- 1Department of Integrated Traditional Chinese and Western Medicine, Affiliated Cancer Hospital of Zhengzhou University, Zhengzhou, China
- 2Department of Oncology, Affiliated Cancer Hospital of Zhengzhou University, Zhengzhou, China
- 3Department of Hepatopancreatobiliary Surgery, Affiliated Cancer Hospital of Zhengzhou University, Zhengzhou, China
- 4Department of Pathology, Affiliated Cancer Hospital of Zhengzhou University, Zhengzhou, China
Macrophage-targeting therapies have become attractive strategies for immunotherapy. Deficiency of MARCO significantly inhibits tumor progression and metastasis in murine models of pancreatic cancer. However, the role of MARCO in patients with pancreatic cancer remains unclear. In the present study, we analyzed tumor-associated macrophage (TAM)-related changes using the Cancer Genome Atlas database. We observed a significant enrichment of M2 macrophages in pancreatic cancer tissues. We found that several pro-tumor markers are increased in cancer tissues, including CD163, CD206, SIRPα, LILRB1, SIGLEC10, AXL, MERTK, and MARCO. Crucially, MARCO is highly or exclusively expressed in pancreatic cancer across many types of solid tumors, suggesting its significant role in pancreatic cancer. Next, we investigated the expression of MARCO in relation to the macrophage marker CD163 in a treatment-naïve pancreatic cancer cohort after surgery (n = 65). MARCO and CD163 were analyzed using immunohistochemistry. We observed increased expression of CD163 and MARCO in pancreatic cancer tissues compared with paracancerous tissues. Furthermore, we observed a large variation in CD163 and MARCO expression in pancreatic cancer tissues among cases, suggesting the heterogeneous expression of these two markers among patients. Correlation to clinical data indicated a strong trend toward worse survival for patients with high CD163 and MARCO macrophage infiltration. Moreover, high CD163 and MARCO expression negatively affected the disease-free survival and overall survival rates of patients with pancreatic cancer. Univariate and multivariate analysis revealed that CD163 and MARCO expression was an independent indicator of pancreatic cancer prognosis. In conclusion, high CD163 and MARCO expression in cancer tissues is a negative prognostic marker for pancreatic cancer after surgery. Furthermore, anti-MARCO may be a novel therapy that is worth studying in depth.
Introduction
Pancreatic cancer remains a highly lethal malignancy and is expected to be the second leading cause of cancer death in the United States within the next 20 to 30 years (1). In the United States, the 5-year survival rate at diagnosis of pancreatic cancer is approximately 10%, and even after surgery it is only approximately 20% (1, 2). Despite improvements in diagnosis and treatment options as well as rapid advances in targeted therapies and immunotherapy, the prognosis of patients with pancreatic cancer remains poor (2). Therefore, an urgent need exists to explore new biomarkers to have a clinically meaningful impact in the screening of patients with high-risk pancreatic cancer.
The tumor microenvironment (TME) has long been of great interest in a wide range of research studies in the field (3–5). Immune cells are a major component of the tumor microenvironment (6). In recent years, as research has progressed, many immune escape mechanisms have been reported, including T cells, tumor-associated macrophages (TAMs), myeloid-derived suppression cells (MDSCs), and natural killer (NK) cells (7–11). Among these immune cells, TAMs constitute the vast majority in the tumor microenvironment, which indicates the possibility that macrophage-targeting therapies are novel and attractive strategies for cancer treatment (12). Crucially, TAMs drive tumor progression through multiple mechanisms, which include increased angiogenesis, immunosuppression, and resistance to therapy (13). However, macrophages can also exhibit a proinflammatory phenotype that kills tumor cells effectively in vitro and in vivo. Antibodies to CD47 or SIRPα, developed against the TAM immunosuppression-related signaling pathway CD47-SIRPα (also known as the “don’t eat me” signal), allow macrophages to regain the ability to phagocytose tumor cells and restore the activity of CD8+ cytotoxic T cells, thereby significantly reducing tumor size and inhibiting tumor metastasis (14). Other preclinical trials targeting the macrophage “don’t eat me” signal have also yielded impressive results through the promotion of tumor cell phagocytosis by macrophages, such as those involving the inhibition of the MHC-I-LILRB1 axis, inhibition of the CD24-SIGLEC-10 axis, and CAR-M cellular immunotherapy (15–17). Despite these exciting findings, the role of macrophages in pancreatic cancer still needs to be fully uncovered.
Recently, studies have demonstrated that the scavenger receptor MARCO is mainly expressed by macrophages, and that higher MARCO expression is associated with the poor prognosis of many types of cancers (18–20). Importantly, preclinical studies have demonstrated that an anti-MARCO antibody inhibits tumor growth and metastasis in 4T1 mammary carcinoma and B16 melanoma mouse models (21). However, the association of decreased MARCO expression by macrophages with tumor progression and poor prognosis in human hepatocellular carcinoma (HCC) was also observed (22). The aforementioned studies suggest that MARCO expression may play distinct roles in different tumor types. Furthermore, in a murine model of pancreatic cancer, Neyen et al. found that MARCO deficiency significantly inhibits tumor progression and metastasis (23). MARCO antibody alters macrophage polarization, enhancing NK cell activation and tumor killing, and the anti-tumor effect of MARCO antibody in combination with PD-1/L1 antibody is more potent in a mouse model of melanoma (24). However, the roles of MARCO-expressing macrophages in human pancreatic cancer remain unclear.
In the present study, we characterized the gene expression profiles from the Cancer Genome Atlas (TCGA) data set (https://portal.gdc.com). Immune cell type analysis indicated that M2 macrophages, monocytes, and uncharacterized cells are significantly different between normal and pancreatic cancer tissues, which suggested that M2 macrophages may contribute more to the progression of pancreatic cancer. We then analyzed many correlated macrophage biomarkers and the results suggested that MARCO-expressing macrophages are highly associated with poor pancreatic cancer prognosis. Furthermore, MARCO expression was analyzed in many solid tumor types. Finally, we analyzed the expression profiles of CD163 and MARCO in both cancer and paracancerous tissues from patients with pancreatic cancer.
Materials and Methods
TCGA Data Analysis
(1) Gene Expression Data Sets
A gene expression data set of pancreatic cancer and normal tissues was acquired from the TCGA database (http://tcga-data.nci.nih.gov/tcga/tcgaAbout.jsp). The data were from 178 pancreatic cancer tissues, 4 paracancerous tissues, and 328 healthy tissues.
(2) Immune Infiltration Estimations
To produce reliable immune infiltration estimations, we used immunedeconv, an R package that integrates six state-of-the-art algorithms (quanTIseq). This R package and all of the aforementioned analysis methods were implemented in R Foundation for Statistical Computing (2020) version 4.0.3 and the software packages ggplot2 and pheatmap as previous described (25).
(3) Analysis of Differential Expression
The limma package (version: 3.40.2) for R was used to study the differential expression of mRNAs. The adjusted P value was analyzed to correct for false positive results in the TCGA or GTEx data set. The defined thresholds for screening for the differential expression of mRNAs were as follows: adjusted P < 0.05 and log (fold change) >1 or log (fold change) < −1. To further confirm the underlying function of potential targets, the data were analyzed using functional enrichment. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis is a practical resource for analytically studying gene functions and associated high-level genome functional information. To better understand the carcinogenesis of mRNAs, the ClusterProfiler package (version: 3.18.0) in R was employed to analyze and enrich the KEGG pathway as previous described (26).
(4) Kaplan–Meier Survival Analysis
For Kaplan–Meier curves, P values and hazard ratios with 95% confidence intervals were generated using log-rank tests and univariate Cox proportional hazards regression. All of the abovementioned analytical methods and R packages were performed using R software version v4.0.3 (R Foundation for Statistical Computing, 2020) as previous described (27); P < 0.05 was considered statistically significant.
Cancer and Paracancerous Tissues From Pancreatic Cancers
Sixty-five patients with pancreatic cancer surgically treated at the Affiliated Cancer Hospital of Zhengzhou University between July 1, 2016 and June 1, 2018 were collected for this study. This study was approved by the Ethics Committee at the Affiliated Cancer Hospital of Zhengzhou University. Clinical parameters were obtained from the records of patients in the same hospital. All methods and procedures associated with this study were conducted in accordance with the Good Clinical Practice guidelines and accorded ethically with the principles of the Declaration of Helsinki and local laws.
