
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 05 November 2021
Sec. Cancer Genetics
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.771099
This article is part of the Research TopicAdvances in Genetics and Molecular Diagnosis in Colorectal, Stomach, and Pancreatic Cancer.View all 11 articles
Background: Previous study implicated that genes of matrix metalloproteinase (MMP) family play an important role in tumor invasion, neoangiogenesis, and metastasis. However, the diverse expression patterns and prognostic values of 24 MMPs in colorectal cancer are yet to be analyzed.
Methods: In this study, by integrating public database and our data, we first investigated the expression levels and protein levels of MMPs in patients with colorectal cancer. Then, by using TCGA and GEO datasets, we evaluated the association of MMPs with clinicopathological parameters and prognosis of colorectal cancer. Finally, by using the cBioPortal online tool, we analyzed the alterations of MMPs and did the network and pathway analyses for MMPs and their nearby genes.
Results: We found that, MMP1, MMP3, MMP7, MMP9–MMP12, and MMP14 were consistently upregulated in public dataset and our samples. Whereas, MMP28 was consistently downregulated in public dataset and our samples. In the clinicopathological analyses, upregulated MMP11, MMP14, MMP16, MMP17, MMP19, and MMP23B were significantly associated with a higher tumor stage. In the survival analyses, upregulated MMP11, MMP14, MMP17, and MMP19 were significantly associated with a shorter progression-free survival (PFS) time and a shorter relapse-free (RFS) time.
Discussion: This study implied that MMP11, MMP14, MMP17, and MMP19 are potential targets of precision therapy for patients with colorectal cancer.
Colorectal cancer (CRC) is the second leading cause of worldwide cancer mortality. It accounts for 9.2% of all cancer deaths according to the Global Cancer Statistics 2020 (1). In the USA, according to the SEER database, those with CRC have an overall 5-year survival rate of ~64%, primarily dependent on pathological stage at diagnosis. CRC patients diagnosed with disease limited to the colon have greater than 90% 5-year survival rate. Five-year survival decreases to ~70% with regional spread, and for patients diagnosed with distant metastases, the 5-year survival rate drops to 12.5% (2). Despite the significant advances in screening and diagnosis, there are limited therapeutic options for patients with advanced disease, which highlight the need for additional tumor molecular markers and prognostic predictors (3).
The human matrix metalloproteinases (MMPs) family belongs to the metzincin superfamily. The main function of MMPs is catalyzing the proteolytic activities and aiding breakdown of the extracellular matrix (ECM) (4). By degrading connective tissue between cells and in the lining of blood vessels, they enable tumor cells to escape from their original location and seed metastases (5). A large body of experimental and clinical evidence has implicated MMPs in tumor invasion, neoangiogenesis, and metastasis (6). Also, from the 1990s to early 2000s, inhibitors of MMPs (MMPI) were studied in various cancer types. However, despite strongly promising preclinical data, all trials failed due to lack of efficacy and severe side effects (7–9). One important reason to explain the failure is that some MMPs have antitumor effects, while the broad-spectrum MMPIs used in the initial trials might block these MMPs and result in tumor progression (10). Recently, with growing knowledge of MMPs in tumor invasion and metastasis and broader roles in cancer biology, narrow-spectrum MMPIs which were safer and more selective were currently being developed (11).
MMPs play complex and distinct roles in CRC. To date, 24 MMPs (MMP1, MMP2, MMP3, MMP4, MMP7, MMP8, MMP9, MMP10, MMP11, MMP12, MMP13, MMP14, MMP15, MMP16, MMP17, MMP19, MMP20, MMP21, MMP23a/MMP23b, MMP24, MMP25, MMP26, MMP27, and MMP28) were identified. For MMP1, Sunami et al. found that the expression of MMP1 was significantly correlated with hematogenous metastasis of colorectal cancer, which were further supported by research made by Shiozawa et al. and Bendardaf et al. (12–14) MMP2 and MMP9 comprise the gelatinase subfamily of MMPs. Marcus et al. found that the concentrations of MMP2 protein expression in tumor tissue were significantly higher than that in tumor-free tissue. In addition, the lymph node status was correlated with the expression of MMP2 in plasma, that is, the expression of MMP2 was significantly increased in patients with lymph node metastasis compared with those without (15). MMP7, also known as matrilysin, is frequently overexpressed in human cancer tissues. Adachi et al. found that the expression of MMP7 correlated significantly with the presence of nodal or distant metastases (16, 17). Another member of the gelatinase subfamily, MMP9, was expressed at significantly higher ratios in the sera of persons with CRC compared with normal controls. Overexpression of p38 gamma MAPK was shown to increase MMP9 transcription, enhancing cell invasion (18). Whereas, TGF-β receptor kinase inhibitors can reduce expression of MMP9 and block CRC metastasis to the liver (19, 20). However, for colitis-associated colon cancer, MMP9 has a protective role and acts as a tumor suppressor (21). MMP12, also called metalloelastase, was reported to be associated with both reduced tumor growth and increased overall survival (22). MMP13, sharing structural homology with MMP1, was reported to be associated with advanced cancer stage, and its overexpression can increase the risk of postoperative relapse (23). In addition to the MMPs mentioned above, MMP3, MMP11, and MMP14 were also found to be highly expressed in malignant tumors as compared with normal tissue (24–26).
