- 1State Key Laboratory of Respiratory Disease, National Clinical Research Centre for Respiratory Disease, Guangzhou Institute of Respiratory Health, First Affiliated Hospital, Guangzhou Medical University, Guangzhou, China
- 2Department of Thoracic Surgery, Hunan Key Laboratory of Early Diagnosis and Precise Treatment of Lung Cancer, The Second Xiangya Hospital of Central South University, Changsha, China
- 3Department of Respiratory Medicine, First Affiliated Hospital of Guangxi Medical University, Nanning, China
- 4Medical Department, Geneplus-Beijing, Beijing, China
- 5Department of Respiratory and Critical Medicine, Zhuhai People’s Hospital (Zhuhai Hospital Affiliated with Jinan University), Zhuhai, China
Background: Epidemiological surveys have suggested that lung cancer has inherited susceptibility and shows familial aggregation. However, the distribution and prevalence of epidermal growth factor receptor (EGFR) germline variants and their roles in lung cancer genetic predisposition in Chinese population remain to be elucidated.
Methods: In this study, EGFR germline and somatic variants were retrospectively reviewed from the next-generation sequencing results of 31,906 patients with lung cancer. Clinical information was also collected for patients with confirmed EGFR germline mutations.
Results: A total of 22 germline EGFR variants were identified in 64 patients with lung cancer, accounting for 0.2% of the total cases studied. Five patients were diagnosed as multiple primary carcinomas. Family history was documented in 31.3% (20/64) of patients, 55% of which were diagnosed as lung cancer. G863D was the most frequent EGFR germline mutation, followed by P848L, D1014N, and K757R. Somatic EGFR-sensitive mutations were identified in 51.6% of patients with germline EGFR mutations. The proportion of L858R mutation, exon 19 deletion, and rare sensitive mutation was 50%, 17.6%, and 32.4%, respectively. D1014N and T790M mutations were common in young patients. The family members of patients with P848L, R776H, V769M, and V774M mutations were more commonly diagnosed with cancers. A total of 19 patients were confirmed to have received EGFR tyrosine kinase inhibitors (TKIs), but the response to EGFR-TKIs differed among patients with different EGFR mutations.
Conclusion: Chinese patients with lung cancer harbored unique and dispersive EGFR germline mutations and showed unique clinical and genetic characteristics, with varied response patterns to EGFR-TKI treatment.
Introduction
Lung cancer is the most common and lethal malignancy in most countries. China reported 733,300 new cases and 610,200 lung cancer deaths in 2015 (1). Tobacco smoking is the greatest risk factor for lung cancer development, with up to 80% of cases attributed to smoking (2). Recently, additional risk factors, including exposure to radon, occupational hazards, biomass fuel, and infectious diseases have been identified as additional risk factors in the carcinogenesis of lung cancer (2).
Epidemiological surveys have further suggested that lung cancer has inherited susceptibility and show familial aggregation (3–6). That is, genetic factors, such as high-frequency single nucleotide polymorphisms with low penetrance and low-frequency pathogenic germline variants with high penetrance, have been confirmed to be related to lung cancer predisposition (7–10). Multiple genome-wide association studies confirmed CHRNA5, TERT, BAT2, and FKBPL as candidate genes associated with lung cancer risk (6, 7). The investigation of pathogenic germline variants mainly focused on epidermal growth factor receptor (EGFR) and other genes commonly related to hereditary tumor syndromes, including ATM, TP53, and BRCA2 (11). There are four well-documented germline mutations in EGFR, including T790M, V843I, R776X, and P848L (12). EGFR T790M is the most frequent mutation in Western countries, with a 0.54% frequency in nonsmokers and 0.34% in patients with nonsquamous nonsmall cell lung cancer (NSCLC) (13, 14). However, the frequency of EGFR T790M germline mutation in Chinese lung cancer patients was 0.0078%, suggesting a distinct germline mutation spectrum among different ethnicities (15). Therefore, the distribution and prevalence of EGFR germline variants and their roles in lung cancer genetic predisposition in Chinese population remain to be elucidated.
In this study, EGFR germline and somatic variants were retrospectively reviewed in 31,906 patients with lung cancer whose tumor tissues or peripheral blood samples were collected to perform a matched tumor-normal next-generation sequencing of 1,021 cancer-related genes. Clinical information was also collected for each patient identified with EGFR germline mutations for comparison.
Methods
Patients and Samples
This study recruited a total of 31,906 Chinese patients with lung cancer who underwent matched tumor-normal next-generation sequencing at Geneplus-Beijing (Beijing, China) from April 2015 to March 2021. Tumor tissues (including formalin-fix paraffin-embedded, frozen, and needle biopsy samples), peripheral blood samples, or effusion samples were obtained from each participant. This study was approved by the Ethics Committee of the First Affiliated Hospital, Guangzhou Medical University (Guangzhou, China) (Approval No. 2020-140). All procedures were conducted in accordance with the Declaration of Helsinki and written informed consent for mutational analysis of genomic DNA (gDNA) and circulating free DNA (cfDNA) was obtained from all participants.
