- 1School of Pharmacy, Yancheng Teachers’ University, Yancheng, China
- 2Department of General Surgery, Xuhui District Central Hospital, Shanghai, China
- 3Physical Examination Centre, Xuhui District Central Hospital, Shanghai, China
Although the association of MEG3 gene rs7158663 polymorphism with cancer susceptibility has been investigated, the findings are inconsistent. The aim of this study was to analyze the association between the rs7158663 polymorphism and cancer susceptibility through a case-control study and meta-analysis. In a case-control study with 430 colorectal cancer (CRC) cases and 445 healthy controls, the rs7158663 polymorphism was genotyped by direct sequencing. STATA software was used to calculate the pooled odds ratio and 95% confidence interval in a meta-analysis including 4,649 cancer cases and 5,590 controls. Both the case-control study and meta-analysis showed that the rs7158663 polymorphism was associated with increased susceptibility to CRC. Individuals carrying the AA or GA genotype were more likely to develop CRC than those carrying the rs7158663 GG genotype. Interestingly, MEG3 expression was significantly lower in colorectal tissues of the AA or GA genotype compared to those of the rs7158663 GG genotype. In addition, the meta-analysis suggested that the rs7158663 polymorphism was also associated with increased susceptibility to breast cancer and gastric cancer. Bioinformatics analysis showed that the rs7158663 A allele contributed to the binding of hsa-miR-4307 and hsa-miR-1265 to MEG3. In conclusion, the current findings suggest that the MEG3 gene rs7158663 polymorphism may serve as a genetic marker for predicting the risk of cancers, such as breast cancer, gastric cancer and CRC. However, the sample size of the current study is still insufficient, especially in the subgroup analysis. Therefore large and well-designed studies are needed to validate our findings.
Introduction
Cancer is one of the most serious public health issues in the world, with approximately 18.1 million new cancer diagnoses and 9.6 million cancer deaths in 2018 (1). Although the precise processes of cancer development and progression are still largely unclear, a growing body of research suggests that genetic predisposition has a substantial influence on the likelihood of individual cancer development (2, 3).
Long non-coding RNA (lncRNA) is a form of RNA transcript that is longer than 200 nucleotides but does not transcribe into protein in cells. LncRNAs play a role in a variety of cell activities, such as cell proliferation, migration, invasion, and angiogenesis, and their dysregulation has been linked to a variety of cancers (4–7). Maternally expressed 3 (MEG3) is one of the most well-studied lncRNAs and is expressed in multiple organs, such as the liver, brain, pancreas, stomach and ovary. However, MEG3 expression is typically suppressed in a variety of cancer tissues (8). Functional studies showed that this lncRNA could regulate the expression of various tumor suppressor genes and oncogenes (9–12). Zhu et al. found that ectopic expression of MEG3 could significantly inhibit proliferation and induce apoptosis in hepatoma cells. MEG3 could function as a tumor suppressor in hepatoma cells through interacting with p53 protein to activate p53-mediated transcriptional activity and influence the expression of partial p53 target genes (9). Dong et al. found that downregulation of MEG3 expression could promote proliferation, migration, and invasion of hepatocellular carcinoma cells by upregulating TGF-β1 expression (10). Zuo et al. demonstrated that MEG3 activated by vitamin D could inhibit glycolysis in colorectal cancer (CRC) via promoting c-Myc degradation (11). Xu et al. found that MEG3 could mediate the miR-149-3p/FOXP3 axis by reducing p53 ubiquitination to exert a suppressive effect on regulatory T cell differentiation and immune escape in esophageal cancer (12). In addition, certain polymorphisms (rs3087918 T>G, rs11160608 A>C, rs4081134 G>A, rs7158663 A>G) within the MEG3 gene are implicated in cancer susceptibility (13–16). For example, MEG3 gene rs3087918 was associated with a decreased risk of breast cancer in a Chinese population (14). MEG3 gene rs11160608 was related to an increased risk of oral squamous cell carcinoma in a Chinese Han population (15). MEG3 gene rs4081134 was significantly associated with a decreased risk of lung cancer in a Northeast Chinese population (16). MEG3 gene rs7158663 is the most interesting polymorphic locus located on the MEG3 transcript. Bioinformatic analysis showed that the rs7158663 polymorphism had the potential to change the local RNA folding structure and affect miRNA-lncRNA interactions, which in turn affected the expression level of miRNA and/or MEG3 (17, 18). Several studies have explored the relationship between this potentially functional polymorphism and cancer susceptibility, but the results are inconsistent and need to be further clarified.
In the current study, we first explored the relationship between the rs7158663 polymorphism and CRC susceptibility using a case-control study, and then analyzed its effect on MEG3 expression in colorectal tissues. In addition, a meta-analysis was conducted to systematically evaluated the relationship between this polymorphism and cancer susceptibility, which would help us to better understand the role of the rs7158663 polymorphism in cancer susceptibility.
Materials And Methods
Sample Collection
Peripheral blood of 430 CRC patients and 445 healthy controls were collected from Shanghai Xuhui District Central Hospital. All participants were genetically unrelated Han Chinese. Diagnosis of CRC patients was histopathologically confirmed. Healthy controls were cancer-free individuals living in the same residential area and seeking routine physical exams. Furthermore, colorectal tissues were obtained from 40 CRC surgery patients who had not received radiochemotherapy before surgery. Written informed consent was obtained from all participants. The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Human Ethics Committee of Shanghai Xuhui District Central Hospital.
Genotyping
TIANamp genomic DNA Kit (Tiangen) was used to isolate genomic DNA from peripheral blood according to the manufacturer’s instructions. Genomic DNA concentration was detected using a NanoDrop spectrophotometer. Direct sequencing was used to detect the genotype of the rs7158663 locus in each individual.
