
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 07 December 2021
Sec. Cancer Metabolism
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.779918
Cancer therapy is moving beyond traditional chemotherapy to include epigenetic approaches. KDM6 demethylases are dynamic regulation of gene expression by histone demethylation in response to diverse stimuli, and thus their dysregulation has been observed in various cancers. In this review, we first briefly introduce structural features of KDM6 subfamily, and then discuss the regulation of KDM6, which involves the coordinated control between cellular metabolism (intrinsic regulators) and tumor microenvironment (extrinsic stimuli). We further describe the aberrant functions of KDM6 in human cancers, acting as either a tumor suppressor or an oncoprotein in a context-dependent manner. Finally, we propose potential therapy of KDM6 enzymes based on their structural features, epigenetics, and immunomodulatory mechanisms, providing novel insights for prevention and treatment of cancers.
The initiation and progression of cancer, traditionally viewed as a set of genetic diseases, have now been considered as a complex cooperation within genetic alterations and epigenetic abnormalities (1). Epigenetics including DNA methylation, histone modification, and chromatin remodeling is involved in the regulation of gene expression and is closely related to multiple human cancers (2). Histone methylation and demethylation play an important role in histone modification (3). The histone H3 lysine 27 (H3K27) methylation status is dominated by histone methyltransferase EZH2 and two lysine demethylases (KDMs), and deregulated H3K27 methylation is often associated with a multitude of cancer types (4, 5).
KDM6 enzymes are capable of removing di-methylated and tri-methylated H3K27, thereby activating or repressing target gene transcription (6). Notably, KDM6 demethylases appear to be highly regulated at the transcriptional level and more susceptible to diverse stimuli like tumor microenvironments, metabolic reactions, differentiation inducers and stress signals (7, 8). There are emerging evidences for deregulation of KDM6 demethylases and important phenotypic consequences in various types of cancer (9–11). However, the precise molecular basis of how these lysine-specific demethylases contribute to oncogenesis has not been extensively pursued. This review summarizes recent advances in understanding structure, alterations and functions of KDM6 histone demethylases in oncogenesis and potential therapeutic targeting of these enzymes, providing a train of thought for the prevention and treatment of cancers.
The KDM6 demethylase subfamily belongs to the Jumonji C (JmjC) domain-containing KDMs, these enzymes require ferrous iron Fe(II), α-ketoglutarate [α-KG, also known as 2-oxoglutarate (2-OG)] and oxygen as co-factors (12). Due to these dependencies, KDM6 demethylases are more responsive to tumor microenvironments and cellular metabolites (10). KDM6 subfamily consists of three distinct members called KDM6A (also known as UTX), KDM6B (also known as JMJD3) and KDM6C (also known as UTY) proteins (13). Whereas KDM6A and KDM6C are respectively located at X and Y chromosomes, KDM6B is found on chromosome 17 (13). The structure of KDM6 members consists of five regions: tetratricopeptide repeat domain (TPR), helical domain, linker region, JmjC domain and a GATA-like zinc finger domain (Figure 1) (12). All three KDM6 members contain JmjC domains at their C termini catalyzing protein lysine demethylation, especially H3K27 dimethylation (H3K27me2) and H3K27 trimethylation (H3K27me3) (10). Of note, demethylase activity of KDM6C is minimal because of a subtle sequence divergence in the JmjC catalytic domain (10). However, in a knock-out mouse model, KDM6C can partially compensate for the demethylase-independent functions of KDM6A (14). The zinc finger domain of KDM6 subfamily is found to have a DNA binding function and plays an important role in the regulatory regions of genes (12, 15). KDM6A and KDM6C, but not KDM6B, have similar N-terminal TPR domains to form scaffold which could mediate protein-protein interactions and the assembly of multiprotein complexes (10). The helical domain of KDMs is anticipated to play roles in specific protein-protein interactions both in the assembly of chromatin-protein and other protein complexes (16). However, to date, the role of the helical domain in the KDM6 subfamily remains unclear, as the amine oxidase like domain serves the active site for substrate and cofactor binding. Whereas enzymatic roles of KDM6A and KDM6B are undisputed in cancer cells, non-enzymatic scaffolding roles for large complexes of all KDM6 members have been recognized (17). In a growing number of cases, mutations affecting these KDM6 response to specific therapies (18, 19).
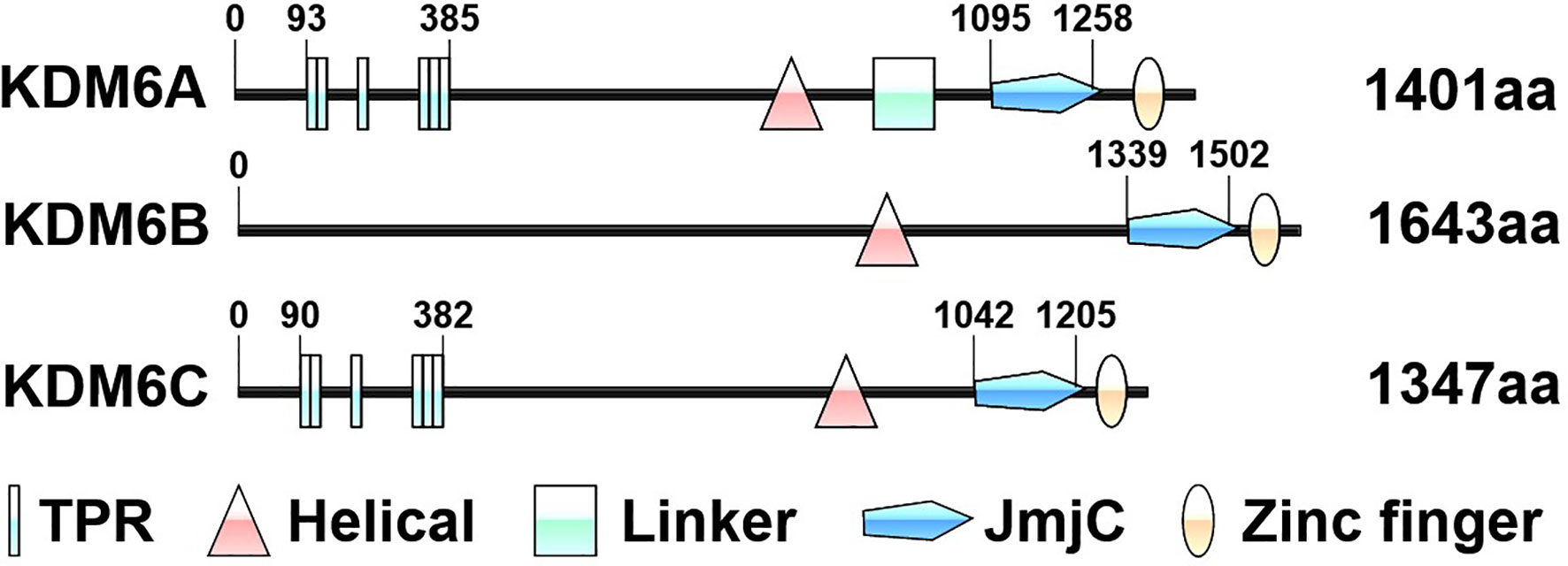
Figure 1 Schematic representation of the protein domains of KDM6 enzymes. The structure of KDM6 members consists of five regions: tetratricopeptide repeat domain (TPR), helical domain, linker region, Jumonji C (JmjC) domain and zinc finger domain. The numbers indicate the amino acid residues. The domains are based on the UniProt database (https://www.uniprot.org/).
Given that KDM6 demethylases are involved in the regulation of a vast array of biological processes via their demethylase dependent or independent functions, their levels and associated activities have to be precisely controlled. However, this area of research has never been addressed. Below is the summary of available reports on the regulation of KDM6 (Figure 2).
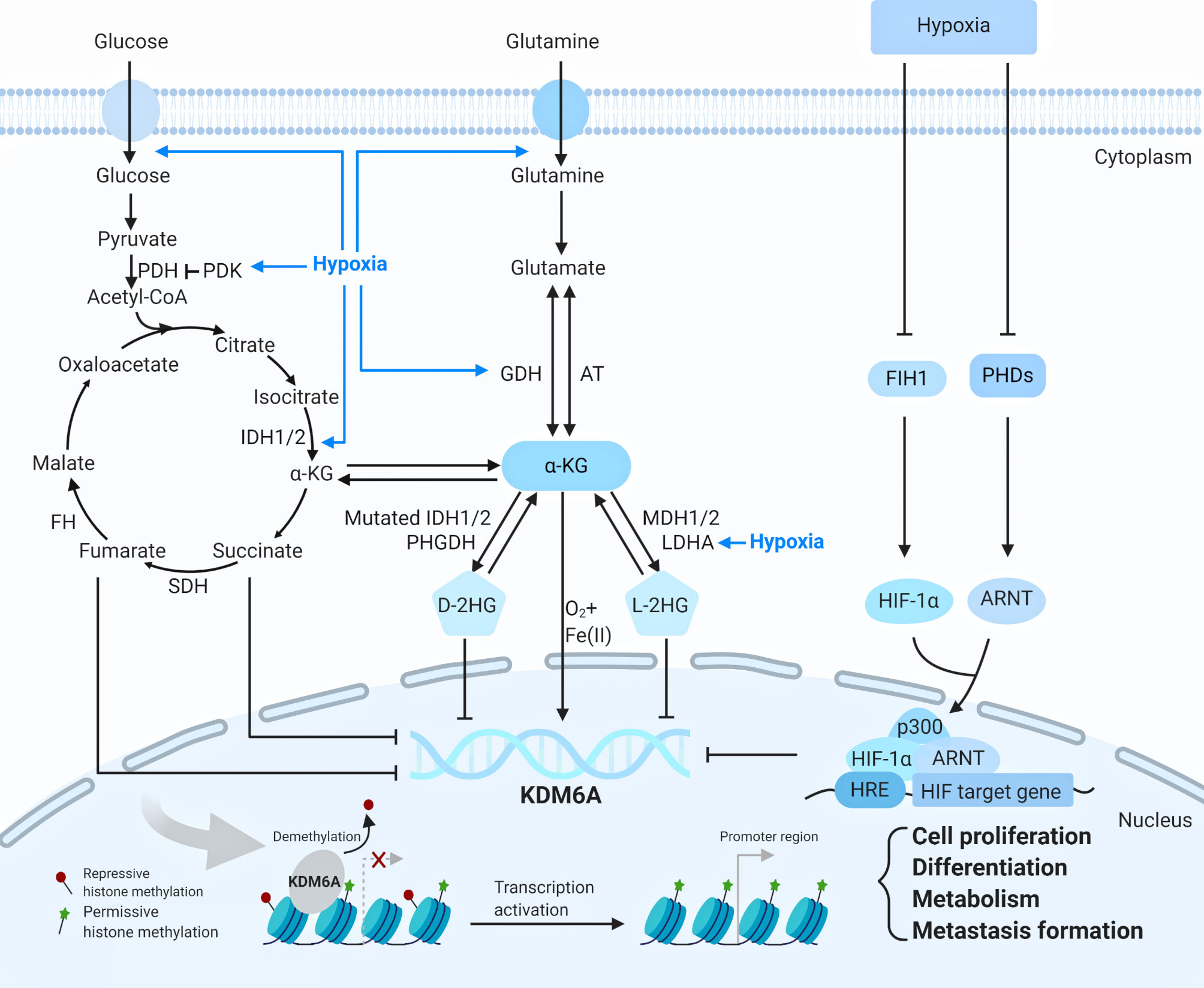
Figure 2 The regulation of KDM6A (Created with BioRender.com). KDM6A is upregulated or downregulated by metabolic reactions and hypoxia through diverse mechanisms. KDM6A is re-cruited to the chromatin and respond to the transcription through influencing cell proliferation, differentiation, metabolism and metastasis formation. ARNT, aryl hydrocarbon nuclear translocator, AT amino transferases, D-2HG D-2-hydroxyglutarate, FH fumarate hydratase, FIH1 factor-inhibiting hypoxia-inducible factor 1, GDH glutamate dehydrogenases, HIF-1α hypoxia-inducible factor-1α, HRE hypoxia response element, IDH1/2 isocitrate dehydrogenases 1 or 2, LDHA lactate dehydrogenase A, L-2HG L-2- hy-droxyglutarate, MDH 1/2 malate dehydrogenase 1 or 2, PDH pyruvate dehydrogenase, PHDs prolyl hydroxylases, PDK pyruvate dehydrogenase kinase, PHGDH D-3-phosphoglycerate, SDH succinate dehydrogenase, α-KG α-ketoglutarate.
