- 1Medizinische Klinik II, University Hospital Frankfurt, Frankfurt, Germany
- 2Dr. Senckenberg Institute of Pathology, University Hospital Frankfurt, Frankfurt, Germany
- 3Wildlab, University Hospital MVZ GmbH, Frankfurt, Germany
- 4Frankfurt Institute for Advanced Studies (FIAS), Frankfurt, Germany
KRAS is one of the most commonly mutated oncogenes in cancer, enabling tumor proliferation and maintenance. After various approaches to target KRAS have failed over the past decades, the first specific inhibitor of the p.G12C mutation of KRAS was recently approved by the FDA after showing promising results in adenocarcinomas of the lung and other solid tumors. Lung cancer, the most common cancer worldwide, is a promising use case for these new therapies, as adenocarcinomas in particular frequently harbor KRAS mutations. However, in squamous cell carcinoma (SCC) of the lung, KRAS mutations are rare and their impact on clinical outcome is poorly understood. In this review, we discuss the current knowledge on the prevalence and prognostic and predictive significance of KRAS mutations in the context of SCC.
Introduction
Lung cancer is the most common cancer worldwide with a high mortality rate (1). Histologically, ~85% of all cases are non-small cell lung cancer (NSCLC), which is subdivided into adenocarcinoma (ADC) and squamous cell carcinoma (SCC), among others, with the latter accounting for ~30% of all NSCLC (2).
In recent decades, the development of immune checkpoint inhibitors (ICI) and agents targeting specific oncogenic driver mutations (EGFR, ALK, ROS, BRAF, MET, RET, ERBB2 and NTRK) has fundamentally changed the therapeutic landscape of NSCLC by prolonging survival rates and reducing toxicities associated with conventional chemotherapy (3). While these driver mutations are common in ADC, they are rare in SCC. Instead, the mutational landscape of SCC consists of genes such as TP53, CDKN2A, PTEN, PIK3CA, KEAP1, MLL2, HLA-A, NFE2L2, NOTCH1, FGFR1-4, and others (4, 5). Unfortunately, there are no approved therapies that target any of these mutations, so chemotherapy and ICI are usually the only therapeutic approaches for advanced-stage SCC patients (6).
The most common driver mutation in NSCLC patients is found in the Kirsten rat sarcoma virus oncogene homolog (KRAS) gene. KRAS mutations are quite common in ADC with an incidence of ~25% in Western countries and ~10% in Asian populations and are associated with past and current tobacco use (7). In recent years, several studies pointed to a variety of effects of KRAS mutations on ADC biology, namely, patterns of metastatic spread, efficacy of chemotherapy, ICI, and EGFR therapy, as described elsewhere (8, 9). In addition, there are less frequently mutated isoforms of KRAS, namely, NRAS (neuroblastoma rat sarcoma viral oncogene homolog) and HRAS (Harvey rat sarcoma viral oncogene homolog) (10). In contrast to ADC, RAS mutations are rare in SCC.
Recently, after decades of unsuccessful efforts, sotorasib, a selective inhibitor of KRAS with codon 12 glycine to cysteine substitutions (p.G12C), has been approved by the U.S. Food and Drug Administration (FDA) after showing promising results in phase I and II trials in NSCLC and other solid tumors (11–13). Adagrasib, another KRAS p.G12C inhibitor, is currently being studied in clinical trials (14). In addition to direct inhibition, there are other approaches to target KRAS. BI 1701963, for example, is an inhibitor of SOS1, which in turn is an important mediator of KRAS activation as a guanine nucleotide exchange factor (15). The farnesyltransferase inhibitor tipifarnib received breakthrough therapy designation from the FDA for HRAS-mutated head and neck SCC (16). Furthermore, combined inhibition of multiple pathways of the KRAS signaling network may provide clinical benefit in patients with KRAS p.G12D-mutated tumors, as shown by preclinical data of the MEK inhibitor trametinib in combination with the ROS1/TRK and SRC/FAK/JAK2 inhibitor repotrectinib (17).
Since the clinical relevance of RAS mutations in the context of SCC is unclear and new therapies are emerging, the current state of knowledge will be discussed here.
Prevalence and Incidence
Cancer Mutation Databases
Cancer mutation databases are one way to estimate the prevalence of KRAS mutations in SCC (Table 1). Probably the largest mutation database is the Catalogue of Somatic Mutations in Cancer (COSMIC), whose data come from over 25,000 manually curated publications totaling over 1.3 million tumor samples (30). At the time of writing, COSMIC provides over 5,000 samples of SCC patients tested for RAS mutations, with mutation rates of 3.5, 1.0, and 0.8% for KRAS, NRAS, and HRAS, respectively. Among patients with KRAS mutations, the most common amino acid substitutions were p.G12D (28%), p.G12C (27%), and p.G12V (16%).
The Cancer Genome Atlas (TCGA) is a curated, open-access database providing comprehensive and likely refined genomic, epigenetic, transcriptomic and proteomic data of more than 11,000 patients with various cancer types (18). TCGA dataset, accessible via the cBioPortal at https://www.cbioportal.org/ (31), currently contains samples from 841 SCC patients with available mutation data. Of these 841 patients, mutations in KRAS, NRAS, and HRAS occurred in 1.3, 0.8, and 2.0% of cases, respectively. Interestingly, only 3 of the 11 samples with KRAS mutations were codon 12 mutations.
