
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Oncol., 31 January 2022
Sec. Pharmacology of Anti-Cancer Drugs
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.809527
This article is part of the Research TopicMetabolism of Anti-Cancer Drugs: Interplay Involving Pharmacology and PharmacokineticsView all 5 articles
 Nicholas R. Powell1
Nicholas R. Powell1 Tyler Shugg1
Tyler Shugg1 Reynold C. Ly1
Reynold C. Ly1 Costantine Albany2
Costantine Albany2 Milan Radovich2
Milan Radovich2 Bryan P. Schneider2
Bryan P. Schneider2 Todd C. Skaar1*
Todd C. Skaar1*Docetaxel therapy occasionally causes severe and life-threatening toxicities. Some docetaxel toxicities are related to exposure, and inter-individual variability in exposure has been described based on genetic variation and drug-drug interactions that impact docetaxel clearance. Cytochrome P450 3A4 (CYP3A4) and CYP3A5 metabolize docetaxel into inactive metabolites, and this is the primary mode of docetaxel clearance. Supporting their role in these toxicities, increased docetaxel toxicities have been found in patients with reduced- or loss-of-function variants in CYP3A4 and CYP3A5. However, since these variants in CYP3A4 are rare, little is known about the safety of docetaxel in patients who are homozygous for the reduced-function CYP3A4 variants. Here we present a case of life-threatening (grade 4) pneumonitis, dyspnea, and neutropenia resulting from a single dose of docetaxel. This patient was (1) homozygous for CYP3A4*22, which causes reduced expression and is associated with increased docetaxel-related adverse events, (2) heterozygous for CYP3A4*3, a rare reduced-function missense variant, and (3) homozygous for CYP3A5*3, a common loss of function splicing defect that has been associated with increased docetaxel exposure and adverse events. The patient also carried functional variants in other genes involved in docetaxel pharmacokinetics that may have increased his risk of toxicity. We identified one additional CYP3A4*22 homozygote that received docetaxel in our research cohort, and present this case of severe hematological toxicity. Furthermore, the one other CYP3A4*22 homozygous patient we identified from the literature died from docetaxel toxicity. This case report provides further evidence for the need to better understand the impact of germline CYP3A variants in severe docetaxel toxicity and supports using caution when treating patients with docetaxel who have genetic variants resulting in CYP3A poor metabolizer phenotypes.
Docetaxel is an antimicrotubular taxane derivative used for treatment of a variety of solid tumors. Due to its cytotoxicity, docetaxel occasionally causes severe and life-threatening toxicities (1). Both the incidence and severity of docetaxel-induced adverse events are related to exposure (2), and significant inter-individual variability in exposure has been described based on genetic variation and drug-drug interactions that impact docetaxel clearance (1, 3).
The drug-metabolizing enzymes and drug transporters that contribute to docetaxel pharmacokinetics are presented in Supplemental Figure S1 (4). It is transported into hepatocytes via solute carrier organic anion transporter 1B3 (SLCO1B3) (5, 6) and excreted into the bile by multiple ATP-binding cassette (ABC) transporters (7). Within hepatocytes, cytochrome P450 3A4 (CYP3A4) and CYP3A5 metabolize docetaxel into inactive metabolites, and this is the primary mode of docetaxel clearance (1, 8). To avoid toxicities, the drug label recommends 50% reduction in dose when co-administered with strong CYP3A inhibitors (1). Similarly, pharmacogenetic evaluations have demonstrated increased docetaxel toxicities in patients with reduced- or loss-of-function variants in CYP3A4 and CYP3A5 (9, 10).
Pharmacogenetic guidelines for docetaxel have not been published yet, in part, due to the need for additional evidence. Here we present two cases that provide further evidence for a role of germline CYP3A genetics in severe docetaxel toxicity.
