
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 03 January 2022
Sec. Gynecological Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.797007
This article is part of the Research Topic Surgical Horizons in Cervical Cancer View all 10 articles
 Sadia Safi1
Sadia Safi1 Yasmin Badshah1
Yasmin Badshah1 Maria Shabbir1*
Maria Shabbir1* Kainat Zahra1
Kainat Zahra1 Khushbukhat Khan1
Khushbukhat Khan1 Erum Dilshad2
Erum Dilshad2 Tayyaba Afsar3
Tayyaba Afsar3 Ali Almajwal3
Ali Almajwal3 Nawaf W. Alruwaili3
Nawaf W. Alruwaili3 Dara Al-disi3
Dara Al-disi3 Mahmoud Abulmeaty3
Mahmoud Abulmeaty3 Suhail Razak3*
Suhail Razak3*Our study aimed to identify the new blood-based biomarkers for the diagnosis and prognosis of cervical cancer. Moreover, the three-dimensional (3D) structure of Kruppel-like factor 9 (KLF9) was also determined in order to better understand its function, and a signaling pathway was constructed to identity its upstream and downstream targets. In the current study, the co-expressions of tumor protein D52 (TPD52), KLF9, microRNA 223 (miR-223), and protein kinase C epsilon (PKCϵ) were evaluated in cervical cancer patients and a possible relation with disease outcome was revealed. The expressions of TPD52, KLF9, miR-223, and PKCϵ were studied in the blood of 100 cervical cancer patients and 100 healthy controls using real-time PCR. The 3D structure of KLF9 was determined through homology modeling via the SWISS-MODEL and assessed using the Ramachandran plot. The predicted 3D structure of KLF9 had a similarity index of 62% with its template (KLF4) with no bad bonds in it. In order to construct a genetic pathway, depicting the crosstalk between understudied genes, STRING analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG), and DAVID software were used. The constructed genetic pathway showed that all the understudied genes are linked to each other and involved in the PI3K/Akt signaling pathway. There was a 23-fold increase in TPD52 expression, a 2-fold increase in miR-223 expression, a 0.14-fold decrease in KLF9 expression, and a 0.05-fold decrease of PKCϵ expression in cervical cancer. In the present study, we observed an association of the expressions of TPD52, KLF9, miR-223, and PKCϵ with tumor stage, metastasis, and treatment status of cervical cancer patients. Elevated expressions of TPD52 and miR-223 and reduced expressions of KLF9 and PKCϵ in peripheral blood of cervical cancer patients may serve as predictors of disease diagnosis and prognosis. Nevertheless, further in vitro and tissue-level studies are required to strengthen their role as potential diagnostic and prognostic biomarkers.
Cervical cancer arises from the cervix in women. It is the fourth most prevalent and the fourth most frequent cause of cancer mortality, with approximately 604,000 new cases and 342,000 causalities all over the world in 2020 (1). Various studies have confirmed the association between genital human papillomavirus (HPV) and cervical cancer. Sexual contact is the key risk factor associated with HPV acquisition. HPV has been recommended as solely the “necessary cause” of cervical cancer (2). Pap smear has been the most widely used cervical cytology screening technique for the past 50 years. However, the Pap smear is far from perfect, and its foremost shortcoming is the possibility of a false-negative result (3). No significant improvements in the Pap test have been made, due to which false-negative results that arise from the Pap test are continuously being reported even now. Laboratory misinterpretations, preparation errors, and improper sampling are the main causes of erroneous negative results (4). Although the basic treatment for cervical cancer is surgery or chemoradiation therapy, patients with advanced-stage tumor have poor disease prognosis with severe side effects. Hence, substitute screening approaches are required in underdeveloped and developing countries (5).
It has been reported that KRAS and phosphoinositide 3-kinases, upon activation via different receptors, e.g., G protein-coupled receptors (GPCRs) and receptor tyrosine kinase (RTK), cause the activation of the major downstream signaling pathways. Various studies have confirmed the interactions of Kruppel-like factor 9 (KLF9), protein kinase C epsilon (PKCϵ), tumor protein D52 (TPD52), and microRNA 223 (miR-223) with the downstream components of these signaling pathways, which eventually lead to carcinogenesis (6–9).
TPD52 (CR542034.1) is situated at the 8q21 chromosome, on an area that is commonly amplified in numerous cancers particularly in humans (10). The primary evidence of the importance of an altered expression of TPD52 in various cancers was obtained from the position of this gene on chromosome 8q, and during the mid-1990s, it became widely understood that the expression of TPD52 increases in certain tumor types, as well as in MYC oncogene. Nevertheless, the role of TPD52 in the onset of cancer is still debatable (11). The expression of TPD52 is upregulated in certain types of cancers, such as breast, prostate, ovarian, and pancreatic cancer, Burkitt’s lymphoma, multiple myeloma, and melanoma (12). On the other hand, the expression of TPD52 is also downregulated in other cancer types such as leiomyosarcoma, papillary renal cell cancer, clear cell renal cell cancer, lung cancer, and liposarcoma. Due to its altered expression in various cancers, it is referred to as a controversial gene (13). Several studies have reported evidence of the role of TPD52 in various signaling pathways of cancers, i.e., in the PI3K/Akt signaling pathway (14), protein kinase B/Akt signaling pathway (15), and nuclear factor-κB transactivation (16).
KLF9 (NM_001206.4) is a regulator of transcription in cellular adhesion, differentiation, and proliferation in the endometrium (17). Irregular expression of KLF9 may contribute toward the onset of several carcinomas and their proliferation (18). KLF9 is known to interact substantially with the Akt pathway. One of the studies validated the involvement of KLF9 in the Akt pathway and indicated that KLF9 substantially inhibits AKT activation and abrogates tumor growth in prostate cancer (19, 20).
PKCϵ (NM_005400.3) is one of the members of the protein kinase C family. Out of 10 isoforms of serine/threonine kinases, PKCϵ is the most widely studied for its contribution to malignant transformation (21). A recent study has revealed the interaction of PKCϵ with Akt, suggesting that the downregulation of PKCϵ causes the inhibition of Akt in breast cancer cells, thus increasing drug efficacy in breast cancer patients (22). The overexpression of PKCϵ has been reported in a wide range of carcinomas, including breast cancer, lung cancer, prostate cancer (23, 24), and brain tumors (25).
Similarly, recent studies have suggested the reduced expression of miR-223 (NC_000023.11) in metastatic and end-stage osteosarcoma patients, indicating the inhibitory role of miR-223 in osteosarcoma. An increased expression of miR-223 revokes atherosclerosis advancement by activating the PI3K/AKT pathway through blockade of TLR4 signaling. Its dysregulation is also associated with aberrant Akt/mTor pathway in various diseases such as myocardial infarction (26), colorectal cancer (27), and pancreatic cancer (28).
Kruppel-like factor (KLF) proteins have been found in diverse species and are known to have evolved by gene duplication (29, 30). However, the structures of all KLFs, except that of KLF4 (PDB ID: 2BWU), remain unpredicted. The prediction of the first ever structure of KLF4 provided new insights toward a better understanding of the molecular basis and functional anatomy of KLF4 and the other members of the KLF family (31) The three-dimensional (3D) structure of proteins helps in understanding their functions and their interactions with their binding partners (32). Our study describes the approaches to identify and determine the conserved domains and 3D structure of KLF9 and the development of a genetic pathway, thus establishing a crosstalk between KLF9 and its upstream and downstream targets. Additionally, although the individual expression status of TPD52, KLF9, miR-223, and PKCϵ has been previously studied in various tumors, no study has investigated the co-expressions of TPD52, KLF9, miR-223, and PKCϵ in any cancer type. Hence, our study also aimed to identify the combined expression patterns of TPD52, KLF9, miR-223, and PKCϵ, and their relationship with clinicopathological features, and to investigate the diagnostic and prognostic value of these genes in cervical cancer patients.
Blood samples were collected only from those patients who gave approval to collect their blood voluntarily in Combined Military Hospital (CMH), Rawalpindi, after approval by the Ethical Committee of Combined Military Hospital and ASAB, National University of Science and Technology, Islamabad, Pakistan. All participants were informed about the study objectives and signed the informed consent. The study protocol was carried out in accordance with the principles of the Declaration of Helsinki (33).
Blood samples were collected from female patients with histologically confirmed diagnosis of localized and/or metastasized carcinoma of the cervix (n = 100) and currently were on chemotherapy, radiotherapy, or chemoradiotherapy. Patients with co-infection of HIV were excluded from our study. The median age of cervical cancer patients was 47.5 years (range, 35–60 years). Furthermore, a control group was also included in the present study, which comprised blood samples from healthy individuals (n = 100), for accurate interpretion of the results.
RNA was extracted from whole blood drawn from peripheral veins of cancer patients using the TriZol reagent (Thermo Fischer Scientific, Waltham, MA, USA). The reaction was conducted on ice to avoid RNA degradation. For cDNA synthesis, 20 μl of the reaction mixture was prepared by adding 1 μl of Oligo dT20 [Random Hexamer, 1 μl dNTP mix (2.5 mM)], <5 μg of RNA, and RNAse-free water up to 10 μl. The reaction mixture was incubated at 65°C in a thermocycler for 5 min. In the next step, 10× reaction buffer (2 μl), 100 mM DTT (1 μl), RNase inhibitor (0.5 μl), and RTase (1 μl) were added into the PCR tube (same) and placed in a thermocycler for 50 min at 42°C and for 10 min at 70°C. The synthesized cDNA was stored at −20°C.
For analysis of the expression of the candidate gene and microRNA (miRNA), real-time PCR was used. Real-time reaction mixture was made by adding 10 μl of Wiz pure qPCR master mix (SYBR), 6 mM of forward and reverse primers, and 10 μg of cDNA with RNAse-free water up to a volume of 20 μl. The conditions for quantitative PCR (qPCR) amplification were 40 cycles with an initial temperature of 95°C for 10 min, which basically activated Hot Start DNA polymerase, followed by 95°C for 15 s and then amplification for 1 min for 61°C, followed by real-time analysis for 45 s at 75°C. The primer sequences and the GC (guanine–cytosine) content are presented in Table 1. The specificity of primers was confirmed by observing the melt curve analysis of qPCR. The reagent and software used for real-time PCR were SYBR Green dye and 7300 SDS software, respectively. For quantifying the gene expression, the 2−ΔΔCT method was performed. Moreover, the Livak method was used for conversion of the cycle threshold (Ct) values, obtained for real-time PCR, into fold change. β-actin was used as a control, and the experiment was performed in triplicate. The Ct values obtained in triplicate for each sample was found to be almost the same, hence confirming the validity of the results.
Statistical analysis was performed with one-way and two-way ANOVA in order to show the relationship of the expressions of TPD52, KLF9, miR-223, and PKCϵ with the different clinicopathological features of cervical cancer. Spearman’s rho correlation was used to test the association of age and the stage of the disease. All these statistical tests were performed using GraphPad Prism 6.0 software. Similarly, GraphPad prism was employed for generating the receiver operating characteristic (ROC) curve.
The 3D structure of KLF9 protein (NP_001197.1) was determined through homology modeling via SWISS-MODEL, a bioinformatics web server. For prediction of the 3D structure of KLF9, the first amino acid sequence of the KLF9 gene was retrieved from the National Center for Biotechnology Information (NCBI) in FASTA format. In order to find the conserved domains and the evolutionary relationships between all the 17 members of the KLF family, multiple sequence alignment was done using Clustal Omega. For a better understanding of the evolutionary histories and conservation of the different members of the KLF family, phylogenetic analysis was performed using MEGA 7. The secondary structure of KLF9 was predicted via different servers, i.e., UCL Bioinformatics Group (34), SPIDER2 (35), and Predict Protein (36). For 3D structure predictions, KLF4 was chosen as a template due to the fact that its structure has already been crystallographically predicted in RCSB-PDB (Research Collaboratory for Structural Bioinformatics Protein Data Bank). Hence, the structure of KLF4 (PDB ID: 2BWU) was taken from RCSB-PDB. After acquistion of the template (KLF4) structure, the sequence of KLF9 in FASTA format was aligned to the crystallographically determined structure of KLF4 via the SWISS-MODEL and a 3D model of KLF9 was generated.
