- 1Department of Hematology, The First Affiliated Hospital of Xiamen University and Institute of Hematology, School of Medicine, Xiamen University, Xiamen, China
- 2Department of Hematology, Key Laboratory of Xiamen for Diagnosis and Treatment of Hematological Malignancy, Xiamen, China
The key factors leading to transformed follicular lymphoma (t-FL) include the aberrations of epigenetic modifiers as early and driving events, especially mutations in the gene encoding for histone acetyltransferase. Therefore, reversal of this phenomenon by histone deacetylase (HDAC) inhibitors is essential for the development of new treatment strategies in t-FL. Several t-FL cell lines were treated with various doses of chidamide and subjected to cell proliferation, apoptosis and cell cycle analyses with CCK-8 assay, Annexin V/PI assay and flow cytometry, respectively. Chidamide dose-dependently inhibited cell proliferation, caused G0/G1 cycle arrest and triggered apoptosis in t-FL cells. In addition, the effects of chidamide on tumor growth were evaluated in vivo in xenograft models. RNA-seq analysis revealed gene expression alterations involving the PI3K-AKT signaling pathway might account for the mechanism underlying the antitumor activity of chidamide as a single agent in t-FL. These findings provide a basis for further clinical exploration of chidamide as a promising treatment for FL.
Introduction
Follicular lymphoma (FL) is the most common indolent lymphoma, accounting for approximately 20% of all non-Hodgkin lymphoma (NHL) cases (1, 2). With the use of current front-line regimens, the majority of FL patients have an initial response to therapy, with 40 to 80% demonstrating complete response (3, 4). However, despite improvement in front-line treatment, conventional therapy for FL is not curative, and approximately 20% of patients still experience either refractoriness or early relapse, which occurs in the first 2 years after diagnosis and treatment by chemoimmunotherapy (5–7). Moreover, such early relapse cases are often chemo-resistant, leading to significantly shorter survival (4, 8). Thus, integrating molecular targeted therapies into current treatment protocols and adjusting conventional treatment to improve survival, without compromising long-term quality of life is urgently needed in FL patients with poor prognosis.
Histone deacetylases (HDACs), which act as “epigenetic erasers”, are known to catalyze the removal of acetyl groups from histones and non-histone proteins, thereby altering the transcription of oncogenes and tumor suppressor genes (9, 10). Aberrant HDAC expression occurs in both solid tumors and hematological cancers, including B-cell lymphoma (11, 12). Prior findings showed that dysregulation of histone acetylation contributes to lymphomagenesis, particularly in GC-derived lymphomas (13, 14). FL, similar to other cancers, has recurrent alterations in genes involved in maintaining chromatin structure and transcription machinery genes (4, 8). For example, somatic mutations or genomic loss in the CREBBP and EP300 genes that encode HATs lead to an imbalance between acetylation and deacetylation, and the occurrence of these mutations is associated with disease relapse and poor prognosis in FL (15–17). Perturbing the balance between histone acetylation and deacetylation, which is tightly regulated by HDACs, is one of the main mechanisms by which epigenetics may be exploited to harness chromatin remodeling (18). In this context, it raises the possibility that HDACs may serve as a potentially attractive therapeutic target in this disease.
According to previous studies, Class I HDACs 1-3 are the most important HDAC enzymes with close associations with the corresponding malignant phenotypes (19, 20). Currently, multiple HDAC inhibitors have been developed, Chidamide is a noteworthy drug that may target specifically subtypes 1, 2 and 3 of Class I and subtype 10 of Class IIb HDACs and lead to increased acetylation of histones H3 and H4, resulting ultimately in the activation of gene transcription (21, 22). Chidamide was first approved by the Chinese FDA for the treatment of relapsed or refractory (R/R) peripheral T cell lymphoma (PTCL) (23, 24). Afterwards, more and more studies have focused on the anti-cancer effects of chidamide in various tumors, including hematological tumors (25–27). However, significant knowledge gaps remain, including the mechanism underlying chidamide’s therapeutic effects. In addition, its clinical utility in FL is currently unclear.
This study evaluated the effects of chidamide in well-characterized transformed follicular lymphoma cell lines and a xenograft model of t-FL. In addition, we analyzed the molecular basis of chidamide’s effects by evaluating gene expression using microarrays in cells treated with the single agent in t-FL: 1) to identify genes and pathways affected by chidamide; 2) to determine biomarkers that could be used in preclinical studies.
Materials and Methods
Cell Lines and Molecules
Established human cell lines derived from t-FL, including RL, DOHH2, SU-DHL4 and Karpas422 cells, were obtained from Cobioer Biotechnology Company (Jiangsu, China). All cell lines were cultured at 37°C in a 5% CO2 incubator in RPMI-1640 (Gibco, CA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, CA, USA), 100 units/ml penicillin and 100 mg/ml streptomycin (Invitrogen, CA, USA). Chidamide (CS055; HBI-8000) was provided by Shenzhen Chipscreen Biosciences (Shenzhen, China) and dissolved in sterile DMSO (Sigma, MO, USA) to produce a 50-mM stock solution stored at -20°C for in vitro experiments and diluted in 0.5% (w/v) CMC-Na suspension for oral gavage.
Cell Viability Assessment
Cytotoxicity was determined with Cell Counting Kit-8 (CCK-8, APExBIO, Texas, USA). Briefly, 3×104 cells/well were seeded in 100μl medium in 96-well plates and treated with various concentrations of chidamide alone for 24, 36 and 48 h. The CCK-8 reagent (10μl/well) was then added and incubated for additional 2 h, after which absorbance at 450 nm was detected on a Bio-Rad microplate reader (Bio-Rad, CA, USA). Data from three independent triplicate experiments were presented as a percentage of viable cells relative to untreated controls. IC50 values were determined with the GraphPad Prism 6 software.
FACS Analysis of the Cell Cycle and Apoptosis
Cells were treated with various concentrations of chidamide for the designated times. Cells were harvested and processed according to the manufacturer’s protocols. For cell cycle analysis, propidium iodide (PI)/RNase staining buffer from BD Pharmingen (556463, New Jersey, USA) was used. Cells were then analyzed on a CytoFlex S flow cytometer (Beckman Coulter, CA, USA). Data analysis was performed with the FlowJo software (San Carlos, CA, USA). Apoptosis was measured with the Annexin V/PI apoptosis detection kit (BD Pharmingen, USA). Cells positive for Annexin V were determined to be apoptotic (28), and were located in the right quadrant of the dot plot. Statistical analysis was performed by ANOVA. P-values below 0.05 in comparison to the control group were considered significant.
Western Blot
Protein extraction, separation and immunoblotting were performed as previously described (29). The following antibodies were used: anti-PDK1 (CA3062, 1:1000, Cell Signaling Technology, MA, USA), anti-P-PDK1 (Ser241) (CA3061, 1:1000, CST), anti-Akt (CA9272, 1:1000, CST), anti-P-Akt (Ser473) (CA4060, 1:1000, CST), anti-P-Akt (Thr308) (CA9275, 1:1000, CST), anti-CDK2 (CA2546, 1:1000, CST), anti-P-CDK2 (Thr160) (CA2561, 1:1000, CST), anti-PARP (CA9532, 1:1000, CST), anti-Cleaved PARP (CA5625, 1:1000, CST), anti-Caspase-3 (CA9662, 1:1000, CST), anti-Cleaved Caspase-3 (CA9661, 1:1000, CST), anti-P27 (CA3698, 1:1000, CST), anti-HDAC1 (CA5356, 1:1000, CST), anti-HDAC2 (CA5113, 1:1000, CST), anti-HDAC3 (CA3949, 1:1000, CST), anti-HDAC10 (ab108934, 1:1000, Abcam, Cambridge, UK), anti-Histone H3 (CA4499, 1:2000, CST), anti-Histone H3/acetyl K27 (ab4729, 1:1000, Abcam), and secondary HRP-linked antibodies (1:2000, Cell Signaling Technology, MA, USA). Anti-GAPDH (CA60004-1-Ig, 1:10000, Proteintech, Suite, USA) was used as a loading control. Blots were then detected using the hypersensitive ECL chemiluminescence kit (NCM Biotech, Suzhou, China) and the Bio-Rad ChemiDoc XRS + detection system (Bio-Rad, CA, USA).
RNA-Sequencing
DOHH2 cells were incubated with chidamide for 24h, followed by total RNA extraction with TRIzol reagent (Invitrogen, NY, USA) according to the manufacturer’s instructions. Totally, 1μg total RNA with RIN above 6.5 was used for subsequent library preparation. Next generation sequencing libraries were constructed according to the manufacturer’s protocol. Then, libraries with different indices were multiplexed and loaded on an Illumina HiSeq instrument according to the manufacturer’s instructions (Illumina, CA, USA). Sequencing was carried out using the 2x150 bp paired-end (PE) configuration; image analysis and base calling were conducted with HiSeq Control Software (HCS) + OLB + GAPipeline-1.6 (Illumina) on the HiSeq instrument. KEGG pathway and GO analyses were performed using the R Studio approach. Differential expression analysis used the DESeq2 Bioconductor package, a model based on negative binomial distribution; adjusted p value (padj)<0.05 indicated differential expression.
In vivo Experiments
All animal procedures were performed in accordance with the guidelines of the Animal Care and Use Committee and Ethics Committee of Xiamen University. DOHH2 cells (200μl of PBS, 1×107 cells/mouse) were inoculated subcutaneously into the back of female CB17/Icr-Prkdcscid/IcrlcoCrl mice (approximately 14-16g of body weight, Xiamen University Laboratory Animal Center, Fujian, China). After 3 days, mice were randomly divided into two groups (8 animals per group), to receive vehicle (PBS with 0.2% methyl cellulose/0.1% Tween 80) and chidamide (10 mg/kg/d), administered by oral gavage for 3 successive weeks, respectively. Tumor size and body weight were measured every two days. Tumor volumes were calculated according to the formula V= (L × W2)/2 [V, volume (mm3); L, length (mm); W, width (mm)]. After euthanasia, tumor tissues were extracted and divided into two parts: one part was frozen at -80°C for protein extraction followed by Western blot, and the other was fixed with 4% paraformaldehyde for hematoxylin and eosin (H&E) staining, immunohistochemistry (IHC) and immunofluorescence (IF). The slides were incubated overnight at 4°C with primary antibodies targeting Ki67 (27309-1-AP, 1:2000, Proteintech, Suite, USA) and PCNA (10205-2-AP, 1:200, Proteintech) antibodies. Subsequently, DAB (DAB-2032, MXB Biotechnologies, Fujian, China) was applied for 5 min at room temperature according to the manufacturer’s instructions. TUNEL-FITC (A111-03, Vazyme Biotech, Jiangsu, China) was applied at room temperature for 20 min, and analysis was performed under a fluorescence microscope (Nikon, Eclipse Ci-L, Japan).