Sample Processing and DNA Extraction
Peripheral blood samples were collected in Streck tubes (Streck, Omaha, NE, USA) and centrifuged within 72 h to separate the plasma from the peripheral blood cells. To detect germline and somatic mutations, gDNA was extracted from the peripheral blood cells and fresh tumor tissues using a QIAamp DNA Blood mini kit (Qiagen, Hilden, Germany). Formalin-fixed, paraffin-embedded (FFPE) DNA was isolated using Maxwell® 16 FFPE Plus LEV DNA purification kit (Qiagen, Hilden, Germany). QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) was used to extract cfDNA from liquid biopsies. DNA extractions were performed according to the manufacturer’s instructions. The DNA concentration was measured using a Qubit fluorometer and Qubit dsDNA HS (high sensitivity) assay kit (Invitrogen, Carlsbad, CA, USA).
Library Preparation, Target Capture, and Sequencing
Sequencing libraries were prepared from ctDNA using KAPA DNA Library preparation kits (Kapa Biosystems, Wilmington, MA, USA), and genomic DNA sequencing libraries were prepared using Illumina’s TruSeq DNA Library preparation kits (Illumina, San Diego, CA, USA). Libraries were hybridized to custom-designed biotinylated oligonucleotide probes (Roche NimbleGen, Madison, WI, USA) targeting cancer-related genes ranging from 16 to 1,021, including but not limited to all driver mutations in lung cancer (EGFR, ALK, ROS1, RET, KRAS, NRAS, TP53, BRAF, ERBB2, and MET).
Sequencing Data Analysis
Terminal adaptor sequences were removed from the raw sequencing data. Subsequently, reads with more than 50% low-quality bases, or more than 50% undefined bases, were discarded. The remaining reads were mapped to the reference human genome (hg19) using the Burrows-Wheel Aligner (BWA). Somatic variants, including single nucleotide variants (SNVs), small insertions and deletions (InDels), copy number alterations (CNAs), and structural variants were assessed. MuTect2 (version 1.1.4) and NChot2 were employed to identify somatic SNVs, while GATK was used to identify small insertions and deletions (indels). CNAs were identified using Contra (v.2.0.8). Structural variants (SVs) were identified using NCsv (an in-house tool). The candidate variants were all manually verified using the Integrative Genomics Viewer.
Clinical and Genetic Data Analysis
All nonsynonymous variants in the coding region of EGFR gene were screened, and variants with frequencies greater than 0.01 in general populations were excluded. A variant was included in the final analysis cohort only when: (i) it was reported to be associated with targeted therapy; (ii) previously documented as a germline variant; or (iii) reported as functional. Clinical characteristics such as age at diagnosis, family history, and treatment history were collected for each patient in the final analysis cohort.
Statistical Analysis
The difference in age at diagnosis between different groups was evaluated using a two-tailed unpaired Mann-Whitney U test. Fisher’s exact test was utilized to assess the differences in other demographic characteristics. Statistical significance was determined at p < 0.05.
Results
Patient Characteristics
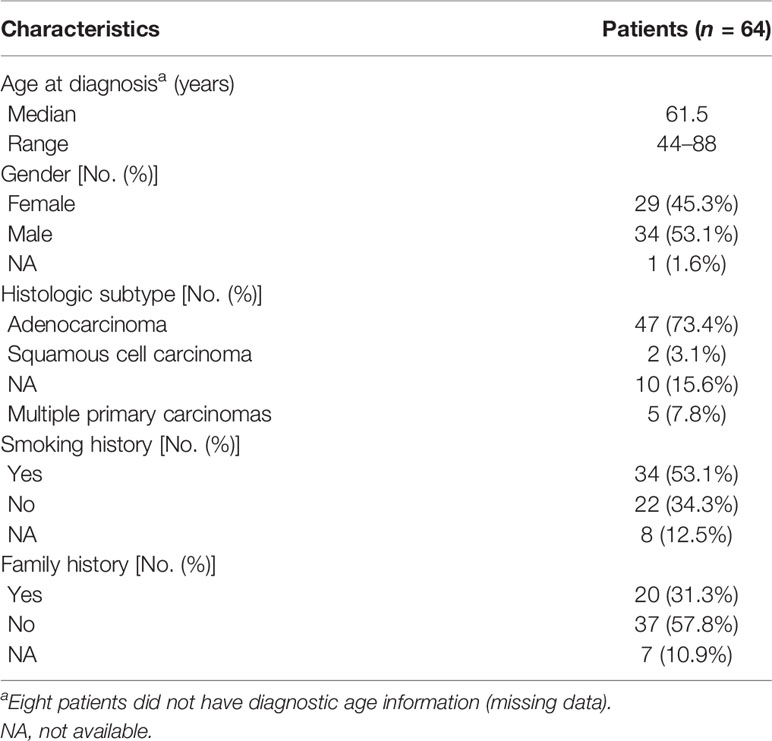
In the final analysis cohort, a total of 22 germline EGFR variants were identified in 64 patients with lung cancer. The prevalence of EGFR germline mutation in Chinese patients with lung cancer was 0.2% (64/31,906), which was higher than that found in a previous study (15). The baseline characteristics are summarized in Table 1. The median age at diagnosis was 61.5 years (range: 44–88 years). Regarding the diagnosis, most patients were classified as adenocarcinoma (73.4%, 47/64), five patients with multiple primary carcinomas, including three patients with double primary lung adenocarcinomas, one patient with mucoepidermoid carcinoma and adenocarcinoma of the lung, and one patient with lung adenocarcinoma and liver cancer. More than half the patients (53.1%, 34/64) were ever smokers, and family history was documented for approximately one-third of the patients (31.3%, 20/64). Among those whose cancer family history was obtained, lung cancer was the most recurrent type among the family members of 55% (11/20) patients.