Patient 1 is a 52-year-old male with metastatic prostate cancer who was administered a single dose of docetaxel 75 mg/m2. Nine days later, he presented to the emergency department with complaints of fever, chills, hemoptysis, and was grade 3 febrile neutropenic. He was discharged on levofloxacin for suspected community-acquired pneumonia. On day 11, he returned to the emergency department and was admitted for acute respiratory distress accompanied by fever, hypoxemia, tachypnea, and tachycardia. On day 12, the respiratory distress worsened and he was moved to the intensive care unit. On day 13, he was intubated due to hypoxic respiratory failure secondary to docetaxel-induced pneumonitis and was started on corticosteroids. Neutropenia progressed to grade 4 by day 16. By day 23, the patient was weaned off respiratory support and discharged. The timeline of events is shown in Figure 1. Noteworthy past medical history includes diagnosis of a pulmonary embolism 14 days prior to docetaxel treatment, which was treated with enoxaparin and considered resolved. Aspartate aminotransferase (AST), alanine aminotransferase (ALT), and total bilirubin were normal throughout the time course. Alkaline Phosphatase was elevated to 162 U/L on day 13. These labs indicate that general liver function was not impaired or diseased at the time of docetaxel administration. The patient was not concomitantly prescribed any CYP3A inhibitors.
The pneumonitis, dyspnea, and neutropenia were grade 4 toxicities caused by one docetaxel dose. Although the patient’s earlier pulmonary embolism may have been a predisposing factor to the pneumonitis, not all patients with a history of pulmonary emboli who receive docetaxel develop life-threatening toxicity; therefore, we investigated potential underlying genetic defects in docetaxel clearance.
Our CYP3A genetic analyses (see Appendix for methods and Table 1 for results) of this patient revealed the following: (1) homozygous CYP3A4*22, which causes reduced expression of the active enzyme (26) and is associated with increased docetaxel-related adverse events (9, 10); (2) heterozygous CYP3A4*3, a rare reduced-function missense variant (22–24); and (3) homozygous CYP3A5*3, a common loss of function splicing defect that has been associated with increased docetaxel exposure and adverse events (9, 10). All genotyping and sequencing was carried out in a phenotype-blind process.