In order to construct a genetic pathway depicting the crosstalk between understudied genes, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used and STRING analysis was performed to study the gene linkage, while the genetic pathway was obtained via DAVID software.
The results of the multiple sequence alignment of KLF9 with the rest of the members of the KLF family via Clustal Omega (37) depicted the conserved domains across all KLF family members. Figure 1 depicts the results of multiple sequence alignment using Clustal Omega. Three tandem C2H2 zinc finger domains, 1, 2, and 3, were found to be conserved throughout the members of the KLF family.
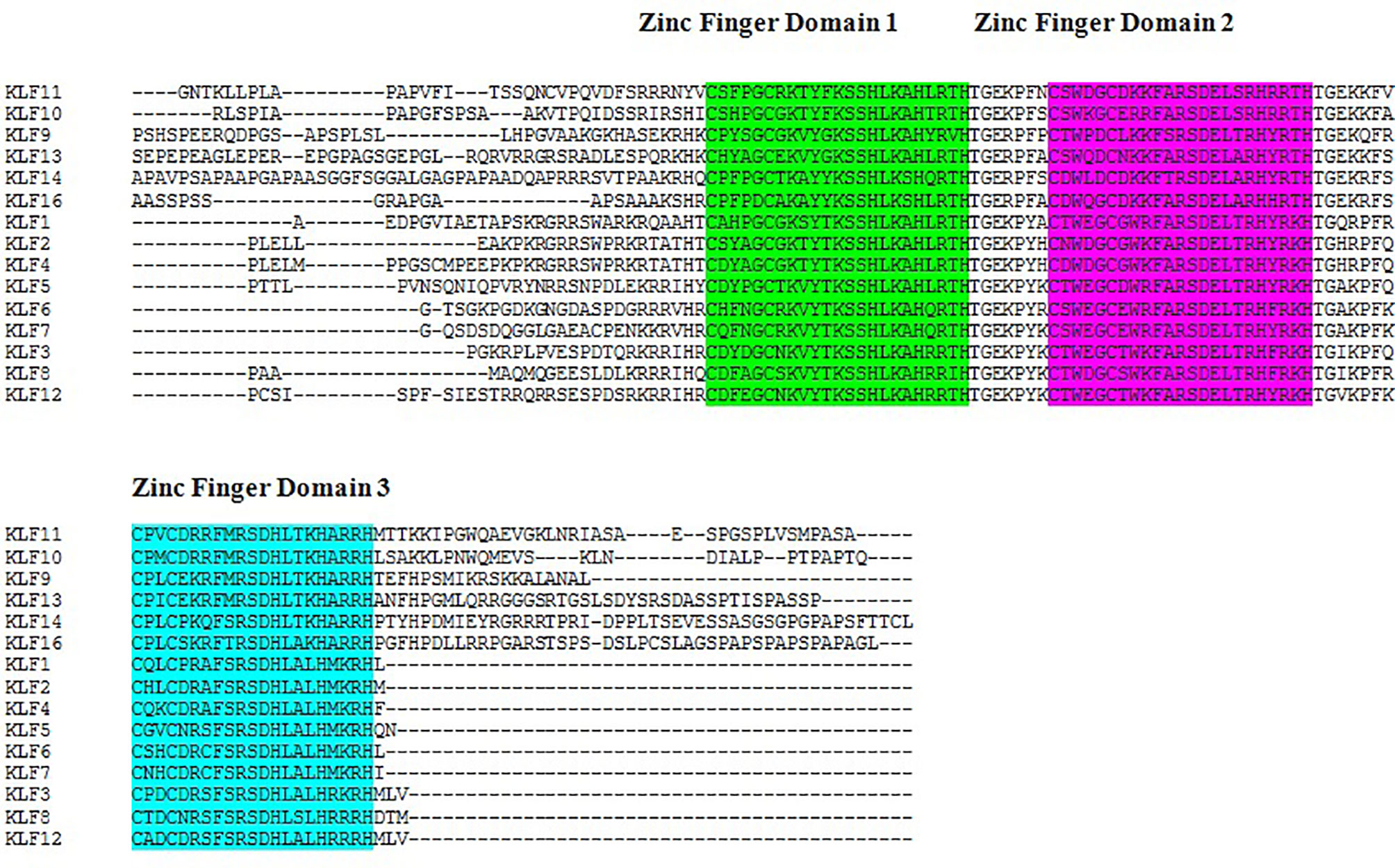
Figure 1 Sequence alignment of Kruppel-like factors depicting the conserved domains obtained from Clustal Omega. Certain sequence alignments have been deleted for formatting. Zinc figure domain 1 (labeled green), zinc figure domain 2 (labeled purple), and zinc figure domain 3 (labeled blue) are highlighted.
Phylogenetic analysis of the KLFs performed by MEGA 7 (38) using the UPGMA (unweighted pair group method with arithmetic mean) phylogenetic tree placed KLF9 in group 3 based on its transcription repression activity (Figure 2). Like in earlier studies, the KLF family members were divided into three groups based on their evolutionary histories, structural characteristics, and binding domains, which help define their functions. Group 1 includes KLF3, KLF8, and KLF12. These members serve as repressors of transcription by mediating interactions with the co-repressors Sin3A and CtBP. Group 2 includes KLF1, KLF2, and KLF4–KLF7. These members act as activators of transcription. Group 3 includes KLF9–KLF11, KLF13, KLF14, and KLF16. These members serve as repressors of transcription by mediating interactions with the co-repressors Sin3A and CtBP (41).
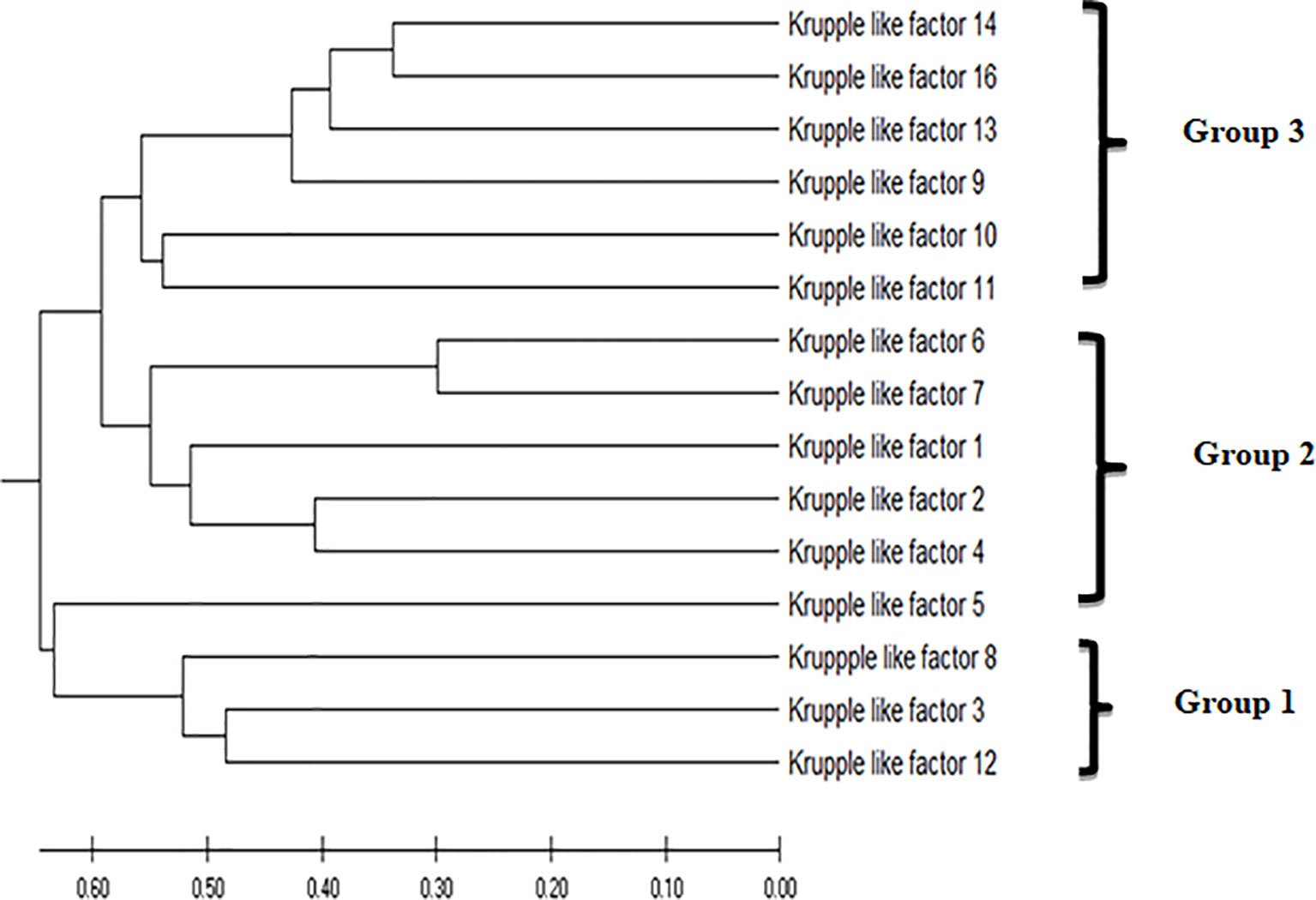
Figure 2 The evolutionary history was inferred using the unweighted pair group method with arithmetic mean (UPGMA). The optimal tree with the sum of branch length = 7.54632578 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (39) and are in the units of the number of amino acid substitutions per site. This analysis involved 15 amino acid sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 602 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (40).
Each member of KLF family, despite having highly conserved consensus sequences at the C-terminal region, has unique functions involved in cellular processes. This is due to great variations in sequences at the N-terminus region of KLFs that mediate interactions with diverse activators and repressors of transcription. The KLF sequences contain conserved motifs, at the N-terminus, comprising CtBP and Sin3A binding sites (41). Co-repressor C-terminal binding protein (CtBP) is a co-repressor of transcription. The main mechanism by which CtBP proteins suppress transcription is by recruiting histone methyl transferases and histone deacetylases (HDACs) to transcriptional complexes, which causes chromatin compaction and transcriptional silencing by the methylation and deacetylation of proteins, respectively (42, 43). KLF3, KLF5, KLF8, and KLF12 contain the conserved motif CtBP binding site. KLF3, KLF8, and KLF12 contain the conserved sequence PXDLS that mediates the interaction between KLFs and CtBP. This interaction facilitates the functions of KLF3 and KLF8 in co-repression and the activity of KLF12 in repressing AP-2α gene expression (44). Sin3A is a protein that functions as a repressor of transcription. It is involved in the recruitment and binding of HDACs (45). KLF9, KLF10, KLF11, KLF13, KLF14, and KLF16 possess binding sites for Sin3A. These KLFs possess the R1 domain that enclose a Sin3-interacting domain (SID), an α-helical hydrophobic structure that meditates binding with the PAH domain of Sin3 proteins (46). It was found that KLF9, KLF10, KLF11, KLF13, KLF14, and KLF16 possess a conserved α-helical motif in their structure, i.e., AA/VXXL, a binding site for Sin3A that facilitates interaction with Sin3A and causes transcriptional repression (47). Unexpectedly, KLF1 possesses no SID, but still interacts with Sin3A and acts as a co-repressor (48).