KDM6A is regulated by metabolic reactions and hypoxia through diverse mechanisms. As KDM6A uses cellular metabolites such as Fe(II), and tricarboxylic acid (TCA) cycle intermediate α-KG and oxygen for their catalytic reactions, its activity is influenced by mutations in the TCA cycle enzymes isocitrate dehydrogenase 1 and 2 (IDH1 and 2), succinate dehydrogenase (SDH), and fumarase hydratase (FH). Glucose and glutamine respectively are transported into cells and both are converted to α-KG, which related to 2-hydroxyglutarate (2-HG) (10). The metabolite 2-HG can be produced as either a D(R)- or L(S)-enantiomer, each of which functions as a potent inhibitor of KDM6A enzyme involved in diverse biologic processes (20). Oncogenic mutations in IDH1 or 2 produce D-2HG, which causes a pathologic blockade in cell differentiation (21). Production of L-2HG instead results from promiscuous substrate usage primarily by lactate dehydrogenase A (LDHA) (Figure 2) (20). Hypoxic tumor microenvironment is closely associated with cellular metabolites (10). Under hypoxia, effective concentration of α-KG is decreased, and the 2-HG level is increased via metabolic reprogramming, thus contributing to the inhibition of α-KG-dependent KDM6A, which is related to cellular heterogeneity, cancer resistance, and progression (20). Interestingly, in the latest study KDM6A was found to be a direct sensor of oxygen (22, 23). In hypoxic conditions, factor-inhibiting hypoxia-inducible factor 1 (FIH1) and prolyl hydroxylases (PHDs) are inactivated, allowing hypoxia-inducible factor-1α (HIF-1α) to dimerize with aryl hydrocarbon nuclear translocator (ARNT; also called HIF-1β) (24). The HIF complex translocates to the nucleus and binds to hypoxia response element (HRE) in HIF target gene promoters (24). KDM6A inactivation via hypoxia promotes hypermethylation of H3K27, preventing transcriptional reprogramming for cellular differentiation (Figure 2) (22, 24).
KDM6A also exerted the function under the stimulation of certain specific proteins and ligands. Recent studies have demonstrated demethylase-independent functions of KDM6A as a scaffold protein that facilitates the binding of other factors that directly regulate transcription. The KDM6A were also found to be associated with estrogen receptor (ER) and retinoic acid receptor (RAR) upon ligand treatments, including estrogens and retinoic acid, which is necessary for cellular differentiation, cancer tumorigenesis and metastasis (13). Although whether KDM6A interacts with ER or RAR directly, or depends on certain complex, remains unclear, these studies have shed light that KDM6A contributes to oncogenesis on the ligand-dependent transcriptional regulation (25, 26).
The role of KDM6B in cell proliferation and differentiation is regulated by growth factors, cytokines and ligands via distinct signaling pathways in human cancers. KDM6B is upregulated via nuclear factor-κB (NF-κB) binding to its promotor region by tumor necrosis factor α (TNFα), and that KDM6B in turn upregulates mitogen-activated protein kinase (MAPK) pathway, thereby promoting cancer cell growth and survival in a catalytically-independent manner (Figure 3) (27, 28). RAS is activated by GDP/GTP exchange stimulated by epidermal growth factor (EGF), and activator protein 1 (AP-1) transcription factors are good candidates for acting as transactivators of KDM6B transcription (29). KDM6B expression is induced by activation of RAS–RAF pathway, and contributes to activate tumor suppressor proteins p16INK4A and p14ARF, which triggers cellular senescence and apoptosis in a demethylase-dependent fashion (Figure 3) (30, 31). Signal transducer and activator of transcription 3 (STAT3) is activated by phosphorylation on receptor-associated Janus kinases (JAKs), and regulates the proliferation and self-renewal ability of cancer stem cells through repression of KDM6B expression (Figure 3) (32). Moreover, like KDM6A, KDM6B interacts with ER or RAR in the presence of estrogens or retinoic acid, and regulates the proliferation and differentiation of cancer cells. However, the mechanisms underlying KDM6B interaction with ER or RAR are unknown (33).
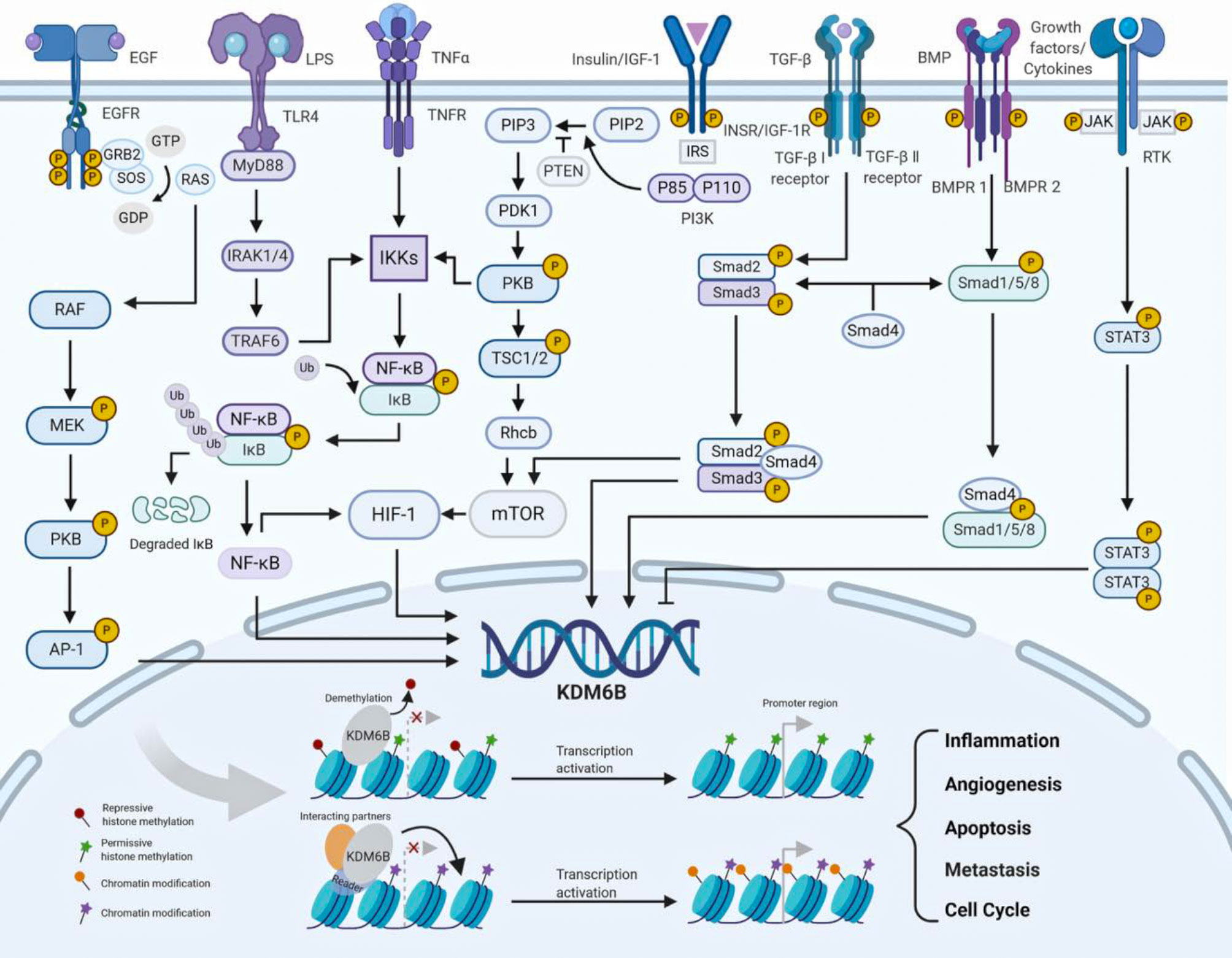
Figure 3 The regulation of KDM6B (Created with BioRender.com). KDM6B is upregulated or downregulated by growth factors, cytokines and cellular stresses via distinct signaling pathways. KDM6B is recruited to the chromatin and involved in a wide range of biological processes, such as angiogenesis, metastasis, cell cycle, apoptosis and inflammation. AP-1 activator protein 1, BMP bone morphogenetic protein, BMPR1 bone morphogenetic protein receptor 1, BMPR2 bone morphogenetic protein receptor 2, EGF epidermal growth factor, EGFR epidermal growth factor receptor, ERK extracellular signal-regulated kinase, GRB2 growth factor receptor bound protein 2, HIF-1 hypoxia inducible factor-1, IGF-1 insulin-like growth factor 1, IGF-1R insulin-like growth factor 1 receptor, IκB inhibitor of nuclear factor-κB, IKKs inhibitor of nuclear factor-κ kinases, INSR insulin receptor, IRAK1/4 interleukin-1 receptor-associated kinase 1/4, IRS insulin receptor substrate, JAK Janus kinase, LPS lipopolysaccharide, MEK MAPK/ERK kinase, MAPK mitogen-activated protein kinase, mTOR mammalian target of rapamycin, MyD88 myeloiddifferentiationfactor88, NF-κB nuclear factor-κB, PI3K phosphoinositide 3-kinase, PIP2 phosphatidylinositol- 3,4-bisphosphate, PIP3 phosphatidylinositol-3,4,5-trisphosphate, PDK1 3-phosphoinositide-dependent protein kinase 1, PTEN phosphatase and tensin homolog deleted on chromosome ten, PKB protein kinase B, RTK receptor tyrosine kinase, SOS son of sevenless, STAT3 signal transducer and activator of transcrip-tion 3 , TGF-β transforming growth factor-β, TLR4 toll-like receptor 4, TNF-α tumor necrosis factor-α, TNFR tumor necrosis factor receptor, TRAF6 TNF receptor-associated factor 6, TSC1/2 tuberous sclerosis complex 1/2.