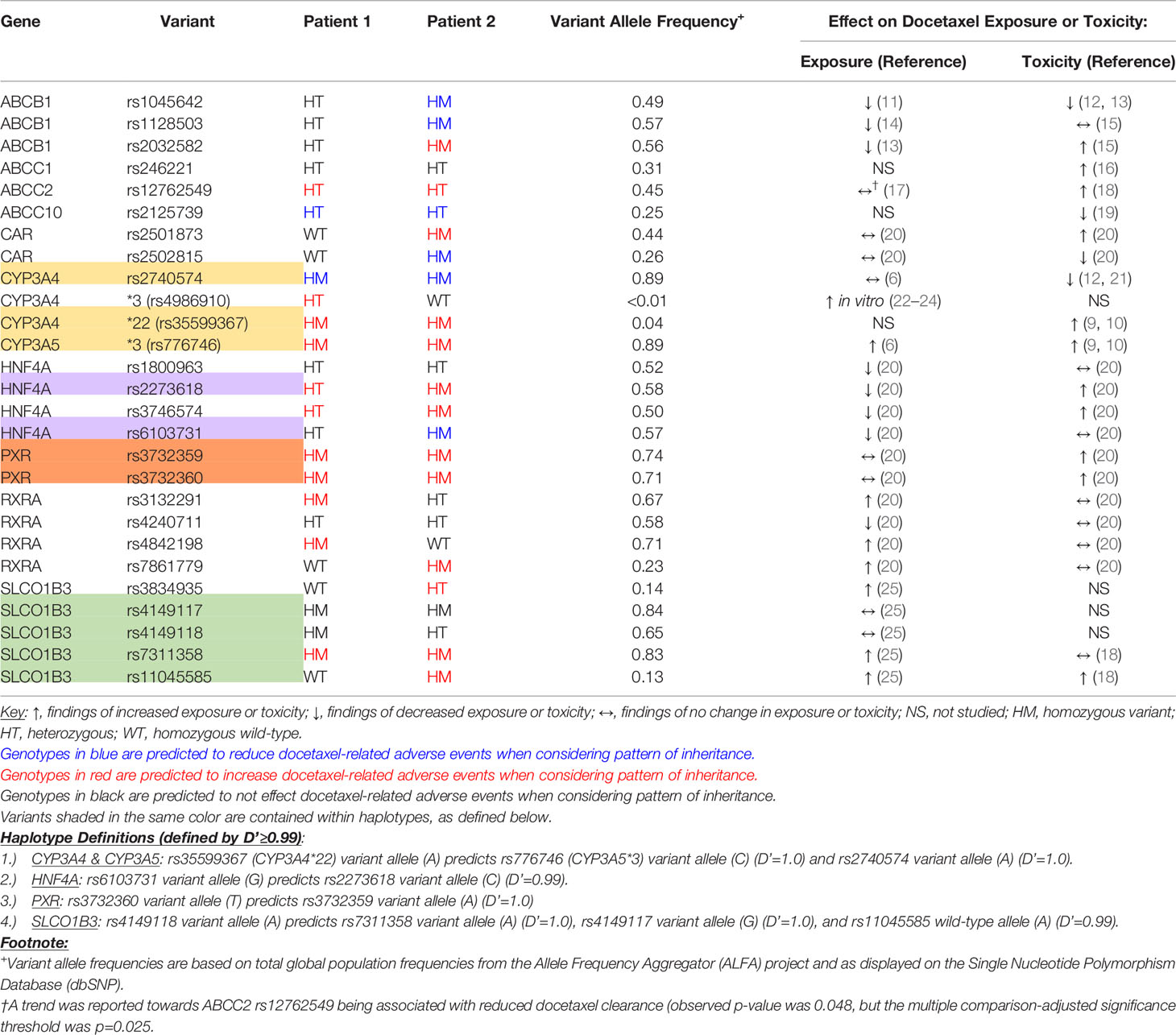
Table 1 Patient status for genetic variants in docetaxel pharmacokinetics-related genes that have previously been associated with exposure or toxicity.
Since CYP3A4*22 is a rare functional variant that was a plausible cause of the docetaxel toxicity, we searched our research cohort for additional CYP3A4*22 homozygotes that received docetaxel. We identified only one additional patient who was homozygous for CYP3A4*22. She was also homozygous for CYP3A5*3, but did not have the CYP3A4*3 variant. She was a 64-year old female breast cancer patient who was treated with docetaxel and cyclophosphamide for a total of 4 cycles. She was also prophylactically treated with granulocyte colony stimulating factor drugs with each cycle and darbepoetin alfa after her 1st and 4th cycle. Over the course of 4 cycles she experienced grade 3 leukopenia, grade 2 anemia, generalized weakness, erythema on her extremities, and numbness in her extremities indicating peripheral neuropathy. We believe her toxicity could be regarded as severe and was caused by docetaxel.
Given the genomic proximity of CYP3A4 and CYP3A5 and the existence of known haplotypes containing functional variants in both genes (27), we conducted linkage disequilibrium (LD) analyses on the these CYP3A variants. Using the global population data from the 1000 Genomes Project, we used LDpair (28) to determine the linkage disequilibrium between CYP3A4*22, CYP3A4*3, and CYP3A5*3. CYP3A4*22 always occurred with CYP3A5*3 (Supplemental Table S1). Also, 62% of the CYP3A4*3 alleles occurred with CYP3A4*22 (Supplemental Table S2). Thus, this is a rare haplotype containing reduced-function CYP3A variants that appears to be associated with docetaxel exposure and toxicity.
In our analysis of other pharmacokinetic genes, both patients were heterozygous for ABCC2 rs12762549, which has been associated with docetaxel-related leukopenia (18). Both patients had variants in HNF4A, PXR, and RXRA that have been weakly associated with increased exposure and/or toxicity, and patient 2 was homozygous for SLCO1B3 rs11045585. All pharmacokinetic variants potentially involved in these cases are reviewed in Table 1. Analysis of pharmacodynamic-related variants using Qiagen Clinical Insights did not identify “pathogenic” or “likely pathogenic” variants associated with observed docetaxel toxicities.