Real-Time Quantitative PCR
Total RNA was isolated from colorectal tissues using the RNAsimple total RNA kit (Tiangen) according to the manufacturer’s instructions. ReverTra Ace qPCR RT Kit (TOYOBO) was used to synthesize cDNA. FastStart Universal SYBR Green Master (Roche) was used to conduct real-time quantitative PCR. MEG3 expression was normalized to the internal control GAPDH. The specific primer sequences are presented in Table S1.
Bioinformatic Analysis
The lncRNASNP online tool (http://bioinfo.life.hust.edu.cn/lncRNASNP) was used to analyze whether the rs7158663 polymorphism affects miRNA binding (19).
Statistical Analysis
Hardy-Weinberg equilibrium (HWE) for the control group was tested by a goodness-of-fit χ2 test. The association of MEG3 gene rs7158663 polymorphism with CRC susceptibility was evaluated using adjusted odds ratios (ORs) with their 95% confidence intervals (CIs). Student’s t-test was used to check the differences for age variable between CRC cases and controls. χ2 test was used to assess the differences in gender variable between CRC cases and controls. The normalized expression levels of MEG3 among different genotypes were compared using one-way ANOVA. All statistical analyses were performed by SAS 9.4 (SAS Institute, Cary, USA). P < 0.05 was defined as the level of significance.
Meta-Analysis
PubMed, CNKI and EMBASE databases were searched based on the following keywords: “Maternally expressed 3 or MEG3”, “polymorphism or variant” and “cancer or carcinoma or malignancy”. The last literature search was conducted on October 7, 2021. The primary inclusion criterion for previous studies was to have sufficient genotype data. If numerous studies had overlapping or duplicate data, only studies with complete data were included. Data from the included studies were extracted independently by two investigators. Disagreements were settled by conversation. The pooled ORs and their 95% CIs were applied to determine the relationship of the rs7158663 polymorphism with cancer susceptibility. The between-study heterogeneity was assessed using Chi-square-based statistic I2 test and Cochran’s Q-test. When I2 > 50% or PH < 0.1, we used the random-effects model to estimate the pooled OR. Otherwise, the fixed-effects model was applied. To assess the quality and consistency of the results, sensitivity analysis was undertaken by removing each study in turn. Begg’s and Egger’s tests were used to assess potential publication bias. Trial sequential analysis (TSA) was conducted in a selected genetic model to assess the statistical reliability of the meta-analysis. TSA was conducted with a 5% risk of type I error and a 20% risk of type II error (20). The statistical analyses were performed by STATA 12.0 (Stata Corporation, College Station, TX, USA).
Results
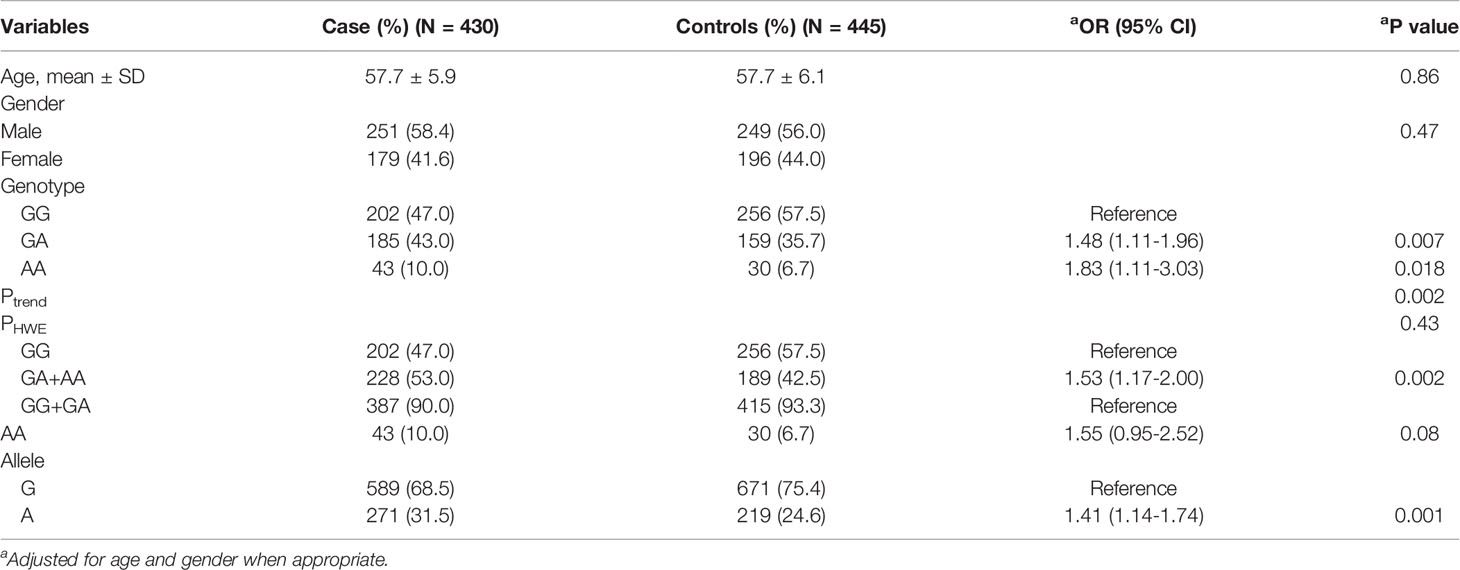
The results of the case-control study are shown in Table 1. There was no statistical difference in the age and gender distribution between the case and control groups (P>0.05). The genotype frequency distribution of the control group was consistent with HWE (PHWE=0.43). There was a significant association between MEG3 gene rs7158663 polymorphism and CRC susceptibility [GA vs. GG: OR=1.48, 95%CI= 1.11-1.96, P=0.007; AA vs. GG: OR=1.83, 95%CI=1.11-3.03, P=0.018; (GA+AA) vs. GG: OR=1.53, 95%CI=1.17-2.00, P=0.002; A vs. G: OR=1.41, 95%CI=1.14-1.74, P=0.001].
