- 1Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran
- 2Student Research Committee, School of Medicine, Shahroud University of Medical Science, Shahroud, Iran
- 3Department of Molecular Medicine and Biotechnology, Faculty of Medicine, Arak University of Medical Sciences, Arak, Iran
- 4Student Research Committee, University of Social Welfare and Rehabilitation Sciences, Tehran, Iran
- 5Department of Pharmacognosy, College of Pharmacy, Hawler Medical University, Erbil, Iraq
- 6Skull Base Research Center, Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 7Institute of Human Genetics, Jena University Hospital, Jena, Germany
- 8Clinical Research Development Unit of Tabriz Valiasr Hospital, Tabriz University of Medical Sciences, Tabriz, Iran
Stress granule (SG) formation is a well-known cellular mechanism for minimizing stress-related damage and increasing cell survival. In addition to playing a critical role in the stress response, SGs have emerged as critical mediators in human health. It seems logical that SGs play a key role in cancer cell formation, development, and metastasis. Recent studies have shown that many SG components contribute to the anti-cancer medications’ responses through tumor-associated signaling pathways and other mechanisms. SG proteins are known for their involvement in the translation process, control of mRNA stability, and capacity to function in both the cytoplasm and nucleus. The current systematic review aimed to include all research on the impact of SGs on the mechanism of action of anti-cancer medications and was conducted using a six-stage methodological framework and the PRISMA guideline. Prior to October 2021, a systematic search of seven databases for eligible articles was performed. Following the review of the publications, the collected data were subjected to quantitative and qualitative analysis. Notably, Bortezomib, Sorafenib, Oxaliplatin, 5-fluorouracil, Cisplatin, and Doxorubicin accounted for the majority of the medications examined in the studies. Overall, this systematic scoping review attempts to demonstrate and give a complete overview of the function of SGs in the mechanism of action of anti-cancer medications by evaluating all research.
Introduction
Stress granules (SG) are, from a higher perspective, a subset of RNP granules. Cellular mRNAs appear in the messenger ribonucleoprotein (mRNP) structure within the cell by being coated with proteins (1). SGs are structured, ranging from 100 to 2000 nm, and present in cytoplasmic foci (2). The word stress in the title of these granules indicates the effect of stress on the formation of SGs. Types of stress can be divided into two categories: conditional such as heat shock, arsenite, and hypoxia (3) and other categories including genotoxic drugs and x-ray (4). Stress granules proteins component can be included in three subgroups of RNA binding proteins, non-RNA binding proteins, and transcription initiation factors (5). Proteomic studies and the study of interprotein interactions of this structural component of SGs indicate a large number of proteins that can be included in the structure of SGs (6, 7). Identification of this protein component is essential when more attention is paid to the mechanism of formation of SGs and their effect on the pathogenesis of various diseases. In general, the stress on the cell is followed by the cessation of one of the most critical cell processes called translation (8). Stopping translation accurately at the initiation stage provides the cell with a supply of resources like the RNA-binding proteins involved in this process to build SGs (5).
Interestingly, the major component of the protein component of SGs is RNA-binding proteins that have two specific domains that predispose to the formation of protein aggregates and the construction of SGs, including prion-like domains (PLDs) and intrinsically disordered domains (IDDs) (9). Among these, proteins such as TIA1, PABP, and G3BP have the most involvement in the structure of SGs (10). These proteins can participate in the formation of SGs in two ways. First, the core structure is formed before forming the outer shell structure, where proteins such as G3BP1 and TIA1 attach to the mRNA in the nucleus and form the mRNP structure. This mRNP is transported to the cytoplasm as a core for the SGs formation, although it can also follow the translation process (11, 12). Then, by increasing the core size and connecting other components, a structure of 200 nm is formed, and by continuing the same process, a liquid-like shell is created by relying on microtubules, and it completes the SG structure. Liquid–liquid phase separation (LLPS) is a thermodynamically driven, reversible event that involves the separation of a liquid into two separate liquid phases with differing solute concentrations (13). Alternatively, the structure of SGs can begin to form during the liquid-liquid phase separation process before the formation of the core structure (14) due to poorly binding untranslated mRNPs (15). Then, with the addition of more untranslated mRNPs and more proteins as SGs protein components, the formation of the structure of SGs follows (16).
It should be noted that the structure of SGs is temporary. Cells under stress use SGs as a strategy to protect the translation process, and as the stress is relieved, the structure of SGs moves toward disassembling (17). Disequilibrium between assembly and disassembly can create the conditions for the pathogenesis of various diseases, from neurodegenerative diseases (18, 19) to autoimmune diseases (20) and cancer (21). SGs are involved in various dimensions of cancer, from formation to progression, metastasis, and response to various forms of treatment (22). Cancer is identified by abnormal cell proliferation with the potential to invade and spread to other parts of the body (23).
SGs are present in many cancers, and their up-reg has been proven in many different tumors, including hepatocellular carcinoma (24), sarcoma (25), pancreatic cancer (26), prostate cancer (27, 28), breast cancer (29), malignant glioma (30). Cancer cells are subjected to various stresses due to overgrowth and overuse of nutrients and the effect of various therapies (31). Cancer cells take advantage of the structural ability of SGs under various stresses to survive (32). This research reviews all the studies in the field of cancer treatment in which traces of SGs have been seen in an attempt to review the progress made in targeting SGs in cancer therapy in order to be able to find ways and means of treating cancer.
Methods
The Overall Framework of the Review
The strategy in this article was established on the basis proposed by Arksey and O’Malley (2005) (33). Later versions of this strategy were developed by Levac et al. (2010) (34) and Colquhoun et al. (2014) (35). This review follows a 5-step framework, including the following steps: classification of the research question, search strategy, study selection, charting the data, Collating, summarizing, and reporting the results. Consultation is the sixth and final step, which is not covered in this article. During the article’s writing, the Preferred Reporting Items for Systematic Reviews and Meta-Analysis Extension for Scoping Reviews (PRISMA-ScR) Checklist (36) is used to consider and observe two critical aspects clarity and transparency.
Classification of the Research Question
The main research question that was developed is as follows:
‘What do studies on the involvement of stress granules in anti-cancer drugs and cancer treatments represent?’
‘What are these anti-cancer medications, and what is their functional mechanism?’
Critical studies are considered to be included in general and comprehensive questions.
Search Strategy
Researchers used PubMed, Scopus, Cochrane, Google Scholar, Embase, Web of Science, and ProQuest to find the articles. The search was not limited by date, language, subject, or type of publication. Review publications were also revised to ensure that related articles were not neglected. For our research on anti-cancer medications and Stress granules, we almost used the following search query: “cancer*” OR “neoplasm*” OR “cyst*” OR “carcinoma*” OR “adenocarcinoma*” OR “neurofibroma*” OR “tumor*” OR “tumour*” OR “malign*” AND “stress granule” OR “stress granules”. Keywords were selected according to background reading and subject headings in PubMed and Embase search engines to have the most coverage on cancer studies. Medical subject heading (MeSH) for the PubMed database and emtree for the Embase database are correctly used in the search. The most recent search was conducted on October 16, 2021. EndNote X8.1 was used to manage the references.
Study Selection
The publications found during the search were screened for Stress granules involving anti-cancer medications in humans, cell lines, and animal models. Journal articles, conference presentations, erratum, conference abstracts, and reports were among the publications screened. Two reviewers (MRA and MSM) independently completed the screening (first only title and abstract, second full-text). At this point, the article titles and abstracts were reviewed using the inclusion and exclusion criteria listed below.
Inclusion Criteria
I. Stress granules involved in anti-cancer medications (any cancer) (all human studies, animal studies, cell culture studies)
II. Articles in English only
III. Original studies
Exclusion Criteria
I. Research on stress granules in diseases other than cancer
II. Languages different from English
III. Non-original studies
IV. Stress granules have been studied using bioinformatics and impractical techniques.
Charting the Data
Following the completion of the final articles that address the research questions, the data-charting was created to organize the study variables using the following headings: author’s name, year of publication, country, type of study, human samples, animal models, cell lines, SG protein components, methods, major findings, and references. Separately, two reviewers (MRA and MSM) extracted data from articles using charts.
Collating, Summarizing, and Reporting the Results
A quantitative and qualitative analysis of the publications’ findings, presented in tables and charts, was performed. The quantitative analysis section reviewed a descriptive numerical summary of the studies’ scope, nature, and distribution. In the qualitative analysis section, the presented data were confirmed in light of the broader context proposed by Levac et al. in a narrative review.
Results
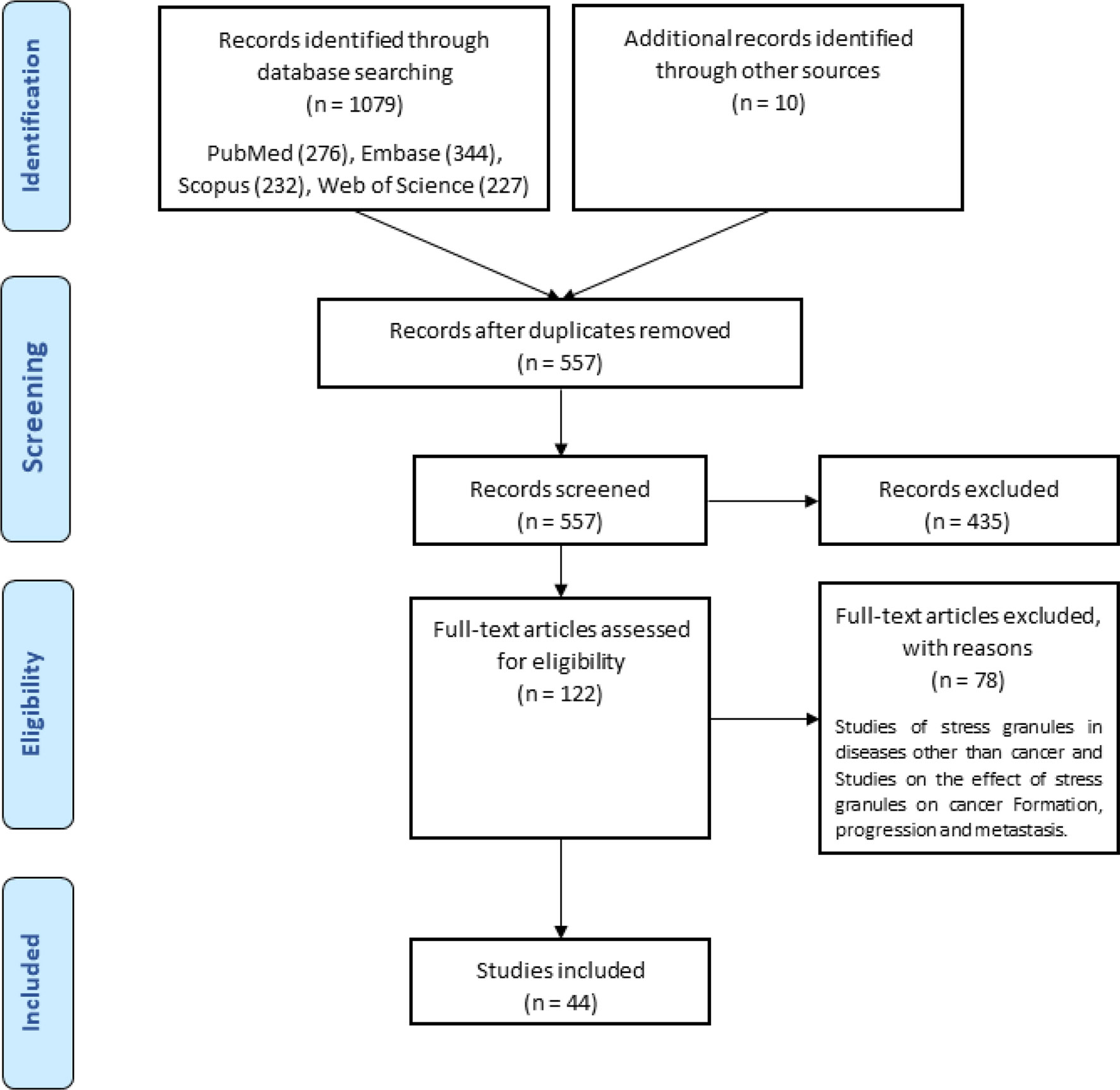
A total of ten hundred and seventy-nine items were returned from a keyword search across seven databases. Meanwhile, ten additional records were discovered through other sources, increasing the total number of articles. Endnote software identified and eliminated 522 duplicate records, bringing the total to 557. Following a review of the article titles and abstracts, 122 publications that addressed the research subject were chosen. Following a study of the entire texts of 122 publications, 44 articles for the charting data stage were included in Table 1. The procedure for discovering relevant articles and research is depicted in Figure 1. Eligible research was published between 2007 and 2021. The percentage of various research is depicted in Figure 1. Meanwhile, cell culture research accounts for the vast majority of studies, accounting for approximately 72.7 percent of all studies (24, 30, 37–43, 45, 47, 49, 50, 52, 53, 55, 57–59, 61–63, 65, 66, 68–73, 75, 76). Following that, cell culture, animal, and tissue specimen studies accounted for 13.6% of studies (26, 29, 48, 60, 64, 76), cell culture and tissue specimen studies accounted for 9.1% of studies (46, 51, 54, 67), and cell culture and animal studies accounted for 2% of the total studies (57, 74). Pancreatic cancer (26, 46), gastric cancer (67, 76), breast cancer (29), sarcoma (64), colorectal cancer samples (54, 60), primary malignant B cells (51), and osteosarcoma (48) were among the human cancer samples utilized in the research. Figure 2 depicts the quantity of each SGs protein component investigated in all investigations. G3BP1 has the greatest rate (16.9%), followed by eIF2α (13.4%), TIA-1 (9.2%), and eIF4G1 and FXR-1 (4.3%). Figure 3 is a schematic image of the proportion of anti-cancer medications utilized in studies in which bortezomib (26, 30, 39, 42, 59, 62, 63, 73) with 14% has the largest share and followed by 5-Fluorouracil (49, 54, 74), cisplatin (43, 48, 58), Oxaliplatin (26, 67, 76), and Sorafenib (24, 57, 62) with 5.3% of all anti-cancer medications used in studies. The number of studies is limited to twelve countries, with the United States accounting for the most with nine, followed by Canada with eight, China with six, South Korea with four, Switzerland, Germany, Brazil, Japan, and Australia with two each, and Italy, France, Poland, the United Kingdom, Argentina, Chile, and Russia with one each.