Immunohistochemistry and Image Analysis
Formalin-fixed and paraffin-embedded sections of pancreatic cancer tissue and paracancerous tissue (3–5 μm thick) were dewaxed and rehydrated. Antigen retrieval was performed by heating the slides in 10 mM Tris buffer with 1 mM EDTA (pH 9) in a streamer for 20 min. Then, endogenous peroxidase activity was inhibited through immersion in 3% H2O2 for 5 min. After washing with Tris-buffered saline containing Tween, endogenous biotin was inhibited through sequential incubation with 0.1% antibiotin protein and 0.01% biotin (Dako, Glostrup, Denmark), respectively, for 10 min at room temperature. Other nonspecific binding sites were blocked with 3% skimmed milk powder for 30 min at room temperature. Sections of pancreatic cancer tissue and paracancerous tissue were incubated with the monoclonal mouse antibody anti-human CD163(abcam, Cat#: ab182422) and MARCO (Bioss, Cat#: bs-2659R) for one night at 4°C. Subsequently, the sections were serially rinsed and incubated with secondary antibodies. Immunohistochemical staining was evaluated independently by two experienced pathologists blinded to the patients’ clinical characteristics and outcomes. The median was selected as the cutoff value for high or low CD163 and MARCO expression.
Follow-Up and Survival Analysis
Disease-free survival (DFS) was calculated from the date of surgery to the time of recurrence or metastasis, and patients alive in a stable state were censored at the time of last contact (28). The overall survival (OS) was calculated from the date of surgery to the time of death, and patients who were alive at the time of last contact were censored (28). The DFS and OS were calculated using the Kaplan–Meier method. After surgery, all of the patients were scheduled for follow-up evaluations at our hospital from the date of initial treatment to the follow-up deadline of September 1, 2021 or to the time of death. Clinical examinations were performed by our oncology specialists every 3 months, including complete blood examinations and chest and abdominal computed tomography scans during the first 2-year period. From years 2 to 5, patients were examined every 6 months. Beyond 5 years, patients were examined every year. If follow-up evaluations revealed metastatic disease and/or local recurrence, other therapies were applied, including conventional therapies (surgery, chemotherapy, and radiotherapy) and immunotherapy.
Statistical Analysis
The GraphPad Prism 9.0 and SPSS 24.0 software packages were used to perform the statistical analyses. The DFS and OS were calculated using the Kaplan–Meier method from the time of surgery. The prognostic factors were analyzed using univariate and multivariable Cox proportional hazards regression models. The χ2 test was used in a prespecified analysis to compare the characteristics of groups. Other data were analyzed using a t test. For all statistical analyses, significance was indicated at a level of P < 0.05.
Results
M2 Macrophages Are Significantly Heterogeneous Among Pancreatic Cancer Patients
To evaluate the significant changes of immune cells in patients with pancreatic cancer, RNA-seq data from the TCGA database were analyzed. Immune infiltration estimations were analyzed using quanTIseq. Figures 1A, B indicate that among the immune cells, M2 macrophages, monocytes, and uncharacterized cells in cancer tissues were significantly different compared with normal tissues. Furthermore, among tumors, the M2 macrophage content in each sample was also distinct, highlighting the heterogeneity in patients with pancreatic cancer patients (Figures 1A, B).
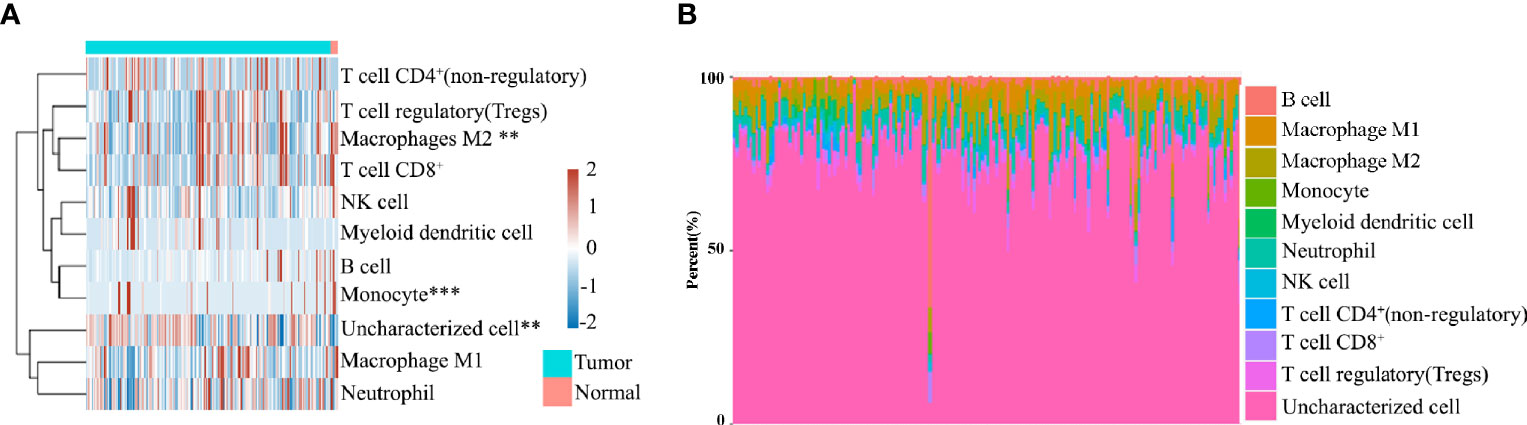
Figure 1 (A) Immune cell score heat map, where different colors represent the expression trend in different samples. **P < 0.01, ***P < 0.001. (B) The percentage abundance of tumor-infiltrating immune cells in each sample, with different colors and different types of immune cells.
Identification of Differentially Expressed Genes and Signal Pathways
Next, we retrieved the transcriptome profiling data of pancreatic cancers from the TCGA database, which comprised 178 pancreatic cancer tissues, and 4 paracancer tissues. A total of 12 039 genes were distinguished as differentially expressed mRNAs, with 11 392 genes being upregulated and 647 genes being downregulated (Figures 2A, B). The enriched KEGG signaling pathways were selected to demonstrate the primary biological actions of major potential mRNAs. The upregulated pathways related to TAMs included the osteoclast differentiation, endocytosis, the chemokine signaling pathway, and cell adhesion molecules (CAMs; Figure 2C). By contrast, no highly relevant downregulated pathways were associated with TAMs (Figure 2D). Taken together, the function of macrophages may play significant roles in pancreatic cancer. Thus, further studies on the roles of TAMs in pancreatic cancer are required.
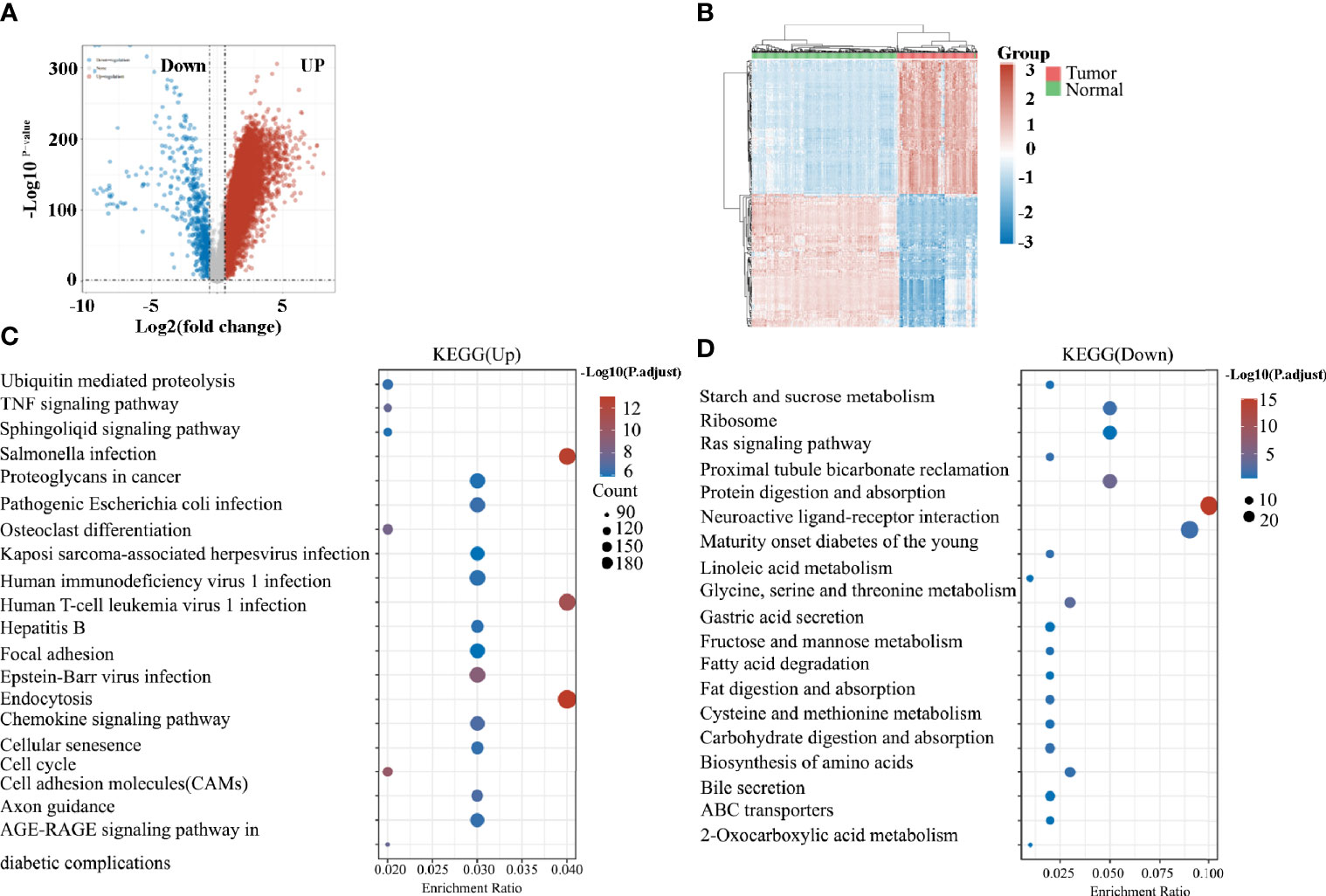
Figure 2 (A) Volcano plots constructed using fold-change values and adjusted P values. The red point in the plot represents the over-expressed mRNAs and the blue point indicates the down-expressed mRNAs with statistical significance. (B) Hierarchical clustering analysis of mRNAs, which were differentially expressed between tumor and normal tissues. (C) The top 20 upregulated KEGG signaling pathways in tumors. (D) The top 20 downregulated KEGG signaling pathways in tumors.