KLF9 contains the conserved hydrophobic motif AAQCL in its amino acid sequence, as shown in Figure 3. It serves as a SID and is able to recruit and bind Sin3A. Sin3A proteins bind HDAC1, HDAC2, and other proteins, probably assembling multi-unit complexes (HDAC1 and HDAC2), altering chromatin compaction and so repressing transcription. A number of studies have justified the presence of such conserved motifs in KLF9 (49).
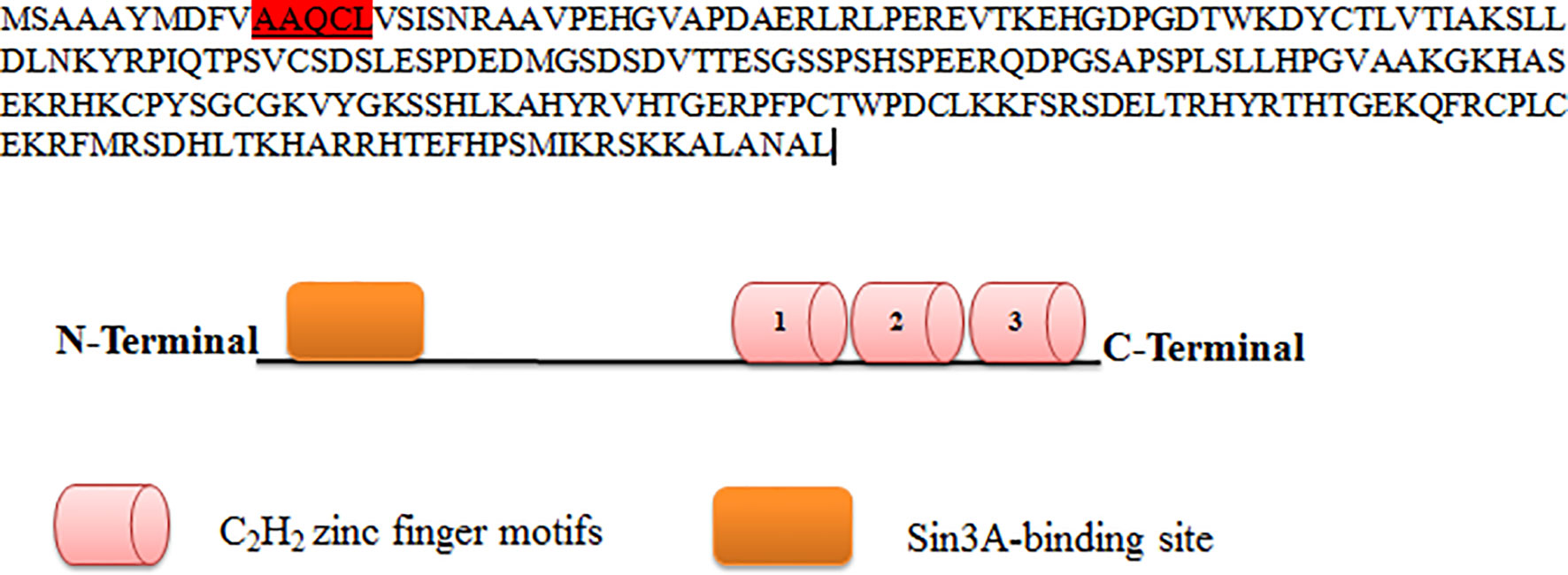
Figure 3 Protein sequence of KLF9. KLF9 is highly homologous to other members of the Kruppel-like factor (KLF) family at carboxy-terminal DNA-binding regions, which contain three C2H2 zinc finger motifs. At the N-terminal region is the Sin3A binding region.
Subcellular localization of KLF9 was found to be inside the nucleus (Figure 4). By modeling the functional domain features and the hidden associations of gene ontology, different servers gave different nuclear signals. Hum-mPLoc 3.0 showed a nuclear signal of 1.88, while DeepLoc-1.0 showed a nuclear signal of 0.99.
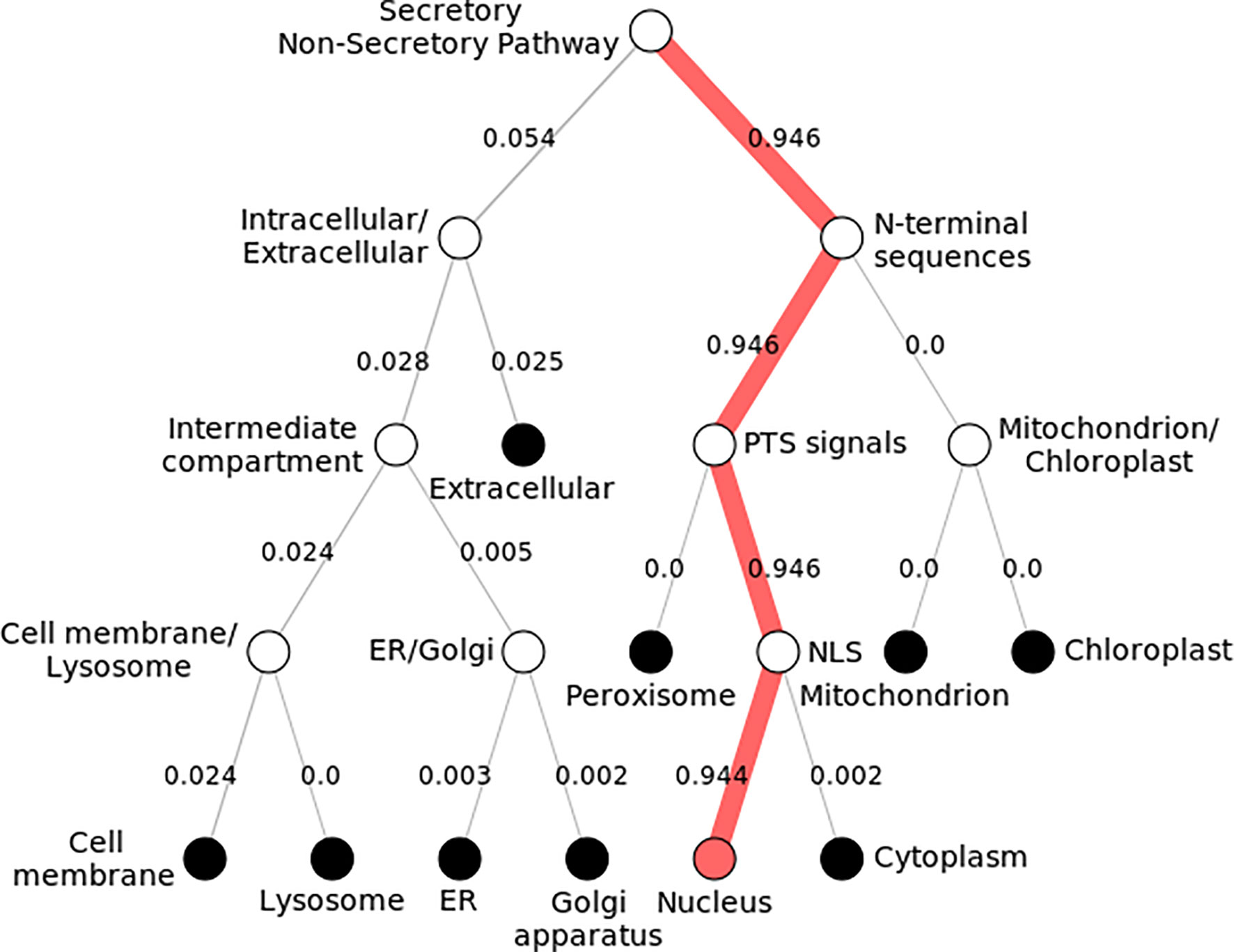
Figure 4 Subcellular localization of KLF9. Pathway following subcellular localization of KLF9 generated by DeepLoc-1.0. Numerous locations are shown, and each follows a distinct pathway and score. The KLF9 protein is localized inside the nucleus (depicted by 0.9 score). It directs toward the nucleus by executing peroxisomal targeting signals (PTS) and nuclear localization signals (NLS).
The similarity index between the structures of the template (KLF4) and target (KLF9) was found to be 62%. The 3D structure of KLF9 is shown in Figure 5A. Using Chimera, the structure of KLF9 obtained via the SWISS-MODEL was superimposed on KLF4 (template) for the anlysis of structural conservation between the target (KLF9) and template (KLF4). The template is labeled red, while target is labeled blue. Figure 5B illustrates the superimposed structures of the template (KLF4) and target (KLF9) proteins. Ramachandran plots were used to analyze the quality of the model obtained via the SWISS-MODEL. These plots were used for visualization of the dihedral angles, i.e., phi (φ) and psi (ψ) angles of the amino acids. It was found that most of the amino acids were found to be lying in favorable regions, i.e., 95.06%, and Ramachadran outliers were 1.23% (A146 PRO). Bad bonds in the structure were 0/721, while bad angles were 16/965. Figure 5C illutrates the Ramachadran plot.
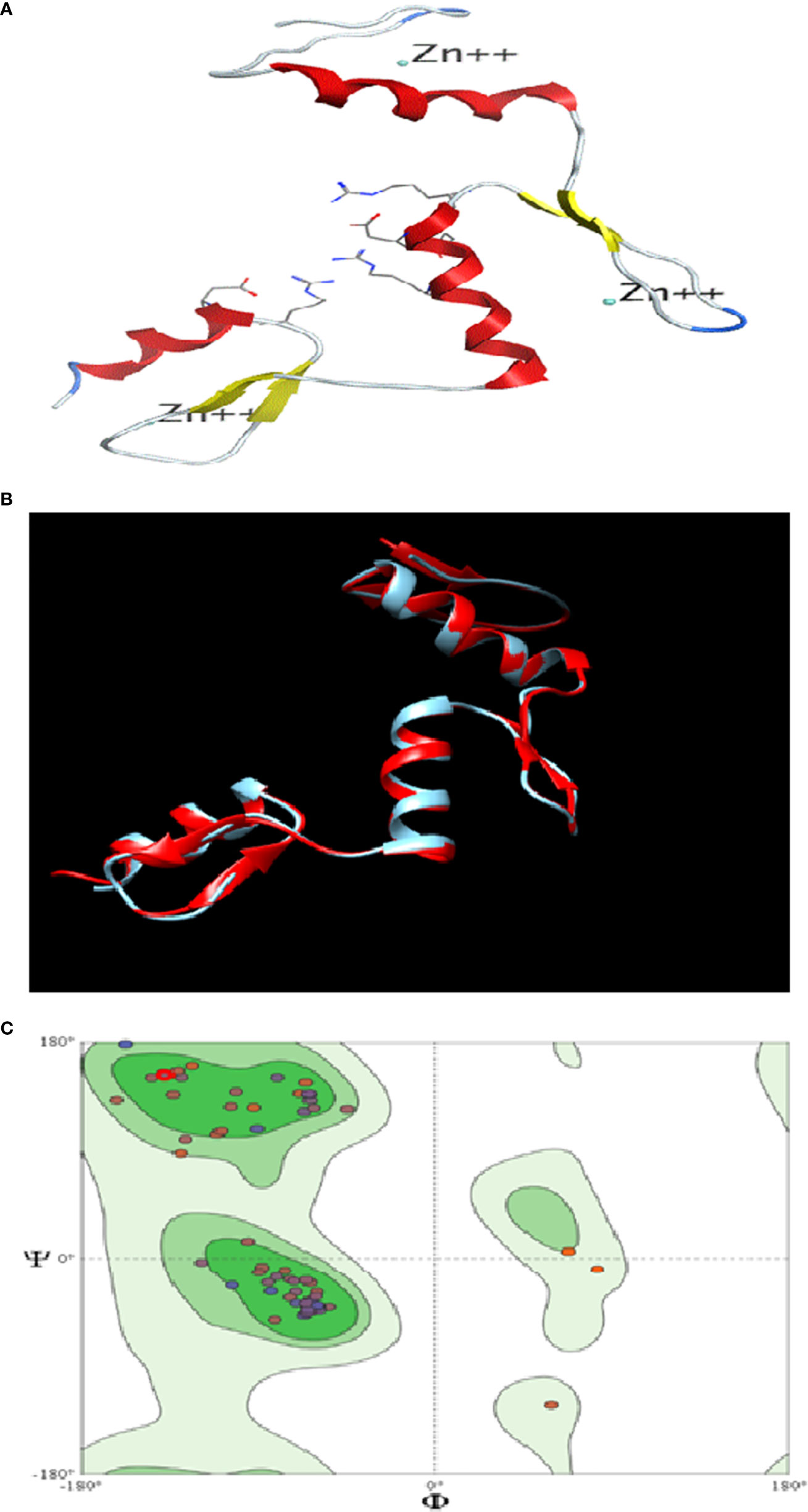
Figure 5 In silico analysis of KLF9. (A) Three-dimensional structure of KLF9. (B) Comparison of the crystallographically determined structure 2bwu (labeled red) and the predicted structure KLF9 (labeled blue) for the analysis of structure conservation. (C) Ramachandran plot analysis determining the quality of the model. Most amino acids (95%) were found in favored regions, showing that the model is of good steriochemical quality.
