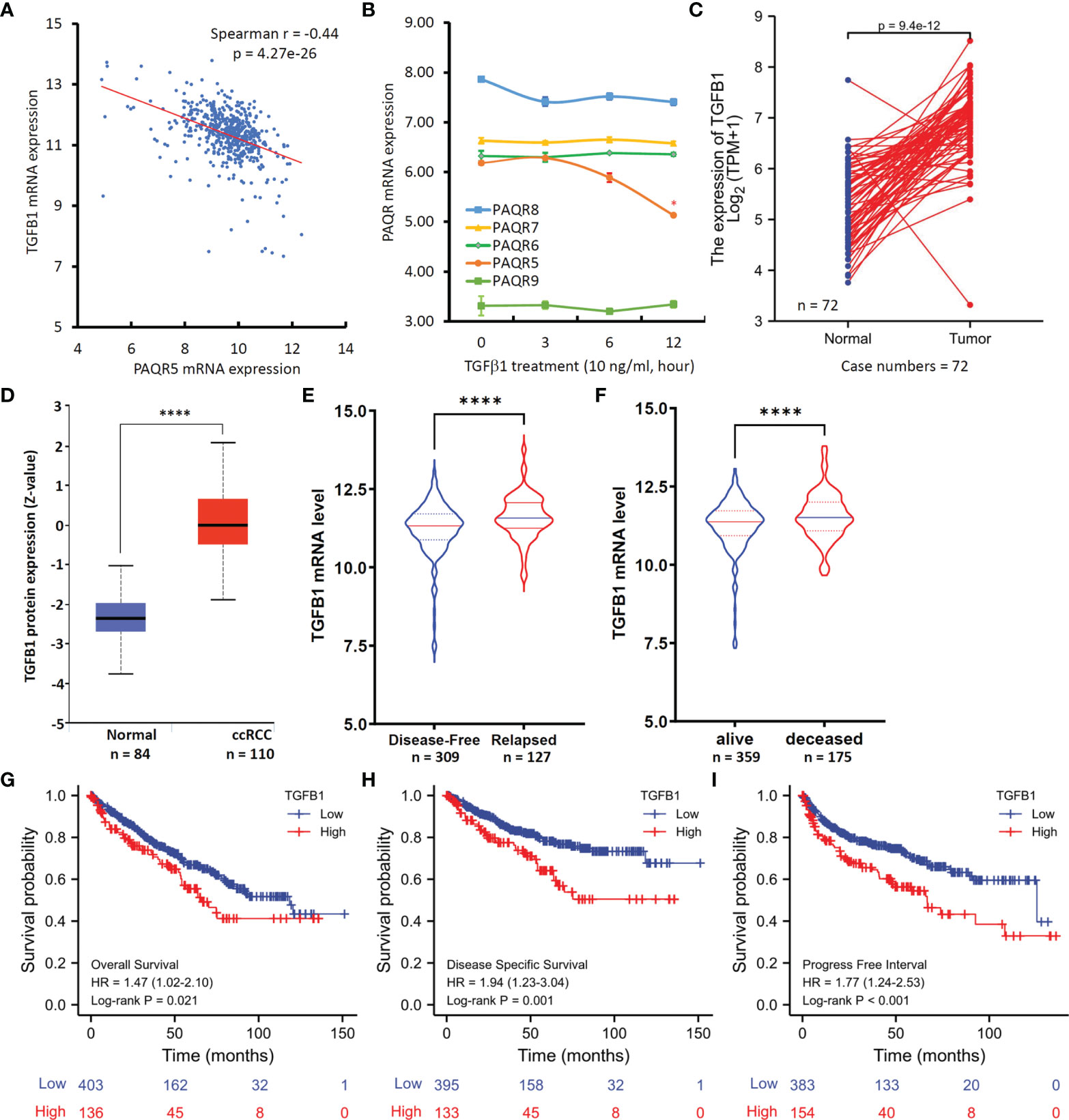
Figure 5 TGFB1 expression inversely correlates with PAQR5 expression in ccRCC tissues. (A) Spearman coefficient analysis was conducted using the TCGA-Firehose Legacy RNAseq dataset downloaded from the cBioportal platform (case number = 534). The mRNA levels were presented as log2(RSEM+1) values. RSEM: RNA-Seq by Expectation-Maximization. (B) Human IOSE cells were treated with TGFβ1 (10 ng/ml) for the indicated time-period. Total RNAs were extracted for gene expression analysis on the Affymetrix Human Genome U133A plus 2.0 GeneChip. The error bar represents the SEM of the mean. The asterisk indicates a significant difference compared to the 0-h control (Student t-test, p < 0.05). (C) TGFB1 expression levels between case-match pairs of normal kidneys and ccRCC tissues in the TCGA dataset. The p-value was derived from Wilcoxon signed-rank test. (D) TGFβ1 protein level in ccRCC and normal kidney tissues using the CPTAC dataset (22). The Quadro-asterisk indicates a significant difference (unpaired t-test, p < 0.0001). (E, F) TGFB1 expression levels were compared between disease-free vs. relapsed or alive vs. deceased patients using the TCGA-Firehose Legacy RNAseq dataset downloaded from the cBioportal platform. The Quadro-asterisk indicates a significant difference (unpaired t-test, p < 0.0001). (G–I) Kaplan-Meier curve analysis was conducted for overall and disease-specific survival, as well as the progression-free interval in ccRCC patients based on TGFB1 expression using the TCGA dataset with a minimum p-value approach (26).