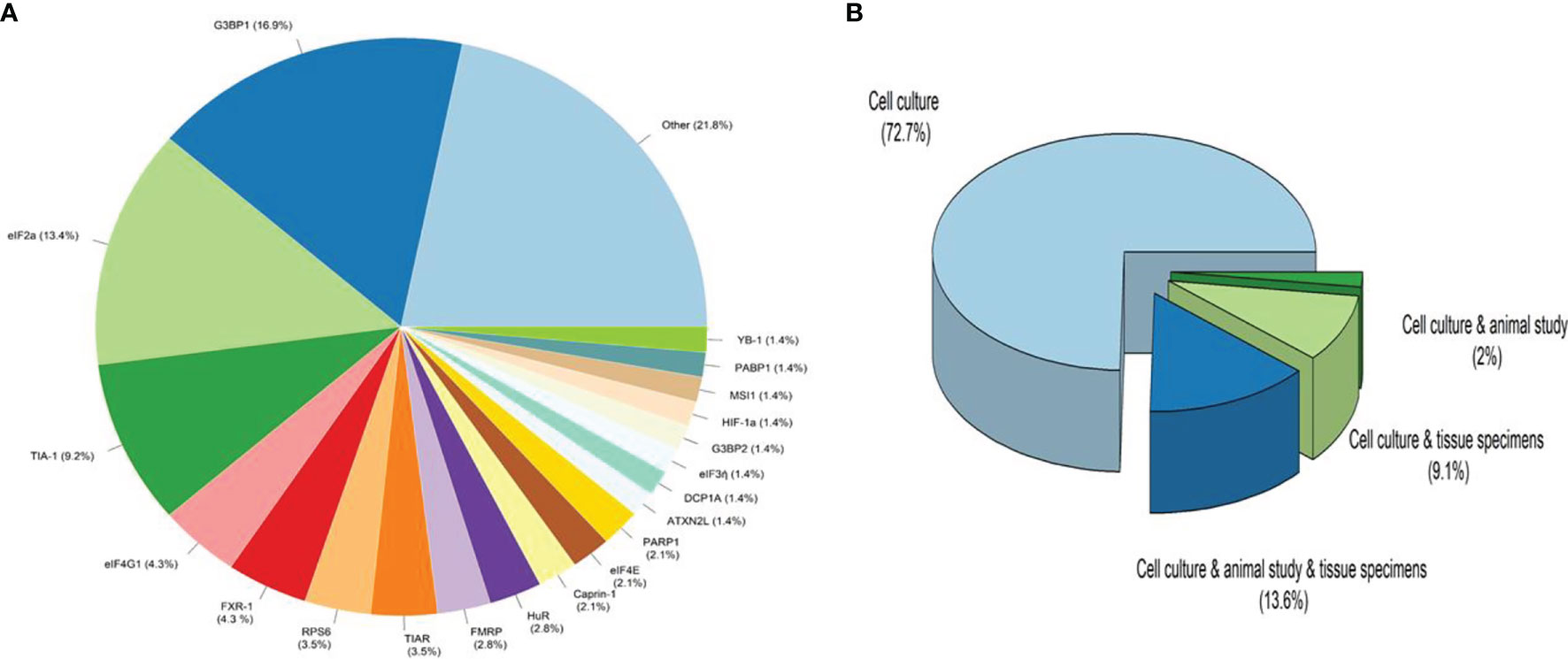
Figure 2 The ratio of Stress Granules protein components and type of studies. (A). Other refers to proteins that have been considered only once in all studies, including CCAR1, DDX3, DDX6, eIF3b, eIF3c, eIF3f, eIF4A1, eIF4D, eIF4E, eIF4G1, FMR1, FMRP, G3BP1, hnRNPA1, hnRNPk, hnRNPA2B1, HSP90a, mTOR, PRMT1, RACK1, RAPTOR, Rbfox2, Sam68, SQSTM1/p62, SRSF1, TDRD3, TIAL1, TTP, USP9X, YWHAZ, ATXN2. (B). Cell culture studies were the most common kind of research, followed by cell culture, animal studies, and tissue specimen studies with the most significant number (13.6 percent in study design), cell culture and tissue specimen studies with 9.1 percent, and cell culture and animal studies with 2% of all studies.
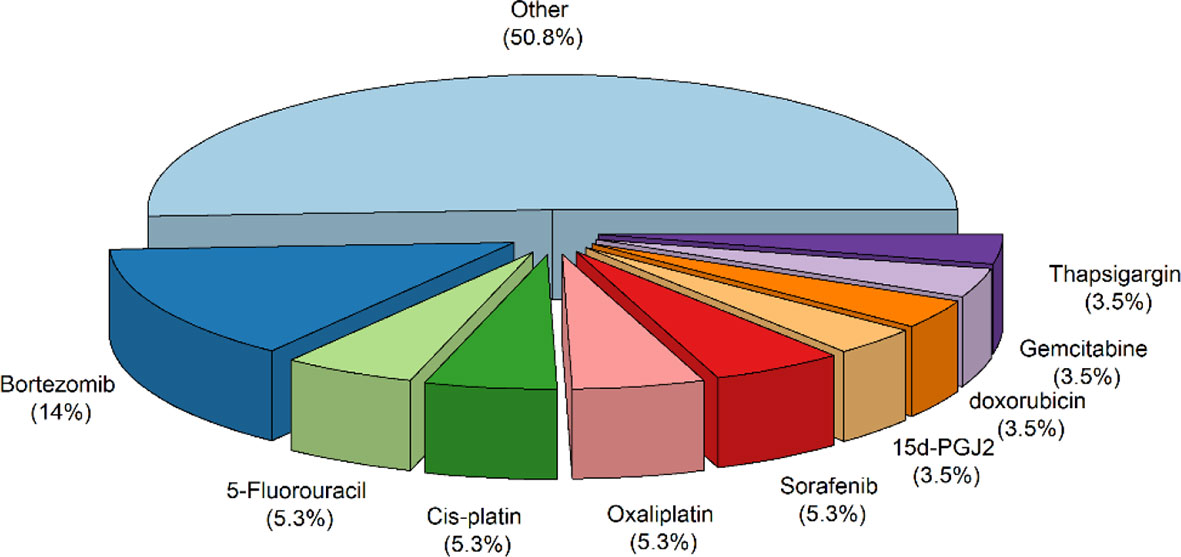
Figure 3 The proportion of anti-cancer medications utilized in studies. Other refers to anti-Cancer medications that have been considered only once in all studies, including Arsenic trioxide, boric acid, c108, camptothecin, Capecitabine, celecoxib, cis-diamminedichloroplatinum, Darinaparsin, docetaxel, fasnall, ibrutinib, Imatinib, lapatinib, Mitoxantrone, Mn3O4, MO-460, morusin, MS-275, nocodazole, Phenethyllisothiocyanate, Psammaplysin F, QLT0267, raloxifene, resveratrol, TAT-RasGAP317–326 (peptide), torkinib, tunicamycin, Verrucarin, Vinca Alkaloid.
Discussion
Stress Granules Branch Off From RNP Granules
RNP granules are non-membrane-bound cellular compartments with high protein and RNA concentrations. Nuclear granules like Cajal bodies, paraspeckles, the nucleolus, and cytoplasmic granules like stress granules and processing bodies fall into this category (77, 78). RNP granules are dynamic in nature and rely on RNA for assembly. As a result, the formation of dynamic RNP granules for the concentration of specific cellular components is a strategy that has been conserved across a wide range of organisms and cellular compartments (79). Among these, P-bodies (80) and SGs (81) are two types of cytoplasmic mRNP granules that form from pools of non-translating mRNA. The P-bodies were discovered during research into the localization of proteins associated with the 5′ to 3′ mRNA decay pathway, and the discovery of mRNA decay mediators in these structures led to the initial hypothesis that P-bodies are cellular sites of mRNA decay (82). SGs were named after dense cytoplasmic bodies formed in chicken embryonic fibroblasts when they were stressed in 1988 (83). SGs are dense bodies made up of RNA and proteins that are found in the cytosol when the cells are under stress (84). Ribonucleoproteins appear in response to various stresses, and their number decreases as the stress fades away and is restricted to SGs being disassembled (22).
SGs in Cancer Treatment
The most challenging aspect of the clinical picture is the use of SGs by cancer cells in response to treatment and chemotherapy. In cancer cells, the equilibrium between assembling and disassembling SGs versus chemotherapy is disrupted, and this imbalance tends to increase the number of SGs. Aside from pathophysiological conditions, numerous studies have linked cancer cell survival to the accumulation of SGs in response to chemotherapy drugs, which can aggravate cancer. EIF2α phosphorylation is the common denominator of the majority of chemotherapeutic agents (85). It is thought that four stress-related kinases phosphorylate eIF2α (17, 18).
Among these are the double-stranded RNA-dependent protein kinase (PKR), PKR-like endoplasmic reticulum kinase (PERK) (86), haem-regulated inhibitor (HRI), and general control nonderepressible 2 (GCN2) (86, 87). Chemotherapy drugs typically stimulate SG accumulation by activating these phosphorylating kinases. Simultaneously, studies show that targeting SGs as anti-stress granule therapy in combination with conventional chemotherapy could provide a new perspective on cancer treatment and has the potential to be recognized as a new treatment through further research.
Chemotherapy Drug Traces in the Induction of SG Assembly
Sorafenib
Sorafenib has shown anti-tumor efficacy in animal models of RCC (88), HCC (89), and DTC (90) by inhibiting tumor proliferation and angiogenicity and promoting tumor death. Although first identified as a Raf inhibitor, it was later shown that sorafenib has several targets, including many protein kinases in the Ras–Raf–MEK–ERK signaling cascade. Sorafenib has the potential to block a variety of oncogenic Ras and Raf mutations, including the BRAF V600E mutant, which is linked to tumor angiogenesis and invasion, as well as the silencing of tumor suppressor genes in a spectrum of cancer types and also inhibits VEGF receptors, platelet-derived growth factor receptor family proteins (PDGFR and Kit), and FMS-related tyrosine kinase 3 (FLT-3) (91), as well as the oncogenic RET kinase (92) and the degradation of the anti-apoptotic myeloid cell leukemia 1 (Mcl-1) protein (93). Sorafenib was approved for use in solid tumors based on these findings. It also reduces the severity of its side effects because it is a potent inhibitor of epoxide hydrolase solution due to the structure of its distributed 1,3-di urea (94).
