- 1Comprehensive Cancer Center, Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Roma, Italy
- 2Section of Oncology, Department of Translational Medicine, Università Cattolica Del Sacro Cuore, Roma, Italy
- 3Section of Oncology, Department of Medicine, University of Verona School of Medicine and Verona University Hospital Trust, Verona, Italy
Non-small cell lung cancer (NSCLC) represents the perfect paradigm of ‘precision medicine’ due to its complex intratumoral heterogeneity. It is truly characterized by a range of molecular alterations that can deeply influence the natural history of this disease. Several molecular alterations have been found over time, paving the road to biomarker-driven therapy and radically changing the prognosis of ‘oncogene addicted’ NSCLC patients. Kirsten rat sarcoma (KRAS) mutations are present in up to 30% of NSCLC (especially in adenocarcinoma histotype) and have been identified decades ago. Since its discovery, its molecular characteristics and its marked affinity to a specific substrate have led to define KRAS as an undruggable alteration. Despite that, many attempts have been made to develop drugs capable of targeting KRAS signaling but, until a few years ago, these efforts have been unsuccessful. Comprehensive genomic profiling and wide-spectrum analysis of genetic alterations have only recently allowed to identify different types of KRAS mutations. This tricky step has finally opened new frontiers in the treatment approach of KRAS-mutant patients and might hopefully increase their prognosis and quality of life. In this review, we aim to highlight the most interesting aspects of (epi)genetic KRAS features, hoping to light the way to the state of art of targeting KRAS in NSCLC.
Introduction: KRAS Mutations
Non-small cell lung cancer (NSCLC) represents one of the most multifaceted tumor based on its complex molecular features. The identification of driver mutations, predominantly identified in the adenocarcinoma histotype, have opened the era of ‘biomarker-driven therapy’ (1). The ‘oncogene addicted’ disease, characterized by a targetable gene alteration, represents a subgroup of NSCLC in which targeted agents have radically changed prognosis and quality of life of these patients (2). With the advent of tumor genotyping, upfront routine testing for targetable biomarker alterations is strongly recommended in all patients with diagnosis of NSCLC to personalize treatment (3). Additional molecular analysis by next generation sequencing (NGS) may be considered as part of broader testing panels, with the final aim of detecting rare druggable alterations to enroll patients in clinical trials. Among gene alterations involved in cancer development, kirsten rat sarcoma (KRAS) represents the most common oncogene driver in human cancer. It has been demonstrated that the wild-type KRAS is a proto-oncogene that is frequently activated during many types of cancer progression (4). Once the KRAS gene mutates, it acquires oncogenic properties that are causally involved in the development of human cancers (5, 6). Such oncogenic forms of the KRAS gene are prevalent in pancreatic carcinomas (>80%), colon carcinomas (40–50%), and lung carcinomas (30–50%), but are also present in biliary tract malignancies, endometrial cancer, cervical cancer, bladder cancer, liver cancer, myeloid leukemia and breast cancer (7). KRAS protein is a small guanine triphosphatase (GTPase) that serves as a binary switch in signal transduction for most receptor tyrosine kinases and plays a key role in regulating cell growth, differentiation and apoptosis by interacting with multiple effectors including mitogen-activated protein kinase (MAPK) (8). Oncogenic KRAS mutations mostly occur in exon 2 at codon 12, less frequently at codon 13 (3-5%) and rarely at exon 3 codon 61 (less than 1%) (9). KRAS signaling begins with the stimulation of a vast array of upstream receptors including receptor tyrosine kinases (RTKs). Adaptor proteins interact with the intracellular domain of activated EGFR and in turn recruit guanine nucleotide exchange factors (GEFs), such as Son of Sevenless (SOS), to the cellular membrane where they can associate with KRAS to promote the switch from inactive GDP to active GTP. KRAS signaling terminates upon the hydrolysis of GTP to GDP by the intrinsic GTPase activity of RAS. Cancer-causing mutations in KRAS drastically impair the GTPase activity, resulting in KRAS proteins that are locked in the active GTP-bound conformation, regardless of the upstream signal. As consequence, these missense mutations cause a constitutive activation of KRAS downstream pathways and promote cell proliferation and tumor development (10). In this review we aim to give an overview on KRAS role in lung cancer pathogenesis and targeting with a special focus on new potential treatments based on epigenetic modulation.
KRAS in Lung Cancer: Clinical Features and Therapeutical Implications Before the Advent of Direct Inhibitors
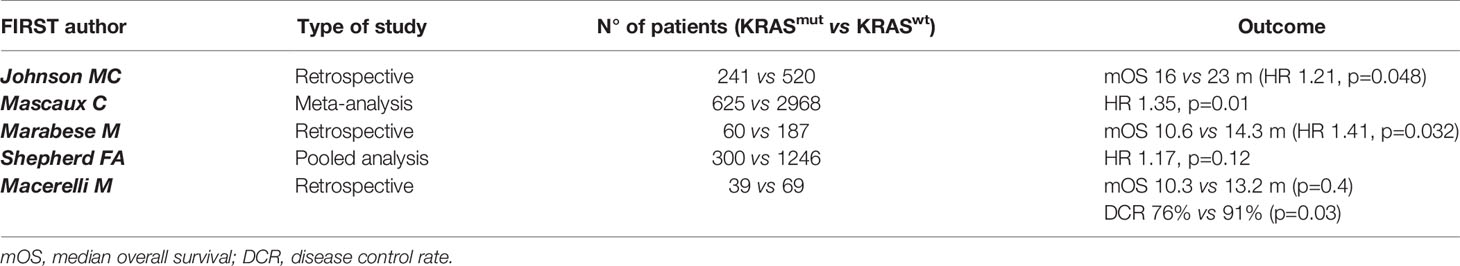
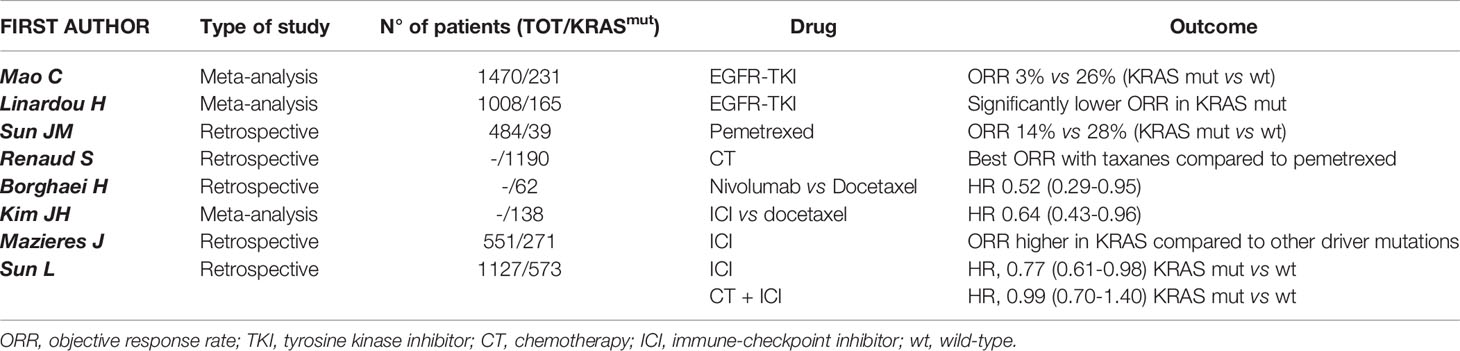
In NSCLC, KRAS mutations occur in 30% of adenocarcinoma and less frequently (about 7%) in squamous-cell lung carcinoma (11). Nevertheless, more recent results suggest that, in most cases, KRAS mutations do not occur in pure squamous-cell lung carcinoma but in mixed histology of adenosquamous or neuroendocrine carcinomas (12, 13). Although most of KRAS-mutant NSCLC are diagnosed in former or active smokers, KRAS mutations can also be detected in never smoker patients with early onset of cancer, thus its mutational state cannot be predicted on the basis of smoking history alone (14). Because of its high frequency in lung adenocarcinoma, several preclinical and clinical investigations have been conducted seeking effective therapeutic approaches targeting KRAS mutations but, for many years, these efforts have been unsuccessful. The deeper knowledge of genomic alterations has led to the identification of different KRAS subtype mutations. Indeed, it has been demonstrated that in NSCLC the most frequent KRAS allelic variations are the p.G12C (39%) and p.G12V (17-18%), resulting from a G:C to T:A transversion as a classical smoking-induced alteration (15). Remarkably, smokers and never smokers have a different spectrum of mutations and codon variants in KRAS. Thus, transition mutations (G12D) are more common in never smokers, whereas transversion mutations (G12C and G12V) are more common in former or current smokers (16). Moreover, KRAS-mutant NSCLC in smoker patients are genomically more complex, with a higher mutational burden and higher frequency of additional mutations in TP53 or STK11 genes compared to never smoker tumors, as result of antigenic exposure and oxidative stress in epithelial cells (17). From a clinical point of view, KRAS-mutant cancers have generally been associated with poorer overall survival (OS) compared to KRAS wild type tumors, especially in the advanced stages (18–20); however, other studies in early (where the benefit of adjuvant chemotherapy is minimal) or advanced stage of KRAS-mutant lung cancer have provided conflicting results, thus the prognostic value of KRAS alteration is still debated (21–23). A systematic review and meta-analysis including 3,620 patients has shown that KRAS mutations confers a significantly worse prognosis in patients with lung adenocarcinoma (19). Furthermore, KRAS mutations also have a predictive value in lung cancer. Indeed, as already outlined, KRAS represents a critical downstream effector on the EGFR pathway. Thus, there is a biologic rationale supporting the hypothesis that KRAS-mutant NSCLC are intrinsically resistant to EGFR TKIs because of a constitutive activation of the kinase that regulates EGFR signaling (24–27). With this regard, several studied have evaluated the efficacy of erlotinib or gefitinib according to KRAS mutational status confirming an association between KRAS mutations and limited activity of EGFR TKIs (28, 29). Several investigators have also hypothesized that KRAS may be a predictive biomarker of response to chemotherapy. Indeed, early preclinical data have shown that KRAS mutations correlate with a greater sensitivity to pemetrexed in NSCLC models (30) probably due to the pemetrexed ability to alter KRAS RNA expression. From a biologic point of view, this result is explained by a strong dependency of KRAS-mutant cells on folate metabolism pathways. Unfortunately, these preclinical results did not translate into an increased clinical activity (31, 32); indeed, a retrospective study conducted on 1,190 KRAS-mutant NSCLC patients treated with upfront platinum-based doublet chemotherapy has demonstrated that pemetrexed was associated with a shorter progression-free interval compared to other agents, such as taxanes (32). Nevertheless, at the ESMO 2021 congress, in the NVALT22 study cisplatin-pemetrexed chemotherapy plus bevacizumab did not improve PFS compared with carboplatin-paclitaxel plus bevacizumab in KRAS mutant patients (33). Concerning other potential targets, Mainardi et al. (34) have found that Src homology region 2 (SHP2), a cytoplasmic Src homology 2 domain containing protein tyrosine phosphatase that regulates several cellular processes, is necessary for KRAS-mutant tumor cell growth in in vivo models of NSCLC. Accordingly, the authors have highlighted that SHP2 inhibition promotes a senescence response in KRAS-mutant NSCLC cells, suggesting a rationale for targeting SHP2 in NSCLC harboring KRAS mutations. Indeed, a recent study has reported that the co-inhibition of SHP2 and KRAS in xenograft models is effectively able to suppress feedback reactivation of KRAS pathway, producing a significant tumor shrinkage (35). With regard to immunotherapy, KRAS mutations have been associated with immune checkpoint inhibitors (ICIs) benefit in NSCLC patients. In particular, a subgroup analysis of Checkmate 057 has shown greater outcomes wit ICIs in KRAS-mutant patients (36). Kim et al. (37) compared in a meta-analysis ICIs treatment and docetaxel in pretreated NSCLC patients, stratifying results by KRAS status. They have found that, in KRAS-mutant NSCLC patients, overall survival (OS) was improved with ICI versus chemotherapy; on the other hand, this benefit was not reported in KRAS wild-type patients. Moreover, through the ImmunoTarget registry, it has been evaluated the sensitivity of ICIs in NSCLC patients with a variety of driver mutations. Accordingly, KRAS mutations resulted in increased immunotherapy benefit compared to other driver mutations (38). Recently, new data are available regarding (chemo)immunotherapy efficacy according to KRAS status. Sun et al. have demonstrated that among patients with PD-L1>50% treated with ICI, KRAS mutant tumors are associated with a superior overall survival compared with KRAS wild type (mOS 21.1 vs 13.6 months, p=0.03). However, among patients treated with chemoimmunotherapy, there was no significant survival difference between patients with KRAS mutant and KRAS wild type status (mOS, 20.0 vs 19.3 months; p=0.93). Moreover, among patients with KRAS mutant status, OS did not differ between those treated with ICI monotherapy and chemoimmunotherapy (mOS, 21.1 vs 20.0 months; p= 0.78). The authors conclude that among patients with PD-L1 expression of 50% or greater NSCLC treated with ICI monotherapy, KRAS wild type is associated with worse survival compared with KRAS mutant patients; in contrast, survival did not differ appreciably between patients with KRAS mutant and KRAS wild type status who were treated with chemoimmunotherapy. Definitely patients with PD-L1>50% and KRAS-mutant NSCLC had favorable survival (mOS ≥20 months) with either ICI monotherapy or chemoimmunotherapy, while patients with KRAS wild type who were treated with ICI monotherapy had worse survival compared with those treated with chemoimmunotherapy (39). With regard to KRAS mutation subtype, another study has shown that treatment outcomes with chemoimmunotherapy are similar in patients with G12C and non-G12C subtypes (40). All these results could have a potential biological rationale because of the correlation between a higher incidence of KRAS mutations in smokers and, as already known, the elevated rate of tumoral DNA mutations and the higher tumor mutational burden (TMB) in smoker patients compared to never-smokers (41, 42). Another important issue is the potential clinical relevance of KRAS mutations in combination with other genetic alterations. In particular, variations in KRAS mutation subtypes have been associated with distinct biological behaviors that may led to different clinical outcomes (43, 44). For example, tumors with KRAS G12C mutations exhibited higher ERK1/2 phosphorylation than those with KRAS G12D (45); to support this data, a recent study using mouse models with KRAS-mutant tumors, demonstrated higher efficacy of the MEK inhibitor selumetinib in KRAS G12C cancers compared with KRAS G12D tumors. Concerning additional alterations, EGFR/ALK/BRAF co-mutations are rare in KRAS-mutant lung adenocarcinoma; concomitant mutations in onco-suppressor genes, especially TP53, STK11 and KEAP1, are more often found (11, 14, 46, 47). Furthermore, additional alterations may affect the immunogenicity of KRAS-mutant tumors. For example, it has been recently demonstrated that tumors carrying KRAS/TP53 mutations have an enhanced tumor inflammation, increased PD-L1 expression and tumor-infiltrating lymphocytes (TILs) and, consequently, there is a remarkable clinical benefit wit ICIs (48). In contrast, additional STK11 mutations in KRAS-mutant NSCLC could decrease immune surveillance through NF-kB pathway modulation. Indeed, these tumors are associated with low TILs infiltration that further reduces immune surveillance (49, 50). Several trials have shown that STK11/LKB1 mutations in KRAS-mutant NSCLC might be predictive of primary resistance to ICIs or chemotherapy as well (51–53). Taken together, all this evidence suggests that KRAS mutations cannot be evaluated as a single entity, but it needs to be placed in the context of the specific tumor features and seen as part of a complex process in cancer development and progression. The deeper understanding of underlying KRAS biology and function has allowed, after many years of failures, to develop agents able to stop the winding path towards KRAS unstoppable tumors (Tables 1, 2).
The Era of KRAS Direct Inhibition
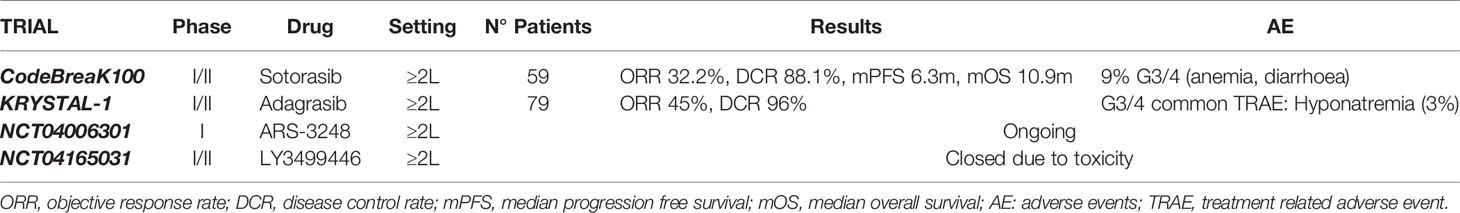
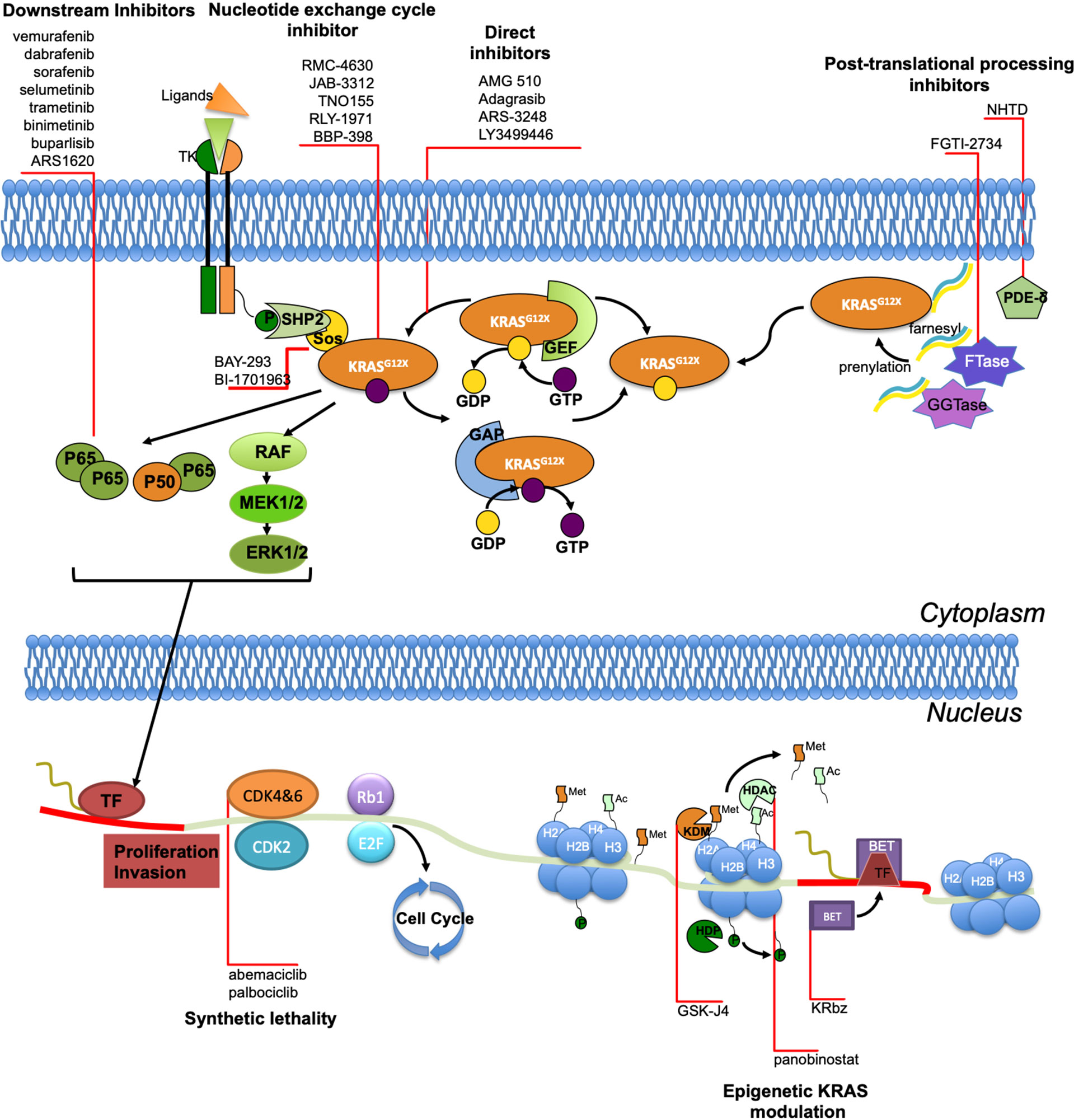
Although KRAS was one of the earliest oncogenic drivers discovered, for decades a therapeutic breakthrough in its targeting has been hampered by several biochemical obstacles (such as the high affinity of GTP for KRAS, the difficulty of finding pockets on KRAS large enough for small-molecule drugs), prompting the increasingly widespread opinion that KRAS was definitely undruggable (54, 55). Therefore, tackling KRAS alterations has been focused on inhibition of its membrane binding (or subcellular localization), identification of lethality partners and on inhibition of its downstream effectors. Concerning direct inhibition, the peculiar crystal structure of the KRAS G12C subtype and the unique reactivity of the cysteine thiol result in the ability to covalently bind the cysteine residue within the region adjacent to the nucleotide-binding pocket, inducing a significant perturbation of protein functional domains and a negative allosteric regulation of RAS signaling in tumor cells (56). This feature can be exploited to create covalent small-molecule inhibitors, binding to KRAS and locking cells in the inactive GDP-bound state (57–59). Thus, various pharmaceutical companies have begun to develop KRAS G12C-specific inhibitors, many of which are being explored in ongoing clinical trials. AMG 510/Sotorasib was the first compound to enter clinical trials with promising results since early trials phases, especially in NSCLC patients. Canon et al. (60) have identified this highly selective molecule inducing tumor regression in preclinical models. In particular, AMG 510 potently inhibits cellular viability in KRAS G12X cell lines and induces tumor regression in xenograft models (60). Furthermore, AMG 510 has shown synergistic growth inhibitory effects when combined with other inhibitors of protein that regulate KRAS activation (such as MEK, AKT, PI3K and members of EGFR family) or with immunotherapy (60). Preliminary results of AMG 510 clinical activity have been presented at the 2019 American Society of Clinical Oncology meeting (61), showing a very promising activity in a cohort of 34 NSCLC patient harboring KRAS G12C mutation achieving a surprising objective response rate (ORR) of 48% and a disease control rate (DCR) of 96%. Regarding its safety profile, the adverse events (AEs) have been manageable with only 9% of grade 3-4 AEs (mostly anaemia and diarrhoea) (62). The final results from the phase I trial have shown a median progression-free survival (PFS) of 6.3 months and a median duration of response of 10.9 months in NSCLC subgroup (59 patients), with a confirmed ORR in 32.2% and DCR in 88.1% of patients (63). Thus, the trial has confirmed the antitumor activity of AMG 510 in pretreated NSCLC patients carrying KRAS G12C mutation, consistent with preliminary results (64). The phase II CodeBreak 100 trial has validated the power of KRAS inhibition with sotorasib in patients with advanced NSCLC. In the phase II trial sotorasib has now shown a durable ORR of 37.1%, a DCR of 80.6%, a median PFS of 6.8 months and a median OS of 12.5 months, in the face of grade 3-4 toxicities rate, mostly diarrhea and hepatic transaminase elevation (11%) (65, 66). These exciting data have led to the fast-track designation of AMG 510 by Food and Drug Administration (FDA). Another cysteine 12-directed covalent inhibitor is MRYX849/adagrasib. Similar to sotorasib, adagrasib is a KRAS G12C inhibitor that has demonstrated a great activity on tumor regression in cell lines and patient-derived xenograft models, as well as in vivo, showing tumor regression in 65% of KRAS G12C mutant preclinical models including different tumor histotypes (67). Indeed, this agent is under investigation in the KRYSTAL trial, a phase I/II multiple expansion cohort trial of MRTX849 in patients with advanced solid tumors carrying KRAS G12C mutation. In this trial, 79 patients were treated with adagrasib 600 mg BID. Overall, the treatment has been well tolerated and most patients has experienced grade 1-2 AEs, such as nausea, diarrhea, vomiting, fatigue and increased ALT. The only commonly reported (>2%) grade 3-4 treatment-related serious AE has been hyponatremia. Among the 51 patients evaluable for clinical activity, the trial has reported an ORR of 45% and DCR of 96% (68). Interestingly, other studies have highlighted that MRTX849 has synergistic effects when combined with EGFR, Src homology 2 domain-containing tyrosinephosphatase-2 (SHP2), mammalian target of rapamycin (mTOR) or cyclin-dependent kinase 4 (CDK4) and CDK6 inhibitors and its efficacy has resulted increased in case of SHP2, MYC or mTOR gene loss (67). A third KRAS G12C covalent inhibitor is JNJ-74699157 (ARS-3248), currently evaluated in a phase I clinical trial enrolling different KRAS G12C mutated tumor types (NCT04006301). A fourth KRAS G12C covalent inhibitor is LY3499446, evaluated in a phase I/II clinical trial (NCT04165031): the study was early terminated due to an unexpected toxicity finding. Concerning pan-RAS inhibitors, many compounds have been tested but the major issue reported is the difficult discrimination between active and inactive KRAS state, with a consequent high off-target activity and a marked toxicity (69). Regarding preclinical evidence, two previous compounds (ARS-853 and ARS-1620) have demonstrated to reduce cell growth and inhibit downstream signaling to MAPK exclusively in tumor cell lines with KRAS G12C mutations (57, 58, 70). Notably, ARS-853 is a potent G12C allele-specific inhibitor that reduces cancer cell proliferation in vitro; its mechanism exploits the need of KRAS G12C nucleotide exchange to switch in the activated status, trapping it in the GDP-RAS state (58, 70). However, ARS-853 has not entered clinical evaluations in KRAS mutant lung cancer yet. Instead, ARS-1620, a covalent compound with high potency and selectivity for KRAS G12C, have shown rapid and sustained tumor regression in vivo (57). The interesting feature of tumor cells harboring KRAS mutations is that they develop, over time, escape mechanisms to break free from the KRAS activation status and undergo adaptative changes to became insusceptible to the drug (71). Instead of relying on the drug-inhibited MEK pathway to proliferate, these adapted cells are capable of using EGFR and aurora kinase downstream signaling cascades to remain in the active form (35). These data may help elucidating why most patients enrolled in clinical trials tend to exhibit partial responses to KRAS inhibitors. For this reason, overcome treatment resistance by developing combination therapies might be a promising approach for the future to create a more durable response to KRAS G12C inhibitors (Table 3).