Statistical Analyses
Statistical analyses were performed with Statistical Product and Service Solutions (SPSS) 21.0 (IBM Corp., Amronk, New York, USA), GraphPad Prism 6 (GraphPad Software, CA, USA) and Microsoft Office Excel (WA, USA). Unpaired Student’s t-test was performed to compare group pairs. Multiple groups were compared by one-way ANOVA, followed by post-hoc Bonferroni test. All quantifications were performed based on at least three independent experiments. P<0.05 was considered statistically significant.
Results
Effect of Chidamide on t-FL Cell Viability
We evaluated the anti-proliferative activity of chidamide in four t-FL cell lines, including RL, DOHH2, SU-DHL4 and Karpas422 cells. As assessed by the CCK-8 assay, after exposure to a series of concentrations for 24, 36 and 48 h, chidamide potently reduced cell viability in all four FL cell lines in a dose-dependent manner (Figure 1). The IC50 values of chidamide for these four cell lines over different treatment periods (Supplementary Table 1) revealed that chidamide inhibition of RL, DOHH2, SU-DHL4 and Karpas422 cells was time-dependent. IC50 values for chidamide-treated DOHH2 cells (9.08± 2.03, 0.85 ± 0.07 and 0.54 ± 0.05 μM, respectively) were close to those obtained in SU-DHL4 cells (4.56± 0.31, 3.17± 0.2 and 1.67± 0.05 μM, respectively) following incubation times of 24, 36 and 48 h. However, for the same incubation times, IC50 values in RL (30.39± 26.45, 7.447 ± 0.87 and 1.87± 0.25 μM, respectively) and Karpas-422 cells 10.92 ± 0.15, 5.10 ± 0.23 and 3.09 ± 0.23 μM, respectively) decreased steeply with increasing drug exposure time (about 16.0 and 3.0-fold from 24 h to 48 h, respectively), and were higher than the corresponding IC50 values recorded for DOHH2 and SU-DHL4 cells. These findings suggested that DOHH2 and SU-DHL4 cells may be more sensitive to chidamide in terms of viability and proliferation.
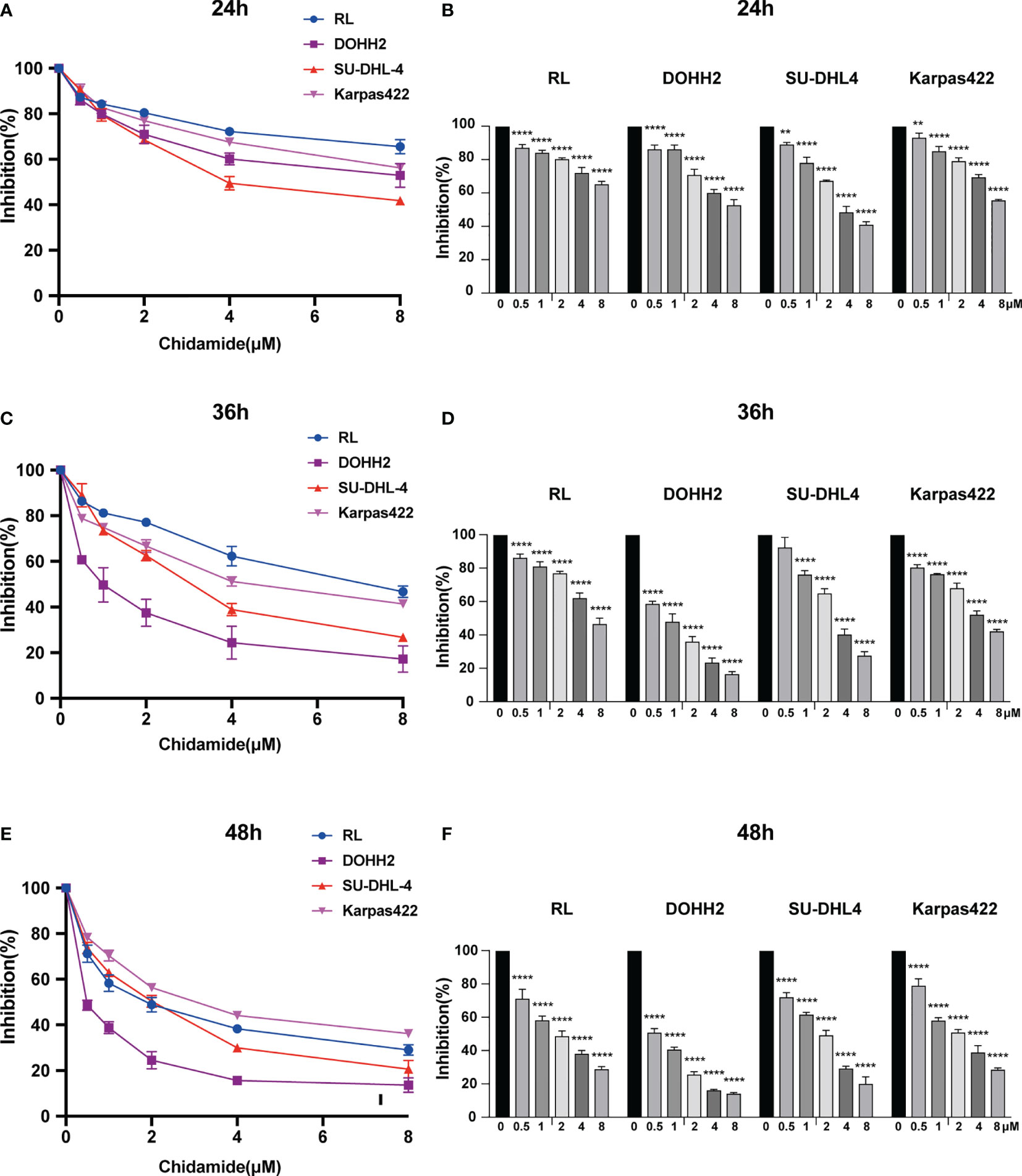
Figure 1 Chidamide inhibits viability in FL cell lines. Human FL RL, DOHH2, SU-DHL4 and Karpas422 cells were exposed to the indicated concentrations of chidamide for (A) 24 h, (C) 36 h or (E) 48 h, after which cell proliferation was measured by the CCK-8 assay. Data (mean ± S.D.) from at least three independent experiments are shown in (B) 24 h, (D) 36 h and (F) 48 h. (**p < 0.01; ****p < 0.0001).
Chidamide Induces Caspase-Dependent Apoptosis in t-FL Cells
To further assess the antitumor effect of chidamide on t-FL cells, flow cytometry after Annexin V/PI staining was performed to examine whether chidamide induces apoptosis in t-FL cells. Four t-FL cell lines were cultured with increasing concentrations of chidamide for 24 and 48 h before apoptosis assessment. Consistent with the CCK-8 assay, dose and time-dependent induction of cell death was evident in both DOHH2 and SU-DHL4 cells, whereas RL and Karpars422 cells showed reduced, although significant induction of apoptosis (Figure 2 and Supplementary Figure 1), further indicating that DOHH2 and SU-DHL4 cells were more sensitive to chidamide and suggesting cell line-specific differences. Thus, besides the reduced cell proliferation, increased cell death might be another factor contributing to chidamide activity in t-FL cells. We next sought to confirm the potential mechanism underlying chidamide’s anti-apoptotic effect. Western blot was carried out to detect the expression of activated caspase-3 and cleaved PARP after chidamide treatment for 12, 24 and 36 h, respectively, in DOHH2 and SU-DHL4 cells. Chidamide treatment for 24 h markedly upregulated cleaved caspase-3 and cleaved PARP (Figure 3). Taken together, these results suggested that chidamide induced apoptosis in FL cells by triggering the caspase dependent pathway.
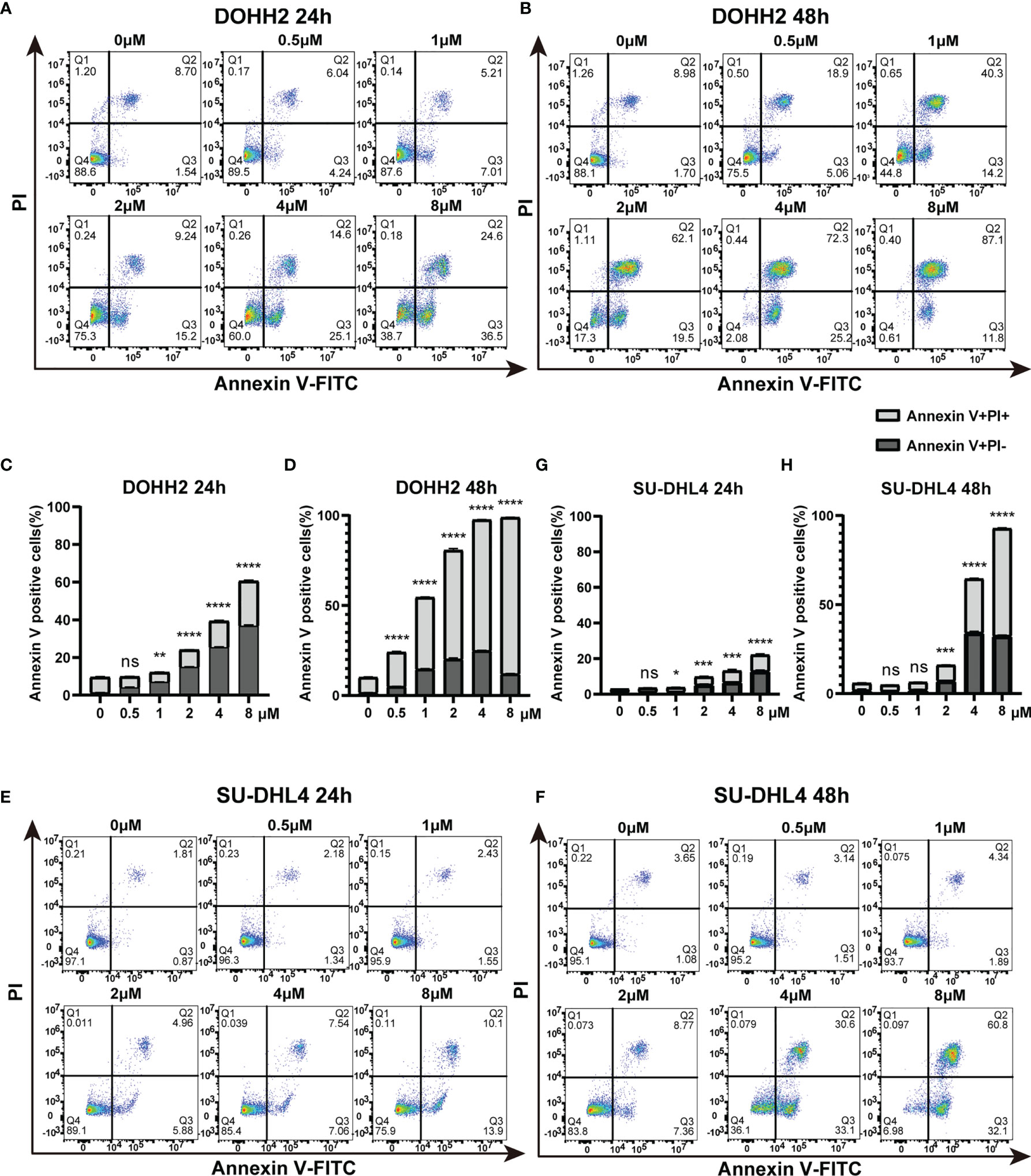
Figure 2 Chidamide induces apoptosis in highly sensitive FL DOHH2 and SU-DHL4 cells. (A, B) DOHH2 and (E, F) SU-DHL4 cells were treated with various doses of chidamide for 24 h or 48 h, after which the percentages of apoptotic cells were examined by Annexin V/PI double staining. The levels of apoptotic cells were remarkably increased at the indicated times of exposure to chidamide in (C, D) DOHH2 and (G, H) SU-DHL4 cells. Data are mean ± S.D. (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; NS, p > 0.05).