In this study, we observed a significantly increased expression of TPD52 (23.8 ± 0.42) in understudied samples of cervical cancer compared to the controls. The expression of KLF9 was found to be downregulated in the blood of cervical cancer patients (0.14 ± 1.6) relative to healthy controls. There was an elevation of miR-223 expression in cervical cancer patients (2.0 ± 1.8) relative to controls. In the case of PKCϵ, its expression was found to be significantly reduced in cervical cancer patients (0.05 ± 5.7). Overall, we found that the expressions of TPD52 and miR-223 were increased 23- and 2-fold in peripheral blood of cervical cancer patients, respectively, whereas expressions of KLF9 and PKCϵ were 0.14- and 0.05-fold reduced in cervical cancer patients relative to healthy individuals (Figure 6).
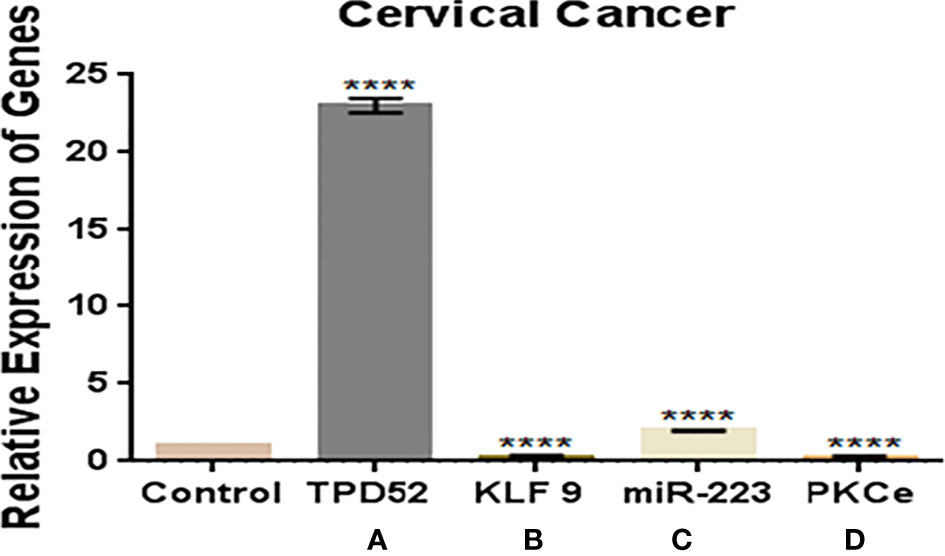
Figure 6 Expressions of TPD52, KLF9, miR-223, and PKCϵ in blood of cervical cancer patients. (A) TPD52 expression was increased 23-fold. (B) KLF9 expression was decreased 0.4-fold. (C) miR-223 expression was increased 2-fold. (D) PKCϵ decreased 0.05-fold. Fold change is plotted on the y-axis and study groups on the x-axis. Illustrative data are presented as the mean ± SEM of triplicate experimentations. Statistical significance was measured by ordinary two-way ANOVA (****p < 0.0001).
The clinicopathological features of cervical cancer patients are shown in Table 2. The relative expressions of TPD52, KLF9, miR-223, and PKCϵ in cervical cancer patients were measured with respect to their clinical features. The fold change and expression status of TPD52, KLF9, miR-223, and PKCϵ for each clinicopathological feature, i.e., low tumor stage groups I–II and advanced tumor stage groups III–IV, distant metastatic vs. non-metastatic group, and treatment status of patients (e.g., chemotherapy, radiotherapy, or chemoradiotherapy), are shown in Table 3. Significant results (p < 0.001) were found between all groups of patients. The expression of TPD52 was found to be significantly higher in the lower tumor stage and non-metastatic groups of patients in comparison to its high expression in the advanced tumor stage and distant metastatic groups of patients (Figures 7A, B). A similar trend was found for miR-223 (Figures 7E, F). In the case of KLF9, its expression was much more significantly reduced in the advanced tumor stage and distant metastatic groups relative to its less reduced expression in the lower tumor stage and non-metastatic groups (Figures 7C, D). On the other hand, for PKCϵ, its expression was much more significantly reduced in the lower tumor stage and non-metastatic groups relative to its less reduced expression in the advanced tumor stage and distant metastatic groups (Figures 7G, H).
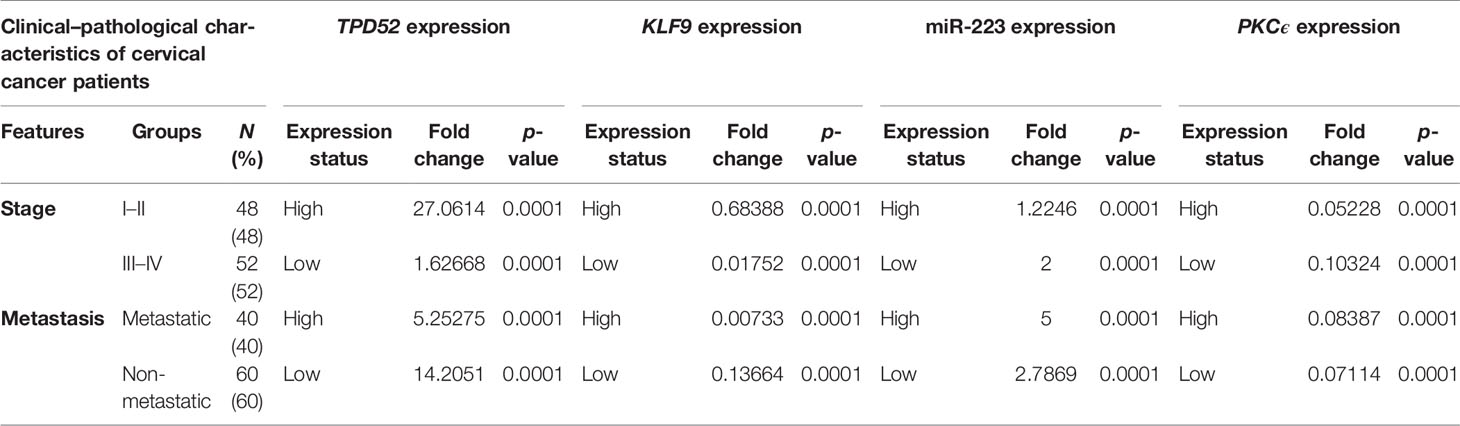
Table 3 Relationship between TPD52, KLF9, PKCϵ, and miR-223 expression and clinicopathological features of cervical cancer.
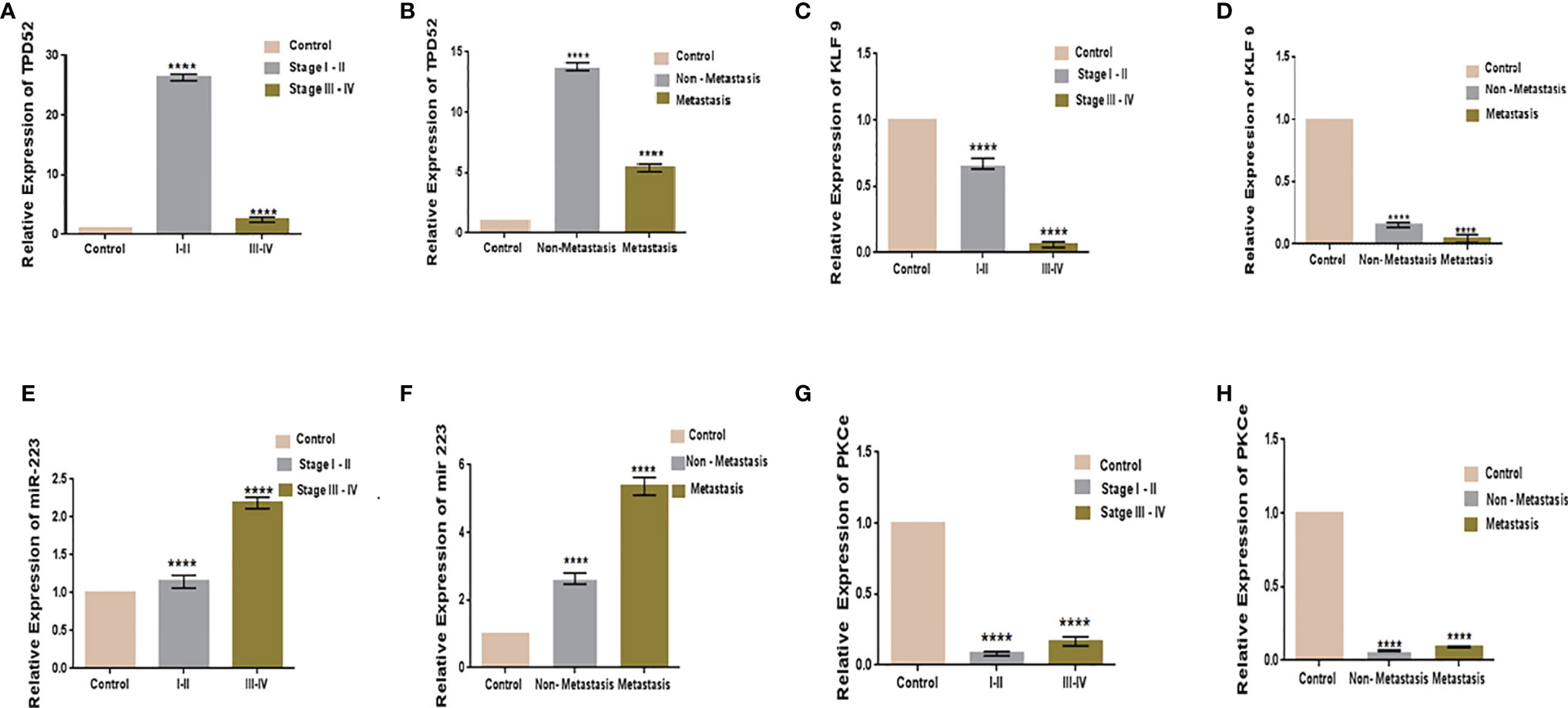
Figure 7 Relative gene expression with clinical features of cervical cancer. Relative TPD52 expression with tumor stage (A) and metastasis (B). Relative KLF9 expression with tumor stage (C) and metastasis (D). Relative miR-223 expression with tumor stage (E) and metastasis (F). Relative PKCϵ expression with tumor stage (G) and metastasis (H). Fold change is plotted on the y-axis and study groups on the x-axis. Illustrative data are presented as the mean ± SEM of triplicate experimentations. Statistical significance was measured by ordinary one-way ANOVA (****p < 0.0001).