The MSK-IMPACT Clinical Sequencing Cohort (MSKCC), in which samples from a total of over 10,000 patients were analyzed, included over 1,500 NSCLC patients. Of all 170 SCC patients, 11 showed a KRAS mutation (6.5%) with the most common being p.G12C (n = 5) followed by p.G12V (n = 2) (19).
Multi-Center Studies
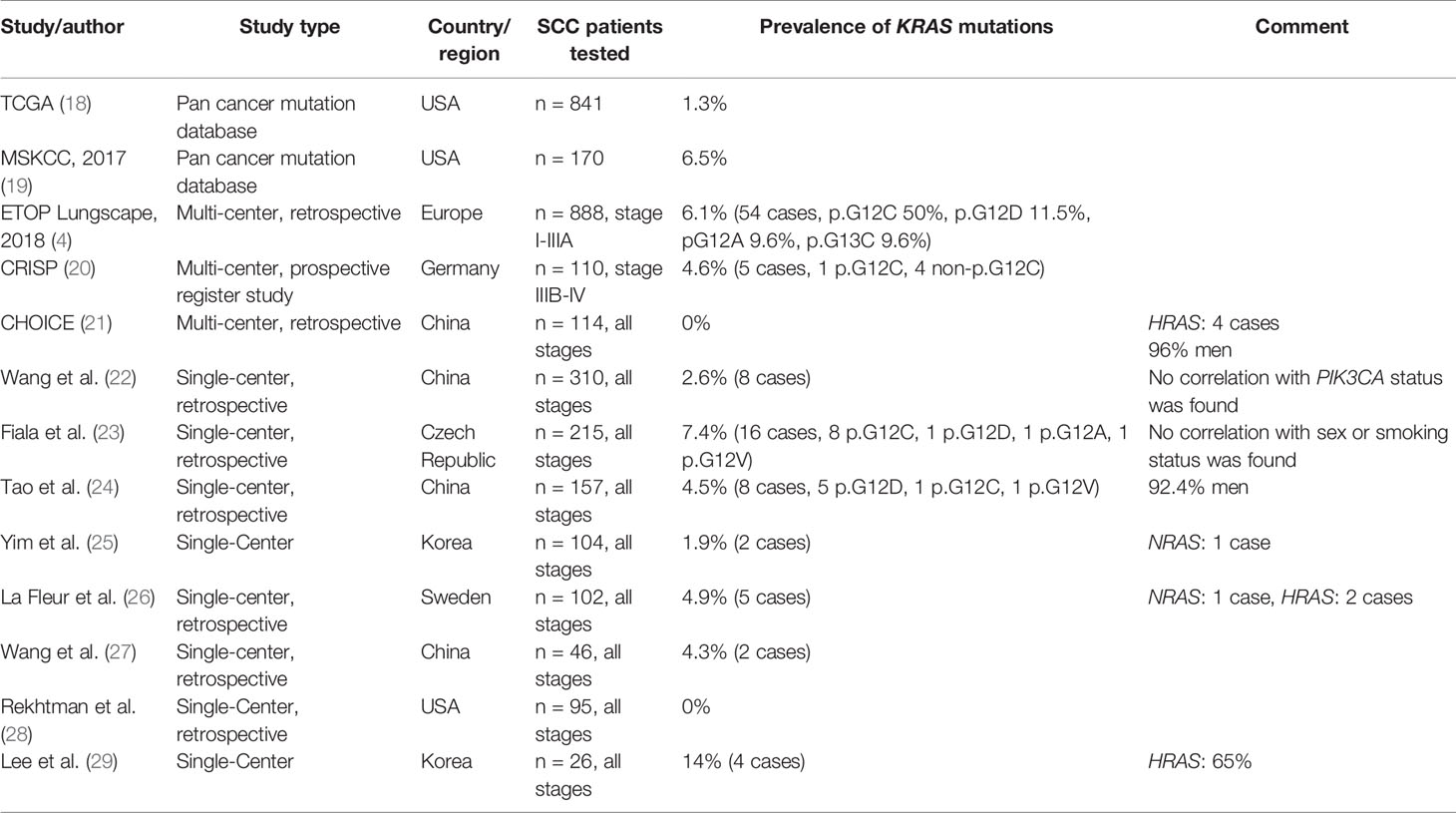
As part of the European Thoracic Oncology Platform’s (ETOP) Lungscape project, PCR multiplex tests were performed on tumor samples of 888 patients of 17 different sites with resected stages I–IIIA SCC. In this cohort, a KRAS mutation was found in 6.1% (4). The most common mutations were p.G12C (50%), p.G12D (11.5%), p.G12A, and p.G13C (9.6% each) (Figure 1). Mutation rates did not differ significantly between current/former smokers and never-smokers (6.3% vs. 5.0%, p >0.99). However, as expected, the group of never-smokers was quite small with three mutations in 60 patients.