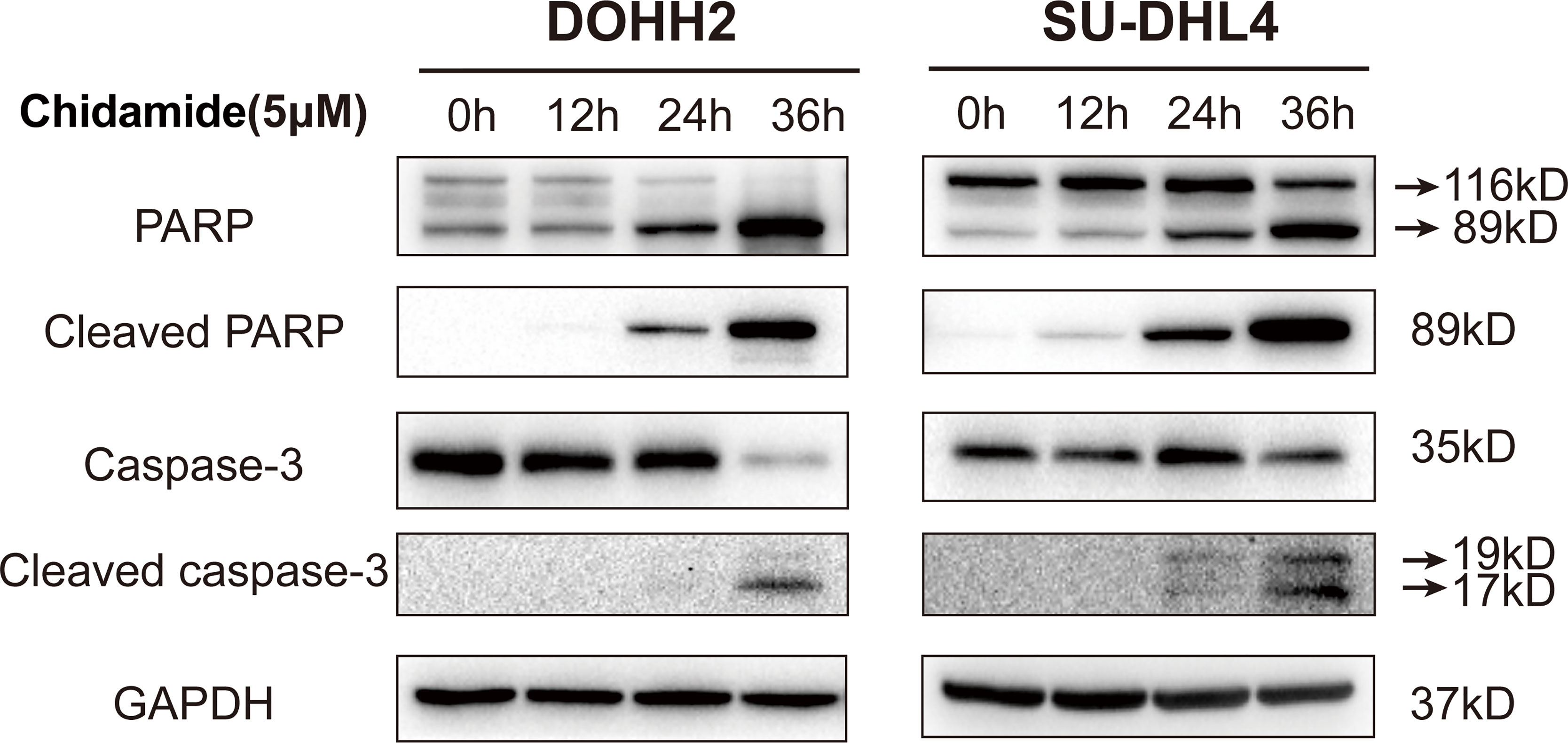
Figure 3 Chidamide induces apoptosis in FL cells by triggering the caspase dependent pathway. Western blot of apoptosis signaling proteins in DOHH2 and SU-DHL4 cells, including PARP, cleaved-PARP, caspase-3 and cleaved caspase-3, exposed to 5μM chidamide for 12h, 24 h and 36 h.
Chidamide Induces Cell Cycle Arrest in the G0/G1 Phase
Aberrant HDAC expression has previously been shown to impair a subset of genes involved in cell cycle regulation. Accordingly, inducing cell cycle arrest may be an underlying mechanism of chidamide’s effect on FL cells. In the above CCK-8 assay, chidamide exhibited a potent inhibitory effect on the proliferation of all four FL cells studied. We further investigated the effect of chidamide on cell cycle distribution in these four FL cell lines. After 24 h of incubation with various concentrations of chidamide, cell cycle analysis by flow cytometry revealed that chidamide induced the accumulation of cells in the sub-G0/G1 phase and reduced cells in the S phase in a concentration dependent manner, with little change in the G2 phase in RL, DOHH2, SU-DHL4 and Karpas422 cells (Figure 4 and Supplementary Figure 2). These data indicated that chidamide inhibited the proliferation of FL cells by inducing cell cycle arrest.
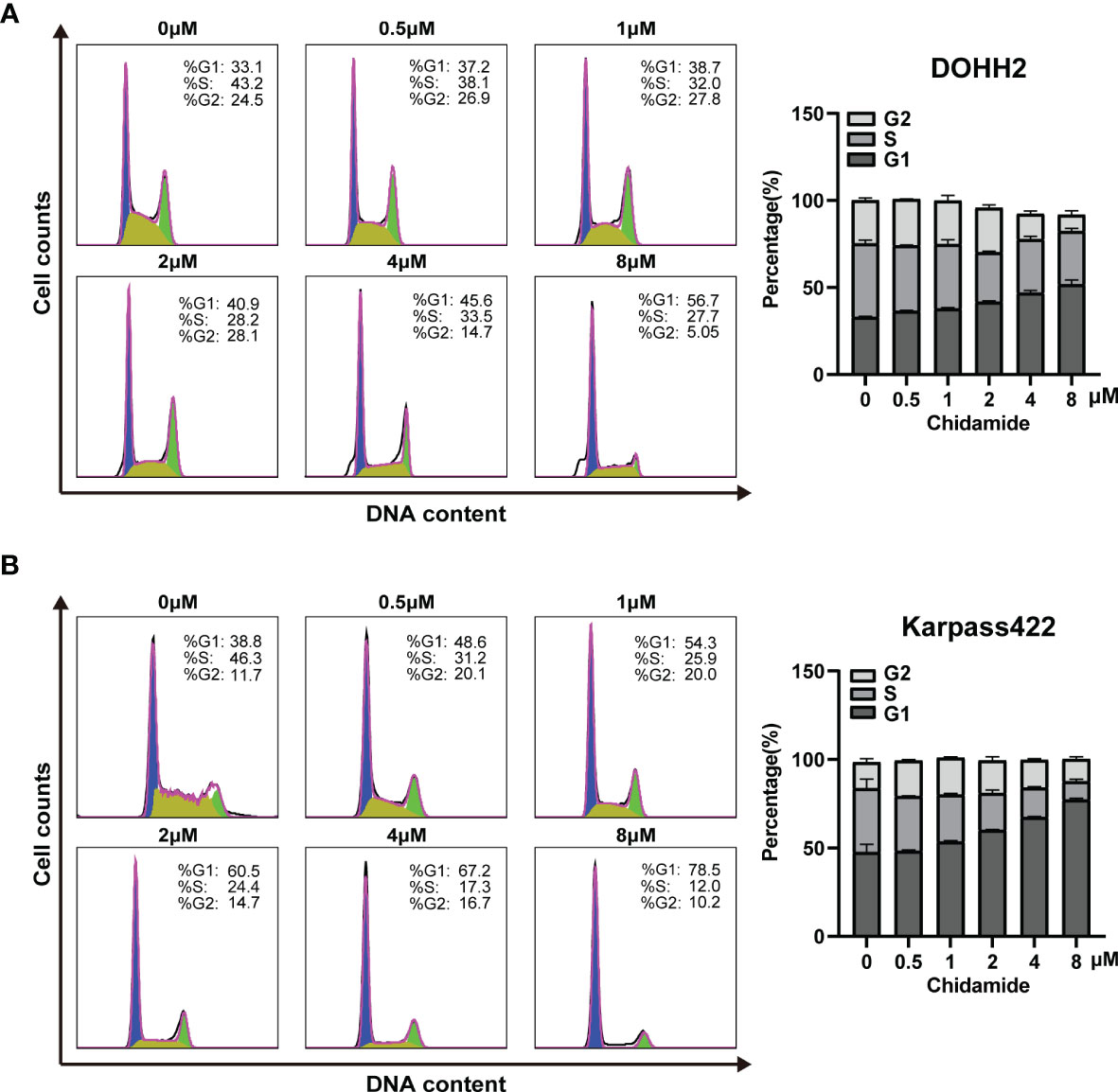
Figure 4 Chidamide induces FL cell cycle arrest in the G0/G1 phase. DOHH2 and Karpas422 cells were treated with chidamide for 24 h at the indicated concentrations, and cell cycle distribution in (A) DOHH2 and (B) Karpas422 cells was analyzed by flow cytometry. Representative flow cytograms are shown in the right panel.