Pathways can lead to sorafenib-treated cancer cells becoming resistant to the drug. The formation of SGs can be considered as a frontier in resistance to sorafenib treatment. It should be noted that sorafenib produces SGs in a variety of cancer cells, including HeLa (cervix), MCF-7 (breast), PC3, and LnCaP (prostate), with a high degree of potency (80%) (24). Resistance to sorafenib chemotherapy occurs through the pathway in which ATF4 and PERK are involved. Sorafenib induces eIF2α phosphorylation by PERK, and this phosphorylation leads to the formation of SGs. Phosphorylated eIF2α, on the other hand, induces preferential induction of ATF4 expression, which promotes cell death (95). Low ATF4 expression is required for resistance to chemotherapy due to its activity in promoting the expression of antioxidant and chaperone genes that contribute to cell survival and growth (96). On the other hand, under the influence of Sorafenib, PERK mediates the formation of SGs by phosphorylation eIF2α. By capturing ATF4 mRNA, SGs have been shown to minimize expression to the extent necessary for survival and resistance to chemotherapy and increase chemotherapy resistance (24). Sorafenib phosphorylates GCN2 to phosphorylate its downstream protein, eIF2α, promoting cell apoptosis (57). Meanwhile, cox2 protein, which is more expressed in sorafenib-treated cells, inhibits the apoptotic activity of cells with its anti-apoptotic function (97); Cox2 mRNA is localized in the structure of SGs. Combination therapy with sorafenib and celecoxib, which inhibits cox2, is better in chemotherapy-resistant cells than treatment with sorafenib alone (57). It was found that reducing the number of SGs could increase the effectiveness of chemotherapy. Psammaplysin F is a marine sponge-derived metabolite that has the ability to reduce the number of SGs and increase the effectiveness of chemotherapy drugs such as Sorafenib and Bortezomib (62).
Bortezomib
Bortezomib is an anti-cancer drug that was made for the first time in 1995, approved by the food and drug administration (FDA) in 2003 to treat multiple myeloma and mantle cell lymphoma (Velcade, PS-341; Millennium Pharmaceuticals, Inc., Cambridge, MA) (98–100). It is a 26S proteasome inhibitor, modified dipeptide boronic acid derived from leucine and phenylalanine. It could inhibit the proteasome reversibly in mammalian cells (101, 102). The proteasome controls protein production and function in normal cells by degrading ubiquitylated proteins and ridding the cell of aberrant or misfolded proteins (103). Clinical and preclinical evidence supports the proteasome’s role in sustaining myeloma cells’ eternal nature, and cell-culture and xenograft data suggest a similar function in solid tumor malignancies. While various processes are believed to be at work, proteasome inhibition may limit the degradation of pro-apoptotic proteins, prompting programmed cell death in cancer cells (99, 104). The 26S proteasome consists of a 20S core complex and 19S regulatory complex, and remarkably, the β-subunits of the 20S core complex have the catalytic function. bortezomib’s binding position is the threonine hydroxyl group in β1-subunit and β5-subunit of the 20S core in the proteasome structure (99, 102). Bortezomib inhibits the chymotrypsin-like activity of the proteasome through the boronic acid group in its binding to the threonine hydroxyl group in the β5-subunit (105, 106).
Bortezomib restraint more than 75% of proteasomes in whole blood samples up to one hour after the dose of bortezomib (99), and additionally, it binds 83% of human plasma proteins (101). Bortezomib has essential activities such as anti-tumor function, growth inhibition, and suppression of apoptosis. On the other hand, bortezomib prevents the progression of the cell cycle in the transition from the G2 phase to the M phase (107) and could influence the NF-κB signaling pathway, leading to anti-apoptotic target genes and expression of anti-apoptotic proteins (108). NOXA is a pro-apoptotic protein that bortezomib provokes in cancer cells (109, 110). Cytochrome P-450 enzymes 3A4, 2D6, 2C19, 2C9, and1A2 are responsible for metabolizing bortezomib through oxidative ways. According to the reports in this metabolization, two isomers from a single metabolite are generated due to bortezomib deboronation, and hydroxylation and deamination occur (101). The metabolization of bortezomib produces more than 30% inactive metabolites (111).
SGs are constituted by provoking bortezomib in cancer cells such as HeLa cells, Calu-I (lung cancer), and Caco (colon cancer) cells, but not all cancer cells like Hs578T breast cancer cells. Under long-term bortezomib (1 M, 10 h) therapy, the synthesis of SGs under stimulation by bortezomib is reversible; therefore, SGs disassemble and partially activate translation; this event occurs independently of eIF2α dephosphorylation. HRI and GCN2 are two kinases responsible for the phosphorylation of eIF2α caused by Bortezomib induction (39). The findings suggest that HRI may promote cancer cell resistance to bortezomib (39, 81). Following HRI reduction, SG formation decreases, and also IF2α phosphorylation is reduced through bortezomib (12). The efficiency of bortezomib was increased by knocking down the HRI in HeLa cells (62). RACK1 or TRAF2 is an apoptotic molecule inactivated by SGs cause to impede cancer cell resistance to bortezomib (39). Flow cytometry analysis shows that cells were treated with bortezomib, which raised the permeability of the plasma membrane. Angiogenesis increased in a conforming in vivo model, U87 cells conditioned culture media under bortezomib for 24 hours. Silencing G3BP1 as an SGs protein component might enhance bortezomib-induced apoptosis (59).
There is arginylated calreticulin in the structure of SGs, and it moves to the plasma membrane, where it can regulate cell death in cells treated with bortezomib. Arginylated calreticulin also acts as an apoptosis promoter (63). Bortezomib’s efficacy for solid tumors is inadequate due to resistance to cell death induction (30); nevertheless, insertion of arginylated calreticulin into the plasma membrane of glioma cells treated with bortezomib can initiate the apoptotic pathway (63). Bortezomib’s cytotoxicity would be increased by inhibiting the development of SGs. Chikungunya virus expressed non-structural protein 3 (nsP3), which might impede the development of SGs by inducing G3BP into cytoplasmic foci. Transfecting nsP3 into cancer cells and then treating them with bortezomib might pave the way for a novel strategy for cancer treatment (73). SGs regulate the production of the anti-apoptotic protein p21WAF1/CIP1; Bortezomib promotes the accumulation of p21 mRNA and its translation. p21WAF1/CIP1 and its regulatory protein CUGBP1 inhibit apoptosis in response to bortezomib therapy (42).
5-Fluorouracil
5-Fluorouracil (5-FU) is a uracil and thymine analog used as an antimetabolite and anti-cancer medicine. In the 1950s, researchers observed that rat hepatoma cells utilize pyrimidine uracil to the biosynthesis of nucleic acid, and this finding showed a clear horizon in cancer treatment (112–114). 5-FU is broadly used for treating solid tumors like breast, gastrointestinal system (colon, rectum, anus, esophagus, pancreas, and stomach), head and neck, and ovary (115). The fluorine atom is placed instead of hydrogen of uracil in the 5-FU structure (113). 5-FU inhibits thymidylate synthase (TS), and its metabolites incorporate into RNA and DNA, hence applying its antineoplastic effect (116). TS is the only enzyme that produces de novo thymidylate to DNA replication and repair (117). Increasing dUTP could result from TS inhibition and 5-FU metabolite FdUTP might become misincorporated into DNA (118, 119). As a result of these occurrences are DNA strand breaks and cell death (116). Thymidine kinase produces thymidylate from thymidine, so it is a potential salvage pathway TS deficit and provides a mechanism for resistance to 5-FU (120). Dihydropyrimidine dehydrogenase (DPD) is the rate-limiting enzyme in 5-FU catabolism that turns 5-FU to dihydro fluorouracil (DHFU). DHFU is expressed in the liver, and as well as more than 80% of consumed 5-FU is generally catabolized in the liver (121).
The enzymes responsible for metabolizing uracil and thymine could also metabolize 5-FU, and the mechanism of entering 5-FU into the cell is the same as for uracil. 5-FU undergoes intracellular transmutation to active metabolites such as fluorodeoxyuridine monophosphate (FdUMP), fluorodeoxyuridine triphosphate (FdUTP), and fluorouridine triphosphate (FUTP) (122). The 5-FU metabolite integrated into RNA then prevents pre-rRNA maturation into rRNA (123, 124), damages post-transcriptional modification of tRNAs (125, 126), and the assembly and activity of snRNA/protein complexes, resulting in pre-mRNA restraint splicing (127). The suppression of pre-rRNA maturation into rRNA by 5-FU therapy leads to a lack of synthesis of functional ribosomes (128). The incorporation of 5-FU metabolite into RNA is a factor in triggering SGs assembly (49).
There is a stemness gene in neuronal and epithelium cells, namely Musashi-1, which is an RNA-binding protein (129). A study indicated that Musashi-1 has a fundamental role in increasing the extension of CD44+ colorectal cancer stem cells and SG formation. Remarkably, when colorectal cancer cell lines are treated with 5-FU, Musashi-1 leads to SGs formation. Musashi-1 interacted with SGs through its C-terminal region. 5-FU stimulated SGs contained Musashi-1 along with G3BP. The C-terminal of Musashi-1 is critical for SGs formation under the induction of 5-FU. Furthermore, they realized that Musashi-1 causes colorectal cancer drug resistance by forming SGs during 5-FU treatment because Musashi-1 prevents apoptosis in colorectal carcinoma cells via the formation of SGs under 5-FU treatment (54). On the other hand, 5-FU could activate PRK (protein kinase RNA-activated), directing to eIF2α phosphorylation (79), thereby forming SG. Based on experiences, 5-FU influences on SGs formation under stress, and SGs become larger. By induction of 5-FU in HeLa cells, SGs include mediator protein RACK1, and disassembly of SGs was affected (49).
Cisplatin
Cisplatin is an anti-cancer medication that is useful in the treatment of a variety of malignancies (130). This compound has the chemical formula cl2H6N2pt, which is essentially insoluble in water but soluble in dimethylpropane and N-dimethylformamide (131). M. Peyron discovered and synthesized cisplatin in 1844. Years later, in 1960, Rosenberg demonstrated that platinum electrolytes might halt cell development (132). Despite its anti-cisplatin function, it produces side effects and difficulties in patients, including nephrotoxicity, ototoxicity, myelosuppression, gastrotoxicity, and allergies (133, 134). These cisplatin adverse effects are most likely caused by the substance’s interaction with the N7 position in purine molecules in DNA or by disrupting the fusion of double-stranded or single-stranded DNA molecules (135). Cisplatin is used to treat several malignancies, including ovarian, testicular, and cervical cancers. However, it is essential to note that in these cancers, tumor cells can develop resistance to cisplatin for a variety of reasons, including reduced cisplatin (DDP) levels in the cell, increased glutathione and glutathione S-transferase activity, accumulation of metallothionein’s in the cell, and improved DNA repair (136). There is widespread agreement that cisplatin enters the cell via passive transport, which lends credence to the idea that DDP cannot be absorbed completely (137).
One study discovered that Cisplatin therapy results in a lower rate of SG production than predicted. The fraction of cells containing SGs is modest, accounting for 5% of total cells. It is unknown what causes reduced SG production in cisplatin-treated cells; Cisplatin may interfere with SG formation. On the other hand, most cisplatin-induced SGs are likely to be undetectable under a microscope and are distinct from those generated with sodium azide or sodium arsenite (138). However, one study revealed that cisplatin had no effect on SG formation and had no effect on eIF2α. It does not cause ER stress and, when combined with other chemotherapeutic medicines such as ThapsiGargin or tunicamycin, can cause apoptosis in cancer cells (43). Remarkably, another study noted that a primary effect of cisplatin is to prevent the translation from progressing by increasing 4E-BP1 dephosphorylation and eIF2α phosphorylation, respectively. It inhibits the production of SGs in a concentration-and time-dependent way by targeting ribosomes. Cisplatin inhibits translation initiation and promotes cytosolic small ribosomal 40S subunit aggregation to impede ribosome interaction in translation complexes (139). Resistance to cisplatin can result in SGs containing caprin1, one of the components that may be integrated into their structure, and cause chemotherapy resistance, prevent cisplatin-induced apoptosis, and promote tumor development (48).
Gemcitabine
Chemotherapy is likely to give significant local control while also prolonging life. However, there is no practical or widely used therapy for advanced or metastatic pancreatic cancer. Gemcitabine, a deoxycytidine nucleoside analog (2′-deoxy-2′,2′-difluorocytidine; dFdC), has demonstrated anti-cancer efficacy against a wide range of malignancies, including pancreatic, lung, and breast cancers. GEM action is dependent on its entrance into cells, where it is immediately phosphorylated by deoxycytidine kinase (DCK), producing monophosphate and diphosphate (dFdCDP) (140, 141). Because of the inhibition of ribonucleotide reductase, diphosphate has an anti-cancer action. Another active GEM metabolite that may be integrated into DNA is the triphosphate metabolite (dFdCTP). The suppression of DNA synthesis is the most significant mode of action of gemcitabine. When dFdCTP is integrated into DNA, it incorporates a single deoxynucleotide, inhibiting chain elongation. This non-terminal location of gemcitabine prevents DNA polymerases from proceeding, a process known as “masked chain termination,” which also prevents gemcitabine removal by DNA repair enzymes (142).