KDM6B is upregulated or downregulated by different signaling pathways, and play essential roles in the tumor microenvironment. It has been previously shown that bone morphogenetic protein 4 (BMP4) activates the Smad signaling cascade and upregulates KDM6B expression as the regulator of balances between self-renewal and differentiation. Interestingly, a positive feedback loop is found between KDM6B and BMP signaling, and KDM6B also could employs BMP and NF-kB pathways to modulate the tumor microenvironment by angiogenesis and tumor associated-macrophage infiltration (Figure 3) (34, 35). Recently, the mechanisms of KDM6B in regulating inflammatory genes have been studied extensively and deeply, supporting the view that KDM6B involves the engagement of an inflammatory tumor microenvironment (36). TNF receptor associated factor 6 (TRAF6) is activated by toll like receptor 4 (TLR4) signaling stimulated by lipopolysaccharides (LPS), and induces phosphorylation of the inhibitor of nuclear factor-κ kinase (IKK) complex, leading to activation of KDM6B via NF-κB pathway (28, 37). Consequently, KDM6B is recruited to promoter regions of inflammatory genes TNF-α and interleukin-6 (IL-6), where the H3K27me3 levels strikingly decrease (Figure 3) (38, 39). KDM6B is upregulated via Smad signaling by activated by transforming growth factor-β (TGF-β), and that KDM6B in turn upregulates SNAI1, which is a master transcription factor in epithelial-mesenchymal transition (EMT) and cancer progression (Figure 3) (40, 41).
The regulation of KDM6B is an important host response against cellular metabolism and environmental stress. KDM6B expression is up-regulated through HIF-1 under hypoxia and normoxia (29). Independently from oxygen levels, HIF-1 is constitutively transcribed and synthesized through a series of signaling events involving IGF-1/PI3K/mTOR, TNF-α/NF-κB and TGF-β/Smad/mTOR pathways (Figure 3) (42–46). Moreover, similar to KDM6A, KDM6B expression is also subjected to different levels of its cosubstrates from cellular metabolism and tumor microenvironments (10). Studies have demonstrated vitamin D metabolite 1α,25-dihydroxyvitamin D (3) [1,25(OH) (2)D (3)] modulates the KDM6B gene promoter and increases the level of KDM6B RNA in human colon cancer cells (47). Regional glutamine deficiency decreases KDM6B expression, which promotes tumor heterogeneity and therapeutic resistance (48). However, the functions of KDM6B in cellular metabolism as well as regulatory mechanisms of above-mentioned stresses remain to be fully studied.
KDM6C is an essential downstream mediator of NKX3.1 for prostate differentiation and cancer (49). Compared with other two paralogs, very little is known as to how KDM6C is regulated under physiological and pathological conditions. Thus, elucidation of these biological processes may help to understand the context dependent role of KDM6C in human cancers.
KDM6 profoundly influences gene expression, based on enzymatic activity in demethylating H3K27me3, and non-enzymatic scaffolding roles for large complexes that open and close chromatin for transcription. KDM6 plays a crucial and dual role in onset and progression of cancers through activating or repressing target gene transcription, which has been recognized to strongly influence cancer risk, prognosis, and therapy resistance.
In recent years, KDM6A is found to be aberrantly expressed in many cancers as a tumor suppressor or promoter, implying its regulatory role in tumor initiation and progression (Table 1). KDM6A is initially a tumor-suppressor of Notch and Rb-dependent tumors in Drosophila (117). Numerous studies have demonstrated that KDM6A is targeted by loss-of-function mutations as a suppressor in various cancers, including acute myeloid leukemia, B cell lymphoma, multiple myeloma, non-small cell lung cancer, squamous-like pancreatic cancer, T-cell acute lymphoblastic leukemia, urothelial cancer, colorectal cancer, medulloblastoma, breast cancer, hepatocellular cancer and esophageal squamous cell cancer (50–58, 60, 61, 67–73, 118).
KDM6A mutations are commonly lost in all stages and subtypes of urothelial cancer (UC), which is classified into non-muscle-invasive and muscle-invasive tumors (62–65). Papillary UC is driven by loss of mutation of KDM6A in growth signaling pathway, especially activating FGFR3 and PIK3CA (119). In muscle-invasive UC, KDM6A deficiency activates multiple immune response genes and causes bladder cancer in cooperation with p53 dysfunction (120). In the TCGA analysis of muscle-invasive UC, KDM6A is found to interact with KMT2C and KMT2D in the compass complex (121, 122). Gender bias in KDM6A inactivation has been reported in some cancer types (58, 60, 66). More KDM6A mutations are present in non-muscle-invasive UC from females than males (123). Molecular mechanisms through which KDM6A suppresses the progression of urothelial cancer depend on the subtype, not only via its H3K27me3 demethylase activity but also via interactions with other epigenetic complexes.
The function of KDM6A is even different among subtypes of one disease, including acute myeloid leukemia, non-small cell lung cancer, squamous-like pancreatic cancer, T-cell acute lymphoblastic leukemia (T-ALL), breast cancer and glioblastoma (25, 50–52, 57–60, 68, 70, 71, 74, 76, 81–84). In human T-ALL, KDM6A acts as a tumor suppressor and is frequently genetically inactivated in a NOTCH1-induced T-ALL model (57). Interestingly, KDM6A controls oncogenic transcription factor TAL1 and functions as an oncogene for leukemia maintenance in human T-ALL (74). Yu and Taube et al. demonstrate that low expression of KDM6A predicts poor survival in breast cancer (70, 71). while Kim et al. show that high expression of KDM6A is associated with poor prognosis (25, 76). In breast cancer, KDM6A is found to be an important factor in mediating EMT (70, 71). KDM6A promotes the proliferation and migration of hormonally responsive breast cancer via feed-forward transcription with Erα (25). The dual roles of KDM6A suggest its context-specific role in tumorigenesis.
KDM6A also has been shown to be overexpressed and exert different roles in multiple human cancers (25, 74, 75, 77, 78, 80). The overexpression of KDM6A changes histone H3 methylation on the promoters of tumor suppressor gene Rb and Rbl2, contributing to the decreased cell proliferation of retinoblastoma (75). Analysis of TCGA database found that high KDM6A is associated with sex-biased by activating immune-related pathways (86). Nevertheless, KDM6A cannot be perceived solely as a tumor suppressor, as KDM6A is found to have close association with genesis and progression of different cancers (25, 74, 77, 78, 80). In advanced stages of prostate cancer, KDM6A is essential for maintenance of androgen receptor (AR) activity despite anti-androgenic therapy (77, 78, 124). A different pro-oncogenic function has been shown in cervical cancer, where increased KDM6A expression appears to support cell cycle activation by p21CIP1 suppression of replication stress (80). KDM6A and NOTCH1 mutations are assocatiated with poor prognosis in metastatic adenoid cystic cancer (85). KDM6A interacting with GATA activates stem-like phenotypes in ovarian cancer cell lines (87).
KDM6B mediates carcinogenic and anti-cancer signaling pathways by directing distinct transcription factors in a context-dependent manner, imposing wide-ranging effects on proliferation, apoptosis, migration, stem cell behavior, EMT, drug resistance and tumor microenvironment (Table 1). In several cancers, KDM6B is downregulated and regarded as a tumor suppressor by counteracting different transcriptional programs. KDM6B is downregulated in high-risk neuroblastomas, and functions as a tumor suppressor through neuronal differentiation by activating NEFM (88). Low expression of KDM6B is an independent predictor of poor prognosis in colorectal cancer by mediating p15INK4B expression (89). KDM6B expressional reduction is found in acute myeloid leukemia, and KDM6B acts as an oncorepressor by activating C/EBPβ-centered transcriptional program (90). Decreased expression of KDM6B enhances aggressiveness of pancreatic cancer through downregulation of C/EBPα (91). KDM6B shows its tumor-suppressive function on squamous cell carcinoma by mediating cell-cycle and proinflammatory genes, such as CCNB1, CDK1, and IL-6 (92).
However, KDM6B is more frequent upregulated and functions as tumor promotor in various cancers. KDM6B upregulates gene expression of cell cycle and proliferation related genes such as HOX, p16INK4A, ELK1, FOS, cyclin D1 and TP53, thereby promoting cell growth and survival of acute myeloid leukemia, cervical cancer, multiple myeloma, prostate cancer, and gastric cancer (27, 93, 95, 97, 99, 125). Overexpression of KDM6B exerts an anti-apoptotic effect in diffuse large B-cell lymphoma and prostate cancer, and respectively activates the transcription of downstream target genes IRF4 and PTEN via distinct mechanisms (75, 96, 98). High expression of KDM6B is correlated with distant metastasis via modulating H3K27me3 of SLUG gene promoter in hepatocellular carcinomas (100). KDM6B could promote the invasion of hepatocellular cancer, non-small cell lung cancer, renal cell cancer and chordoma through upregulation of EMT-specific genes, such as TIAM1, E-cadherin, SNAI1 and TBXT (101, 112, 113, 115, 116). Breast cancer cells acquire drug resistance to PI3K inhibitor via upregulation of KDM6B-mediated IGFBP5 expression (105). It has been reported that KDM6B impacts migration and invasion of ovarian cancer by regulating TGF-β1 and HER2 (102, 103). KDM6B regulates the differentiation of cancer stem cells via activating distinct target genes, and may contribute to the pathogenesis of T-ALL, acute myeloid leukemia, breast cancer, Hodgkin’s Lymphoma and Glioblastoma (27, 83, 94, 104, 106–108). In colorectal cancer and melanoma, KDM6B expression is elevated and positively correlated with several targets of CXCL9, CXCL10, STC1 and CCL2, which affects the tumor immune microenvironment by T cells and macrophage infiltration (109–111). In chronic myelomonocytic leukemia, KDM6B overexpression results in activation of innate immune genes, such as S100A9, TLR and C3, indicating that KDM6B is involved in tumor immune microenvironment (114).