Several TAM-Related Molecules Are Significantly Increased in Patients With Pancreatic Cancer
We then analyzed the expression of molecules that play a crucial role in the function of TAMs. Consistent with the increased enrichment of M2 macrophages, the M2 markers of CD163 and CD206 were significantly increased in pancreatic cancer compared with controls (Figure 3A). Macrophage-mediated phagocytosis was inhibited by numerous “don’t eat me” signaling pathways, which mainly included CD47/SIRPα, CD24/SIGLEC10, and MHC-I/LILRB1 (15, 17, 29). Noteworthily, the expression of SIRPα, SIGLEC10, and LILRB1 also increased in cancer tissues (Figure 3B). The AXL receptor tyrosine kinase (RTK) was implicated in the proliferation and invasion of many cancers, particularly in pancreatic ductal adenocarcinoma (PDAC) (30). Consistent with previous studies (30, 31), AXL and TIMD4 increased in patients with pancreatic cancer (Figure 3C). MARCO is a pattern recognition receptor that belongs to the class A scavenger receptor family (32). Figure 3D reveals that MARCO expression in pancreatic cancer is significantly increased.
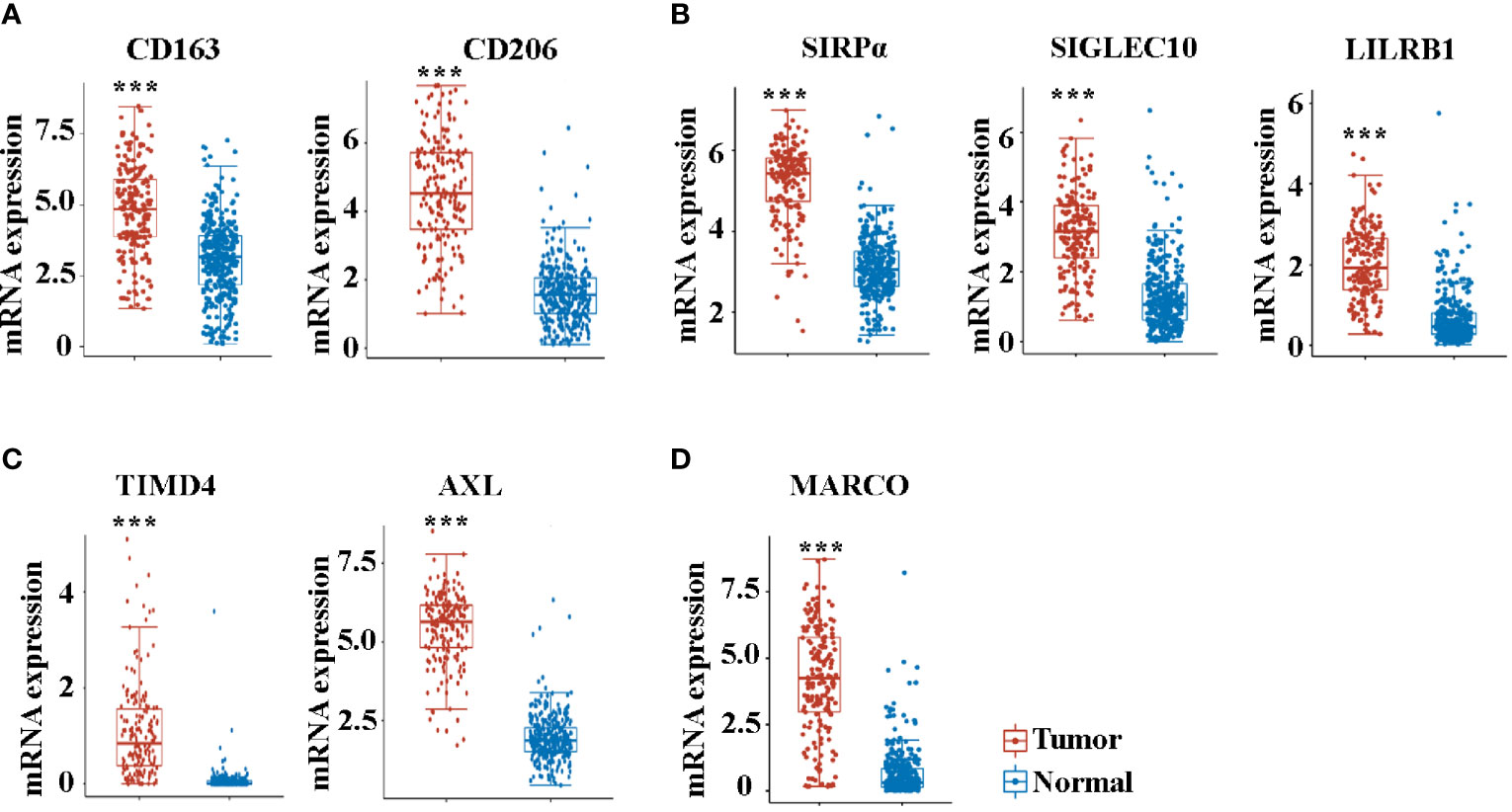
Figure 3 The mRNA expression of CD163 (A), CD206 (A), SIRPα (B), SIGLEC10 (B), LILRB1 (B), TIMD4 (C), AXL (C), and MARCO (D) between cancer tissues and paracancer tissues in pancreatic cancer patients from the TCGA database. ***P < 0.001.
Prognostic Roles of TAM-Related Molecules
Kaplan–Maier survival analysis with log-rank tests was also used to compare the difference in survival between high and low expression of TAM-related molecules. Noteworthily, the analysis revealed that patients exhibited a similar survival time based on the expression levels of CD163, CD206, SIRPα, SIGLEC10, LILRB1, and TIMD4 (Figures 4A–C). Previous studies have firmly demonstrated that the activation of the AXL RTK is associated with poor outcomes in PDAC (30, 33). Consistent with this, AXL hi (Figure 4C) patients exhibited a trend in shorter OS (P = 0.118). Although MARCO hi (Figure 4D) patients revealed a trend in shorter OS, no significant difference existed (P = 0.0518).