KRAS Indirect Inhibition
Due to the long-standing belief that KRAS was undruggable for the above-mentioned reasons, a large branch of research has been conducted on indirect inhibition. These strategies, which turned out to be mainly unsuccessful, were aimed at KRAS expression (including epigenetic modulation), processing and downstream signaling inhibition.
Inhibitors of the Nucleotide Exchange Cycle
Guanine nucleotide exchange factors (GEFs), such as SOS, are essential to promote KRAS signaling by catalyzing the exchange of GDP with GTP, thus resulting in the active KRAS-GTP form; GEF inhibitors are a field of particular interest because their activity is not dependent on the type of KRAS mutation, resulting in pan-RAS inhibition. Several small molecules inhibiting SOS have been identified, although only one has made its way to the clinical experimentation (72). BAY-293 is a SOS1 inhibitor which showed a high affinity for its target but weak activity on cell proliferation in vitro (73). BI-1701963 is another SOS1 inhibitor with promising pre-clinical activity, currently tested in a phase I trial, alone or in combination with trametinib, in solid tumors harboring KRAS mutation (NCT04111458).
An interesting new role has been proposed for miR-148a-3p, a tumor suppressor microRNA which controls cell proliferation by reducing the expression of SOS2; in KRAS-mutated cells this microRNA is commonly lost, representing a possible new target in KRAS-mutant NSCLC (74).
Src homology region 2 (SH2)-containing protein tyrosine phosphatase 2 (SHP2) is essential for the activation of KRAS (and other RTKs) signaling, in fact previous studies identified SHP2 as a scaffold protein that links SOS to GRB-2 enhancing the nucleotide exchange cycle (75). Different SHP2 inhibitors are currently under investigation in clinical trials, alone or in combination with other agents; as SHP2 is also implicated in acquired resistance to KRAS G12C inhibitors, there is a strong rationale in combining KRAS and SHP2 inhibitors (71). RMC-4630 has shown promising activity as a single agent in an ongoing phase I clinical trial (NCT03634982), with a DCR of 71% in the cohort of 7 patients with NSCLC and KRAS G12C mutation; RMC-4630 is also being tested in a phase Ib/II clinical in combination with cobimetinib/osimertinib (NCT03989115); JAB-3068 and JAB-3312 are SHP2 allosteric inhibitors investigated in a phase I/II clinical trial (respectively NCT03518554, NCT03565003 and NCT04121286, NCT04045496); TNO155 is being explored as a single agent or with an anti-EGFR (EGF816) in a phase I clinical trial (NCT03114319), in combination with adagrasib in a phase I/II clinical trial (NCT04330664) and in combination with spartalizumab (anti-PD-1) or ribociclib in a phase Ib trial (NCT04000529); RLY-1971 and BBP-398 as single agents are under evaluation in phase I trials (NCT04252339 and NCT04528836).
Inhibitors of RAS Post-Translational Processing
KRAS undergoes several post-translational modifications in order to become active (76). The first step is the prenylation of the carboxyl terminal by the addition of a farnesyl tail, mediated by farnesyltransferase (FTase) or geranylgeranyltransferase (GGTase). Although an interesting preclinical activity of FTase inhibitors (FTIs), first-generation FTIs tipifarnib and lonafarnib, and second-generation salirasib, have not shown significant activity in NSCLC patients (77–79). A possible mechanism of intrinsic resistance to FTIs may be the alternative prenylation of KRAS by GGTase; preclinical data regarding the activity of FGTI-2734, a dual FTase and GGTase inhibitor, on KRAS mutant cell lines (including a NSCLC line) are promising (80). Another potential target involved in KRAS post-translational modifications comprehends phosphodiesterase-δ (PDE-δ), a prenyl-binding protein that transports prenylated KRAS to the cellular membrane; NHTD is a PDE-δ inhibitor with promising in vitro and in vivo activity on mouse xenografts (81).
Inhibitors of Downstream Signaling
Targeting KRAS intracellular pathways has not scored interesting results due to the redundancy and crosstalk of downstream signaling. The inhibition of RAF/MEK/ERK pathway using selective BRAF inhibitors, such as vemurafenib or dabrafenib, whose activity is well established in tumors carrying BRAF V600E mutation, is not effective in KRAS mutant tumors. These drugs can effectively bind only to BRAF monomers, but in KRAS mutant tumors the MAPK pathway is activated by RAF dimers, which are not inhibited by vemurafenib or dabrafenib; in addition, these TKIs cause an upregulation in ERK phosphorylation and activation mediated by CRAF (the so called “MAPK paradox”) (82). On the other hand, nonselective RAF inhibitors like sorafenib (multi-TKI inhibitor of RAF, VEGFR and PDGFR amongst others), did not show a relevant clinical activity in subgroups of KRAS mutant NSCLC: in the MISSION phase III trial sorafenib was tested against placebo in pretreated NSCLC patients, and although a significantly longer PFS, OS was similar between the two arms both in the overall population (8.2 vs 8.3 months; HR, 0.99, p=0.49) and in the KRAS mutant population (6.4 vs 5.1 months; HR, 0.76, p=0.279) (83); in the BATTLE-1 and BATTLE-2 trials, 8-weeks DCR was non statistically different with sorafenib according to KRAS status, although in the latter study RAF and MEK inhibitors showed a better clinical activity than EGFR inhibitor erlotinib in KRAS mutant patients (84, 85). MEK inhibitors have been used alone or in combination with chemotherapy, based on preclinical data of enhanced activity, with disappointing results up to now. The randomized phase III SELECT-1 trial has compared selumetinib plus docetaxel versus docetaxel in 510 patients with KRAS mutant pretreated NSCLC, showing no significant difference in mPFS (3.9 vs 2.8 months, p=0.44) and mOS (8.7 vs 7.9 months, p=0.64) (86). Trametinib has not demonstrated a better clinical activity compared to docetaxel in a randomized phase II study conducted on KRAS mutant NSCLC patients (87); trametinib has also been evaluated in phase I/Ib trials in combination with docetaxel or pemetrexed in patients with NSCLC, showing a favorable toxicity profile and a slightly better activity on patients with KRAS mutation compared to KRAS wild-type patients (88, 89). Binimetinib has been tested in the first-line setting in the phase IB SAKK 19/16 study, in combination with platinum-based chemotherapy, in patient with KRAS mutant NSCLC; the addition of binimetinib has not shown early signs of clinical activity (90). Several trials are being conducted using MEK inhibitors alone or in combination with other agents. A preclinical study by Lee et al. has shown that in murine models of KRAS G12D/p53- lung cancer, the combination of trametinib with anti-PD-1 or anti-PD-L1 immune-checkpoint inhibitors has synergistic anti-tumor effect; this might be explained by an immunomodulatory effect of trametinib that, according to the authors, might be determined by the depletion of PMN-MDSC from the tumor microenvironment (91). ERK inhibitors have shown little activity as monotherapy, so they are currently under investigation mostly in combination with other drugs (e.g. ICIs or other targeted agents) in phase I basket trials (NCT03745989; NCT04418167; NCT03415126).
PI3K-AKT-mTOR is another downstream pathway whose signaling is started by the phosphorylation of PI3K by means of KRAS-GTP; it has important interconnections with RAF-MEK-ERK pathway, in fact the inhibition one of them causes a compensating hyperactivation of the other (92). This might be the reason why single agents directed against this pathway have proven to be mostly unsuccessful. In the phase II BASALT-1 trial, buparlisib, a pan-PI3K inhibitor, was tested in a cohort of 63 patients with pretreated NSCLC harboring a PI3K pathway activation, including 3 patients with concomitant KRAS alterations; the study was closed due to futility, with a 12-week PFS of 23.3% and 20% in squamous and non-squamous histology (93). After the disappointing results of everolimus in unselected patients with pretreated NSCLC (RR<5%), a phase II trial with ridaforolimus was conducted on 79 patients with pretreated KRAS mutated NSCLC, obtaining similar outcomes (RR 1%) (94). More emphasis should be given to combination therapies which include PI3K pathway inhibitors, as it is involved in resistance mechanisms to direct KRAS inhibitors: preclinical studies have demonstrated that the addition of PI3Ki to ARS1620, a selective RAS G12C inhibitor, was effective in cell lines and in xenografts after the failure of ARS1620 monotherapy (95). Dual inhibition of MAPK and PI3K pathways has shown synergistic effects on cell proliferation in preclinical models (96–98), but in the clinical setting alarming signs of toxicity have been reported (99, 100).