EGFR Germline and Somatic Mutations
In our cohort, the mutation spectrum of EGFR germline mutations was considerably different from another study evaluating Chinese cancer patients (Figure 1A) (15). G863D, identified in nine of our patients (14.1%), was the most frequent EGFR germline mutation, followed by P848L (10.9%), D1014N (10.9%), K757R (9.4%), V897A (7.8%), and R831H (6.3%). EGFR-T790M, the dominant EGFR germline mutation in Western countries, was only present in two cases in our cohort. Most mutations (86.4%, 19/22) occurred within the tyrosine kinase domain, except for A647T (one case), V689M (one case), and D1014N (seven cases).
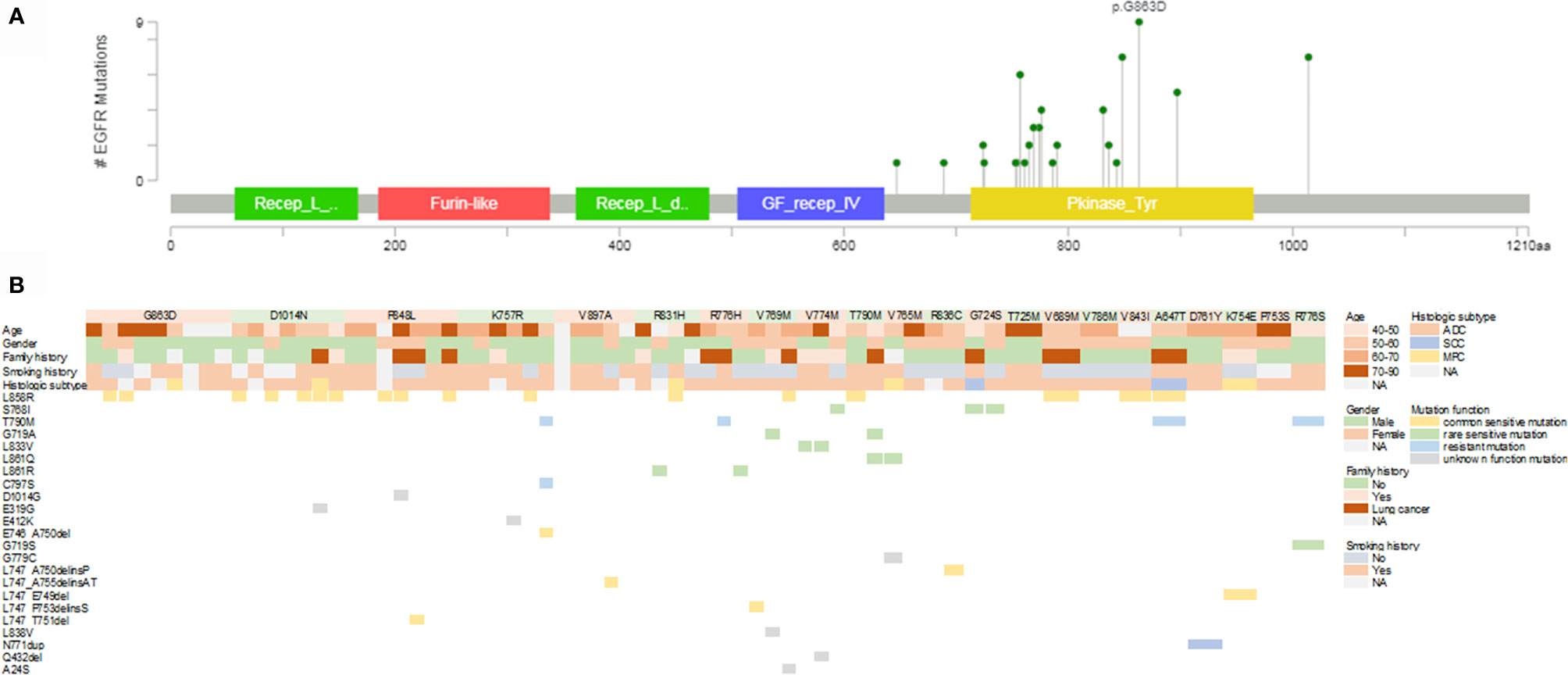
Figure 1 Mutational landscape of germline and somatic EGFR gene mutations. (A) Lollipop plot of the distribution of EGFR germline mutations. (B) Mutational profiles of patients with germline EGFR mutations. EGFR, epidermal growth factor receptor; ADC, adenocarcinoma; SCC, squamous cell carcinoma; MPC, multiple primary carcinoma; NA, not available.
A total of 46 EGFR somatic mutations were concurrently identified in 36 patients with EGFR germline mutations (Figure 1B). EGFR-sensitive mutations were identified in 51.6% (33/64) of patients with germline EGFR mutations. EGFR L858R was the most common mutation, with a detection rate of 26.6% (17/64). The distribution of deletion or deletion-insertion mutations in EGFR exon 19 was dispersive, accounting for 9.4% of the cases (6/64). Rare sensitive mutations, including S768I, G719A, L861Q/R, L833V, and G719S were found in 17.2% of patients (11/64). EGFR-resistant mutations were also identified in six cases, including three cases with T790M mutation, one with T790M and C797S mutations, and one with N771dup mutation.
