- 1Department of Microbiology, Guilin Medical University, Guilin, China
- 2Key Laboratory of Tumor Immunology and Microenvironmental Regulation, Guilin Medical University, Guilin, China
- 3Department of Physiology, Guilin Medical University, Guilin, China
Background: Hepatocellular carcinoma (HCC) is one of the most common malignant tumors, the pathogenesis of which remains unclear. Mediator complex subunit 19 (MED19), a subunit of the Mediator complex, is a multi-protein co-activator necessary for DNA transcription factors to induce RNA polymerase II transcription. In the current study, we aimed to study the role of MED19 in HCC and elucidate its mechanism.
Methods: MED19 expression in HCC tissues was determined. The relationship between MED19 and the clinical prognosis was explored. The influence of MED19 on HCC cell viability, migration, invasion, and apoptosis was studied. The expression of AKT/mTOR pathway genes and proteins was detected by qRT-PCR and western blot. The correlation between MED19 and immune infiltration was investigated.
Results: MED19 was upregulated in HCC tissues compared with tumor-adjacent tissues, and was associated with a poor prognosis. Furthermore, high MED19 expression was correlated with race, gender, etc. Knockdown of MED19 inhibited cell proliferation, migration, invasion, and promoted apoptosis. Knockdown of MED19 decreased p-AKT and p-mTOR protein expression. Additionally, the downstream effectors of the AKT/mTOR pathway, p70S6K1 and 4EBP1, were affected by MED19. Notably, MED19 expression was positively correlated with the infiltration levels of B cells, CD4+ T cells, CD8+ T cells, macrophages, etc.
Conclusion: MED19 is significantly upregulated in HCC tissues and cells. MED19 may promote the progression of HCC in vitro and may be related to immune infiltration. Together, our data show that MED19 could be considered as a new possible biomarker as well as a novel therapeutic target for HCC.
Introduction
Hepatocellular carcinoma (HCC) is the most common primary malignancy of hepatocytes and serves as the third leading cause of cancer-related death worldwide (1).The treatment of HCC is limited. At present, surgery is the main treatment strategy for early HCC; however, following surgery recurrence and metastasis are common. Patients with advanced HCC are prescribed sorafenib, a specific molecular targeting drug, but drug resistance and side effects have become serious problems (2, 3). Furthermore, the prognosis of HCC is very poor with the 5-year survival rate reported at less than 5% (4). Therefore, researchers and clinicians must elucidate the molecular mechanisms of the occurrence, development, invasion, and metastasis of HCC to find and develop new therapeutic targets.
Mediator is an evolutionarily conserved multi-protein complex (5, 6). As an important part of the transcription mechanism of eukaryotes, the Mediator complex participates in gene expression and mediates the interaction between different proteins (7–9). In humans, mutations, or changes in the Mediator complex have a wide-ranging impact on the occurrence and development of a variety of diseases including cancer.
In 2003, MED19, or lung cancer metastasis-associated protein 1, was cloned from lung large carcinoma and found to be an important part of the Mediator complex (10–12). MED19 is mainly confined to the nuclear region of the cell (https://www.proteinatlas.org/ENSG00000156603-MED19/cell). Further, several studies have shown that MED19 plays a key role in malignant tumor growth by regulating signals related to cell growth, differentiation, cell cycle, and apoptosis (13–16). Zhang et al. found that the expression of MED19 in breast cancer tissues was significantly higher than that in adjacent tissues (17, 18). Further, MED19 promoted breast cancer cell proliferation through the EGFR/MEK/ERK signaling pathway (19). In addition, studies have reported that the expression of MED19 was positively correlated with the expression of bone morphogenetic protein 2 (BMP2) in bladder urothelial carcinoma bone metastasis and invasion (20). MED19 transcription can activate the expression of endogenous Tspan8, and regulate the adhesion and invasion of melanoma in a Tspan8-dependent manner (21). As a member of the Mediator complex, MED19 plays a key role in the activation and inhibition of tumor signal transduction and transcriptional regulation and has a role in the induction of other developmental diseases. Zou et al. (2011) reported that the inhibition of MED19 reduced HCC cellular proliferation, induced cell-cycle arrest, and suppressed tumor formation (22). However, the specific mechanism explaining how MED19 affects the occurrence and development of HCC is unclear. Therefore, an in-depth understanding of the biological function of MED19 and its mechanism of action in HCC might be helpful in the identification of potential targets for clinical treatment.
In the present study, we show that MED19 is upregulated in HCC tissues and that MED19 upregulation was closely correlated with a poor prognosis. Furthermore, MED19 knockdown was observed to modulate the migration, invasion, and apoptosis of HCC cells, and may promote the occurrence and development of HCC through the AKT/mTOR pathway. Additionally, MED19 was correlated with tumor immune infiltration. Together, this study shows that MED19 plays an important oncogenic role in the occurrence and development of HCC, and could be considered as a newpossible biomarker as well as a novel therapeutic target for HCC.
Materials and Methods
Public Datasets Analysis
Tumor Immune Estimation Resource (TIMER) 2.0 is a comprehensive resource for the systematic assessment of diverse cancer types. Using the TCGA database, TIMER 2.0 explores the differential gene expression between tumors and normal tissues, the correlation between gene expression and clinical results, and the analysis of tumor immune infiltration (23). Gene Expression Profiling Interactive Analysis (GEPIA) is a web server based on the visual analysis of the TCGA database (24). It provides several key interactive customization features, such as differential expression analysis, patient survival analysis, and related gene detection, etc. UALCAN is a comprehensive, user-friendly, and interactive web resource for analyzing cancer data (25). LinkedOmics is a platform that can access, analyze and compare cancer multi-omics data within and across tumor types (26). In the current study, the TIMER 2.0 database was used to analyze immune infiltration and the expression profile of MED19 in different types of human cancers. Additionally, using the GEPIA datasets, the survival analysis of MED19 in HCC was evaluated. Using the UALCAN dataset, factors related to MED19 transcription in HCC were investigated. Using the LinkedOmics datasets, genes in HCC that are related to MED19 were assessed and used to draw heat maps. Finally, the relationship between MED19 and several key HCC regulators, including AKT and mTOR was assessed.
Immunohistochemistry
Human HCC tissue specimens and adjacent non-tumor tissues were obtained from patients who underwent surgical hepatectomy. All patients were sourced from the Second Affiliated Hospital of Guilin Medical University, China and signed informed consent. Immunohistochemical staining analyses were performed using formalin-fixed paraffin-embedded tissue sections. The sections were deparaffinized, rehydrated, and incubated in EDTA at 120°C for 5 minutes for antigen repair. After incubating with 3% H2O2 at room temperature for 15 minutes, the sections were sealed with fetal bovine serum and incubated with primary antibody overnight. Goat anti-rabbit antibody coupled with HRP was used for immune detection. Finally, the immune complex was displayed with chromogenic substrate, and the sections were re-stained with hematoxylin. In order to reduce the non-specific binding of antibody, titration was used to optimize the concentration. The diagnosis of the liver cancer samples was verified by pathologists.
Cell Lines and Cell Culture
The human hepatocellular carcinoma cell lines HepG2 and Huh7 are generated by our laboratory. The HepG2 and Huh7 cells were maintained in DMEM high glucose medium (Gibco; Thermo Fisher Scientific) containing 10% serum (South American Fetal Bovine, EXCELL).
Plasmid Construction and Cell Transfection
The MED19 knockdown plasmid was designed by Genechem, Shanghai, China. The sh-MED19 sequence was as follows: sense 5’-CAGTACTCTTTCAATCCTAT-3’, irrelevant nucleotides not targeting any annotated human genes were used as the negative control (sh-NC): sense 5’-TTCTCCGAACGTGTCACGT-3’. Cell transfection with plasmids was conducted using Lipofectamine 2000 (Invitrogen, USA) in accordance with the manufacturer’s instructions.
Cell Proliferation Analysis
Cell proliferation was measured by the MTT assay (Solarbio, China). Briefly, the cells were transfected with the sh-MED19 plasmid or corresponding negative control and were then seeded into 96- well plates (5×103 cells/well), and cultured at 37°C for 0 h, 24 h, 48 h, 72 h. After incubation, 20 ul of the MTT solution was added to each well and incubated for 4 h at each time point at 37°C. The MTT solution was aspirated and 200ul DMSO (Solarbio, China) was added to each well to dissolve the formazan crystals. Cellular proliferation in each well was quantified by measuring optical density using an EPOCH 2 microplate spectrophotometer at a wavelength of 490 nm. The transfected cells were used for cell colony formation assay. Approximately, 100-1000 cells were added per well in a six-well plate. The colonies were then fixed with 4% paraformaldehyde, stained with crystal violet solution and counted.