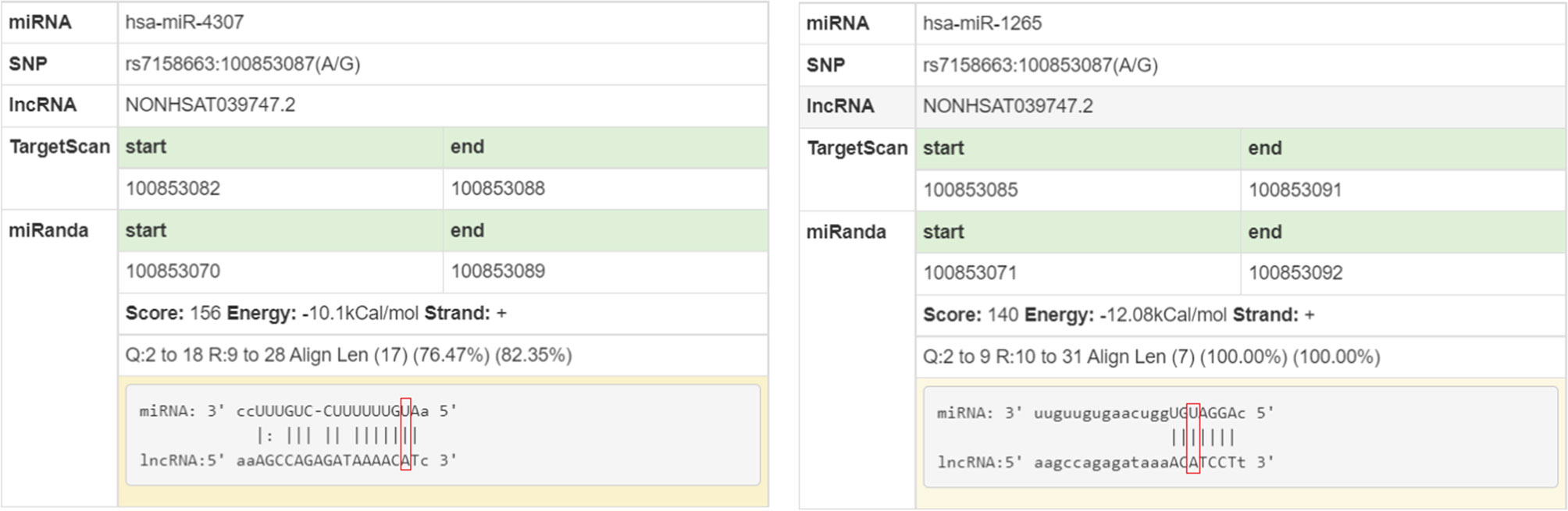
Genotype-tissue expression results showed that the rs7158663 polymorphism was significantly associated with the expression of MEG3 in colorectal tissues. MEG3 expression was significantly lower in colorectal tissues of the AA or GA genotype compared to those of the rs7158663 GG genotype (Figure 1). The results of bioinformatics analysis showed that the rs7158663 A allele contributed to the binding of hsa-miR-4307 and hsa-miR-1265 to MEG3 (Figure 2).
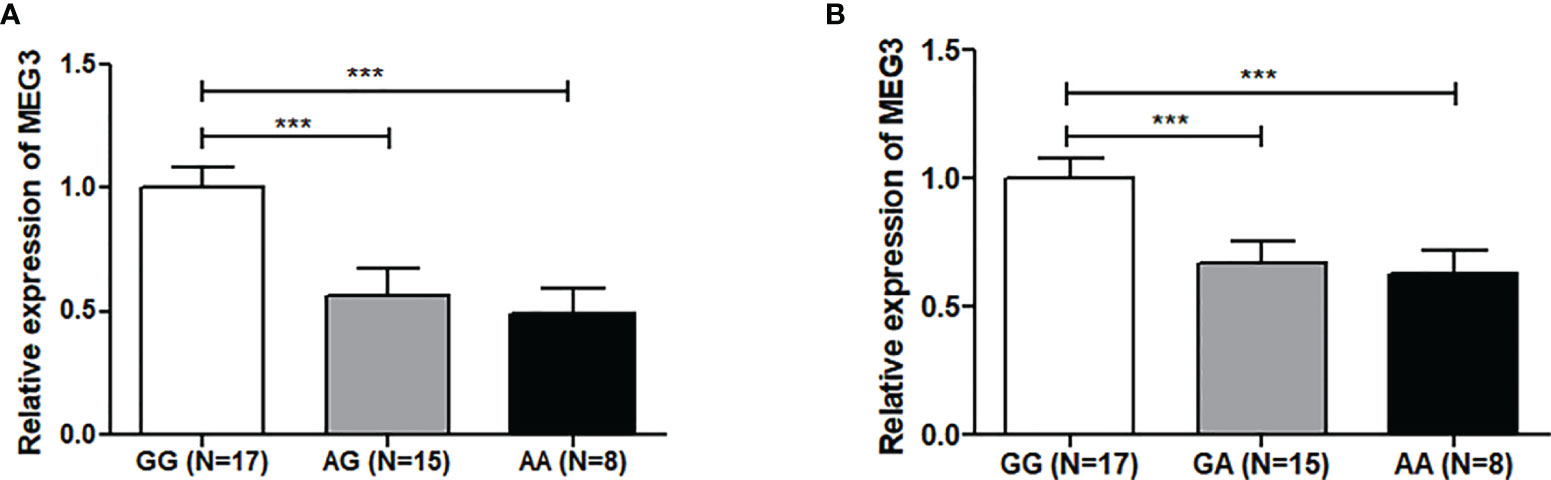
Figure 1 Relationship between rs7158663 genotype and MEG3 expression in CRC tissues (A) and normal paracancerous tissues (B). ***p < 0.001.