Clinical and Genetic Feature Comparison Among Patients With Different Germline Mutations
We also investigated the possible differences in clinical and genetic characteristics among patients harboring different germline mutations. Compared with the age of patients with P848L or K757R germline mutation, that at diagnosis among patients with D1014N was significantly lower (median: 57 years for D1014N, 65.5 years for P848L, 66 years for K757R, p = 0.014 and 0.046, respectively). No significant differences were observed when comparing other groups (Figure 2A). Owing to the small number of patients in certain germline mutation groups, only those with ≥3 patients were included in the comparative analysis. More than three-quarters of patients with G863D, D1014N, and K757R were males. Whereas, all the patients with V769M (n = 3) and R836C (n = 2) were females. For the former, the differences were statistically significant (p = 0.045, 0.033, and 0.048, respectively) (Figure 2B). More than half of the patients with P848L, R776H, V769M, and V774M had cancer family history (including lung cancer). In addition, the percentage of lung cancer family history was higher than 50% in patients with P848L, R776H, and V769M (Figure 2C). All the patients with V769M mutation were never smokers, which was significantly different from patients with D1014N and K757R mutation (p = 0.048 and 0.029, respectively) (Figure 2D). Majority of patients were diagnosed as adenocarcinoma, with other subtypes dispersedly distributed across several groups. Multiple primary carcinomas were found in patients with G863D, D1014N, R831H, V765M, and K754E (Figure 2E). More than half of the patients with D1014N, P848L, and V769M harbored somatic exon 19 deletion or L858R mutations. Rare EGFR-sensitive mutations were frequently found in patients with V769M and V774M, but not in patients with G863D, D1014N, P848L, K757R, and V897A (Figure 2F).
Figure 2 Comparison of clinical and genetic features among patients with different EGFR germline mutations. (A) Age at diagnosis (years). (B) Gender. (C) Cancer family history. (D) Smoking history. (E) Histologic subtype. (F) Somatic EGFR-sensitive mutations. ADC, adenocarcinoma; SCC, squamous cell carcinoma; MPC, multiple primary carcinoma.
Patient Response to EGFR-TKIs
EGFR tyrosine kinase inhibitors (TKIs) were administered to patients with or without somatic EGFR-sensitive mutations. A total of 19 patients were confirmed to receive EGFR-TKIs; the survival information from their medical records is summarized in Table 2. Among patients with somatic L858R mutation (P1-P8), those with P848L (P4), V769M (P6), and K757R (P5) received the short duration of treatment (DOT) with EGFR-TKIs (2, 3, and 6 months, respectively). Only one patient with solely germline P848L mutation did not respond to EGFR-TKIs (P18). However, one patient with germline P848L mutation and somatic exon 19 deletion mutation had a DOT of 10 months (P9). Different from P5, one patient with germline K757R mutation and somatic exon 19 deletion mutation had a durable response to gefitinib and osimertinib (P10). A similar finding was observed in patients with V769M mutation, which showed that one patient with germline V769M mutation and somatic exon 19 deletion mutation had a DOT of 17 months for gefitinib (P11). Patients with germline D1014N (P1 and P2)/V843I (P8) mutation and somatic L858R mutation also responded well to EGFR-TKIs, with a DOT longer than 1 year. Patients with exon 19 deletion somatic mutation and R836C (P12)/K754E (P13) had a modest DOT with EGFR-TKIs. Germline T790M mutations were identified in two patients (P7 with L858R somatic mutation and P15 with somatic L861Q and G719A mutations). P7 showed a modest DOT for icotinib combined with two cycles of chemotherapy, but a durable DOT for the osimertinib group. The DOT for icotinib for P15 was 15 months. One patient with somatic primary T790M mutation and germline R776H had a DOT of 5 months for osimertinib (P17). In addition, patients with somatic S768I and germline V774M mutations (P14) and germline R831H were sensitive to EGFR-TKIs.