Cell Migration and Invasion Assay
Cell migratory ability was assessed using the scratch assay. After transfection with the targeted plasmid, 106 cells were seeded in a six-well plate with DMEM containing 2% serum. After adhering to the well, the cells were scratched with a 10 ul pipette tip. The migration distance was measured at 0 h, 24 h and 48 h, and the migration capacity was calculated. Cell invasion was measured in a 24-well plate and transwell chamber covered with Matrigel (CORNING, USA). The transfected cells were resuspended in serum-free media and counted. The upper chamber was inoculated with 105 cells per well, and 10% serum media was added to the lower chamber of the 24-well plate as an inducer to trigger cell invasion. After 36 h, the bottom of the chamber was fixed with 4% paraformaldehyde, stained with 1% crystal violet and counted (100X).
Cell Apoptosis Assay
After cell transfection for 24 h, the 6-well plate was redigested to observe the transfection efficiency. A sterile cover glass was placed in the 6-well plate in advance. The cells were then added to the 6 well-plate and cultured overnight to a density of approximately 50-80%. Following the culture period, the cells were fixed with 0.5 ml fixative solution for 10 min, stained with 0.5 ml Hoechst 33258 staining solution for 5 min, washed with anti-fluorescence quenching solution, and observed under a fluorescence microscope. Normal cell nuclei appeared blue, apoptotic nuclei were densely stained, or fragmented and densely stained, and whitish.
Flow Cytometry for Apoptosis Detection
After 24 h of cell transfection to observe the transfection efficiency, apoptosis was measured using the Annexin V-Phycoerytirin (Annexin V-PE) cell apoptosis detection kit (C1065, Beyotime Biotechnology, China). These data were acquired by flow cytometry (Thermo Fisher, USA) and analyzed by Flowjo software.
qRT-PCR
Total RNA was extracted using the TRIZOL. RNA was reverse transcribed to cDNA using the qRT-PCR kit (Thermo Scientific, USA) according to the manufacturer’s protocol. The PCR cycling conditions were as follows: 40 cycles with pre-denaturation at 95°C for 15 min, denaturation at 95°C for 10 s, and annealing and extension at 60°C for 32 s.
The primers used in this study were synthesized by Wuhan Genecreate Biological Engineering Co., Ltd, and were as follows: MED19-F: CTGACAGGCAGCACGAATCT, MED19-R: CTCCTTCACCTTCTTCCCACA; AKT-F: TACTCTTTCCAGACCCACGAC, AKT-R: AGGTTCTCCAGCTTGAGGTC; mTOR-F: CGCTGTCATCCCTTTATCGAC, mTOR-R: CAGAGTCAAGTGGTCATAGTCCG; 4EBP1-F: CTCACCTGTACCAAAACACC, 4EBP1-R: CCCGCTTATCTTCTGGGCTA; p70S6K1-F: GTGCTGTGGATTGGTGGAGT, p70S6K1-R: GAGGTAGGGAGGCAAATTGAG; GAPDH-F: AGAAGGCTGGGGCTCATTTG, GAPDH-R: AGGGGCCATCCACAGTCTTC. All samples were normalized to internal controls and fold changes were calculated based on relative quantification (2-ΔΔCt).
Western Blot
48 h after cell transfection, the protein samples were extracted with high-efficiency RIPA cell lysis buffer and protease inhibitor (100:1). Then, the protein samples were electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel, then transferred to polyvinylidene fluoride (PVDF) membrane and blocked with 5% skimmed milk powder for 2 h. The samples were incubated with the primary antibody overnight. All antibodies used in this study are shown in Table 1. The membrane was washed three times, incubated with the corresponding enzyme-labeled secondary antibodies for 1 h, including horseradish peroxidase (HRP) – conjugated anti-rabbit (cat. no. 111-035-003, 1:10000; Jackson ImmunoResearch, USA), and horseradish peroxidase (HRP) – conjugated anti-mouse (cat. no. 115-035-003, 1:10000; Jackson ImmunoResearch, USA). The signals of the protein bands were analyzed using ChemiDoc XRS+ biomolecular imaging system (BIO-RAD, USA). Analysis using Image J software.
Statistical Analysis
All in vitro experiments were performed in triplicate. All statistical calculations and analyses were performed using GraphPad Prism 8.0.2 software (GraphPad Software, San Diego, CA, USA). The data were presented as mean ± SD. The comparison between the two groups was performed using by unpaired Student’s t -test (for parametric data). P values <0.05 were considered statistically different.
Results
MED19 Was Up-Regulated in Hepatocellular Carcinoma
In this study, the carcinogenic effect of MED19 during HCC was investigated. First, we analyzed the expression patterns of MED19 in different tumor and non-tumor tissues. Using TIMER 2.0, MED19 was shown to be highly expressed for bladder urothelial carcinoma (BLCA), breast cancer (BRCA), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD) and other types in the TCGA project (Figure 1A). Based on the GEPIA dataset, matching TCGA normal and GTEx data, MED19 expression was found to be significantly higher in HCC tissues relative to other non-tumor tissues (Figure 1B). Based on the TCGA dataset, MED19 expression was highest in stage III HCC tissues (Figure 1C). The high expression of MED19 was associated with overall survival and disease-free survival, suggesting a poor prognosis (Figures 1D, E). In addition, within the clinical specimens collected, MED19 was highly expressed in cancer tissues compared with paracancerous tissues (Figure 1F). The level of MED19 transcription was significantly higher in HCC patients relative to healthy subjects in the subgroup analysis based on gender, age, race, tumor grade, etc. (Figure 2). Therefore, the expression of MED19 may serve as a potential diagnostic indicator in HCC.
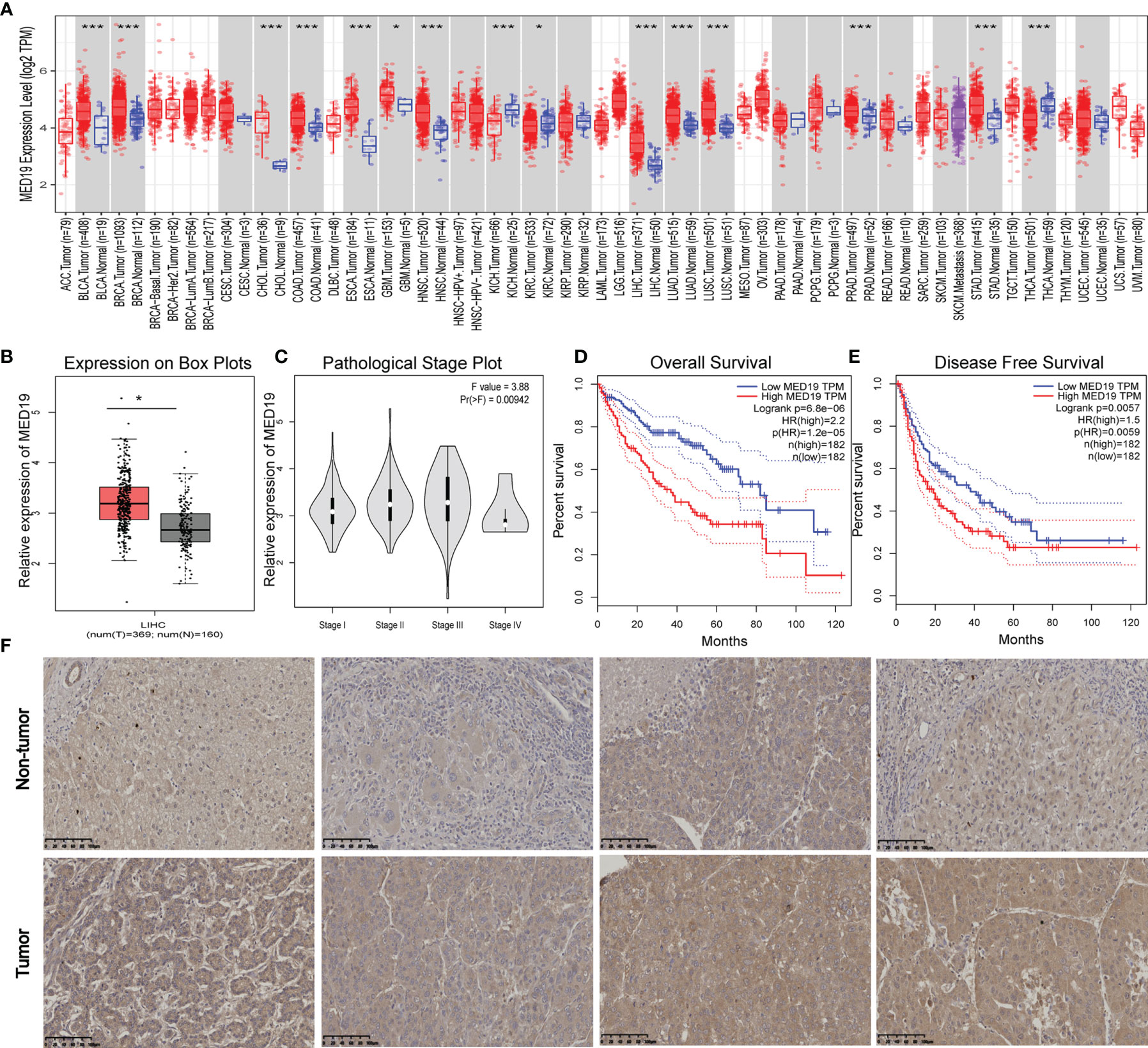
Figure 1 MED19 is highly expressed in HCC. (A) Expression of MED19 gene in different cancers or specific cancer subtypes. (B) MED19 was upregulated in LIHC samples compared with normal tissues. (C) Based on the TCGA data, the expression of the MED19 at different pathological stages of LIHC. (D, E) The higher expression of MED19 was associated with a shorter overall survival time and disease-free survival time of HCC. (F) IHC images of HCC tissues showed that the expression of MED19 in HCC tissues was higher than that in adjacent non-tumor tissues. *P < 0.05; ***P < 0.001.