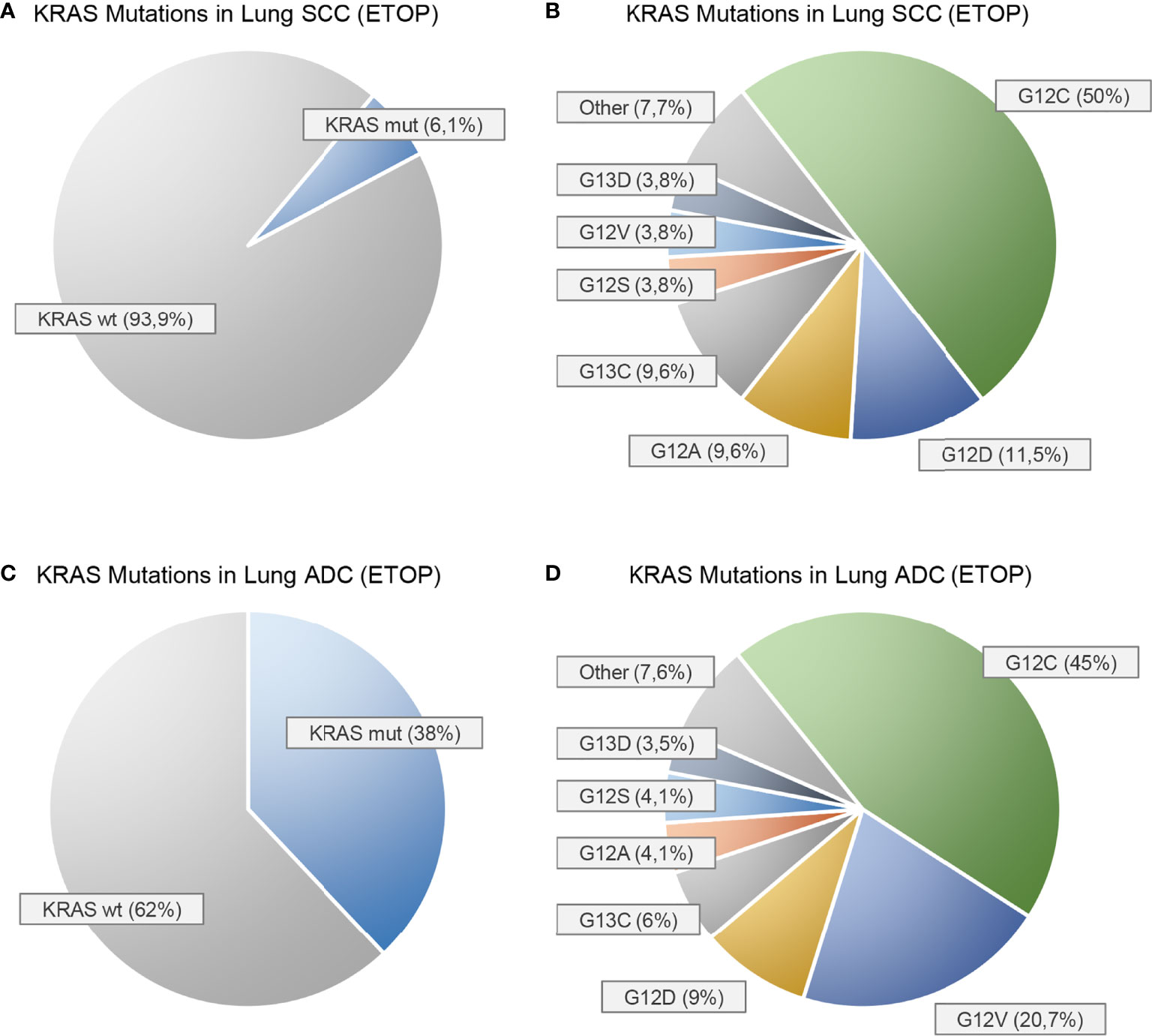
Figure 1 Prevalence of KRAS mutations in squamous cell carcinomas (A, B) and adenocarcinomas (C, D) of the lung as reported by the European Thoracic Oncology Platform (ETOP) Lungscape iBiobank [Ref. (4)]. (A, C) show the proportion of KRAS mutated (mut) and KRAS wild-type (wt) patients. (B, D) show the frequencies of the individual KRAS mutations.
An analysis from the Germany-wide prospective CRISP registry with more than 150 participating centers reported a KRAS mutation rate of 4.6% in 109 SCC patients (20). In contrast, no KRAS mutation was detected in 114 SCC patients at 6 hospitals in China by whole-exome sequencing in the retrospective CHOICE study. However, HRAS mutations occurred in 3.5% of cases (21).
Single-Center Studies
A study of 310 SCC patients from China revealed 8 cases of KRAS mutations (2.6%) that did not correlate with PIK3CA mutation status (22). In another work on 157 Chinese patients with early-stage SCC, KRAS mutations were detected in 4.5% of all cases (5 of 8 patients had a p.G12D mutation, 1 p.G12C, 1 p.G12V, and 1 other). Male patients were particularly prevalent in this study, accounting for 92.4% (24). Another study from China with 46 SCC patients reported a prevalence of KRAS mutations of 4.3% (27).
In a retrospective single-center study from the Czech Republic with a total of 215 patients with advanced SCC, a KRAS mutation was described in 7.4% (23). Approximately 68.8% of these mutations were in codon 12, with p.G12C being the most common mutation (50%). As in the ETOP study, no correlation with sex or smoking status was observed. In a study of 102 Swedish SCC patients, mutation rates of 4.9, 1.0, and 2.0% were described for KRAS, NRAS, and HRAS, respectively (26). In contrast, another retrospective study examined 95 resected SCC and found no KRAS mutation (28).
A retrospective study from Korea, in which whole-exome sequencing of 104 SCC samples was performed, revealed 2 KRAS mutations and 1 NRAS mutation (25). In contrast, another Korean study using only 26 SCC found 14% KRAS and 65% HRAS mutations (29). The paper gave no indication of a cause for these very high mutation rates compared with other publications.
Estimated Incidence
Using mutation prevalence data of the COSMIC and TCGA databases and cancer incidence data from the American Cancer Society, Prior et al. estimated an incidence of 2,794 new KRAS-mutated SCC cases per year (782 for KRAS p.G12C) in the United States (32). This is roughly equivalent to an incidence of 0.85 per 100,000 inhabitants per year (0.23 per 100,000 inhabitants per year for KRAS p.G12C mutant SCC).