Synthetic Lethality
Synthetic lethal screenings are used to identify, in different cell lines, genes that are indispensable for cell survival and proliferation. In KRAS mutated cell lines, previous studies have shown that selective inhibition of BCL-XL, CDK4, XPO1, GATA2, or NF-κB results in cell death, according to the principle of synthetic lethality (101–105). Abemaciclib, a CDK4/6 inhibitor, has been investigated in the JUNIPER trial, a phase III study in KRAS mutated pretreated NSCLC, and it has not improved OS compared to erlotinib, the control arm (7.4 vs 7.8 months, HR 0.968, p=0.77) (106). Although palbociclib, another CDK4/6i, hasn’t shown relevant clinical activity in patients with pretreated NSCLC harboring cell cycle gene alterations (107), preclinical data suggest that the combination of palbociclib and MEKi has synergistic antitumoral and radiosensitizing effects on KRAS mutated NSCLC cell lines (108, 109); different clinical trials are evaluating this combination in the clinical setting (NCT03170206; NCT02022982). Based on synthetic lethality, NCT02079740 is a phase Ib/II trial investigating safety and tolerability of the combination of the BCL inhibitor navitoclax and trametinib, while NCT03095612 is a phase I/II trial evaluating selinexor, an inhibitor of exportin-1 (XPO1), in combination with docetaxel in previously treated KRAS mutant NSCLC. NF-κB is a key transcription factor for KRAS mutated NSCLC; its activity is downregulated by the proteasome inhibitor bortezomib, which restores the function of IκB (NF-κB inhibitor). In a phase II trial, 16 patients with previously treated NSCLC harboring KRAS G12D mutation were treated with bortezomib, but results were disappointing: only 1 partial response was seen, mPFS was 1 month and mOS 13 months. To note, the patient that obtained a partial response had a special histotype (invasive mucinous adenocarcinoma) (110).
New Potential Approaches
Although the recent discovery of direct inhibitors, KRAS mutated NSCLC remains a challenging disease. Several mechanisms of resistance to direct inhibitors have been identified, both on-target (acquired point mutations or KRAS amplification) and off-target (activating mutations in RAF/MEK/ERK pathway, oncogenic fusions of other oncogenes such as ALK and RET or histological transitions) (111). Different strategies to improve the results obtained so far are under investigation, for example the use of KRAS direct inhibitors in combination with immunotherapy in the phase II KRYSTAL-7 trial (NCT04613596) and in the phase Ib CodeBreak 101 trial (NCT04185883). Recently, preliminary results about two combination arms of the CodeBreak 101 trial were presented at the AACR-NCI-EORTC 2021 Virtual International Conference on Molecular Targets and Cancer Therapeutics. In 33 pretreated patients with KRAS-mutant NSCLC, sotorasib was combined with two different doses of afatinib (20 mg in cohort one and 30 mg in cohort two), showing respectively an ORR of 20% and 35% and a DCR of 70% and 74%; the most common grade 3 AE was diarrhea. Sotorasib and trametinib combination was tested in 18 patients with heavily pretreated NSCLC, achieving early signs of activity: DCR was 87% in patients not previously treated with a KRAS G12C inhibitor; the most common AEs were diarrhea, nausea, vomiting and skin reactions (112). Other fields of research are exploring new potential indirect targets, for example p65BTK, an isoform of Bruton’s tyrosine kinase expressed in more than 50% NSCLC samples that acts as a downstream effector of RAF/MEK/ERK pathway in KRAS mutated cell lines; this important oncogene, which up to now has been known to be expressed only in hematopoietic cells, could be targetable by BTK inhibitors like ibrutinib (113). A further important open issue is targeting non-G12C KRAS mutations; for instance, KRAS G12V mutation, although more common in colorectal cancers (CRC) or pancreatic adenocarcinomas, accounts for about 6% of NSCLC mutations. Direct KRAS G12V inhibitors are still in the preclinical setting, but they showed promising results in patient-derived xenograft models of NSCLC, CRC and pancreatic cancers, obtaining even complete tumor regressions (114). The deeper understanding of the metabolic reprogramming in KRAS-mutant cells has allowed to identify additional vulnerabilities. Kerr and colleagues showed that KRAS G12D copy gain, which is a common step during tumor progression, not only induces a glycolytic switch in lung cancer models, but also enhances glutathione (GSH) biosynthesis, translating into lower ROS levels and increased resistance to oxidative stress; treating KRAS-mutant NSCLC cell lines with the glucose analogue 2-deoxy-D-glucose and with BSO (an inhibitor of the synthesis of GSH) resulted in a dramatic apoptosis, confirming their dependency on glucose and GSH (115). The upregulation of fatty acid synthetase (FASN) is also frequent in KRAS-mutant cancer cells in order to provide a large amount of lipids, which are necessary to store energy and build cell membranes (116); indeed, FASN inhibitor TVB-2640 was tested (alone or in combination with a taxane) in a phase Ib trial in solid tumors including a cohort of pretreated NSCLC patients: a higher activity was seen in patients harboring a KRAS mutation, with a median time to progression of 22 weeks vs 5 weeks (p<0.02) (117). Lastly, LKB1 mutations are found simultaneously with KRAS mutation in about 10% of NSCLC and seems to confer susceptibility to phenformin, a mitochondrial inhibitor and analogue of the antidiabetic drug metformin. This might be explained because LKB1 is a kinase responsible for the phosphorylation of AMPK under condition of energy stress (low intracellular ATP levels), leading to the control of cell growth; in the absence of LKB1, the depletion of ATP induced by phenformin results in apoptosis due to the inability of these cells to recognize the state of energy deficiency (118).
Epigenetic KRAS Modulation
Epigenetic modulation of gene expression is an important regulatory process in biology (119) and its alterations contribute to cancer development and progression. Gene regulation occurs in the context of packaging of DNA into an organizing structure, the nucleosome, composed of a DNA strand wound around a core of eight histone proteins (120). Each histone protein has a N-terminal tail extended outward through the DNA strand. Amino acid residues on the histone tail are modified by post-translational methylation, acetylation and phosphorylation (121). On the contrary, deacetylation, demethylation, and dephosphorylation of histones work decreasing access of transcription factors to promoter regions (122). These post-translational modifications can alter the secondary structure of the histone protein tails, increasing the distance between DNA strands and histones and facilitating the access to transcription factors to gene promoter regions (123). Therefore, it is clear that epigenetic regulation is strongly correlated with gene expression and, potentially, with cancer development. In particular, epigenetic modification concur to lung cancer spreading (124–126) and some data available have shown that there is an interaction between KRAS mutations and epigenetic changes (127). For example, histone deacetylase inhibitors have shown the ability of blocking KRAS signalling through gene transcription inhibition and by promoting apoptosis process. Indeed, promising results have been achieved for the treatment of several tumours (122). Notably, Panobinostat has been evaluated in KRAS-mutant NSCLC A549 cell line, demonstrating a significant reduction of cell proliferation (128). Yamada et al. (129) have also evaluated the synergistic efficacy of MEK plus a histone deacetylase inhibitor, demonstrating that this combination has activity in KRAS-mutant lung cancer cells and might be consider for a promising novel therapeutic approach for patients with NSCLC harbouring KRAS mutations. Liu et al. (127) have demonstrated that KRAS mutations increase telomerase activity and length using RAS/MEK pathway activation. In vitro experiments have shown that there is a clear correlation between chemoresistance in KRAS-mutant tumors and telomerase length; as consequence, targeting telomerase/telomere may represent a potential therapeutic strategy for patients carrying KRAS mutations. Unluckily, imetelstat, a telomerase inhibitor, has been tested in a phase II study as maintenance treatment and has failed to improve PFS in patients with advanced NSCLC, only showing a trend toward an improved PFS and OS in patients with shorter telomere (130). Hong et al. (124) have speculated that GSK-J4, the histone H3K27 demethylase inhibitor, is able to induce oxidative and metabolic stress in cancer cell lines when a KRAS mutation is detected, linking the role of oncogenic KRAS in the metabolic stress response to GSK-J4 sensitivity. The trial results suggest GSK-J4 as a potential treatment option for cancer patients with KRAS mutations. Indeed, previous studies have shown that KRAS mutant cancers are more sensitive to oxidative stress (131, 132); moreover, a recent study has exhibited that in NSCLC with KRAS mutations there is an altered expression of genes involved in metabolism upon glutamine deprivation, confirming the relationship between KRAS mutations and oxidative stress susceptibility (133). Sunaga et al. (134) have used RNA interference (RNAi) to knock down the KRAS-mutant cells transcript. They have demonstrated that this mechanism reduces cell proliferation through MAPK pathway downregulation, but cancer progression is not completely abolished due to escape pathways using EGFR, STAT3 and Akt phosphorylation. These findings reveal that inhibition of KRAS signaling is effective, but it could be not enough for a such complex and sophisticated issue (134). Another study has examined the inhibitory growth effects of an anti-KRAS ribozyme adenoviral vector (KRbz-ADV) in in vivo and in vitro NSCLC models. In this study, KRbz-ADV has reduced NSCLC cell growth, determining tumor shrinkage in vitro and inducing complete xenograft regression in 70% of cases after repeated intratumoral injection of KRbz-ADV in vivo (135). A further epigenetic regulation is mediated by bromodomain and extraterminal (BET) proteins that bind acetylated histones and recruit transcription factors to active promoters and enhancers. For this reason, agents targeting BET proteins have emerged as potential therapeutic strategies (136) and have been tested in hematopoietic and solid tumors, showing a significant MYC suppression in cancer cells (137). Interestingly, BET inhibitors have demonstrated to suppress KRAS pathway, in particular in NF1-mutant tumors, by suppressing KRAS-driven transcriptional output (138); as consequence, MEK and ERK downstream appear silenced. Therefore, Guerra et al. have demonstrated that BET inhibitors might enhance the efficacy of MEK inhibitors in KRAS-mutant cancers (139). They have also identified a subset of lung cancer harbouring KRAS mutations. Specifically, several tumors express high level of the homeobox gene HOXC10. HOX genes are development controllers often overexpressed in cancer and, on the contrary, are normally expressed in the corresponding tissue during non-tumoral development (139, 140). In the case of lung tissues, HOX genes are not expressed at all during lung development, on the other hand HOXC10 appears overexpressed in NSCLC. In this study, the authors have shown that HOXC10 is a biomarker of response to MEK/BET inhibitors and regulates expression of pre-replication complex proteins in conjunction with MEK. Thus, MEK/BET inhibitor can suppress KRAS output and HOXC10 to trigger replication defects and induce cell death (139). Fabrizio et al. have investigated the role of methylation of Kelch-like ECH-associated protein cytosine-guanine dinucleotide (KEAP1CpGs); they have found that this methylation shows a significant inverse correlation with the KEAP1 transcript levels. Notably, these results were limited to the KRAS wild-type squamous cell carcinoma and adenocarcinoma, whereas in adenocarcinoma histotype the effect of epigenetic silencing of KEAP1 was also observed in the EGFR mutated subpopulation. In conclusion, the study has revealed that the epigenetic regulation of KEAP1 expression has different features in KRAS and EGFR settings of NSCLC, suggesting KEAP1 methylation as a predictive marker for response to anti-EGFR agents in oncogene addicted disease. Moreover, the correlation between epigenetic features and histotype underlies an interplay with lung cancer pathogenesis and development (141). Despite the many obstacles and its intrinsic complexity, the intriguing scenario of epigenetic targeting has led to many efforts trying to find the weak spot of KRAS-mutant tumors, although in this regard there is still a long way to go.