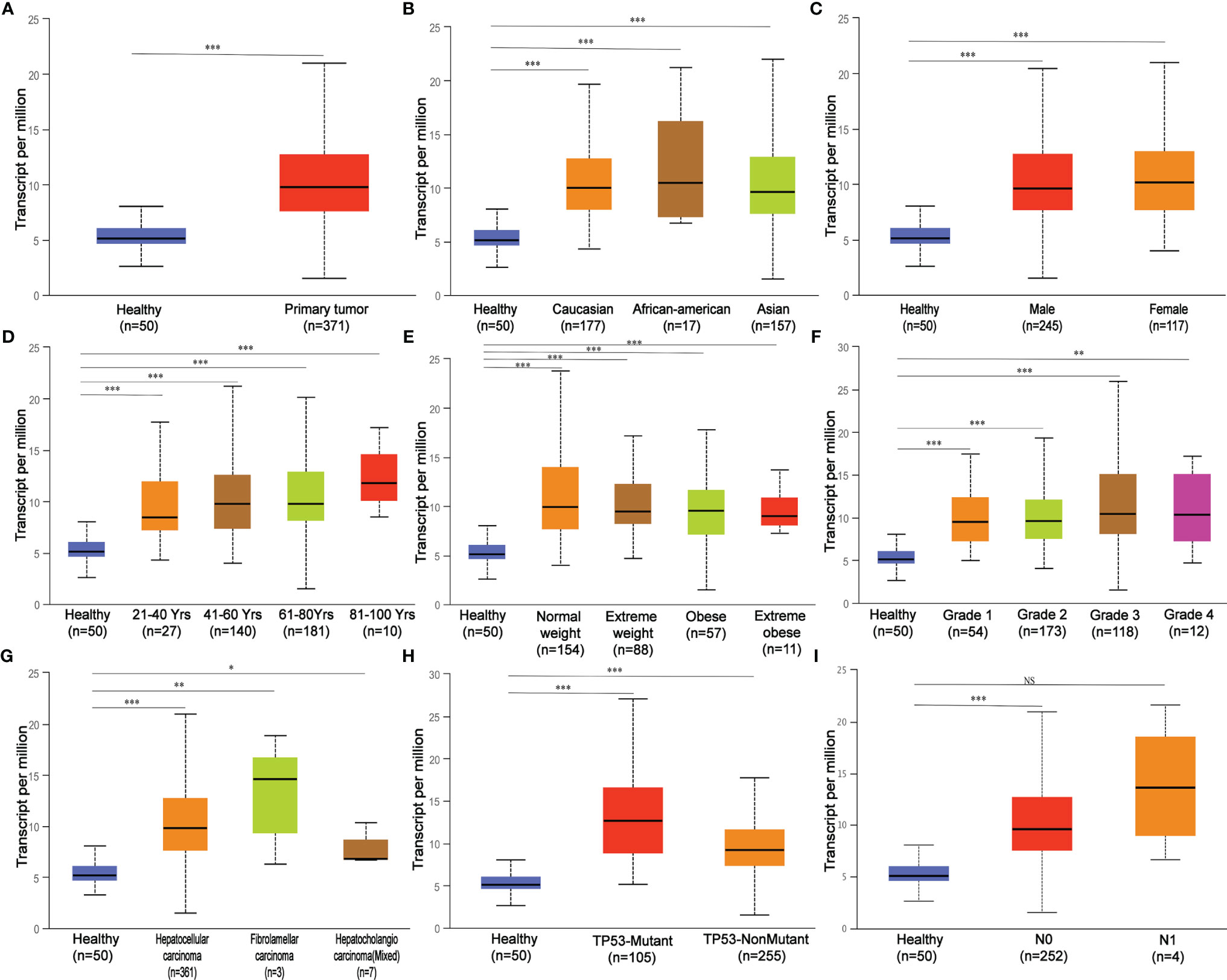
Figure 2 MED19 transcription in subgroups of patients with hepatocellular carcinoma stratified based on gender, age, and other criteria. (A) Boxplot showing relative expression of MED19 in healthy and LIHC samples. (B) Boxplot showing relative expression of MED19 in healthy individuals of any ethnicity or LIHC patients of Caucasian, African-American or Asian ethnicity. (C) Boxplot showing relative expression of MED19 in healthy individuals of either gender or male or female LIHC patients. (D) Boxplot showing relative expression of MED19 in healthy individuals of any age or LIHC patients aged 21-40, 40-60, 61-80, or 81-100 yr. (E) Boxplot showing relative expression of MED19 in healthy individuals of any weight or LIHC patients weighted normal, extreme, obese, or extreme obese. (F) Boxplot showing relative expression of MED19 in healthy individuals or LIHC patients with grade 1, 2, 3 or 4 tumors. (G) Boxplot showing relative expression of MED19 in healthy individuals or LIHC patients of hepatocellular carcinoma, fibrolamellar carcinoma, or hepatocholangio carcinoma (mixed). (H) Boxplot showing relative expression of MED19 in healthy individuals or LIHC patients with TP53-mutant or TP53- nonmutant. (I) Boxplot showing relative expression of MED19 in healthy individuals or LIHC patients with no regional lymph node metastasis or metastases in 1 to 3 axillary lymph nodes. *P < 0.05; **P < 0.01; ***P < 0.001; NS, nonsignificant.
MED19 Knockdown Inhibited Proliferation and Promoted Apoptosis of HCC Cell
In the current study, we used HepG2 and Huh7 cells as knockdown models to explore the potential biological function of MED19. shRNA was transfected into cells within a plasmid vector. The transfection efficiency was evaluated by fluorescence microscope and knockdown efficiency was evaluated by western blot and qRT-PCR (Figures 3A, B). The proliferation ability of Huh7 and HepG2 cells was detected by the MTT and colony formation assays. The downregulation of MED19 significantly inhibited cell proliferation (Figure 3C) and colony formation efficiency (Figure 3D). Due to a higher transfection efficiency of HepG2 relative to Huh7, fewer HepG2 colonies were observed. In addition, we found that the apoptotic ability of HCC cells increased significantly after MED19 was knockdown by Hoechst and flow cytometry (Figures 3E, F).
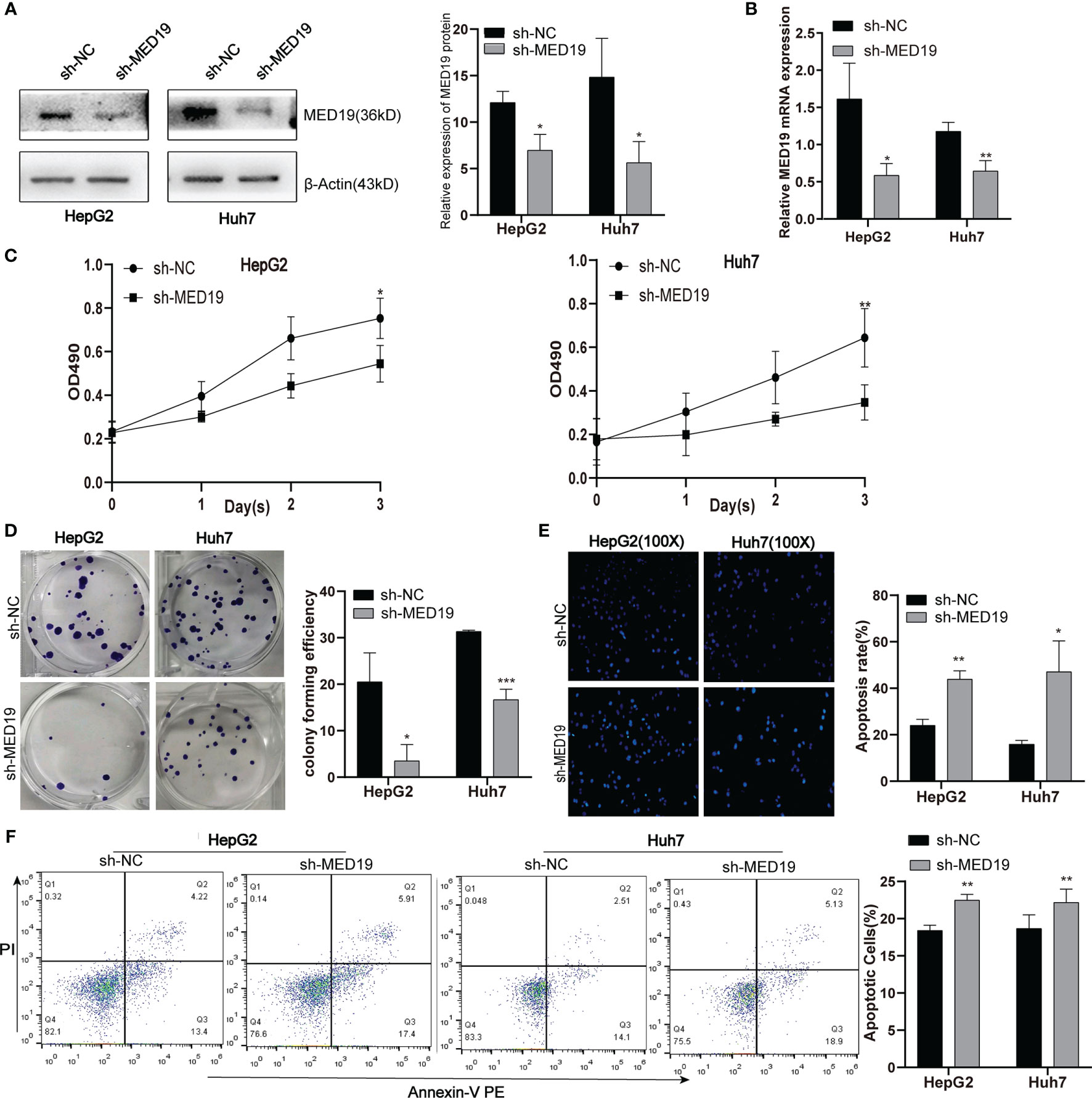
Figure 3 MED19 knockdown inhibited HCC cell proliferation and promoted cell apoptosis. (A, B) Relative MED19 RNA and protein levels in HepG2 and Huh7 cells after transfection with sh-MED19 or sh-NC plasmid. (C) The viability of HepG2 and Huh7 cells in the sh-MED19 and sh-NC group. (D) Influence of MED19 knockdown on clone formation of HepG2 and Huh7 cells. (E) The apoptosis situation of HepG2 and Huh7 cells induced by Hoechst 33258. (F) The apoptosis situation of HepG2 and Huh7 cells by flow cytometry. *P < 0.05; **P < 0.01; ***P < 0.001. Perform three independent replicates. Statistical analysis is performed using an unpaired t -test. All results are expressed as mean ± SD.