We also found that the expression of TPD52 was lowest in patients undergoing chemoradiotherapy relative to its higher expression in patients receiving a combination of chemotherapy and radiotherapy (Figure 8A). A similar trend was followed in the expression profile of miR-223 (Figure 8C), whereas for KLF9 and PKCϵ, patients undergoing chemoradiotherapy showed higher expressions relative to patients on chemotherapy and radiotherapy, where their expressions were significantly reduced (Figures 8B, D). However, it is to be noted that the expressions of TPD52 and miR-223 were higher relative to healthy controls and that the expressions of KLF9 and PKCϵ were lower in comparison to healthy controls in each group of patients.
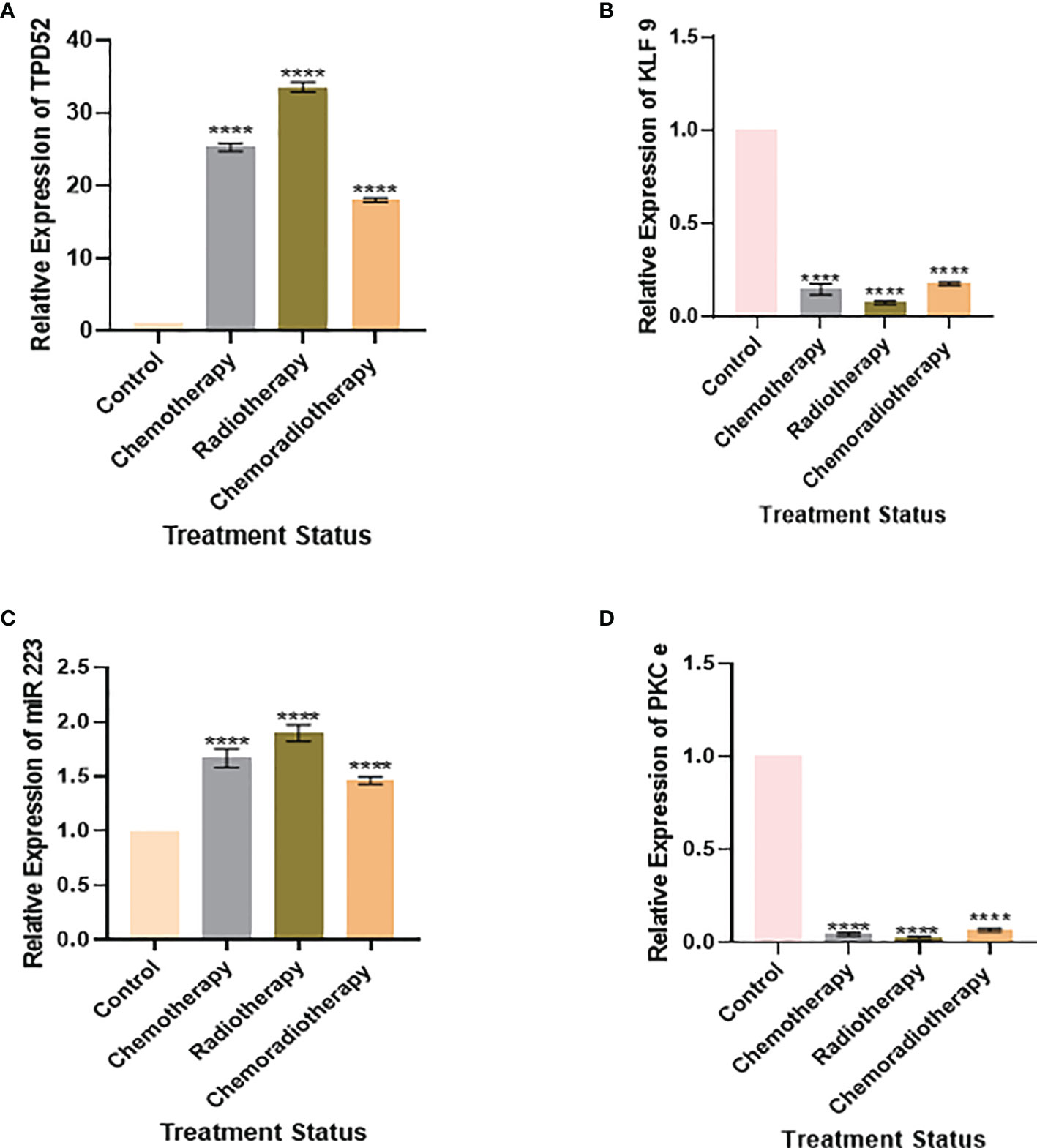
Figure 8 Relative gene expression with treatment status. (A) Relative TPD52 expression. (B) Relative KLF9 expression. (C) Relative miR-223 expression. (D) Relative PKCϵ expression. Illustrative data are presented as the mean ± SEM of triplicate experimentations. Fold change is plotted on the y-axis and study groups on the x-axis. Statistical significance was measured by ordinary one-way ANOVA (****p < 0.0001).
For verification of the relationship between these blood-based biomarkers (TPD52, KLF9, miR-223, and PKCϵ) and cervical cancer, ROC curves were generated (Figure 9). The area under the ROC curve (AUC) was calculated and 95% confidence intervals were determined.
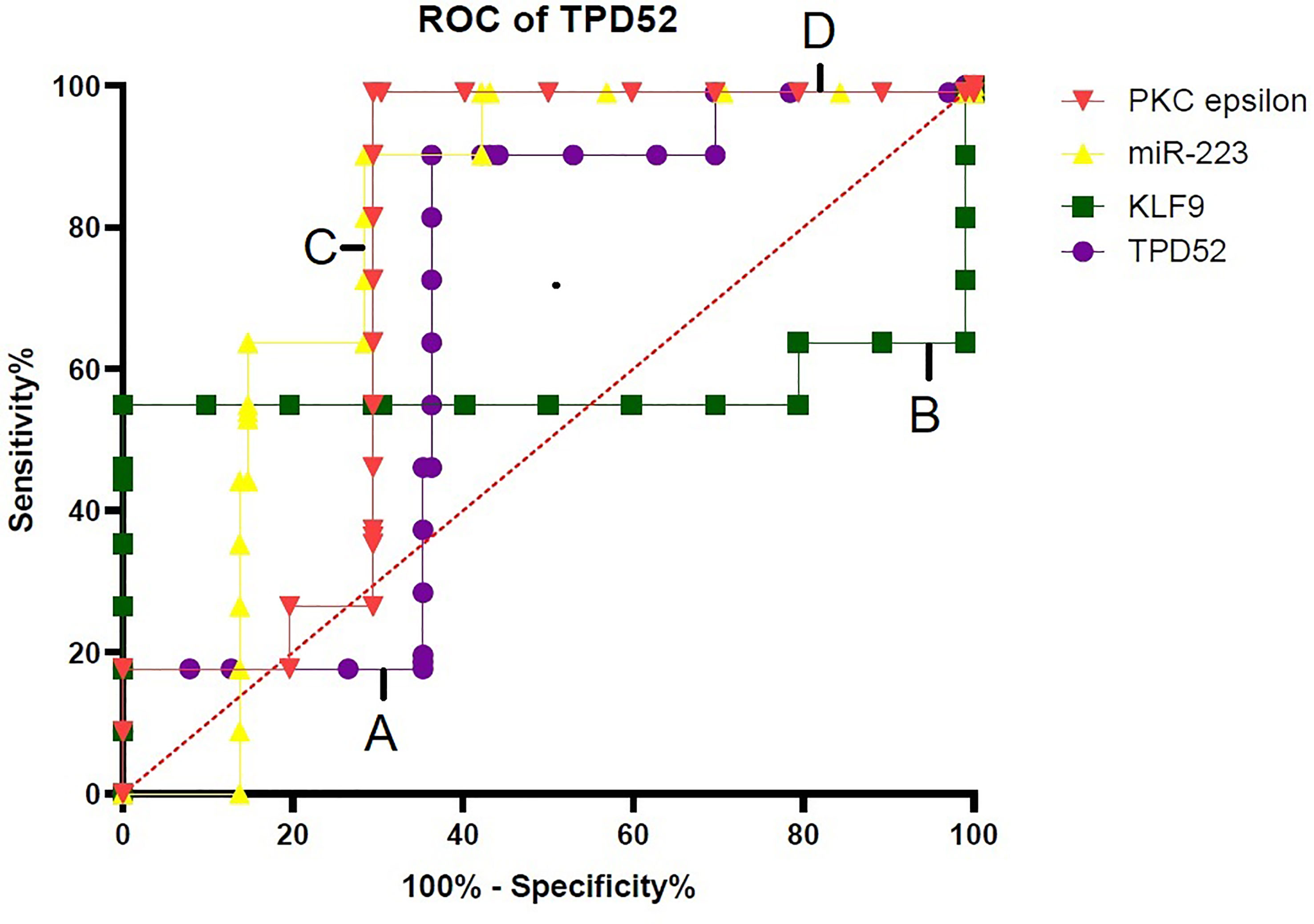
Figure 9 Specificity of TPD52, KLF9, miR-223, and PKCϵ in the diagnosis of cervical cancer. Receiver operating characteristic (ROC) curve for TPD52, KLF9, miR-223, and PKCe predicted high risk of cervical cancer. (A) TPD52: area under the ROC curve (AUC) = 0.6685 and 95% confidence interval (CI) = 0.5880–0.7490. (B) KLF9: AUC = 0.5706, 95%CI = 0.4775–0.6638. (C) miR-223: AUC = 0.7884, 95%CI = 0.7184–0.8583. (D) PKCϵ: AUC = 0.7595, 95%CI = 0.6852–0.8338.
The association between patient’s age and cancer stage is shown in Figure 10. Participants diagnosed with stage IV were significant older than those in the early stages. Furthermore, age showed a significant positive correlation with stage (r = 0.503, p < 0.001).
Tumors arising in the genital tract of females were found to be the fourth most frequent set of malignancies among females. The absence of screening methods, diagnostic techniques, and treatment and the lack of proper knowledge are the leading causes of cervical cancer incidences (50). The late diagnosis of the illness results in increased mortality rates (51). Although various screening techniques are being used for the diagnosis of cervical cancer, the death rates in developing states continue to be high, i.e., 87%. Pap smear is currently used for screening cervical neoplasia at an early stage. However, the false-negative results that are often produced by the Pap test is one of its major drawbacks (50). Hence, discovering the biological and molecular mechanisms of tumor progression and identifying diagnostic biomarkers have become essential in cancer research studies.
With improvements in technology, there has been a significant increase in the structure determination of numerous proteins. Still, the prediction of protein structures remains a challenging task. However, certain theoretical models can be used to assess the topological characteristics of proteins. The 3D structure of protein helps in understanding their functions and their interactions with their binding partners. Homology modeling can help in predicting low-resolution structures. Hence, in this study, the 3D structure of KLF9 was predicted via the SWISS-MODEL Workspace. The template used for 3D structure predictions was KLF4. The server used for the visualization of the 3D structures was Chimera. The similarity index between the structure of a template (KLF4) and a target (KLF9) was known to be 62%, and no bad bonds were found in the predicted structure. This study also predicted the possible crosstalk of KLF9 with TPD52, miR-223, and PKCϵ. KEGG and STRING were used to determine gene linkage with neighboring genes, while DAVID 6.8 was used to dig out the biological meaning from a large set of genes.