Prognostic and Predictive Value
Because KRAS mutations are rare in SCC, there are few data on the impact of mutations on patient outcomes. Therefore, pooled analyses are a promising tool to find correlations. To our knowledge, there are two such pooled cohorts of interventional trials.
Zer et al. evaluated data from four double-blind, placebo-controlled National Cancer Institute of Canada Clinical Trials Group (NCIC CTG) trials of EGFR tyrosine kinase inhibitors (TKIs) (33). The four studies were BR.21 (erlotinib in second- and third-line stages III-IV NSCLC) (34), BR19 (adjuvant gefitinib in completely resected stages IB-IIIA NSCLC) (35), BR.26 (dacomitinib in stages IIIB-IV NSCLC after first-line chemotherapy and EGFR TKI) (36), and TOPICAL (first-line erlotinib in advanced stage NSCLC unsuitable for chemotherapy) (29). All studies enrolled patients regardless of their EGFR mutational status. In the retrospective analyses, KRAS and EGFR mutation status was determined for all four studies. The pooled analysis with a total of 1,362 patients included 385 SCC cases in which KRAS mutations were present in 6% (45% p.G12C/p.G12V; 32% p.G12D/p.G12S; and 20% p.G12A/p.G12R). Although no significant impact on overall survival (OS) was found, supplemental data showed that KRAS mutation tended to have a negative impact on OS (hazard ratio [HR] 1.288, 95% confidence interval [CI] 0.68–2.43) for the 177 evaluable patients.
The other pooled analysis of prospective trials was performed by Shepherd et al. (37). This study included four trials evaluating adjuvant chemotherapy versus observation in patients with resected early-stage NSCLC, including JBR.10 (vinorelbine plus cisplatin in stages IB-II resected NSCLC) (38), IALT (cisplatin-containing doublet in stages I–III resected NSCLC) (39), ANITA (vinorelbine plus cisplatin in stages IB–IIIA NSCLC) (40), and CALGB 9633 (paclitaxel plus carboplatin in stage IB resected NSCLC) (41). Of the 707 evaluable SCC patients, 6% had a KRAS mutation. In this analysis, OS tended to be worse in the presence of a KRAS mutation with a HR of 1.41 (95% CI, 0.89–2.23). No difference was observed between patients in the treatment and observation groups.
Apart from these two studies, there are mainly retrospectively collected data. Although there are plenty of studies examining the impact of RAS on survival in ADC, data on RAS mutant SCC are seldom reported. A systematic review from 2004 by Mascaux et al. comprised 4 retrospective studies reporting survival data on RAS mutant SCC (42). With a mutation rate of 7.1% in 280 patients, OS did not significantly differ between mutant and wild-type group with a HR of 1.49 (95% CI, 0.88–2.52). It can be assumed that most patients included in this review only received surgery and/or radio-/chemotherapy.
In a recent paper Xu et al. studied 64 Chinese patients with advanced-stage SCC who underwent ICI (43). Overall, the seven patients (10.9%) with a KRAS mutation had a higher overall response rate than wild-type patients (71.4% vs. 22.8%, p = 0.009). Yet, progression-free survival (PFS, HR 0.62, 95% CI 0.26–1.48) and OS (HR 0.68, 95% CI 0.2–2.3) did not differ significantly.
However, in another retrospective single-center study by Fiala et al. of 223 Czech SCC patients treated with EGFR TKI (EGFR wild-type and mutant), OS differed significantly between KRAS mutant (7.4%) and wild-type patients with a median survival of 5.7 vs. 8.2 months (p = 0.039) (23). Anyway, PFS was not significantly different between the groups. Of note, only univariate analysis was done in this study diminishing its validity.
In contrast, Tao et al. reported 157 Chinese SCC patients, 4.5% of whom had a KRAS mutation. OS and DFS were not significantly different between groups with a HR of 0.91 (95% CI 0.22–3.76) and 0.56 (0.14–2.28), respectively (24).
Discussion
Controversies Over KRAS Status Related to SCC
As pointed out, KRAS mutations in SCC are rare and appear to range from 1 to 7% in most publications (Table 1). It does not seem to differ significantly between Western and Eastern populations, as is the case with ADC (7).
The low prevalence has led some authors to suggest that there are no KRAS-mutated SCC and that reported cases may be misclassified ADC or adenosquamous carcinoma (ADSQC) (28). A phenomenon supporting this hypothesis might be ADC to SCC transition (AST). Data from genetically engineered mouse models (44, 45) show that the loss of LKB1/STK11 of KRAS mutated ADC can lead to an ADC to SCC transition (AST) as reviewed in (46). Besides mutational events, transcriptional reprogramming was also shown to be a potential driver of AST (47). As a consequence, cases of KRAS mutated SCC (or ADSQC) might in part be the result of an AST with very early onset. In line with this, KRAS mutations do occur in both the squamous and the glandular components of ADSQC (48, 49).