MED19 Knockdown Inhibited HCC Cell Migration and Invasion
To explore the effect of MED19 on cell migration and invasion, a transwell experiment was performed. The number of cells that crossed the chamber in the sh-MED19 group was significantly lower than the sh-NC group (Figures 4A, D). The invasion experiment was performed in a chamber containing Matrigel. The knockdown of MED19 inhibited the invasion ability of HepG2 and Huh7 cells (Figures 4B, E). Wound-healing experiments further confirmed the influence of MED19 expression on the migration ability of HCC cells. The migration rate of HCC cells in the sh-MED19 group was markedly lower than that in the sh-NC group (Figures 4C, F). Together, these data suggested that MED19 may act as an oncogene and promotes migration and invasion in HCC.
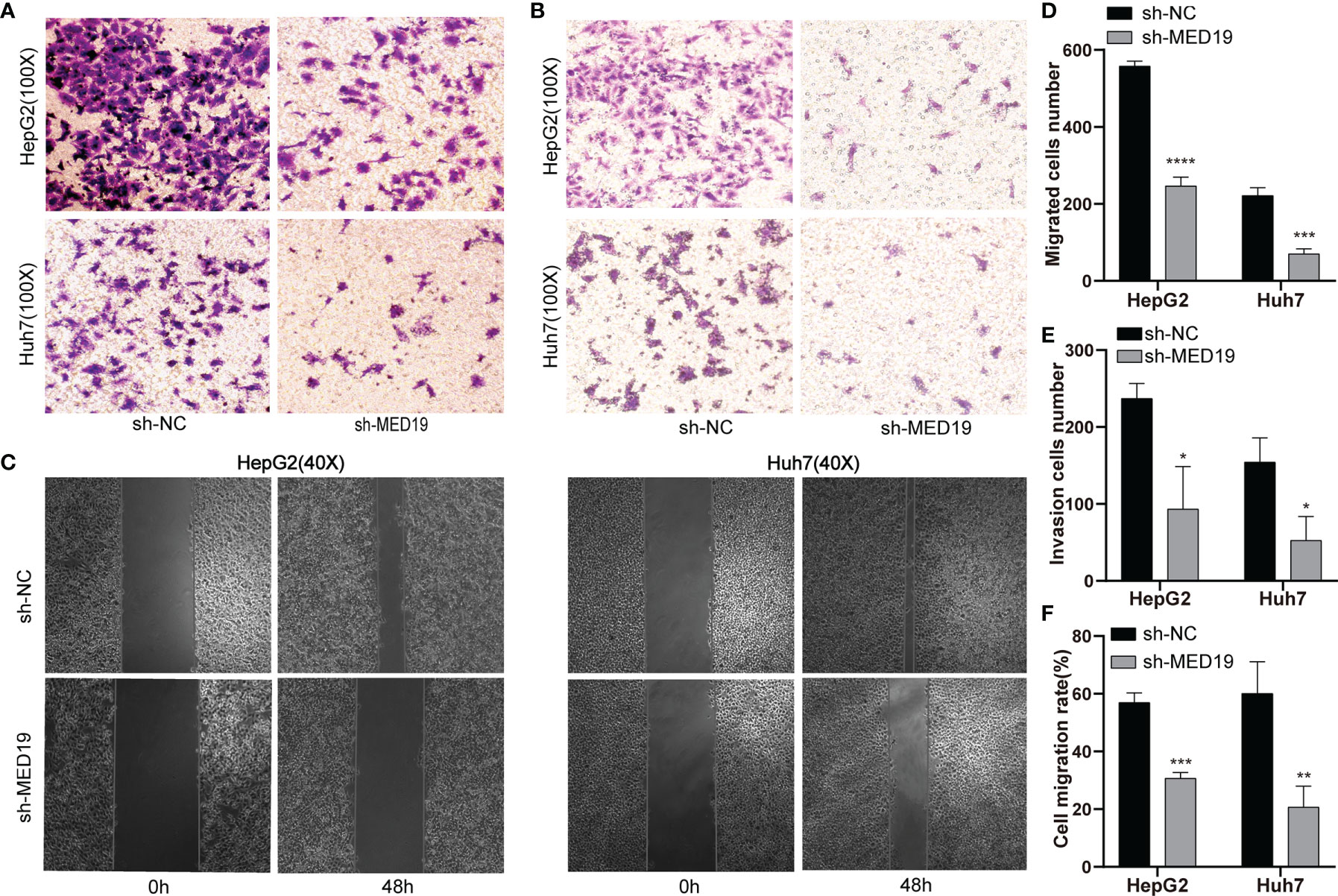
Figure 4 MED19 knockdown inhibited the migration and invasion of HCC cells in vitro. (A, D) Transwell assay was used to detect the migration of HCC cells between the sh-NC and the sh-MED19 group. (B, E) Transwell assay was used to detect the invasion of HCC cells between the sh-NC and sh-MED19 groups. (C, F) Wound-healing was utilized to evaluate the effect of MED19 knockdown on the migration of HCC cells. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. Perform three independent replicates. Statistical analysis is performed using an unpaired t -test. All results are expressed as mean ± SD.
MED19 Knockdown Inhibited the Activation of the AKT/mTOR Signaling Pathway In Vitro
To further explore the functional role of MED19 in cell proliferation, migration, invasion, and apoptosis, we aimed to identify the potential mechanism of MED19 in HCC cells. Initially, data from the TCGA database indicated that MED19 was positively correlated with AKT and mTOR (Figures 5A-C). To further explore the potential relationship between MED19 and other genes related to the AKT/mTOR signaling pathway (AKT, p-AKT, mTOR, p-mTOR, 4EBP1, p-4EBP1, p70S6K1, p-p70S6K1), proteins expression was determined by western blot. Western blot analysis revealed decreased p-AKT, p-mTOR, p-p70S6K1 protein expression, and increased p-4EBP1 expression in MED19-downregulated cells (Figures 5D, F). Together, these data indicate that the activity of the AKT/mTOR pathway was decreased in MED19-depleted HCC cells. However, after treatment with noval AKT activator, SC79, the activation of the AKT/mTOR signaling pathway in HCC was partially restored (Figures 5E, G). This suggests that SC79 increased the phosphorylation of AKT and mTOR in MED19 knocked down HCC cells, thus further supporting the hypothesis that the AKT/mTOR pathway is the molecular target of MED19 in HCC cells.