Transcriptional Signature of Chidamide’s Effects in t-FL Cells
To obtain a global profile of the transcriptional changes after chidamide treatment, we performed genome-wide gene expression (GEP) on the most sensitive cell line (DOHH2 cells) treated with DMSO or chidamide (5µM) for 24 h. Totally, 4114 and 2095 genes in DOHH2 cells were significantly upregulated and downregulated (log2FC≥1, P<0.05) by chidamide, respectively (Figure 5A). KEGG analysis revealed that chidamide affected several important biological processes, including DNA replication, MAPK signaling, PI3K/AKT signaling and cell cycle regulation (Figures 5B, C). The downregulated transcripts mainly comprised HDAC, P53 and CDK2 or genes involved in the PI3K/AKT pathway (Figure 5D). In this context, Western blot was performed to validate the target specificity of chidamide on HDAC1, 2, 3 and 10. As shown in Figure 6, exposure of DOHH2 and SU-DHL4 cells to chidamide resulted in time-dependent downregulation of HDAC1, 2, 3 and 10, thereby causing hyper-acetylation of histones H3 and H4. Chidamide also upregulated p27 and downregulated phosph-CDK2 (Thr160) in a time-dependent manner. However, Western blot detected no expression change for P27 in RL and Karpas-422 cells (Supplementary Figure 3), which might be partially responsible for their lower sensitivity to chidamide. Taken together, these results suggested that the mechanism underlying the anti-proliferative activity of chidamide might involve downregulation of molecules related to the PI3K kinase pathway and cell cycle arrest in t-FL cells.
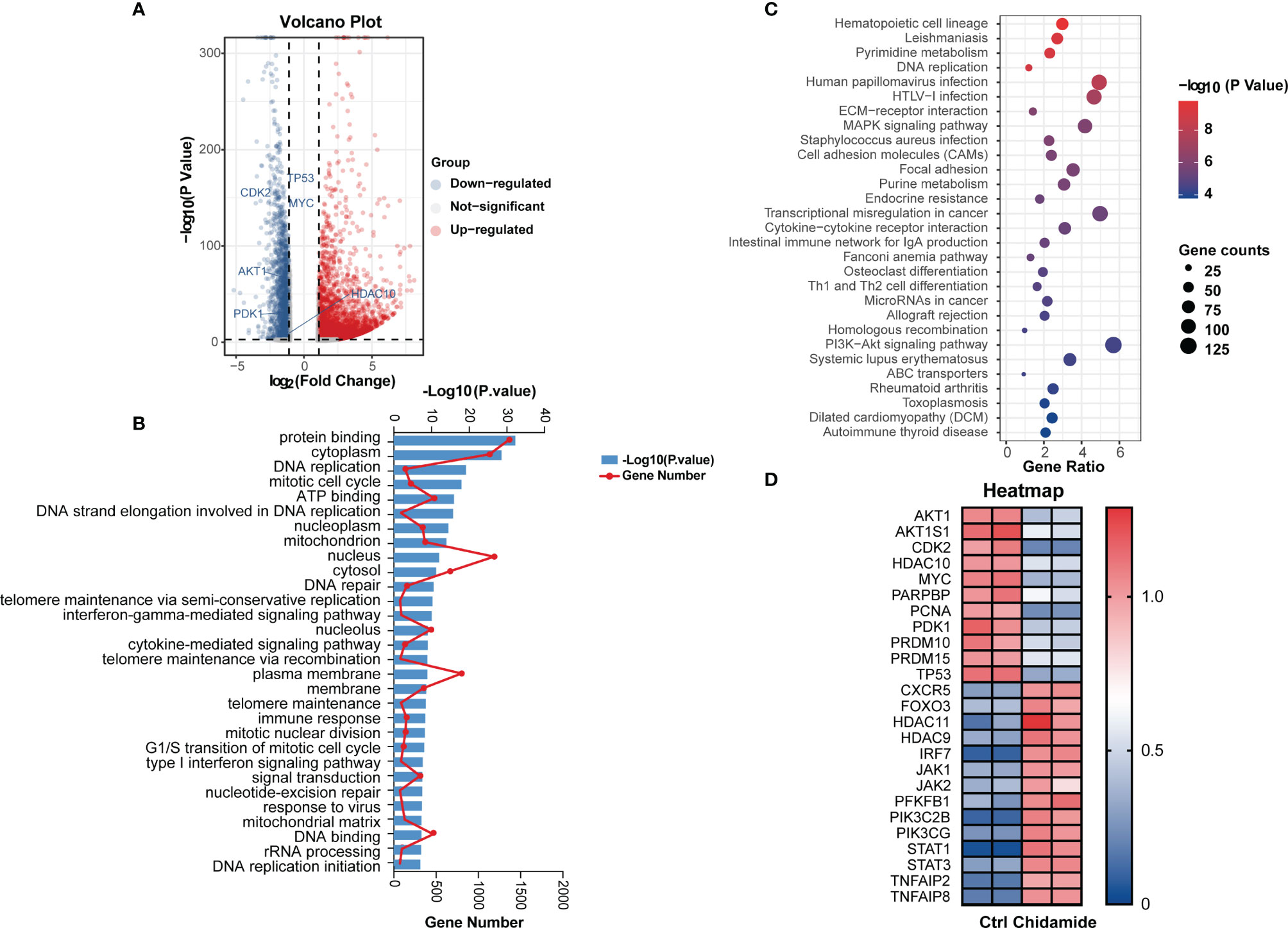
Figure 5 Effects of chidamide in genome-wide gene expression and relative pathway in DOHH2 cells. RNA-seq was performed to profile genome-wide gene expression in DOHH2 cells treated with 5μM chidamide for 24 h. (A) Volcano plot depicting 4114 and 2095 genes significantly upregulated and downregulated compared with the DMSO control groups, respectively. (B) Gene Ontology (GO) enrichment analysis of DEGs was performed by Gene Set Enrichment Analysis (GSEA). Pathways affected by chidamide in the DOHH2 cell line. (C) Differential signaling pathways in KEGG pathway enrichment analysis showing genes involved in PI3K/AKT signaling were significantly enriched. (D) Heat map of differentially expressed genes in DOHH2 cells in response to chidamide for 24 h.
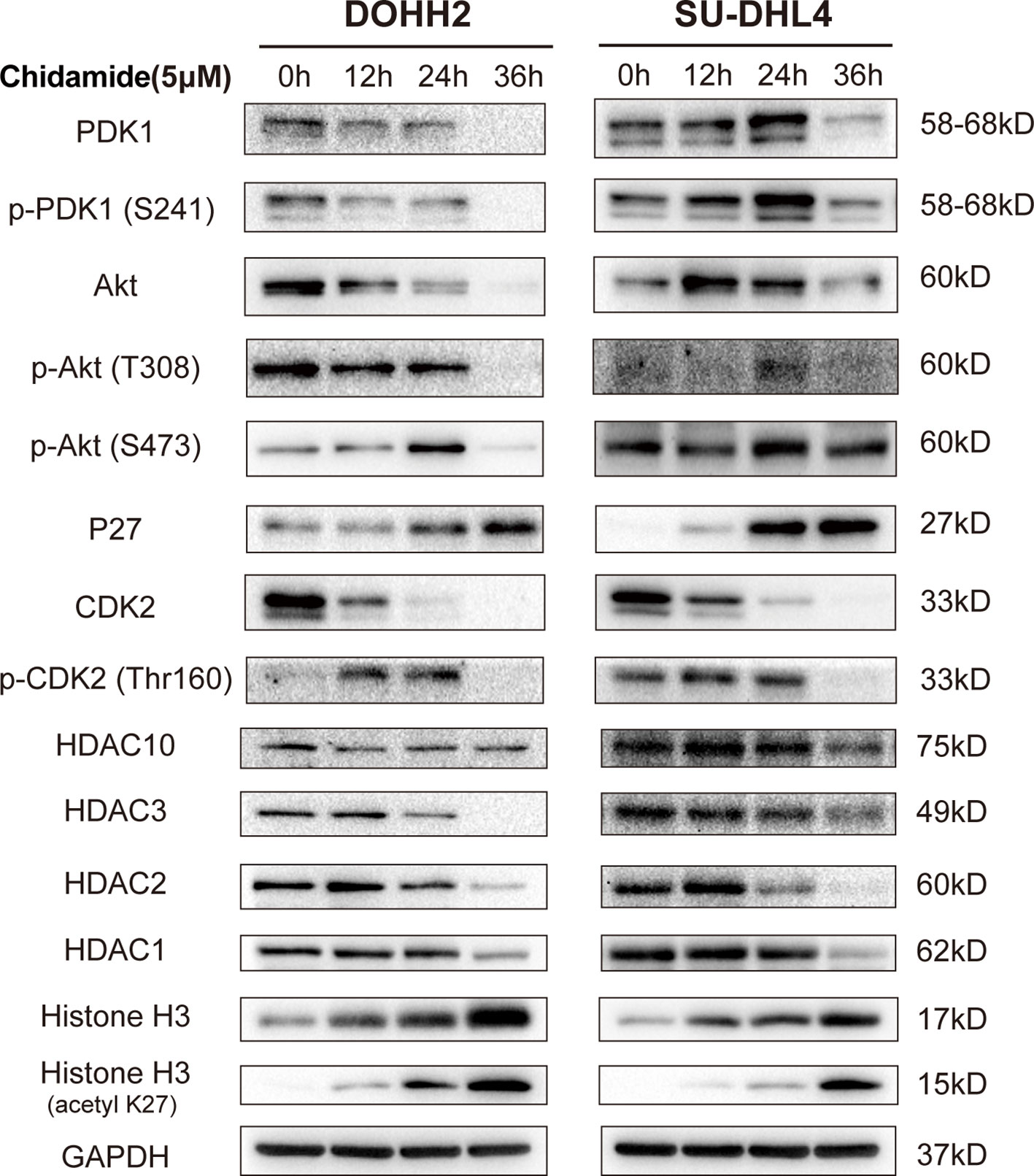
Figure 6 Differential gene and protein expression in FL cells after chidamide treatment. DOHH2 and SU-DHL4 cells were treated with 5μM chidamide for the indicated times. The levels of differential genes were determined by Western blot.