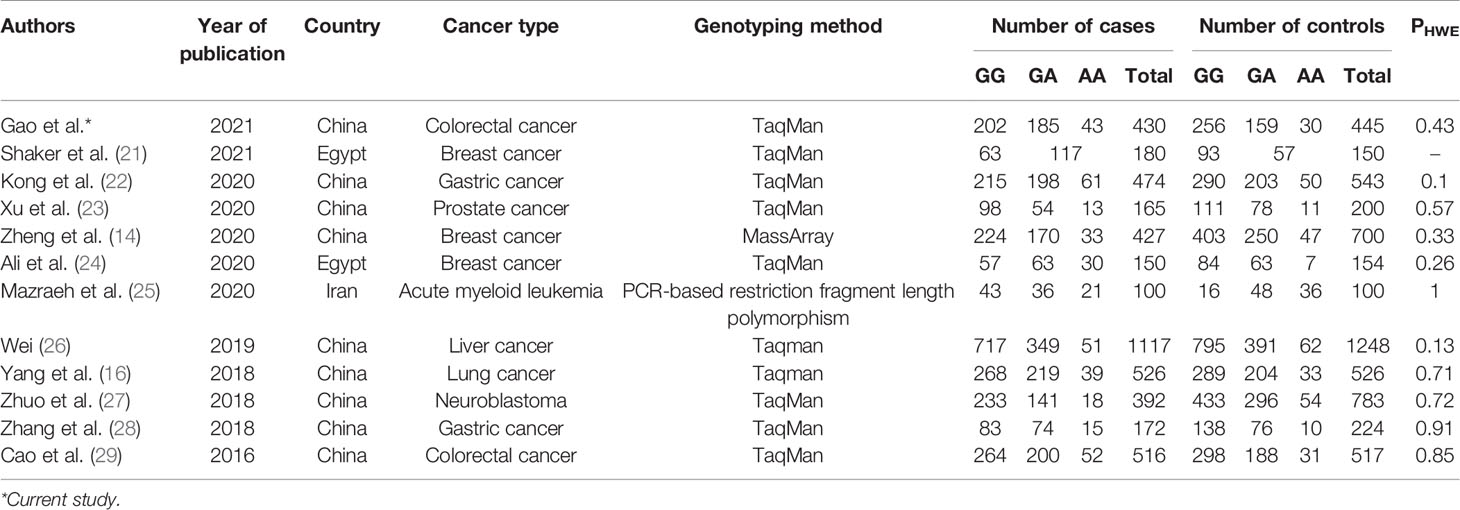
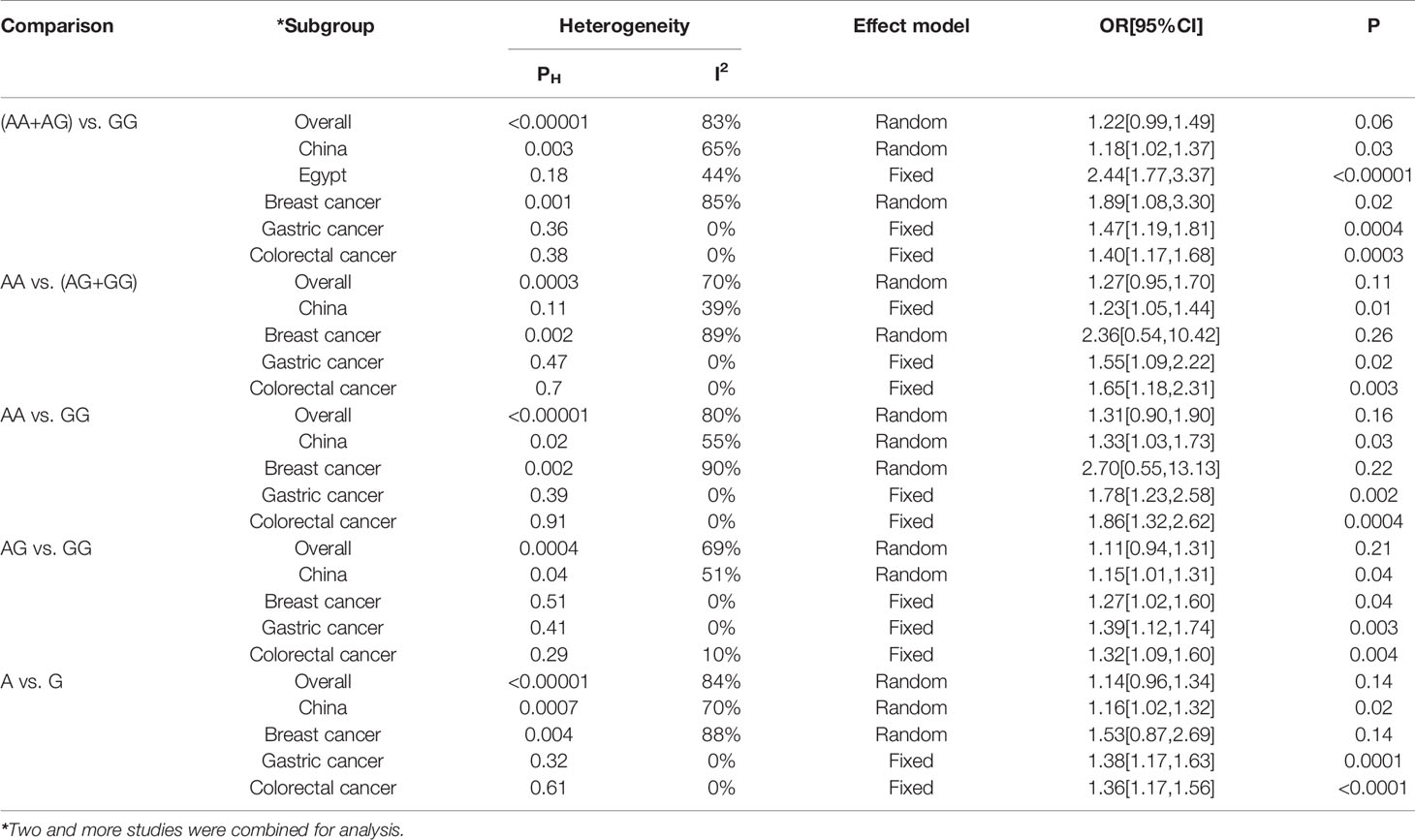
Based on database searches, a total of 11 case-control studies exploring the association of the rs7158663 polymorphism with cancer susceptibility were included in the current meta-analysis (Figure S1). The relevant studies were published between 2016 and 2021. The current meta-analysis combined our results included 4,649 cancer cases and 5,590 controls (Table 2). The overall combined analysis showed that the rs7158663 polymorphism was not associated with cancer susceptibility (Table 3). However, the country-based stratified analysis showed that the rs7158663 polymorphism was associated with cancer susceptibility in the Chinese population under (AA+AG) vs. GG, AA vs. (AG+GG), AA vs. GG, AG vs. GG, and A vs. G models, and in the Egyptians under (AA+AG) vs. GG model. The stratified analysis based on cancer type showed that the rs7158663 polymorphism was associated with susceptibility to breast cancer under (AA+AG) vs. GG and AG