The observations presented in this case report implicate the reduced-function variants in CYP3A in the severe docetaxel-induced toxicities. First, CYP3A4 is the main route of docetaxel detoxification and elimination; second, the CYP3A4*22 variant reduces CYP3A4-mediated drug clearance; third, it is a rare variant (0.2% of people are homozygous for CYP3A4*22) (29); fourth, the only two patients in our research cohort that were CYP3A4*22 homozygotes and received docetaxel developed severe toxicity; and fifth, the patient that also had the rare CYP3A4*3 variant (variant allele frequency < 0.01) had even greater toxicities. Both patients were also homozygous for the CYP3A5*3 variant, which causes loss of CYP3A5 function. Previous studies have linked increased docetaxel exposure to toxicity, which provides mechanistic plausibility. Since docetaxel is cleared by the time these toxicities occur, and it would not be appropriate to rechallenge the patient with docetaxel, it is not feasible to obtain plasma docetaxel concentrations after the toxicities have occurred; this is a limitation of this observation. Also, we found no concurrent medications that inhibit CYP3A activity, suggesting it may be a genetic cause and we did not find any variants linked to pharmacodynamic sensitivity to docetaxel toxicities.
We searched the literature for reports of additional CYP3A4*22 homozygous patients who received docetaxel, and found only one report by Sim et al.; that patient died from typhlitis induced by a single dose of docetaxel (9). Thus, we are aware of a total of 3 cases of CYP3A4*22 homozygotes that were treated with docetaxel; all three experienced severe toxicities, one of which was fatal and one of which was life-threatening.
Several studies have reported the association of heterozygous CYP3A4*22 carriers with docetaxel-induced toxicities. Gastric cancer patients who were CYP3A4*22 carriers had higher rates of toxicity (58%) compared to non-carriers who were homozygous for CYP3A5*3 (41%) and non-carriers heterozygous for CYP3A5*3 (17%) (10). Increased docetaxel-induced adverse events were also observed in breast cancer patients who were CYP3A4*22 carriers and CYP3A5*3 homozygotes (9). Most of those toxicities were less severe than those we report in the current study, as would be expected in carriers of only one loss of function CYP3A4*22 allele. A role of CYP3A metabolism in docetaxel toxicities is also supported by the FDA label recommendations to avoid the concomitant use of strong CYP3A4 inhibitors (1).
Additional factors may also contribute to the toxicities. For example, the two patients in this report also had genetic variants in SLCO1B3 and ABCC2 transporters, which have been associated with docetaxel toxicities (18). They also had genetic variants in additional CYP regulatory genes that have been weakly associated with docetaxel toxicities (20). Patient 2 was homozygous for variants in ABCB1 that have been associated with reduced exposure, potentially compensating for the loss in function of CYP3A (11–15). The previous pulmonary embolism in patient 1 may also have contributed to the pneumonitis. Considered together, the final toxicity phenotypes are likely composed of the net effect of all these genetic and environmental factors.
Taxane-induced pneumonitis has been speculated to occur through an immune-mediated delayed hypersensitivity reaction. Although patient 1’s pneumonitis could have been allergic in nature, evidence also supports the possibility of it being caused by increased docetaxel exposure. Docetaxel-induced interstitial pneumonitis is estimated to occur in about 1 to 5% of patients and is known to be dose-dependent, occurring more frequently among those treated with 100mg/m2 vs. 60mg/m2 (2.2% vs. 0.7%) (30); but, allergic reactions are not typically dose-dependent. The patient had grade 4 neutropenia, a symptom frequently observed in patients with genetic variants resulting in increased docetaxel exposure (10), but not typically observed in docetaxel hypersensitivity reactions (31). Also, immune-mediated hypersensitivity often requires sensitization periods; however, in patient 1 and in some other case reports, the toxicity occurs after a single dose (32, 33). It is possible that the reduced clearance may sustain single-dose docetaxel exposure long enough to allow for allergic sensitization to occur. There may also be genetic variability in immune function genes that result in increased susceptibility to docetaxel toxicities, although those have yet to be identified.