As previously described, the relationship between MMPs and the prognosis of human CRC was only partly reported. By integrating state-of-art databases, we conducted a systematical analysis for all 24 human MMPs. Differential expression analyses were implemented in public database and our samples. Prognosis analyses were evaluated in The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets. Pathway and network analyses were further used to investigate the mechanisms underlying them. To the best of our knowledge, this is among the first bioinformatic analyses to comprehensively evaluate all 24 MMPs in CRC.
This study was approved by the Academic Committee of Sun Yat-Sen University, and it was conducted according to the principles expressed in the Declaration of Helsinki.
Oncomine is an online cancer microarray database (https://www.oncomine.org/resource/login.html). Gene expression array datasets from Oncomine were used to analyze the transcription levels of MMPs in different cancers. Differential gene expression analyses of all MMPs were implemented between cancer samples and normal controls. p-value was calculated using Student’s t-test. Cutoffs of p-value and fold change were 0.01 and 1.5, respectively.
Gene Expression Profiling Interactive Analysis (GEPIA) is an interactive web server which was developed by Tang et al. (27) By using a standard processing pipeline, they analyzed the RNA sequencing expression data of 9,736 tumors and 8,587 normal samples. GEPIA provides customizable tumor/normal differential expression analysis, profiling according to cancer types. Cutoff of p-value and fold change used in GEPIA were 0.01 and 2, respectively.
All fresh frozen tissues were archived from The Sixth Affiliated Hospital of Sun Yat-Sen University. The related protocol of human sample usage and the informed consent was approved by the Ethical Review Board of the The Sixth Affiliated Hospital of Sun Yat-Sen University.
Total RNA was extracted from the tumor and normal tissues of 12 patients using Total RNA Kit (Vazyme, China) according to the manufacturer’s instruction. Detailed information of these 12 patients can be found in Supplementary Table S1. For cDNA synthesis, 1 μg total RNA was reverse-transcribed into cDNA by Hiscript@ III RT Super Mix with gDNA wiper (Vazyme, Nanjing, China). Quantitative PCR reaction was then performed using 2×SYBR mix (Vazyme, China) and the reaction was run on Applied Biosystems 7500 Real-time PCR system. The Ct values obtained from different samples were compared using the 2-ΔΔCt method. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as internal reference genes. Sequence information of all used primers is listed in Supplementary Table S2.
UALCAN is a comprehensive, user-friendly, and interactive web resource for analyzing cancer OMICS data. It is built on PERL-CGI with high-quality graphics using JavaScript and CSS (http://ualcan.path.uab.edu/index.html) (28). Using UALCAN, we evaluated the protein level of MMPs in cancer tissue and normal tissue of colorectal cancer patients.
For preparation of protein extracts, 12 pairs of cancer and adjacent normal tissues were crushed with a mortar under ice cold conditions and lysed with RIPA lysis buffer together with protease inhibitors. Cells were collected and lysed with RIPA lysis buffer together with protease inhibitors. After centrifugation at 12,000 rpm at 4°C for 20 min, supernatants were collected and protein concentration was determined using the Pierce™ BCA protein assay (Thermo, Waltham, MA, USA). Proteins were separated by electrophoresis on a 10% SDS-polyacrylamide gel, electroblotted onto a PVDF membrane, and blocked by 5% nonfat dry milk for 1 h. Membranes were then washed in TBST three times for 5 min and then incubated with anti-MMP1 (Abcam, Cambridge, MA, USA), anti-MMP2 (Abcam), anti-MMP3 (Abcam, USA), anti-MMP7 (Abcam, USA), anti-MMP8 (Abcam, USA), anti-MMP9 (Invitrogen, Waltham, MA, USA), anti-MMP11 (Bioss, Beijing, China), anti-MMP12 (Abcam, USA), anti-MMP14 (Abcam, USA), anti-MMP17 (Abcam, USA), anti-MMP19 (Bioss, China), anti-MMP28 (Abcam, USA), anti-Collagen (Abcam), anti-TIMP2 (Bioss, China), or anti-GAPDH (Abcam). Subsequently, the membranes were washed with PBST and incubated for 1 h with goat anti-rabbit IgG (Abcam). Finally, membranes were washed three times and immunoreactivity was determined by using a Chemi DOC™ XRS+ system (BioRad Laboratories, Hercules, CA, USA).