vs. GG models, and gastric cancer under (AA+AG) vs. GG, AA vs. (AG+GG), AA vs. GG, AG vs. GG, and A vs. G models, and colorectal cancer under (AA+AG) vs. GG, AA vs. (AG+GG), AA vs. GG, AG vs. GG, and A vs. G models.
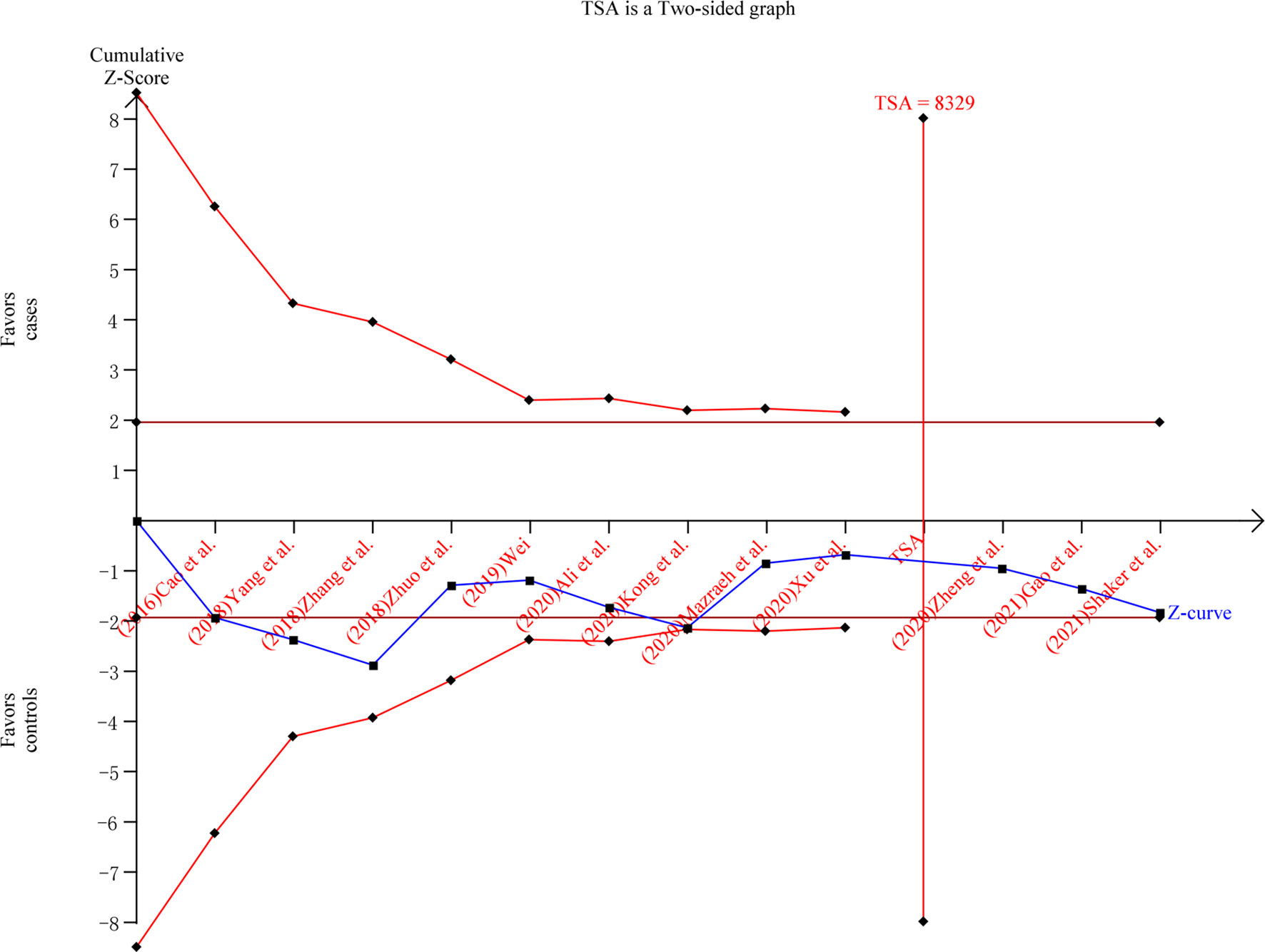
The sensitivity analysis showed that after Mazraeh’s study was removed, the overall combined results were significantly altered under all comparison models (Figure S2). After Zhuo’s study was removed, the overall combined results were significantly altered under (AA+AG) vs. GG, and AA vs. (AG+GG) models. After Xu’s study was removed, the overall combined results were significantly altered under (AA+AG) vs. GG model. Begg’s and Egger’s tests showed no publication bias in the current meta-analysis (Table 4). TSA was conducted in the (AA+AG) vs. GG model. The result showed that the cumulative Z-curve (blue line) has crossed the required information sizes (n=8,329) (Figure 3), which indicated that the cumulative evidence was adequate in the overall analysis.