To understand if TGFβ1 is an upstream regulator for PAQR5 expression, we analyzed gene expression profiles in TGFβ1-treated cells using a microarray dataset (NCBI GDS2795) (24). As shown in Figure 5B, PAQR5 expression was significantly reduced in human immortalized ovarian surface epithelial (IOSE) cells after TGFβ1 treatment for up to 12 hours. However, TGFβ1 treatment had no significant effect on other PAQR genes. These data verify TGFβ1 as an upstream suppressor of PAQR5 gene expression.
We then assessed the clinical significance of TGFβ1 expression in ccRCC patients. Compared to normal kidney tissues, TGFB1 expression at the mRNA and protein levels was significantly increased in ccRCC tissues (Figures 5C, D). These data are in line with previous reports of increased TGFβ1 plasma level in RCC patients compared to non-malignant controls (32–34). Further analysis revealed that TGFB1 expression was significantly higher in relapsed or deceased ccRCC patients compared to relapse-free or alive patients (Figures 5E, F). In addition, higher TGFB1 expression at the mRNA level was significantly associated with poor overall (Figure 5G) and disease-specific survival outcomes (Figure 5H), as well as shorter progression-free intervals (Figure 5I) compared to patients with lower TGFB1 expression. These data are consistent with a previous prognosis study using TGFβ1 protein level as the biomarker for ccRCC patients (35).
In this study, our analysis revealed that the PAQR5 gene was predominantly expressed in normal kidney tissue when compared to other organs, and was significantly downregulated in ccRCC tissues, as assessed at the mRNA and protein levels. PAQR5 downregulation was associated with tumor progression, including tumor stage, cancer grade, lymph node invasion, and distal metastasis. PAQR5 expression was significantly reduced in ccRCC tissues from relapsed or deceased cases. In addition, patients with lower PAQR5 expression had an inferior survival outcome than patients with higher PAQR5 expression. DNA methylation analysis indicated that PAQR5 promoter methylation in ccRCC tissues was significantly higher than in normal kidney tissues and correlated with tumor stage and cancer grade in ccRCC tissues, indicating that DNA methylation could be a potential mechanism for PAQR5 downregulation. In addition, PAQR5 expression was negatively correlated with TGFB1 expression, which was significantly increased in ccRCC tissues and associated with a poor prognosis. TGFβ1 treatment in human cancer cells significantly suppressed PAQR5 expression. As far as the authors are aware, this is the first report about the expression of PAQR genes in human kidney cancers.
Current literature showed that mPRs were involved in cancer progression of the breast, ovarian, prostate, brain, bladder, and uterine (10). Altered expression levels of mPR genes were associated with disease prognosis in breast, endometrial, bladder, and prostate cancers (12–15, 36, 37). Specifically, PAQR6 gene upregulation or copy number gain was associated with an unfavorable prognosis in the prostate, bladder, and endometrial cancers (13, 14, 36). In contrast, an increased expression of PAQR5/8 genes was associated with a favorable prognosis in endometrial cancers (12). This study found that PAQR5 expression was significantly reduced in RCC tissues compared to normal kidney tissues. However, PAQR5 expression correlated considerably with all pathological and clinical parameters, including tumor stage, cancer grade, lymph node invasion, distal metastasis, disease relapse, and survival outcomes only in ccRCC but not in pRCC or ChRCC. These results suggest that PAQR5 potentially plays a tumor-suppressive role in ccRCC development and progression, although further mechanistic study is needed to elucidate PAQR5 function in ccRCC progression.