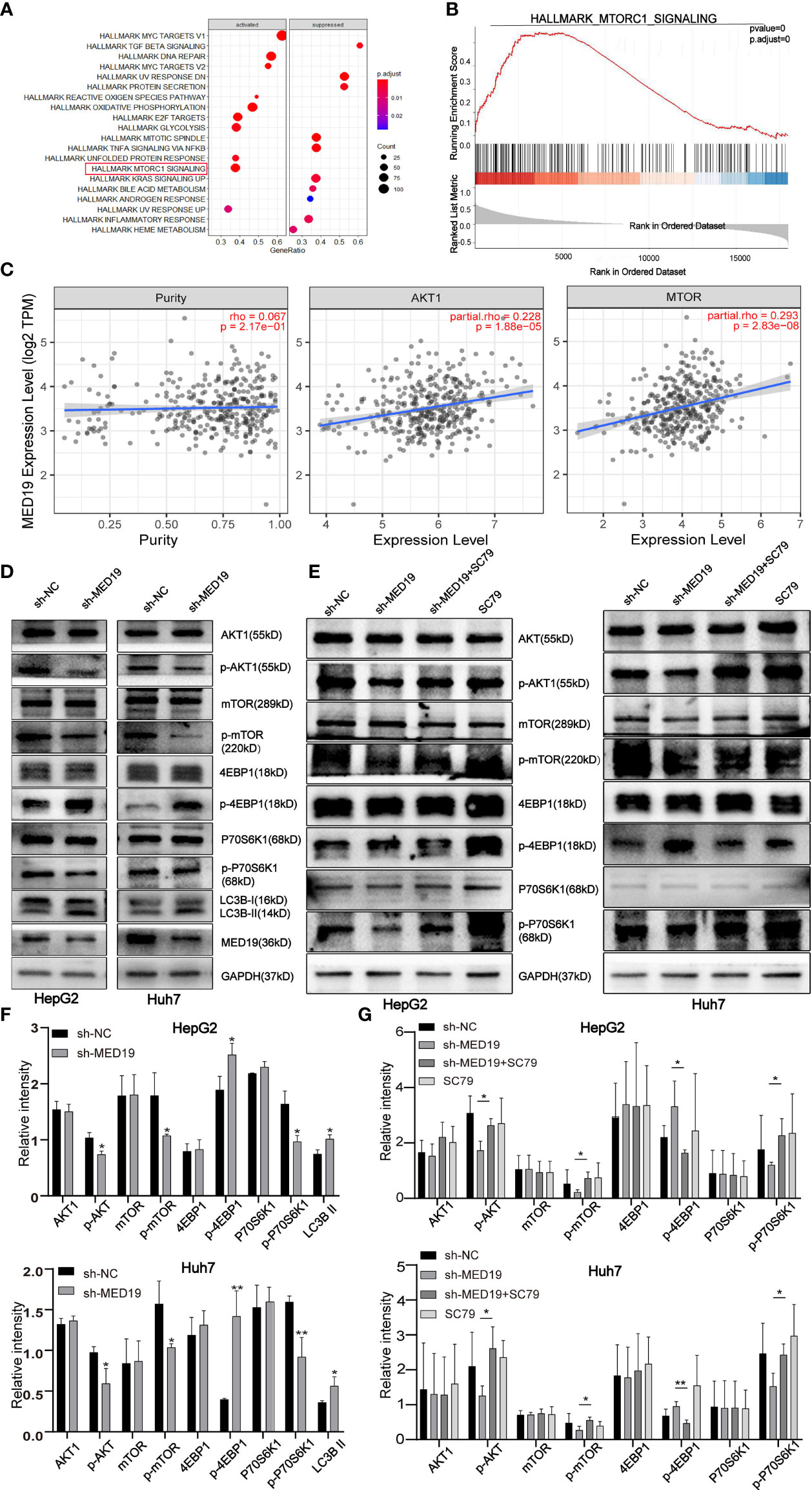
Figure 5 MED19 knockdown inhibited AKT/mTOR signaling pathway. (A, B) Pathway enrichment analysis of MED19 in HCC. (C) Co-expression analysis of MED19 and AKT/mTOR pathway genes. (D, F) The protein expression of AKT, p-AKT, mTOR, p-mTOR, p70S6K1, p-p70S6K1, 4EBP1, p-4EBP1, and LC3B I/II in HepG2 and Huh7 cells from the sh-MED19 and sh-NC groups. (E, G) SC79 enhances the expression of p-AKT, p-mTOR, and p-p70S6K1 but inhibits the expression of p-4EBP1. *P < 0.05; **P < 0.01. Perform three independent replicates. Statistical analysis is performed using an unpaired t -test. All results are expressed as mean ± SD.
Previous studies have reported that abnormal AKT/mTOR signals were closely related to autophagy (27, 28). Based on this, we speculate that MED19 may reduce autophagy in HCC cells. Therefore, WB was used to determine the expression level of key autophagy-related proteins, LC3B-I and LC3B-II. The protein level of LC3B-II in the MED19 knockdown group was higher than the corresponding control group (Figures 5D, F). In general, the downregulation of MED19 inhibited the proliferation, migration, invasion, and apoptosis of HCC cells through the AKT/mTOR signaling pathway and may be related to autophagy. In addition, using the LinkedOmics database, we found that MED19 in HCC was related to many genes. In Figure 6, we highlight genes that are positively and negatively related to MED19 in HCC (Figures 6A-C).
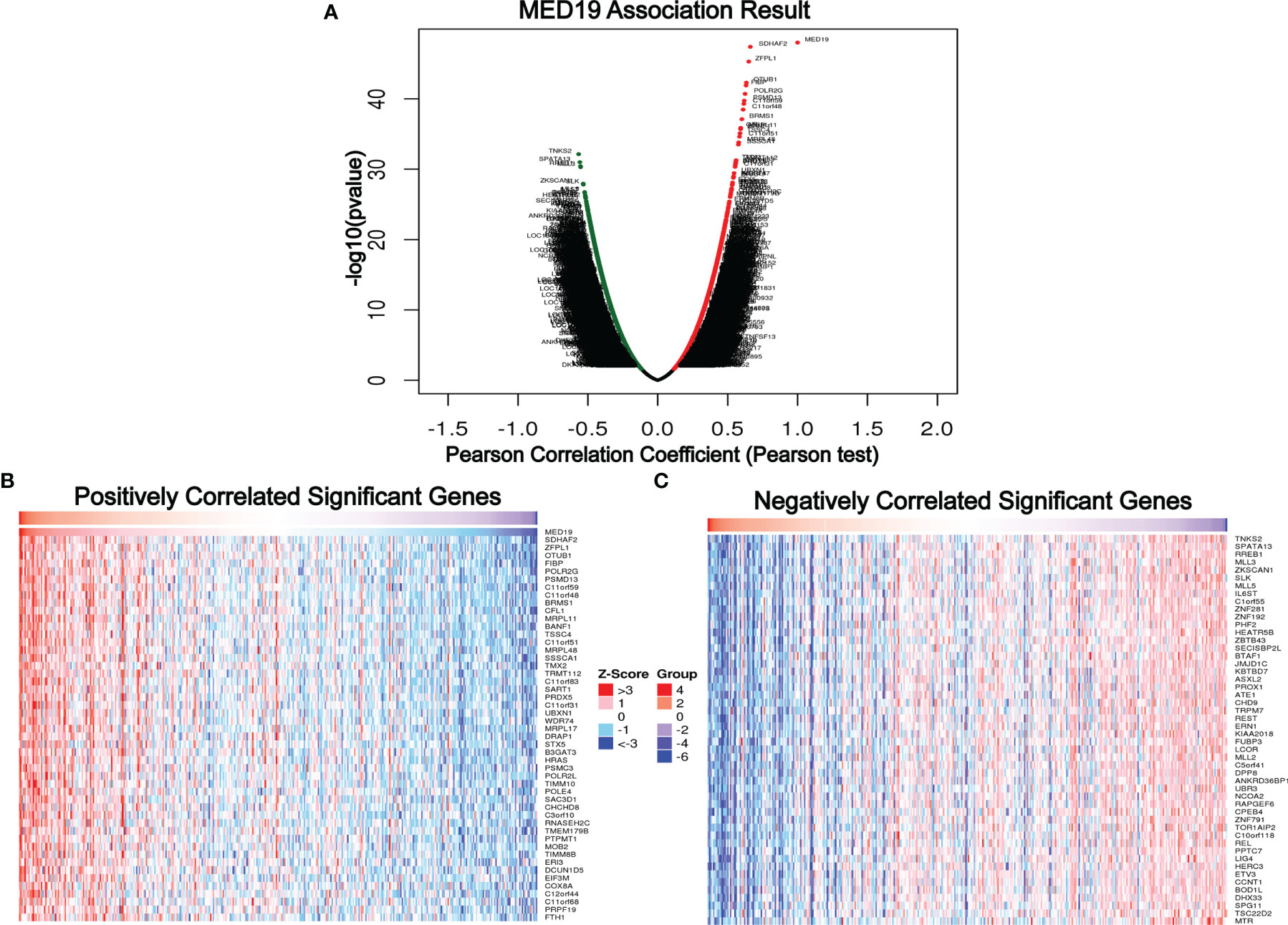
Figure 6 Genes differentially expressed in correlation with MED19 in hepatocellular carcinoma. (A) A Pearson test was used to analyze correlations between MED19 and genes differentially expressed in HCC. In the volcano map, red indicates positively correlated genes and green indicates negatively correlated genes. (B, C) Heat maps showing genes positively and negatively correlated with MED19 in HCC (TOP 50).