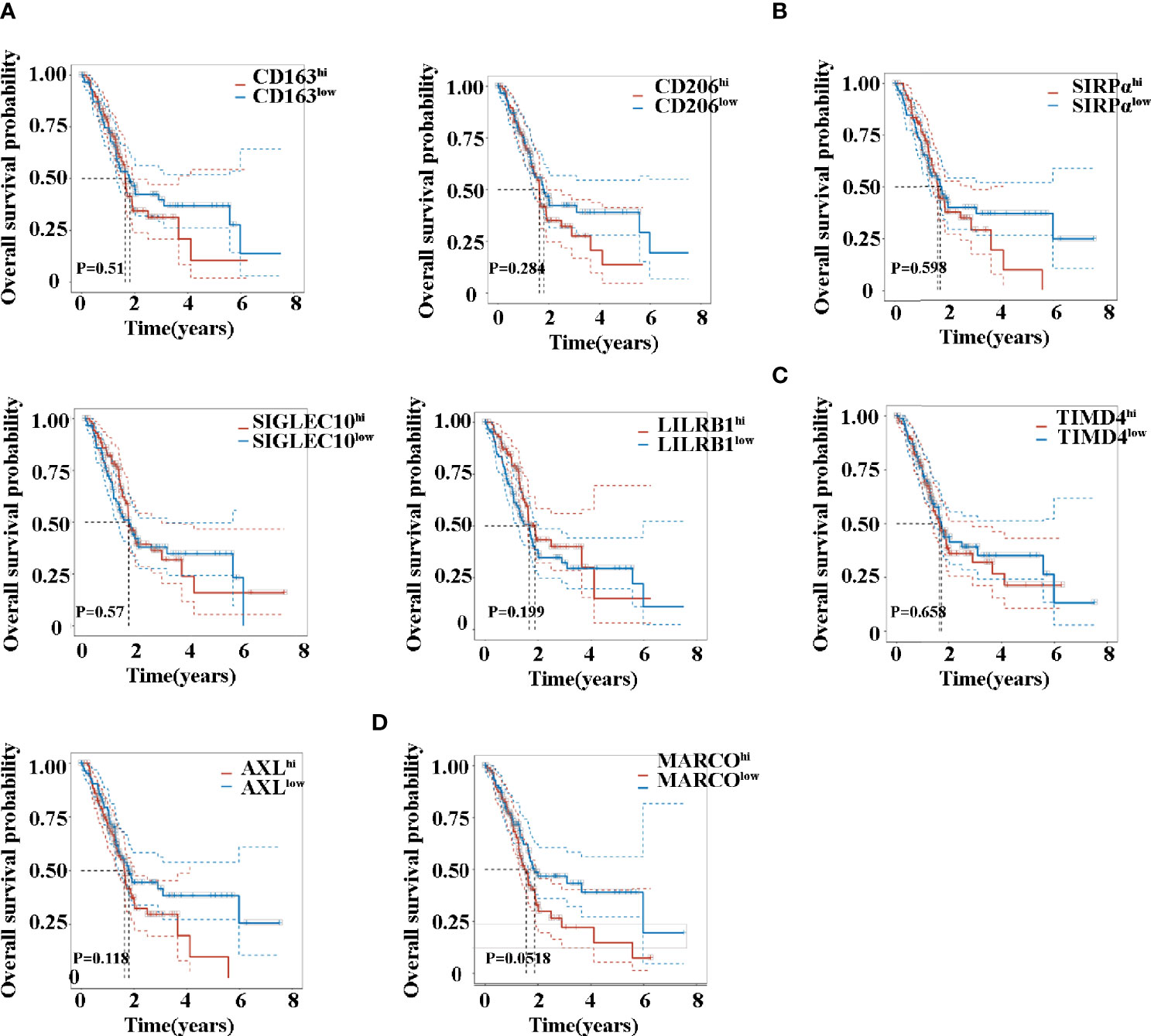
Figure 4 Overall survival time of patients with pancreatic cancer between high and low expression of CD163 (A), CD206 (A), SIRPα (B), SIGLEC10 (B), LILRB1 (B), TIMD4 (C), AXL (C), and MARCO (D).
MARCO Is Highly or Exclusively Expressed in Pancreatic Cancer Across Many Types of Solid Tumors
Next, we compared the expression of MARCO between tumor tissues and control tissues across solid tumors. Noteworthily, a majority of solid tumors exhibited decreased MARCO expression compared with the controls, including bladder cancer, colon cancer, cholangiocarcinoma, lung cancer, HCC, ovarian cancer, prostate cancer, breast cancer, adrenocortical carcinoma, esophageal cancer, gastric cancer, endometrial cancer, and uterine sarcoma (Figure 5B). Although the expression of MARCO in testicular cancer, cervical cancer, melanoma, thyroid cancer, brain cancer, and renal cancer increased, there were fewer fold changes than in pancreatic cancer (Figure 5A). Additionally, the expression of MARCO in sarcoma, squamous carcinoma of the head and neck, and thoracic cancer did not change significantly (Figures 5C). Taken together, these data indicated that MARCO expression may play distinct roles in different cancer types. Furthermore, the highest expression of MARCO in pancreatic cancer suggested that MARCO may play a significant role in this type of cancer.
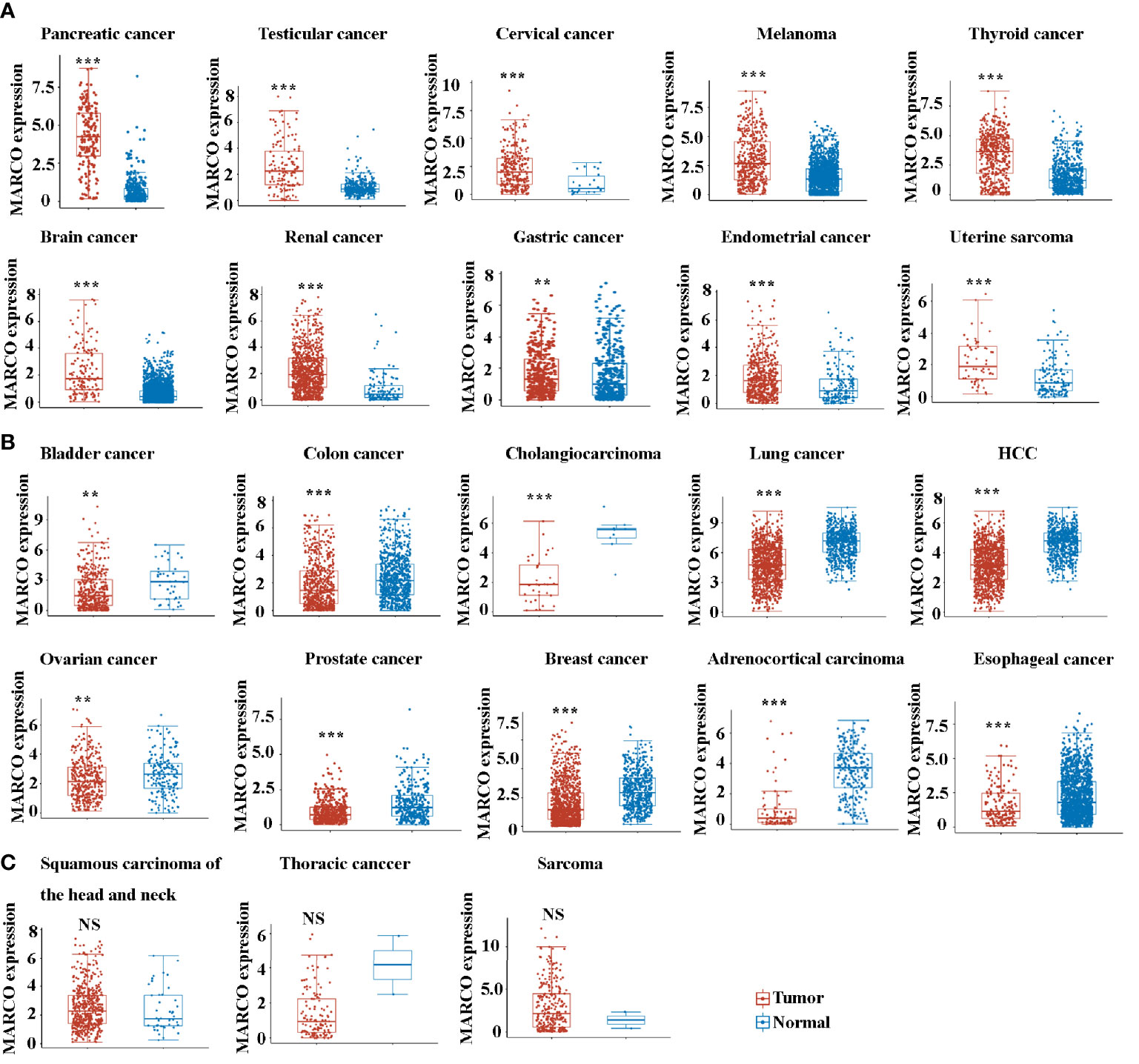
Figure 5 (A) The up-regulated mRNA expression of MARCO across many types of solid tumors from the TCGA database. (B) The down-regulated mRNA expression of MARCO across many types of solid tumors from the TCGA database. (C) The unchanged mRNA expression of MARCO across many types of solid tumors from the TCGA database. **P < 0.01, ***P < 0.001. NS means no significant.