On the other hand, gemcitabine mediates PERK- eIF2α phosphorylation and suppresses translation at the cellular level (1, 143). In response to various stress events, the eukaryotic initiation factor 2 (eIF2) subunit is phosphorylated at serine 51, triggering the Integrated Stress Response (ISR) (144). Resistance to gemcitabine chemotherapy is achieved in this way: Phosphorylated eIF2α (p-eIF2α) significantly reduces translation initiation and total protein synthesis, enabling cellular resources to be conserved. Furthermore, p-eIF2α promotes the preferential translation of specific mRNAs, most notably ATF4, whose overexpression increases the genes’ expression involved in oxidative stress (OS), metabolism, and nutrition absorption (145, 146). Thus, p-eIF2α gene reprogramming helps cells recover from stress-induced damage, increasing apoptosis in response to moderate stress and enabling survival in response to chronic stress (146, 147). In addition to inhibiting translation, phosphorylated eIF2α causes the cell to produce more SGs (15). In the sorafenib treatment, it was established that SGs promote chemotherapy resistance via suppressing ATF4 expression (24). Treatment with gemcitabine also maintains ATF4 preferred expression, which may contribute to chemotherapy resistance (26). After gemcitabine therapy, it was discovered in pancreatic epithelial cells that if eIF3f, a component of SGs, is knocked down, the gemcitabine-resistant cell becomes sensitive to this chemotherapeutic agent (46). On the other hand, gemcitabine can improve the sensitivity of other chemotherapy medicines by blocking glutamine metabolism (74).
Oxaliplatin
Oxaliplatin is a third-generation cisplatin analog that has demonstrated promising therapeutic results in colon cancer patients resistant to cisplatin. Oxaliplatin is used in combination with other medicines, such as 5-fluorouracil with leucovorin, to achieve response rates of up to 60%, and the inclusion of irinotecan to enhance pancreatic cancer therapy (148). Oxaliplatin has been linked to several different modes of action. Oxaliplatin, like other platinum-based compounds, causes cytotoxicity primarily through DNA damage. Apoptosis in cancer cells can be induced by the development of DNA lesions, the halt of DNA synthesis, the inhibition of RNA synthesis, and the activation of immunologic responses. Oxaliplatin also has synergistic effects with other cytotoxic medicines, although the underlying processes are less well known (149).
Oxaliplatin resistance, like cisplatin resistance, is obtained by a variety of mechanisms, including lower drug uptake and/or greater efflux of the drug, intracellular sequestration, decreased DNA adduct production, improved DNA repair, or increased adduct tolerance, and decreased sensitivity to platinum DNA adducts (150–152). The overall effect of oxaliplatin absorption and outflow is cellular accumulation. The human copper transporter hCTR1, as well as the organic cation transporters OCT1, 2, and 3, can all facilitate oxaliplatin absorption (SLC22A1-3) (153, 154). P-type ATPases, particularly ATP7A and ATP7B, appear to have a functional role in oxaliplatin efflux or sequestration (155, 156). The production of platinum-DNA adducts may be reduced as a result of decreased oxaliplatin transport. Differences in platinum DNA adducts and downstream signaling may explain the activity in colon tumors that are inherently resistant to cisplatin (157).
Resistance to Oxaliplatin may be connected to SGs. ATXN2L, as an SG component, contributed to the recurrence and development of Gastric Cancer (GC), even when treated with Oxaliplatin. ATXN2L expression was increased by EGF and its downstream PI3K/Akt signaling. On the one hand, ATXN2L overexpression aids migration and invasion through EMT. ATXN2L, on the other hand, aiding SGs assembly during oxaliplatin-induced stress. ATXN2L overexpression resulted in intrinsic and acquired oxaliplatin resistance. In turn, oxaliplatin-resistant cell lines expressed more ATXN2L as well as EGF and EGFR. These findings formed a positive feedback loop connecting EGF, ATXN2L, and oxaliplatin resistance because Oxaliplatin had previously been demonstrated to increase PI3K/Akt signaling in a compensatory way. ATXN2L might be utilized as a prognostic and therapeutic target in GC, primarily if oxaliplatin-based chemotherapy is applied (67).
Doxorubicin
Doxorubicin is a commonly used anti-cancer medication; typical indications include hematological (such as leukemia and lymphoma, including both Hodgkin’s and non-lymphoma) Hodgkin’s and solid organ malignancies (such as breast cancer, thyroid cancer, sarcoma, osteosarcoma, Kaposi’s sarcoma, and others) (158–160). It is regarded as one of the frontline medicines in many chemotherapy regimens since it is a time-tested anti-cancer agent. Chemotherapeutic regimens, including Doxorubicin (anthracyclines), are superior to regimens that do not contain anthracyclines in studies (161, 162). The two most widely hypothesized and effective mechanisms related to doxorubicin action are damage to cell membrane DNA and other cellular proteins caused by free radical production and intercalation into the cellular DNA, resulting in failure of DNA repair mediated primarily by topoisomerase IIa (163). Doxorubicin is transformed to the unstable intermediate metabolite semiquinone, which is unstable and is converted back to Doxorubicin throughout the process, generating reactive oxygen species (ROS). These free radicals cause extensive cellular damage, including lipid peroxidation, cell membrane degradation, DNA damage, and the induction of apoptosis (164).
One set of genes is responsible for free radical production (NADH dehydrogenase, NO synthase, and xanthine oxidase). In contrast, the other set is responsible for free radical deactivation (NADH dehydrogenase, NO synthase, and xanthine oxidase) (antioxidants, namely glutathione peroxidase, superoxide dismutase, and catalase) (165, 166). According to the second hypothesized mode of action, when Doxorubicin enters the target cell’s nucleus, it intercalates with the host DNA and targets TOP2A (167). TOP2A is in charge of separating entangled DNA, as well as temporarily generating and eventually repairing double-strand DNAs (double-strand breaks [DSB]) (72). Doxorubicin slows the repair process by interfering with the function of TOP2A, resulting in the formation of a significant number of DSBs (168). The presence of DSBs triggers the apoptotic pathway (caspase-dependent) by activating the p53 and FOXO3 genes. The ratio of anti-apoptotic to pro-apoptotic members of the Bcl2 protein family has changed (169). Other suggested modes of action for Doxorubicin include the inhibition of DNA and RNA synthesis as well as the promotion of mitochondrial ROS generation, which triggers the death cascade (163). Furthermore, Doxorubicin has the ability to activate p53, a tumor suppressor that tries to protect cells from specific tumorigenic changes (170).
Although a variety of stressors have been identified as happening in the tumor microenvironment, including local hyperthermia, UV, ionizing radiation exposure, ER stress, oxidative stress, genotoxic stress, and chemo-toxic and inflammatory stress, oxidative stress best depicts the prevalent phenomena surrounding the tumor tissues. Aside from the oxidative stress caused by hypoxia and ATO treatments, Doxorubicin significantly increases ER stress and pro-apoptotic processes that promote the development of SGs (1). In particular, Doxorubicin increases the number of SGs by directly affecting phosphorylation on eIF2α (53). In a study on the fission yeast Schizosaccharomyces pombe, the effect of Doxorubicin on SGs was further studied, and it was found that in this Schizosaccharomyces pombe, Doxorubicin, along with heat, causes the formation of SGs from a non- eIF2α -independent pathway but is targeted. The formation of SGs decreases the sensitivity of cells to Doxorubicin (171).
Conclusion
SGs have evolved into one of cancer cells’ primary stress-response mechanisms. SGs allow cancer cells to go through the most challenging phases of their development process on account of their structural capabilities. Many studies have shown that SGs have a role in cancer treatment and responsiveness to anti-cancer medications. A general point of agreement is that SGs are involved in and play a critical role in various pathways in various malignancies. On the other hand, the impact of SGs on cell cycle regulatory factors and critical elements implicated in cancer cell proliferation is utilized as a biased mechanism. Utilizing the capabilities of SGs in the process of chemotherapy resistance (Figure 4), as well as the existence of more SGs in cells receiving chemotherapeutic drugs, is associated with cancer at the following critical stages. Much research has been conducted on the effects of SGs on anti-cancer medications. The goal of this research was to offer a comprehensive review to conclude this subject. Overall, this research may pave the way for future investigations on SGs in treating malignancies and offer a roadmap to lead these studies.
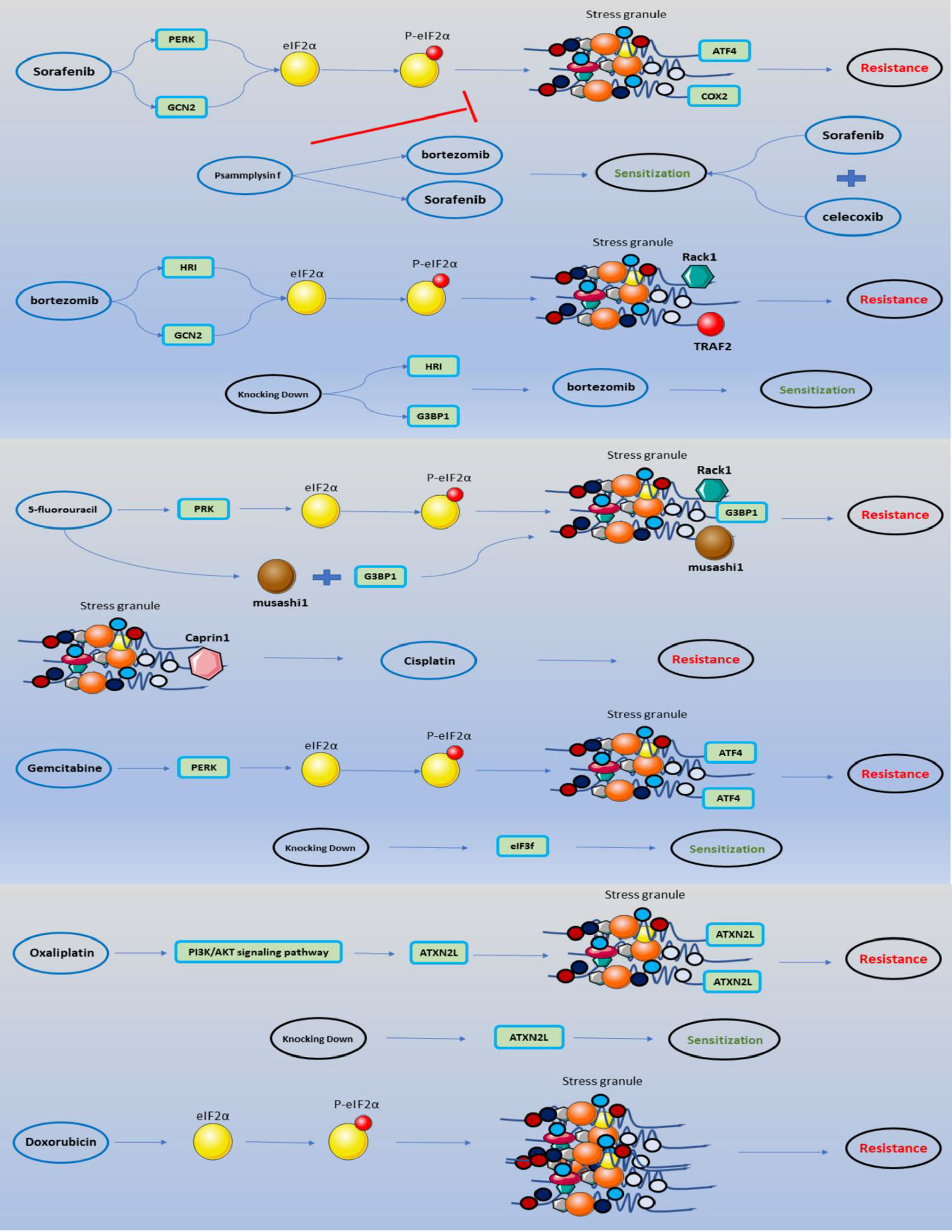
Figure 4 SGs involved in anti-cancer medications mechanism of actions. The impact of anti-cancer medications on the development of SGs through eIF2α phosphorylation is depicted in a schematic. Accumulation of SGs with particular features leads to chemoresistance, which may be anticipated by enhancing the sensitivity of specific medications by combining specific pharmaceuticals or knocking down a portion of the protein components of SGs.
Author Contributions
MA, HS, and MR wrote the draft and revised it. MT designed and supervised the study. BH, MM, MP, EG, and MH contributed in data collection and designing the tables and figures. All authors contributed to the article and approved the submitted version.