Gene linkage analysis via KEGG and STRING is shown in Figure 11. Our genetic pathway depicted that all the understudied genes are linked to each other and are involved in the Akt pathway. The pathway obtained via DAVID software depicted that PKCϵ is found is upstream to the Ras/Raf pathway and bridges the activation of this pathway by GPCRs. Some studies have also described the involvement of PKCϵ in the Ras/Raf pathway and have revealed that PKCϵ activates GPCR coupled Ras/Raf pathway and helps in the remodeling of cardiomyocytes (24). We also found that the regulation of PKCϵ by the STAT3 gene (signal transducer and activator of transcription 3) stimulates the activity of cyclin D in the nucleus via activation of o-myc (family of transcription regulatory genes), which leads to enhanced cell cycle progression. A regulatory link of PKCϵ with STAT3 has also been established in prostate adenocarcinoma (52). A few studies also depicted the activation of STAT3 via Rho kinases, which validates our results (53). Moreover, TPD52 also activates STAT3. A recent study has ascertained the activation of STAT3 via TPD52 (16). Hence, the transcriptional activity of STAT3 is regulated by PKCϵ, TPD52, and Rho-kinases. PKCϵ involvement was also found in the Rho signaling pathway, which eventually leads to metastasis. According to a recent study, PKCϵ also facilitates metastasis in breast cancer by activating Rho-GTPases (54). Our genetic pathway showed that Rho-GTPases are found downstream of PKCϵ, and ERK phosphorylation in the Ras/Raf pathway occurs due to the activation of a downstream target of PKCϵ (Rho GTPases). Our finding is in agreement with the previously published report by Pan et al. (55), who also found the same phosphorylation mechanism of ERK in the Ras/Raf pathway. Our genetic pathway also depicted the involvement of PKCϵ in the Akt pathway. We found that PKCϵ is located upstream of TPD52, and both of these genes activate the Akt pathway, which promotes tumor proliferation and invasion. The role of PKCϵ in Akt activation, by phosphorylating Akt at serine 473, has already been established (56). Akt is known to regulate proliferation and the cell cycle by targeting cyclin D1, p21, p53, and p27 (57). Forkhead box O (FOXO) is a transcription factor that serves as a downstream target of Akt (protein kinase B). Akt inhibits FOXO by phosphorylating it, and hence promoting cell survival, growth, and proliferation. TPD52 and PKCϵ block the transcriptional activity of FOXO, activate cyclin D, and inactivate p27 (regulator of the cell cycle), leading to enhanced cellular proliferation. According to Zhang et al. (58), the PI3K/Akt signaling pathway inactivates FOXO and, hence, cause the downregulation of cell cycle controls, i.e., CDKI and p27. Our results manifested that the decreased expression of KLF9 inhibits the progesterone growth hormones (progesterone receptor gene, PGR), which in return directly blocks FOXO and ultimately promotes tumorigenesis. Pabona et al. (59) validates our finding by demonstrating KLF9 as a regulator of PGR. Loss of KLF9 leads to the inhibition of PGR and FOXO signaling, hence leading to oncogenesis and tumor invasion in endometrial cells. The genetic pathway constructed in the current study also proposes that the increased expression of miR-223 causes the activation of STMN1 and inhibition of FOXO. In gastric cancer, overexpression of miR-223 also leads to a reduced expression of FOXO and the inhibition of cyclin D, p21, and p27 (60). Moreover, miR-223 is also involved in the activation of phosphatidylinositol 3-kinase (PI3K), which in return produces phosphatidylinositol triphosphate (PIP3) in the cell membrane. PIP3 activates Akt signaling. Zhu et al. (8) also reported on the role of overexpressed miR-223 in the activation of Akt and onset of tumorigenesis in cervical cancer.
This study also aimed to identify new biomarkers and critical genes linked to the prognosis and diagnosis of cervical cancer. In our study, we have measured the co-expressions of TPD52, KLF9, miR-223, and PKCϵ in cervical cancer. Expression dysregulation of the biomarkers PKCϵ, TPD52, miR-223, and KLF9 was determined by comparing the expression fold change with the expression profile of the healthy group. Previously, numerous studies that determine the expressions of biomarkers in patient blood using real-time-PCR were conducted. For instance, the prognostic significance of KLF7 was studied in tongue cancer (61). The plasma levels of several miRNAs, such as miR-218, miR-223, miR-7, miR30, and miR-21, were studied in hepatocellular carcinoma and gastric and ovarian cancer (62–64). Recently, the relative expressions of matrix metalloproteinases (MMPs) in blood of breast cancer patients were investigated to determine their role in cancer progression (65), hence indicating their possible application in disease prognosis. The current study also evaluated the mRNA expression of these molecules in blood of cervical cancer patients and provided a foundation for conducting an in-depth, proteome-level analysis in vitro and in vivo. The outcome of the current study indicated the prognostic significance of these molecules for cervical cancer. The diagnostic specificity of these biomarkers was also determined through ROC curve analysis. However, further evaluation on a larger cohort size and at the protein level is required to determine its clinical significance.
Earlier, the role of understudied genes had been independently studied in various tumors, which confirmed the involvement of these genes in cancer, metastasis, and expansion and in resistance to therapy. To the best of our knowledge, the co-expression of these genes in cervical cancer has not been studied yet. We observed an increased expression of TPD52 in cervical cancer patients relative to healthy controls who have very low levels of the TPD52 gene in their blood. Various studies reported the upregulation of TPD52 expression in quite a few cancers, such as breast, prostate, and pancreatic cancer, Burkitt’s lymphoma, multiple myeloma, and melanoma (12). On the other hand, the expression of TPD52 is downregulated in some cancers, such as papillary renal cell cancer, lung cancer, and liposarcoma (13). In the case of KLF9, we observed its significantly reduced expression in cervical cancer patients relative to healthy controls. Similar downregulation of KLF9 has been reported in endometrium cancer, where its downregulation is linked to estrogen-mediated growth control (66). The reduced expression of KLF9 has also been reported in breast cancer, human colorectal tumors, and hepatocellular carcinoma (67). Various studies have discovered that expression profiling of various circulating miRNAs in the blood may probably be used in therapeutic interventions and in identifying different tumor types (68). We have found an upregulation of miR-223 in cervical cancer patients relative to the healthy individuals. According to a recent study, the expression of miR-223 is significantly elevated in gastric adenocarcinoma cells. The upregulation of miR-223 encouraged cell proliferation and reduced apoptosis in gastric adenocarcinoma cells, while the downregulation of miR-223 expression has been linked to various cancer subtypes, including leukemia and gastric, esophageal, and colorectal cancer (69). In the case of PKCϵ, we observed its reduced expression in cervical cancer patients relative to healthy controls who had significantly high levels of this gene in their blood. On the contrary, an upregulation of PKCϵ has been reported in a large number of carcinomas, including breast, lung, and prostate cancer (70). Various reports have confirmed the role of this gene as an oncogene and its involvement in tumor metastasis (55). Our study found that the expression of TPD52 was upregulated in the advanced-stage tumor group (1.62 ± 0.4) and in the distant metastatic group of patients (5.25 ± 0.42) relative to lower stage tumor and non-metastatic groups, where its expression levels were increased 27.0 ± 1.68- and 14.2 ± 1.68-fold, respectively. Hence, TPD52 may serve as a potent early diagnostic biomarker in cervical cancer. A recent study has reported the decreased expression of TPD52 in tumorous tissues of hepatic cellular carcinoma (HCC) in comparison to healthy tissues. Further correlation analysis exposed that the reduced expression of TPD52 in HCC was suggestively linked to advanced stage tumor, signifying that a reduced TPD52 expression may promote tumor metastasis (71). These results are inconsistent with our study. Furthermore, in the case of KLF9, we observed its reduced expression in advanced tumor stage (0.01 ± 1.6) and in distant metastasis (0.007 ± 1.39). A downregulated expression of KLF9 was suggestively found in the lower stage tumor group (0.68 ± 1.6) and the non-metastatic group (0.13 ± 1.82). Our result is encouragingly inconsistent with recent findings that point to the fact that a reduced expression of KLF9 is linked to poor survival and prognosis in pancreatic ductal adenocarcinoma and leads to tumor metastasis (9). Our study found that the expression of miR-223 was increased in the advanced tumor stage (2.07 ± 3.9) and distant metastasis (5.8 ± 4.25) groups, while its expression was decreased in the lower tumor stage group (1.2 ± 43.9) and the non-metastatic group (2.7 ± 4.5). Further studies have revealed that miR-223 plays a significant part in the metastasis of cervical cancer. The upregulation of miR-223 promotes metastasis in cervical cancer cells (72). These results validate our results showing that the increased expression of miR-223 in cervical cancer patients causes metastasis and poor prognosis. The expression of PKCϵ was much more downregulated in the advanced tumor stage (0.10 ± 5.8) and distant metastasis (0.08 ± 6.36) groups relative to the lower tumor stage group and the non-metastatic group, where its expression was reduced 0.05 ± 6.0- and 0.07 ± 5.87-fold, respectively. According to recent studies, PKCϵ causes tumor metastasis to the bone by promoting translation increase and causes osteosarcoma metastasis (73). These findings contradict our study as PKCϵ inhibited metastasis in cervical cancer. The contradictory results may be due to the different cancer types.
Our study also discovered the effect of treatment on the expression profiles of understudied genes. It was found that patients undergoing chemoradiotherapy showed better prognosis. In the case of TPD52, patients undergoing chemoradiotherapy showed the lowest expression (18.52 ± 1.84) relative to patients on chemotherapy (26.2 ± 1.5) and radiotherapy (34.7 ± 1.83). Likewise, patients on chemoradiotherapy showed the lowest miR-223 expression (1.51 ± 3.8) relative to patients undergoing chemotherapy (1.76 ± 3.7) and radiotherapy (2.03 ± 4.2). These results show patients’ response to treatment and indicate that chemoradiotherapy has better prognosis, while radiotherapy is linked to poor prognosis in cervical cancer. During treatment expression profiling, KLF9 and PKCϵ were found to be slightly less reduced in patients treated with chemoradiotherapy, who showed better prognosis, relative to chemotherapy and radiotherapy. The expression patterns of KLF9 in patients undergoing chemotherapy and radiotherapy were found to be 0.10 ± 0.60 and 0.08 ± 1.85, respectively. In the case of PKCϵ, these were found to be 0.06 ± 0.1 and 0.04 ± 6.48, respectively. Hence, it was deduced that chemoradiotherapy is linked to better survival of cervical cancer patients.
To further validate our findings, Spearman’s rho correlation was used to test the association of age and the stage of the disease. The association of age and stage of the disease was found in line with the frequency found in the literature in adults (74) and children (75). However, some studies showing evidence of a relationship between age and cancer in adults (76) have reported that cancer does not have to be a consequence of old age.
All the involved genes and miRNAs in our study are known to be implicated in various cancer signaling pathways, such as the PI3K/Akt, nuclear factor-κB, Wnt/β-catenin, and Ras signaling pathways. Hence, these genes and miRNAs may serve as potential diagnostic and prognostic biomarkers. Moreover, these genes can further be investigated as targets for anticancer therapy.
In the present study, we identified the conserved domains and the 3D structure of KLF9 and developed a genetic pathway establishing the crosstalk between KLF9 and its upstream and downstream targets. Moreover, upregulation of the expressions of TPD52 and miR-223 and downregulation of the expressions of KLF9 and PKCϵ were found in peripheral blood of cervical cancer patients. Altered expressions of these genes have been found to be related to tumor progression. Alterations in the expression levels of the understudied genes in cervical cancer may serve as a potential circulating biomarker for cancer diagnosis and prognosis. Hence, understanding the functions, signaling pathways, and genetic networks of TPD52, KLF9, miR-223, and PKCϵ may synergistically reveal the mechanisms of disease progression and serve as a target for inhibitors, therefore assisting in the development of effective anticancer therapy.