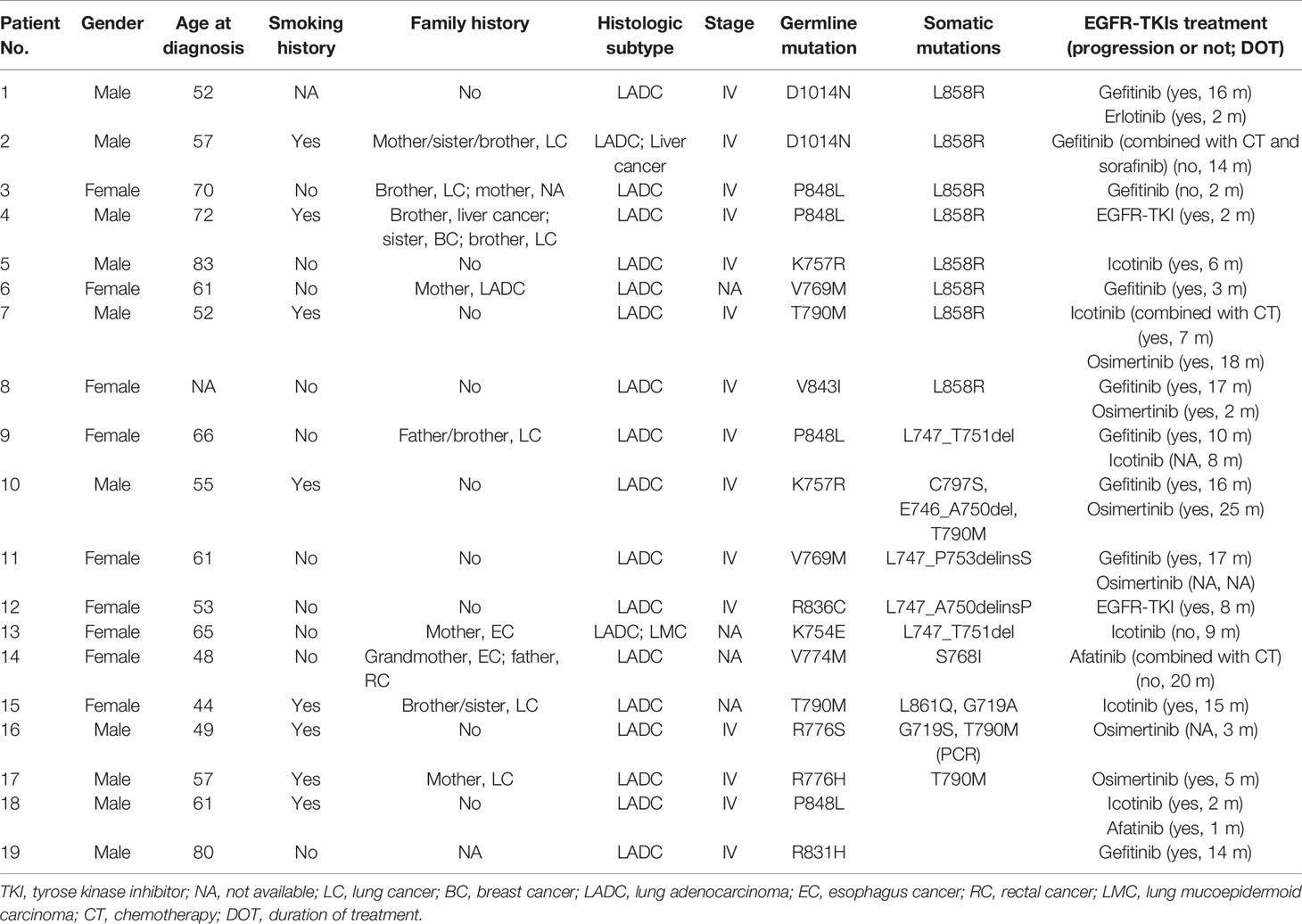
Table 2 The clinical and genetic features and patients’ response to EGFR-TKIs in patients with germline EGFR mutations.
Discussion
In this study, we identified 22 EGFR germline mutations in 64 out of 31,906 Chinese patients with lung cancer. The prevalence of EGFR germline mutations was 0.2%. The median age at diagnosis in our cohort was similar to that of the general Chinese population, which suggests that new lung cancer cases occur most frequently in individuals aged 60–74 years (1). The proportion of patients with multiple primary cancers in our study was higher than that reported (0.4%–2.4%) in the general Chinese population (16). In addition, the proportion of patients with cancer family history was remarkably high in our cohort. Unfortunately, none of the family members with cancer underwent genetic testing to confirm the presence of the corresponding germline mutations. These findings suggest that genetic susceptibility may play a role in the development of lung cancer. Germline EGFR mutations may not contribute to early onset of lung cancer. However, germline mutation analysis should be considered for patients with multiple primary carcinomas or cancer family history.
Our study revealed a unique EGFR germline mutation profile in Chinese patients with lung cancer. G863D, the most frequent EGFR germline mutation in our cohort, has not been previously reported as a germline mutation. Additionally, R836C, V897A, A647T, V689M, T725M, D761Y, R776S, V765M, V774M, P753S, and K754E were also reported for the first time as germline mutations in this large-scale, retrospective study. Somatic EGFR mutation rate in our study was 51.6%, similar to the 50.2% reported by the PIONEER study of Chinese patients with lung adenocarcinoma (17). However, the distribution of EGFR somatic mutations differed from that found by another study. In this study, the proportion of L858R, exon 19 deletion, and rare sensitive mutation in patients with somatic EGFR mutations was 50%, 17.6%, and 32.4%, respectively. In a previous study, L858R, exon 19 deletion, and other mutations accounted for 40%–45%, 45%, and 10% of EGFR mutations, respectively (18). Our study found unique clinical features for patients harboring different germline mutations. D1014N and T790M mutations were common in young patients. The family members of patients with P848L, R776H, V769M, and V774M more commonly suffered from various cancers. The distributions of EGFR somatic mutations among patients with different germline mutations were also different. Future studies should confirm whether the unique distribution of EGFR somatic mutations may influence the efficacy of EGFR-TKIs in patients with germline EGFR mutations.