The individual mPR protein was shown to localize on the plasma membrane and other organelle membranes (10, 38). Immunostaining studies have demonstrated that PAQR7/8 was mainly localized at the plasma membrane, while PAQR5 was localized in the cytoplasm and nucleus in normal endometrium (12). Our results showed that PAQR5 protein was expressed in the cytoplasm of renal tubular cells but not in the glomerulus. In ccRCC tissues, PAQR5 protein staining was primarily lost in the cytoplasm with weak signals at the plasma membrane. We speculate that PAQR5 protein loss might be due to PAQR5 gene downregulation in ccRCC tissues.
The TGFβ1 protein is a multi-functional cytokine, and its role has been implicated in human cancers (39), including enhancement of proliferative and metastatic potential in human RCC cells (40). Increased TGFB1 mRNA or TGFβ1 protein level in cancer tissue or patient plasma was identified as a prognostic factor for rapid progression and poor survival outcome (32–35). Our analysis discovered a strong and inverse correlation between PAQR5 and TGFB1 in ccRCC tissues. Similar to PAQR5, TGFB1 expression was also associated with rapid disease progression and unfavorable survival outcomes. TGFβ1 treatment suppressed PAGR5 expression but did not affect other PAQR genes in human immortalized epithelial cells. It is postulated that increased TGFB1 expression in ccRCC tissues is a potential mediator for PAQR5 downregulation, although further mechanistic study is warranted.
To date, very few reports studied the biological functions of mPR proteins in cancer biology, especially their roles in different types of human cancers after progesterone stimulation. It has been shown that mPR proteins might work either with G-proteins, growth factor receptors, or alone to stimulate diverse intracellular signaling pathways including MAPK, JNK, PI3K, NF-κB modulate cellular function (10). Further studies are needed to understand progesterone-stimulated mPR signaling in renal function or cancer cell behavior.
PAQR5 expression was significantly reduced in ccRCC tissues compared to normal tissues, as verified at the protein levels. PAQR5 downregulation was significantly associated with clinical and pathological parameters in ccRCC patients, representing a novel prognostic factor of rapid disease progression and poor survival outcome. PAQR5 downregulation was accompanied by increased promoter methylation, indicating a potential mechanism for reduced gene expression. PAQR5 expression was strongly and inversely correlated with TGFB1 expression in ccRCC tissues. TGFβ1 treatment specifically reduced PAQR5 expression, representing a distinct mechanism for PAQR5 modulation.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
RZ and BL designed the study. CT, XY, MY, and BL analyzed the bioinformatics data. WL and SY performed the IHC experiments. BL and QS performed the statistical analysis and generated figures and tables. RZ, QS, and BL drafted the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We were grateful for the English editing service from the Uniwin SCI LLC.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.827344/full#supplementary-material
Supplementary Figure S1 | Gene expression profiles in normal tissues. The cDNA microarray dataset generated from 65 tissue types (19) was analyzed for the gene expression profiles. The figure panels were created on the Oncomine platform. The most predominant tissue types were highlighted in red font.
Supplementary Figure S2 | PAQR5 expression in pan-cancer comparison.
Supplementary Table S1 | The relationship of PAQR5 expression with clinicopathological parameters in pRCC patients.
Supplementary Table S2 | The relationship of PAQR5 expression with clinicopathological parameters in ChRCC patients.
Supplementary Table S3 | Spearman correlation analysis of PAQR5 expression in the whole transcriptome.
Supplementary Table S4 | Gene-list enrichment analysis of PAQR5-related genes.