MARCO Is Highly Expressed in Patients With Pancreatic Cancer
Subsequently, we sought to assess the role of MARCO in pancreatic cancer using immunohistochemistry (IHC). In total, 65 cancerous tissues and paracancerous tissues were included. The tissues were taken from 43 male and 22 female patients with a median age of 63.5 years (range = 41-74 years). The characteristics of patients with pancreatic cancer from the Affiliated Cancer Hospital of Zhengzhou University are presented in Table 1. Before starting the immunohistochemical staining, we first confirmed the paracancerous and cancerous tissues using HE staining. Figure 6A presents representative HE images of paracancerous and cancerous tissues. Then, CD163 and MARCO expression in both paracancerous and cancerous tissues were examined using IHC. A comparison of CD163 expression between paracancerous and cancerous tissues revealed significantly higher expression in cancerous tissues than in paracancerous tissues (Figures 6B, C). The expression of MARCO was also detected in paracancerous and cancerous tissues, and our data demonstrated that MARCO expression significantly increased in cancerous tissues compared with paracancerous tissues (Figures 6D, E).
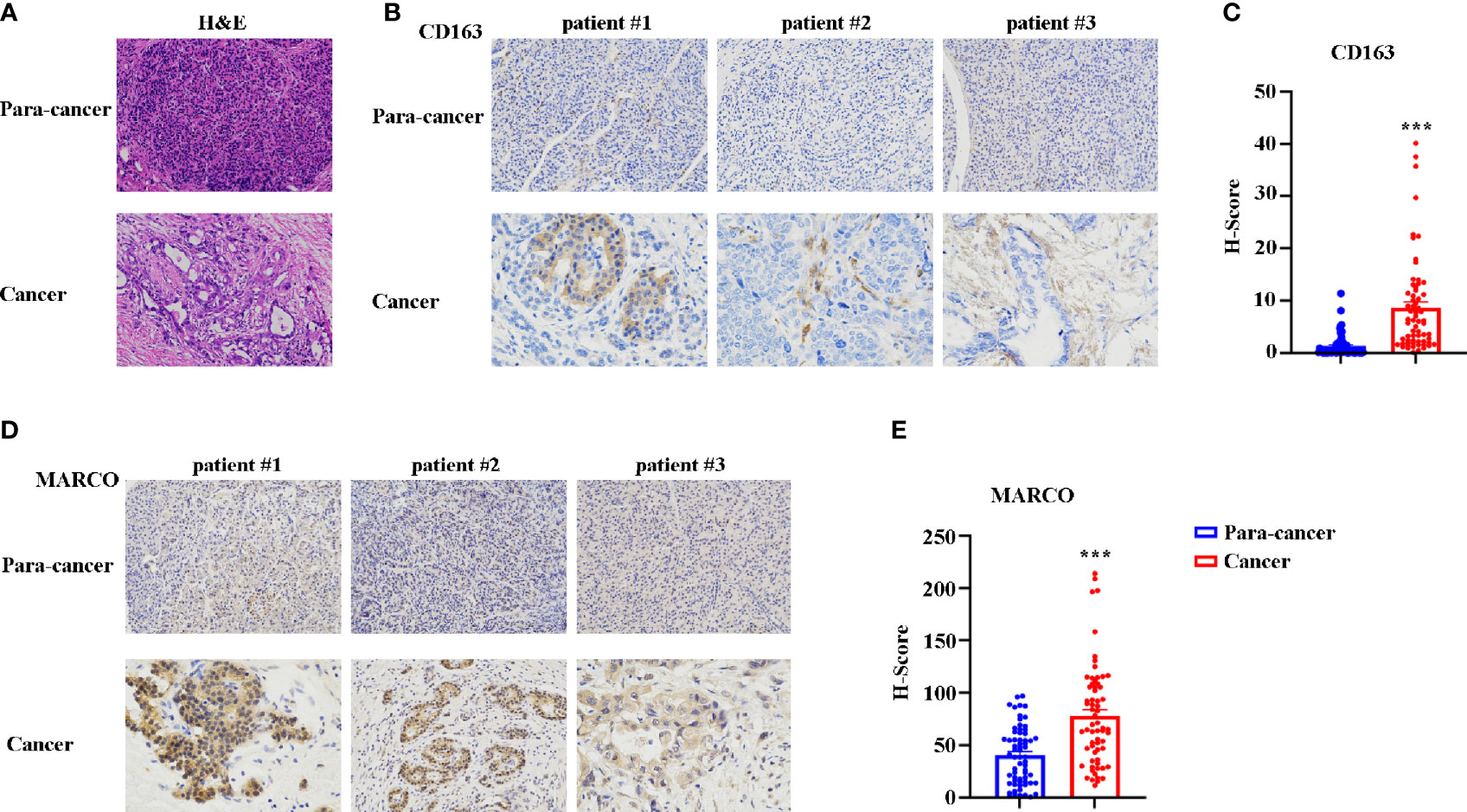
Figure 6 The expression of CD163 and MARCO in pancreatic cancer examined using immunohistochemistry (IHC). (A) Representative HE staining of cancer tissue and paracancer tissue in pancreatic cancer. (B) Representative IHC image of CD163 in pancreatic cancer tissues and paracancer tissues. (C) Quantitative analysis of CD163 expression between pancreatic cancer tissues and paracancer tissues. (D) Representative IHC image of MARCO in pancreatic cancer tissues and paracancer tissues. (E) Quantitative analysis of MARCO expression between pancreatic cancer tissues and paracancer tissues. ***P < 0.001.
Relationship of MARCO With Clinicopathologic Features of Pancreatic Cancer Patients
CD163 and MARCO expression were diverse in each sample (Figures 6C, E). We calculated the H-score of CD163 expression and divided the patients into high expression (CD163 hi) and low expression (CD163 low) subgroups according to a cutoff value of a mean H-score. We also classified the cohort into two subgroups according to the mean H-score of MARCO in tumor tissues. High CD163 expression was positively correlated with high ECOG PS (P = 0.002), high TNM stage (P = 0.000), and low histopathological grading (P = 0.006; Table 2). Similarly, high MARCO expression in tumor tissues was positively correlated with high ECOG PS (P = 0.002), high TNM stage (P = 0.002), and low histopathological grading (P = 0.010; Table 2).
Prognostic Implication of MARCO in Patients With Pancreatic Cancer
Until the last follow-up, 55 patients died. CD163 hi patients with pancreatic cancer had a shorter DFS (4.5 months vs 12.0 months, P = 0.0043) and OS (11.0 months vs 24.0 months, P = 0.0018) than 163 low patients (Figures 7A, B). Similarly, patients with pancreatic cancer with high MARCO expression in tumor tissues had a shorter DFS (3.0 months vs 15.5 months, P = 0.0004) and OS (13.0 months vs 24.0 months, P = 0.0003) than MARCO low patients (Figures 7C, D). Furthermore, we analyzed the combined role of CD163 and MARCO in pancreatic cancer. Patients with pancreatic cancer with high expression of both CD163 and MARCO had the shortest DFS (3.0 months vs 10.0 months vs 19.0 months, P = 0.0146 and P = 0.0004, receptively) and OS (7.5 months vs 22.0 months vs 34.5 months, P = 0.0004 and P = 0.0004, respectively) among patients with high expression of only CD163 or MARCO and patients with low expression of both CD163 and MARCO (Figures 7E, F). A univariate analysis indicated that high ECOG PS, larger tumor size, high TNM stage, and low histopathological grading were risk factors for both DFS and OS (Table 3). Noteworthily, CD163 and MARCO expression as well as the combined expression of CD163 and MARCO in tumor tissues were also correlated with DFS and OS (Table 3). These risk factors revealed by the univariate analysis were adopted as covariates in a multivariate Cox proportional hazards model. Noteworthily, high expression of CD163 and MARCO as well as their combined expression were still independent prognostic indicators for both DFS and OS (Table 4).