In their very thorough study, Rekthman et al. reviewed all six cases of KRAS mutated SCC known to their department by repeating immunohistochemistry (IHC) and morphological examination (28). Two of the cases were reclassified as ADSQC after detection of glandular components in other specimens of the same tumor (preoperative biopsy vs either metastatic/recurrent site or same-site resection). Another three cases turned out to be poorly differentiated ADC, defined as ΔNp63-negativity/TTF1-positivity in IHC (50). The last case consisted of two distinct cell populations: the first one ΔNp63-positive/TTF1-negative (~5%) and the second one ΔNp63-negative/TTF1-positive (~95%). Morphologically, this tumor was predominantly squamous with ~5% glands. As this is suggestive for an ADSQC but does not match the World Health Organization definition of ADSQC, which requires more than 10% of each component (51), the authors classified this tumor as “biphasic tumor with <10% glands”. Overall, the authors concluded that KRAS-mutated SCC may in some cases represent misclassified ADSQC or poorly differentiated ADC.
Indeed, morphological differentiation between ADC and SCC can be difficult in some cases, and small sample sizes and poor differentiation are known diagnostic pitfalls (52). Therefore, it is very likely, that at least some KRAS-mutant SCC described in the literature are actually misclassified ADC or ADSQC. However, given the low reported frequency of KRAS mutations, the sample size of Rekthman et al. (28) appears too small for definitive conclusions.
Given the continuity with which KRAS mutations are described in the literature, it is unlikely that there are no KRAS-mutant SCC. Nevertheless, we believe it is reasonable to closely review cases of KRAS-mutated SCC in collaboration between clinicians and pathologists.
Lack of Data and Potential Bias
In recent years, many studies have attempted to correlate outcome parameters with the presence of specific KRAS mutations associated with ADC, including prognostic value, predictive value (e.g., with EGFR TKI, ICI, etc.), patterns of metastatic spread, and other aspects, as described elsewhere (8, 9). In any case, these results cannot be simply extrapolated to SCC.
Clearly, the rarity of KRAS mutations in SCC complicates the ability to conduct such studies. However, in our review of the literature, even if larger cohorts were studied, we noticed a tendency to either not stratify patient outcomes with KRAS-mutated NSCLC by histology, or, at most, to mention them in the supplementary information (4, 21, 26).
Also, the fact that the College of American Pathologists (CAP), the International Association for the Study of Lung Cancer (IASLC), the Association for Molecular Pathology (AMP), and the European Society of Clinical Oncology (ESMO) do not recommend KRAS mutation testing in SCC (52, 53) may introduce bias in retrospective studies.
Conclusion
As mentioned earlier, new drugs that directly or indirectly target mutant KRAS are in development or have been approved recently (8, 9). Although KRAS is much more commonly mutated in ADC, a small proportion of 1–7% of SCC patients may also benefit from these new therapies. However, the biological effects of KRAS mutations are highly dependent on the tumor cell of origin (54) and there is limited evidence in SCC. Whether the outcome of KRAS-directed therapy is different in ADC, SCC, and other solid tumors is yet unknown. Therefore, we welcome the fact that the ongoing studies of sotorasib and adagrasib include NSCLC with non-adenocarcinomatous histology (NCT04303780, NCT04685135). One patient with SCC has already been enrolled in the CodeBreaK100 phase II study of sotorasib (11).
Against the background that lung cancer is the most common malignancy worldwide, the incidence of KRAS-mutated SCC may seem low with roughly 0.85 per 100,000 inhabitants per year. Nevertheless, it represents a relevant subgroup in an increasingly segmented clinical and therapeutic landscape.
Author Contributions
FA and MS contributed to conception and review of the literature. FA wrote the first draft of the manuscript. SW and PW wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