Due to point substitutions affecting substrate binding, KDM6C, the Y-chromosome homolog of KDM6A, has markedly lower demethylase activity. Some studies confirm the frequent co-occurrence of KDM6A and KDM6C, and identify KDM6C as a demethylase independent tumor suppressor in acute myeloid leukemia and squamous-like pancreatic carcinoma (52, 58). Compared with other KDM6 subfamily, the function of KDM6C in human cancers needs further investigated.
The diversity of interacting transcription factors explains why KDM6 acts as a pro-oncogenic or tumor-suppressive factor in different tumor types. Notably, functional roles of KDM6 subfamily in tumorigenesis were found to be highly cell type-specific and pathologic context-dependent. Thus, taking into consideration the dual roles of KDM6 in different cellular contexts would make a better approach in overcoming tumor progression. In the same type of cancer, differential roles and associations of KDM6 subfamily in cancer should be further discussed in the future.
Since KDM6 demethylases are overexpressed in many cancers and contribute to cancer development, KDM6 family members have attracted considerable interest as targets for enzymatic inhibition. Most KDM6 demethylase inhibitors target the active site of JmjC domain via binding competitively with α-KG and chelating Fe(II) residue (126). Unfortunately, these agents are broad-spectrum inhibitors and also effect on other members of JmjC domain-containing family. Due to this reason, use of these compounds might result in significant cytotoxicity (127). Recently, some compounds inhibit a more restrictive subset of KDMs, with KDM6A being among the targets. MC3324, a dual KDM1A-KDM6A inhibitor, regulates drug resistance in ERα-positive breast cancer (128). Metformin has also been recognized as a catalytic inhibitor of KDM6A, and is currently in phase II clinical trials of bladder cancer (NCT03379909) (Table 2) (129). To date, the most described selective inhibitors of KDM6A and KDM6B are GSK-J1 and cell-active ethyl ester prodrug GSK-J4 (141). The synergistic inhibition of KDM6B and KDM1A by GSK-J1 and TCP is efficient in head and neck squamous cell carcinoma in vivo (130). GSK-J4 are relatively unspecific as it also inhibits KDM5 subfamily in vitro (142). GSK-J4 has nevertheless been proposed for application in several cancer types, particularly in vivo of acute myeloid leukemia, breast cancer, TAL1 positive T-ALL, colorectal cancer and osteosarcoma (Table 2) (74, 78, 93, 99, 106, 131–137). These indicate that GSK-J4 might be a promising treatment for clinical cancer therapy. The main obstacle of this treatment is achieving specific inhibition of key demethylases thus reducing possible toxicity or side effects. Inhibition of KDM6A and KDM6B might provide benefit in certain circumstances, but inhibition might produce deleterious responses. Based on intensive studies across a broad spectrum of tumors, the functional roles of KDM6A and KDM6B were inconsistent in the same type of cancer. If ablation of KDM6A or KDM6B increases cancer cell metastasis, GSK-J4 treatment might result in increased tumor growth instead. Therefore, development of highly specific small-molecule inhibitors of KDM6 subfamily will provide novel targeted therapeutic options. In addition, targeting non-catalytic functions of KDM6 will require chemical probes and inhibitors targeting protein-protein interactions, and may be an emerging avenue.
KDM6A, a known tumor suppressor, is frequently inactivated in cancers (143). Given that the imbalance between KDM6A and EZH2 expression appears to sensitize many cancers to epigenetic inhibitors, resetting epigenetic balance could be useful in combination approaches (13). In multiple myeloma and bladder cancer, KDM6A loss confers sensitivity to inhibitors of the EZH2 methyltransferase, while KDM6A loss in pancreatic cancers sensitizes to bromodomain and extra-terminal (BET) and histone deacetylase (HDAC) inhibitors (Table 2) (55, 58, 138–140). In several cancers, KDM6B is downregulated and regarded as a tumor suppressor, suggesting there are no effects of KDM6 inhibitors in these cellular contexts (88–92). KDM6B loss conferring sensitivity to inhibitors of EZH2, BET and HDAC should be investigated in future. However, mechanistic details of how loss of KDM6 stimulates enhancer activation are lacking.
Because some epigenetic regulators have shown potent immunomodulatory activity, combination with immune checkpoint inhibitors (ICIs) could be promising (144). Notably, KDM6 demethylases regulates multiple innate and adaptive immune responses (145, 146). Furthermore, in bladder cancer KDM6A loss activates cytokine and chemokine pathways, and these cells are more sensitive to combined inhibition of IL-6 and CCL2 (Table 2) (120). A number of trials combining epigenetic inhibitors with ICIs are in progress, and need to be explored (147).
Because functional roles of KDM6 subfamily were highly cell type-specific and pathologic context-dependent, it is essential to consider different cellular functions of KDM6 subfamily when targeting their highly related enzymatic domains. Therefore, developing a deeper understanding of differential roles and associations of KDM6 subfamily in cancer will provide precision therapeutic strategies. In addition, development of highly specific small-molecule inhibitors of KDM6 subfamily will be of utmost important in modern molecular oncology.
The regulation of KDM6 is complex and intriguing, and involves the coordinated control between cellular metabolism (intrinsic regulators) and tumor microenvironment (extrinsic stimuli). A better understanding of regulatory mechanisms of KDM6 demethylases in cancer progression will guide provide novel targeted therapeutic options for epigenetic inhibitors.
Due to potent immunomodulatory activities of KDM6, ICIs should be taken into consideration when targeting the KDM6 demethylases. Considering the discussed effects KDM6 exerts on both innate and adaptive immune cells, it is essential to understand how targeting the KDM6 demethylases would affect tumor immune microenvironment. Development of combining epigenetic inhibitors with ICIs will complement the existing arsenal of epigenetic drugs.
JC, SL, JZ, and JF performed paper searches and wrote the manuscript. CH, WS, and WW designed the study, revised the manuscript and made the decision to submit. All authors contributed to the article and approved the submitted version.
This work was supported by National Natural Science Foundation of China (81802963, 81972261, 81901660), Zhejiang Provincial Natural Science Foundation of China (LY18H160046), Zhejiang Medical Science Foundation (2018KY531), Lin He’s New Medicine and Clinical Translation Academician Workstation Research Fund (18331215).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AP-1: Activator protein 1
AR: Androgen receptor
ARNT: Aryl hydrocarbon nuclear translocator
BET: Bromodomain and extra-terminal
BMP4: Bone morphogenetic protein 4
CCL2: C-C chemokine ligand 2
CDK1: Cyclin-dependent kinase 1
C/EBPβ: CCAAT/enhancer-binding protein beta
EGF: Epidermal growth factor
EMT: Epithelial-mesenchymal transition
ER: Estrogen receptor
EZH2: Enhancer of zeste homolog 2
FGFR3: Fibroblast growth factor receptor 3
FH: Fumarase hydratase
FIH1: Factor-inhibiting hypoxia-inducible factor 1
HDAC: Histone deacetylase
HER2: Human epidermal growth factor receptor 2
HIF-1α: Hypoxia-inducible factor-1α
HRE: Hypoxia response element
H3K27: Histone H3 lysine 27
H3K27me2: H3K27 dimethylation
H3K27me3: H3K27 trimethylation
ICIs: Immune checkpoint inhibitors
IDH1 and 2: Isocitrate dehydrogenase 1 and 2
IKK: Inhibitor of nuclear factor-κ kinase
IL-6: Interleukin-6
IRF4: Interferon regulatory factor 4
JAKs: Janus kinases
JmjC: Jumonji C
JMJD3: Jumonji domain-containing protein D3
KDM: Lysine demethylase
LDHA: Lactate dehydrogenase A
LPS: Lipopolysaccharides
MAPK: Mitogen-activated protein kinase
NEFM: Neurofilament medium
NF-κB: Nuclear factor-κB
PHDs: Prolyl hydroxylases
PTEN: Phosphatase and tensin homolog deleted on chromosome ten
RAR: Retinoic acid receptor
SDH: Succinate dehydrogenase
STAT3: Signal transducer and activator of transcription 3
T-ALL: T-cell acute lymphoblastic leukemia
TAL1: T-cell acute leukemia protein 1
TCA: Tricarboxylic acid
TCGA: The Cancer Genome Atlas
TGF-β: Transforming growth factor-β
TLR4: Toll like receptor 4
TNFα: Tumor necrosis factor α
TPR: Tetratricopeptide repeat domain
TRAF6: TNF receptor associated factor 6
UC: Urothelial cancer
UTX: Ubiquitously transcribed tetratricopeptide repeat X
UTY: Ubiquitously transcribed tetratricopeptide repeat Y
1,25(OH) (2)D (3): 1α,25-dihydroxyvitamin D (3)
2-HG: 2-hydroxyglutarate
2-OG: 2-oxoglutarate
α-KG: α-ketoglutarate
1. Sharma S, Kelly TK, Jones PA. Epigenetics in Cancer. Carcinogenesis (2010) 31:27–36. doi: 10.1073/pnas.96.25.14523
2. Højfeldt JW, Agger K, Helin K. Histone Lysine Demethylases as Targets for Anticancer Therapy. Nat Rev Drug Discov (2013) 12:917–30. doi: 10.1038/nrd4154
3. Jambhekar A, Dhall A, Shi Y. Roles and Regulation of Histone Methylation in Animal Development. Nat Rev Mol Cell Biol (2019) 20:625–41. doi: 10.1038/s41580-019-0151-1
4. Dawson MA, Kouzarides T. Cancer Epigenetics: From Mechanism to Therapy. Cell (2012) 150:12–27. doi: 10.1016/j.cell.2012.06.013
5. Jones PA, Baylin SB. The Epigenomics of Cancer. Cell (2007) 128:683–92. doi: 10.1016/j.cell.2007.01.029
6. Arcipowski KM, Martinez CA, Ntziachristos P. Histone Demethylases in Physiology and Cancer: A Tale of Two Enzymes, JMJD3 and UTX. CurrOpin Genet Dev (2016) 36:59–67. doi: 10.1016/j.gde.2016.03.010
7. Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC Domain-Containing UTX and JMJD3 as Histone H3 Lysine 27 Demethylases. Proc Natl Acad Sci U S A (2007) 104:18439–44. doi: 10.1073/pnas.0707292104
8. Chen X, Xiao X, Guo F. Roles of H3K27me3 Demethylase JMJD3 in Inflammation and Cancers. J Rare Dis Res Treat (2019) 4(1):71–6.