The original contributions presented in the study are included in the article/supplementary materials. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved. This experimental protocol for the use of human was approved (ref. no. IRB-110) by the Ethical Committee of Combined Military Hospital and ASAB, NUST. The patients/participants provided written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
MS, SS, KZ, YB, KK, NA and SR designed and conceived the study and analyzed the results. ED, SR, TA, MS, KK and AA conceived an initial part of the study, performed the experiment and histology, and helped in compiling the results. MS, KZ, and SS performed experiments. MS, SR, ED, AA, NA and TA helped in writing the results. SR, TA, DD, and AA wrote the paper with input from all other authors. MS, KZ, SR, YB, DD, SS, TA, KK, NA and AA made a substantial contribution in the interpretation of data and revised the manuscript for intellectual content. All authors read and approved the final manuscript.
We are grateful to the Deanship of Scientific Research at King Saud University for funding this research through Research Group Project no. RGP-193. Furthermore, we are grateful to the Department of Healthcare Biotechnology, Atta-ur-Rahman School of Applied Biosciences, National University of Sciences and Technology, Islamabad, Capital, Pakistan, and Higher Education Commission for their funding through grant no. 10067. The funding body has no role in designing the study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to the Deanship of Scientific Research at King Saud University for its funding of this research through Research Group number RGP-193.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
2. Castellsague X, Bosch FX, Munoz N. Environmental Co-Factors in HPV Carcinogenesis. Virus Res (2002) 89:191–9. doi: 10.1016/S0168-1702(02)00188-0
3. Mittal M, Kulkarni C, Aggarwal P. Comparison Between Manual Liquid Based Cytology and Conventional Pap Smear for Evaluation of Cervical Lesions and It’s Histopathological Correlation in Cases of Epithelial Abnormalities. J Med Sci Clin Res (2018) 6:117–22. doi: 10.18535/jmscr/v6i11.22
4. Haridas S, Subashini P. A Survey of Different Methods for Automated Diagnosis of Cervical Cancer in Pap-Smear Image. J Math Comput Sci (2021) 11:6829–56. doi: 10.28919/jmcs/6264
5. W.H. Organization. WHO Guidelines for Screening and Treatment of Precancerous Lesions for Cervical Cancer Prevention. Geneva:World Health Organization (2013).
6. Zhao Z, Liu H, Hou J, Li T, Du X, Zhao X, et al. Tumor Protein D52 (TPD52) Inhibits Growth and Metastasis in Renal Cell Carcinoma Cells Through the PI3K/Akt Signaling Pathway. Oncol Res Featuring Preclin Clin Cancer Ther (2017) 25:773–9. doi: 10.3727/096504016X14774889687280
7. Irnaten M, Duff A, Clark A, O’Brien C. Intra-Cellular Calcium Signaling Pathways (PKC, RAS/RAF/MAPK, PI3K) in Lamina Cribrosa Cells in Glaucoma. J Clin Med (2021) 10:62. doi: 10.3390/jcm10010062
8. Zhu X, Shen H, Yin X, Yang M, Wei H, Chen Q, et al. Macrophages Derived Exosomes Deliver miR-223 to Epithelial Ovarian Cancer Cells to Elicit a Chemoresistant Phenotype. J Exp Clin Cancer Res (2019) 38:81. doi: 10.1186/s13046-019-1095-1
9. Fan X, Wang X, Zhang J, Ma X, Mao Z. Krüppel-Like Transcription Factor 9 Is Down Regulated in Pancreatic Ductal Adenocarcinoma. SAJ Cancer Sci (2018) 5:103.
10. Ito C, Mukudai Y, Itose M, Kato K, Motohashi H, Shimane T, et al. Tumor Proteins D52 and D54 Have Opposite Effects on the Terminal Differentiation of Chondrocytes. BioMed Res Int (2017) 2017:180–191. doi: 10.1155/2017/6014278
11. Shehata M, Weidenhofer J, Thamotharampillai K, Hardy JR, Byrne JA. Tumor Protein D52 Overexpression and Gene Amplification in Cancers From a Mosaic of Microarrays. Crit Rev™ Oncogene (2008) 14:33–55. doi: 10.1615/CritRevOncog.v14.i1.30
12. Largo C, Alvarez S, Saez B, Blesa D, Martin-Subero JI, González-García I, et al. Identification of Overexpressed Genes in Frequently Gained/Amplified Chromosome Regions in Multiple Myeloma. Haematologica (2006) 91:184–91. doi: 10.1186/1755-8166-7-24
13. Tennstedt P, Bölch C, Strobel G, Minner S, Burkhardt L, Grob T, et al. Patterns of TPD52 Overexpression in Multiple Human Solid Tumor Types Analyzed by Quantitative PCR. Int J Oncol (2014) 44:609–15. doi: 10.3892/ijo.2013.2200
14. Guo H, German P, Bai S, Barnes S, Guo W, Qi X, et al. The PI3K/AKT Pathway and Renal Cell Carcinoma. J Genet Genomics (2015) 42:343–53. doi: 10.1016/j.jgg.2015.03.003
15. Sourbier C, Lindner V, Lang H, Agouni A, Schordan E, Danilin S, et al. The Phosphoinositide 3-Kinase/Akt Pathway: A New Target in Human Renal Cell Carcinoma Therapy. Cancer Res (2006) 66:5130–42. doi: 10.1158/0008-5472.CAN-05-1469
16. Dasari C, Yaghnam DP, Walther R, Ummanni R. Tumor Protein D52 (Isoform 3) Contributes to Prostate Cancer Cell Growth via Targeting Nuclear Factor-κb Transactivation in LNCaP Cells. Tumor Biol (2017) 39:1010428317698382. doi: 10.1177/1010428317698382
17. Simmen FA, Su Y, Xiao R, Zeng Z, Simmen RC. The Krüppel-Like Factor 9 (KLF9) Network in HEC-1-A Endometrial Carcinoma Cells Suggests the Carcinogenic Potential of Dys-Regulated KLF9 Expression. Reprod Biol Endocrinol (2008) 6:41. doi: 10.1186/1477-7827-6-41
18. Ying M, Sang Y, Li Y, Guerrero-Cazares H, Quinones-Hinojosa A, Vescovi AL, et al. Krüppel-Like Family of Transcription Factor 9, a Differentiation-Associated Transcription Factor, Suppresses Notch1 Signaling and Inhibits Glioblastoma-Initiating Stem Cells. Stem Cells (2011) 29:20–31. doi: 10.1002/stem.561
19. Ye S, Zhang D, Cheng F, Wilson D, Mackay J, He K, et al. Wnt/β-Catenin and LIF–Stat3 Signaling Pathways Converge on Sp5 to Promote Mouse Embryonic Stem Cell Self-Renewal. J Cell Sci (2016) 129:269–76. doi: 10.1242/dev.135210
20. Jeong J-W. In Search of Molecular Mechanisms in Endometriosis. Michigan:Oxford University Press (2014).
21. Gorin MA, Pan Q. Protein Kinase Cϵ: An Oncogene and Emerging Tumor Biomarker. Mol Cancer (2009) 8:9. doi: 10.1186/1476-4598-8-9
22. Chauvin L, Goupille C, Blanc C, Pinault M, Domingo I, Guimaraes C, et al. Long Chain N-3 Polyunsaturated Fatty Acids Increase the Efficacy of Docetaxel in Mammary Cancer Cells by Downregulating Akt and Pkcϵ/δ-Induced ERK Pathways. Biochim Biophys Acta (BBA)-Mol Cell Biol Lipids (2016) 1861:380–90. doi: 10.1016/j.bbalip.2016.01.012
23. Wheeler DL, Reddig PJ, Ness KJ, Leith CP, Oberley TD, Verma AK. Overexpression of Protein Kinase C-ϵ in the Mouse Epidermis Leads to a Spontaneous Myeloproliferative-Like Disease. Am J Pathol (2005) 166:117–26. doi: 10.1016/S0002-9440(10)62237-7
24. Sharif TR, Sharif M. Overexpression of Protein Kinase C Epsilon in Astroglial Brain Tumor Derived Cell Lines and Primary Tumor Samples. Int J Oncol (1999) 15:237–80. doi: 10.3892/ijo.15.2.237
25. Xiao H, Goldthwait DA, Mapstone T. The Identification of Four Protein Kinase C Isoforms in Human Glioblastoma Cell Lines: PKC Alpha, Gamma, Epsilon, and Zeta. J Neurosurge (1994) 81:734–40. doi: 10.3171/jns.1994.81.5.0734
26. Xiaoyu L, Wei Z, Ming Z, Guowei J. Anti-Apoptotic Effect of MiR-223-3p Suppressing PIK3C2A in Cardiomyocytes From Myocardial Infarction Rat Through Regulating PI3K/Akt Signaling Pathway. Cardiovasc Toxicol (2021) 21(8):1–14. doi: 10.1007/s12012-021-09658-x
27. Liu Z, Ma T, Duan J, Liu X, Liu L. MicroRNA−223−Induced Inhibition of the FBXW7 Gene Affects the Proliferation and Apoptosis of Colorectal Cancer Cells via the Notch and Akt/mTOR Pathways. Mol Med Rep (2021) 23:1–1. doi: 10.3892/mmr.2020.11662
28. Wang J, Bai X, Song Q, Fan F, Hu Z, Cheng G, et al. miR-223 Inhibits Lipid Deposition and Inflammation by Suppressing Toll-Like Receptor 4 Signaling in Macrophages. Int J Mol Sci (2015) 16:24965–82. doi: 10.3390/ijms161024965
29. Black AR, Black JD, Azizkhan-Clifford J. Sp1 and Krüppel-Like Factor Family of Transcription Factors in Cell Growth Regulation and Cancer. J Cell Physiol (2001) 188:143–60. doi: 10.1002/jcp.1111
30. Dang DT, Pevsner J, Yang VW. The Biology of the Mammalian Krüppel-Like Family of Transcription Factors. Int J Biochem Cell Biol (2000) 32:1103–21. doi: 10.1016/S1357-2725(00)00059-5
31. Schuetz A, Nana D, Rose C, Zocher G, Milanovic M, Koenigsmann J, et al. The Structure of the Klf4 DNA-Binding Domain Links to Self-Renewal and Macrophage Differentiation. Cell Mol Life Sci (2011) 68:3121–31. doi: 10.1007/s00018-010-0618-x
32. Berruyer P, Lelli M, Conley MP, Silverio DL, Widdifield CM, Siddiqi G, et al. Three-Dimensional Structure Determination of Surface Sites. J Am Chem Soc (2017) 139:849–55. doi: 10.1021/jacs.6b10894
33. W.M. Association. World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects (2008). Available at: http://www.wma.net/e/policy/b3.htm.
34. Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable Web Services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res (2013) 41:W349–57. doi: 10.1093/nar/gkt381
35. Yang Y, Heffernan R, Paliwal K, Lyons J, Dehzangi A, Sharma A, et al. Spider2: A Package to Predict Secondary Structure, Accessible Surface Area, and Main-Chain Torsional Angles by Deep Neural Networks. Predict Protein Secondary Structure Springer (2017) pp:55–63. doi: 10.1007/978-1-4939-6406-2_6
36. Rost B, Yachdav G, Liu J. The Predictprotein Server. Nucleic Acids Res (2004) 32:W321–6. doi: 10.1093/nar/gkh377
37. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol Syst Biol (2011) 7:539–48. doi: 10.1038/msb.2011.75
38. Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol (2016) 33:1870–4. doi: 10.1093/molbev/msw054
39. Liberles D, Teufel A. Evolution and Structure of Proteins and Proteomes. Basel:Multidisciplinary Digital Publishing Institute (2018).