AR, androgen receptor; BSA, bovine serum albumin; ChRCC, chromophobe RCC; ccRCC, clear cell RCC; CPTAC, clinical proteomic tumor analysis consortium; ER, estrogen receptor; IOSE, immortalized ovarian surface epithelial; KEGG, kyoto encyclopedia of genes and genomes; KICH, kidney chromophobe cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney papillary carcinoma; MAPK, mitogen-activated protein kinase; mPR, membrane-bound progesterone receptors; mTOR, mammalian target of rapamycin; PI3K, phosphoinositide 3-kinase; PAQR, progesterone and adipoQ receptor; PGRMC1/2, progesterone receptor membrane component 1/2; PGR, progesterone receptor; RCC, renal cell carcinoma; pRCC, papillary RCC SEM, standard error of the mean; TGFB1, transforming growth factor beta 1; TCGA, the cancer genome atlas; TBS-T, tris-buffered saline with tween-20.
1. Porta C, Cosmai L, Leibovich BC, Powles T, Gallieni M, Bex A. The Adjuvant Treatment of Kidney Cancer: A Multidisciplinary Outlook. Nat Rev Nephrol (2019) 15(7):423–33. doi: 10.1038/s41581-019-0131-x
2. Concolino G, Marocchi A, Conti C, Tenaglia R, Di Silverio F, Bracci U. Human Renal Cell Carcinoma as a Hormone-Dependent Tumor. Cancer Res (1978) 38(11 Pt 2):4340–4.
3. Concolino G, Di Silverio F, Marocchi A, Bracci U. Renal Cancer Steroid Receptors: Biochemical Basis for Endocrine Therapy. Eur Urol (1979) 5(2):90–3. doi: 10.1159/000473074
4. Czarnecka AM, Niedzwiedzka M, Porta C, Szczylik C. Hormone Signaling Pathways as Treatment Targets in Renal Cell Cancer (Review). Int J Oncol (2016) 48(6):2221–35. doi: 10.3892/ijo.2016.3460
5. Langner C, Ratschek M, Rehak P, Schips L, Zigeuner R. Steroid Hormone Receptor Expression in Renal Cell Carcinoma: An Immunohistochemical Analysis of 182 Tumors. J Urol (2004) 171(2 Pt 1):611–4. doi: 10.1097/01.ju.0000108040.14303.c2
6. Batra S, Colleen S, Grundsell H, Hakansson L. Progesterone and Estradiol Concentration in Human Kidney and Renal Cell Carcinoma. J Steroid Biochem (1979) 11(5-6):1535–7. doi: 10.1016/0022-4731(79)90344-3
7. Satomi Y, Takai S, Kondo I, Fukushima S, Furuhata A. Postoperative Prophylactic Use of Progesterone in Renal Cell Carcinoma. J Urol (1982) 128(5):919–22. doi: 10.1016/s0022-5347(17)53276-0
8. Ahmed T, Benedetto P, Yagoda A, Watson RC, Scher HI, Herr HW, et al. Estrogen, Progesterone, and Androgen-Binding Sites in Renal Cell Carcinoma. Observations Obtained in Phase II Trial of Flutamide. Cancer (1984) 54(3):477–81. doi: 10.1002/1097-0142(19840801)54:3<477::aid-cncr2820540316>3.0.co;2-k
9. Zhu Y, Bond J, Thomas P. Identification, Classification, and Partial Characterization of Genes in Humans and Other Vertebrates Homologous to a Fish Membrane Progestin Receptor. Proc Natl Acad Sci USA (2003) 100(5):2237–42. doi: 10.1073/pnas.0436133100
10. Valadez-Cosmes P, Vazquez-Martinez ER, Cerbon M, Camacho-Arroyo I. Membrane Progesterone Receptors in Reproduction and Cancer. Mol Cell Endocrinol (2016) 434:166–75. doi: 10.1016/j.mce.2016.06.027
11. Pru JK, Clark NC. PGRMC1 and PGRMC2 in Uterine Physiology and Disease. Front Neurosci (2013) 7:168. doi: 10.3389/fnins.2013.00168
12. Sinreih M, Knific T, Thomas P, Frkovic Grazio S, Rizner TL. Membrane Progesterone Receptors Beta and Gamma Have Potential as Prognostic Biomarkers of Endometrial Cancer. J Steroid Biochem Mol Biol (2018) 178:303–11. doi: 10.1016/j.jsbmb.2018.01.011