Funding
The research protocol was approved and supported by a grant (grant number: 68047) from Student Research Committee, Tabriz University of Medical Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ivanov P, Kedersha N, Anderson P. Stress Granules and Processing Bodies in Translational Control. Cold Spring Harbor Perspect Biol (2019) 11(5):a032813. doi: 10.1101/cshperspect.a032813
2. Moser JJ, Fritzler MJ. Cytoplasmic Ribonucleoprotein (RNP) Bodies and Their Relationship to GW/P Bodies. Int J Biochem Cell Biol (2010) 42(6):828–43. doi: 10.1016/j.biocel.2009.11.018
3. Zeng W-j, Lu C, Shi Y, Wu C, Chen X, Li C, et al. Initiation of Stress Granule Assembly by Rapid Clustering of IGF2BP Proteins Upon Osmotic Shock. Biochim Biophys Acta (BBA) - Mol Cell Res (2020) 1867:118795. doi: 10.1016/j.bbamcr.2020.118795
4. Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of Stress Granules Inhibits Apoptosis by Suppressing Stress-Responsive MAPK Pathways. Nat Cell Biol (2008) 10(11):1324–32. doi: 10.1038/ncb1791
5. Cao X, Jin X, Liu B. The Involvement of Stress Granules in Aging and Aging-Associated Diseases. Aging Cell (2020) 19(4):e13136. doi: 10.1111/acel.13136
6. Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell (2016) 164(3):487–98. doi: 10.1016/j.cell.2015.12.038
7. Marmor-Kollet H, Siany A, Kedersha N, Knafo N, Rivkin N, Danino YM, et al. Spatiotemporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis. Mol Cell (2020) 80(5):876–91.e6. doi: 10.1016/j.molcel.2020.10.032
8. Mateju D, Eichenberger B, Voigt F, Eglinger J, Roth G, Chao JA. Single-Molecule Imaging Reveals Translation of mRNAs Localized to Stress Granules. Cell (2020) 183(7):1801–12.e13. doi: 10.1016/j.cell.2020.11.010
9. Malinovska L, Kroschwald S, Alberti S. Protein Disorder, Prion Propensities, and Self-Organizing Macromolecular Collectives. Biochim Biophys Acta (2013) 1834(5):918–31. doi: 10.1016/j.bbapap.2013.01.003
10. Bley N, Lederer M, Pfalz B, Reinke C, Fuchs T, Glaß M, et al. Stress Granules are Dispensable for mRNA Stabilization During Cellular Stress. Nucleic Acids Res (2015) 43(4):e26. doi: 10.1093/nar/gku1275
11. Fasken MB, Corbett AH. Process or Perish: Quality Control in mRNA Biogenesis. Nat Struct Mol Biol (2005) 12(6):482–8. doi: 10.1038/nsmb945
12. Anderson P, Kedersha N. Stress Granules: The Tao of RNA Triage. Trends Biochem Sci (2008) 33(3):141–50. doi: 10.1016/j.tibs.2007.12.003
13. Chernov KG, Barbet A, Hamon L, Ovchinnikov LP, Curmi PA, Pastré D. Role of Microtubules in Stress Granule Assembly: Microtubule Dynamical Instability Favors the Formation of Micrometric Stress Granules in Cells. J Biol Chem (2009) 284(52):36569–80. doi: 10.1074/jbc.M109.042879
14. Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, et al. Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Mol Cell (2015) 57(5):936–47. doi: 10.1016/j.molcel.2015.01.013
15. Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. Distinct Stages in Stress Granule Assembly and Disassembly. eLife (2016) 5:e18413. doi: 10.7554/eLife.18413
16. Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell (2015) 60(2):208–19. doi: 10.1016/j.molcel.2015.08.018
17. Aulas A, Fay MM, Lyons SM, Achorn CA, Kedersha N, Anderson P, et al. Stress-Specific Differences in Assembly and Composition of Stress Granules and Related Foci. J Cell Sci (2017) 130(5):927–37. doi: 10.1242/jcs.199240
18. Wolozin B, Ivanov P. Stress Granules and Neurodegeneration. Nat Rev Neurosci (2019) 20(11):649–66. doi: 10.1038/s41583-019-0222-5
19. Asadi MR, Sadat Moslehian M, Sabaie H, Jalaiei A, Ghafouri-Fard S, Taheri M, et al. Stress Granules and Neurodegenerative Disorders: A Scoping Review. Front Aging Neurosci (2021) 13:650740. doi: 10.3389/fnagi.2021.650740
20. Yoshinaga M, Takeuchi O. RNA Binding Proteins in the Control of Autoimmune Diseases. Immunol Med (2019) 42(2):53–64. doi: 10.1080/25785826.2019.1655192
21. Hamidi H, Ivaska J. Every Step of the Way: Integrins in Cancer Progression and Metastasis. Nat Rev Cancer (2018) 18(9):533–48. doi: 10.1038/s41568-018-0038-z
22. Asadi MR, Rahmanpour D, Moslehian MS, Sabaie H, Hassani M, Ghafouri-Fard S, et al. Stress Granules Involved in Formation, Progression and Metastasis of Cancer: A Scoping Review. Front Cell Dev Biol (2021) 9:745394. doi: 10.3389/fcell.2021.745394
23. Fabbri L, Chakraborty A, Robert C, Vagner S. The Plasticity of mRNA Translation During Cancer Progression and Therapy Resistance. Nat Rev Cancer (2021) 21(9):558–77. doi: 10.1038/s41568-021-00380-y
24. Adjibade P, St-Sauveur VG, Quevillon Huberdeau M, Fournier MJ, Savard A, Coudert L, et al. Sorafenib, a Multikinase Inhibitor, Induces Formation of Stress Granules in Hepatocarcinoma Cells. Oncotarget (2015) 6(41):43927–43. doi: 10.18632/oncotarget.5980
25. Somasekharan SP, El-Naggar A, Leprivier G, Cheng H, Hajee S, Grunewald TG, et al. YB-1 Regulates Stress Granule Formation and Tumor Progression by Translationally Activating G3BP1. J Cell Biol (2015) 208(7):913–29. doi: 10.1083/jcb.201411047
26. Grabocka E, Bar-Sagi D. Mutant KRAS Enhances Tumor Cell Fitness by Upregulating Stress Granules. Cell (2016) 167(7):1803–13.e12. doi: 10.1016/j.cell.2016.11.035
27. Takayama KI, Suzuki T, Fujimura T, Takahashi S, Inoue S. Association of USP10 With G3BP2 Inhibits P53 Signaling and Contributes to Poor Outcome in Prostate Cancer. Mol Cancer Res MCR (2018) 16(5):846–56. doi: 10.1158/1541-7786.MCR-17-0471
28. Vellky JE, McSweeney ST, Ricke EA, Ricke WA. RNA-Binding Protein DDX3 Mediates Posttranscriptional Regulation of Androgen Receptor: A Mechanism of Castration Resistance. Proc Natl Acad Sci USA (2020) 117(45):28092–101. doi: 10.1073/pnas.2008479117
29. Zhang Y, Yue C, Krichevsky AM, Garkavtsev I. Repression of the Stress Granule Protein G3BP2 Inhibits Immune Checkpoint Molecule PD-L1. Mol Oncol (2021). doi: 10.1002/1878-0261.12915
30. Vilas-Boas Fde A, da Silva AM, de Sousa LP, Lima KM, Vago JP, Bittencourt LF, et al. Impairment of Stress Granule Assembly via Inhibition of the Eif2alpha Phosphorylation Sensitizes Glioma Cells to Chemotherapeutic Agents. J Neuro-Oncol (2016) 127(2):253–60. doi: 10.1007/s11060-015-2043-3
31. Liu Y, Li Q, Zhou L, Xie N, Nice EC, Zhang H, et al. Cancer Drug Resistance: Redox Resetting Renders a Way. Oncotarget (2016) 7(27):42740–61. doi: 10.18632/oncotarget.8600
32. Protter DSW, Parker R. Principles and Properties of Stress Granules. Trends Cell Biol (2016) 26(9):668–79. doi: 10.1016/j.tcb.2016.05.004
33. Arksey H, O'Malley L. Scoping Studies: Towards a Methodological Framework. Int J Soc Res Method (2005) 8(1):19–32. doi: 10.1080/1364557032000119616
34. Levac D, Colquhoun H, O'Brien KK. Scoping Studies: Advancing the Methodology. Implementation Sci (2010) 5(1):69. doi: 10.1186/1748-5908-5-69
35. Colquhoun HL, Levac D, O'Brien KK, Straus S, Tricco AC, Perrier L, et al. Scoping Reviews: Time for Clarity in Definition, Methods, and Reporting. J Clin Epidemiol (2014) 67(12):1291–4. doi: 10.1016/j.jclinepi.2014.03.013
36. Tricco AC, Lillie E, Zarin W, O'Brien KK, Colquhoun H, Levac D, et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann Internal Med (2018) 169(7):467–73. doi: 10.7326/M18-0850
37. Kim WJ, Kim JH, Jang SK. Anti-Inflammatory Lipid Mediator 15d-PGJ2 Inhibits Translation Through Inactivation of Eif4a. EMBO J (2007) 26(24):5020–32. doi: 10.1038/sj.emboj.7601920
38. Busà R, Geremia R, Sette C. Genotoxic Stress Causes the Accumulation of the Splicing Regulator Sam68 in Nuclear Foci of Transcriptionally Active Chromatin. Nucleic Acids Res (2010) 38(9):3005–18. doi: 10.1093/nar/gkq004
39. Fournier MJ, Gareau C, Mazroui R. The Chemotherapeutic Agent Bortezomib Induces the Formation of Stress Granules. Cancer Cell Int (2010) 10:12. doi: 10.1186/1475-2867-10-12
40. Kalra J, Sutherland BW, Stratford AL, Dragowska W, Gelmon KA, Dedhar S, et al. Suppression of Her2/neu Expression Through ILK Inhibition Is Regulated by a Pathway Involving TWIST and YB-1. Oncogene (2010) 29(48):6343–56. doi: 10.1038/onc.2010.366
41. Annibaldi A, Dousse A, Martin S, Tazi J, Widmann C. Revisiting G3BP1 as a RasGAP Binding Protein: Sensitization of Tumor Cells to Chemotherapy by the RasGAP 317-326 Sequence Does Not Involve G3BP1. PloS One (2011) 6(12):e29024. doi: 10.1371/journal.pone.0029024
42. Gareau C, Fournier MJ, Filion C, Coudert L, Martel D, Labelle Y, et al. P21(WAF1/CIP1) Upregulation Through the Stress Granule-Associated Protein CUGBP1 Confers Resistance to Bortezomib-Mediated Apoptosis. PloS One (2011) 6(5):e20254. doi: 10.1371/journal.pone.0020254
43. Martins I, Kepp O, Schlemmer F, Adjemian S, Tailler M, Shen S, et al. Restoration of the Immunogenicity of Cisplatin-Induced Cancer Cell Death by Endoplasmic Reticulum Stress. Oncogene (2011) 30(10):1147–58. doi: 10.1038/onc.2010.500
44. Mason TA, Kolobova E, Liu J, Roland JT, Chiang C, Goldenring JR. Darinaparsin Is a Multivalent Chemotherapeutic Which Induces Incomplete Stress Response With Disruption of Microtubules and Shh Signaling. PloS One (2011) 6(11):e27699. doi: 10.1371/journal.pone.0027699
45. Woldemichael GM, Turbyville TJ, Vasselli JR, Linehan WM, McMahon JB. Lack of a Functional VHL Gene Product Sensitizes Renal Cell Carcinoma Cells to the Apoptotic Effects of the Protein Synthesis Inhibitor Verrucarin a. Neoplasia (New York NY) (2012) 14(8):771–7. doi: 10.1593/neo.12852
46. Wen F, Zhou R, Shen A, Choi A, Uribe D, Shi J. The Tumor Suppressive Role of Eif3f and Its Function in Translation Inhibition and rRNA Degradation. PloS One (2012) 7(3):e34194. doi: 10.1371/journal.pone.0034194
47. Fournier MJ, Coudert L, Mellaoui S, Adjibade P, Gareau C, Côté MF, et al. Inactivation of the Mtorc1-Eukaryotic Translation Initiation Factor 4E Pathway Alters Stress Granule Formation. Mol Cell Biol (2013) 33(11):2285–301. doi: 10.1128/MCB.01517-12