PW has received consulting fees and honoraria (private/institutional) for lectures by Bayer, Janssen-Cilag, Novartis, Roche, MSD, Astellas Pharma, Bristol-Myers Squibb, Thermo Fisher Scientific, Molecular Health, Sophia Genetics, Qiagen, Eli Lilly, Myriad, Hedera Dx, and Astra Zeneca. MS has received consulting fees and honoraria for lectures by Novartis, BMS, Roche, Lilly, Boehringer-Ingelheim, Pfizer, MSD, Astra-Zeneca, Celgene, AbbVie, Takeda, Janssen-Cilag, and Tesaro. JS has received consulting fees and honoraria for lectures by Novartis, BMS, Leo Pharma, Oncopeptides, Astra-Zeneca, Roche, Takeda, Boehringer-Ingelheim, Amgen, and Pfizer.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor declared a past co-authorship with one of the authors MS.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ADC, Lung adenocarcinoma; ADSQC, Adenosquamous carcinoma; AMP, Association for Molecular Pathology; AST, Adenocarcinoma to squamous cell carcinoma transition; CAP, College of American Pathologists; CI, Confidence interval; COSMIC, Catalogue of Somatic Mutations in Cancer; EGFR, Epidermal growth factor receptor; ESMO, European Society of Clinical Oncology; ETOP, European Thoracic Oncology Platform; FDA, U.S. Food and Drug Administration; HR, Hazard ratio; HRAS, Harvey rat sarcoma viral oncogene homolog; IASLC, International Association for the Study of Lung Cancer; ICI, Immune checkpoint inhibitor; IHC, Immunohistochemistry; KRAS, Kirsten rat sarcoma viral oncogene homolog; LKB1, Liver kinase B1; MSKCC, MSK-IMPACT Clinical Sequencing Cohort; NCIC CTG, National Cancer Institute of Canada Clinical Trials Group; NRAS, Neuroblastoma rat sarcoma viral oncogene homolog; NSCLC, Non-small cell lung cancer; OS, Overall survival; p.G12C, Codon 12 glycine to cysteine substitution; PCR , Polymerase chain reaction; PFS, Progression-free survival; RAS, Rat sarcoma viral oncogene homolog; SCC, Lung squamous cell carcinoma; STK11, Serine/threonine kinase 11; TCGA, The Cancer Genome Atlas; TKI, Tyrosine kinase inhibitor.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71:7–33. doi: 10.3322/caac.21654
2. Heist RS, Sequist LV, Engelman JA. Genetic Changes in Squamous Cell Lung Cancer: A Review. J Thorac Oncol (2012) 7:924–33. doi: 10.1097/JTO.0b013e31824cc334
3. Herbst RS, Morgensztern D, Boshoff C. The Biology and Management of Non-Small Cell Lung Cancer. Nature (2018) 553:446–54. doi: 10.1038/nature25183
4. Kerr KM, Dafni U, Schulze K, Thunnissen E, Bubendorf L, Hager H, et al. Prevalence and Clinical Association of Gene Mutations Through Multiplex Mutation Testing in Patients With NSCLC: Results From the ETOP Lungscape Project. Ann Oncol (2018) 29:200–8. doi: 10.1093/annonc/mdx629
5. Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct Patterns of Somatic Genome Alterations in Lung Adenocarcinomas and Squamous Cell Carcinomas. Nat Genet (2016) 48:607–16. doi: 10.1038/ng.3564
6. C. G. A. R. Network. Comprehensive Genomic Characterization of Squamous Cell Lung Cancers. Nature (2012) 489:519–25. doi: 10.1038/nature11404
7. Dearden S, Stevens J, Wu YL, Blowers D. Mutation Incidence and Coincidence in Non Small-Cell Lung Cancer: Meta-Analyses by Ethnicity and Histology (Mutmap). Ann Oncol (2013) 24:2371–6. doi: 10.1093/annonc/mdt205
8. Friedlaender A, Drilon A, Weiss GJ, Banna GL, Addeo A. KRAS as a Druggable Target in NSCLC: Rising Like a Phoenix After Decades of Development Failures. Cancer Treat Rev (2020) 85:101978. doi: 10.1016/j.ctrv.2020.101978
9. Uras IZ, Moll HP, Casanova E. Targeting KRAS Mutant Non-Small-Cell Lung Cancer: Past, Present and Future. Int J Mol Sci (2020) 21:4325. doi: 10.3390/ijms21124325
10. Hobbs GA, Der CJ, Rossman KL. RAS Isoforms and Mutations in Cancer at a Glance. J Cell Sci (2016) 129:1287–92. doi: 10.1242/jcs.182873
11. Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, et al. Sotorasib for Lung Cancers With KRAS P.G12C Mutation. N Engl J Med (2021) 384:2371–81. doi: 10.1056/NEJMoa2103695
13. Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, et al. KRAS G12C Inhibition With Sotorasib in Advanced Solid Tumors. N Engl J Med (2020) 383:1207–17. doi: 10.1056/NEJMoa1917239
14. Jaenne PA, Rybkin II, Spira AI, Riely GJ, Papadopoulos KP, Sabari JK, et al. KRYSTAL-1: Activity and Safety of Adagrasib (MRTX849) in Advanced/ Metastatic Non–Small-Cell Lung Cancer (NSCLC) Harboring KRAS G12C Mutation. Eur J Cancer. (2020) 138(suppl. 2):S1–S2. doi: 10.1016/S0959-8049(20)31076-5
15. Gort E, Johnso ML, Hwang JJ, Pant S, Dünzinger U, Riemann K, et al. A phase I, Open-Label, Dose-Escalation Trial of BI 1701963 as Monotherapy and in Combination With Trametinib in Patients With KRAS Mutated Advanced or Metastatic Solid Tumors. J Clin Oncol (2020) 38(15_suppl). doi: 10.1200/JCO.2020.38.15_suppl.TPS3651
16. Ho AL, Brana I, Haddad R, Bauman J, Bible K, Oosting S, et al. Tipifarnib in Head and Neck Squamous Cell Carcinoma With HRAS Mutations. J Clin Oncol (2021) 39:1856–64. doi: 10.1200/JCO.20.02903
17. Lee NV, Deng W, Zhai D, Rodon L, Parra A, Cowell J, et al. Abstract 1104: Repotrectinib Increases Effectiveness of MEK Inhibitors in KRAS Mutant Cancer Models, in: AACR Annual Meeting 2021, Philadelphia. (2021) 81:1104. doi: 10.1158/1538-7445.AM2021-1104
18. Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): An Immeasurable Source of Knowledge. Contemp Oncol (Pozn) (2015) 19:A68–77. doi: 10.5114/wo.2014.47136
19. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational Landscape of Metastatic Cancer Revealed From Prospective Clinical Sequencing of 10,000 Patients. Nat Med (2017) 23:703–13. doi: 10.1038/nm.4333
20. Sebastian M, Eberhardt WEE, Hoffknecht P, Metzenmacher M, Wehler T, Kokowski K, et al. KRAS G12C-Mutated Advanced Non-Small Cell Lung Cancer: A Real-World Cohort From the German Prospective, Observational, Nation-Wide CRISP Registry (AIO-TRK-0315). Lung Cancer (2021) 154:51–61. doi: 10.1016/j.lungcan.2021.02.005
21. Zhang XC, Wang J, Shao GG, Wang Q, Qu X, Wang B, et al. Comprehensive Genomic and Immunological Characterization of Chinese Non-Small Cell Lung Cancer Patients. Nat Commun (2019) 10:1772. doi: 10.1038/s41467-019-09762-1
22. Wang L, Hu H, Pan Y, Wang R, Li Y, Shen L, et al. PIK3CA Mutations Frequently Coexist With EGFR/KRAS Mutations in Non-Small Cell Lung Cancer and Suggest Poor Prognosis in EGFR/KRAS Wildtype Subgroup. PloS One (2014) 9:e88291. doi: 10.1371/journal.pone.0088291
23. Fiala O, Pesek M, Finek J, Benesova L, Bortlicek Z, Minarik M, et al. Gene Mutations in Squamous Cell NSCLC: Insignificance of EGFR, KRAS and PIK3CA Mutations in Prediction of EGFR-TKI Treatment Efficacy. Anticancer Res (2013) 33:1705–11. doi: 10.1016/S0169-5002(13)70293-9
24. Tao D, Han X, Zhang N, Lin D, Wu D, Zhu X, et al. Genetic Alteration Profiling of Patients With Resected Squamous Cell Lung Carcinomas. Oncotarget (2016) 7:36590–601. doi: 10.18632/oncotarget.9096
25. Kim Y, Hammerman PS, Kim J, Yoon JA, Lee Y, Sun JM, et al. Integrative and Comparative Genomic Analysis of Lung Squamous Cell Carcinomas in East Asian Patients. J Clin Oncol (2014) 32:121–8. doi: 10.1158/1538-7445.AM2014-1526
26. La Fleur L, Falk-Sörqvist E, Smeds P, Berglund A, Sundström M, Mattsson JS, et al. Mutation Patterns in a Population-Based Non-Small Cell Lung Cancer Cohort and Prognostic Impact of Concomitant Mutations in KRAS and TP53 or STK11. Lung Cancer (2019) 130:50–8. doi: 10.1016/j.lungcan.2019.01.003
27. Wang L, Hu H, Pan Y, Wang R, Li Y, Shen L, et al. Assessment of Nine Driver Gene Mutations in Surgically Resected Samples From Patients With Non-Small-Cell Lung Cancer. Cancer Manag Res (2020) 12:4029–38. doi: 10.2147/CMAR.S250822
28. Rekhtman N, Paik PK, Arcila ME, Tafe LJ, Oxnard GR, Moreira AL, et al. Clarifying the Spectrum of Driver Oncogene Mutations in Biomarker-Verified Squamous Carcinoma of Lung: Lack of EGFR/KRAS and Presence of PIK3CA/AKT1 Mutations. Clin Cancer Res (2012) 18:1167–76. doi: 10.1158/1078-0432.CCR-11-2109
29. Lee HY, Lee SH, Won JK, Lee DS, Kwon NJ, Choi SM, et al. Analysis of Fifty Hotspot Mutations of Lung Squamous Cell Carcinoma in Never-Smokers. J Korean Med Sci (2017) 32:415–20. doi: 10.3346/jkms.2017.32.3.415
30. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: The Catalogue of Somatic Mutations In Cancer. Nucleic Acids Res (2019) 47:D941–7. doi: 10.1093/nar/gky1015
31. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The Cbio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov (2012) 2:401–4. doi: 10.1158/2159-8290.CD-12-0095
32. Prior IA, Hood FE, Hartley JL. The Frequency of Ras Mutations in Cancer. Cancer Res (2020) 80:2969–74. doi: 10.1158/0008-5472.CAN-19-3682
33. Zer A, Ding K, Lee SM, Goss GD, Seymour L, Ellis PM, et al. Pooled Analysis of the Prognostic and Predictive Value of KRAS Mutation Status and Mutation Subtype in Patients With Non-Small Cell Lung Cancer Treated With Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors. J Thorac Oncol (2016) 11:312–23. doi: 10.1016/j.jtho.2015.11.010
34. Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, et al. Erlotinib in Previously Treated Non-Small-Cell Lung Cancer. N Engl J Med (2005) 353:123–32. doi: 10.1056/NEJMoa050753
35. Goss GD, O'Callaghan C, Lorimer I, Tsao MS, Masters GA, Jett J, et al. Gefitinib Versus Placebo in Completely Resected Non-Small-Cell Lung Cancer: Results of the NCIC CTG BR19 Study. J Clin Oncol (2013) 31:3320–6. doi: 10.1200/JCO.2013.51.1816
36. Ellis PM, Shepherd FA, Millward M, Perrone F, Seymour L, Liu G, et al. Dacomitinib Compared With Placebo in Pretreated Patients With Advanced or Metastatic Non-Small-Cell Lung Cancer (NCIC CTG BR.26): A Double-Blind, Randomised, Phase 3 Trial. Lancet Oncol (2014) 15:1379–88. doi: 10.1016/S1470-2045(14)70472-3
37. Shepherd FA, Domerg C, Hainaut P, Jänne PA, Pignon JP, Graziano S, et al. Pooled Analysis of the Prognostic and Predictive Effects of KRAS Mutation Status and KRAS Mutation Subtype in Early-Stage Resected Non-Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. J Clin Oncol (2013) 31:2173–81. doi: 10.1200/JCO.2012.48.1390