9. Gažová I, Lengeling A, Summers KM. Lysine Demethylases KDM6A and UTY: The X and Y of Histone Demethylation. Mol Genet Metab (2019) 127:31–44. doi: 10.1016/j.ymgme.2019.04.012
10. Chang S, Yim S, Park H. The Cancer Driver Genes IDH1/2, JARID1C/ KDM5C, and UTX/ KDM6A: Crosstalk Between Histone Demethylation and Hypoxic Reprogramming in Cancer Metabolism. Exp Mol Med (2019) 51:1–17. doi: 10.1038/s12276-019-0230-6
11. Walport LJ, Hopkinson RJ, Vollmar M, Madden SK, Gileadi C, Oppermann U, et al. Human UTY(KDM6C) is a Male-Specific Nϵ-Methyl Lysyl Demethylase. J Biol Chem (2014) 289:18302–13. doi: 10.1074/jbc.M114.555052
12. Sengoku T, Yokoyama S. Structural Basis for Histone H3 Lys 27 Demethylation by UTX/KDM6A. Genes Dev (2011) 25:2266–77. doi: 10.1101/gad.172296.111
13. Wang L, Shilatifard A. UTX Mutations in Human Cancer. Cancer Cell (2019) 35:168–76. doi: 10.1016/j.ccell.2019.01.001
14. Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY Demonstrate Histone Demethylase-Independent Function in Mouse Embryonic Development. PLoS Genet (2012) 8:e1002964. doi: 10.1371/journal.pgen.1002964
15. Jones SE, Olsen L, Gajhede M. Structural Basis of Histone Demethylase KDM6B Histone 3 Lysine 27 Specificity. Biochemistry (2018) 57:585–92. doi: 10.1021/acs.biochem.7b01152
16. Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, et al. Solution Structure of the SWIRM Domain of Human Histone Demethylase LSD1. Structure (2006) 14:457–68. doi: 10.1016/j.str.2005.12.004
17. Johansson C, Tumber A, Che K, Cain P, Nowak R, Gileadi C, et al. The Roles of Jumonji-Type Oxygenases in Human Disease. Epigenomics (2014) 6:89–120. doi: 10.2217/epi.13.79
18. Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX Play a Demethylase-Independent Role in Chromatin Remodeling to Regulate T-Box Family Member-Dependent Gene Expression. Mol Cell (2010) 40:594–605. doi: 10.1016/j.molcel.2010.10.028
19. Yin X, Yang S, Zhang M, Yue Y. The Role and Prospect of JMJD3 in Stem Cells and Cancer. BioMed Pharmacother (2019) 118:109384. doi: 10.1016/j.biopha.2019.109384
20. Intlekofer AM, Dematteo RG, Venneti S, Finley LW, Lu C, Judkins AR, et al. Hypoxia Induces Production of L-2-Hydroxyglutarate. Cell Metab (2015) 22:304–11. doi: 10.1016/j.cmet.2015.06.023
21. Intlekofer AM, Wang B, Liu H, Shah H, Carmona-Fontaine C, Rustenburg AS, et al. L-2-Hydroxyglutarate Production Arises From Noncanonical Enzyme Function at Acidic Ph. Nat Chem Biol (2017) 13:494–500. doi: 10.1038/nchembio.2307
22. Chakraborty AA, Laukka T, Myllykoski M, Ringel AE, Booker MA, Tolstorukov MY, et al. Histone Demethylase KDM6A Directly Senses Oxygen to Control Chromatin and Cell Fate. Science (2019) 363:1217–22. doi: 10.1126/science.aaw1026
23. Batie M, Frost J, Frost M, Wilson JW, Schofield P, Rocha S. Hypoxia Induces Rapid Changes to Histone Methylation and Reprograms Chromatin. Science (2019) 363:1222–6. doi: 10.1126/science.aau5870
24. Gallipoli P, Huntly BJP. Histone Modifiers are Oxygen Sensors. Science (2019) 363:1148–9. doi: 10.1126/science.aaw8373
25. Xie G, Liu X, Zhang Y, Li W, Liu S, Chen Z, et al. UTX Promotes Hormonally Responsive Breast Carcinogenesis Through Feed-Forward Transcription Regulation With Estrogen Receptor. Oncogene (2017) 36:5497–511. doi: 10.1038/onc.2017.157
26. Rocha-Viegas L, Villa R, Gutierrez A, Iriondo O, Shiekhattar R, Croce LD. Role of UTX in Retinoic Acid Receptor-Mediated Gene Regulation in Leukemia. Mol Cell Biol (2014) 34:3765–75. doi: 10.1128/mcb.00839-14
27. Ohguchi H, Harada T, Sagawa M, Kikuchi S, Tai YT, Richardson PG, et al. KDM6B Modulates MAPK Pathway Mediating Multiple Myeloma Cell Growth and Survival. Leukemia (2017) 31:2661–9. doi: 10.1038/leu.2017.141
28. De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natolil G. The Histone H3 Lysine-27 Demethylase Jmjd3 Links Inflammation to Inhibition of Polycomb-Mediated Gene Silencing. Cell (2007) 130:1083–94. doi: 10.1016/j.cell.2007.08.019
29. Kim JH, Sharma A, Dhar SS, Lee SH, Gu B, Chan CH, et al. UTX and MLL4 Coordinately Regulate Transcriptional Programs for Cell Proliferation and Invasiveness in Breast Cancer Cells. Cancer Res (2014) 74:1705–17. doi: 10.1158/0008-5472.CAN-13-1896
30. Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenführ M, et al. Histone Demethylase JMJD3 Contributes to Epigenetic Control of INK4a/ARF by Oncogenic RAS. Genes Dev (2009) 23:1177–82. doi: 10.1101/gad.511109
31. Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, et al. The H3K27me3 Demethylase JMJD3 Contributes to the Activation of the INK4A-ARF Locus in Response to Oncogene- and Stress-Induced Senescence. Genes Dev (2009) 23:1171–6. doi: 10.1101/gad.510809
32. Sherry-Lynes MM, Sengupta S, Kulkarni S, Cochran BH. Regulation of the JMJD3 (KDM6B) Histone Demethylase in Glioblastoma Stem Cells by STAT3. PLoS One (2017) 12:e0174775. doi: 10.1371/journal.pone.0174775
33. Pinton G, Nilsson S, Moro L. Targeting Estrogen Receptor Beta (Erβ) for Treatment of Ovarian Cancer: Importance of KDM6B and SIRT1 for Erβ Expression and Functionality. Oncogenesis (2018) 7:15. doi: 10.1038/s41389-018-0027-9
34. Park WY, Hong BJ, Lee J, Choi C, Kim MY. H3K27 Demethylase JMJD3 Employs the NF-κb and BMP Signaling Pathways to Modulate the Tumor Microenvironment and Promote Melanoma Progression and Metastasis. Cancer Res (2016) 76:161–70. doi: 10.1158/0008-5472.CAN-15-0536
35. Fei T, Xia K, Li Z, Zhou B, Zhu S, Chen H, et al. Genome-Wide Mapping of SMAD Target Genes Reveals the Role of BMP Signaling in Embryonic Stem Cell Fate Determination. Genome Res (2010) 20:36–44. doi: 10.1101/gr.092114.109
36. Greten FR, Grivennikov SI. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity (2019) 51:27–41. doi: 10.1016/j.immuni.2019.06.025
37. Saccani S, Natoli G. Dynamic Changes in Histone H3 Lys 9 Methylation Occurring at Tightly Regulated Inducible Inflammatory Genes. Genes Dev (2002) 16:2219–24. doi: 10.1101/gad.232502
38. Przanowski P, Dabrowski M, Ellert-Miklaszewska A, Kloss M, Mieczkowski J, Kaza B, et al. The Signal Transducers Stat1 and Stat3 and Their Novel Target Jmjd3 Drive the Expression of Inflammatory Genes in Microglia. J Mol Med (Berl) (2014) 92:239–54. doi: 10.1007/s00109-013-1090-5
39. Min Y, Kim MJ, Lee S, Chun E, Lee KY. Inhibition of TRAF6 Ubiquitin-Ligase Activity by PRDX1 Leads to Inhibition of NFKB Activation and Autophagy Activation. Autophagy (2018) 14:1347–58. doi: 10.1080/15548627.2018.1474995
40. Ramadoss S, Chen X, Wang CY. Histone Demethylase KDM6B Promotes Epithelial-Mesenchymal Transition. J Biol Chem (2012) 287:44508–17. doi: 10.1074/jbc.M112.424903
41. David CJ, Massagué J. Contextual Determinants of Tgfβ Action in Development, Immunity and Cancer. Nat Rev Mol Cell Biol (2018) 19:419–35. doi: 10.1038/s41580-018-0007-0
42. Jiang BH, Jiang G, Zheng JZ, Lu Z, Hunter T, Vogt PK. Phosphatidylinositol 3-Kinase Signaling Controls Levels of Hypoxia-Inducible Factor 1. Cell Growth Differ (2001) 12:363–9. doi: 10.1073/pnas.040560897
43. Stiehl DP, Jelkmann W, Wenger RH, Hellwig-Bürgel T. Normoxic Induction of the Hypoxia-Inducible Factor 1alpha by Insulin and Interleukin-1beta Involves the Phosphatidylinositol 3-Kinase Pathway. FEBS Lett (2002) 512:157–62. doi: 10.1016/s0014-5793(02)02247-0
44. Treins C, Giorgetti-Peraldi S, Murdaca J, Semenza GL, Obberghen EV. Insulin Stimulates Hypoxia-Inducible Factor 1 Through a Phosphatidylinositol 3-Kinase/Target of Rapamycin-Dependent Signaling Pathway. J Biol Chem (2002) 277:27975–81. doi: 10.1074/jbc.M204152200
45. Van Uden P, Kenneth NS, Rocha S. Regulation of Hypoxia-Inducible Factor-1alpha by NF-Kappab. Biochem J (2008) 412:477–84. doi: 10.1042/BJ20080476
46. Rozen-Zvi B, Hayashida T, Hubchak SC, Hanna C, Platanias LC, Schnaper HW. TGF-β/Smad3 Activates Mammalian Target of Rapamycin Complex-1 to Promote Collagen Production by Increasing HIF-1α Expression. Am J Physiol Renal Physiol (2013) 305:F485–94. doi: 10.1152/ajprenal.00215.2013
47. Pereira F, Barbáchano A, Silva J, Bonilla F, Campbell MJ, Muñoz A, et al. KDM6B/JMJD3 Histone Demethylase is Induced by Vitamin D and Modulates its Effects in Colon Cancer Cells. Hum Mol Genet (2011) 20:4655–65. doi: 10.1093/hmg/ddr399
48. Pan M, Reid MA, Lowman XH, Kulkarni RP, Tran TQ, Liu X, et al. Regional Glutamine Deficiency in Tumours Promotes Dedifferentiation Through Inhibition of Histone Demethylation. Nat Cell Biol (2016) 18:1090–101. doi: 10.1038/ncb3410
49. Dutta A, Le Magnen C, Mitrofanova A, Ouyang X, Califano A, Abate-Shen C. Identification of an NKX3.1-G9a-UTY Transcriptional Regulatory Network That Controls Prostate Differentiation. Science (2016) 352:1576–80. doi: 10.1126/science.aad9512
50. Stief SM, Hanneforth AL, Weser S, Mattes R, Carlet M, Liu WH, et al. Loss of KDM6A Confers Drug Resistance in Acute Myeloid Leukemia. Leukemia (2020) 34:50–62. doi: 10.1038/s41375-019-0497-6
51. Greif PA, Hartmann L, Vosberg S, Stief SM, Mattes R, Hellmann I, et al. Evolution of Cytogenetically Normal Acute Myeloid Leukemia During Therapy and Relapse: An Exome Sequencing Study of 50 Patients. Clin Cancer Res (2018) 24:1716–26. doi: 10.1158/1078-0432.CCR-17-2344
52. Gozdecka M, Meduri E, Mazan M, Tzelepis K, Dudek M, Knights AJ, et al. UTX-Mediated Enhancer and Chromatin Remodeling Suppresses Myeloid Leukemogenesis Through Noncatalytic Inverse Regulation of ETS and GATA Programs. Nat Genet (2018) 50:883–94. doi: 10.1038/s41588-018-0114-z
53. Li X, Zhang Y, Zheng L, Liu M, Chen CD, Jiang H. UTX is an Escape From X-Inactivation Tumor-Suppressor in B Cell Lymphoma. Nat Commun (2018) 9:2720. doi: 10.1038/s41467-018-05084-w
54. Licht JD. Disordered Histone Methylation in Hematological Malignancies the Case of UTX/KDM6A. Trans Am Clin Climatol Assoc (2018) 129:24–36.