The response to EGFR-TKIs differed among patients with different somatic and germline EGFR mutations. Multiple preclinical studies have suggested that P848L mutation is not a sensitive type (19, 20). The progression-free survival of patients with somatic and germline P848L mutation using erlotinib was 78 days and 4 months, respectively (21, 22). In our study, patients with P848L alone or combined with L858R somatic mutation did not respond to EGFR-TKIs. However, germline P848L combined with exon 19 deletion was sensitive to gefitinib and icotinib. V769M mutation has previously shown controversial and more insensitive efficacy to EGFR-TKIs (23–25), which influenced the effectiveness of EGFR-TKIs in patients with somatic L858R mutations but not in patients with exon 19 deletion mutations. A similar response pattern was observed for patients with K757R mutation, which previously showed more sensitive efficacy to EGFR-TKIs (15, 22). Similar to the previous favorable efficacy of gefitinib in one patient with germline D1014N and somatic L858R mutations (15), two patients in our study also showed good response to EGFR-TKIs. Previous studies suggested that R836C showed inconsistent responses to gefitinib in two cases (26, 27). In our study, modest survival was observed in one patient with germline R836C and somatic exon 19 deletion mutations. In our study, K754E, less sensitive to erlotinib than wild-type EGFR (28), showed modest sensitivity in a patient with concurrent somatic exon 19 deletion mutation. Multiple previous studies have reported the durable response to both first- and third-generation EGFR-TKIs in patients with germline T790M and somatic-sensitive mutations (29–31), which was also observed in our study. Patients with R776H and known sensitive mutations showed sensitivity to EGFR-TKIs such as gefitinib and erlotinib (32, 33). In our study, one patient with germline R776H and somatic T790M mutation showed modest sensitivity to osimertinib. Several studies demonstrated that V774M showed modest sensitivity to EGFR-TKIs (22, 34). In this study, afatinib combined with chemotherapy greatly prolonged survival time for the patient with germline V774M and somatic S768I mutation. R831H was reported to be a ligand-dependent activating mutation with sensitivity to erlotinib (35). One patient with germline R831H mutation responded well to gefitinib treatment.
We recognized several potential limitations in our study. Owing to the low prevalence of EGFR germline mutations in lung cancer patients, this study is a retrospective descriptive study. Only two patients (P18 and P19) with EGFR germline mutations received EGFR-TKI treatment; hence, we could not evaluate the efficacy difference among patients with (n = 17) or without (n = 2) EGFR somatic mutations. Therefore, we could not determine whether EGFR germline mutations should be regarded as driver mutations.
In conclusion, a small number of Chinese patients with lung cancer harbored unique and dispersive EGFR germline mutations, which may be related to their second primary carcinomas and cancer family history. Patients with different germline EGFR mutations showed unique clinical and genetic characteristics and variant response patterns to EGFR-TKIs treatment.
Data Availability Statement
The data analyzed in this study is subject to the following licenses/restrictions: This study recruited a total of 31,906 Chinese patients with lung cancer who underwent matched tumor-normal next-generation sequencing (NGS) at Geneplus-Beijing (Beijing, China) between April 2015 and March 2021. Requests to access these datasets should be directed to “Xin Yi, https://www.geneplus.org.cn”.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the First Affiliated Hospital, Guangzhou Medical University (Guangzhou, China) (Approval No. 2020-140). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
XL, MC, and CZ designed the study. MP wrote and edited the manuscript, which was approved by all the authors. QC, HD, and JD evaluated and examined the data. OL and YW performed the data collection. MY and RC analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Natural Science Foundation of Changsha (No. kq2014001); State Key Laboratory of Respiratory Disease-The open project (SKLRD-OP-202111); Zhongnanshan Medical Foundation of Guangdong Province (ZNSA-2020003); Fundamental and Applied Fundamental Research Project of City-School (Institute) Joint Funding Project, Guangzhou Science and Technology Bureau (202102010357); and Wu Jieping Medical Foundation (320.6750.2020-19-8).
Conflict of Interest