Conclusions
NSCLC is a heterogeneous disease, in which the only histological classification is largely inadequate to completely define its intrinsic complexity. Molecular markers have dramatically reshaped NSCLC treatment, but KRAS mutations still represent a challenge for physicians and patients. Recently, promising compounds have opened the doors to a new era in which KRAS targeting is possible. The most promising therapeutic approaches are represented by KRAS G12C direct inhibitors, AMG 510 and MRTX849, that have shown encouraging preliminary results in ongoing clinical trials. Actually, the impressive preclinical and clinical activity showed by AMG 510, has led to the fast-track designation by the FDA, representing one of the major breakthrough in the lung cancer research of the last years (142). Other treatment strategies including the inhibition of specific pathways, such as RAF/MEK/ERK, are under investigation as well (NCT03745989; NCT04418167; NCT03415126). The role of ICIs in KRAS mutant NSCLC treatment is also under consideration, alone or in combination with targeted therapy approaches (NCT04613596). Also epigenetic deregulation has been increasingly recognized as one of the major mechanisms of gene expression modulation in lung cancer. Furthermore, some data support the hypothesis that there could be an interplay between KRAS epigenetic regulation and lung cancer pathogenesis (141), suggesting a sophisticated balance between genomic and epigenomic features and lung cancer development. However, although much progress has been made in KRAS-mutant NSCLC treatment, clinical trials have shown that outcomes cannot be given for granted even when the same subtype mutation occurs. This limited efficacy underlies a relevant molecular heterogeneity among KRAS-mutant lung cancer, with such an innate or acquired resistance to targeted therapy, requiring further investigations. Overcoming mechanisms of resistance, implementing gene expression profiling in routine practice, tailoring treatment strategies and extending pharmacological approaches to KRAS mutations other than G12C currently represent the biggest challenge to be addressed for the near future. On the whole, the historical undruggable KRAS alteration is likely to be targeted and a significant turning point has been reached, allowing to offer a range of opportunities in the growing individualized treatment paradigm for KRAS-mutant NSCLC patients (Figure 1).
Author Contributions
MF, AS, and EB, conceived the original idea of the article, drafting, and writing the paper. CC, SP, LB, MM, and GP revised the scientific content of specific sections of the manuscript and participated in drafting specific section of the paper. EV, MD, IS, FM, AV, GV, and ED, participated in the critical revision of the paper. MM, GT, and EB participated in the critical revision and editing of the manuscript. GT and EB conceived the original idea and provided critical revision of the manuscript as well as the final approval of the version to publish. All authors contributed to the article and approved the submitted version.
Funding
EB is supported by Institutional funds of Università Cattolica del Sacro Cuore (UCSC-project D1-2019-2021). EB is currently supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) under Investigator Grant (IG) No. IG20583. GT is supported by AIRC under IG No. IG18599. CC is supported by AIRC Grant “Luigi Bonatti and Anna Maria Bonatti Rocca”.
Conflict of Interest
EB received speakers’ and travels’ fee from MSD, Astra-Zeneca, Pfizer, Eli-Lilly, BMS, Novartis and Roche. EB received institutional research grants from Astra-Zeneca, Roche.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Mayekar MK, Bivona TG. Current Landscape of Targeted Therapy in Lung Cancer. Clin Pharmacol Ther (2017) 102(5):757–64. doi: 10.1002/cpt.810
2. Ferrara MG, di Noia V, D’argento E, Vita E, Damiano P, Cannella A, et al. Oncogene-Addicted Non-Small-Cell Lung Cancer: Treatment Opportunities and Future Perspectives. Cancers (2020) 12(5):1196–217. doi: 10.3390/cancers12051196
3. Planchard D, Popat S, Kerr K, Novello S, Smit EF, Faivre-Finn C, et al. Metastatic Non-Small Cell Lung Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann Oncol (2018) 29(Suppl 4):iv192–237. doi: 10.1093/annonc/mdy275
4. Zhang Z, Wang Y, Vikis HG, Johnson L, Liu G, Li J, et al. Wildtype Kras2 can Inhibit Lung Carcinogenesis in Mice. Nat Genet (2001) 29(1):25–33. doi: 10.1038/ng721
5. McCoy MS, Toole JJ, Cunningham JM, Chang EH, Lowy DR, Weinberg RA. Characterization of a Human Colon/Lung Carcinoma Oncogene. Nature (1983) 302(5903):79–81. doi: 10.1038/302079a0
6. Kranenburg O. The KRAS Oncogene: Past, Present, and Future. Biochim Biophys Acta - Rev Cancer (2005) 1756(2):81–2. doi: 10.1016/j.bbcan.2005.10.001
7. Hajdúch M, Jančík S, Drábek J, Radzioch D. Clinical Relevance of KRAS in Human Cancers. J Biomed Biotechnol (2010) 2010. doi: 10.1155/2010/150960.
8. D’Arcangelo M, Cappuzzo F. K-Ras Mutations in Non-Small-Cell Lung Cancer: Prognostic and Predictive Value. ISRN Mol Biol (2012) 2012:1–8. doi: 10.5402/2012/837306
9. Ghimessy A, Radeczky P, Laszlo V, Hegedus B, Renyi-Vamos F, Fillinger J, et al. Current Therapy of KRAS-Mutant Lung Cancer. Cancer Metastasis Rev (2020) 39(4):1159–77. doi: 10.1007/s10555-020-09903-9
10. Westcott PMK, To MD. The Genetics and Biology of KRAS in Lung Cancer. Chin J Cancer (2013) 32(2):63–70. doi: 10.5732/cjc.012.10098
11. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer Statistics, 2008. CA: A Cancer J Clin (2008) 58(2):71–96. doi: 10.3322/CA.2007.0010
12. Rekhtman N, Paik PK, Arcila ME, Tafe LJ, Oxnard GR, Moreira AL, et al. Clarifying the Spectrum of Driver Oncogene Mutations in Biomarker-Verified Squamous Carcinoma of Lung: Lack of EGFR/KRAS and Presence of PIK3CA/AKT1 Mutations. Clin Cancer Res (2012) 18(4):1167–76. doi: 10.1158/1078-0432.CCR-11-2109
13. Simbolo M, Barbi S, Fassan M, Mafficini A, Ali G, Vicentini C, et al. Gene Expression Profiling of Lung Atypical Carcinoids and Large Cell Neuroendocrine Carcinomas Identifies Three Transcriptomic Subtypes With Specific Genomic Alterations. J Thorac Oncol (2019) 14(9):1651–61. doi: 10.1016/j.jtho.2019.05.003
14. Riely GJ, Kris MG, Rosenbaum D, Marks J, Li A, Chitale DA, et al. Frequency and Distinctive Spectrum of KRAS Mutations in Never Smokers With Lung Adenocarcinoma. Clin Cancer Res (2008) 14(18):5731–4. doi: 10.1158/1078-0432.CCR-08-0646
15. Ferrer I, Zugazagoitia J, Herbertz S, John W, Paz-Ares L, Schmid-Bindert G. KRAS-Mutant Non-Small Cell Lung Cancer: From Biology to Therapy. Lung Cancer (2018) 124:53–64. doi: 10.1016/j.lungcan.2018.07.013
16. Dogan S, Shen R, Ang DC, Johnson ML, D’Angelo SP, Paik PK, et al. Molecular Epidemiology of EGFR and KRAS Mutations in 3,026 Lung Adenocarcinomas: Higher Susceptibility of Women to Smoking-Related KRAS-Mutant Cancers. Clin Cancer Res (2012) 18(22):6169–77. doi: 10.1158/1078-0432.CCR-11-3265
17. Chapman AM, Sun KY, Ruestow P, Cowan DM, Madl AK. Lung Cancer Mutation Profile of EGFR, ALK, and KRAS: Meta-Analysis and Comparison of Never and Ever Smokers. Lung Cancer (2016) 102:122–34. doi: 10.1016/j.lungcan.2016.10.010
18. Johnson ML, Sima CS, Chaft J, Paik PK, Pao W, Kris MG, et al. Association of KRAS and EGFR Mutations With Survival in Patients With Advanced Lung Adenocarcinomas. Cancer (2013) 119(2):356–62. doi: 10.1002/cncr.27730
19. Mascaux C, Iannino N, Martin B, Paesmans M, Berghmans T, Dusart M, et al. The Role of RAS Oncogene in Survival of Patients With Lung Cancer: A Systematic Review of the Literature With Meta-Analysis. Br J Cancer (2005) 92(1):131–9. doi: 10.1038/sj.bjc.6602258
20. Marabese M, Ganzinelli M, Garassino MC, Shepherd FA, Piva S, Caiola E, et al. KRAS Mutations Affect Prognosis of Non-Small-Cell Lung Cancer Patients Treated With First-Line Platinum Containing Chemotherapy. Oncotarget (2015) 6(32):34014–22. doi: 10.18632/oncotarget.5607
21. Shepherd FA, Domerg C, Hainaut P, Jänne PA, Pignon JP, Graziano S, et al. Pooled Analysis of the Prognostic and Predictive Effects of KRAS Mutation Status and KRAS Mutation Subtype in Early-Stage Resected Non-Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. J Clin Oncol (2013) 31(17):2173–81. doi: 10.1200/JCO.2012.48.1390
22. Macerelli M, Caramella C, Faivre L, Besse B, Planchard D, Polo V, et al. Does KRAS Mutational Status Predict Chemoresistance in Advanced Non-Small Cell Lung Cancer (NSCLC)? Lung Cancer (Amsterdam Netherlands) (2014) 83(3):383–8. doi: 10.1016/j.lungcan.2013.12.013
23. Bria E, Gralla RJ, Raftopoulos H, Cuppone F, Milella M, Sperduti I, et al. Magnitude of Benefit of Adjuvant Chemotherapy for Non-Small Cell Lung Cancer: Meta-Analysis of Randomized Clinical Trials. Lung Cancer (2009) 63(1):50–7. doi: 10.1016/j.lungcan.2008.05.002
24. Giaccone G, Ruiz MG, le Chevalier T, Thatcher N, Smit E, Rodriguez JA, et al. Erlotinib for Frontline Treatment of Advanced Non-Small Cell Lung Cancer: A Phase II Study. Clin Cancer Res (2006) 12(20 PART 1):6049–55. doi: 10.1158/1078-0432.CCR-06-0260
25. Jackman DM, Yeap BY, Lindeman NI, Fidias P, Rabin MS, Temel J, et al. Phase II Clinical Trial of Chemotherapy-Naïve Patients ≥ 70 Years of Age Treated With Erlotinib for Advanced Non-Small-Cell Lung Cancer. J Clin Oncol (2007) 25(7):760–6. doi: 10.1200/JCO.2006.07.5754
26. Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, et al. KRAS Mutations and Primary Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib. PLoS Med (2005) 2(1):0057–61. doi: 10.1371/journal.pmed.0020017
27. Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD, et al. KRAS Mutation Is an Important Predictor of Resistance to Therapy With Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small Cell Lung Cancer. Clin Cancer Res (2007) 13(10):2890–6. doi: 10.1158/1078-0432.CCR-06-3043
28. Mao C, Qiu LX, Liao RY, Du FB, Ding H, Yang WC, et al. KRAS Mutations and Resistance to EGFR-TKIs Treatment in Patients With Non-Small Cell Lung Cancer: A Meta-Analysis of 22 Studies. Lung Cancer (2010) 69(3):272–8. doi: 10.1016/j.lungcan.2009.11.020
29. Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, et al. Assessment of Somatic K-RAS Mutations as a Mechanism Associated With Resistance to EGFR-Targeted Agents: A Systematic Review and Meta-Analysis of Studies in Advanced Non-Small-Cell Lung Cancer and Metastatic Colorectal Cancer. Lancet Oncol (2008) 9(10):962–72. doi: 10.1016/S1470-2045(08)70206-7
30. Moran DM, Trusk PB, Pry K, Paz K, Sidransky D, Bacus SS. KRAS Mutation Status Is Associated With Enhanced Dependency on Folate Metabolism Pathways in Non-Small Cell Lung Cancer Cells. Mol Cancer Ther (2014) 13(6):1611–24. doi: 10.1158/1535-7163.MCT-13-0649
31. Sun JM, Hwang DW, Ahn JS, Ahn MJ, Park K. Prognostic and Predictive Value of KRAS Mutations in Advanced Non-Small Cell Lung Cancer. PLoS One (2013) 8(5):e64816–24. doi: 10.1371/journal.pone.0064816
32. Renaud S, Guerrera F, Seitlinger J, Reeb J, Voegeli AC, Legrain M, et al. KRAS-Specific Amino Acid Substitutions Are Associated With Different Responses to Chemotherapy in Advanced Non–small-Cell Lung Cancer. Clin Lung Cancer (2018) 19(6):e919–31. doi: 10.1016/j.cllc.2018.08.005
33. Dingemans AC, Ernst S, Mellema W, Burgers S, Staal-van den Brekel J, Hendriks LEL, et al. A Randomized Phase III Study Comparing Cisplatin-Pemetrexed (Cis-Pem) With Carboplatin (C)-Paclitaxel (P)-Bevacizumab (B) in Chemotherapy Naïve Pat. OncologyPRO.