By integrating TCGA dataset and standardized survival endpoints defined by Liu et al. recently, we performed clinicopathological and survival analyses (29). Nonparametric Kruskal-Wallis test was used to evaluate the association of American Joint Committee on Cancer (AJCC) stage of colorectal cancer (stage I, stage II, stage III, and stage IV) with the expression of MMPs. Four kinds of survival analyses were implemented, including overall survival (OS), disease-specific survival (DSS), disease-free survival (DFS) also called disease-free interval (DFI), and progression-free survival (PFS) also called progression-free interval (PFI). Disease-free survival is a concept used to describe the period after a successful treatment during which there are no signs and symptoms of the disease that was treated. In addition, by using the GEO dataset GSE39582, we did a relapse-free survival (RFS) analyses (30). As MMP4, MMP23A/MMP23B were not included in the GSE39582 dataset, only 22 MMPs were analyzed in the RFS analyses. Samples were split into two groups by median expression (high vs. low expression), and Kaplan-Meier plot were depicted (denoted with log rank p-value). Hazard ratio (HR) and 85% confidence intervals (CIs) were calculated by multivariate Cox regression adjusting the effect of age at diagnosis and sex.
TCGA collected many types of data for each of over 20,000 tumor and normal samples (31). The colorectal cancer dataset, including data from 640 cases with pathology reports, was selected for further analyses of MMPs using cBioPortal (http://www.cbioportal.org/). The genomic profiles included mutations, putative copy number alterations (CNAs) from genomic identification of significant targets in cancer (GISTIC), mRNA expression Z scores (RNA-seq v.2 RSEM), and protein expression Z scores (reversed-phase protein array (RPPA)). Coexpression and network were calculated according to the cBioPortal’s online instructions. By using the expression data in TCGA, we also calculated the correlation of MMPs with each other and several cancer-associated genes, including MYC, TP53, cyclin-D, as well as CDK4/6. The correlation coefficient was calculated using Spearman’s method.
HCT116 were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA), penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in a humidified CO2 (5%) atmosphere. MMP11, MMP14, MMP17, MMP19, small interfering RNA (siRNA), and nontargeting siRNA (si-control) were purchased from Ribobio (Guangzhou, China) and used at 20 mM. Opti-MEM transfection media and Lipo3000 (Invitrogen) were used to transfect the cells once they reached 50% confluency. Knockdown was assessed by Western blotting after 48 h of transfection. Sequence information of all used primers is listed in Supplementary Table S3.
By using the Oncomine database, we did a Pan-cancer differential gene expression analyses for all MMPs. As shown in Figure 1, MMP1–MMP4, MMP7–MMP14, and MMP24 were significantly upregulated in colorectal cancer samples, while MMP15, MMP17, MMP19, and MMP24–MMP28 were significantly downregulated in colorectal cancer samples. Detailed performance of each MMP in Oncomine database can be found in Supplementary Tables S4, S5.