Correlation Between MED19 Expression and Immune Cells Infiltration and Markers of Different Subsets in HCC
In the process of tumor invasion, immune cells, as an important part of the tumor microenvironment (TME), are closely related to the occurrence and development of cancer (29–31). Cancer immunotherapy utilizes engineered auto-immune cells to eliminate tumor cells. Therefore, understanding the infiltrating immune cells in the TME is essential for deciphering the mechanism of immunotherapy, defining predictive biomarkers, and identifying new therapeutic targets. It is reported that cancer-associated fibroblasts in the tumor microenvironment matrix are involved in regulating the function of various tumor-infiltrating immune cells (32, 33). Studies have found that the expression of tumor-related genes is related to the infiltration level of CD4+ T cells, CD8+ T cells, macrophages, etc. (29, 34–40). The immune cells analyzed in HCC tissues included CD8+ T cells, B cells, tumor-associated macrophages (TAMs), monocytes, M1 and M2 macrophages, neutrophils, and natural killer (NK) cells. To explore the role of MED19 in immune responses within the HCC microenvironment, TIMER 2.0 was used to assess the potential relationship between the infiltration level of different immune cells and the expression of the MED19 gene in HCC. Because tumor purity affects the analysis of immune cell infiltration, the correlation analysis of tumor purity has been adjusted. These results indicate that the expression of MED19 in HCC was significantly correlated with the increased expression of marker genes in B cells, CD8+, CD4+, myeloid dendritic cells, macrophage, and neutrophils (Figure 7). Together, these data indicate that high MED19 expression creates an immunosuppressive microenvironment supports HCC progression. Therefore, as a potential target of HCC, MED19 may be beneficial for future immunotherapy.
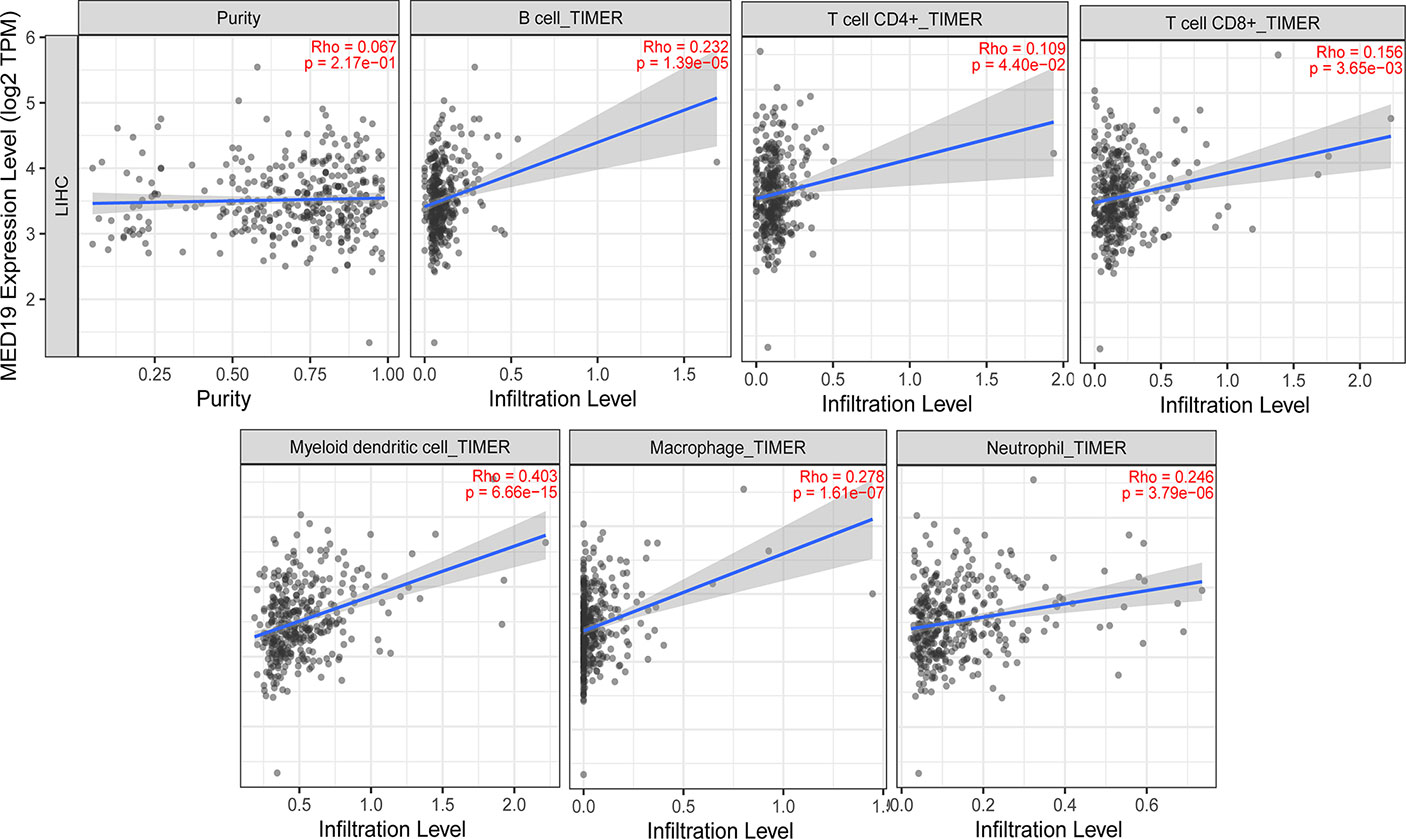
Figure 7 Correlation analysis between the expression of MED19 and the level of immune cells infiltration in HCC. After tumor purity adjustment, MED19 expression in HCC was positively correlated with infiltration levels of B cells, CD4+ T cells, CD8+ T cells, myeloid dendritic cells, macrophages, and neutrophils.
Discussion
MED19 is crucial in stabilizing Mediator’s complex’s transcriptional regulation processes (41–43). Numerous studies have shown that MED19 plays a role in tumor growth, migration, invasion, and apoptosis of various cancer types. In this study, we observed that the expression of MED19 in HCC tissue was increased compared with normal liver tissue, which was confirmed by IHC.
The high expression of MED19 in HCC patients indicates a poor prognosis, suggesting that the expression of MED19 may play an important role in HCC metastasis. Cui et al. found that inhibiting the expression of MED19 inhibited the proliferation and tumorigenesis of human prostate cancer cells (44, 45), and also inhibited tumor growth and metastasis in colorectal cancer (46). MED19 knockdown inhibited the proliferation and migration of bladder cancer cells by down-regulating the WNT/β-catenin signaling pathway (47). Given these results, through a series of experiments, we found that knocking down MED19 significantly reduced the ability of migration, invasion, proliferation, and colony-formation of HCC cells. In addition, MED19 knockdown significantly increased the proportion of apoptotic HCC cells. Together, our data showed that MED19 was related to the tumorigenesis and development of HCC, which may be a related oncogene of HCC.
Tumor immunotherapy based on immune infiltration is a current research hotspot and which still requires more investigation and optimization. Immune cells influence the tumor microenvironment and affect tumor progression and metastasis (18, 37, 48, 49). Based on many findings, we evaluated the correlation between the abnormal expression of MED19 in HCC and immune infiltrating cells. The results showed that the expression of MED19 was significantly correlated with the expression of B cells, CD8+, CD4+ and other cell-related genes.
Finally, based on the role of MED19 in the proliferation, migration, and invasion of HCC, we explored the mechanism of MED19 in the AKT/mTOR signaling pathway. The AKT/mTOR signaling pathway plays an active role in promoting tumor invasion and metastasis (50–53). Here, we show that MED19 knockdown reduced the expression of p-AKT, p-mTOR and p-P70S6K1, but increased the expression of p-4EBP1. Additionally, SC79, an AKT agonist, partially restored the expression of p-AKT, p-mTOR, p-p70S6K1, and p-4EBP1. In summary, MED19 knockdown inhibited the proliferation, migration, and invasion of HCC through the AKT/mTOR signaling pathway.
In obesity studies, adipose tissue inflammation is a key process that promotes cancer (54, 55). The tumor-promoting effect of obesity alters the level of the microenvironment and inflammatory Mediators and affects the level and function of immune infiltrating cells. Dean et al. found that MED19 regulates adipogenesis and participates in the process of fat metabolism by mediating PPAR-γ (56). In addition, MED19 binds to GATA transcription factors and regulates GATA-driven genes together with MED1 (57).
Our study showed that MED19 affects HCC oncogenesis through the AKT/mTOR pathway and may be related to autophagy. Based on this result, further experiments to determine other factors in this possible pathway are urgently needed. Primarily, these studies should aim to further determine the correlation between MED19 and autophagy. Secondly, the effect of MED19 supplementation on HCC cyclin protein Cyclin D1/B1 and apoptosis protein Bax, Bcl-2, etc. should be further elucidated. Thirdly, in vivo tumor formation experiments should be elaborated upon and the effect of MED19 supplementation on HCC should be well defined in vivo and in vitro. Lastly, future work should explore the role and mechanism of the MED19-gene(s) axis in HCC.