34. Mainardi S, Mulero-Sánchez A, Prahallad A, Germano G, Bosma A, Krimpenfort P, et al. SHP2 Is Required for Growth of KRAS-Mutant Non-Small-Cell Lung Cancer In Vivo Letter. Nat Med (2018) 24(7):961–7. doi: 10.1038/s41591-018-0023-9
35. Ryan MB, de la Cruz FF, Phat S, Myers DT, Wong E, Shahzade HA, et al. Vertical Pathway Inhibition Overcomes Adaptive Feedback Resistance to KrasG12C Inhibition. Clin Cancer Res (2020) 26(7):1617–43. doi: 10.1158/1078-0432.CCR-19-3523
36. Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab Versus Docetaxel in Advanced Nonsquamous Non–Small-Cell Lung Cancer. N Engl J Med (2015) 373(17):1627–39. doi: 10.1056/NEJMoa1507643
37. Kim JH, Kim HS, Kim BJ. Prognostic Value of KRAS Mutation in Advanced Non-Small-Cell Lung Cancer Treated With Immune Checkpoint Inhibitors: A Metaanalysis and Review. Oncotarget (2017) 8(29):48248–52. doi: 10.18632/oncotarget.17594
38. Mazieres J, Drilon A, Lusque A, Mhanna L, Cortot AB, Mezquita L, et al. Immune Checkpoint Inhibitors for Patients With Advanced Lung Cancer and Oncogenic Driver Alterations: Results From the IMMUNOTARGET Registry. Ann Oncol (2019) 30(8):1321–8. doi: 10.1093/annonc/mdz167
39. Sun L, Hsu M, Cohen RB, Langer CJ, Mamtani R, Aggarwal C. Association Between KRAS Variant Status and Outcomes With First-Line Immune Checkpoint Inhibitor-Based Therapy in Patients With Advanced Non-Small-Cell Lung Cancer. JAMA Oncol (2021) 7(6):937–9. doi: 10.1001/jamaoncol.2021.0546
40. Arbour KC, Ricciuti B, Rizvi H, Hellmann MD, Yu HA, Ladanyi M, et al. Chemo-Immunotherapy Outcomes of KRAS-G12C Mutant Lung Cancer Compared to Other Molecular Subtypes of KRAS-Mutant Lung Cancer. J Clin Oncol (2021) 39(15_suppl):9088–8. doi: 10.1200/JCO.2021.39.15_suppl.9088
41. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Mutational Landscape Determines Sensitivity to PD-1 Blockade in Non-Small Cell Lung Cancer. Science (2015) 348(6230):124–8. doi: 10.1126/science.aaa1348
42. Addeo A, Banna GL, Weiss GJ. Tumor Mutation Burden - From Hopes to Doubts. JAMA Oncol (2019) 5(7):934–5. doi: 10.1001/jamaoncol.2019.0626
43. Kim ES, Hirsh V, Mok T, Socinski MA, Gervais R, Wu YL, et al. Gefitinib Versus Docetaxel in Previously Treated Non-Small-Cell Lung Cancer (INTEREST): A Randomised Phase III Trial. Lancet (2008) 372(9652):1809–18. doi: 10.1016/S0140-6736(08)61758-4
44. Ciuleanu T, Stelmakh L, Cicenas S, Miliauskas S, Grigorescu AC, Hillenbach C, et al. Efficacy and Safety of Erlotinib Versus Chemotherapy in Second-Line Treatment of Patients With Advanced, Non-Small-Cell Lung Cancer With Poor Prognosis (TITAN): A Randomised Multicentre, Open-Label, Phase 3 Study. Lancet Oncol (2012) 13(3):300–8. doi: 10.1016/S1470-2045(11)70385-0
45. Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, et al. American Society of Clinical Oncology Provisional Clinical Opinion: Testing for KRAS Gene Mutations in Patients With Metastatic Colorectal Carcinoma to Predict Response to Anti-Epidermal Growth Factor Receptor Monoclonal Antibody Therapy. J Clin Oncol (2009) 27(12):2091–6. doi: 10.1200/JCO.2009.21.9170
46. Rodenhuis S, van de Wetering ML, Mooi WJ, Evers SG, van Zandwijk N, Bos JL. Mutational Activation of the K- Ras Oncogene. N Engl J Med (1987) 317(15):929–35. doi: 10.1056/NEJM198710083171504
47. Forbes S, Clements J, Dawson E, Bamford S, Webb T, Dogan A, et al. Cosmic 2005. Br J Cancer (2006) 94(2):318–22. doi: 10.1038/sj.bjc.6602928
48. Dong ZY, Zhong WZ, Zhang XC, Su J, Xie Z, Liu SY, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res (2017) 23(12):3012–24. doi: 10.1158/1078-0432.CCR-16-2554
49. Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-Occurring Genomic Alterations Define Major Subsets of KRAS-Mutant Lung Adenocarcinoma With Distinct Biology, Immune Profiles, and Therapeutic Vulnerabilities. Cancer Discov (2015) 5(8):861–78. doi: 10.1158/2159-8290.CD-14-1236
50. Schabath MB, Welsh EA, Fulp WJ, Chen L, Teer JK, Thompson ZJ, et al. Differential Association of STK11 and TP53 With KRAS Mutation-Associated Gene Expression, Proliferation and Immune Surveillance in Lung Adenocarcinoma. Oncogene (2016) 35(24):3209–16. doi: 10.1038/onc.2015.375
51. Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov (2018) 8(7):822–35. doi: 10.1158/2159-8290.CD-18-0099
52. Moro M, Caiola E, Ganzinelli M, Zulato E, Rulli E, Marabese M, et al. Metformin Enhances Cisplatin-Induced Apoptosis and Prevents Resistance to Cisplatin in Co-Mutated KRAS/LKB1 NSCLC. J Thorac Oncol (2018) 13(11):1692–704. doi: 10.1016/j.jtho.2018.07.102
53. Koyama S, Akbay EA, Li YY, Aref AR, Skoulidis F, Herter-Sprie GS, et al. STK11/LKB1 Deficiency Promotes Neutrophil Recruitment and Proinflammatory Cytokine Production to Suppress T-Cell Activity in the Lung Tumor Microenvironment. Cancer Res (2016) 76(5):999–1008. doi: 10.1158/0008-5472.CAN-15-1439
54. Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the Undruggable RAS: Mission Possible? Nat Rev Drug Discov (2014) 13(11):828–51. doi: 10.1038/nrd4389
55. Lindsay CR, Jamal-Hanjani M, Forster M, Blackhall F. KRAS: Reasons for Optimism in Lung Cancer. Eur J Cancer (2018) 99:20–7. doi: 10.1016/j.ejca.2018.05.001
56. Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) Inhibitors Allosterically Control GTP Affinity and Effector Interactions. Nature (2013) 503(7477):548–51. doi: 10.1038/nature12796
57. Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, et al. Targeting KRAS Mutant Cancers With a Covalent G12C-Specific Inhibitor. Cell (2018) 172(3):578–89.e17. doi: 10.1016/j.cell.2018.01.006
58. Lito P, Solomon M, Li LS, Hansen R, Rosen N. Cancer Therapeutics: Allele-Specific Inhibitors Inactivate Mutant KRAS G12C by a Trapping Mechanism. Science (2016) 351(6273):604–8. doi: 10.1126/science.aad6204
59. Gehringer M, Laufer SA. Emerging and Re-Emerging Warheads for Targeted Covalent Inhibitors: Applications in Medicinal Chemistry and Chemical Biology. J Med Chem (2019) 62(12):5673–724. doi: 10.1021/acs.jmedchem.8b01153
60. Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The Clinical KRAS(G12C) Inhibitor AMG 510 Drives Anti-Tumour Immunity. Nature (2019) 575(7781):217–23. doi: 10.1038/s41586-019-1694-1
61. Fakih M, O’Neil B, Price TJ, Falchook GS, Desai J, Kuo J, et al. Phase 1 Study Evaluating the Safety, Tolerability, Pharmacokinetics (PK), and Efficacy of AMG 510, a Novel Small Molecule KRAS G12C Inhibitor, in Advanced Solid Tumors. J Clin Oncol (2019) 37(15_suppl):3003–3. doi: 10.1200/JCO.2019.37.15_suppl.3003
62. Govindan R, Fakih MG, Price TJ, Falchook GS, Desai J, Kuo JC, et al. Phase I Study of AMG 510, a Novel Molecule Targeting KRAS G12C Mutant Solid Tumours. Ann Oncol (2019) 30:v163–4. doi: 10.1093/annonc/mdz244.008
63. Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, et al. KRAS G12C Inhibition With Sotorasib in Advanced Solid Tumors. N Engl J Med (2020) 383(13):1207–17. doi: 10.1056/NEJMoa1917239
64. Salgia R, Pharaon R, Mambetsariev I, Nam A, Sattler M. The Improbable Targeted Therapy: KRAS as an Emerging Target in Non-Small Cell Lung Cancer (NSCLC). Cell Rep Med (2021) 2(1):1207–17. doi: 10.1016/j.xcrm.2020.100186
65. Sotorasib Edges Closer to Approval. Cancer Discov (2021) 11(5):OF2. doi: 10.1158/2159-8290.CD-NB2021-0309
66. Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, et al. Sotorasib for Lung Cancers With KRAS P.G12C Mutation. N Engl J Med (2021) 384(25):2371–81. doi: 10.1056/NEJMoa2103695
67. Hallin J, Engstrom LD, Hargi L, Calinisan A, Aranda R, Briere DM, et al. The KRASG12C Inhibitor MRTX849 Provides Insight Toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov (2020) 10(1):54–71. doi: 10.1158/2159-8290.CD-19-1167
68. Jänne PA, Rybkin II, Spira AI, Riely GJ, Papadopoulos KP, Sabari JK, et al. KRYSTAL-1: Activity and Safety of Adagrasib (MRTX849) in Advanced/ Metastatic Non–Small-Cell Lung Cancer (NSCLC) Harboring KRAS G12C Mutation. Eur J Cancer (2020) 138:S1–2. doi: 10.1158/2159-8290
69. Welsch ME, Kaplan A, Chambers JM, Stokes ME, Bos PH, Zask A, et al. Multivalent Small-Molecule Pan-RAS Inhibitors. Cell (2017) 168(5):878–89.e29. doi: 10.1016/j.cell.2017.02.006
70. Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, et al. Selective Inhibition of Oncogenic KRAS Output With Small Molecules Targeting the Inactive State. Cancer Discov (2016) 6(3):316–29. doi: 10.1158/2159-8290.CD-15-1105
71. Xue JY, Zhao Y, Aronowitz J, Mai TT, Vides A, Qeriqi B, et al. Rapid Non-Uniform Adaptation to Conformation-Specific KRAS(G12C) Inhibition. Nature (2020) 577(7790):421–5. doi: 10.1038/s41586-019-1884-x
72. Kessler D, Gerlach D, Kraut N, McConnell DB. Targeting Son of Sevenless 1: The Pacemaker of KRAS. Curr Opin Chem Biol (2021) 62:109–18. doi: 10.1016/j.cbpa.2021.02.014
73. Hillig RC, Sautier B, Schroeder J, Moosmayer D, Hilpmann A, Stegmann CM, et al. Discovery of Potent SOS1 Inhibitors That Block RAS Activation via Disruption of the RAS–SOS1 Interaction. Proc Natl Acad Sci USA (2019) 116(7):2551–60. doi: 10.1073/pnas.1812963116
74. Xie Q, Yu Z, Lu Y, Fan J, Ni Y, Ma L. microRNA-148a-3p Inhibited the Proliferation and Epithelial–Mesenchymal Transition Progression of Non-Small-Cell Lung Cancer via Modulating Ras/MAPK/Erk Signaling. J Cell Physiol (2019) 234(8):12786–99. doi: 10.1002/jcp.27899
75. Dance M, Montagner A, Salles JP, Yart A, Raynal P. The Molecular Functions of Shp2 in the Ras/Mitogen-Activated Protein Kinase (ERK1/2) Pathway. Cell Signalling (2008) 20(3):453–9. doi: 10.1016/j.cellsig.2007.10.002
76. Friday BB, Adjei AA. K-Ras as a Target for Cancer Therapy. Biochim Biophys Acta - Rev Cancer (2005) 1756(2):127–44. doi: 10.1016/j.bbcan.2005.08.001
77. Kim ES, Kies MS, Fossella FV, Glisson BS, Zaknoen S, Statkevich P, et al. Phase II Study of the Farnesyltransferase Inhibitor Lonafarnib With Paclitaxel in Patients With Taxane-Refractory/Resistant Nonsmall Cell Lung Carcinoma. Cancer (2005) 104(3):561–9. doi: 10.1002/cncr.21188