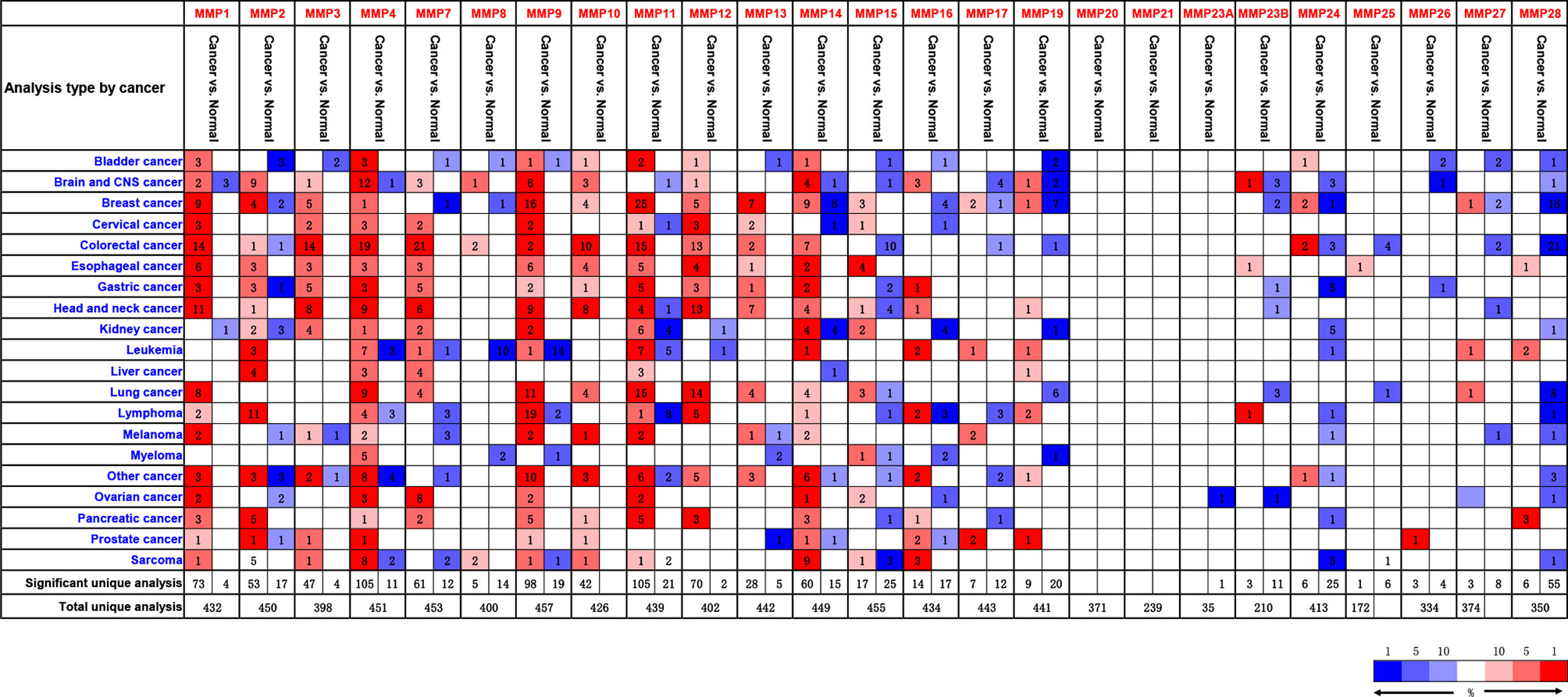
Figure 1 The transcription levels of MMPs in different types of cancers (Oncomine). Upregulated records are highlighted in red, while downregulated records are highlighted in blue. The number in each block means the number of unique analyses, which were fully described in Supplementary Tables S4, S5.
We then used GEPIA to compare the expression level of all MMPs between colorectal tumor tissue and normal tissue. As shown in Figure 2, we found that MMP1, MMP3, MMP7, MMP9–MMP12, and MMP14 were significantly upregulated in tumor tissue, while MMP28 was significantly downregulated in tumor tissue.
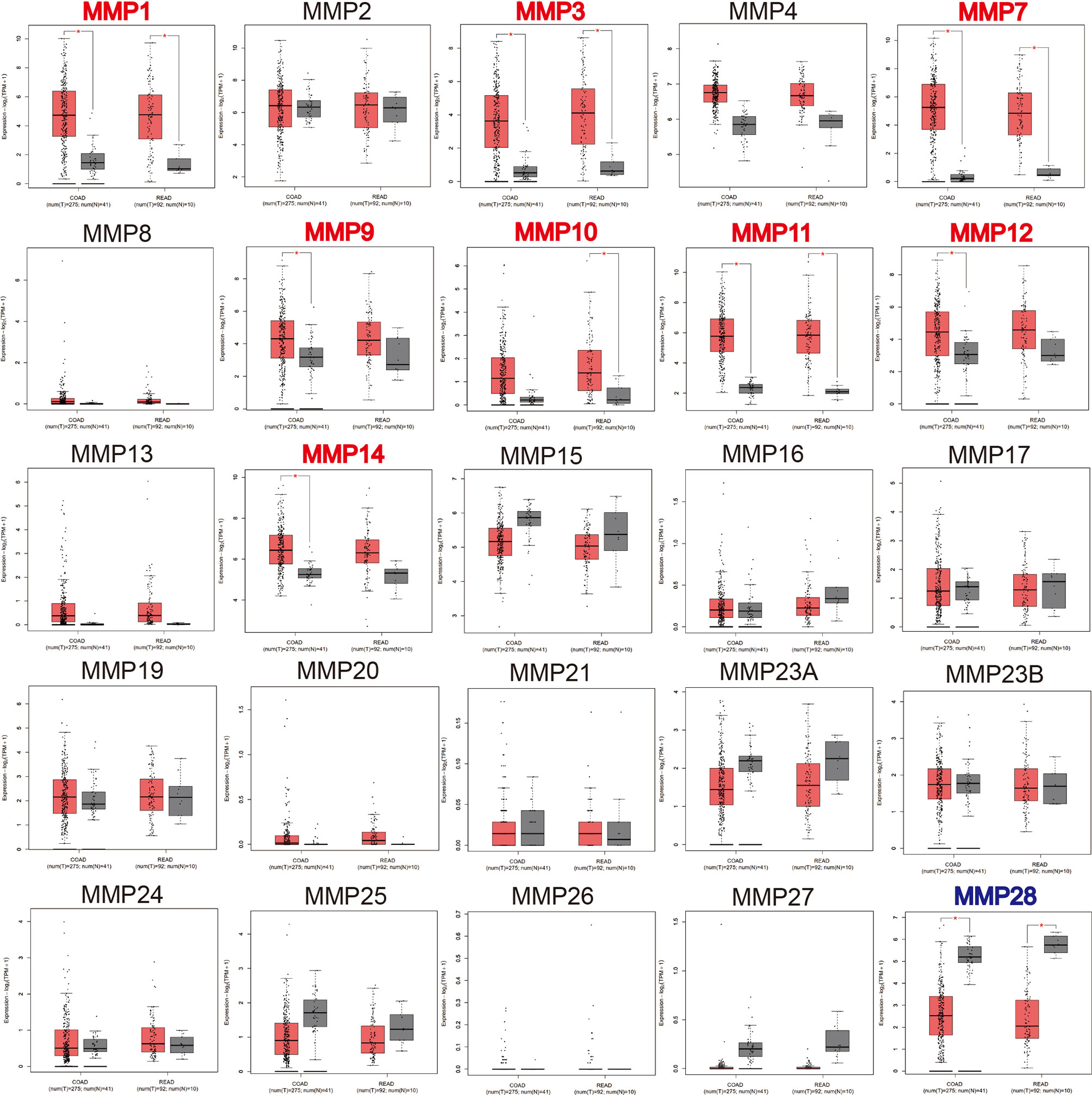
Figure 2 The transcription levels of MMPs in colorectal cancer tissue and normal tissue (GEPIA). Significant records are denoted by the red asterisk on top of the boxplot. The titles of upregulated records are highlighted in red, while the titles of downregulated records are highlighted in blue.