Authors MY and RC were employed by Geneplus-Beijing.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer Statistics in China, 2015. CA Cancer J Clin (2016) 66(2):115–32. doi: 10.3322/caac.21338
2. Corrales L, Rosell R, Cardona AF, Martín C, Zatarain-Barrón ZL, Arrieta O. Lung Cancer in Never Smokers: The Role of Different Risk Factors Other Than Tobacco Smoking. Crit Rev Oncol Hematol (2020) 148:102895. doi: 10.1016/j.critrevonc.2020.102895
3. Lin H, Huang YS, Yan HH, Yang XN, Zhong WZ, Ye HW, et al. A Family History of Cancer and Lung Cancer Risk in Never-Smokers: A Clinic-Based Case-Control Study. Lung Cancer (2015) 89(2):94–8. doi: 10.1016/j.lungcan.2015.05.017
4. Gaughan EM, Cryer SK, Yeap BY, Jackman DM, Costa DB. Family History of Lung Cancer in Never Smokers With Non-Small-Cell Lung Cancer and its Association With Tumors Harboring EGFR Mutations. Lung Cancer (2013) 79(3):193–7. doi: 10.1016/j.lungcan.2012.12.002
5. Cannon-Albright LA, Carr SR, Akerley W. Population-Based Relative Risks for Lung Cancer Based on Complete Family History of Lung Cancer. J Thorac Oncol (2019) 14(7):1184–91. doi: 10.1016/j.jtho.2019.04.019
6. Ding X, Chen Y, Yang J, Li G, Niu H, He R, et al. Characteristics of Familial Lung Cancer in Yunnan-Guizhou Plateau of China. Front Oncol (2018) 8:637. doi: 10.3389/fonc.2018.00637
7. Jin G, Zhu M, Yin R, Shen W, Liu J, Sun J, et al. Low-Frequency Coding Variants at 6p21.33 and 20q11.21 Are Associated With Lung Cancer Risk in Chinese Populations. Am J Hum Genet (2015) 96(5):832–40. doi: 10.1016/j.ajhg.2015.03.009
8. Liu M, Liu X, Suo P, Gong Y, Qu B, Peng X, et al. The Contribution of Hereditary Cancer-Related Germline Mutations to Lung Cancer Susceptibility. Transl Lung Cancer Res (2020) 9(3):646–58. doi: 10.21037/tlcr-19-403
9. McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, et al. Large-Scale Association Analysis Identifies New Lung Cancer Susceptibility Loci and Heterogeneity in Genetic Susceptibility Across Histological Subtypes. Nat Genet (2017) 49(7):1126–32. doi: 10.1038/ng.3892
10. Zanetti KA, Wang Z, Aldrich M, Amos CI, Blot WJ, Bowman ED, et al. Genome-Wide Association Study Confirms Lung Cancer Susceptibility Loci on Chromosomes 5p15 and 15q25 in an African-American Population. Lung Cancer (2016) 98:33–42. doi: 10.1016/j.lungcan.2016.05.008
11. Parry EM, Gable DL, Stanley SE, Khalil SE, Antonescu V, Florea L, et al. Germline Mutations in DNA Repair Genes in Lung Adenocarcinoma. J Thorac Oncol (2017) 12(11):1673–8. doi: 10.1016/j.jtho.2017.08.011
12. Yamamoto H, Yatabe Y, Toyooka S. Inherited Lung Cancer Syndromes Targeting Never Smokers. Transl Lung Cancer Res (2018) 7(4):498–504. doi: 10.21037/tlcr.2018.06.01
13. Girard N, Lou E, Azzoli CG, Reddy R, Robson M, Harlan M, et al. Analysis of Genetic Variants in Never-Smokers With Lung Cancer Facilitated by an Internet-Based Blood Collection Protocol: A Preliminary Report. Clin Cancer Res (2010) 16(2):755–63. doi: 10.1158/1078-0432.CCR-09-2437
14. Hu Y, Alden RS, Odegaard JI, Fairclough SR, Chen R, Heng J, et al. Discrimination of Germline EGFR T790M Mutations in Plasma Cell-Free DNA Allows Study of Prevalence Across 31,414 Cancer Patients. Clin Cancer Res (2017) 23(23):7351–9. doi: 10.1158/1078-0432.CCR-17-1745
15. Lu S, Yu Y, Li Z, Yu R, Wu X, Bao H, et al. EGFR and ERBB2 Germline Mutations in Chinese Lung Cancer Patients and Their Roles in Genetic Susceptibility to Cancer. J Thorac Oncol (2019) 14(4):732–6. doi: 10.1016/j.jtho.2018.12.006
16. Li M, Xie M. Advances In Research On Multiple Primary Malignant Tumors. China Oncol (2017) 27(2):156–60. doi: 10.19401/j.cnki.1007-3639.2017.02.013
17. Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, et al. A Prospective, Molecular Epidemiology Study of EGFR Mutations in Asian Patients With Advanced Non-Small-Cell Lung Cancer of Adenocarcinoma Histology (PIONEER). J Thorac Oncol (2014) 9(2):154–62. doi: 10.1097/JTO.0000000000000033
18. Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal Growth Factor Receptor Mutations in Lung Cancer. Nat Rev Cancer (2007) 7(3):169–81. doi: 10.1038/nrc2088
19. Han B, Zhou X, Zhang RX, Zang WF, Chen ZY, Song HD, et al. Mutations of the Epidermal Growth Factor Receptor Gene in NSCLC Patients. Oncol Lett (2011) 2(6):1233–7. doi: 10.3892/ol.2011.366
20. de Gunst MM, Gallegos-Ruiz MI, Giaccone G, Rodriguez JA. Functional Analysis of Cancer-Associated EGFR Mutants Using a Cellular Assay With YFP-Tagged EGFR Intracellular Domain. Mol Cancer (2007) 6:56. doi: 10.1186/1476-4598-6-56