55. Ezponda T, Dupéré-Richer D, Will CM, Small EC, Varghese N, Patel T, et al. UTX/KDM6A Loss Enhances the Malignant Phenotype of Multiple Myeloma and Sensitizes Cells to EZH2 Inhibition. Cell Rep (2017) 21:628–40. doi: 10.1016/j.celrep.2017.09.078
56. Terashima M, Ishimura A, Wanna-Udom S, Suzuki T. Epigenetic Regulation of Epithelial-Mesenchymal Transition by KDM6A Histone Demethylase in Lung Cancer Cells. BiochemBioph Res Co (2017) 490:1407–13. doi: 10.1016/j.bbrc.2017.07.048
57. Wu Q, Tian Y, Zhang J, Tong X, Huang H, Li S, et al. In Vivo CRISPR Screening Unveils Histone Demethylase UTX as an Important Epigenetic Regulator in Lung Tumorigenesis. Proc Natl Acad Sci U S A (2018) 115:E3978–86. doi: 10.1073/pnas.1716589115
58. Andricovich J, Perkail S, Kai Y, Casasanta N, Peng W, Tzatsos A. Loss of KDM6A Activates Super-Enhancers to Induce Gender-Specific Squamous-Like Pancreatic Cancer and Confers Sensitivity to BET Inhibitors. Cancer Cell (2018) 33:512–26.e8. doi: 10.1016/j.ccell.2018.02.003
59. Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic Analyses Identify Molecular Subtypes of Pancreatic Cancer. Nature (2016) 531:47–52. doi: 10.1038/nature16965
60. Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, et al. The H3K27me3 Demethylase UTX Is a Gender-Specific Tumor Suppressor in T-Cell Acute Lymphoblastic Leukemia. Blood (2015) 125:13–21. doi: 10.1182/blood-2014-05-577270
61. Schulz WA, Lang A, Koch J, Greife A. The Histone Demethylase UTX/KDM6A in Cancer: Progress and Puzzles. Int J Cancer (2019) 145:614–20. doi: 10.1002/ijc.32116
62. Audenet F, Isharwal S, Cha EK, Donoghue MTA, Drill FN, Ostrovnaya I, et al. Clonal Relatedness and Mutational Differences Between Upper Tract and Bladder Urothelial Carcinoma. Clin Cancer Res (2019) 25:967–76. doi: 10.1158/1078-0432.CCR-18-2039
63. Lang A, Whongsiri P, Yilmaz M, Lautwein T, Petzsch P, Greife A, et al. Knockdown of UTX/KDM6A Enriches Precursor Cell Populations in Urothelial Cell Cultures and Cell Lines. Cancers (2020) 12(4):1023. doi: 10.3390/cancers12041023
64. Li R, Du Y, Chen Z, Xu D, Lin T, Jin S, et al. Macroscopic Somatic Clonal Expansion in Morphologically Normal Human Urothelium. Science (2020) 370:82–9. doi: 10.1126/science.aba7300
65. Lang A, Yilmaz M, Hader C, Murday S, Kunz X, Wagner N, et al. Contingencies of UTX/KDM6A Action in Urothelial Carcinoma. Cancers (2019) 11(4):481. doi: 10.3390/cancers11040481
66. Kaneko S, Li X. X Chromosome Protects Against Bladder Cancer in Females via a KDM6A-Dependent Epigenetic Mechanism. Sci Adv (2018) 4:eaar5598. doi: 10.1126/sciadv.aar5598
67. Zhou Z, Zhang HS, Liu Y, Zhang ZG, Du GY, Li H, et al. Loss of TET1 Facilitates DLD1 Colon Cancer Cell Migration via H3K27me3-Mediated Down-Regulation of E-Cadherin. J Cell Physiol (2018) 233:1359–69. doi: 10.1002/jcp.26012
68. Zha L, Cao Q, Cui X, Li F, Liang H, Xue B, et al. Epigenetic Regulation of E-Cadherin Expression by the Histone Demethylase UTX in Colon Cancer Cells. Med Oncol (2016) 33:21. doi: 10.1007/s12032-016-0734-z
69. Yi J, Shi X, Xuan Z, Wu J. Histone Demethylase UTX/KDM6A Enhances Tumor Immune Cell Recruitment, Promotes Differentiation and Suppresses Medulloblastoma. Cancer Lett (2021) 499:188–200. doi: 10.1016/j.canlet.2020.11.031
70. Taube JH, Sphyris N, Johnson KS, Reisenauer KN, Nesbit TA, Joseph R, et al. The H3K27me3-Demethylase KDM6A is Suppressed in Breast Cancer Stem-Like Cells, and Enables the Resolution of Bivalency During the Mesenchymal-Epithelial Transition. Oncotarget (2017) 8:65548–65. doi: 10.18632/oncotarget.19214
71. Yu W, Huang W, Yang Y, Qiu R, Zeng Y, Hou Y, et al. GATA3 Recruits UTX for Gene Transcriptional Activation to Suppress Metastasis of Breast Cancer. Cell Death Dis (2019) 10:832. doi: 10.1038/s41419-019-2062-7
72. Kodama T, Newberg JY, Kodama M, Rangel R, Yoshihara K, Tien JC, et al. Transposon Mutagenesis Identifies Genes and Cellular Processes Driving Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma. Proc Natl Acad Sci U S A (2016) 113:E3384–93. doi: 10.1073/pnas.1606876113
73. Li Y, Yang J, Zhang X, Liu H, Guo J. KDM6A Suppresses Hepatocellular Carcinoma Cell Proliferation by Negatively Regulating the TGF-Beta/SMAD Signaling Pathway. Exp Ther Med (2020) 20:2774–82. doi: 10.3892/etm.2020.9000
74. Benyoucef A, Palii CG, Wang C, Porter CJ, Chu A, Dai F, et al. UTX Inhibition as Selective Epigenetic Therapy Against TAL1-Driven T-Cell Acute Lymphoblastic Leukemia. Genes Dev (2016) 30:508–21. doi: 10.1101/gad.276790.115
75. Terashima M, Ishimura A, Yoshida M, Suzuki Y, Sugano S, Suzuki T. The Tumor Suppressor Rb and its Related Rbl2 Genes Are Regulated by Utx Histone Demethylase. BiochemBiophys Res Commun (2010) 399:238–44. doi: 10.1016/j.bbrc.2010.07.061
76. Lu H, Xie Y, Tran L, Lan J, Yang Y, Murugan NL, et al. Chemotherapy-Induced S100A10 Recruits KDM6A to Facilitate OCT4-Mediated Breast Cancer Stemness. J Clin Invest (2020) 130:4607–23. doi: 10.1172/JCI138577
77. Rathkopf D, Scher HI. Androgen Receptor Antagonists in Castration-Resistant Prostate Cancer. Cancer J (2013) 19:43–9. doi: 10.1097/PPO.0b013e318282635a
78. Morozov VM, Li Y, Clowers MM, Ishov AM. Inhibitor of H3K27 Demethylase JMJD3/UTX GSK-J4 is a Potential Therapeutic Option for Castration Resistant Prostate Cancer. Oncotarget (2017) 8:62131–42. doi: 10.18632/oncotarget.19100
79. Wang F, Zhu Y, Wang F, Wang Y, Dong BJ, Wang N, et al. Wnt/beta-Catenin Signaling Contributes to Prostate Cancer Heterogeneity Through Reciprocal Suppression of H3K27 Trimethylation. BiochemBiophys Res Commun (2020) 527:242–9. doi: 10.1016/j.bbrc.2020.04.112
80. Soto DR, Barton C, Munger K, McLaughlin-Drubin ME. KDM6A Addiction of Cervical Carcinoma Cell Lines is Triggered by E7 and Mediated by P21cip1 Suppression of Replication Stress. PLoS Pathog (2017) 13:e1006661. doi: 10.1371/journal.ppat.1006661
81. Biswas M, Chatterjee SS, Boila LD, Chakraborty S, Banerjee D, Sengupta A. MBD3/NuRD Loss Participates With KDM6A Program to Promote DOCK5/8 Expression and Rac GTPase Activation in Human Acute Myeloid Leukemia. FASEB J (2019) 33:5268–86. doi: 10.1096/fj.201801035R
82. Kim J, Lee SH, Jang JH, Kim MS, Lee EH, Kim YZ. Increased Expression of the Histone H3 Lysine 4 Methyltransferase MLL4 and the Histone H3 Lysine 27 Demethylase UTX Prolonging the Overall Survival of Patients With Glioblastoma and a Methylated MGMT Promoter. J Neurosurg (2017) 126:1461–71. doi: 10.3171/2016.4.JNS1652
83. Liau BB, Sievers C, Donohue LK, Gillespie SM, Flavahan WA, Miller TE, et al. Adaptive Chromatin Remodeling Drives Glioblastoma Stem Cell Plasticity and Drug Tolerance. Cell Stem Cell (2017) 20:233–46.e7. doi: 10.1016/j.stem.2016.11.003
84. Leng X, Wang J, An N, Wang X, Sun Y, Chen Z. Histone 3 Lysine-27 Demethylase KDM6A Coordinates With KMT2B to Play an Oncogenic Role in NSCLC by Regulating H3k4me3. Oncogene (2020) 39:6468–79. doi: 10.1038/s41388-020-01449-y
85. Ho AS, Ochoa A, Jayakumaran G, Zehir A, Mayor CV, Tepe J, et al. Genetic Hallmarks of Recurrent/Metastatic Adenoid Cystic Carcinoma. J Clin Invest (2019) 129:4276–89. doi: 10.1172/JCI128227