We further validated the expression level of MMPs in 12 colorectal cancer patients which were recruited from our hospital (including seven patients with colon cancer and five patients with rectal cancer, detailed information can be found in Supplementary Table S1) and measured the expression level of 24 MMPs in their tumor tissue and adjacent normal tissue by quantitative real-time polymerase chain reaction (qRT-PCR). As shown in Figure 3, we found that MMP1, MMP3, MMP7, MMP9-MMP12, and MMP14 were significantly upregulated in tumor tissue, while MMP15–MMP17, MMP19–MMP21, MMP23A, MMP23B, and MMP25–MMP28 were significantly downregulated in tumor tissue.
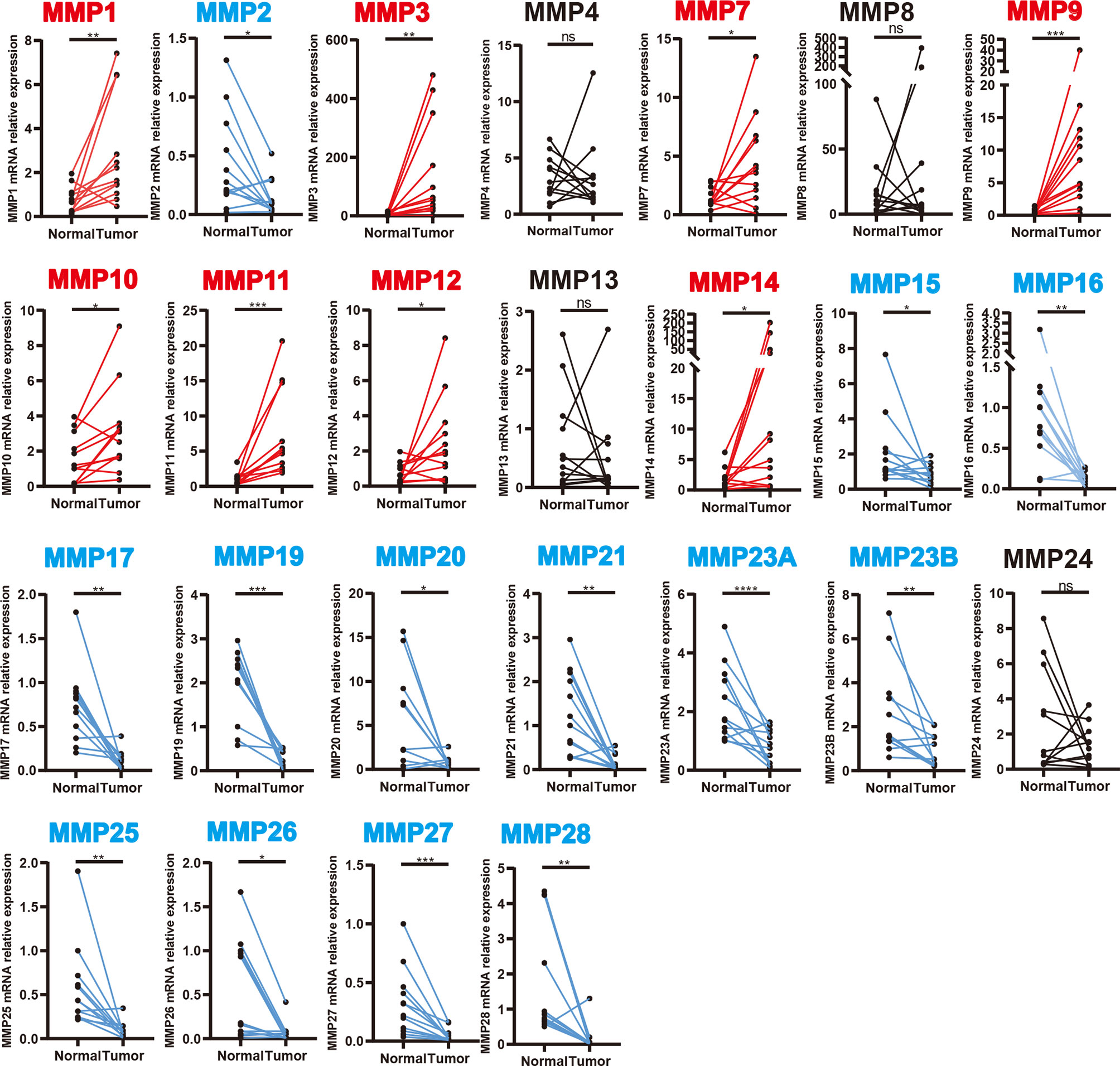
Figure 3 Transcription levels of MMPs in colorectal cancer tissue and adjacent normal tissue (12 colorectal cancer patients in our hospital). Significant records are denoted by the red asterisk on top of the plot (nsp > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001). Upregulated records are highlighted in red, while downregulated records are highlighted in blue.
In summary, MMP1, MMP3, MMP7, MMP9–MMP12, and MMP14 were consistently upregulated in Oncomine, GEPIA, and our samples. Thus, MMP28 was consistently downregulated in Oncomine, GEPIA, and our samples.
By using the UALCAN database, we further evaluated the protein levels of MMPs in patients with colorectal cancer. As some proteins were not included in UALCAN, we can only do the analyses for MMP1, MMP2, MMP3, MMP7, MMP8, MMP9, MMP12, MMP14, and MMP28. As shown in Figure 4A, the protein level of MMP1, MMP2, MMP3, MMP7, MMP8, MMP9, MMP12, and MMP14 in colorectal tumor tissue were significantly higher than that in normal tissue, while the protein level of MMP28 in tumor was significantly lower than that in normal tissue.