78. Wong NS, Meadows KL, Rosen LS, Adjei AA, Kaufmann SH, Morse MA, et al. A Phase I Multicenter Study of Continuous Oral Administration of Lonafarnib (SCH 66336) and Intravenous Gemcitabine in Patients With Advanced Cancer. Cancer Invest (2011) 29(9):617–25. doi: 10.3109/07357907.2011.621912
79. Adjei AA, Mauer A, Bruzek L, Marks RS, Hillman S, Geyer S, et al. Phase II Study of the Farnesyl Transferase Inhibitor R115777 in Patients With Advanced Non-Small-Cell Lung Cancer. J Clin Oncol (2003) 21(9):1760–6. doi: 10.1200/JCO.2003.09.075
80. Kazi A, Xiang S, Yang H, Chen L, Kennedy P, Ayaz M, et al. Dual Farnesyl and Geranylgeranyl Transferase Inhibitor Thwarts Mutant KRAS-Driven Patient-Derived Pancreatic Tumors. Clin Cancer Res (2019) 25(19):5984–96. doi: 10.1158/1078-0432.CCR-18-3399
81. Leung ELH, Luo LX, Li Y, Liu ZQ, Li LL, Shi DF, et al. Identification of a New Inhibitor of KRAS-Pdeδ Interaction Targeting KRAS Mutant Nonsmall Cell Lung Cancer. Int J Cancer (2019) 145(5):1334–45. doi: 10.1002/ijc.32222
82. Moore AR, Rosenberg SC, McCormick F, Malek S. RAS-Targeted Therapies: Is the Undruggable Drugged? Nat Rev Drug Discov (2020) 19(8):533–52. doi: 10.1038/s41573-020-0068-6
83. Paz-Ares L, Hirsh V, Zhang L, de Marinis F, Yang JCH, Wakelee HA, et al. Monotherapy Administration of Sorafenib in Patients With Non-Small Cell Lung Cancer (MISSION) Trial: A Phase III, Multicenter, Placebo-Controlled Trial of Sorafenib in Patients With Relapsed or Refractory Predominantly Nonsquamous Non-Small-Cell Lung Canc. J Thorac Oncol (2015) 10(12):1745–53. doi: 10.1097/JTO.0000000000000693
84. Blumenschein GR, Saintigny P, Liu S, Kim ES, Tsao AS, Herbst RS, et al. Comprehensive Biomarker Analysis and Final Efficacy Results of Sorafenib in the BATTLE Trial. Clin Cancer Res (2013) 19(24):6967–75. doi: 10.1158/1078-0432.CCR-12-1818
85. Papadimitrakopoulou V, Jack Lee J, Wistuba II, Tsao AS, Fossella FV, Kalhor N, et al. The BATTLE-2 Study: A Biomarker-Integrated Targeted Therapy Study in Previously Treated Patients With Advanced Non-Small-Cell Lung Cancer. J Clin Oncol (2016) 34(30):3638–47. doi: 10.1200/JCO.2015.66.0084
86. Jänne PA, van den Heuvel MM, Barlesi F, Cobo M, Mazieres J, Crinò L, et al. Selumetinib Plus Docetaxel Compared With Docetaxel Alone and Progression-Free Survival in Patients With KRAS-Mutant Advanced Non-Small Cell Lung Cancer: The SELECT-1 Randomized Clinical Trial. JAMA - J Am Med Assoc (2017) 317(18):1844–53. doi: 10.1001/jama.2017.3438
87. Blumenschein GR, Smit EF, Planchard D, Kim DW, Cadranel J, de Pas T, et al. A Randomized Phase II Study of the MEK1/MEK2 Inhibitor Trametinib (GSK1120212) Compared With Docetaxel in KRAS-Mutant Advanced Non-Small-Cell Lung Cancer (NSCLC). Ann Oncol (2015) 26(5):894–901. doi: 10.1093/annonc/mdv072
88. Gandara DR, Leighl N, Delord JP, Barlesi F, Bennouna J, Zalcman G, et al. A Phase 1/1b Study Evaluating Trametinib Plus Docetaxel or Pemetrexed in Patients With Advanced Non–Small Cell Lung Cancer. J Thorac Oncol (2017) 12(3):556–66. doi: 10.1016/j.jtho.2016.11.2218
89. Gandara DR, Hiret S, Blumenschein GR, Barlesi F, Delord J-P, Madelaine J, et al. Oral MEK1/MEK2 Inhibitor Trametinib (GSK1120212) in Combination With Docetaxel in KRAS -Mutant and Wild-Type (WT) Advanced Non-Small Cell Lung Cancer (NSCLC): A Phase I/Ib Trial. J Clin Oncol (2013) 31(15_suppl):8028–8. doi: 10.1200/jco.2013.31.15_suppl.8028
90. Froesch P, Mark M, Rothschild SI, Li Q, Godar G, Rusterholz C, et al. Binimetinib, Pemetrexed and Cisplatin, Followed by Maintenance of Binimetinib and Pemetrexed in Patients With Advanced Non-Small Cell Lung Cancer (NSCLC) and KRAS Mutations. The Phase 1b SAKK 19/16 Trial. Lung Cancer (2021) 156:91–9. doi: 10.1016/j.lungcan.2021.04.002
91. Lee JW, Zhang Y, Eoh KJ, Sharma R, Sanmamed MF, Wu J, et al. The Combination of MEK Inhibitor With Immunomodulatory Antibodies Targeting Programmed Death 1 and Programmed Death Ligand 1 Results in Prolonged Survival in Kras/p53-Driven Lung Cancer. J Thorac Oncol (2019) 14(6):1046–60. doi: 10.1016/j.jtho.2019.02.004
92. Wee S, Jagani Z, Kay XX, Loo A, Dorsch M, Yao YM, et al. PI3K Pathway Activation Mediates Resistance to MEK Inhibitors in KRAS Mutant Cancers. Cancer Res (2009) 69(10):4286–93. doi: 10.1158/0008-5472.CAN-08-4765
93. Vansteenkiste JF, Canon JL, de Braud F, Grossi F, de Pas T, Gray JE, et al. Safety and Efficacy of Buparlisib (BKM120) in Patients With PI3K Pathway-Activated Non-Small Cell Lung Cancer: Results From the Phase II BASALT-1 Study. J Thorac Oncol (2015) 10(9):1319–27. doi: 10.1097/JTO.0000000000000607
94. Riely GJ, Brahmer JR, Planchard D, Crinò L, Doebele RC, Mas Lopez LA, et al. A Randomized Discontinuation Phase II Trial of Ridaforolimus in Non-Small Cell Lung Cancer (NSCLC) Patients With KRAS Mutations. J Clin Oncol (2012) 30(15_suppl):7531–1. doi: 10.1200/jco.2012.30.15_suppl.7531
95. Misale S, Fatherree JP, Cortez E, Li C, Bilton S, Timonina D, et al. KRAS G12C NSCLC Models Are Sensitive to Direct Targeting of KRAS in Combination With PI3K Inhibition. Clin Cancer Res (2019) 25(2):796–807. doi: 10.1158/1078-0432.CCR-18-0368
96. Broutin S, Stewart A, Thavasu P, Paci A, Bidart JM, Banerji U. Insights Into Significance of Combined Inhibition of MEK and M-TOR Signalling Output in KRAS Mutant Non-Small-Cell Lung Cancer. Br J Cancer (2016) 115(5):549–52. doi: 10.1038/bjc.2016.220
97. Dogan Turacli I, Ozkan AC, Ekmekci A. The Comparison Between Dual Inhibition of mTOR With MAPK and PI3K Signaling Pathways in KRAS Mutant NSCLC Cell Lines. Tumor Biol (2015) 36(12):9339–45. doi: 10.1007/s13277-015-3671-0
98. Simmons BH, Lee JH, Lalwani K, Giddabasappa A, Snider BA, Wong A, et al. Combination of a MEK Inhibitor at Sub-MTD With a PI3K/mTOR Inhibitor Signiwcantly Suppresses Growth of Lung Adenocarcinoma Tumors in KrasG12D-LSL Mice. Cancer Chemother Pharmacol (2012) 70(2):213–20. doi: 10.1007/s00280-012-1899-6
99. LoRusso P, Shapiro G, Pandya SS, Kwak EL, Jones C, Belvin M, et al. A First-In-Human Phase Ib Study to Evaluate the MEK Inhibitor GDC-0973, Combined With the Pan-PI3K Inhibitor GDC-0941, in Patients With Advanced Solid Tumors. J Clin Oncol (2012) 30(15_suppl):2566–6. doi: 10.1200/jco.2012.30.15_suppl.2566
100. Tolcher AW, Bendell JC, Papadopoulos KP, Burris HA, Patnaik A, Jones SF, et al. A Phase IB Trial of the Oral MEK Inhibitor Trametinib (GSK1120212) in Combination With Everolimus in Patients With Advanced Solid Tumors. Ann Oncol (2015) 26(1):58–64. doi: 10.1093/annonc/mdu482
101. Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM, et al. Synthetic Lethal Interaction of Combined BCL-XL and MEK Inhibition Promotes Tumor Regressions in KRAS Mutant Cancer Models. Cancer Cell (2013) 23(1):121–8. doi: 10.1016/j.ccr.2012.11.007
102. Puyol M, Martín A, Dubus P, Mulero F, Pizcueta P, Khan G, et al. A Synthetic Lethal Interaction Between K-Ras Oncogenes and Cdk4 Unveils a Therapeutic Strategy for Non-Small Cell Lung Carcinoma. Cancer Cell (2010) 18(1):63–73. doi: 10.1016/j.ccr.2010.05.025
103. Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, et al. The GATA2 Transcriptional Network Is Requisite for RAS Oncogene-Driven Non-Small Cell Lung Cancer. Cell (2012) 149(3):642–55. doi: 10.1016/j.cell.2012.02.059
104. Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA Interference Reveals That Oncogenic KRAS-Driven Cancers Require Tbk1. Nature (2009) 462(7269):108–12. doi: 10.1038/nature08460
105. Kim J, McMillan E, Kim HS, Venkateswaran N, Makkar G, Rodriguez-Canales A, et al. XPO1-Dependent Nuclear Export Is a Druggable Vulnerability in KRAS-Mutant Lung Cancer. Nature (2016) 538(7623):114–7. doi: 10.1038/nature19771
106. Goldman JW, Mazieres J, Barlesi F, Dragnev KH, Koczywas M, Göskel T, et al. A Randomized Phase III Study of Abemaciclib Versus Erlotinib in Patients With Stage IV Non-Small Cell Lung Cancer With a Detectable KRAS Mutation Who Failed Prior Platinum-Based Therapy: JUNIPER. Front Oncol (2020) 10. doi: 10.3389/fonc.2020.578756
107. Edelman MJ, Redman MW, Albain KS, McGary EC, Rafique NM, Petro D, et al. SWOG S1400C (Nct02154490)—A Phase II Study of Palbociclib for Previously Treated Cell Cycle Gene Alteration–Positive Patients With Stage IV Squamous Cell Lung Cancer (Lung-MAP Substudy). J Thorac Oncol (2019) 14(10):1853–9. doi: 10.1016/j.jtho.2019.06.027
108. Tao Z, le Blanc JM, Wang C, Zhan T, Zhuang H, Wang P, et al. Coadministration of Trametinib and Palbociclib Radiosensitizes KRAS-Mutant Non-Small Cell Lung Cancers In Vitro and In Vivo. Clin Cancer Res (2016) 22(1):122–33. doi: 10.1158/1078-0432.CCR-15-0589
109. Zhou J, Zhang S, Chen X, Zheng X, Yao Y, Lu G, et al. Palbociclib, a Selective CDK4/6 Inhibitor, Enhances the Effect of Selumetinib in RAS-Driven Non-Small Cell Lung Cancer. Cancer Lett (2017) 408:130–7. doi: 10.1016/j.canlet.2017.08.031
110. Drilon A, Schoenfeld AJ, Arbour KC, Litvak A, Ni A, Montecalvo J, et al. Exceptional Responders With Invasive Mucinous Adenocarcinomas: A Phase 2 Trial of Bortezomib in Patients With KRAS G12D-Mutant Lung Cancers. Cold Spring Harbor Mol Case Stud (2019) 5(2):122–33. doi: 10.1101/mcs.a003665
111. Awad MM, Liu S, Rybkin II, Arbour KC, Dilly J, Zhu VW, et al. Acquired Resistance to KRAS G12C Inhibition in Cancer. N Engl J Med (2021) 384(25):2382–93. doi: 10.1056/NEJMoa2105281
112. Amgen Announces New LUMAKRASTM (Sotorasib) Combination Data From Phase 1b CodeBreaK 101 StudyPatients With KRAS G12C-Mutated Cancers At AACR-NCI-EORTC 2021| Amgen. Available at: https://www.amgen.com/newsroom/press-releases/2021/10/amgen-announces-new-lumakras-sotorasib-combination-data-from-phase-1b-codebreak-101-study-in-patients-with-kras-g12cmutated-cancers-at-aacrncieortc-2021.