48. Sabile AA, Arlt MJ, Muff R, Husmann K, Hess D, Bertz J, et al. Caprin-1, a Novel Cyr61-Interacting Protein, Promotes Osteosarcoma Tumor Growth and Lung Metastasis in Mice. Biochim Biophys Acta (2013) 1832(8):1173–82. doi: 10.1016/j.bbadis.2013.03.014
49. Kaehler C, Isensee J, Hucho T, Lehrach H, Krobitsch S. 5-Fluorouracil Affects Assembly of Stress Granules Based on RNA Incorporation. Nucleic Acids Res (2014) 42:1–13. doi: 10.1093/nar/gku264
50. Podszywalow-Bartnicka P, Wolczyk M, Kusio-Kobialka M, Wolanin K, Skowronek K, Nieborowska-Skorska M, et al. Downregulation of BRCA1 Protein in BCR-ABL1 Leukemia Cells Depends on Stress-Triggered TIAR-Mediated Suppression of Translation. Cell Cycle (Georgetown Tex) (2014) 13(23):3727–41. doi: 10.4161/15384101.2014.965013
51. Yeomans A, Lemm E, Wilmore S, Cavell BE, Valle-Argos B, Krysov S, et al. PEITC-Mediated Inhibition of mRNA Translation Is Associated With Both Inhibition of Mtorc1 and Increased Eif2α Phosphorylation in Established Cell Lines and Primary Human Leukemia Cells. Oncotarget (2016) 7(46):74807–19. doi: 10.18632/oncotarget.11655
52. Henderson KA, Kobylewski SE, Yamada KE, Eckhert CD. Boric Acid Induces Cytoplasmic Stress Granule Formation, Eif2α Phosphorylation, and ATF4 in Prostate DU-145 Cells. Biometals an Int J Role Metal Ions Biol Biochem Med (2015) 28(1):133–41. doi: 10.1007/s10534-014-9809-5
53. Szaflarski W, Fay MM, Kedersha N, Zabel M, Anderson P, Ivanov P. Vinca Alkaloid Drugs Promote Stress-Induced Translational Repression and Stress Granule Formation. Oncotarget (2016) 7(21):30307–22. doi: 10.18632/oncotarget.8728
54. Chiou G-Y, Yang T-W, Huang C-C, Tang C-Y, Yen J-Y, Tsai M-C, et al. Musashi-1 Promotes a Cancer Stem Cell Lineage and Chemoresistance in Colorectal Cancer Cells. Sci Rep (2017) 7. doi: 10.1038/s41598-017-02057-9
55. Narayanan N, Wang Z, Li L, Yang Y. Arginine Methylation of USP9X Promotes its Interaction With TDRD3 and Its Anti-Apoptotic Activities in Breast Cancer Cells. Cell Discovery (2017) 3:16048. doi: 10.1038/celldisc.2016.48
56. Chen H-Y, Lin L-T, Wang M-L, Tsai K-L, Huang P-I, Yang Y-P, et al. Musashi-1 Promotes Chemoresistant Granule Formation by PKR/Eif2α Signalling Cascade in Refractory Glioblastoma. Biochim Biophys Acta (BBA) - Mol Basis Dis (2018) 1864:1850–61. doi: 10.1016/j.bbadis.2018.02.017
57. Chen W, Zhao X, Cao W, Diao W, Guo H. MP72-03 Sorafenib-triggered Stress Granules Promote Resistance in Renal Cancer Cells By Recruiting Anti-apoptotic Cyclooxygenase 2. J Urol (2018) 199:e952–e3. doi: 10.1016/j.juro.2018.02.2287
58. Timalsina S, Arimoto-Matsuzaki K, Kitamura M, Xu X, Wenzhe Q, Ishigami-Yuasa M, et al. Chemical Compounds That Suppress Hypoxia-Induced Stress Granule Formation Enhance Cancer Drug Sensitivity of Human Cervical Cancer HeLa Cells. J Biochem (2018) 164(5):381–91. doi: 10.1093/jb/mvy062
59. Bittencourt LFF, Negreiros-Lima GL, Sousa LP, Silva AG, Souza IBS, Ribeiro R, et al. G3BP1 Knockdown Sensitizes U87 Glioblastoma Cell Line to Bortezomib by Inhibiting Stress Granules Assembly and Potentializing Apoptosis. J Neuro-Oncol (2019) 144(3):463–73. doi: 10.1007/s11060-019-03252-6
60. Choi S, Sa M, Cho N, Kim K, Park S-H. Rbfox2 Dissociation From Stress Granules Suppresses Cancer Progression. Exp Mol Med (2019) 51:49. doi: 10.1038/s12276-019-0246-y
61. Shi Q, Zhu Y, Ma J, Chang K, Ding D, Bai Y, et al. Prostate Cancer-Associated SPOP Mutations Enhance Cancer Cell Survival and Docetaxel Resistance by Upregulating Caprin1-Dependent Stress Granule Assembly. Mol Cancer (2019) 18(1):170. doi: 10.1186/s12943-019-1096-x
62. Christen KE, Davis RA, Kennedy D. Psammaplysin F Increases the Efficacy of Bortezomib and Sorafenib Through Regulation of Stress Granule Formation. Int J Biochem Cell Biol (2019) 112:24–38. doi: 10.1016/j.biocel.2019.04.008
63. Comba A, Bonnet LV, Goitea VE, Hallak ME, Galiano MR. Arginylated Calreticulin Increases Apoptotic Response Induced by Bortezomib in Glioma Cells. Mol Neurobiol (2019) 56(3):1653–64. doi: 10.1007/s12035-018-1182-x
64. El-Naggar AM, Somasekharan SP, Wang Y, Cheng H, Negri GL, Pan M, et al. Class I HDAC Inhibitors Enhance YB-1 Acetylation and Oxidative Stress to Block Sarcoma Metastasis. EMBO Rep (2019) 20(12):e48375. doi: 10.15252/embr.201948375
65. Fuentes-Villalobos F, Farkas C, Riquelme-Barrios S, Armijo ME, Soto-Rifo R, Pincheira R, et al. DISC1 Promotes Translation Maintenance During Sodium Arsenite-Induced Oxidative Stress. Biochim Biophys Acta Gene Regul Mech (2019) 1862(6):657–69. doi: 10.1016/j.bbagrm.2019.05.001
66. Kashiwagi S, Fujioka Y, Kondo T, Satoh AO, Yoshida A, Fujioka M, et al. Localization of BCR-ABL to Stress Granules Contributes to Its Oncogenic Function. Cell Structure Funct (2019) 44(2):195–204. doi: 10.1247/csf.19033
67. Lin L, Li X, Pan C, Lin W, Shao R, Liu Y, et al. ATXN2L Upregulated by Epidermal Growth Factor Promotes Gastric Cancer Cell Invasiveness and Oxaliplatin Resistance. Cell Death Dis (2019) 10(3):173. doi: 10.1038/s41419-019-1362-2
68. Soung NK, Kim HM, Asami Y, Kim DH, Cho Y, Naik R, et al. Mechanism of the Natural Product Moracin-O Derived MO-460 and Its Targeting Protein Hnrnpa2b1 on HIF-1α Inhibition. Exp Mol Med (2019) 51(2):1–14. doi: 10.1038/s12276-018-0200-4
69. Adjibade P, Simoneau B, Ledoux N, Gauthier WN, Nkurunziza M, Khandjian EW, et al. Treatment of Cancer Cells With Lapatinib Negatively Regulates General Translation and Induces Stress Granules Formation. PloS One (2020) 15(5):e0231894. doi: 10.1371/journal.pone.0231894
70. Amen T, Kaganovich D. Fasnall Induces Atypically Transient Stress Granules Independently of FASN Inhibition. iScience (2020) 23(10):101550. doi: 10.1016/j.isci.2020.101550
71. Attwood K, Robichaud A, Westhaver L, Castle E, Brandman D, Balgi A, et al. Raloxifene Prevents Stress Granule Dissolution, Impairs Translational Control and Promotes Cell Death During Hypoxia in Glioblastoma Cells. Cell Death Dis (2020) 11:989. doi: 10.1038/s41419-020-03159-5
72. Illarionova NB, Morozova KN, Petrovskii DV, Sharapova MB, Romashchenko AV, Troitskii SY, et al. 'Trojan-Horse' Stress-Granule Formation Mediated by Manganese Oxide Nanoparticles. Nanotoxicology (2020) 14(10):1432–44. doi: 10.1080/17435390.2020.1856433
73. Lu X, Alam U, Willis C, Kennedy D. Role of Chikungunya Nsp3 in Regulating G3BP1 Activity, Stress Granule Formation and Drug Efficacy. Arch Med Res (2020) 52:48–57. doi: 10.1016/j.arcmed.2020.10.002
74. Mukhopadhyay S, Goswami D, Adiseshaiah PP, Burgan W, Yi M, Guerin TM, et al. Undermining Glutaminolysis Bolsters Chemotherapy While NRF2 Promotes Chemoresistance in KRAS-Driven Pancreatic Cancers. Cancer Res (2020) 80(8):1630–43. doi: 10.1158/0008-5472.CAN-19-1363
75. Park Y-J, Choi D, Cho SW, Han J, Yang S, Choi C. Stress Granule Formation Attenuates RACK1-Mediated Apoptotic Cell Death Induced by Morusin. Int J Mol Sci (2020) 21:5360. doi: 10.3390/ijms21155360
76. Zhao J, Fu X, Chen H, Min L, Sun J, Yin J, et al. G3BP1 Interacts With YWHAZ to Regulate Chemoresistance and Predict Adjuvant Chemotherapy Benefit in Gastric Cancer. Br J Cancer (2021) 124(2):425–36. doi: 10.1038/s41416-020-01067-1
77. Anderson P, Kedersha N. RNA Granules. J Cell Biol (2006) 172(6):803–8. doi: 10.1083/jcb.200512082
78. Biamonti G. Nuclear Stress Bodies: A Heterochromatin Affair? Nat Rev Mol Cell Biol (2004) 5(6):493–8. doi: 10.1038/nrm1405
79. Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, et al. Stress Granules and Processing Bodies Are Dynamically Linked Sites of mRNP Remodeling. J Cell Biol (2005) 169(6):871–84. doi: 10.1083/jcb.200502088
80. Parker R, Sheth U. P Bodies and the Control of mRNA Translation and Degradation. Mol Cell (2007) 25(5):635–46. doi: 10.1016/j.molcel.2007.02.011
81. Buchan JR, Parker R. Eukaryotic Stress Granules: The Ins and Outs of Translation. Mol Cell (2009) 36(6):932–41. doi: 10.1016/j.molcel.2009.11.020
82. Collier NC, Heuser J, Levy MA, Schlesinger MJ. Ultrastructural and Biochemical Analysis of the Stress Granule in Chicken Embryo Fibroblasts. J Cell Biol (1988) 106(4):1131–9. doi: 10.1083/jcb.106.4.1131
83. Gutierrez-Beltran E, Moschou PN, Smertenko AP, Bozhkov PV. Tudor Staphylococcal Nuclease Links Formation of Stress Granules and Processing Bodies With mRNA Catabolism in Arabidopsis. Plant Cell (2015) 27(3):926–43. doi: 10.1105/tpc.114.134494
84. Kedersha N, Ivanov P, Anderson P. Stress Granules and Cell Signaling: More Than Just a Passing Phase? Trends Biochem Sci (2013) 38:494–506. doi: 10.1016/j.tibs.2013.07.004
85. Gao X, Jiang L, Gong Y, Chen X, Ying M, Zhu H, et al. Stress Granule: A Promising Target for Cancer Treatment. Br J Pharmacol (2019) 176(23):4421–33. doi: 10.1111/bph.14790
86. Hamanaka RB, Bennett BS, Cullinan SB, Diehl JA. PERK and GCN2 Contribute to Eif2alpha Phosphorylation and Cell Cycle Arrest After Activation of the Unfolded Protein Response Pathway. Mol Biol Cell (2005) 16(12):5493–501. doi: 10.1091/mbc.e05-03-0268
87. Holcik M, Sonenberg N. Translational Control in Stress and Apoptosis. Nat Rev Mol Cell Biol (2005) 6(4):318–27. doi: 10.1038/nrm1618
88. Chang YS, Adnane J, Trail PA, Levy J, Henderson A, Xue D, et al. Sorafenib (BAY 43-9006) Inhibits Tumor Growth and Vascularization and Induces Tumor Apoptosis and Hypoxia in RCC Xenograft Models. Cancer Chemother Pharmacol (2007) 59(5):561–74. doi: 10.1007/s00280-006-0393-4
89. Liu L, Cao Y, Chen C, Zhang X, McNabola A, Wilkie D, et al. Sorafenib Blocks the RAF/MEK/ERK Pathway, Inhibits Tumor Angiogenesis, and Induces Tumor Cell Apoptosis in Hepatocellular Carcinoma Model PLC/PRF/5. Cancer Res (2006) 66(24):11851–8. doi: 10.1158/0008-5472.CAN-06-1377
90. Salvatore G, De Falco V, Salerno P, Nappi TC, Pepe S, Troncone G, et al. BRAF is a Therapeutic Target in Aggressive Thyroid Carcinoma. Clin Cancer Res an Off J Am Assoc Cancer Res (2006) 12(5):1623–9. doi: 10.1158/1078-0432.CCR-05-2378
91. Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, et al. BAY 43-9006 Exhibits Broad Spectrum Oral Antitumor Activity and Targets the RAF/MEK/ERK Pathway and Receptor Tyrosine Kinases Involved in Tumor Progression and Angiogenesis. Cancer Res (2004) 64(19):7099–109. doi: 10.1158/0008-5472.CAN-04-1443
92. Carlomagno F, Anaganti S, Guida T, Salvatore G, Troncone G, Wilhelm SM, et al. BAY 43-9006 Inhibition of Oncogenic RET Mutants. J Natl Cancer Inst (2006) 98(5):326–34. doi: 10.1093/jnci/djj069
93. Yu C, Bruzek LM, Meng XW, Gores GJ, Carter CA, Kaufmann SH, et al. The Role of Mcl-1 Downregulation in the Proapoptotic Activity of the Multikinase Inhibitor BAY 43-9006. Oncogene (2005) 24(46):6861–9. doi: 10.1038/sj.onc.1208841
94. Liu JY, Park SH, Morisseau C, Hwang SH, Hammock BD, Weiss RH. Sorafenib Has Soluble Epoxide Hydrolase Inhibitory Activity, Which Contributes to Its Effect Profile In Vivo. Mol Cancer Ther (2009) 8(8):2193–203. doi: 10.1158/1535-7163.MCT-09-0119
95. Jiang HY, Wek RC. Phosphorylation of the Alpha-Subunit of the Eukaryotic Initiation Factor-2 (Eif2alpha) Reduces Protein Synthesis and Enhances Apoptosis in Response to Proteasome Inhibition. J Biol Chem (2005) 280(14):14189–202. doi: 10.1074/jbc.M413660200
96. Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, et al. ER-Stress-Induced Transcriptional Regulation Increases Protein Synthesis Leading to Cell Death. Nat Cell Biol (2013) 15(5):481–90. doi: 10.1038/ncb2738
97. Hao Y, Zhang J, Shan G, Zhang N, Jin W, Nan K. Establishment of Optimal Regulatory Network of Colorectal Cancer Based on P42.3 Protein. Saudi J Biol Sci (2017) 24(8):1781–6. doi: 10.1016/j.sjbs.2017.11.012
98. Kane RC, Bross PF, Farrell AT, Pazdur R. Velcade: U.S. FDA Approval for the Treatment of Multiple Myeloma Progressing on Prior Therapy. Oncologist (2003) 8(6):508–13. doi: 10.1634/theoncologist.8-6-508
99. Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the First Proteasome Inhibitor Anticancer Drug: Current Status and Future Perspectives. Curr Cancer Drug Targets (2011) 11(3):239–53. doi: 10.2174/156800911794519752
100. Adams J. Development of the Proteasome Inhibitor PS-341. Oncologist (2002) 7(1):9–16. doi: 10.1634/theoncologist.7-1-9
101. Bross PF, Kane R, Farrell AT, Abraham S, Benson K, Brower ME, et al. Approval Summary for Bortezomib for Injection in the Treatment of Multiple Myeloma. Clin Cancer Res an Off J Am Assoc Cancer Res (2004) 10(12 Pt 1):3954–64. doi: 10.1158/1078-0432.CCR-03-0781
102. Berkers CR, Verdoes M, Lichtman E, Fiebiger E, Kessler BM, Anderson KC, et al. Activity Probe for In Vivo Profiling of the Specificity of Proteasome Inhibitor Bortezomib. Nat Methods (2005) 2(5):357–62. doi: 10.1038/nmeth759
103. Dikic I. Proteasomal and Autophagic Degradation Systems. Annu Rev Biochem (2017) 86:193–224. doi: 10.1146/annurev-biochem-061516-044908
104. Adams J, Palombella VJ, Sausville EA, Johnson J, Destree A, Lazarus DD, et al. Proteasome Inhibitors: A Novel Class of Potent and Effective Antitumor Agents. Cancer Res (1999) 59(11):2615–22.
105. Crawford LJ, Walker B, Ovaa H, Chauhan D, Anderson KC, Morris TC, et al. Comparative Selectivity and Specificity of the Proteasome Inhibitors BzLLLCOCHO, PS-341, and MG-132. Cancer Res (2006) 66(12):6379–86. doi: 10.1158/0008-5472.CAN-06-0605
106. Chen D, Dou QP. The Ubiquitin-Proteasome System as a Prospective Molecular Target for Cancer Treatment and Prevention. Curr Protein Pept Sci (2010) 11(6):459–70. doi: 10.2174/138920310791824057
107. Ling YH, Liebes L, Jiang JD, Holland JF, Elliott PJ, Adams J, et al. Mechanisms of Proteasome Inhibitor PS-341-Induced G(2)-M-Phase Arrest and Apoptosis in Human non-Small Cell Lung Cancer Cell Lines. Clin Cancer Res an Off J Am Assoc Cancer Res (2003) 9(3):1145–54.
108. Adams J. The Proteasome: A Suitable Antineoplastic Target. Nat Rev Cancer (2004) 4(5):349–60. doi: 10.1038/nrc1361
109. Qin JZ, Ziffra J, Stennett L, Bodner B, Bonish BK, Chaturvedi V, et al. Proteasome Inhibitors Trigger NOXA-Mediated Apoptosis in Melanoma and Myeloma Cells. Cancer Res (2005) 65(14):6282–93. doi: 10.1158/0008-5472.CAN-05-0676
110. Fernández Y, Verhaegen M, Miller TP, Rush JL, Steiner P, Opipari AW Jr., et al. Differential Regulation of Noxa in Normal Melanocytes and Melanoma Cells by Proteasome Inhibition: Therapeutic Implications. Cancer Res (2005) 65(14):6294–304. doi: 10.1158/0008-5472.CAN-05-0686
111. Schwartz R, Davidson T. Pharmacology, Pharmacokinetics, and Practical Applications of Bortezomib. Oncol (Williston Park NY) (2004) 18(14 Suppl 11):14–21.
112. Rutman RJ, Cantarow A, Paschkis KE. Studies in 2-Acetylaminofluorene Carcinogenesis. III. The Utilization of Uracil-2-C14 by Preneoplastic Rat Liver and Rat Hepatoma. Cancer Res (1954) 14(2):119–23.
113. Wohlhueter RM, McIvor RS, Plagemann PG. Facilitated Transport of Uracil and 5-Fluorouracil, and Permeation of Orotic Acid Into Cultured Mammalian Cells. J Cell Physiol (1980) 104(3):309–19. doi: 10.1002/jcp.1041040305
114. Parker WB, Cheng YC. Metabolism and Mechanism of Action of 5-Fluorouracil. Pharmacol Ther (1990) 48(3):381–95. doi: 10.1016/0163-7258(90)90056-8
115. Zhang N, Yin Y, Xu SJ, Chen WS. 5-Fluorouracil: Mechanisms of Resistance and Reversal Strategies. Molecules (Basel Switzerland) (2008) 13(8):1551–69. doi: 10.3390/molecules13081551
116. Longley DB, Harkin DP, Johnston PG. 5-Fluorouracil: Mechanisms of Action and Clinical Strategies. Nat Rev Cancer (2003) 3(5):330–8. doi: 10.1038/nrc1074
117. Sommer H, Santi DV. Purification and Amino Acid Analysis of an Active Site Peptide From Thymidylate Synthetase Containing Covalently Bound 5-Fluoro-2'-Deoxyuridylate and Methylenetetrahydrofolate. Biochem Biophys Res Commun (1974) 57(3):689–95. doi: 10.1016/0006-291X(74)90601-9
118. Aherne GW, Hardcastle A, Raynaud F, Jackman AL. Immunoreactive dUMP and TTP Pools as an Index of Thymidylate Synthase Inhibition; Effect of Tomudex (ZD1694) and a Nonpolyglutamated Quinazoline Antifolate (CB30900) in L1210 Mouse Leukaemia Cells. Biochem Pharmacol (1996) 51(10):1293–301. doi: 10.1016/0006-2952(96)00035-4
119. Mitrovski B, Pressacco J, Mandelbaum S, Erlichman C. Biochemical Effects of Folate-Based Inhibitors of Thymidylate Synthase in MGH-U1 Cells. Cancer Chemother Pharmacol (1994) 35(2):109–14. doi: 10.1007/BF00686631
120. Grem JL, Fischer PH. Enhancement of 5-Fluorouracil's Anticancer Activity by Dipyridamole. Pharmacol Ther (1989) 40(3):349–71. doi: 10.1016/0163-7258(89)90084-3
121. Schilsky RL. Biochemical and Clinical Pharmacology of 5-Fluorouracil. Oncol (Williston Park NY) (1998) 12(10 Suppl 7):13–8.
122. Hashimoto Y, Yoshida Y, Yamada T, Aisu N, Yoshimatsu G, Yoshimura F, et al. Current Status of Therapeutic Drug Monitoring of 5-Fluorouracil Prodrugs. Anticancer Res (2020) 40(8):4655–61. doi: 10.21873/anticanres.14464
123. Focaccetti C, Bruno A, Magnani E, Bartolini D, Principi E, Dallaglio K, et al. Effects of 5-Fluorouracil on Morphology, Cell Cycle, Proliferation, Apoptosis, Autophagy and ROS Production in Endothelial Cells and Cardiomyocytes. PloS One (2015) 10(2):e0115686. doi: 10.1371/journal.pone.0115686
124. Ghoshal K, Jacob ST. Specific Inhibition of Pre-Ribosomal RNA Processing in Extracts From the Lymphosarcoma Cells Treated With 5-Fluorouracil. Cancer Res (1994) 54(3):632–6.
125. Santi DV, Hardy LW. Catalytic Mechanism and Inhibition of tRNA (Uracil-5-)Methyltransferase: Evidence for Covalent Catalysis. Biochemistry (1987) 26(26):8599–606. doi: 10.1021/bi00400a016
126. Negrei C, Hudita A, Ginghina O, Galateanu B, Voicu SN, Stan M, et al. Colon Cancer Cells Gene Expression Signature As Response to 5- Fluorouracil, Oxaliplatin, and Folinic Acid Treatment. Front Pharmacol (2016) 7(172). doi: 10.3389/fphar.2016.00172
127. Zhao X, Yu YT. Incorporation of 5-Fluorouracil Into U2 snRNA Blocks Pseudouridylation and pre-mRNA Splicing In Vivo. Nucleic Acids Res (2007) 35(2):550–8. doi: 10.1093/nar/gkl1084
128. Therizols G, Bash-Imam Z, Panthu B, Machon C, Vincent A, Nait-Slimane S, et al. Alteration of Ribosome Function Upon 5-Fluorouracil Treatment Favours Cancer Cell Drug-Tolerance. bioRxiv (2020) 2020.06.04.131201. doi: 10.1101/2020.06.04.131201
129. Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, et al. Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell (2017) 168(6):1028–40.e19. doi: 10.1016/j.cell.2017.02.027
130. D'Errico M, Parlanti E, Teson M, Degan P, Lemma T, Calcagnile A, et al. The Role of CSA in the Response to Oxidative DNA Damage in Human Cells. Oncogene (2007) 26(30):4336–43. doi: 10.1038/sj.onc.1210232
131. Cini R, Fanizzi FP, Intini FP, Maresca L, Natile G. Platinum Amides From Platinum Nitriles: X-Ray Crystal Structures of the Unbridged Dinuclear Compounds Bis[Bis(1-Imino-1-Hydroxy-2,2-Dimethylpropane)Dichloroplatinum(II)] and Bis[Bis(1-Imino-1-Hydroxy-2,2-Dimethylpropane)(1-Amino-1-Oxo-2,2-Dimethylpropane)Dichloroplatinum(II)]. J Am Chem Soc (1993) 115(12):5123–31. doi: 10.1021/ja00065a025
132. Kelland L. The Resurgence of Platinum-Based Cancer Chemotherapy. Nat Rev Cancer (2007) 7(8):573–84. doi: 10.1038/nrc2167
133. Hartmann J, Fels L, Knop S, Stolte H, Kanz L, Bokemeyer C. A Randomized Trial Comparing the Nephrotoxicity of Cisplatin/Ifosfamide-Based Combination Chemotherapy With or Without Amifostine in Patients With Solid Tumors. Investigational New Drugs (2000) 18:281–9. doi: 10.1023/A:1006490226104
134. Hartmann JT, Lipp HP. Toxicity of Platinum Compounds. Expert Opin Pharmacother (2003) 4(6):889–901. doi: 10.1517/14656566.4.6.889
135. Baik MH, Friesner RA, Lippard SJ. Theoretical Study of Cisplatin Binding to Purine Bases: Why Does Cisplatin Prefer Guanine Over Adenine? J Am Chem Soc (2003) 125(46):14082–92. doi: 10.1021/ja036960d
136. Gale GR, Morris CR, Atkins LM, Smith AB. Binding of an Antitumor Platinum Compound to Cells as Influenced by Physical Factors and Pharmacologically Active Agents. Cancer Res (1973) 33(4):813–8.