Discussion
Recent studies have shown that certain genetic variants on lncRNA genes may be associated with cancer risk (30–32). These genetic variants contain the rs7158663 polymorphism on the MEG3 gene. For instance, Cao et al. found that rs7158663 AA genotype had significantly higher CRC risk than GG genotype, which was consistent with our results. The further stratified analysis revealed that the elevated risk was strongly associated with people with age ≤ 60 and a family history of cancer. However, there was no link found between the rs7158663 polymorphism and CRC site or stage (29). Both Zhang et al. and Kong et al. found that individuals carrying the rs7158663 AG+AA genotype or A allele had a significantly increased risk of gastric cancer (22, 28). However, some studies suggested that the rs7158663 polymorphism was not associated with cancer risk. For instance, Wei found that the rs7158663 polymorphism was not associated with hepatocarcinogenesis (26). Yang et al. found that the rs7158663 polymorphism was not associated with susceptibility to lung cancer (16). Zhuo et al. found that the rs7158663 polymorphism was not linked with neuroblastoma susceptibility, regardless of whether it was corrected for age and gender (27). These inconsistent results forced us to clarify the relationship between the rs7158663 polymorphism and cancer susceptibility by meta-analysis. By combining two and more studies for analysis, we found that the rs7158663 polymorphism was not associated with overall cancer susceptibility. However, the country-based stratified analysis showed that the rs7158663 polymorphism was associated with cancer susceptibility in the Chinese population under (AA+AG) vs. GG, AA vs. (AG+GG), AA vs. GG, AG vs. GG, and A vs. G models, and in the Egyptians under (AA+AG) vs. GG model. The stratified analysis based on cancer type showed that the rs7158663 polymorphism was associated with susceptibility to breast cancer under (AA+AG) vs. GG and AG vs. GG models, and gastric cancer under (AA+AG) vs. GG, AA vs. (AG+GG), AA vs. GG, AG vs. GG, and A vs. G models, and colorectal cancer under (AA+AG) vs. GG, AA vs. (AG+GG), AA vs. GG, AG vs. GG, and A vs. G models. There was no publication bias in the current meta-analysis, and TSA suggested that the sample size in the overall combined analysis was adequate. However, the sensitivity analysis results suggested that the current meta-analysis results were not sufficiently stable. Therefore we needed more studies to confirm the current findings.
MEG3 could inhibit the malignant phenotype of many cancers including gastric, breast and colorectal cancers (33–35). The current study found that the rs7158663 polymorphism could affect MEG3 expression in colorectal tissues. MEG3 expression was significantly lower in colorectal tissues of the AA or GA genotype compared to those of the rs7158663 GG genotype. Bioinformatics analysis showed that the rs7158663 A allele contributed to the binding of hsa-miR-4307 and hsa-miR-1265 to MEG3. Therefore, we speculated that the rs7158663 polymorphism may affect an individual’s susceptibility to CRC by influencing the regulation of MEG3 expression by miRNAs.
Although the current study has yielded some meaningful results, some shortcomings needed to be pointed out. Due to the insufficient sample size of the current case-control study and the unavailability of some clinical data, we did not further analyze the relationship between the rs7158663 polymorphism and the clinicopathological features. In addition, we did not consider the effect of the rs7158663 polymorphism interaction with environmental factors on cancer susceptibility.