Conclusion
In summary, the data provided in this article show that MED19, as an oncogene, plays an important role in the proliferation, migration, and invasion of HCC cells through the AKT/mTOR signaling pathway, and may be related to autophagy. Therefore, MED19, as a potential biomarker for HCC diagnosis, may represent a potential therapeutic target for HCC treatment. Further efforts and investigations are needed to clarify the tumor-promoting mechanism of MED19.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author Contributions
All authors participated in the design and performed of the experiments. YTZ wrote the article. YTZ and PQ contributed to the bioinformatics analysis. XX, ML, and HH analyzed the data. YLZ and JY directed the study and modified the article.
Funding
This work was supported by the National Natural Science Foundation of China [NSFC, No. 82160517], the Scientific Research and Technology Development Program of Guangxi [grant number AD18281009, AD18281010], and Thousands of Young and Middle-aged Backbone Teachers in Guangxi colleges and Universities Training Plan.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of Hepatocellular Carcinoma. Hepatology (2021) 73(Suppl 1):4–13. doi: 10.1002/hep.31288
2. Gordan JD, Kennedy EB, Abou-Alfa GK, Beg MS, Brower ST, Gade TP, et al. Systemic Therapy for Advanced Hepatocellular Carcinoma: ASCO Guideline. J Clin Oncol (2020) 38(36):4317–45. doi: 10.1200/JCO.20.02672
3. Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, et al. N(6)-Methyladenosine-Modified CircRNA-SORE Sustains Sorafenib Resistance in Hepatocellular Carcinoma by Regulating Beta-Catenin Signaling. Mol Cancer (2020) 19(1):163. doi: 10.1186/s12943-020-01281-8
4. Feng GS, Hanley KL, Liang Y, Lin X. Improving the Efficacy of Liver Cancer Immunotherapy: The Power of Combined Preclinical and Clinical Studies. Hepatology (2021) 73(Suppl 1):104–14. doi: 10.1002/hep.31479
5. Abdella R, Talyzina A, Chen S, Inouye CJ, Tjian R, He Y. Structure of the Human Mediator-Bound Transcription Preinitiation Complex. Science (2021) 372(6537):52–6. doi: 10.1126/science.abg3074
6. Chen X, Yin X, Li J, Wu Z, Qi Y, Wang X, et al. Structures of the Human Mediator and Mediator-Bound Preinitiation Complex. Science (2021) 372(6546). doi: 10.1126/science.abg0635
7. Clark AD, Oldenbroek M, Boyer TG. Mediator Kinase Module and Human Tumorigenesis. Crit Rev Biochem Mol Biol (2015) 50(5):393–426. doi: 10.3109/10409238.2015.1064854
8. Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei PJ, Burlingame AL, et al. Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Cell (2016) 166(6):1411–1422 e16. doi: 10.1016/j.cell.2016.08.050
9. Soutourina J. Transcription Regulation by the Mediator Complex. Nat Rev Mol Cell Biol (2018) 19(4):262–74. doi: 10.1038/nrm.2017.115
10. Chen L, Liang Z, Tian Q, Li C, Ma X, Zhang Y, et al. Overexpression of LCMR1 Is Significantly Associated With Clinical Stage in Human NSCLC. J Exp Clin Cancer Res (2011) 30:18. doi: 10.1186/1756-9966-30-18
11. Xu Y, Liang Z, Li C, Yang Z, Chen L. LCMR1 Interacts With DEK to Suppress Apoptosis in Lung Cancer Cells. Mol Med Rep (2017) 16(4):4159–64. doi: 10.3892/mmr.2017.7095
12. Rosenblum-Vos LS, Rhodes L, Evangelista CC Jr, Boayke KA, Zitomer RS. The ROX3 Gene Encodes an Essential Nuclear Protein Involved in CYC7 Gene Expression in Saccharomyces Cerevisiae. Mol Cell Biol (1991) 11(11):5639–47. doi: 10.1128/mcb.11.11.5639-5647.1991
13. Ding XF, Huang GM, Shi Y, Li JA, Fang XD. Med19 Promotes Gastric Cancer Progression and Cellular Growth. Gene (2012) 504(2):262–7. doi: 10.1016/j.gene.2012.04.033
14. Liu Y, Tao X, Fan L, Jia L, Gu C, Feng Y. Knockdown of Mediator Complex Subunit 19 Inhibits the Growth of Ovarian Cancer. Mol Med Rep (2012) 6(5):1050–6. doi: 10.3892/mmr.2012.1065
15. Li XH, Fang DN, Zeng CM. Knockdown of MED19 by Short Hairpin RNA-Mediated Gene Silencing Inhibits Pancreatic Cancer Cell Proliferation. Cancer Biother Radiopharm (2011) 26(4):495–501. doi: 10.1089/cbr.2010.0863
16. Wang T, Hao L, Feng Y, Wang G, Qin D, Gu G. Knockdown of MED19 by Lentivirus-Mediated shRNA in Human Osteosarcoma Cells Inhibits Cell Proliferation by Inducing Cell Cycle Arrest in the G0/G1 Phase. Oncol Res (2011) 19(5):193–201. doi: 10.3727/096504011x12970940207760
17. Zhang X, Fan Y, Liu B, Qi X, Guo Z, Li L. Med19 Promotes Breast Cancer Cell Proliferation by Regulating CBFA2T3/HEB Expression. Breast Cancer (2017) 24(3):433–41. doi: 10.1007/s12282-016-0722-3
18. Wang Z, Yang C, Li L, Jin X, Zhang Z, Zheng H, et al. Tumor-Derived HMGB1 Induces CD62L(dim) Neutrophil Polarization and Promotes Lung Metastasis in Triple-Negative Breast Cancer. Oncogenesis (2020) 9(9):82. doi: 10.1038/s41389-020-00267-x
19. Zhang X, Gao D, Fang K, Guo Z, Li L. Med19 Is Targeted by miR-101-3p/miR-422a and Promotes Breast Cancer Progression by Regulating the EGFR/MEK/ERK Signaling Pathway. Cancer Lett (2019) 444:105–15. doi: 10.1016/j.canlet.2018.12.008
20. Wen H, Feng CC, Ding GX, Meng DL, Ding Q, Fang ZJ, et al. Med19 Promotes Bone Metastasis and Invasiveness of Bladder Urothelial Carcinoma via Bone Morphogenetic Protein 2. Ann Diagn Pathol (2013) 17(3):259–64. doi: 10.1016/j.anndiagpath.2012.11.004
21. Agaesse G, Barbollat-Boutrand L, Sulpice E, Bhajun R, El Kharbili M, Berthier-Vergnes O, et al. A Large-Scale RNAi Screen Identifies LCMR1 as a Critical Regulator of Tspan8-Mediated Melanoma Invasion. Oncogene (2017) 36(35):5084. doi: 10.1038/onc.2017.195
22. Zou SW, Ai KX, Wang ZG, Yuan Z, Yan J, Zheng Q. The Role of Med19 in the Proliferation and Tumorigenesis of Human Hepatocellular Carcinoma Cells. Acta Pharmacol Sin (2011) 32(3):354–60. doi: 10.1038/aps.2010.223
23. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for Analysis of Tumor-Infiltrating Immune Cells. Nucleic Acids Res (2020) 48(W1):W509–14. doi: 10.1093/nar/gkaa407
24. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res (2017) 45(W1):W98–W102. doi: 10.1093/nar/gkx247
25. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (2017) 19(8):649–58. doi: 10.1016/j.neo.2017.05.002
26. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing Multi-Omics Data Within and Across 32 Cancer Types. Nucleic Acids Res (2018) 46(D1):D956–63. doi: 10.1093/nar/gkx1090
27. Zhao H, Zhang X, Wang M, Lin Y, Zhou S. Stigmasterol Simultaneously Induces Apoptosis and Protective Autophagy by Inhibiting Akt/mTOR Pathway in Gastric Cancer Cells. Front Oncol (2021) 11:629008. doi: 10.3389/fonc.2021.629008
28. Yu F, Ma R, Liu C, Zhang L, Feng K, Wang M, et al. SQSTM1/p62 Promotes Cell Growth and Triggers Autophagy in Papillary Thyroid Cancer by Regulating the AKT/AMPK/mTOR Signaling Pathway. Front Oncol (2021) 11:638701. doi: 10.3389/fonc.2021.638701
29. Acharya N, Madi A, Zhang H, Klapholz M, Escobar G, Dulberg S, et al. Endogenous Glucocorticoid Signaling Regulates CD8(+) T Cell Differentiation and Development of Dysfunction in the Tumor Microenvironment. Immunity (2020) 53(3):658–671 e6. doi: 10.1016/j.immuni.2020.08.005