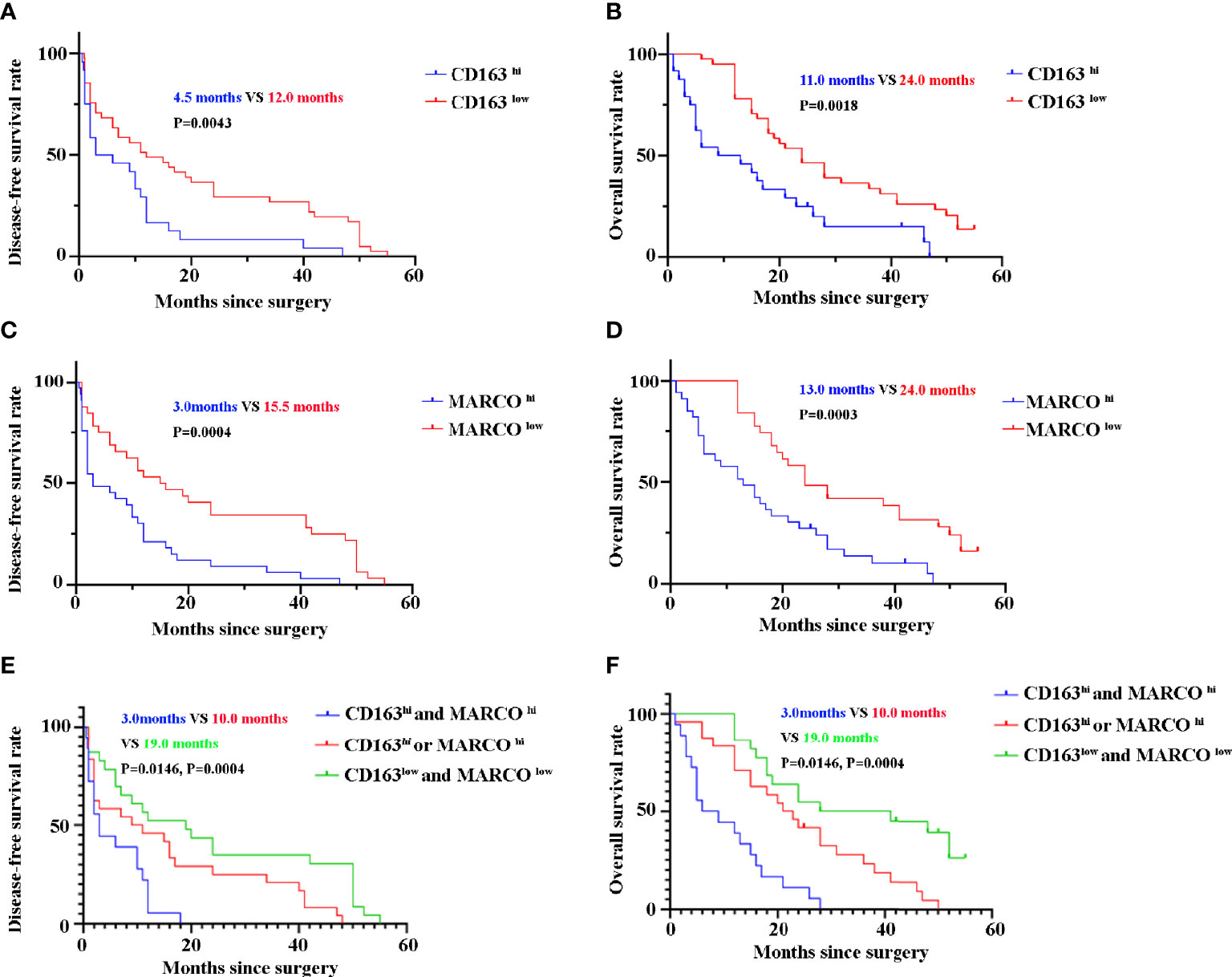
Figure 7 The disease-free survival (DFS) and overall survival (OS) of pancreatic cancer patients between high and low expression of CD163 and MARCO. (A) DFS curve of patients with pancreatic cancer between CD163 hi and CD163 low. (B) OS curve of patients with pancreatic cancer between CD163 hi and CD163 low. (C) DFS curve of patients with pancreatic cancer between MARCO hi and MARCO low. (D) OS curve of patients with pancreatic cancer between MARCO hi and MARCO low. (E) DFS curve of patients with pancreatic cancer between CD163hi MARCO hi and others. (F) OS curve of patients with pancreatic cancer between CD163hi MARCO hi and others.
Discussion
Research advances in PD-1/PD-L1 immune negative regulatory signaling pathways have driven tremendous advances in cancer immunotherapy. However, clinical studies have demonstrated that only 25-30% of tumors suppress the immune response through the PD-1/PD-L1 pathway, whereas others escape the immune response through a different molecular pathway or mechanism (34). Therefore, the inhibition of the PD-1/PD-L1 pathway alone may not be sufficient. The search for mechanisms or immunosuppressive pathways other than PD-1/PD-L1 is of great clinical value. TAMs control the tumor microenvironment and shape anti-tumor responses, affecting the clinical response rate in many cancers, including pancreatic cancer (12, 35–37). In the present study, we analyzed pancreatic cancer data from previous TCGA bulk RNA-seq data and determined that M2 macrophages are significantly enriched in tumor tissues. The significantly increased M2 markers of CD163 and CD206 also confirmed this conclusion. The upregulated KEGG pathways in tumor tissues include the osteoclast differentiation, endocytosis, the chemokine signaling pathway, and CAMs, which are correlated to TAMs (38–43). Significantly, these pathways are correlated to tumor progression (38–43). However, the survival analysis indicated that no significant differences exist based on the expression levels of both CD163 and CD206. It is worth emphasizing that this analysis was based on the mRNA levels of CD163 and CD206; mRNA expression does not always represent protein expression. In previous studies that have used IHC or IF, CD163 hi or CD206 hi has been significantly correlated with shorter survival (44, 45). In the present study, we also confirmed the protein expression of CD163 in patients with pancreatic cancer using IHC, which exhibited increased expression in tumor tissues. Furthermore, patients with high CD163 expression exhibited shorter DFS and OS. The univariate and multivariate analyses also revealed that high CD163 expression is an independent prognostic marker.
Many studies have supported TAMs as being M2-like macrophages; however, experimental evidence suggests that TAMs are not only a unique and distinct M2 myeloid population but also that they share M1 and M2 signature polarization (46, 47). “Don’t eat me” signaling pathways, such as CD47-SIRPα, the MHC-I-LILRB1 axis, and the CD24-SIGLEC-10 axis can be expressed by all types of macrophages (48) and play crucial roles in inhibiting macrophage phagocytosis of tumor cells in numerous cancer types (13, 15, 17, 29). This study found SIRPα, LILRB1, and SIGLEC-10 to be increased in pancreatic cancer tissues. However, patients exhibited a similar survival time based on the expression levels of SIRPα, SIGLEC10, and LILRB1. Further studies are required to identify the roles of these markers in patients with pancreatic cancer. In general, AXL and TIMD4 function as two phagocytosis-related molecules in normal homeostasis (49, 50). Recently, AXL and TIMD4 have also been reported to be positively correlated to the poor progression of many cancers (31, 51–53). The present study found the mRNA expression levels of AXL and TIMD4 to be increased. Furthermore, TIMD4 was not found to be an independent prognostic marker in the survival analysis. By contrast, high AXL expression has a tread to be an independent poor prognostic marker in pancreatic cancer. Previous studies have also confirmed that AXL is critical in the progression and metastasis of pancreatic cancer (30, 33). Inhibiting AXL has extended survival, reduced primary and metastatic burden, and enhanced sensitivity to gemcitabine in pancreatic mouse models (30, 33). These data reveal that high AXL expression is a negative prognostic marker in pancreatic cancer.
MARCO-expressing macrophages are present in the tumor microenvironment in human breast cancer, metastatic melanoma, periampullary adenocarcinoma of the intestine, and non-small-cell lung cancer (18, 19, 32, 54). In the majority of cancers, macrophage expression of MARCO is correlated with an immunosuppressive phenotype (32). By contrast, decreased MARCO expression by macrophages is correlated with tumor progression and poor prognosis in HCC (22). The present study found that a majority of solid tumors have decreased MARCO expression compared with controls, including bladder cancer, colon cancer, cholangiocarcinoma, lung cancer, HCC, ovarian cancer, prostate cancer, breast cancer, adrenocortical carcinoma, esophageal cancer, gastric cancer, endometrial cancer, and uterine sarcoma. Although the expression of MARCO in testicular cancer, cervical cancer, melanoma, thyroid cancer, brain cancer, and renal cancer increase, there are fewer fold changes than in pancreatic cancer. Additionally, the expression of MARCO in sarcoma, squamous carcinoma of the head and neck, and thoracic cancer does not change significantly. These data suggest that MARCO expression may play a more significant role in pancreatic cancer compared with other tumor types. Further studies are required to uncover the roles of MARCO in distinct tumor types.
Although MARCO deficiency significantly inhibited tumor progression and metastasis in a pancreatic mouse model (23), the role of MARCO in human pancreatic cancer has not been previously reported. In the present study, we first confirmed that MARCO expression increased in pancreatic cancer tissues compared with paracancerous control tissues using IHC. Critically, patients with pancreatic cancer with high MARCO expression exhibited shorter DFS and OS. The univariate and multivariate analyses also demonstrated that high MARCO expression is an independent prognostic marker in pancreatic cancer. Furthermore, high expression of both CD163 and MARCO was associated with the shortest DFS and OS. The univariate and multivariate analyses confirmed that combined CD163 and MARCO expression is an independent prognostic marker in pancreatic cancer. Therefore, we first determined that MARCO expression is a biomarker that predicts the progression of pancreatic cancer.