86. Emran AA, Nsengimana J, Punnia-Moorthy G, Schmitz U, Gallagher SJ, Newton-Bishop J, et al. Study of the Female Sex Survival Advantage in Melanoma-A Focus on X-Linked Epigenetic Regulators and Immune Responses in Two Cohorts. Cancers (2020) 12(8):2082. doi: 10.3390/cancers12082082
87. Chen HJ, Huang RL, Liew PL, Su PH, Chen LY, Weng YC, et al. GATA3 as a Master Regulator and Therapeutic Target in Ovarian High-Grade Serous Carcinoma Stem Cells. Int J Cancer (2018) 143:3106–19. doi: 10.1002/ijc.31750
88. Yang L, Zha Y, Ding J, Ye B, Liu M, Yan C, et al. Histone Demethylase KDM6B has an Anti-Tumorigenic Function in Neuroblastoma by Promoting Differentiation. Oncogenesis (2019) 8:3. doi: 10.1038/s41389-018-0112-0
89. Tokunaga R, Sakamoto Y, Nakagawa S, Miyake K, Izumi D, Kosumi K, et al. The Prognostic Significance of Histone Lysine Demethylase JMJD3/KDM6B in Colorectal Cancer. Ann Surg Oncol (2016) 23:678–85. doi: 10.1245/s10434-015-4879-3
90. Yu SH, Zhu KY, Chen J, Liu XZ, Xu PF, Zhang W, et al. JMJD3 Facilitates C/EBPbeta-Centered Transcriptional Program to Exert Oncorepressor Activity in AML. Nat Commun (2018) 9:3369. doi: 10.1038/s41467-018-05548-z
91. Yamamoto K, Tateishi K, Kudo Y, Sato T, Yamamoto S, Miyabayashi K, et al. Loss of Histone Demethylase KDM6B Enhances Aggressiveness of Pancreatic Cancer Through Downregulation of C/EBPalpha. Carcinogenesis (2014) 35:2404–14. doi: 10.1093/carcin/bgu136
92. Al Labban D, Jo SH, Ostano P, Saglietti C, Bongiovanni M, Panizzon R, et al. Notch-Effector CSL Promotes Squamous Cell Carcinoma by Repressing Histone Demethylase KDM6B. J Clin Invest (2018) 128:2581–99. doi: 10.1172/JCI96915
93. Li Y, Zhang M, Sheng M, Zhang P, Chen Z, Xing W, et al. Therapeutic Potential of GSK-J4, a Histone Demethylase KDM6B/JMJD3 Inhibitor, for Acute Myeloid Leukemia. J Cancer Res Clin (2018) 144:1065–77. doi: 10.1007/s00432-018-2631-7
94. Mallaney C, Ostrander EL, Celik H, Kramer AC, Martens A, Kothari A, et al. Kdm6b Regulates Context-Dependent Hematopoietic Stem Cell Self-Renewal and Leukemogenesis. Leukemia (2019) 33:2506–21. doi: 10.1038/s41375-019-0462-4
95. McLaughlin-Drubin ME, Park D, Munger K. Tumor Suppressor P16ink4a is Necessary for Survival of Cervical Carcinoma Cell Lines. Proc Natl Acad Sci U S A (2013) 110:16175–80. doi: 10.1073/pnas.1310432110
96. Zhang Y, Shen L, Stupack DG, Bai N, Xun J, Ren G, et al. JMJD3 Promotes Survival of Diffuse Large B-Cell Lymphoma Subtypes via Distinct Mechanisms. Oncotarget (2016) 7:29387–99. doi: 10.18632/oncotarget.8836
97. Cao Z, Shi X, Tian F, Fang Y, Wu JB, Mrdenovic S, et al. KDM6B is an Androgen Regulated Gene and Plays Oncogenic Roles by Demethylating H3K27me3 at Cyclin D1 Promoter in Prostate Cancer. Cell Death Dis (2021) 12:2. doi: 10.1038/s41419-020-03354-4
98. Daures M, Idrissou M, Judes G, Rifaï K, Penault-Llorca K, Bignon YJ, et al. A New Metabolic Gene Signature in Prostate Cancer Regulated by JMJD3 and EZH2. Oncotarget (2018) 9:23413–25. doi: 10.18632/oncotarget.25182
99. Xu Z, Xia Y, Xiao Z, Jia Y, Li L, Jin Y, et al. Comprehensive Profiling of JMJD3 in Gastric Cancer and Its Influence on Patient Survival. Sci Rep (2019) 9:868. doi: 10.1038/s41598-018-37340-w
100. Tang B, Qi G, Tang F, Yuan S, Wang Z, Liang X, et al. Aberrant JMJD3 Expression Upregulates Slug to Promote Migration, Invasion, and Stem Cell-Like Behaviors in Hepatocellular Carcinoma. Cancer Res (2016) 76:6520–32. doi: 10.1158/0008-5472.CAN-15-3029
101. Wang W, Wang Q, Huang DB, Sun QK, Wu SS, Zhao YJ, et al. Tumor-Associated Mesenchymal Stem Cells Promote Hepatocellular Carcinoma Metastasis via a DNM3OS/KDM6B/TIAM1 Axis. Cancer Lett (2021) 503:19–31. doi: 10.1016/j.canlet.2021.01.011
102. Liang S, Yao Q, Wei D, Liu M, Geng F, Wang Q, et al. KDM6B Promotes Ovarian Cancer Cell Migration and Invasion by Induced Transforming Growth Factor-Beta1 Expression. J Cell Biochem (2019) 120:493–506. doi: 10.1002/jcb.27405
103. Mo J, Wang L, Huang X, Lu B, Zou C, Wei L, et al. Multifunctional Nanoparticles for Co-Delivery of Paclitaxel and Carboplatin Against Ovarian Cancer by Inactivating the JMJD3-HER2 Axis. Nanoscale (2017) 9:13142–52. doi: 10.1039/c7nr04473a
104. Jin Q, Martinez CA, Arcipowski KM, Zhu Y, Gutierrez-Diaz BT, Wang KK, et al. USP7 Cooperates With NOTCH1 to Drive the Oncogenic Transcriptional Program in T-Cell Leukemia. Clin Cancer Res (2019) 25:222–39. doi: 10.1158/1078-0432.CCR-18-1740
105. Wang W, Lim KG, Feng M, Bao Y, Lee PL, Cai Y, et al. KDM6B Counteracts EZH2-Mediated Suppression of IGFBP5 to Confer Resistance to PI3K/AKT Inhibitor Treatment in Breast Cancer. Mol Cancer Ther (2018) 17:1973–83. doi: 10.1158/1535-7163.MCT-17-0802
106. Yan N, Xu L, Wu X, Zhang L, Fei X, Cao Y, et al. GSKJ4, an H3K27me3 Demethylase Inhibitor, Effectively Suppresses the Breast Cancer Stem Cells. Exp Cell Res (2017) 359:405–14. doi: 10.1016/j.yexcr.2017.08.024
107. Xun J, Wang D, Shen L, Gong J, Gao R, Du L, et al. JMJD3 Suppresses Stem Cell-Like Characteristics in Breast Cancer Cells by Downregulation of Oct4 Independently of Its Demethylase Activity. Oncotarget (2017) 8:21918–29. doi: 10.18632/oncotarget.15747
108. Anderton JA, Bose S, Vockerodt M, Vrzalikova K, Wei W, Kuo M, et al. The H3K27me3 Demethylase, KDM6B, Is Induced by Epstein-Barr Virus and Over-Expressed in Hodgkin's Lymphoma. Oncogene (2011) 30:2037–43. doi: 10.1038/onc.2010.579
109. Nagarsheth N, Peng D, Kryczek I, Wu K, Li W, Zhao E, et al. PRC2 Epigenetically Silences Th1-Type Chemokines to Suppress Effector T-Cell Trafficking in Colon Cancer. Cancer Res (2016) 76:275–82. doi: 10.1158/0008-5472.CAN-15-1938
110. Park WY, Hong BJ, Lee J, Choi C, Kim MY. H3K27 Demethylase JMJD3 Employs the NF-kappaB and BMP Signaling Pathways to Modulate the Tumor Microenvironment and Promote Melanoma Progression and Metastasis. Cancer Res (2016) 76:161–70. doi: 10.1158/0008-5472.CAN-15-0536
112. Zhang Y, Hua PY, Jin CY, Li JD, Zhang GX, Wang B. JMJD3 Enhances Invasiveness and Migratory Capacity of Non-Small Cell Lung Cancer Cell via Activating EMT Signaling Pathway. Eur Rev Med Pharmacol Sci (2019) 23:4784–92. doi: 10.26355/eurrev_201906_18063
113. Lachat C, Bruyère D, Etcheverry A, Aubry M, Mosser J, Warda W, et al. EZH2 and KDM6B Expressions Are Associated With Specific Epigenetic Signatures During EMT in Non Small Cell Lung Carcinomas. Cancers (2020) 12(12):3649. doi: 10.3390/cancers12123649
114. Wei Y, Zheng H, Bao N, Jiang S, Bueso-Ramos CE, Khoury J, et al. KDM6B Overexpression Activates Innate Immune Signaling and Impairs Hematopoiesis in Mice. Blood Adv (2018) 2:2491–504. doi: 10.1182/bloodadvances.2018024166
115. Xia M, Yao L, Zhang Q, Wang F, Mei H, Guo X, et al. Long Noncoding RNA HOTAIR Promotes Metastasis of Renal Cell Carcinoma by Up-Regulating Histone H3K27 Demethylase JMJD3. Oncotarget (2017) 8:19795–802. doi: 10.18632/oncotarget.15047
116. Cottone L, Cribbs AP, Khandelwal G, Wells G, Ligammari L, Philpott M, et al. Inhibition of Histone H3K27 Demethylases Inactivates Brachyury (TBXT) and Promotes Chordoma Cell Death. Cancer Res (2020) 80:4540–51. doi: 10.1158/0008-5472.CAN-20-1387
117. Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, Gupta R, et al. The H3K27me3 Demethylase dUTX Is a Suppressor of Notch- and Rb-Dependent Tumors in Drosophila. Mol Cell Biol (2010) 30:2485–97. doi: 10.1128/MCB.01633-09