30. Huang S, Song Z, Zhang T, He X, Huang K, Zhang Q, et al. Identification of Immune Cell Infiltration and Immune-Related Genes in the Tumor Microenvironment of Glioblastomas. Front Immunol (2020) 11:585034. doi: 10.3389/fimmu.2020.585034
31. Iyengar NM, Gucalp A, Dannenberg AJ, Hudis CA. Obesity and Cancer Mechanisms: Tumor Microenvironment and Inflammation. J Clin Oncol (2016) 34(35):4270–6. doi: 10.1200/JCO.2016.67.4283
32. Barrett RL, Pure E. Cancer-Associated Fibroblasts and Their Influence on Tumor Immunity and Immunotherapy. Elife (2020) 9. doi: 10.7554/eLife.57243
33. Biffi G, Tuveson DA. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol Rev (2021) 101(1):147–76. doi: 10.1152/physrev.00048.2019
34. Chow A, Zhou W, Liu L, Fong MY, Champer J, Van Haute D, et al. Macrophage Immunomodulation by Breast Cancer-Derived Exosomes Requires Toll-Like Receptor 2-Mediated Activation of NF-kappaB. Sci Rep (2014) 4:5750. doi: 10.1038/srep05750
35. DeNardo DG, Ruffell B. Macrophages as Regulators of Tumour Immunity and Immunotherapy. Nat Rev Immunol (2019) 19(6):369–82. doi: 10.1038/s41577-019-0127-6
36. Gibson JT, Orlandella RM, Turbitt WJ, Behring M, Manne U, Sorge RE, et al. Obesity-Associated Myeloid-Derived Suppressor Cells Promote Apoptosis of Tumor-Infiltrating CD8 T Cells and Immunotherapy Resistance in Breast Cancer. Front Immunol (2020) 11:590794. doi: 10.3389/fimmu.2020.590794
37. Jones LM, Broz ML, Ranger JJ, Ozcelik J, Ahn R, Zuo D, et al. STAT3 Establishes an Immunosuppressive Microenvironment During the Early Stages of Breast Carcinogenesis to Promote Tumor Growth and Metastasis. Cancer Res (2016) 76(6):1416–28. doi: 10.1158/0008-5472.CAN-15-2770
38. Krishna S, Lowery FJ, Copeland AR, Bahadiroglu E, Mukherjee R, Jia L, et al. Stem-Like CD8 T Cells Mediate Response of Adoptive Cell Immunotherapy Against Human Cancer. Science (2020) 370(6522):1328–34. doi: 10.1126/science.abb9847
39. Li L, Yu R, Cai T, Chen Z, Lan M, Zou T, et al. Effects of Immune Cells and Cytokines on Inflammation and Immunosuppression in the Tumor Microenvironment. Int Immunopharmacol (2020) 88:106939. doi: 10.1016/j.intimp.2020.106939
40. Matsuzaki J, Tsuji T, Luescher IF, Shiku H, Mineno J, Okamoto S, et al. Direct Tumor Recognition by a Human CD4(+) T-Cell Subset Potently Mediates Tumor Growth Inhibition and Orchestrates Anti-Tumor Immune Responses. Sci Rep (2015) 5:14896. doi: 10.1038/srep14896
41. Boube M, Hudry B, Immarigeon C, Carrier Y, Bernat-Fabre S, Merabet S, et al. Drosophila Melanogaster Hox Transcription Factors Access the RNA Polymerase II Machinery Through Direct Homeodomain Binding to a Conserved Motif of Mediator Subunit Med19. PloS Genet (2014) 10(5):e1004303. doi: 10.1371/journal.pgen.1004303
42. Ding N, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Boyer TG. MED19 and MED26 Are Synergistic Functional Targets of the RE1 Silencing Transcription Factor in Epigenetic Silencing of Neuronal Gene Expression. J Biol Chem (2009) 284(5):2648–56. doi: 10.1074/jbc.M806514200
43. Myers LC, Kornberg RD. Mediator of Transcriptional Regulation. Annu Rev Biochem (2000) 69:729–49. doi: 10.1146/annurev.biochem.69.1.729
44. Cui X, Xu D, Lv C, Qu F, He J, Chen M, et al. Suppression of MED19 Expression by shRNA Induces Inhibition of Cell Proliferation and Tumorigenesis in Human Prostate Cancer Cells. BMB Rep (2011) 44(8):547–52. doi: 10.5483/bmbrep.2011.44.8.547
45. Yu S, Wang Y, Yuan H, Zhao H, Lv W, Chen J, et al. Knockdown of Mediator Complex Subunit 19 Suppresses the Growth and Invasion of Prostate Cancer Cells. PloS One (2017) 12(1):e0171134. doi: 10.1371/journal.pone.0171134
46. He GY, Hu JL, Zhou L, Zhu XH, Xin SN, Zhang D, et al. The FOXD3/miR-214/MED19 Axis Suppresses Tumour Growth and Metastasis in Human Colorectal Cancer. Br J Cancer (2016) 115(11):1367–78. doi: 10.1038/bjc.2016.362
47. Yuan H, Yu S, Cui Y, Men C, Yang D, Gao Z, et al. Knockdown of Mediator Subunit Med19 Suppresses Bladder Cancer Cell Proliferation and Migration by Downregulating Wnt/beta-Catenin Signalling Pathway. J Cell Mol Med (2017) 21(12):3254–63. doi: 10.1111/jcmm.13229
48. Bruni D, Angell HK, Galon J. The Immune Contexture and Immunoscore in Cancer Prognosis and Therapeutic Efficacy. Nat Rev Cancer (2020) 20(11):662–80. doi: 10.1038/s41568-020-0285-7
49. Sui S, An X, Xu C, Li Z, Hua Y, Huang G, et al. An Immune Cell Infiltration-Based Immune Score Model Predicts Prognosis and Chemotherapy Effects in Breast Cancer. Theranostics (2020) 10(26):11938–49. doi: 10.7150/thno.49451
50. Yi J, Zhu J, Wu J, Thompson CB, Jiang X. Oncogenic Activation of PI3K-AKT-mTOR Signaling Suppresses Ferroptosis via SREBP-Mediated Lipogenesis. Proc Natl Acad Sci USA (2020) 117(49):31189–97. doi: 10.1073/pnas.2017152117
51. Duan Y, Haybaeck J, Yang Z. Therapeutic Potential of PI3K/AKT/mTOR Pathway in Gastrointestinal Stromal Tumors: Rationale and Progress. Cancers (Basel) (2020) 12(10). doi: 10.3390/cancers12102972
52. Yang R, Wang M, Zhang G, Bao Y, Wu Y, Li X, et al. E2F7-EZH2 Axis Regulates PTEN/AKT/mTOR Signalling and Glioblastoma Progression. Br J Cancer (2020) 123(9):1445–55. doi: 10.1038/s41416-020-01032-y
53. Gao W, Guo H, Niu M, Zheng X, Zhang Y, Xue X, et al. Circpard3 Drives Malignant Progression and Chemoresistance of Laryngeal Squamous Cell Carcinoma by Inhibiting Autophagy Through the PRKCI-Akt-mTOR Pathway. Mol Cancer (2020) 19(1):166. doi: 10.1186/s12943-020-01279-2
54. Fosam A, Perry RJ. Current Mechanisms in Obesity and Tumor Progression. Curr Opin Clin Nutr Metab Care (2020) 23(6):395–403. doi: 10.1097/MCO.0000000000000690
55. Moraes JA, Encarnacao C, Franco VA, Xavier Botelho LG, Rodrigues GP, Ramos-Andrade I, et al. Adipose Tissue-Derived Extracellular Vesicles and the Tumor Microenvironment: Revisiting the Hallmarks of Cancer. Cancers (Basel) (2021) 13(13). doi: 10.3390/cancers13133328
56. Dean JM, He A, Tan M, Wang J, Lu D, Razani B, et al. MED19 Regulates Adipogenesis and Maintenance of White Adipose Tissue Mass by Mediating PPARgamma-Dependent Gene Expression. Cell Rep (2020) 33(1):108228. doi: 10.1016/j.celrep.2020.108228
Keywords: hepatocellular carcinoma, MED19, AKT/mTOR signaling pathway, proliferation/migration/invasion, tumor immune infiltration
Citation: Zhang Y, Qin P, Xu X, Li M, Huang H, Yan J and Zhou Y (2022) Mediator Complex Subunit 19 Promotes the Development of Hepatocellular Carcinoma by Regulating the AKT/mTOR Signaling Pathway. Front. Oncol. 11:792285. doi: 10.3389/fonc.2021.792285
Received: 10 October 2021; Accepted: 10 November 2021;
Published: 03 January 2022.
Edited by:
João Pessoa, University of Coimbra, PortugalReviewed by:
Pushkar Malakar, Gandhi Institute of Technology and Management (GITAM), IndiaJing Liu, Kunming University of Science and Technology, China
Copyright © 2022 Zhang, Qin, Xu, Li, Huang, Yan and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yali Zhou, emhvdXlhbGlAZ2xtYy5lZHUuY24=; Jianguo Yan, eWFuamlhbmd1b0BnbG1jLmVkdS5jbg==
 Yuting Zhang1,2
Yuting Zhang1,2 Jianguo Yan
Jianguo Yan Yali Zhou
Yali Zhou