13. Yang M, Li JC, Tao C, Wu S, Liu B, Shu Q, et al. PAQR6 Upregulation Is Associated With AR Signaling and Unfavorite Prognosis in Prostate Cancers. Biomolecules (2021) 11(9):1383. doi: 10.3390/biom11091383
14. Cai Z, Chen H, Bai J, Zheng Y, Ma J, Cai X, et al. Copy Number Variations of CEP63, FOSL2 and PAQR6 Serve as Novel Signatures for the Prognosis of Bladder Cancer. Front Oncol (2021) 11:674933. doi: 10.3389/fonc.2021.674933
15. Li B, Lin Z, Liang Q, Hu Y, Xu WF. PAQR6 Expression Enhancement Suggests a Worse Prognosis in Prostate Cancer Patients. Open Life Sci (2018) 13:511–7. doi: 10.1515/biol-2018-0061
16. Pang Y, Dong J, Thomas P. Characterization, Neurosteroid Binding and Brain Distribution of Human Membrane Progesterone Receptors Delta and {Epsilon} (Mprdelta and mPR{Epsilon}) and Mprdelta Involvement in Neurosteroid Inhibition of Apoptosis. Endocrinology (2013) 154(1):283–95. doi: 10.1210/en.2012-1772
17. Paul MR, Pan TC, Pant DK, Shih NN, Chen Y, Harvey KL, et al. Genomic Landscape of Metastatic Breast Cancer Identifies Preferentially Dysregulated Pathways and Targets. J Clin Invest (2020) 130(8):4252–65. doi: 10.1172/JCI129941
18. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia (2004) 6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2
19. Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, et al. Gene Expression Analyses Reveal Molecular Relationships Among 20 Regions of the Human CNS. Neurogenetics (2006) 7(2):67–80. doi: 10.1007/s10048-006-0032-6
20. Cancer Genome Atlas Research Network. Comprehensive Genomic Characterization Defines Human Glioblastoma Genes and Core Pathways. Nature (2008) 455(7216):1061–8. doi: 10.1038/nature07385
21. Sun A, Tawfik O, Gayed B, Thrasher JB, Hoestje S, Li C, et al. Aberrant Expression of SWI/SNF Catalytic Subunits BRG1/BRM Is Associated With Tumor Development and Increased Invasiveness in Prostate Cancers. Prostate (2007) 67(2):203–13. doi: 10.1002/pros.20521
22. Chen F, Chandrashekar DS, Varambally S, Creighton CJ. Pan-Cancer Molecular Subtypes Revealed by Mass-Spectrometry-Based Proteomic Characterization of More Than 500 Human Cancers. Nat Commun (2019) 10(1):5679. doi: 10.1038/s41467-019-13528-0
23. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (2017) 19(8):649–58. doi: 10.1016/j.neo.2017.05.002
24. Qin H, Chan MW, Liyanarachchi S, Balch C, Potter D, Souriraj IJ, et al. An Integrative ChIP-Chip and Gene Expression Profiling to Model SMAD Regulatory Modules. BMC Syst Biol (2009) 3:73. doi: 10.1186/1752-0509-3-73
25. Gyorffy B. Survival Analysis Across the Entire Transcriptome Identifies Biomarkers With the Highest Prognostic Power in Breast Cancer. Comput Struct Biotechnol J (2021) 19:4101–9. doi: 10.1016/j.csbj.2021.07.014
26. Menyhart O, Nagy A, Gyorffy B. Determining Consistent Prognostic Biomarkers of Overall Survival and Vascular Invasion in Hepatocellular Carcinoma. R Soc Open Sci (2018) 5(12):181006. doi: 10.1098/rsos.181006
27. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell (2018) 173(2):400–16.e11. doi: 10.1016/j.cell.2018.02.052
28. Yamashita K, Hosoda K, Nishizawa N, Katoh H, Watanabe M. Epigenetic Biomarkers of Promoter DNA Methylation in the New Era of Cancer Treatment. Cancer Sci (2018) 109(12):3695–706. doi: 10.1111/cas.13812