Chidamide Affects the PI3K/PDK1/AKT Signaling Pathway in t-FL Cells
Chidamide negatively regulated transcripts encoding MYC- and P53-regulated genes, as well as PI3K/AKT signaling pathway effectors, including HDAC10, AKT1, CDK2, MYC, PARPBP, PCNA, PDK1, PRDM10 and PRDM15 in DOHH2 cells (Figure 5D). The inhibitory effect of chidamide on the PI3K/AKT signaling pathway was further confirmed at the protein level. Western blot showed a marked reduction in the expression of PDK1 and phospho-AKT (Ser473 or Thr308) in both DOHH2 and SU-DHL4 cells after chidamide treatment for 36 h (Figure 6). In contrast, chidamide did not affect the expression of phospho-AKT (Ser473) in RL and Karpas-422 cells (Supplementary Figure 3). Thus, we speculated that chidamide might promote apoptosis and suppress proliferation in part by disrupting the PI3K/PDK1/AKT signaling pathway in t-FL cells.
Chidamide Has Antitumor Activity in a FL Tumor Xenograft Model
Finally, the anti-tumor activity of chidamide was examined in a CB17/Icr-Prkdcscid/IcrlcoCrl mouse xenograft model bearing DOHH2 cells. In this study, DOHH2 cells (107) were injected subcutaneously into the back of mice, which were randomly divided into the vehicle control and chidamide groups. Vehicle or chidamide (10 mg/kg/day) was orally given continuously daily for three weeks (Figure 7A). After chidamide treatment for 13 days, compared with the control group, mouse tumors showed obvious growth inhibition with no fatal toxicity (Figures 7B, C). Although a temporary body weight loss was observed at the beginning of chidamide administration, it was recovered after a short period of time. Tumors were collected from 5 mice randomly selected per group at the study endpoint (Day 22). As shown in Figure 7D, chidamide treatment resulted in a marked reduction of tumor burden, reflected by decreased volume and weight of tumor masses, compared with the vehicle control (Figures 7E, F). Besides, chidamide significantly prolonged survival in the treatment group compared with vehicle treated animals (Figure 7G).
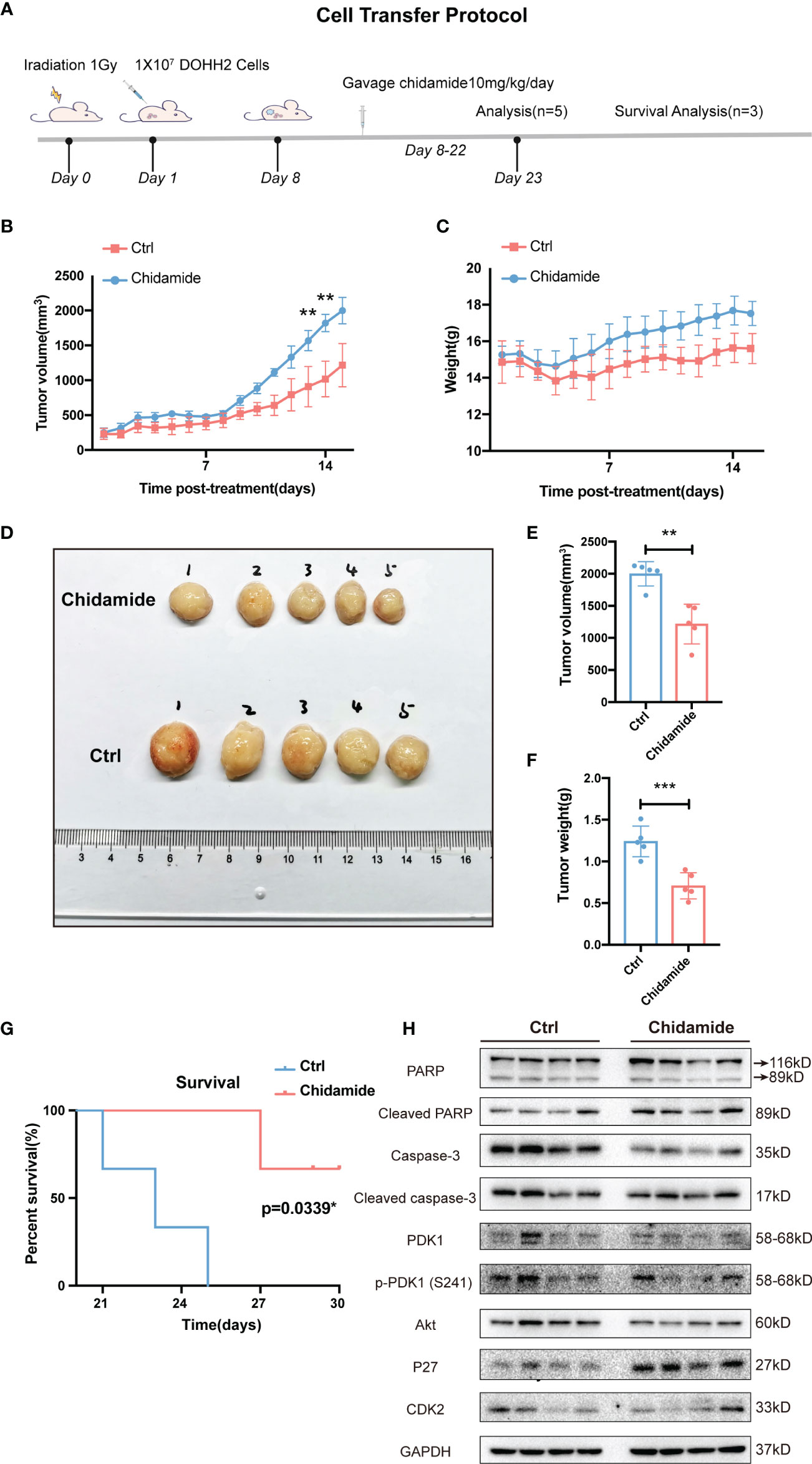
Figure 7 Effect of chidamide on tumor growth in xenograft mouse models. (A) Cell injection protocol in a FL tumor xenograft model. Tumor volumes (B) and body weights (C) of mice were measured daily and presented as mean ± S.D. (D) Images of tumors from DOHH2-bearing xenograft mice after the indicated treatments (n=10). Tumor volumes (E) and weights (F) in the control and chidamide groups were compared to evaluate the treatment response to chidamide. (G) Kaplan Meier overall survival (OS) curves of tumor-bearing xenograft mice. (H) Chidamide suppressed the PDK1-Akt-P27-CDK2 signaling pathway in vivo. The protein levels of PDK1, AKT, P27, CDK2, PARP, cleaved-PARP, caspase3 and cleaved-caspase-3 were determined by Western blot. (**p < 0.01; ***p < 0.001).
The tumor tissues obtained from chidamide treated animals displayed obvious nuclear shrinkage as shown by H&E staining (Figure 8A). Apoptosis in tumor tissues was detected by the TUNEL assay. The number of apoptotic cells was markedly increased in chidamide treated tumors (Figures 8B, C), in agreement with in vitro findings. Immunohistochemical staining revealed that Ki-67 and PCNA levels were decreased in tumor tissues from chidamide treated mice (Figures 8D–F). Compared with the vehicle control group, chidamide treated mice showed significantly reduced p-PDK1 expression, and markedly increased expression of P27, cleaved caspase-3 and cleaved PARP (Figure 7H). These results were consistent with in vitro findings. Taken together, these data suggested that chidamide effectively inhibited t-FL tumorigenesis and development in vivo.
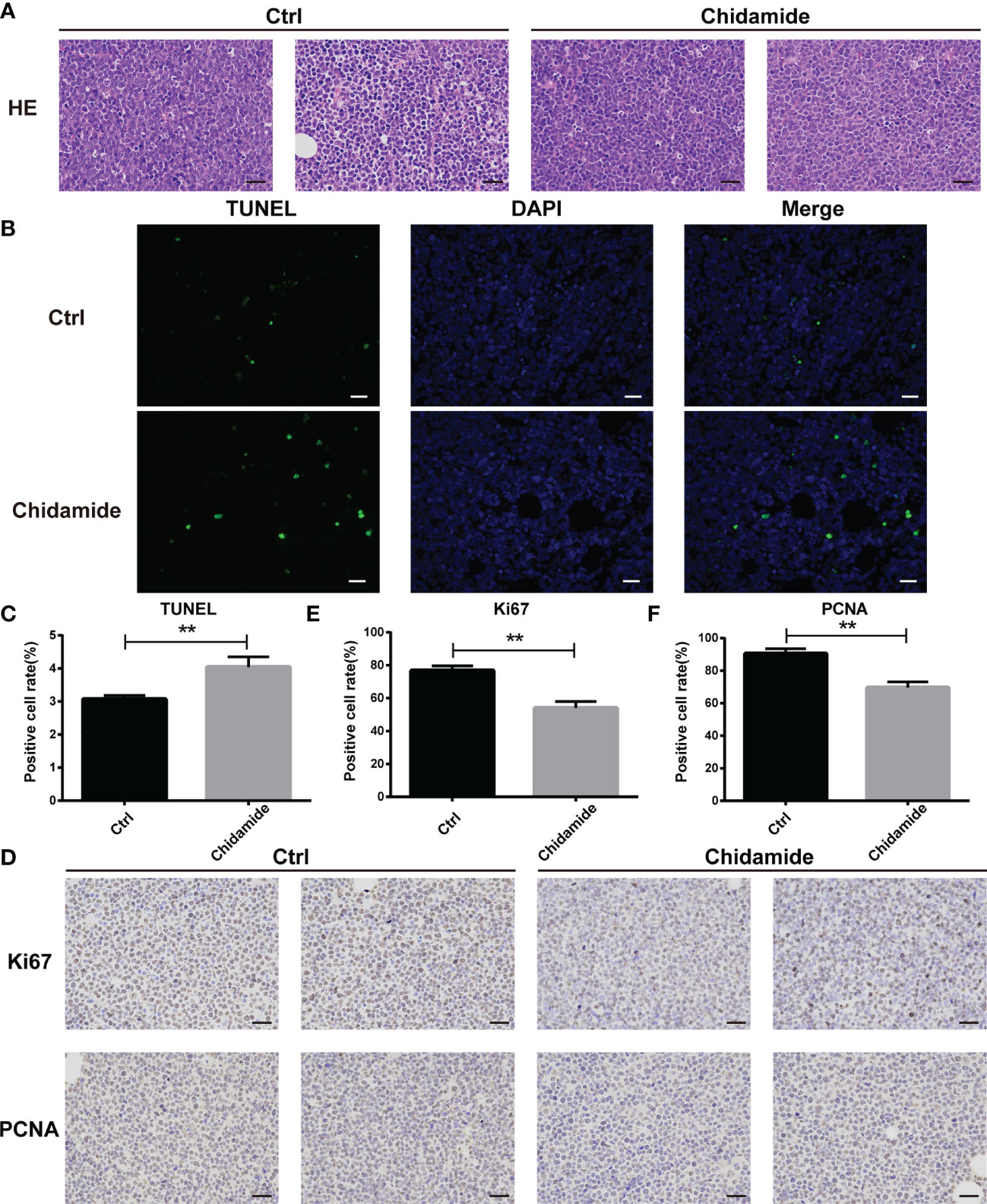
Figure 8 Inhibition of tumor growth in vivo by chidamide. (A) Tumor samples collected after treatment with vehicle/control or chidamide were fixed, sliced and stained with H&E. (B) Representative images of immunofluorescent TUNEL staining performed on serial sections of the tumors are shown; the corresponding statistical results are shown in (C). Images were acquired under a Nikon microscope (original magnification, ×400). (D) The expression levels of Ki67 and PCNA were detected by immunohistochemical staining. (E, F) Data represent three independent experiments, and are mean ± S.D. (**p < 0.01).