Our study has several limitations that require consideration. Although our IHC approach revealed that CD163 and MARCO expression in pancreatic cancer significantly increases and high CD163 and MARCO expression is correlated with poor prognosis, a more detailed understanding of MARCO+ macrophages is required, which can be obtained using more experimental methods such as Sc-RNA-seq, RNA-seq, and multiplex immunofluorescence. Nevertheless, the expression of CD163 and MARCO is still a good predictor of pancreatic cancer prognosis. Furthermore, anti-MARCO therapy may be a novel approach that is worth studying in depth. Lastly, independent studies are required to confirm the findings of this study.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics Statement
This study was reviewed and approved by the Ethics Committee at the Affiliated Cancer Hospital of Zhengzhou University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
BS and JC performed the experiment, wrote the manuscript, and finished the figures and tables. TH, XW, QL, and QX read and edited the manuscript. SL designed experiments, read and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Key Research Project of Henan Province (32211041).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all our authors listed in this manuscript.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71:7–33. doi: 10.3322/caac.21654
2. Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic Cancer. Lancet (2020) 395:2008–20. doi: 10.1016/S0140-6736(20)30974-0
3. Gajewski TF, Meng Y, Blank C, Brown I, Kacha A, Kline J, et al. Immune Resistance Orchestrated by the Tumor Microenvironment. Immunol Rev (2006) 213:131–45. doi: 10.1111/j.1600-065X.2006.00442.x
4. Ingber DE. Can Cancer be Reversed by Engineering the Tumor Microenvironment? Semin Cancer Biol (2008) 18:356–64. doi: 10.1016/j.semcancer.2008.03.016
5. Achard C, Surendran A, Wedge M-E, Ungerechts G, Bell J, Ilkow CS. Lighting a Fire in the Tumor Microenvironment Using Oncolytic Immunotherapy. EBioMedicine (2018) 31:17–24. doi: 10.1016/j.ebiom.2018.04.020
6. Gajewski TF, Schreiber H, Fu Y-X. Innate and Adaptive Immune Cells in the Tumor Microenvironment. Nat Immunol (2013) 14:1014–22. doi: 10.1038/ni.2703
7. Beatty GL, Gladney WL. Immune Escape Mechanisms as a Guide for Cancer Immunotherapy. Clin Cancer Res an Off J Am Assoc Cancer Res (2015) 21:687–92. doi: 10.1158/1078-0432.CCR-14-1860
8. Tang S, Ning Q, Yang L, Mo Z, Tang S. Mechanisms of Immune Escape in the Cancer Immune Cycle. Int Immunopharmacol (2020) 86:106700. doi: 10.1016/j.intimp.2020.106700
9. Zhou J, Tang Z, Gao S, Li C, Feng Y, Zhou X. Tumor-Associated Macrophages: Recent Insights and Therapies. Front Oncol (2020) 10:188. doi: 10.3389/fonc.2020.00188
10. Nagaraj S, Gabrilovich DI. Tumor Escape Mechanism Governed by Myeloid-Derived Suppressor Cells. Cancer Res (2008) 68:2561–3. doi: 10.1158/0008-5472.CAN-07-6229
11. Lim SA, Kim J, Jeon S, Shin MH, Kwon J, Kim T-J, et al. Defective Localization With Impaired Tumor Cytotoxicity Contributes to the Immune Escape of NK Cells in Pancreatic Cancer Patients. Front Immunol (2019) 10:496. doi: 10.3389/fimmu.2019.00496
12. Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-Associated Macrophages as Treatment Targets in Oncology. Nat Rev Clin Oncol (2017) 14:399–416. doi: 10.1038/nrclinonc.2016.217
13. Cassetta L, Pollard JW. Targeting Macrophages: Therapeutic Approaches in Cancer. Nat Rev Drug Discov (2018) 17:887–904. doi: 10.1038/nrd.2018.169
14. Kuo TC, Chen A, Harrabi O, Sockolosky JT, Zhang A, Sangalang E, et al. Targeting the Myeloid Checkpoint Receptor Sirpα Potentiates Innate and Adaptive Immune Responses to Promote Anti-Tumor Activity. J Hematol Oncol (2020) 13:160. doi: 10.1186/s13045-020-00989-w
15. Barkal AA, Brewer RE, Markovic M, Kowarsky M, Barkal SA, Zaro BW, et al. CD24 Signalling Through Macrophage Siglec-10 Is a Target for Cancer Immunotherapy. Nature (2019) 572:392–6. doi: 10.1038/s41586-019-1456-0
16. Klichinsky M, Ruella M, Shestova O, Lu XM, Best A, Zeeman M, et al. Human Chimeric Antigen Receptor Macrophages for Cancer Immunotherapy. Nat Biotechnol (2020) 38:947–53. doi: 10.1038/s41587-020-0462-y
17. Barkal AA, Weiskopf K, Kao KS, Gordon SR, Rosental B, Yiu YY, et al. Engagement of MHC Class I by the Inhibitory Receptor LILRB1 Suppresses Macrophages and is a Target of Cancer Immunotherapy Article. Nat Immunol (2018) 19:76–84. doi: 10.1038/s41590-017-0004-z
18. Jeremiasen M, Borg D, Hedner C, Svensson M, Nodin B, Leandersson K, et al. Tumor-Associated CD68+, CD163+, and MARCO+ Macrophages as Prognostic Biomarkers in Patients With Treatment-Naïve Gastroesophageal Adenocarcinoma. Front Oncol (2020) 10:534761. doi: 10.3389/fonc.2020.534761
19. La Fleur L, Boura VF, Alexeyenko A, Berglund A, Pontén V, Mattsson JSM, et al. Expression of Scavenger Receptor MARCO Defines a Targetable Tumor-Associated Macrophage Subset in Non-Small Cell Lung Cancer. Int J Cancer (2018) 143:1741–52. doi: 10.1002/ijc.31545
20. Chen AX, Gartrell RD, Zhao J, Upadhyayula PS, Zhao W, Yuan J, et al. Single-Cell Characterization of Macrophages in Glioblastoma Reveals MARCO as a Mesenchymal Pro-Tumor Marker. Genome Med (2021) 13:88. doi: 10.1186/s13073-021-00906-x
21. Georgoudaki AM, Prokopec KE, Boura VF, Hellqvist E, Sohn S, Östling J, et al. Reprogramming Tumor-Associated Macrophages by Antibody Targeting Inhibits Cancer Progression and Metastasis. Cell Rep (2016) 15:2000–11. doi: 10.1016/j.celrep.2016.04.084
22. Sun H, Song J, Weng C, Xu J, Huang M, Huang Q, et al. Association of Decreased Expression of the Macrophage Scavenger Receptor MARCO With Tumor Progression and Poor Prognosis in Human Hepatocellular Carcinoma. J Gastroenterol Hepatol (2017) 32:1107–14. doi: 10.1111/jgh.13633
23. Neyen C, Plüddemann A, Mukhopadhyay S, Maniati E, Bossard M, Gordon S, et al. Macrophage Scavenger Receptor a Promotes Tumor Progression in Murine Models of Ovarian and Pancreatic Cancer. J Immunol (2013) 190:3798–805. doi: 10.4049/jimmunol.1203194
24. Eisinger S, Sarhan D, Boura VF, Ibarlucea-Benitez I, Tyystjärvi S, Oliynyk G, et al. Targeting a Scavenger Receptor on Tumor-Associated Macrophages Activates Tumor Cell Killing by Natural Killer Cells. Proc Natl Acad Sci USA (2020) 117:32005–16. doi: 10.1073/pnas.2015343117
25. Sturm G, Finotello F, Petitprez F, Zhang JD, Baumbach J, Fridman WH, et al. Comprehensive Evaluation of Transcriptome-Based Cell-Type Quantification Methods for Immuno-Oncology. Bioinformatics (2019) 35:i436–45. doi: 10.1093/bioinformatics/btz363
26. Yu G, Wang LG, Han Y, He QY. Clusterprofiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omi A J Integr Biol (2012) 16:284–7. doi: 10.1089/omi.2011.0118
27. Lin W, Wu S, Chen X, Ye Y, Weng Y, Pan Y, et al. Characterization of Hypoxia Signature to Evaluate the Tumor Immune Microenvironment and Predict Prognosis in Glioma Groups. Front Oncol (2020) 10:796. doi: 10.3389/fonc.2020.00796