137. Sharp SY, Rogers PM, Kelland LR. Transport of Cisplatin and Bis-Acetato-Ammine-Dichlorocyclohexylamine Platinum(IV) (JM216) in Human Ovarian Carcinoma Cell Lines: Identification of a Plasma Membrane Protein Associated With Cisplatin Resistance. Clin Cancer Res an Off J Am Assoc Cancer Res (1995) 1(9):981–9.
138. Wang S, Kwon SH, Su Y, Dong Z. Stress Granules are Formed in Renal Proximal Tubular Cells During Metabolic Stress and Ischemic Injury for Cell Survival. Am J Physiol Renal Physiol (2019) 317(1):F116–f23. doi: 10.1152/ajprenal.00139.2019
139. Pietras P, Aulas A, Fay MM, Leśniczak M, Lyons SM, Szaflarski W, et al. Mechanisms of Translation Inhibition and Suppression of Stress Granule Formation by Cisplatin. bioRxiv (2021). doi: 10.1101/2021.07.20.452628
140. Plunkett W, Huang P, Xu YZ, Heinemann V, Grunewald R, Gandhi V. Gemcitabine: Metabolism, Mechanisms of Action, and Self-Potentiation. Semin Oncol (1995) 22(4 Suppl 11):3–10.
141. Heinemann V, Quietzsch D, Gieseler F, Gonnermann M, Schönekäs H, Rost A, et al. Randomized Phase III Trial of Gemcitabine Plus Cisplatin Compared With Gemcitabine Alone in Advanced Pancreatic Cancer. J Clin Oncol Off J Am Soc Clin Oncol (2006) 24(24):3946–52. doi: 10.1200/JCO.2005.05.1490
142. Huang P, Chubb S, Hertel LW, Grindey GB, Plunkett W. Action of 2',2'-Difluorodeoxycytidine on DNA Synthesis. Cancer Res (1991) 51(22):6110–7.
143. Palam LR, Gore J, Craven KE, Wilson JL, Korc M. Integrated Stress Response Is Critical for Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma. Cell Death Dis (2015) 6(10):e1913. doi: 10.1038/cddis.2015.264
144. Wek RC, Jiang HY, Anthony TG. Coping With Stress: Eif2 Kinases and Translational Control. Biochem Soc Trans (2006) 34(Pt 1):7–11. doi: 10.1042/BST0340007
145. Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, et al. Regulated Translation Initiation Controls Stress-Induced Gene Expression in Mammalian Cells. Mol Cell (2000) 6(5):1099–108. doi: 10.1016/S1097-2765(00)00108-8
146. Lu PD, Jousse C, Marciniak SJ, Zhang Y, Novoa I, Scheuner D, et al. Cytoprotection by Pre-Emptive Conditional Phosphorylation of Translation Initiation Factor 2. EMBO J (2004) 23(1):169–79. doi: 10.1038/sj.emboj.7600030
147. Marciniak SJ, Ron D. Endoplasmic Reticulum Stress Signaling in Disease. Physiol Rev (2006) 86(4):1133–49. doi: 10.1152/physrev.00015.2006
148. Carrato A, Gallego J, Díaz-Rubio E. Oxaliplatin: Results in Colorectal Carcinoma. Crit Rev Oncol/Hematol (2002) 44(1):29–44. doi: 10.1016/S1040-8428(01)00192-5
149. Alcindor T, Beauger N. Oxaliplatin: A Review in the Era of Molecularly Targeted Therapy. Curr Oncol (Toronto Ont) (2011) 18(1):18–25. doi: 10.3747/co.v18i1.708
150. Johnson SW, Laub PB, Beesley JS, Ozols RF, Hamilton TC. Increased Platinum-DNA Damage Tolerance is Associated With Cisplatin Resistance and Cross-Resistance to Various Chemotherapeutic Agents in Unrelated Human Ovarian Cancer Cell Lines. Cancer Res (1997) 57(5):850–6.
151. Virag P, Fischer-Fodor E, Perde-Schrepler M, Brie I, Tatomir C, Balacescu L, et al. Oxaliplatin Induces Different Cellular and Molecular Chemoresistance Patterns in Colorectal Cancer Cell Lines of Identical Origins. BMC Genomics (2013) 14:480. doi: 10.1186/1471-2164-14-480
152. Martinez-Balibrea E, Martínez-Cardús A, Ginés A, Ruiz de Porras V, Moutinho C, Layos L, et al. Tumor-Related Molecular Mechanisms of Oxaliplatin Resistance. Mol Cancer Ther (2015) 14(8):1767–76. doi: 10.1158/1535-7163.MCT-14-0636
153. Yonezawa A, Masuda S, Yokoo S, Katsura T, Inui K. Cisplatin and Oxaliplatin, But Not Carboplatin and Nedaplatin, Are Substrates for Human Organic Cation Transporters (SLC22A1-3 and Multidrug and Toxin Extrusion Family). J Pharmacol Exp Ther (2006) 319(2):879–86. doi: 10.1124/jpet.106.110346
154. Holzer AK, Manorek GH, Howell SB. Contribution of the Major Copper Influx Transporter CTR1 to the Cellular Accumulation of Cisplatin, Carboplatin, and Oxaliplatin. Mol Pharmacol (2006) 70(4):1390–4. doi: 10.1124/mol.106.022624
155. Samimi G, Safaei R, Katano K, Holzer AK, Rochdi M, Tomioka M, et al. Increased Expression of the Copper Efflux Transporter ATP7A Mediates Resistance to Cisplatin, Carboplatin, and Oxaliplatin in Ovarian Cancer Cells. Clin Cancer Res an Off J Am Assoc Cancer Res (2004) 10(14):4661–9. doi: 10.1158/1078-0432.CCR-04-0137
156. Martinez-Balibrea E, Martínez-Cardús A, Musulén E, Ginés A, Manzano JL, Aranda E, et al. Increased Levels of Copper Efflux Transporter ATP7B Are Associated With Poor Outcome in Colorectal Cancer Patients Receiving Oxaliplatin-Based Chemotherapy. Int J Cancer (2009) 124(12):2905–10. doi: 10.1002/ijc.24273
157. Zdraveski ZZ, Mello JA, Farinelli CK, Essigmann JM, Marinus MG. MutS Preferentially Recognizes Cisplatin- Over Oxaliplatin-Modified DNA. J Biol Chem (2002) 277(2):1255–60. doi: 10.1074/jbc.M105382200
158. Al-Malky HS, Al Harthi SE, Osman AM. Major Obstacles to Doxorubicin Therapy: Cardiotoxicity and Drug Resistance. J Oncol Pharm Pract Off Publ Int Soc Oncol Pharm Practitioners (2020) 26(2):434–44. doi: 10.1177/1078155219877931
159. Gabizon A, Shmeeda H, Barenholz Y. Pharmacokinetics of Pegylated Liposomal Doxorubicin: Review of Animal and Human Studies. Clin Pharmacokinetics (2003) 42(5):419–36. doi: 10.2165/00003088-200342050-00002
160. Morabito A, Gattuso D, Stani SC, Fanelli M, Ferraù F, De Sio L, et al. Safety and Activity of the Combination of Pegylated Liposomal Doxorubicin and Weekly Docetaxel in Advanced Breast Cancer. Breast Cancer Res Treat (2004) 86(3):249–57. doi: 10.1023/B:BREA.0000036898.45123.e9
161. Singal PK, Li T, Kumar D, Danelisen I, Iliskovic N. Adriamycin-Induced Heart Failure: Mechanism and Modulation. Mol Cell Biochem (2000) 207(1-2):77–86. doi: 10.1023/A:1007094214460
162. Novitzky N, Thomas V, Abrahams L, du Toit C, McDonald A. Increasing Dose Intensity of Anthracycline Antibiotics Improves Outcome in Patients With Acute Myelogenous Leukemia. Am J Hematol (2004) 76(4):319–29. doi: 10.1002/ajh.20120
163. Gewirtz DA. A Critical Evaluation of the Mechanisms of Action Proposed for the Antitumor Effects of the Anthracycline Antibiotics Adriamycin and Daunorubicin. Biochem Pharmacol (1999) 57(7):727–41. doi: 10.1016/S0006-2952(98)00307-4
164. Sinha BK, Mimnaugh EG. Free Radicals and Anticancer Drug Resistance: Oxygen Free Radicals in the Mechanisms of Drug Cytotoxicity and Resistance by Certain Tumors. Free Radical Biol Med (1990) 8(6):567–81. doi: 10.1016/0891-5849(90)90155-C
165. Pawłowska J, Tarasiuk J, Wolf C, Paine M, Borowski E. Differential Ability of Cytostatics From Anthraquinone Group to Generate Free Radicals in Three Enzymatic Systems: NADH Dehydrogenase, NADPH Cytochrome P450 Reductase, and Xanthine Oxidase. Oncol Res (2003) 13:245–52. doi: 10.3727/096504003108748294
166. Fogli S, Nieri P, Breschi MC. The Role of Nitric Oxide in Anthracycline Toxicity and Prospects for Pharmacologic Prevention of Cardiac Damage. FASEB J Off Publ Fed Am Societies Exp Biol (2004) 18(6):664–75. doi: 10.1096/fj.03-0724rev
167. Frank NE, Cusack BJ, Talley TT, Walsh GM, Olson RD. Comparative Effects of Doxorubicin and a Doxorubicin Analog, 13-Deoxy, 5-Iminodoxorubicin (GPX-150), on Human Topoisomerase Iiβ Activity and Cardiac Function in a Chronic Rabbit Model. Invest New Drugs (2016) 34(6):693–700. doi: 10.1007/s10637-016-0388-x
168. Chen SH, Chan NL, Hsieh TS. New Mechanistic and Functional Insights Into DNA Topoisomerases. Annu Rev Biochem (2013) 82:139–70. doi: 10.1146/annurev-biochem-061809-100002
169. Roos WP, Kaina B. DNA Damage-Induced Cell Death: From Specific DNA Lesions to the DNA Damage Response and Apoptosis. Cancer Lett (2013) 332(2):237–48. doi: 10.1016/j.canlet.2012.01.007
170. Lotem J, Peled-Kamar M, Groner Y, Sachs L. Cellular Oxidative Stress and the Control of Apoptosis by Wild-Type P53, Cytotoxic Compounds, and Cytokines. Proc Natl Acad Sci USA (1996) 93(17):9166–71. doi: 10.1073/pnas.93.17.9166
Keywords: stress granule, anti-cancer medication, bortezomib, sorafenib, oxaliplatin, 5-fluorouracil, cisplatin, doxorubicin
Citation: Asadi MR, Moslehian MS, Sabaie H, Poornabi M, Ghasemi E, Hassani M, Hussen BM, Taheri M and Rezazadeh M (2021) Stress Granules in the Anti-Cancer Medications Mechanism of Action: A Systematic Scoping Review. Front. Oncol. 11:797549. doi: 10.3389/fonc.2021.797549
Received: 18 October 2021; Accepted: 08 December 2021;
Published: 24 December 2021.
Edited by:
Peng Zhao, The University of Texas Health Science Center at San Antonio, United StatesReviewed by:
Amin Safa, Complutense University of Madrid, SpainRezvan Noroozi, Jagiellonian University, Poland
Copyright © 2021 Asadi, Moslehian, Sabaie, Poornabi, Ghasemi, Hassani, Hussen, Taheri and Rezazadeh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, TW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==; Maryam Rezazadeh, UmV6YXphZGVobUB0YnptZWQuYWMuaXI=
 Mohammad Reza Asadi
Mohammad Reza Asadi Marziyeh Sadat Moslehian1
Marziyeh Sadat Moslehian1 Hani Sabaie
Hani Sabaie Marziye Poornabi
Marziye Poornabi Mehdi Hassani
Mehdi Hassani Mohammad Taheri
Mohammad Taheri