Discussion
Transformed follicular lymphoma (t-FL) is considered a disease predominantly caused by several epigenetic aberrations (e.g., mutations affecting the epigenetic modifiers KMT2D, EZH2, CREBBP and MEF2B) rather than sequential acquisition of genetic aberrations (8, 18). Thus, targeting enzymes involved in the regulation of DNA methylation and histone modifications might be critical for developing more effective treatment strategies in t-FL. In this regard, histone deacetylases (HDACs) represent the most widely studied therapeutic targets, with HDAC inhibitors (HDACi) including chidamide, vorinostat and romidepsin, approved for use in the therapy of cutaneous T-cell lymphoma (23, 30). However, clinical practice has not yet delivered desirable results by applying HDACi in the treatment of t-FL. Furthermore, the biological effects of HDACi in t-FL remain unclear, as well as the identification of response mechanisms. Taking these observations into consideration, we evaluated the activity and underlying mechanism of a benzamide-type selective HDAC inhibitor, chidamide, in preclinical models of t-FL cells.
HDACs have been demonstrated to play a crucial role in the pathogenesis of lymphoma (31, 32). Gil et al. reported that aberrant expression of HDAC9 could lead to lymphoproliferative disorders, including germinal center (GC) and post-GC lymphomas (33). In follicular lymphoma (FL) and diffuse large B cell lymphoma (DLBCL), Bcl6 recruits HDAC3 to repress transcription and trigger B cell lymphoma (34). All these data strongly suggest that HDACs are promising therapeutic targets for GC lymphomas. Furthermore, HDAC inhibitors have been reported to induce Bcl6 downregulation in GC lymphomas including DLBCL (35). Herein, we demonstrated that chidamide had an anticancer effect as a single agent in several t-FL cell lines. At clinically achievable concentrations, chidamide showed cell type- and dose-dependent cytotoxicity in t-FL cells at 24 h, with IC50 values ranging from 4.5µM to 30µM, and varied responses to chidamide among different cells. In DOHH2 and SU-DHL4 cells, which were relatively more sensitive to chidamide, a higher degree of apoptosis induced by chidamide was found as strongly evidenced by the activation of caspase-3. This is likely due to the cell origins or unknown differences of their genetic differentiation.
Since chidamide inhibited cell proliferation and induced apoptosis in t-FL cells, we further investigated whether chidamide regulates cell cycle progression, which is one of the main mechanisms by which HDACi induce tumor cell death. In this context, previous studies have shown that G1 arrest appears to be a common response to chidamide in various tumor cells (36–38). Thus, cell cycle regulators, including cyclins and CDK inhibitors (e.g., p21 and p27), may be tightly controlled by chidamide (27, 36, 39). In general, p27 is known to control G1 length and cell cycle exit by inhibiting the kinase activity of CDK2 bound to cyclin E, thereby causing the dephosphorylation of retinoblastoma protein (Rb), which blocks E2F activity in the transcription of genes required for G1/S transition (40, 41). In this study, microarray analysis of chidamide-treated t-FL cells pointed to cell cycle arrest in the G1 phase. Upon chidamide treatment, CDK2 was found to be specifically inhibited in DOHH2 and SU-DHL4 cells accompanied by p27 activation. However, obvious changes of p27 expression were not observed in RL and Karpas-422 cells, in disagreement with the anti-proliferative phenotype as well as G1 arrest in these cells, indicating that p27-mediated inhibition of CDK2 might partially contribute to chidamide-induced cell cycle arrest. On the other hand, we showed that chidamide treatment simultaneously caused the accumulation of histone H3 acetylation and the activation of p27 and cell cycle exit in DOHH2 and SU-DHL4 cells. The present findings indicate that the mechanism by which chidamide induces G1 arrest by inhibiting HDACs is cell-specific.
The integration of gene expression profiling and sensitivity in cancer cells allowed the identification of functional pathways that might predict the response to chidamide (27, 42). DOHH2 cells with the highest sensitivity to chidamide had high expression levels of genes involved in the PI3K/PDK1/AKT pathway, which is regulated in several human carcinomas, including lymphoid malignancies (43, 44). In addition, an important biological effect of chidamide, both in vitro and in vivo, was the downregulation of the PI3K/AKT pathway. Several studies have indicated that PI3K/AKT pathway downregulation is a relevant mechanism of chidamide’s effects in various cancer cell lines (27, 45, 46). The PI3K/AKT pathway was shown to be activated in human cancers by oncogenic mutations of the PIK3CA gene encoding the catalytic subunit p110a (47). In lymphoid malignancies, including DLBCL (48), MCL (49) and FL (50), hyperactivation of AKT is due to its enhanced activation/phosphorylation at Serine 473 (Ser473). In accordance with apoptosis and caspase-3 activation observed at 24 to 48 h, AKT phosphorylation at ser473 was inhibited in both DOHH2 and SU-DHL4 cells by chidamide treatment. Taken together, these findings suggested that chidamide modulated PI3K/AKT signaling, which is known to be involved in cell proliferation, cell cycle regulation, apoptosis and tumor development (Figure 9).
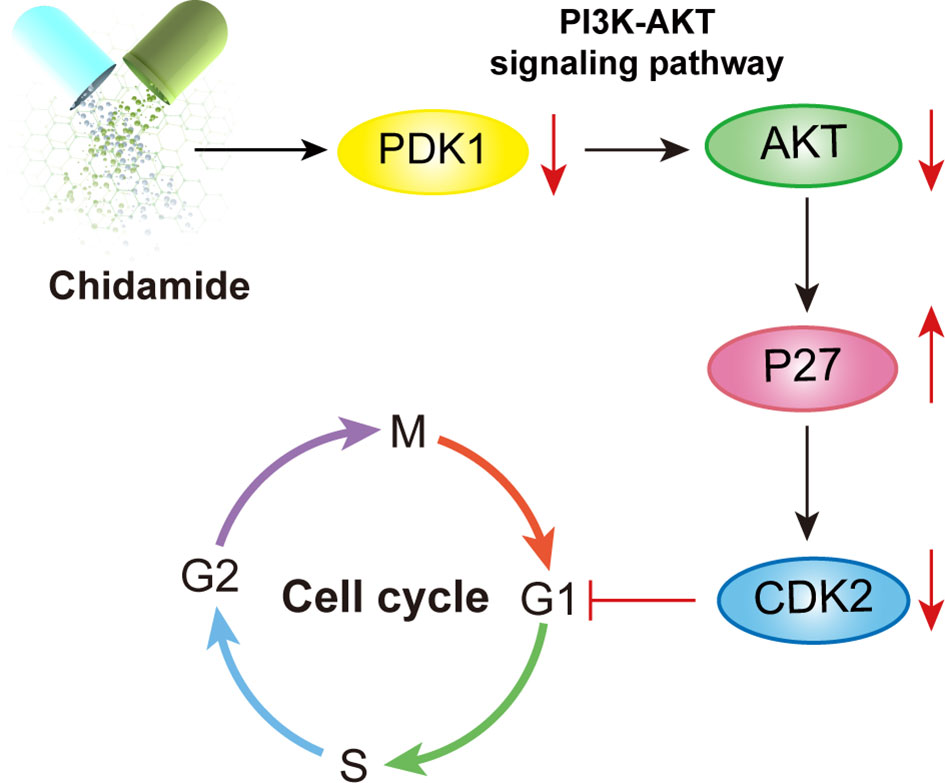
Figure 9 Illustration of the mechanism underlying chidamide induced follicular lymphoma growth inhibition. Chidamide exerts antitumor activity toward FL both in vitro and in vivo, potentially by targeting the PDK1-Akt-P27-CDK2 pathway and triggering caspase dependent apoptosis, finally markedly blocking cells in the G0/G1 phase.
As the activity of chidamide, as monotherapy at the doses tested in this study was modest, understanding potential biomarkers that are predictive of response is very important for the design of future clinical trials (51). In addition, emerging data from in vitro studies indicate that HDAC inhibitors, such as chidamide, may have improved activity when used in combination therapy (52–54). To this end, several recent studies have provided a strong preclinical rationale for combination with chemotherapy, immunotherapy, or molecular targeted therapy, paving the way for possible studies in selected populations (53–55). In our previous work, we revealed that ABT-199 (25) or MLL-menin inhibitor (56) has a synergistic inhibitory effect on acute myeloid leukemia cells when combined with chidamide. Combination with a demethylating agent also showed benefit in diffuse large B cell lymphomas (DLBCLs) (53). In addition, chidamide could increase PD-L1 expression in the tumor microenvironment, and preclinical studies have demonstrated synergy between chidamide, and PD-1 blockade in solid tumors (55, 57). A single-arm-phase II study is therefore currently ongoing to evaluate the activity of chidamide in combination with sintilimab in relapsed or refractory peripheral T-cell lymphomas (58). It is possible that chidamide administration in the context of these combination strategies could further enhance the killing of tumor cells.