In conclusion, our case report adds additional evidence to support the role of pharmacokinetic genetic variants in docetaxel toxicity. Since we are not aware of any cases where docetaxel was safely given to cancer patients with two reduced-function CYP3A4 alleles, we propose that extreme caution should be used when treating those patients with docetaxel.
This report contains original work that was presented as a poster at the Pharmacogenomics Global Research Network – American Society of Human Genetics (PGRN-ASHG) Conference, October, 2021.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by Indiana University Institutional Review Board. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
NP, TS, RL, and TCS wrote manuscript and performed clinical/genetic investigation. CA, MR, and BS performed clinical/genetic investigation. All authors contributed to the article and approved the submitted version.
This study was supported with funding from NIH Grant R35GM131812 (TCS), T32GM008425 (NP), the Monogrammed Medicine Program of the Vera Bradley Center for Breast Cancer Research (TCS), and the Indiana University Grand Challenge Precision Health Initiative (TCS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Emily Nelson, Casey Bales, Sheryl Lynch, and Lori Ann Smallwood for their help collecting patient data, the Indiana University Pharmacogenomics Laboratory for genotyping and sanger sequencing, and the Center for Medical Genomics at Indiana University for performing next generation sequencing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.809527/full#supplementary-material
1. U.S. Food and Drug Administration. Prescribing Information for Taxotere (2020). Available at: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/020449s084lbl.pdf.
2. Kenmotsu H, Tanigawara Y. Pharmacokinetics, Dynamics and Toxicity of Docetaxel: Why the Japanese Dose Differs From the Western Dose. Cancer Sci (2015) 106(5):497–504. doi: 10.1111/cas.12647
3. Hirth J, Watkins PB, Strawderman M, Schott A, Bruno R, Baker LH. The Effect of an Individual's Cytochrome CYP3A4 Activity on Docetaxel Clearance. Clin Cancer Res (2000) 6(4):1255–8.
4. Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, et al. Pharmacogenomics Knowledge for Personalized Medicine. Clin Pharma Therapeutics (2012) 92(4):414–7. doi: 10.1038/clpt.2012.96
5. Lee HH, Leake BF, Teft W, Tirona RG, Kim RB, Ho RH. Contribution of Hepatic Organic Anion-Transporting Polypeptides to Docetaxel Uptake and Clearance. Mol Cancer Ther (2015) 14(4):994–1003. doi: 10.1158/1535-7163.Mct-14-0547
6. Baker SD, Verweij J, Cusatis GA, van Schaik RH, Marsh S, Orwick SJ, et al. Pharmacogenetic Pathway Analysis of Docetaxel Elimination. Clin Pharmacol Ther (2009) 85(2):155–63. doi: 10.1038/clpt.2008.95
7. van Waterschoot RA, Lagas JS, Wagenaar E, Rosing H, Beijnen JH, Schinkel AH. Individual and Combined Roles of CYP3A, P-Glycoprotein (MDR1/ABCB1) and MRP2 (ABCC2) in the Pharmacokinetics of Docetaxel. Int J Cancer (2010) 127(12):2959–64. doi: 10.1002/ijc.25279
8. Engels FK, Ten Tije AJ, Baker SD, Lee CK, Loos WJ, Vulto AG, et al. Effect of Cytochrome P450 3A4 Inhibition on the Pharmacokinetics of Docetaxel. Clin Pharmacol Ther (2004) 75(5):448–54. doi: 10.1016/j.clpt.2004.01.001