29. Shinawi T, Hill VK, Krex D, Schackert G, Gentle D, Morris MR, et al. DNA Methylation Profiles of Long- and Short-Term Glioblastoma Survivors. Epigenetics (2013) 8(2):149–56. doi: 10.4161/epi.23398
30. Men C, Chai H, Song X, Li Y, Du H, Ren Q. Identification of DNA Methylation Associated Gene Signatures in Endometrial Cancer via Integrated Analysis of DNA Methylation and Gene Expression Systematically. J Gynecol Oncol (2017) 28(6):e83. doi: 10.3802/jgo.2017.28.e83
31. Tokumoto T, Tokumoto M, Oshima T, Shimizuguchi K, Fukuda T, Sugita E, et al. Characterization of Multiple Membrane Progestin Receptor (mPR) Subtypes From the Goldfish Ovary and Their Roles in the Induction of Oocyte Maturation. Gen Comp Endocrinol (2012) 177(1):168–76. doi: 10.1016/j.ygcen.2012.03.005
32. Hegele A, Varga Z, von Knobloch R, Heidenreich A, Kropf J, Hofmann R. TGF-Beta1 in Patients With Renal Cell Carcinoma. Urol Res (2002) 30(2):126–9. doi: 10.1007/s00240-002-0245-6
33. Junker U, Knoefel B, Nuske K, Rebstock K, Steiner T, Wunderlich H, et al. Transforming Growth Factor Beta 1 Is Significantly Elevated in Plasma of Patients Suffering From Renal Cell Carcinoma. Cytokine (1996) 8(10):794–8. doi: 10.1006/cyto.1996.0105
34. Kim CS, Kim Y, Kwon T, Yoon JH, Kim KH, You D, et al. Regulatory T Cells and TGF-Beta1 in Clinically Localized Renal Cell Carcinoma: Comparison With Age-Matched Healthy Controls. Urol Oncol (2015) 33(3):113 e19–25. doi: 10.1016/j.urolonc.2014.11.004
35. Lebdai S, Verhoest G, Parikh H, Jacquet SF, Bensalah K, Chautard D, et al. Identification and Validation of TGFBI as a Promising Prognosis Marker of Clear Cell Renal Cell Carcinoma. Urol Oncol (2015) 33(2):69 e11–8. doi: 10.1016/j.urolonc.2014.06.005
36. Kunitomi H, Kobayashi Y, Wu RC, Takeda T, Tominaga E, Banno K, et al. LAMC1 Is a Prognostic Factor and a Potential Therapeutic Target in Endometrial Cancer. J Gynecol Oncol (2020) 31(2):e11. doi: 10.3802/jgo.2020.31.e11
37. Xie M, Zhu X, Liu Z, Shrubsole M, Varma V, Mayer IA, et al. Membrane Progesterone Receptor Alpha as a Potential Prognostic Biomarker for Breast Cancer Survival: A Retrospective Study. PloS One (2012) 7(4):e35198. doi: 10.1371/journal.pone.0035198
38. Foster H, Reynolds A, Stenbeck G, Dong J, Thomas P, Karteris E. Internalisation of Membrane Progesterone Receptor-Alpha After Treatment With Progesterone: Potential Involvement of a Clathrin-Dependent Pathway. Mol Med Rep (2010) 3(1):27–35. doi: 10.3892/mmr_00000214
39. Wang J, Xiang H, Lu Y, Wu T. Role and Clinical Significance of TGFbeta1 and TGFbetaR1 in Malignant Tumors (Review). Int J Mol Med (2021) 47(4):55. doi: 10.3892/ijmm.2021.4888
Keywords: PAQR5, TGFβ1, renal cancer, patient survival, disease progression
Citation: Tao C, Liu W, Yan X, Yang M, Yao S, Shu Q, Li B and Zhu R (2022) PAQR5 Expression Is Suppressed by TGFβ1 and Associated With a Poor Survival Outcome in Renal Clear Cell Carcinoma. Front. Oncol. 11:827344. doi: 10.3389/fonc.2021.827344
Received: 01 December 2021; Accepted: 31 December 2021;
Published: 20 January 2022.
Edited by:
Dong-Hua Yang, St. John’s University, United StatesReviewed by:
Chang-Deng Hu, Purdue University, United StatesCopyright © 2022 Tao, Liu, Yan, Yang, Yao, Shu, Li and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Runzhi Zhu, cnVuemhpemh1QHpqdS5lZHUuY24=; Benyi Li, YmxpQGt1bWMuZWR1; Qiang Shu, c2h1cWlhbmdAemp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.