21. Prim N, Legrain M, Guerin E, Mennecier B, Weingertner N, Voegeli AC, et al. Germ-Line Exon 21 EGFR Mutations, V843I and P848L, in Nonsmall Cell Lung Cancer Patients. Eur Respir Rev (2014) 23(133):390–2. doi: 10.1183/09059180.00009313
22. Klughammer B, Brugger W, Cappuzzo F, Ciuleanu T, Mok T, Reck M, et al. Examining Treatment Outcomes With Erlotinib in Patients With Advanced Non-Small Cell Lung Cancer Whose Tumors Harbor Uncommon EGFR Mutations. J Thorac Oncol (2016) 11(4):545–55. doi: 10.1016/j.jtho.2015.12.107
23. Hellmann MD, Hayashi T, Reva B, Yu HA, Riely GJ, Adusumilli PS, et al. Identification and Functional Characterization of EGFR V769M, a Novel Germline Variant Associated With Multiple Lung Adenocarcinomas. JCO Precis Oncol (2017) 1:PO.16.00019. doi: 10.1200/PO.16.00019
24. Landi L, Cappuzzo F. Irreversible EGFR-TKIs: Dreaming Perfection. Transl Lung Cancer Res (2013) 2(1):40–9. doi: 10.3978/j.issn.2218-6751.2012.12.05
25. Xu C, Wang W-X, Fang M, Zhuang W, Lin G, Chen X-H, et al. Clinical Efficacy of Icotinib in Patients With Advanced Non-Small Cell Lung Cancer Harboring EGFR Exon 18, 20 and 21 Uncommon Mutations. J Clin Oncol (2017) 35(15_suppl):e14050–0. doi: 10.1200/JCO.2017.35.15_suppl.e14050
26. Hsieh MH, Fang YF, Chang WC, Kuo HP, Lin SY, Liu HP, et al. Complex Mutation Patterns of Epidermal Growth Factor Receptor Gene Associated With Variable Responses to Gefitinib Treatment in Patients With Non-Small Cell Lung Cancer. Lung Cancer (2006) 53(3):311–22. doi: 10.1016/j.lungcan.2006.06.005
27. Mompradé E, Arriola E, Martínez-Avilés L. Two Rare Exon 21 EGFR Mutations in Patients Treated With Gefitinib. J Thorac Oncol (2013) 8(4):e36–7. doi: 10.1097/JTO.0b013e318286cf9e
28. Berger AH, Brooks AN, Wu X, Shrestha Y, Chouinard C, Piccioni F, et al. High-Throughput Phenotyping of Lung Cancer Somatic Mutations. Cancer Cell (2016) 30(2):214–28. doi: 10.1016/j.ccell.2016.06.022
29. Tibaldi C, Giovannetti E, Vasile E, Boldrini L, Gallegos-Ruiz MI, Bernardini I, et al. Inherited Germline T790M Mutation and Somatic Epidermal Growth Factor Receptor Mutations in Non-Small Cell Lung Cancer Patients. J Thorac Oncol (2011) 6(2):395–6. doi: 10.1097/JTO.0b013e3182059a6f
30. Bell DW, Gore I, Okimoto RA, Godin-Heymann N, Sordella R, Mulloy R, et al. Inherited Susceptibility to Lung Cancer may be Associated With the T790M Drug Resistance Mutation in EGFR. Nat Genet (2005) 37(12):1315–6. doi: 10.1038/ng1671
31. Ma W, Gong J, Shan J, Lewis D, Xiao W, Moore EH, et al. Safety and Efficacy of Osimertinib in the Treatment of a Patient With Metastatic Lung Cancer and Concurrent Somatic EGFR L858R and Germline EGFR T790M Mutations. JCO Precis Oncol (2018) 2):1–7. doi: 10.1200/PO.18.00076
32. Kobayashi S, Canepa HM, Bailey AS, Nakayama S, Yamaguchi N, Goldstein MA, et al. Compound EGFR Mutations and Response to EGFR Tyrosine Kinase Inhibitors. J Thorac Oncol (2013) 8(1):45–51. doi: 10.1097/JTO.0b013e3182781e35
33. Wu SG, Chang YL, Hsu YC, Wu JY, Yang CH, Yu CJ, et al. Good Response to Gefitinib in Lung Adenocarcinoma of Complex Epidermal Growth Factor Receptor (EGFR) Mutations With the Classical Mutation Pattern. Oncologist (2008) 13(12):1276–84. doi: 10.1634/theoncologist.2008-0093
34. Woo HS, Ahn HK, Lee HY, Park I, Kim YS, Hong J, et al. Epidermal Growth Factor Receptor (EGFR) Exon 20 Mutations in Non-Small-Cell Lung Cancer and Resistance to EGFR-Tyrosine Kinase Inhibitors. Invest New Drugs (2014) 32(6):1311–5. doi: 10.1007/s10637-014-0146-x
Keywords: genetic features, EGFR, treatment, Chinese lung cancer patient, germline mutations
Citation: Lin X, Peng M, Chen Q, Yuan M, Chen R, Deng H, Deng J, Liu O, Weng Y, Chen M and Zhou C (2021) Identification of the Unique Clinical and Genetic Features of Chinese Lung Cancer Patients With EGFR Germline Mutations in a Large-Scale Retrospective Study. Front. Oncol. 11:774156. doi: 10.3389/fonc.2021.774156
Received: 11 September 2021; Accepted: 23 October 2021;
Published: 16 November 2021.
Edited by:
Tao Jiang, Shanghai Pulmonary Hospital, ChinaReviewed by:
Shaodong Hong, Sun Yat-Sen University Cancer Center (SYSUCC), ChinaJian Zhang, Southern Medical University, China
Copyright © 2021 Lin, Peng, Chen, Yuan, Chen, Deng, Deng, Liu, Weng, Chen and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengzhi Zhou, ZG9jdG9yemN6QDE2My5jb20=; Mingjiu Chen, Y2hlbm1pbmdqaXVAY3N1LmVkdS5jbg==; Yuqing Weng, MTQ1NTI1OTgxNEBxcS5jb20=
†These authors have contributed equally to this work and share first authorship
 Xinqing Lin
Xinqing Lin Muyun Peng
Muyun Peng Quanfang Chen3†
Quanfang Chen3† Mingming Yuan
Mingming Yuan Rongrong Chen
Rongrong Chen Chengzhi Zhou
Chengzhi Zhou