9. Sim S, Bergh J, Hellström M, Hatschek T, Xie H. Pharmacogenetic Impact of Docetaxel on Neoadjuvant Treatment of Breast Cancer Patients. Pharmacogenomics (2018) 19(16):1259–68. doi: 10.2217/pgs-2018-0080
10. Meulendijks D, Rozeman EA, Cats A, Sikorska K, Joerger M, Deenen MJ, et al. Pharmacogenetic Variants Associated With Outcome in Patients With Advanced Gastric Cancer Treated With Fluoropyrimidine and Platinum-Based Triplet Combinations: A Pooled Analysis of Three Prospective Studies. Pharmacogenomics J (2017) 17(5):441–51. doi: 10.1038/tpj.2016.81
11. Fajac A, Gligorov J, Rezai K, Lévy P, Lévy E, Selle F, et al. Effect of ABCB1 C3435T Polymorphism on Docetaxel Pharmacokinetics According to Menopausal Status in Breast Cancer Patients. Br J Cancer (2010) 103(4):560–6. doi: 10.1038/sj.bjc.6605789
12. Kus T, Aktas G, Kalender ME, Demiryurek AT, Ulasli M, Oztuzcu S, et al. Polymorphism of CYP3A4 and ABCB1 Genes Increase the Risk of Neuropathy in Breast Cancer Patients Treated With Paclitaxel and Docetaxel. Onco Targets Ther (2016) 9:5073–80. doi: 10.2147/ott.S106574
13. Zhong J, Guo Z, Fan L, Zhao X, Zhao B, Cao Z, et al. ABCB1 Polymorphism Predicts the Toxicity and Clinical Outcome of Lung Cancer Patients With Taxane-Based Chemotherapy. Thorac Cancer (2019) 10(11):2088–95. doi: 10.1111/1759-7714.13184
14. Bosch TM, Huitema AD, Doodeman VD, Jansen R, Witteveen E, Smit WM, et al. Pharmacogenetic Screening of CYP3A and ABCB1 in Relation to Population Pharmacokinetics of Docetaxel. Clin Cancer Res (2006) 12(19):5786–93. doi: 10.1158/1078-0432.Ccr-05-2649
15. Tulsyan S, Chaturvedi P, Singh AK, Agarwal G, Lal P, Agrawal S, et al. Assessment of Clinical Outcomes in Breast Cancer Patients Treated With Taxanes: Multi-Analytical Approach. Gene (2014) 543(1):69–75. doi: 10.1016/j.gene.2014.04.004
16. Huang S, Yang J, Fu F, Wang C, Guo X, He B, et al. Clinical and Genetic Risk Factors for the Prediction of Hepatotoxicity Induced by a Docetaxel, Epirubicin and Cyclophosphamide Regimen in Breast Cancer Patients. Pharmacogenomics (2021) 22(2):87–98. doi: 10.2217/pgs-2020-0080
17. Lewis LD, Miller AA, Owzar K, Bies RR, Markova S, Jiang C, et al. The Relationship of Polymorphisms in ABCC2 and SLCO1B3 With Docetaxel Pharmacokinetics and Neutropenia: CALGB 60805 (Alliance). Pharmacogenet Genomics (2013) 23(1):29–33. doi: 10.1097/FPC.0b013e32835b16d8
18. Kiyotani K, Mushiroda T, Kubo M, Zembutsu H, Sugiyama Y, Nakamura Y. Association of Genetic Polymorphisms in SLCO1B3 and ABCC2 With Docetaxel-Induced Leukopenia. Cancer Sci (2008) 99(5):967–72. doi: 10.1111/j.1349-7006.2008.00765.x
19. Sone K, Oguri T, Uemura T, Takeuchi A, Fukuda S, Takakuwa O, et al. Genetic Variation in the ATP Binding Cassette Transporter ABCC10 Is Associated With Neutropenia for Docetaxel in Japanese Lung Cancer Patients Cohort. BMC Cancer (2019) 19(1):246. doi: 10.1186/s12885-019-5438-2
20. Chew SC, Lim J, Singh O, Chen X, Tan EH, Lee EJ, et al. Pharmacogenetic Effects of Regulatory Nuclear Receptors (PXR, CAR, Rxrα and HNF4α) on Docetaxel Disposition in Chinese Nasopharyngeal Cancer Patients. Eur J Clin Pharmacol (2014) 70(2):155–66. doi: 10.1007/s00228-013-1596-3