28. Sargent DJ, Wieand S, Benedetti J, Labianca R, Haller DG, Shepherd LE, et al. Disease-Free Survival (DFS) vs. Overall Survival (OS) as a Primary Endpoint for Adjuvant Colon Cancer Studies: Individual Patient Data From 12,915 Patients on 15 Randomized Trials. J Clin Oncol (2004) 22:3502. doi: 10.1200/jco.2004.22.90140.3502
29. Michaels AD, Newhook TE, Adair SJ, Morioka S, Goudreau BJ, Nagdas S, et al. CD47 Blockade as an Adjuvant Immunotherapy for Resectable Pancreatic Cancer. Clin Cancer Res an Off J Am Assoc Cancer Res (2018) 24:1415–25. doi: 10.1158/1078-0432.CCR-17-2283
30. Du W, Phinney NZ, Huang H, Wang Z, Westcott J, Toombs JE, et al. AXL is a Key Factor for Cell Plasticity and Promotes Metastasis in Pancreatic Cancer. Mol Cancer Res (2021) 19:1412–21. doi: 10.1158/1541-7786.MCR-20-0860
31. Yano H, Motoshima T, Ma C, Pan C, Yamada S, Nakayama T, et al. The Significance of TIMD4 Expression in Clear Cell Renal Cell Carcinoma. Med Mol Morphol (2017) 50:220–6. doi: 10.1007/s00795-017-0164-9
32. La Fleur L, Botling J, He F, Pelicano C, Zhou C, He C, et al. Targeting MARCO and IL37R on Immunosuppressive Macrophages in Lung Cancer Blocks Regulatory T Cells and Supports Cytotoxic Lymphocyte Function. Cancer Res (2021) 81:956–67. doi: 10.1158/0008-5472.can-20-1885
33. Ludwig KF, Du W, Sorrelle NB, Wnuk-Lipinska K, Topalovski M, Toombs JE, et al. Small-Molecule Inhibition of Axl Targets Tumor Immune Suppression and Enhances Chemotherapy in Pancreatic Cancer. Cancer Res (2018) 78:246–55. doi: 10.1158/0008-5472.CAN-17-1973
34. Xin Yu J, Hodge JP, Oliva C, Neftelinov ST, Hubbard-Lucey VM, Tang J. Trends in Clinical Development for PD-1/PD-L1 Inhibitors. Nat Rev Drug Discov (2020) 19:163–4. doi: 10.1038/d41573-019-00182-w
35. Noy R, Pollard JW. Tumor-Associated Macrophages: From Mechanisms to Therapy. Immunity (2014) 41:49–61. doi: 10.1016/j.immuni.2014.06.010
36. Yang S, Liu Q, Liao Q. Tumor-Associated Macrophages in Pancreatic Ductal Adenocarcinoma: Origin, Polarization, Function, and Reprogramming. Front Cell Dev Biol (2020) 8:607209. doi: 10.3389/fcell.2020.607209
37. Lin Y, Xu J, Lan H. Tumor-Associated Macrophages in Tumor Metastasis: Biological Roles and Clinical Therapeutic Applications. J Hematol Oncol (2019) 12:1–16. doi: 10.1186/s13045-019-0760-3
38. Binnemars-Postma K, Storm G, Prakash J. Nanomedicine Strategies to Target Tumor-Associated Macrophages. Int J Mol Sci (2017) 18(5):979. doi: 10.3390/ijms18050979
39. Li D, Ji H, Niu X, Yin L, Wang Y, Gu Y, et al. Tumor-Associated Macrophages Secrete CC-Chemokine Ligand 2 and Induce Tamoxifen Resistance by Activating PI3K/Akt/Mtor in Breast Cancer. Cancer Sci (2020) 111:47–58. doi: 10.1111/cas.14230
40. Laoui D, Van Overmeire E, De Baetselier P, Van Ginderachter JA, Raes G. Functional Relationship Between Tumor-Associated Macrophages and Macrophage Colony-Stimulating Factor as Contributors to Cancer Progression. Front Immunol (2014) 5:489. doi: 10.3389/fimmu.2014.00489
41. Fend L, Accart N, Kintz J, Cochin S, Reymann C, Le Pogam F, et al. Therapeutic Effects of Anti-CD115 Monoclonal Antibody in Mouse Cancer Models Through Dual Inhibition of Tumor-Associated Macrophages and Osteoclasts. PloS One (2013) 8:e73310. doi: 10.1371/journal.pone.0073310
42. Samineni S, Zhang Z, Shively JE. Carcinoembryonic Antigen-Related Cell Adhesion Molecule 1 Negatively Regulates Granulocyte Colony-Stimulating Factor Production by Breast Tumor-Associated Macrophages That Mediate Tumor Angiogenesis. Int J Cancer (2013) 133:394–407. doi: 10.1002/ijc.28036
43. Francavilla C, Maddaluno L, Cavallaro U. The Functional Role of Cell Adhesion Molecules in Tumor Angiogenesis. Semin Cancer Biol (2009) 19:298–309. doi: 10.1016/j.semcancer.2009.05.004
44. Väyrynen SA, Zhang J, Yuan C, Väyrynen JP, Dias Costa A, Williams H, et al. Composition, Spatial Characteristics, and Prognostic Significance of Myeloid Cell Infiltration in Pancreatic Cancer. Clin Cancer Res an Off J Am Assoc Cancer Res (2021) 27:1069–81. doi: 10.1158/1078-0432.CCR-20-3141
45. Pan Y, Lu F, Fei Q, Yu X, Xiong P, Yu X, et al. Single-Cell RNA Sequencing Reveals Compartmental Remodeling of Tumor-Infiltrating Immune Cells Induced by Anti-CD47 Targeting in Pancreatic Cancer. J Hematol Oncol (2019) 12:1–18. doi: 10.1186/s13045-019-0822-6
46. Allavena P, Sica A, Garlanda C, Mantovani A. The Yin-Yang of Tumor-Associated Macrophages in Neoplastic Progression and Immune Surveillance. Immunol Rev (2008) 222:155–61. doi: 10.1111/j.1600-065X.2008.00607.x
47. Chávez-Galán L, Olleros ML, Vesin D, Garcia I. Much More Than M1 and M2 Macrophages, There are Also CD169(+) and TCR(+) Macrophages. Front Immunol (2015) 6:263. doi: 10.3389/fimmu.2015.00263
48. Vaeteewoottacharn K, Kariya R, Pothipan P, Fujikawa S, Pairojkul C, Waraasawapati S, et al. Attenuation of CD47-Sirpα Signal in Cholangiocarcinoma Potentiates Tumor-Associated Macrophage-Mediated Phagocytosis and Suppresses Intrahepatic Metastasis. Transl Oncol (2019) 12:217–25. doi: 10.1016/j.tranon.2018.10.007
49. Li W, Guo R, Song Y, Jiang Z. Erythroblastic Island Macrophages Shape Normal Erythropoiesis and Drive Associated Disorders in Erythroid Hematopoietic Diseases. Front Cell Dev Biol (2021) 8:613885. doi: 10.3389/fcell.2020.613885
50. Yoshida H, Kawane K, Koike M, Mori Y, Uchiyama Y, Nagata S. Phosphatidylserine-Dependent Engulfment by Macrophages of Nuclei From Erythroid Precursor Cells. Nature (2005) 437:754–8. doi: 10.1038/nature03964
51. Zhu C, Wei Y, Wei X. AXL Receptor Tyrosine Kinase as a Promising Anti-Cancer Approach: Functions, Molecular Mechanisms and Clinical Applications. Mol Cancer (2019) 18:153. doi: 10.1186/s12943-019-1090-3
52. Davra V, Kumar S, Geng K, Calianese D, Mehta D, Gadiyar V, et al. Axl and Mertk Receptors Cooperate to Promote Breast Cancer Progression by Combined Oncogenic Signaling and Evasion of Host Antitumor Immunity. Cancer Res (2021) 81:698–712. doi: 10.1158/0008-5472.CAN-20-2066
53. Baghdadi M, Yoneda A, Yamashina T, Nagao H, Komohara Y, Nagai S, et al. TIM-4 Glycoprotein-Mediated Degradation of Dying Tumor Cells by Autophagy Leads to Reduced Antigen Presentation and Increased Immune Tolerance. Immunity (2013) 39:1070–81. doi: 10.1016/j.immuni.2013.09.014
Keywords: MARCO, tumor-associated macrophage, pancreatic cancer, prognosis, CD163
Citation: Shi B, Chu J, Huang T, Wang X, Li Q, Gao Q, Xia Q and Luo S (2021) The Scavenger Receptor MARCO Expressed by Tumor-Associated Macrophages Are Highly Associated With Poor Pancreatic Cancer Prognosis. Front. Oncol. 11:771488. doi: 10.3389/fonc.2021.771488
Received: 06 September 2021; Accepted: 12 October 2021;
Published: 29 October 2021.
Edited by:
Mark De Ridder, Vrije University Brussel, BelgiumCopyright © 2021 Shi, Chu, Huang, Wang, Li, Gao, Xia and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suxia Luo, emx5eWx1b3N1eGlhMDM2MUB6enUuZWR1LmNu
†These authors have contributed equally to this work
 Bian Shi
Bian Shi Junfeng Chu2†
Junfeng Chu2† Qingxin Xia
Qingxin Xia Suxia Luo
Suxia Luo