113. Grassilli E, Cerrito MG, Bonomo S, Giovannoni R, Conconi D, Lavitrano M. P65btk Is a Novel Biomarker and Therapeutic Target in Solid Tumors. Front Cell Dev Biol (2021) 9. doi: 10.3389/fcell.2021.690365
114. Koltun E, Cregg J, Rice MA, Whalen DM, Freilich R, Jiang J, et al. Abstract 1260: First-In-Class, Orally Bioavailable KRASG12V(ON) Tri-Complex Inhibitors, as Single Agents and in Combinations, Drive Profound Anti-Tumor Activity in Preclinical Models of KRASG12V Mutant Cancers. Cancer Res (2021) 81(13 Supplement):1260–0. doi: 10.1158/1538-7445.AM2021-1260
115. Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras Copy Number Defines Metabolic Reprogramming and Therapeutic Susceptibilities. Nature (2016) 531(7592):110. doi: 10.1038/nature16967
116. Bartolacci C, Padanad M, Andreani C, Melegari M, Rindhe S, George K, et al. Fatty Acid Synthase Is a Therapeutic Target in Mutant KRAS Lung Cancer. J Thorac Oncol (2017) 12(8):S1538. doi: 10.1016/j.jtho.2017.06.030
117. Falchook G, Infante J, Arkenau HT, Patel MR, Dean E, Borazanci E, et al. First-In-Human Study of the Safety, Pharmacokinetics, and Pharmacodynamics of First-In-Class Fatty Acid Synthase Inhibitor TVB-2640 Alone and With a Taxane in Advanced Tumors. EClinicalMedicine (2021) 34:100797. doi: 10.1016/j.eclinm.2021.100797
118. Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, et al. LKB1 Inactivation Dictates Therapeutic Response of Non-Small Cell Lung Cancer to the Metabolism Drug Phenformin. Cancer Cell (2013) 23(2):143–58. doi: 10.1016/j.ccr.2012.12.008
119. Jones PA, Baylin SB. The Epigenomics of Cancer. Cell (2007) 128(4):683–92. doi: 10.1016/j.cell.2007.01.029
120. Luger K. Structure and Dynamic Behavior of Nucleosomes. Curr Opin Genet Dev (2003) 13(2):127–35. doi: 10.1016/S0959-437X(03)00026-1
121. Strahl BD, Allis CD. The Language of Covalent Histone Modifications. Nature (2000) 403(6765):41–5. doi: 10.1038/47412
122. Lane AA, Chabner BA. Histone Deacetylase Inhibitors in Cancer Therapy. J Clin Oncol (2009) 27(32):5459–68. doi: 10.1200/JCO.2009.22.1291
123. Gregory PD, Wagner K, Hörz W. Histone Acetylation and Chromatin Remodeling. Exp Cell Res (2001) 265(2):195–202. doi: 10.1006/excr.2001.5187
124. Park WY, Hong BJ, Lee J, Choi C, Kim MY. H3K27 Demethylase JMJD3 Employs the NF-κb and BMP Signaling Pathways to Modulate the Tumor Microenvironment and Promote Melanoma Progression and Metastasis. Cancer Res (2016) 76(1):161–70. doi: 10.1158/0008-5472.CAN-15-0536
125. Dawson MA, Kouzarides T. Cancer Epigenetics: From Mechanism to Therapy. Cell (2012) 150(1):12–27. doi: 10.1016/j.cell.2012.06.013
126. Ansari J, Shackelford RE, El-Osta H. Epigenetics in Non-Small Cell Lung Cancer: From Basics to Therapeutics. Trans Lung Cancer Res (2016) 5(2):155–71. doi: 10.21037/tlcr.2016.02.02
127. Liu W, Yin Y, Wang J, Shi B, Zhang L, Qian D, et al. Kras Mutations Increase Telomerase Activity and Targeting Telomerase Is a Promising Therapeutic Strategy for Kras-Mutant NSCLC. Oncotarget (2017) 8(1):179–90. doi: 10.18632/oncotarget.10162
128. Greve G, Schiffmann I, Pfeifer D, Pantic M, Schüler J, Lübbert M. The Pan-HDAC Inhibitor Panobinostat Acts as a Sensitizer for Erlotinib Activity in EGFR-Mutated and -Wildtype Non-Small Cell Lung Cancer Cells. BMC Cancer (2015) 15(1):155–71. doi: 10.1186/s12885-015-1967-5
129. Yamada T, Amann JM, Tanimoto A, Taniguchi H, Shukuya T, Timmers C, et al. Histone Deacetylase Inhibition Enhances the Antitumor Activity of a MEK Inhibitor in Lung Cancer Cells Harboring RAS Mutations. Mol Cancer Ther (2018) 17(1):17–25. doi: 10.1158/1535-7163.MCT-17-0146
130. Chiappori AA, Kolevska T, Spigel DR, Hager S, Rarick M, Gadgeel S, et al. A Randomized Phase II Study of the Telomerase Inhibitor Imetelstat as Maintenance Therapy for Advanced Non-Small-Cell Lung Cancer. Ann Oncol (2015) 26(2):354–62. doi: 10.1093/annonc/mdu550
131. Iskandar K, Rezlan M, Yadav SK, Foo CHJ, Sethi G, Qiang Y, et al. Synthetic Lethality of a Novel Small Molecule Against Mutant KRAS-Expressing Cancer Cells Involves AKT-Dependent ROS Production. Antioxidants Redox Signaling (2016) 24(14):781–94. doi: 10.1089/ars.2015.6362
132. Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, et al. Oncogenic K-Ras Decouples Glucose and Glutamine Metabolism to Support Cancer Cell Growth. Mol Syst Biol (2011) 7:354–62. doi: 10.1038/msb.2011.56
133. Gwinn DM, Lee AG, Briones-Martin-del-Campo M, Conn CS, Simpson DR, Scott AI, et al. Oncogenic KRAS Regulates Amino Acid Homeostasis and Asparagine Biosynthesis via ATF4 and Alters Sensitivity to L-Asparaginase. Cancer Cell (2018) 33(1):91–107.e6. doi: 10.1016/j.ccell.2017.12.003
134. Sunaga N, Shames DS, Girard L, Peyton M, Larsen JE, Imai H, et al. Knockdown of Oncogenic KRAS in Non-Small Cell Lung Cancers Suppresses Tumor Growth and Sensitizes Tumor Cells to Targeted Therapy. Mol Cancer Ther (2011) 10(2):336–46. doi: 10.1158/1535-7163.MCT-10-0750
135. Zhang YA, Nemunaitis J, Scanlon KJ, Tong AW. Anti-Tumorigenic Effect of a K-Ras Ribozyme Against Human Lung Cancer Cell Line Heterotransplants in Nude Mice. Gene Ther (2000) 7(23):2041–50. doi: 10.1038/sj.gt.3301331
136. Stathis A, Bertoni F. BET Proteins as Targets for Anticancer Treatment. Cancer Discov (2018) 8(1):24–36. doi: 10.1158/2159-8290.CD-17-0605
137. Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET Bromodomain Inhibition as a Therapeutic Strategy to Target C-Myc. Cell (2011) 146(6):904–17. doi: 10.1016/j.cell.2011.08.017
138. de Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, et al. PRC2 Loss Amplifies Ras-Driven Transcription and Confers Sensitivity to BRD4-Based Therapies. Nature (2014) 514(7521):247–51. doi: 10.1038/nature13561
139. Guerra SL, Maertens O, Kuzmickas R, de Raedt T, Adeyemi RO, Guild CJ, et al. A Deregulated HOX Gene Axis Confers an Epigenetic Vulnerability in KRAS-Mutant Lung Cancers. Cancer Cell (2020) 37(5):705–19.e6. doi: 10.1016/j.ccell.2020.03.004
140. Abate-Shen C. Deregulated Homeobox Gene Expression in Cancer: Cause or Consequence? Nat Rev Cancer (2002) 2(10):777–85. doi: 10.1038/nrc907
141. Fabrizio FP, Mazza T, Castellana S, Sparaneo A, Muscarella LA. Epigenetic Scanning of Keap1 Cpg Sites Uncovers New Molecular-Driven Patterns in Lung Adeno and Squamous Cell Carcinomas. Antioxidants (2020) 9(9):1–19. doi: 10.3390/antiox9090904
Keywords: KRAS, lung cancer, mutations, precision medicine, targeted therapy
Citation: Ferrara MG, Stefani A, Pilotto S, Carbone C, Vita E, Di Salvatore M, D’Argento E, Sparagna I, Monaca F, Valente G, Vitale A, Piro G, Belluomini L, Milella M, Tortora G and Bria E (2021) The Renaissance of KRAS Targeting in Advanced Non-Small-Cell Lung Cancer: New Opportunities Following Old Failures. Front. Oncol. 11:792385. doi: 10.3389/fonc.2021.792385
Received: 10 October 2021; Accepted: 06 December 2021;
Published: 23 December 2021.
Edited by:
Idris Bahce, Academic Medical Center, NetherlandsReviewed by:
Roberto Ferrara, National Cancer Institute Foundation (IRCCS), ItalyChristopher Gerard Azzoli, Brown University, United States
Copyright © 2021 Ferrara, Stefani, Pilotto, Carbone, Vita, Di Salvatore, D’Argento, Sparagna, Monaca, Valente, Vitale, Piro, Belluomini, Milella, Tortora and Bria. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emilio Bria, ZW1pbGlvLmJyaWFAdW5pY2F0dC5pdA==
†These authors share first authorship
‡These authors share last authorship
 Miriam Grazia Ferrara
Miriam Grazia Ferrara Alessio Stefani
Alessio Stefani Sara Pilotto3
Sara Pilotto3 Carmine Carbone
Carmine Carbone Ettore D’Argento
Ettore D’Argento Ileana Sparagna
Ileana Sparagna Geny Piro
Geny Piro Lorenzo Belluomini
Lorenzo Belluomini Michele Milella
Michele Milella Emilio Bria
Emilio Bria