Conclusions
The current study results suggest that the MEG3 gene rs7158663 polymorphism is associated with susceptibility to a variety of cancers, such as breast cancer, gastric cancer and CRC. However, large and well-designed studies are still needed to validate our findings.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by Shanghai Xuhui District Central Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
XG carried out the molecular genetic studies, did the literature search and the statistical analysis, and wrote the paper. XL performed biochemistry tests. XW were responsible for the acquisition of data. SZ participated in study design and coordination and helped to draft the manuscript. XG and SZ interpreted the data and were responsible for the manuscript preparation. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We gratefully acknowledge the cooperation of the patients and normal control individuals involved in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.796774/full#supplementary-material
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Gao X, Wang X. HMGA2 Rs968697 T > C Polymorphism Is Associated With the Risk of Colorectal Cancer. Nucleosides Nucleotides Nucleic Acids (2021) 40(8):821–8. doi: 10.1080/15257770.2021.1952596
3. Ni W, Wang X, Sun Y, Gao X. Meta-Analysis of the Association Between MALAT1 Rs619586 a>G Polymorphism and Cancer Risk. J Int Med Res (2020) 48:300060520941969. doi: 10.1177/0300060520941969
4. Qi ZY, Wang LL, Qu XL. lncRNA LINC00355 Acts as a Novel Biomarker and Promotes Glioma Biological Activities via the Regulation of miR-1225/FNDC3B. Dis Markers (2021) 2021:1683129. doi: 10.1155/2021/1683129
5. Feng Y, Wei G, Zhang L, Zhou H, Wang W, Guo P, et al. LncRNA DARS-AS1 Aggravates the Growth and Metastasis of Hepatocellular Carcinoma via Regulating the miR-3200-5p-Cytoskeleton Associated Protein 2 (CKAP2) Axis. Bioengineered (2021) 12(1):8217–32. doi: 10.1080/21655979.2021.1982272
6. Fu C, Xin J, Zhang W, Lai J, Huang Z. LINC00992 Exerts Oncogenic Activities in Prostate Cancer via Regulation of SOX4. Exp Cell Res (2021) 408(1):112855. doi: 10.1016/j.yexcr.2021.112855
7. Xu L, Liu C, Ye Z, Wu C, Ding Y, Huang J. Overexpressed LINC00467 Promotes the Viability and Proliferation Yet Inhibits Apoptosis of Gastric Cancer Cells via Raising ITGB3 Level. Tissue Cell (2021) 73:101644. doi: 10.1016/j.tice.2021.101644
8. Ghafouri-Fard S, Taheri M. Maternally Expressed Gene 3 (MEG3): A Tumor Suppressor Long Non Coding RNA. BioMed Pharmacother (2019) 118:109129. doi: 10.1016/j.biopha.2019.109129
9. Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J, et al. Long Noncoding RNA MEG3 Interacts With P53 Protein and Regulates Partial P53 Target Genes in Hepatoma Cells. PloS One (2015) 10(10):e0139790. doi: 10.1371/journal.pone.0139790
10. Dong H, Zhang Y, Xu Y, Ma R, Liu L, Luo C, et al. Downregulation of Long Non-Coding RNA MEG3 Promotes Proliferation, Migration, and Invasion of Human Hepatocellular Carcinoma Cells by Upregulating TGF-β1. Acta Biochim Biophys Sin (Shanghai) (2019) 51(6):645–52. doi: 10.1093/abbs/gmz046
11. Zuo S, Wu L, Wang Y, Yuan X. Long Non-Coding RNA MEG3 Activated by Vitamin D Suppresses Glycolysis in Colorectal Cancer via Promoting C-Myc Degradation. Front Oncol (2020) 10:274. doi: 10.3389/fonc.2020.00274
12. Xu QR, Tang J, Liao HY, Yu BT, He XY, Zheng YZ, et al. Long Non-Coding RNA MEG3 Mediates the miR-149-3p/FOXP3 Axis by Reducing P53 Ubiquitination to Exert a Suppressive Effect on Regulatory T Cell Differentiation and Immune Escape in Esophageal Cancer. J Transl Med (2021) 19(1):264. doi: 10.1186/s12967-021-02907-1
13. Dong X, Gao W, Lv X, Wang Y, Wu Q, Yang Z, et al. Association Between lncRNA GAS5, MEG3, and PCAT-1 Polymorphisms and Cancer Risk: A Meta-Analysis. Dis Markers (2020) 2020:6723487. doi: 10.1155/2020/6723487
14. Zheng Y, Wang M, Wang S, Xu P, Deng Y, Lin S, et al. LncRNA MEG3 Rs3087918 was Associated With a Decreased Breast Cancer Risk in a Chinese Population: A Case-Control Study. BMC Cancer (2020) 20(1):659. doi: 10.1186/s12885-020-07145-0
15. Hou Y, Zhang B, Miao L, Ji Y, Yu Y, Zhu L, et al. Association of Long Non-Coding RNA MEG3 Polymorphisms With Oral Squamous Cell Carcinoma Risk. Oral Dis (2019) 25(5):1318–24. doi: 10.1111/odi.13103
16. Yang Z, Li H, Li J, Lv X, Gao M, Bi Y, et al. Association Between Long Noncoding RNA MEG3 Polymorphisms and Lung Cancer Susceptibility in Chinese Northeast Population. DNA Cell Biol (2018) 37(10):812–20. doi: 10.1089/dna.2018.4277
17. Cao X, Zhuang S, Hu Y, Xi L, Deng L, Sheng H, et al. Associations Between Polymorphisms of Long Non-Coding RNA MEG3 and Risk of Colorectal Cancer in Chinese. Oncotarget (2016) 7(14):19054–9. doi: 10.18632/oncotarget.7764