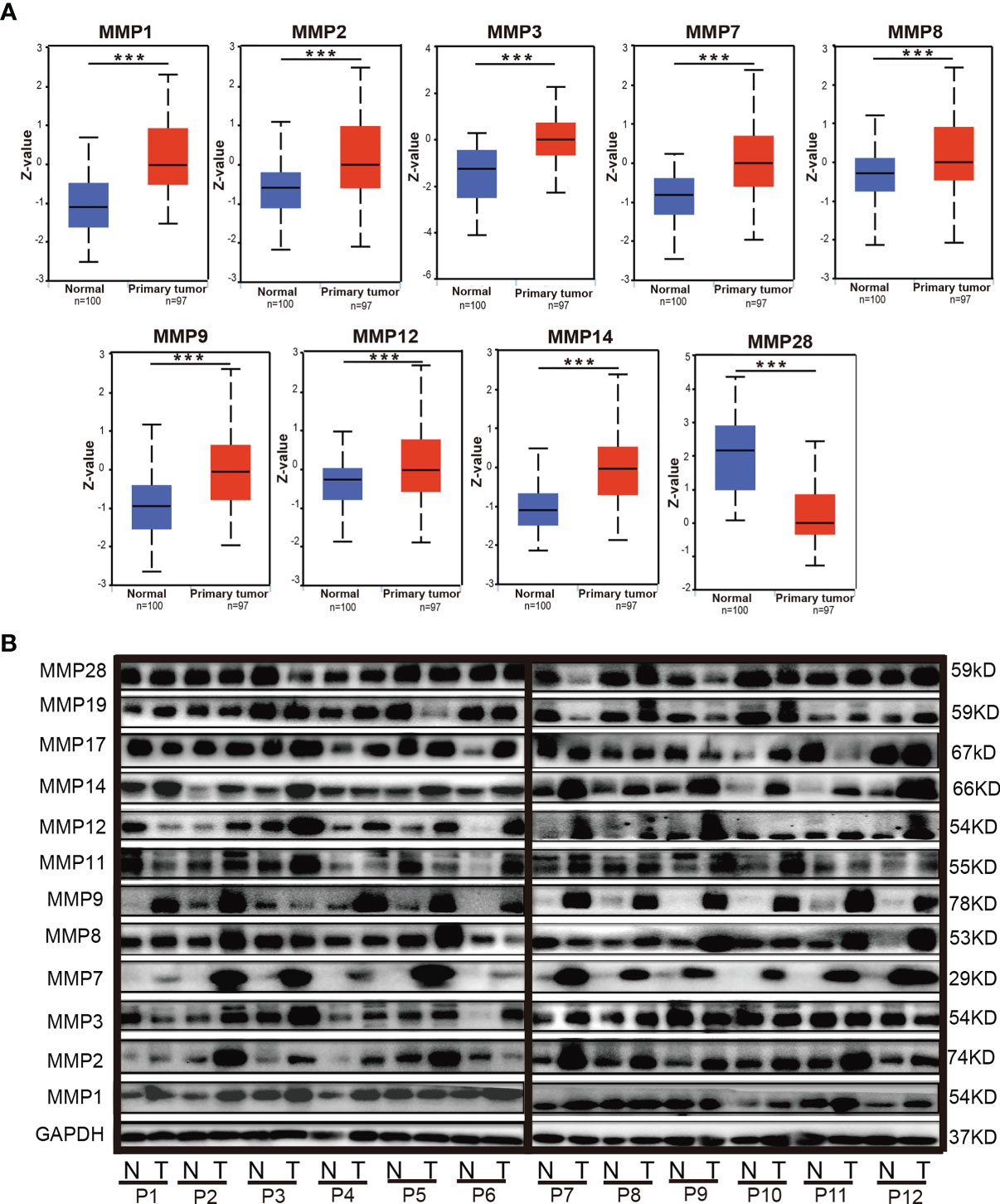
Figure 4 The protein levels of MMPs in colorectal cancer tissue and normal tissue. (A) The protein levels of MMPs in UALCAN database; significant records are denoted by the red asterisk on top of the boxplot (***p < 0.001). (B) The protein levels of MMPs in our 12 samples, which were measured by Western blotting (WB).
We also evaluated the protein level of MMPs in our patients and measured the expression level of MMP1–MMP3, MMP7–MMP9, MMP11, MMP12, MMP14, MMP17, MMP19, and MMP28 in their tumor tissue and adjacent normal tissue by Western blot. As shown in Figure 4B, we found that the protein level of MMP2, MMP7, MMP9, MMP12, and MMP14 in the tumor tissue were basically higher than that in the normal tissue.
By using the TCGA dataset, we analyzed the association of MMP expression with the AJCC stage of colorectal cancer. As shown in Figure 5, MMP11, MMP14, MMP16, MMP17, MMP19, and MMP23b were positively correlated with the tumor stage, that is, the mRNA levels of MMPs in patients with higher tumor stage were always high. Detailed information can be seen in Supplementary Table S6. Take MMP14 as an example, the mean expression level (log2(normalized count of MMP14)) were 8.43, 8.56, 8.74, and 8.72 for stage I, stage II, stage III, and stage IV patients, respectively (p = 0.007).