In summary, this study demonstrated the importance of HDACs in the progression and transformation of FL, and provided a critical link between epigenetic changes and increased FL aggressiveness. We also provided evidence that chidamide exerts anticancer effects by inducing G1 arrest and apoptosis via PI3K/PDK1/AKT signaling pathway inactivation. Furthermore, in t-FL cells relatively sensitive to chidamide-induced apoptosis, chidamide caused significant changes in the transcriptome profile, providing a compelling rationale for chidamide as an effective single-agent in aggressive FL. The data presented here provide the basis for further exploration of chidamide in combination therapies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: SAMN22127095.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee and Ethics Committee of Xiamen University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
Conception and design: BX, JZ, and MZ. Development of methodology: MZ, JT, GP, YJ, and HZ. Analysis and interpretation of data: GP, QL, QC, and LF. Technical support: MD, BX, and JZ. Writing, review, and/or revision of the manuscript: JZ and MZ. Study supervision: BX and JZ. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Scientific Foundation of China (No. 82170180,81770126, 81800163), Fujian Natural Science Foundation of China (No. 2020J011246), Xiamen Municipal Bureau of Science and Technology (No. 3502Z20209003) and Lymphoma Research Fund of Chinese Anti-Cancer Association (No. CORP-117).
Conflict of Interest:
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.780118/full#supplementary-material
Supplementary Figure 1 | Chidamide induces apoptosis in relatively de novo resistant FL RL and Karpas422 cells. Representative flow cytograms depicting RL (A, B) and Karpas422 (E, F) cells exposed for 24 h or 48 h to chidamide. Data represent three independent experiments examining RL (C, D) and Karpas422 (G, H) cells, and are mean ± S.D. (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; NS: p > 0.05)
Supplementary Figure 2 | Chidamide induces G0/G1 arrest in FL cell lines. Representative flow cytograms of SU-DHL4 (A) and RL (B) cells treated for 24 h with chidamide.
Supplementary Figure 3 | Effect of chidamide on protein expression in resistant FL RL and Karpas422 cell lines. RL (A) and Karpas422 (B) cells were treated with 5μM chidamide for the indicated times. The levels of phospho-AKT (Ser473) and P27 were determined by Western blot.
Supplementary Table 1 | IC50 values of chidamide in FL cell lines.
Abbreviations
t-FL, transformed follicular lymphoma; DLBCL, Diffuse large B-cell lymphoma; HDACs, Histone deacetylases; HATs, histone acetyltransferases; NHL, Non-Hodgkin lymphoma; R/R, Relapsed or refractory; FBS, Fetal bovine serum; CCK-8, Cell Counting Kit-8; qRT-PCR, Real-time quantitative PCR; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PI, Propidium iodide; SPSS, Statistical Product and Service Solutions; GSEA, gene set enrichment analysis.
References
1. Teras LR, DeSantis CE, Cerhan JR, Morton LM, Jemal A, Flowers CR. 2016 US Lymphoid Malignancy Statistics by World Health Organization Subtypes. CA Cancer J Clin (2016) 66:443–59. doi: 10.3322/caac.21357
2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71:7–33. doi: 10.3322/caac.21654
3. Solal-Celigny P, Roy P, Colombat P, White J, Armitage JO, Arranz-Saez R, et al. Follicular Lymphoma International Prognostic Index. Blood (2004) 104:1258–65. doi: 10.1182/blood-2003-12-4434
4. Freedman A, Jacobsen E. Follicular Lymphoma: 2020 Update on Diagnosis and Management. Am J Hematol (2020) 95:316–27. doi: 10.1002/ajh.25696
5. Maurer MJ, Bachy E, Ghesquieres H, Ansell SM, Nowakowski GS, Thompson CA, et al. Early Event Status Informs Subsequent Outcome in Newly Diagnosed Follicular Lymphoma. Am J Hematol (2016) 91:1096–101. doi: 10.1002/ajh.24492
6. Casulo C, Byrtek M, Dawson KL, Zhou X, Farber CM, Flowers CR, et al. Early Relapse of Follicular Lymphoma After Rituximab Plus Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone Defines Patients at High Risk for Death: An Analysis From the National LymphoCare Study. J Clin Oncol (2015) 33:2516–22. doi: 10.1200/JCO.2014.59.7534
7. Seymour JF, Marcus R, Davies A, Gallop-Evans E, Grigg A, Haynes A, et al. Association of Early Disease Progression and Very Poor Survival in the GALLIUM Study in Follicular Lymphoma: Benefit of Obinutuzumab in Reducing the Rate of Early Progression. Haematologica (2019) 104:1202–8. doi: 10.3324/haematol.2018.209015
8. Carbone A, Roulland S, Gloghini A, Younes A, von Keudell G, Lopez-Guillermo A, et al. Follicular Lymphoma. Nat Rev Dis Primers (2019) 5:83. doi: 10.1038/s41572-019-0132-x
9. Quintas-Cardama A, Santos FP, Garcia-Manero G. Histone Deacetylase Inhibitors for the Treatment of Myelodysplastic Syndrome and Acute Myeloid Leukemia. Leukemia (2011) 25:226–35. doi: 10.1038/leu.2010.276
10. Bolden JE, Peart MJ, Johnstone RW. Anticancer Activities of Histone Deacetylase Inhibitors. Nat Rev Drug Discov (2006) 5:769–84. doi: 10.1038/nrd2133
11. Korfi K, Ali S, Heward JA, Fitzgibbon J. Follicular Lymphoma, a B Cell Malignancy Addicted to Epigenetic Mutations. Epigenetics (2017) 12:370–7. doi: 10.1080/15592294.2017.1282587
12. Wang X, Waschke BC, Woolaver RA, Chen SMY, Chen Z, Wang JH. HDAC Inhibitors Overcome Immunotherapy Resistance in B-Cell Lymphoma. Protein Cell (2020) 11:472–82. doi: 10.1007/s13238-020-00694-x
13. Lue JK, Prabhu SA, Liu Y, Gonzalez Y, Verma A, Mundi PS, et al. Precision Targeting With EZH2 and HDAC Inhibitors in Epigenetically Dysregulated Lymphomas. Clin Cancer Res (2019) 25:5271–83. doi: 10.1158/1078-0432.CCR-18-3989
14. Cogan JC, Liu Y, Amengual JE. Hypomethylating Agents in Lymphoma. Curr Treat Options Oncol (2020) 21:61. doi: 10.1007/s11864-020-00761-9
15. Desmots F, Roussel M, Pangault C, Llamas-Gutierrez F, Pastoret C, Guiheneuf E, et al. Pan-HDAC Inhibitors Restore PRDM1 Response to IL21 in CREBBP-Mutated Follicular Lymphoma. Clin Cancer Res (2019) 25:735–46. doi: 10.1158/1078-0432.CCR-18-1153
16. Zhang J, Vlasevska S, Wells VA, Nataraj S, Holmes AB, Duval R, et al. The CREBBP Acetyltransferase Is a Haploinsufficient Tumor Suppressor in B-Cell Lymphoma. Cancer Discov (2017) 7:322–37. doi: 10.1158/2159-8290.CD-16-1417
17. Meyer SN, Scuoppo C, Vlasevska S, Bal E, Holmes AB, Holloman M, et al. Unique and Shared Epigenetic Programs of the CREBBP and EP300 Acetyltransferases in Germinal Center B Cells Reveal Targetable Dependencies in Lymphoma. Immunity (2019) 51:535–47.e9. doi: 10.1016/j.immuni.2019.08.006
18. Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent Mutation of Histone-Modifying Genes in Non-Hodgkin Lymphoma. Nature (2011) 476:298–303. doi: 10.1038/nature10351
19. West AC, Johnstone RW. New and Emerging HDAC Inhibitors for Cancer Treatment. J Clin Invest (2014) 124:30–9. doi: 10.1172/JCI69738
20. Inoue S, Mai A, Dyer MJ, Cohen GM. Inhibition of Histone Deacetylase Class I But Not Class II is Critical for the Sensitization of Leukemic Cells to Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand-Induced Apoptosis. Cancer Res (2006) 66:6785–92. doi: 10.1158/0008-5472.CAN-05-4563
21. Ning ZQ, Li ZB, Newman MJ, Shan S, Wang XH, Pan DS, et al. Chidamide (CS055/HBI-8000): A New Histone Deacetylase Inhibitor of the Benzamide Class With Antitumor Activity and the Ability to Enhance Immune Cell-Mediated Tumor Cell Cytotoxicity. Cancer Chemother Pharmacol (2012) 69:901–9. doi: 10.1007/s00280-011-1766-x
22. Dong M, Ning ZQ, Xing PY, Xu JL, Cao HX, Dou GF, et al. Phase I Study of Chidamide (CS055/HBI-8000), a New Histone Deacetylase Inhibitor, in Patients With Advanced Solid Tumors and Lymphomas. Cancer Chemother Pharmacol (2012) 69:1413–22. doi: 10.1007/s00280-012-1847-5
23. Shi Y, Jia B, Xu W, Li W, Liu T, Liu P, et al. Chidamide in Relapsed or Refractory Peripheral T Cell Lymphoma: A Multicenter Real-World Study in China. J Hematol Oncol (2017) 10:69. doi: 10.1186/s13045-017-0439-6
24. Shi Y, Dong M, Hong X, Zhang W, Feng J, Zhu J, et al. Results From a Multicenter, Open-Label, Pivotal Phase II Study of Chidamide in Relapsed or Refractory Peripheral T-Cell Lymphoma. Ann Oncol (2015) 26:1766–71. doi: 10.1093/annonc/mdv237
25. Chen K, Yang Q, Zha J, Deng M, Zhou Y, Fu G, et al. Preclinical Evaluation of a Regimen Combining Chidamide and ABT-199 in Acute Myeloid Leukemia. Cell Death Dis (2020) 11:778. doi: 10.1038/s41419-020-02972-2
26. Guan XW, Wang HQ, Ban WW, Chang Z, Chen HZ, Jia L, et al. Novel HDAC Inhibitor Chidamide Synergizes With Rituximab to Inhibit Diffuse Large B-Cell Lymphoma Tumour Growth by Upregulating CD20. Cell Death Dis (2020) 11:20. doi: 10.1038/s41419-019-2210-0
27. Wang H, Liu YC, Zhu CY, Yan F, Wang MZ, Chen XS, et al. Chidamide Increases the Sensitivity of Refractory or Relapsed Acute Myeloid Leukemia Cells to Anthracyclines via Regulation of the HDAC3 -AKT-P21-CDK2 Signaling Pathway. J Exp Clin Cancer Res (2020) 39:278. doi: 10.1186/s13046-020-01792-8
28. Pan R, Ruvolo V, Mu H, Leverson JD, Nichols G, Reed JC, et al. Synthetic Lethality of Combined Bcl-2 Inhibition and P53 Activation in AML: Mechanisms and Superior Antileukemic Efficacy. Cancer Cell (2017) 32:748–60.e6. doi: 10.1016/j.ccell.2017.11.003
29. Zha J, Bi S, Deng M, Chen K, Shi P, Feng L, et al. Disulfiram/copper Shows Potent Cytotoxic Effects on Myelodysplastic Syndromes via Inducing Bip-Mediated Apoptosis and Suppressing Autophagy. Eur J Pharmacol (2021) 902:174107. doi: 10.1016/j.ejphar.2021.174107
30. Shah. Safety RR. And Tolerability of Histone Deacetylase (HDAC) Inhibitors in Oncology. Drug Saf (2019) 42:235–45. doi: 10.1007/s40264-018-0773-9
31. Lee SH, Yoo C, Im S, Jung JH, Choi HJ, Yoo J. Expression of Histone Deacetylases in Diffuse Large B-Cell Lymphoma and Its Clinical Significance. Int J Med Sci (2014) 11:994–1000. doi: 10.7150/ijms.8522
32. Chen IC, Sethy B, Liou JP. Recent Update of HDAC Inhibitors in Lymphoma. Front Cell Dev Biol (2020) 8:576391. doi: 10.3389/fcell.2020.576391
33. Gil VS, Bhagat G, Howell L, Zhang J, Kim CH, Stengel S, et al. Deregulated Expression of HDAC9 in B Cells Promotes Development of Lymphoproliferative Disease and Lymphoma in Mice. Dis Model Mech (2016) 9:1483–95. doi: 10.1242/dmm.023366
34. Hopken UE. Targeting HDAC3 in CREBBP-Mutant Lymphomas Counterstrikes Unopposed Enhancer Deacetylation of B-Cell Signaling and Immune Response Genes. Cancer Discov (2017) 7:14–6. doi: 10.1158/2159-8290.CD-16-1285
35. Kim SJ, Kim UJ, Yoo HY, Choi YJ, Kang KW. Anti-Cancer Effects of CKD-581, a Potent Histone Deacetylase Inhibitor Against Diffuse Large B-Cell Lymphoma. Int J Mol Sci (2020) 21:4377. doi: 10.3390/ijms21124377
36. Wu YF, Ou CC, Chien PJ, Chang HY, Ko JL, Wang BY. Chidamide-Induced ROS Accumulation and miR-129-3p-Dependent Cell Cycle Arrest in Non-Small Lung Cancer Cells. Phytomedicine (2019) 56:94–102. doi: 10.1016/j.phymed.2018.09.218
37. Liu Z, Chen J, Wang H, Ding K, Li Y, de Silva A, et al. Chidamide Shows Synergistic Cytotoxicity With Cytarabine via Inducing G0/G1 Arrest and Apoptosis in Myelodysplastic Syndromes. Am J Transl Res (2017) 9:5631–42.