38. Winton T, Livingston R, Johnson D, Rigas J, Johnston M, Butts C, et al. Vinorelbine Plus Cisplatin vs. Observation in Resected Non-Small-Cell Lung Cancer. N Engl J Med (2005) 352:2589–97. doi: 10.1056/NEJMoa043623
39. Arriagada R, Bergman B, Dunant A, Le Chevalier T, Pignon JP, Vansteenkiste J. Cisplatin-Based Adjuvant Chemotherapy in Patients With Completely Resected Non-Small-Cell Lung Cancer. N Engl J Med (2004) 350:351–60. doi: 10.1056/NEJMoa031644
40. Douillard JY, Rosell R, De Lena M, Carpagnano F, Ramlau R, Gonzáles-Larriba JL, et al. Adjuvant Vinorelbine Plus Cisplatin Versus Observation in Patients With Completely Resected Stage IB-IIIA Non-Small-Cell Lung Cancer (Adjuvant Navelbine International Trialist Association [ANITA]): A Randomised Controlled Trial. Lancet Oncol (2006) 7:719–27. doi: 10.1016/S1470-2045(06)70804-X
41. Strauss GM, Herndon JE, Maddaus MA, Johnstone DW, Johnson EA, Harpole DH, et al. Adjuvant Paclitaxel Plus Carboplatin Compared With Observation in Stage IB Non-Small-Cell Lung Cancer: CALGB 9633 With the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol (2008) 26:5043–51. doi: 10.1200/JCO.2008.16.4855
42. Mascaux C, Iannino N, Martin B, Paesmans M, Berghmans T, Dusart M, et al. The Role of RAS Oncogene in Survival of Patients With Lung Cancer: A Systematic Review of the Literature With Meta-Analysis. Br J Cancer (2005) 92:131–9. doi: 10.1038/sj.bjc.6602258
43. Xu Y, Li H, Huang Z, Chen K, Yu X, Sheng J, et al. Predictive Values of Genomic Variation, Tumor Mutational Burden, and PD-L1 Expression in Advanced Lung Squamous Cell Carcinoma Treated With Immunotherapy. Transl Lung Cancer Res (2020) 9:2367–79. doi: 10.21037/tlcr-20-1130
44. Han X, Li F, Fang Z, Gao Y, Fang R, Yao S, et al. Transdifferentiation of Lung Adenocarcinoma in Mice With Lkb1 Deficiency to Squamous Cell Carcinoma. Nat Commun (2014) 5:3261. doi: 10.1038/ncomms4261
45. Zhang H, Fillmore Brainson C, Koyama S, Redig AJ, Chen T, Li S, et al. Lkb1 Inactivation Drives Lung Cancer Lineage Switching Governed by Polycomb Repressive Complex 2. Nat Commun (2017) 8:14922. doi: 10.1038/ncomms15901
46. Pan Y, Han H, Labbe KE, Zhang H, Wong KK. Recent Advances in Preclinical Models for Lung Squamous Cell Carcinoma. Oncogene (2021) 40:2817–29. doi: 10.1038/s41388-021-01723-7
47. Quintanal-Villalonga A, Taniguchi H, Zhan YA, Hasan MM, Chavan SS, Meng F, et al. Multi-Omic Analysis of Lung Tumors Defines Pathways Activated in Neuroendocrine Transformation. Cancer Discov (2021). doi: 10.1101/2020.12.02.408476
48. Tochigi N, Dacic S, Nikiforova M, Cieply KM, Yousem SA. Adenosquamous Carcinoma of the Lung: A Microdissection Study of KRAS and EGFR Mutational and Amplification Status in a Western Patient Population. Am J Clin Pathol (2011) 135:783–9. doi: 10.1309/AJCP08IQZAOGYLFL
49. Wang R, Pan Y, Li C, Zhang H, Garfield D, Li Y, et al. Analysis of Major Known Driver Mutations and Prognosis in Resected Adenosquamous Lung Carcinomas. J Thorac Oncol (2014) 9:760–8. doi: 10.1097/JTO.0b013e3182a406d1
50. Pelosi G, Fabbri A, Bianchi F, Maisonneuve P, Rossi G, Barbareschi M, et al. Δnp63 (P40) and Thyroid Transcription Factor-1 Immunoreactivity on Small Biopsies or Cellblocks for Typing Non-Small Cell Lung Cancer: A Novel Two-Hit, Sparing-Material Approach. J Thorac Oncol (2012) 7:281–90. doi: 10.1097/JTO.0b013e31823815d3
51. Travis W. Thoracic Tumours. In: WHO Classification of Tumours, vol. 5. Lyon: World Health Organization (2021).
52. Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med (2018) 142:321–46. doi: 10.5858/arpa.2017-0388-CP
53. Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, et al. Recommendations for the Use of Next-Generation Sequencing (NGS) for Patients With Metastatic Cancers: A Report From the ESMO Precision Medicine Working Group. Ann Oncol (2020) 31:1491–505. doi: 10.1016/j.annonc.2020.07.014
Keywords: lung squamous cell carcinoma (LUSC), NSCLC, RAS, KRAS, review
Citation: Acker F, Stratmann J, Aspacher L, Nguyen NTT, Wagner S, Serve H, Wild PJ and Sebastian M (2021) KRAS Mutations in Squamous Cell Carcinomas of the Lung. Front. Oncol. 11:788084. doi: 10.3389/fonc.2021.788084
Received: 01 October 2021; Accepted: 22 November 2021;
Published: 15 December 2021.
Edited by:
Matthias Scheffler, University Hospital of Cologne, GermanyReviewed by:
Federica Pezzuto, University of Padua, ItalyJohannes Brägelmann, University Hospital of Cologne, Germany
Copyright © 2021 Acker, Stratmann, Aspacher, Nguyen, Wagner, Serve, Wild and Sebastian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fabian Acker, ZmFiaWFuLmFja2VyQGtndS5kZQ==
 Fabian Acker
Fabian Acker Jan Stratmann1
Jan Stratmann1 Peter J. Wild
Peter J. Wild Martin Sebastian
Martin Sebastian