40. Kumar S, Stecher G, Li M, Knyaz C, Tamura K, MEGA X. Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol Biol Evol (2018) 35:1547–9. doi: 10.1093/molbev/msy096
41. Shao M, Ge G-Z, Liu W-J, Xiao J, Xia H-J, Fan Y, et al. Characterization and Phylogenetic Analysis of Krüppel-Like Transcription Factor (KLF) Gene Family in Tree Shrews (Tupaia Belangeri Chinensis). Oncotarget (2017) 8:16325. doi: 10.18632/oncotarget.13883
42. Bellesis AG, Jecrois AM, Hayes JA, Schiffer CA, Royer WE. Assembly of Human C-Terminal Binding Protein (CtBP) Into Tetramers. J Biol Chem (2018) 293:9101–12. doi: 10.1074/jbc.RA118.002514
43. Chinnadurai G. Transcriptional Regulation by C-Terminal Binding Proteins. Int J Biochem Cell Biol (2007) 39:1593–607. doi: 10.1016/j.biocel.2007.01.025
44. McConnell BB, Yang VW. Mammalian Krüppel-Like Factors in Health and Diseases. Physiol Rev (2010) 90:1337–81. doi: 10.1152/physrev.00058.2009
45. Silverstein RA, Ekwall K. Sin3: A Flexible Regulator of Global Gene Expression and Genome Stability. Curr Genet (2005) 47:1–17. doi: 10.1007/s00294-004-0541-5
46. Li X, Zhang B, Wu Q, Ci X, Zhao R, Zhang Z, et al. Interruption of KLF5 Acetylation Converts Its Function From Tumor Suppressor to Tumor Promoter in Prostate Cancer Cells. Int J Cancer (2015) 136:536–46. doi: 10.1002/ijc.29028
47. Zhang J-S, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R. A Conserved α-Helical Motif Mediates the Interaction of Sp1-Like Transcriptional Repressors With the Corepressor Msin3a. Mol Cell Biol (2001) 21:5041–9. doi: 10.1128/MCB.21.15.5041-5049.2001
48. Chen X, Bieker JJ. Unanticipated Repression Function Linked to Erythroid Krüppel-Like Factor. Mol Cell Biol (2001) 21:3118–25. doi: 10.1128/MCB.21.9.3118-3125.2001
49. Oktaba K, Gutiérrez L, Gagneur J, Girardot C, Sengupta AK, Furlong EE, et al. Dynamic Regulation by Polycomb Group Protein Complexes Controls Pattern Formation and the Cell Cycle in Drosophila. Dev Cell (2008) 15:877–89. doi: 10.1016/j.devcel.2008.10.005
50. Imam SZ, Rehman F, Zeeshan MM, Maqsood B, Asrar S, Fatima N, et al. Perceptions and Practices of a Pakistani Population Regarding Cervical Cancer Screening. Asian Pac J Cancer Prev (2008) 9:42–4.
51. Bhaumik S. India has World’s Highest Number of Cervical Cancer Deaths. BMJ: Br Med J (2013) 346:346–52. doi: 10.1136/bmj.f3108
52. Hafeez BB, Zhong W, Weichert J, Dreckschmidt NE, Jamal MS, Verma AK. Genetic Ablation of PKC Epsilon Inhibits Prostate Cancer Development and Metastasis in Transgenic Mouse Model of Prostate Adenocarcinoma. Cancer Res (2011) 71:2318–27. doi: 10.1158/0008-5472.CAN-10-4170
53. Aziz MH, Manoharan HT, Verma AK. Protein Kinase Cϵ, Which Sensitizes Skin to Sun’s UV Radiation–Induced Cutaneous Damage and Development of Squamous Cell Carcinomas, Associates With Stat3. Cancer Res (2007) 67:1385–94. doi: 10.1158/0008-5472.CAN-06-3350
54. Urtreger AJ, Kazanietz MG, de Kier Joffé EDB. Contribution of Individual PKC Isoforms to Breast Cancer Progression. IUBMB Life (2012) 64:18–26. doi: 10.1002/iub.574
55. Pan Q, Bao LW, Teknos TN, Merajver SD. Targeted Disruption of Protein Kinase Cϵ Reduces Cell Invasion and Motility Through Inactivation of RhoA and RhoC GTPases in Head and Neck Squamous Cell Carcinoma. Cancer Res (2006) 66:9379–84. doi: 10.1158/0008-5472.CAN-06-2646
56. Zhang J, Baines CP, Zong C, Cardwell EM, Wang G, Vondriska TM, et al. Functional Proteomic Analysis of a Three-Tier Pkcϵ-Akt-eNOS Signaling Module in Cardiac Protection. Am J Physiology-Heart Circulatory Physiol (2005) 288:H954–61. doi: 10.1152/ajpheart.00756.2004
57. Dansen TB, Burgering BM. Unravelling the Tumor-Suppressive Functions of FOXO Proteins. Trends Cell Biol (2008) 18:421–9. doi: 10.1016/j.tcb.2008.07.004
58. Zhang X, Tang N, Hadden TJ, Rishi AK. Akt, FoxO and Regulation of Apoptosis. Biochim Biophys Acta (BBA)-Mol Cell Res (2011) 1813:1978–86. doi: 10.1016/j.bbamcr.2011.03.010
59. Pabona JMP, Simmen FA, Nikiforov MA, Zhuang D, Shankar K, Velarde MC, et al. Krüppel-Like Factor 9 and Progesterone Receptor Coregulation of Decidualizing Endometrial Stromal Cells: Implications for the Pathogenesis of Endometriosis. J Clin Endocrinol Metab (2012) 97:E376–92. doi: 10.1210/jc.2011-2562
60. Wu L, Li H, Jia CY, Cheng W, Yu M, Peng M, et al. MicroRNA-223 Regulates FOXO1 Expression and Cell Proliferation. FEBS Lett (2012) 586:1038–43. doi: 10.1016/j.febslet.2012.02.050
61. Lyu J, Wang J, Miao Y, Xu T, Zhao W, Bao T, et al. KLF7 Is Associated With Poor Prognosis and Regulates Migration and Adhesion in Tongue Cancer. FEBS Lett (2021) 586(7):1038–43. doi: 10.1111/odi.13767
62. Laios A, O’Toole S, Flavin R, Martin C, Kelly L, Ring M, et al. Potential Role of miR-9 and miR-223 in Recurrent Ovarian Cancer. Mol Cancer (2008) 7:1–14. doi: 10.1186/1476-4598-7-35
63. Bhattacharya S, Steele R, Shrivastava S, Chakraborty S, Di Bisceglie AM, RB R. Serum miR-30e and miR-223 as Novel Noninvasive Biomarkers for Hepatocellular Carcinoma. Am J Pathol (2016) 186:242–7. doi: 10.1016/j.ajpath.2015.10.003
64. Li B-s, Zhao Y-l, Guo G, Li W, Zhu E-d, Luo X, et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as Novel Potential Biomarkers for Gastric Cancer Detection. Plos One (2012) 1439(1):678–82. doi: 10.1371/journal.pone.0041629
65. Eiro N, Cid S, Aguado N, Fraile M, de Pablo N, Fernández B, et al. MMP1 and MMP11 Expression in Peripheral Blood Mononuclear Cells Upon Their Interaction With Breast Cancer Cells and Fibroblasts. Int J Mol Cancer (2021) 22:371. doi: 10.3390/ijms22010371
66. Heard ME, Simmons CD, Simmen FA, Simmen RC. Krüppel-Like Factor 9 Deficiency in Uterine Endometrial Cells Promotes Ectopic Lesion Establishment Associated With Activated Notch and Hedgehog Signaling in a Mouse Model of Endometriosis. Endocrinology (2014) 155:1532–46. doi: 10.1210/en.2013-1947
67. Diakiw SM, Perugini M, Kok CH, Engler GA, Cummings N, To LB, et al. Methylation of KLF 5 Contributes to Reduced Expression in Acute Myeloid Leukaemia and Is Associated With Poor Overall Survival. Br J Haematol (2013) 161:884–8. doi: 10.1111/bjh.12295
68. Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in Body Fluids—the Mix of Hormones and Biomarkers. Nat Rev Clin Oncol (2011) 8:467. doi: 10.1038/nrclinonc.2011.76
69. Croce CM, Calin GA. miRNAs, Cancer, and Stem Cell Division. Cell (2005) 122:6–7. doi: 10.1016/j.cell.2005.06.036
70. Ishiguro H, Akimoto K, Nagashima Y, Kojima Y, Sasaki T, Ishiguro-Imagawa Y, et al. Apkcλ/ι Promotes Growth of Prostate Cancer Cells in an Autocrine Manner Through Transcriptional Activation of Interleukin-6. Proc Natl Acad Sci (2009) 106:16369–74. doi: 10.1073/pnas.0907044106
71. Wang Y, Chen C-L, Pan Q-Z, Wu Y-Y, Zhao J-J, Jiang S-S, et al. Decreased TPD52 Expression Is Associated With Poor Prognosis in Primary Hepatocellular Carcinoma. Oncotarget (2016) 7:6323. doi: 10.18632/oncotarget.6319
72. Ding J, Zhao Z, Song J, Luo B, Huang L. MiR-223 Promotes the Doxorubicin Resistance of Colorectal Cancer Cells via Regulating Epithelial–Mesenchymal Transition by Targeting FBXW7. Acta Biochim Biophys Sin (2018) 50:597–604. doi: 10.1093/abbs/gmy040
73. Hu H-J, Deng X-W, Li R-X, Chen D-W, Xue C. Inhibition of Protein Kinase C Activity Inhibits Osteosarcoma Metastasis. Arch Med Sci: AMS (2019) 15:1028. doi: 10.5114/aoms.2018.79450
74. Alibhai SM, Krahn MD, Fleshner NE, Cohen MM, Tomlinson GA, Naglie G. The Association Between Patient Age and Prostate Cancer Stage and Grade at Diagnosis. BJU Int (2004) 94:303–6. doi: 10.1111/j.1464-410X.2004.04883.x
75. Federico SM, Chen X, Easton J, Wu J, Mao S, Liu Y, et al. Association of Age at Diagnosis and Stage of Disease With ATRX Mutations in Neuroblastoma. Am Soc Clin Oncol (2016), 187–200. doi: 10.1038/nrc3526
Keywords: cervical cancer, microRNA 223, PKCϵ, PI3K/Akt signaling pathway, Ramachandran plots, KLF9
Citation: Safi S, Badshah Y, Shabbir M, Zahra K, Khan K, Dilshad E, Afsar T, Almajwal A, Alruwaili NW, Al-disi D, Abulmeaty M and Razak S (2022) Predicting 3D Structure, Cross Talks, and Prognostic Significance of KLF9 in Cervical Cancer. Front. Oncol. 11:797007. doi: 10.3389/fonc.2021.797007
Received: 18 October 2021; Accepted: 30 November 2021;
Published: 03 January 2022.
Edited by:
Alberto Farolfi, Istituto Scientifico Romagnolo per lo Studio e il Trattamento dei Tumori (IRCCS), ItalyReviewed by:
Tang Youyong, Southern Medical University, ChinaCopyright © 2022 Safi, Badshah, Shabbir, Zahra, Khan, Dilshad, Afsar, Almajwal, Alruwaili, Al-disi, Abulmeaty and Razak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suhail Razak, c21hcmF6aUBrc3UuZWR1LnNh; cnVoYWlsMTIzNDVAeWFob28uY29t; Maria Shabbir, bXNoYWJiaXJAYXNhYi5udXN0LmVkdS5waw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.