38. Lin L, Que Y, Lu P, Li H, Xiao M, Zhu X, et al. Chidamide Inhibits Acute Myeloid Leukemia Cell Proliferation by lncRNA VPS9D1-AS1 Downregulation via MEK/ERK Signaling Pathway. Front Pharmacol (2020) 11:569651. doi: 10.3389/fphar.2020.569651
39. He J, Chen Q, Gu H, Chen J, Zhang E, Guo X, et al. Therapeutic Effects of the Novel Subtype-Selective Histone Deacetylase Inhibitor Chidamide on Myeloma-Associated Bone Disease. Haematologica (2018) 103:1369–79. doi: 10.3324/haematol.2017.181172
40. Sun C, Wang G, Wrighton KH, Lin H, Songyang Z, Feng XH, et al. Regulation of P27(Kip1) Phosphorylation and G1 Cell Cycle Progression by Protein Phosphatase PPM1G. Am J Cancer Res (2016) 6:2207–20.
41. Rao S, Lowe M, Herliczek TW, Keyomarsi K. Lovastatin Mediated G1 Arrest in Normal and Tumor Breast Cells Is Through Inhibition of CDK2 Activity and Redistribution of P21 and P27, Independent of P53. Oncogene (1998) 17:2393–402. doi: 10.1038/sj.onc.1202322
42. van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene Expression Profiling Predicts Clinical Outcome of Breast Cancer. Nature (2002) 415:530–6. doi: 10.1038/415530a
43. Blachly JS, Baiocchi RA. Targeting PI3-Kinase (PI3K), AKT and mTOR Axis in Lymphoma. Br J Haematol (2014) 167:19–32. doi: 10.1111/bjh.13065
44. Nepstad I, Hatfield KJ, Gronningsaeter IS, Reikvam H. The PI3K-Akt-mTOR Signaling Pathway in Human Acute Myeloid Leukemia (AML) Cells. Int J Mol Sci (2020) 21:2907. doi: 10.3390/ijms21082907
45. Liang S, Zhou X, Cai D, Rodrigues-Lima F, Wang L. Chidamide Inhibits Cell Proliferation via the PI3K/AKT Pathway in K562 Cells Based on Network Pharmacology and Experimental Validation. Curr Pharm Des (2021) 27:2990–8. doi: 10.2174/1381612827666210701152250
46. Zhang N, Liang C, Song W, Tao D, Yao J, Wang S, et al. Antitumor Activity of Histone Deacetylase Inhibitor Chidamide Alone or in Combination With Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Icotinib in NSCLC. J Cancer (2019) 10:1275–87. doi: 10.7150/jca.28570
47. Noorolyai S, Shajari N, Baghbani E, Sadreddini S, Baradaran B. The Relation Between PI3K/AKT Signalling Pathway and Cancer. Gene (2019) 698:120–8. doi: 10.1016/j.gene.2019.02.076
48. Paul J, Soujon M, Wengner AM, Zitzmann-Kolbe S, Sturz A, Haike K, et al. Simultaneous Inhibition of PI3Kdelta and PI3Kalpha Induces ABC-DLBCL Regression by Blocking BCR-Dependent and -Independent Activation of NF-kappaB and AKT. Cancer Cell (2017) 31:64–78. doi: 10.1016/j.ccell.2016.12.003
49. Rudelius M, Pittaluga S, Nishizuka S, Pham TH, Fend F, Jaffe ES, et al. Constitutive Activation of Akt Contributes to the Pathogenesis and Survival of Mantle Cell Lymphoma. Blood (2006) 108:1668–76. doi: 10.1182/blood-2006-04-015586
50. Hu N, Wang F, Sun T, Xu Z, Zhang J, Bernard D, et al. Follicular Lymphoma-Associated BTK Mutations are Inactivating Resulting in Augmented AKT Activation. Clin Cancer Res (2021) 27:2301–13. doi: 10.1158/1078-0432.CCR-20-3741
51. Zhang MC, Fang Y, Wang L, Cheng S, Fu D, He Y, et al. Clinical Efficacy and Molecular Biomarkers in a Phase II Study of Tucidinostat Plus R-CHOP in Elderly Patients With Newly Diagnosed Diffuse Large B-Cell Lymphoma. Clin Epigenet (2020) 12:160. doi: 10.1186/s13148-020-00948-9
52. Ding N, You A, Tian W, Gu L, Deng D. Chidamide Increases the Sensitivity of Non-Small Cell Lung Cancer to Crizotinib by Decreasing C-MET mRNA Methylation. Int J Biol Sci (2020) 16:2595–611. doi: 10.7150/ijbs.45886
53. Wang X, Wang D, Ding N, Mi L, Yu H, Wu M, et al. The Synergistic Anti-Tumor Activity of EZH2 Inhibitor SHR2554 and HDAC Inhibitor Chidamide Through ORC1 Reduction of DNA Replication Process in Diffuse Large B Cell Lymphoma. Cancers (Basel) (2021) 13:4249. doi: 10.3390/cancers13174249
54. Cao L, Zhao S, Yang Q, Shi Z, Liu J, Pan T, et al. Chidamide Combined With Doxorubicin Induced P53-Driven Cell Cycle Arrest and Cell Apoptosis Reverse Multidrug Resistance of Breast Cancer. Front Oncol (2021) 11:614458. doi: 10.3389/fonc.2021.614458
55. Tu K, Yu Y, Wang Y, Yang T, Hu Q, Qin X, et al. Combination of Chidamide-Mediated Epigenetic Modulation With Immunotherapy: Boosting Tumor Immunogenicity and Response to PD-1/PD-L1 Blockade. ACS Appl Mater Interfaces (2021) 13:39003–17. doi: 10.1021/acsami.1c08290
56. Ye J, Zha J, Shi Y, Li Y, Yuan D, Chen Q, et al. Co-Inhibition of HDAC and MLL-Menin Interaction Targets MLL-Rearranged Acute Myeloid Leukemia Cells via Disruption of DNA Damage Checkpoint and DNA Repair. Clin Epigenet (2019) 11:137. doi: 10.1186/s13148-019-0723-0
57. Que Y, Zhang XL, Liu ZX, Zhao JJ, Pan QZ, Wen XZ, et al. Frequent Amplification of HDAC Genes and Efficacy of HDAC Inhibitor Chidamide and PD-1 Blockade Combination in Soft Tissue Sarcoma. J Immunother Cancer (2021) 9:e001696. doi: 10.1136/jitc-2020-001696
Keywords: transformed-follicular lymphoma (t-FL), chidamide, HDAC, PI3K/AKT signaling, epigenetic antitumor therapy
Citation: Zhong M, Tan J, Pan G, Jiang Y, Zhou H, Lai Q, Chen Q, Fan L, Deng M, Xu B and Zha J (2021) Preclinical Evaluation of the HDAC Inhibitor Chidamide in Transformed Follicular Lymphoma. Front. Oncol. 11:780118. doi: 10.3389/fonc.2021.780118
Received: 20 September 2021; Accepted: 09 November 2021;
Published: 03 December 2021.
Edited by:
Stefano Luminari, University of Modena and Reggio Emilia, ItalyReviewed by:
Daniele Derudas, Ospedale Oncologico Armando Businco, ItalyKen Young, Duke University, United States
Xiaoyang Li, Ocean University of China, China
Copyright © 2021 Zhong, Tan, Pan, Jiang, Zhou, Lai, Chen, Fan, Deng, Xu and Zha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Zha, bGN3XzEwMDZAMTYzLmNvbQ==; Bing Xu, eHViaW5nemhhbmdqaWFuQDEyNi5jb20=
†These authors have contributed equally to this work
 Mengya Zhong1,2†
Mengya Zhong1,2† Bing Xu
Bing Xu Jie Zha
Jie Zha