118. Li SH, Lu HI, Huang WT, Tien WY, Lan YC, Lin WC, et al. The Prognostic Significance of Histone Demethylase UTX in Esophageal Squamous Cell Carcinoma. Int J Mol Sci (2018) 19(1):297. doi: 10.3390/ijms19010297
119. Barrows D, Feng L, Carroll TS, Allis CD. Loss of UTX/KDM6A and the Activation of FGFR3 Converge to Regulate Differentiation Gene-Expression Programs in Bladder Cancer. Proc Natl Acad Sci U S A (2020) 117:25732–41. doi: 10.1073/pnas.2008017117
120. Kobatake K, Ikeda KI, Nakata Y, Yamasaki N, Ueda T, Kanai A, et al. Kdm6a Deficiency Activates Inflammatory Pathways, Promotes M2 Macrophage Polarization, and Causes Bladder Cancer in Cooperation With P53 Dysfunction. Clin Cancer Res (2020) 26:2065–79. doi: 10.1158/1078-0432.CCR-19-2230
121. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the Cbioportal. Sci Signal (2013) 6:pl1. doi: 10.1126/scisignal.2004088
122. Cerami E, Gao J, Dogrusoz U, Aksoy BA, Gross BE, Sumer SO, et al. The Cbio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov (2012) 2:401–4. doi: 10.1158/2159-8290.CD-12-0095
123. Hurst CD, Alder O, Platt FM, Droop A, Stead LF, Burns JE, et al. Genomic Subtypes of Non-Invasive Bladder Cancer With Distinct Metabolic Profile and Female Gender Bias in KDM6A Mutation Frequency. Cancer Cell (2017) 32:701–15.e7. doi: 10.1016/j.ccell.2017.08.005
124. Wang F, Zhu Y, Wang F, Wang Y, Dong BJ, Wang N, et al. Wnt/β-Catenin Signaling Contributes to Prostate Cancer Heterogeneity Through Reciprocal Suppression of H3K27 Trimethylation. BiochemBiophys Res Commun (2020) 527:242–9. doi: 10.1016/j.bbrc.2020.04.112
125. Rejlova K, Musilova A, Kramarzova KS, Zaliova M, Fiser K, Alberich-Jorda M, et al. Low HOX Gene Expression in PML-Rarα-Positive Leukemia Results From Suppressed Histone Demethylation. Epigenetics (2018) 13:73–84. doi: 10.1080/15592294.2017.1413517
126. Tricarico R, Nicolas E, Hall MJ, Golemis EA. X- and Y-Linked Chromatin-Modifying Genes as Regulators of Sex-Specific Cancer Incidence and Prognosis. Clin Cancer Res (2020) 26:5567–78. doi: 10.1158/1078-0432.CCR-20-1741
127. Jambhekar A, Anastas JN, Shi Y. Histone Lysine Demethylase Inhibitors. Cold Spring HarbPerspect Med (2017) 77(1):a026484. doi: 10.1101/cshperspect.a026484
128. Benedetti R, Dell'Aversana C, De Marchi T, Rotili D, Liu NQ, Novakovic B, et al. Inhibition of Histone Demethylases LSD1 and UTX Regulates Erα Signaling in Breast Cancer. Cancers (2019) 11(12):2027. doi: 10.3390/cancers11122027
129. Cuyàs E, Verdura S, Llorach-Pares L, Fernández-Arroyo S, Luciano-Mateo F, Cabré N, et al. Metformin Directly Targets the H3K27me3 Demethylase KDM6A/UTX. Aging Cell (2018) 17:e12772. doi: 10.1111/acel.12772
130. Zhang W, Cheng J, Diao P, Wang D, Zhang W, Jiang H, et al. Therapeutically Targeting Head and Neck Squamous Cell Carcinoma Through Synergistic Inhibition of LSD1 and JMJD3 by TCP and GSK-J1. Br J Cancer (2020) 122:528–38. doi: 10.1038/s41416-019-0680-6
131. Sui A, Xu Y, Li Y, Hu Q, Wang Z, Zhang H, et al. The Pharmacological Role of Histone Demethylase JMJD3 Inhibitor GSK-J4 on Glioma Cells. Oncotarget (2017) 8:68591–8. doi: 10.18632/oncotarget.19793
132. Cregan S, Breslin M, Roche G, Wennstedt S, MacDonagh L, Albadri C, et al. Kdm6a and Kdm6b: Altered Expression in Malignant Pleural Mesothelioma. Int J Oncol (2017) 50:1044–52. doi: 10.3892/ijo.2017.3870
133. Mathur R, Sehgal L, Havranek O, Köhrer S, Khashab T, Jain N, et al. Inhibition of Demethylase KDM6B Sensitizes Diffuse Large B-Cell Lymphoma to Chemotherapeutic Drugs. Haematologica (2017) 102:373–80. doi: 10.3324/haematol.2016.144964
134. Sakaki H, Okada M, Kuramoto K, Takeda H, Watarai H, Suzuki S, et al. GSKJ4, A Selective Jumonji H3K27 Demethylase Inhibitor, Effectively Targets Ovarian Cancer Stem Cells. Anticancer Res (2015) 35:6607–14.
135. Lochmann TL, Powell KM, Ham J, Floros KV, Heisey DAR, Kurupi RIJ, et al. Targeted Inhibition of Histone H3K27 Demethylation Is Effective in High-Risk Neuroblastoma. Sci Transl Med (2018) 10(441):eaao4680. doi: 10.1126/scitranslmed.aao4680
136. Jiang Y, Li F, Gao B, Ma M, Chen M, Wu Y, et al. KDM6B-Mediated Histone Demethylation of LDHA Promotes Lung Metastasis of Osteosarcoma. Theranostics (2021) 11:3868–81. doi: 10.7150/thno.53347
137. Zhang J, Ying Y, Li M, Wang M, Huang X, Jia M, et al. Targeted Inhibition of KDM6 Histone Demethylases Eradicates Tumor-Initiating Cells via Enhancer Reprogramming in Colorectal Cancer. Theranostics (2020) 10:10016–30. doi: 10.7150/thno.47081
138. Ler LD, Ghosh S, Chai X, Thike AA, Heng HL, Siew EY, et al. Loss of Tumor Suppressor KDM6A Amplifies PRC2-Regulated Transcriptional Repression in Bladder Cancer and can be Targeted Through Inhibition of EZH2. Sci Transl Med (2017) 9(378):eaai8312. doi: 10.1126/scitranslmed.aai8312
139. Ramakrishnan S, Granger V, Rak M, Hu Q, Attwood K, Aquila L, et al. Inhibition of EZH2 Induces NK Cell-Mediated Differentiation and Death in Muscle-Invasive Bladder Cancer. Cell Death Differ (2019) 26:2100–14. doi: 10.1038/s41418-019-0278-9
140. Watanabe S, Shimada S, Akiyama Y, Ishikawa Y, Ogura T, Ogawa K, et al. Loss of KDM6A Characterizes a Poor Prognostic Subtype of Human Pancreatic Cancer and Potentiates HDAC Inhibitor Lethality. Int J Cancer (2019) 145:192–205. doi: 10.1002/ijc.32072
141. Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, et al. A Selective Jumonji H3K27 Demethylase Inhibitor Modulates the Proinflammatory Macrophage Response. Nature (2012) 488:404–8. doi: 10.1038/nature11262
142. Heinemann B, Nielsen JM, Hudlebusch HR, Boesen T, Lees MJ, Larsen DV, et al. Inhibition of Demethylases by GSK-J1/J4. Nature (2014) 514:E1–2. doi: 10.1038/nature13688
143. Van der Meulen J, Speleman F, Van Vlierberghe P. The H3K27me3 Demethylase UTX in Normal Development and Disease. Epigenetics (2014) 9:658–68. doi: 10.4161/epi.28298
144. Topper MJ, Vaz M, Marrone KA, Brahmer JR, Baylin SB. The Emerging Role of Epigenetic Therapeutics in Immuno-Oncology. Nat Rev Clin Oncol (2020) 17:75–90. doi: 10.1038/s41571-019-0266-5
145. Cribbs AP, Terlecki-Zaniewicz S, Philpott M, Baardman J, Ahern D, Lindow M, et al. Histone H3K27me3 Demethylases Regulate Human Th17 Cell Development and Effector Functions by Impacting on Metabolism. Proc Natl Acad Sci U S A (2020) 117:6056–66. doi: 10.1073/pnas.1919893117
146. Cribbs A, Hookway ES, Wells G, Lindow M, Obad S, Oerum H, et al. Inhibition of Histone H3K27 Demethylases Selectively Modulates Inflammatory Phenotypes of Natural Killer Cells. J Biol Chem (2018) 293:2422–37. doi: 10.1074/jbc.RA117.000698
Keywords: histone demethylase, KDM6, cancer, epigenetics, cancer therapy
Citation: Hua C, Chen J, Li S, Zhou J, Fu J, Sun W and Wang W (2021) KDM6 Demethylases and Their Roles in Human Cancers. Front. Oncol. 11:779918. doi: 10.3389/fonc.2021.779918
Received: 21 September 2021; Accepted: 17 November 2021;
Published: 07 December 2021.
Edited by:
Tuuli Käämbre, National Institute of Chemical Physics and Biophysics, EstoniaReviewed by:
Adam Paul Cribbs, University of Oxford, United KingdomCopyright © 2021 Hua, Chen, Li, Zhou, Fu, Sun and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijian Sun, d2VpamlhbnN1bkB3bXUuZWR1LmNu; Wenqian Wang, d2FuZ3dlbnFpYW5Ad211LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.