21. Bosó V, Herrero MJ, Santaballa A, Palomar L, Megias JE, de la Cueva H, et al. SNPs and Taxane Toxicity in Breast Cancer Patients. Pharmacogenomics (2014) 15(15):1845–58. doi: 10.2217/pgs.14.127
22. Liu Q, Ou-Yang QG, Lin QM, Lu XR, Ma YQ, Li YH, et al. Effects of 27 CYP3A4 Protein Variants on Saxagliptin Metabolism In Vitro. Fundam Clin Pharmacol (2021). doi: 10.1111/fcp.12693
23. Zhou XY, Hu XX, Wang CC, Lu XR, Chen Z, Liu Q, et al. Enzymatic Activities of CYP3A4 Allelic Variants on Quinine 3-Hydroxylation In Vitro. Front Pharmacol (2019) 10:591. doi: 10.3389/fphar.2019.00591
24. Kumondai M, Gutiérrez Rico EM, Hishinuma E, Ueda A, Saito S, Saigusa D, et al. Functional Characterization of 40 CYP3A4 Variants by Assessing Midazolam 1'-Hydroxylation and Testosterone 6β-Hydroxylation. Drug Metab Dispos (2021) 49(3):212–20. doi: 10.1124/dmd.120.000261
25. Chew SC, Sandanaraj E, Singh O, Chen X, Tan EH, Lim WT, et al. Influence of SLCO1B3 Haplotype-Tag SNPs on Docetaxel Disposition in Chinese Nasopharyngeal Cancer Patients. Br J Clin Pharmacol (2012) 73(4):606–18. doi: 10.1111/j.1365-2125.2011.04123.x
26. Wang D, Sadee W. CYP3A4 Intronic SNP Rs35599367 (CYP3A4*22) Alters RNA Splicing. Pharmacogenet Genomics (2016) 26(1):40–3. doi: 10.1097/fpc.0000000000000183
27. Plummer SJ, Conti DV, Paris PL, Curran AP, Casey G, Witte JS. CYP3A4 and CYP3A5 Genotypes, Haplotypes, and Risk of Prostate Cancer. Cancer Epidemiol Biomarkers Prev (2003) 12(9):928–32.
28. Machiela MJ, Chanock SJ. LDlink: A Web-Based Application for Exploring Population-Specific Haplotype Structure and Linking Correlated Alleles of Possible Functional Variants. Bioinformatics (2015) 31(21):3555–7. doi: 10.1093/bioinformatics/btv402
29. Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: The NCBI Database of Genetic Variation. Nucleic Acids Res (2001) 29(1):308–11. doi: 10.1093/nar/29.1.308
30. Harvey V, Mouridsen H, Semiglazov V, Jakobsen E, Voznyi E, Robinson BA, et al. Phase III Trial Comparing Three Doses of Docetaxel for Second-Line Treatment of Advanced Breast Cancer. J Clin Oncol (2006) 24(31):4963–70. doi: 10.1200/jco.2005.05.0294
31. Syrigou E, Dannos I, Kotteas E, Makrilia N, Tourkantonis I, Dilana K, et al. Hypersensitivity Reactions to Docetaxel: Retrospective Evaluation and Development of a Desensitization Protocol. Int Arch Allergy Immunol (2011) 156(3):320–4. doi: 10.1159/000324454
32. Read WL, Mortimer JE, Picus J. Severe Interstitial Pneumonitis Associated With Docetaxel Administration. Cancer (2002) 94(3):847–53. doi: 10.1002/cncr.10263
Keywords: DOCETAXEL, case report, CYP3A4, pneumonitis, pharmacogenetics
Citation: Powell NR, Shugg T, Ly RC, Albany C, Radovich M, Schneider BP and Skaar TC (2022) Life-Threatening Docetaxel Toxicity in a Patient With Reduced-Function CYP3A Variants: A Case Report. Front. Oncol. 11:809527. doi: 10.3389/fonc.2021.809527
Received: 05 November 2021; Accepted: 17 December 2021;
Published: 31 January 2022.
Edited by:
Haishu Lin, Shenzhen Technology University, ChinaReviewed by:
Umamaheswaran Gurusamy, University of California, San Francisco, United StatesCopyright © 2022 Powell, Shugg, Ly, Albany, Radovich, Schneider and Skaar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Todd C. Skaar, dHNrYWFyQGl1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.