18. Ghaedi H, Zare A, Omrani MD, Doustimotlagh AH, Meshkani R, Alipoor S, et al. Genetic Variants in Long Noncoding RNA H19 and MEG3 Confer Risk of Type 2 Diabetes in an Iranian Population. Gene (2018) 675:265–71. doi: 10.1016/j.gene.2018.07.002
19. Gong J, Liu W, Zhang J, Miao X, Guo AY. lncRNASNP: A Database of SNPs in lncRNAs and their Potential Functions in Human and Mouse. Nucleic Acids Res (2015) 43:D181–6. doi: 10.1093/nar/gku1000
20. Wetterslev J, Thorlund K, Brok J, Gluud C. Trial Sequential Analysis may Establish When Firm Evidence Is Reached in Cumulative Meta-Analysis. J Clin Epidemiol (2008) 61(1):64–75. doi: 10.1016/j.jclinepi.2007.03.013
21. Shaker O, Ayeldeen G, Abdelhamid A. The Impact of Single Nucleotide Polymorphism in the Long Non-Coding MEG3 Gene on microRNA-182 and microRNA-29 Expression Levels in the Development of Breast Cancer in Egyptian Women. Front Genet (2021) 12:683809. doi: 10.3389/fgene.2021.683809
22. Kong X, Yang S, Liu C, Tang H, Chen Y, Zhang X, et al. Relationship Between MEG3 Gene Polymorphism and Risk of Gastric Cancer in Chinese Population With High Incidence of Gastric Cancer. Biosci Rep (2020) 40(11):BSR20200305. doi: 10.1042/BSR20200305
23. Xu B, Zhang M, Liu C, Wang C, You Z, Wang Y, et al. Association of Long Non-Coding RNA MEG3 Polymorphisms and Risk of Prostate Cancer in Chinese Han Population. Urol J (2020) 18(2):176–80. doi: 10.22037/uj.v16i7.5585
24. Ali MA, Shaker OG, Alazrak M, AbdelHafez MN, Khalefa AA, Hemeda NF, et al. Association Analyses of a Genetic Variant in Long Non-Coding RNA MEG3 With Breast Cancer Susceptibility and Serum MEG3 Expression Level in the Egyptian Population. Cancer Biomark (2020) 28(1):49–63. doi: 10.3233/CBM-191072
25. Mazraeh SA, Gharesouran J, Fard SG, Ketab FNG, Hosseinzadeh H, Moradi M, et al. Association Between WT1 and MEG3 Polymorphisms and Risk of Acute Myeloid Leukemia. Meta Gene (2020) 23:100636. doi: 10.1016/j.mgene.2019.100636
26. Wei ZM. Analysis of the Effect of Genetic Variants of MEG3-P53-MDM2-PTEN Molecular Pathway on Hepatocarcinogenesis. Master's degree dissertation. (2019). doi: 10.27690/d.cnki.ggdyk.2019.000060
27. Zhuo ZJ, Zhang R, Zhang J, Zhu J, Yang T, Zou Y, et al. Associations Between lncRNA MEG3 Polymorphisms and Neuroblastoma Risk in Chinese Children. Aging (Albany NY) (2018) 10(3):481–91. doi: 10.18632/aging.101406
28. Zhang Q, Ai L, Dai YG. Association Between Polymorphism of Long Non-Coding RNA Maternally Expressed Gene 3 and Risk of Gastric Cancer. Chin J Bases Clinics Gen Surg (2018) 25:1323–6. doi: 10.7507/1007-9424.201805074
29. Cao X, Zhuang S, Hu Y, Xi L, Deng L, Sheng H, et al. Associations Between Polymorphisms of Long Non-Coding RNA MEG3 and Risk of Colorectal Cancer in Chinese. Oncotarget (2016) 7(14):19054–9. doi: 10.18632/oncotarget.7764
30. Du P, Li G, Zhu J, Luo Y, Fu N, Li Y. Association of lncRNA PRNCR1 Polymorphisms With Cancer Susceptibility: A Meta-Analysis of the Current Literature. J Genet (2021) 100:19. doi: 10.1007/s12041-021-01269-3
31. Li HN, Deng N, Zhao X, Liu J, He T, Ding XW. Contributions of HOTAIR Polymorphisms to the Susceptibility of Cancer. Int J Clin Oncol (2021) 26(6):1022–38. doi: 10.1007/s10147-021-01884-1
32. Li N, Cui Z, Huang D, Gao M, Li S, Song M, et al. Association of LINC00673 Rs11655237 Polymorphism With Cancer Susceptibility: A Meta-Analysis Based on 23,478 Subjects. Genomics (2020) 112(6):4148–54. doi: 10.1016/j.ygeno.2020.07.015
33. Wu X, Li J, Ren Y, Zuo Z, Ni S, Cai J. MEG3 can Affect the Proliferation and Migration of Colorectal Cancer Cells Through Regulating miR-376/PRKD1 Axis. Am J Transl Res (2019) 11:5740–51.
34. Jiao J, Zhang S. Long Non−Coding RNA MEG−3 Suppresses Gastric Carcinoma Cell Growth, Invasion and Migration via EMT Regulation. Mol Med Rep (2019) 20(3):2685 –93. doi: 10.3892/mmr.2019.10515
Keywords: MEG3, polymorphism, rs7158663, cancer, susceptibility
Citation: Gao X, Li X, Zhang S and Wang X (2021) The Association of MEG3 Gene rs7158663 Polymorphism With Cancer Susceptibility. Front. Oncol. 11:796774. doi: 10.3389/fonc.2021.796774
Received: 17 October 2021; Accepted: 23 November 2021;
Published: 09 December 2021.
Edited by:
Walter Hernán Pavicic, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Macrina Beatriz Silva Cázares, Autonomous University of San Luis Potosí, MexicoQing Chun Zhao, Shenyang Pharmaceutical University, China
Copyright © 2021 Gao, Li, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueren Gao, Z2FveHJAeWN0dS5lZHUuY24=
 Xueren Gao
Xueren Gao Xianyang Li
Xianyang Li Shulong Zhang 2and
Shulong Zhang 2and  Xiaoting Wang3
Xiaoting Wang3