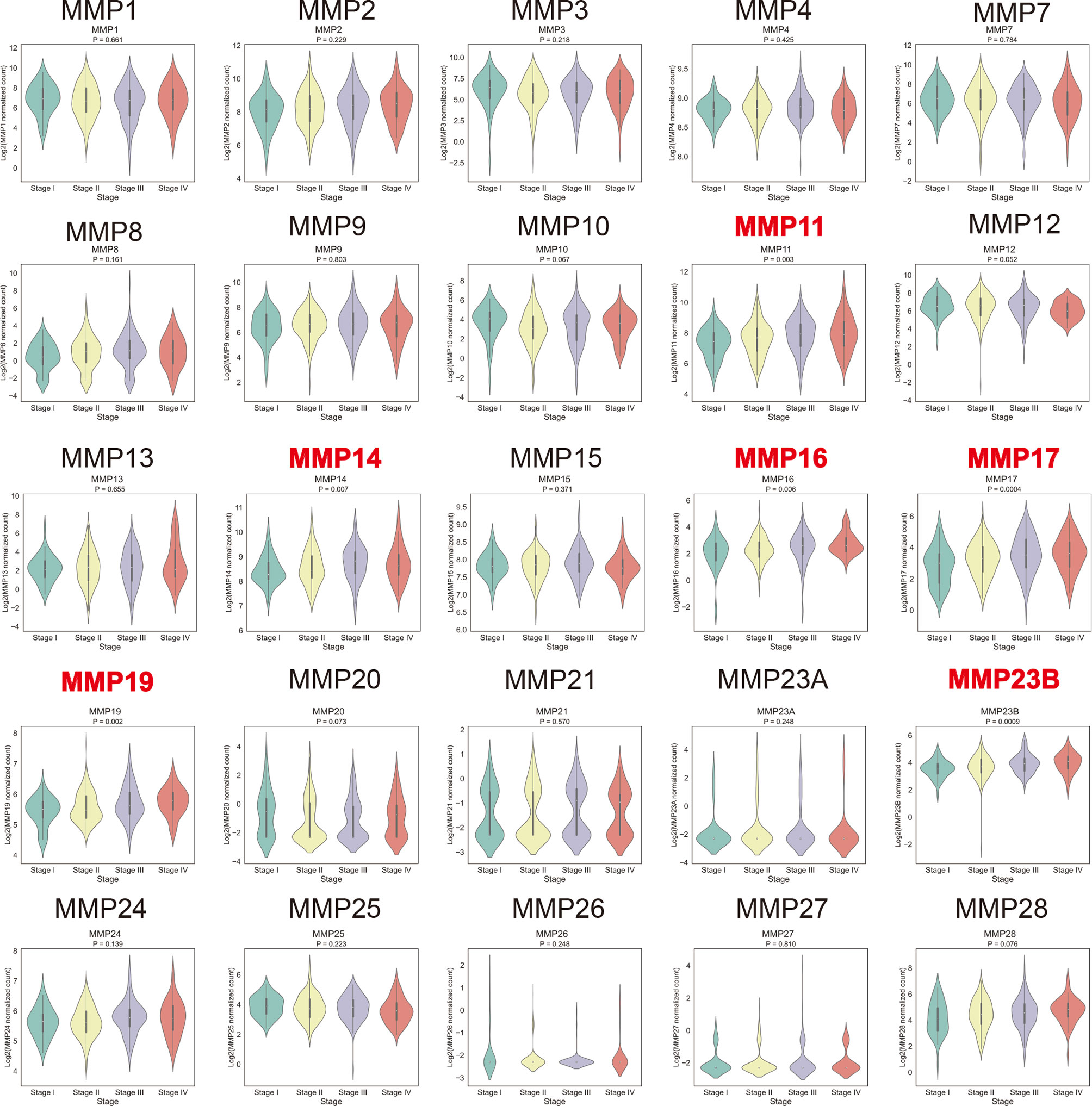
Figure 5 Association of the mRNA levels of MMPs with tumor stage of patients with colorectal cancer. The titles of upregulated records are highlighted in red, while the titles of downregulated records are highlighted in blue.
By integrating the TCGA data and four standardized survival endpoints defined by Liu et al. in 2018, we further performed the OS, DSS, DFS, and PFS analyses for all MMPs (Supplementary Figures S1–S4; Table 1). In the OS analyses, upregulated MMP11, MMP16, MMP17, MMP19, and MMP23B were significantly associated with a shorter overall survival time (Table 1; Supplementary Figure S1); in the DSS analyses, upregulated MMP14, MMP16, MMP17, MMP19, and MMP23B were significantly associated with a shorter disease-specific survival time (Table 1; Supplementary Figure S2); in the PFS analyses, upregulated MMP11, MMP14, MMP16, MMP17, MMP19, and MMP23B were significantly associated with a shorter progression-free time (Table 1; Supplementary Figure S3); and in the DFS analyses, downregulated MMP1, MMP3, MMP9, and MMP12 were significantly associated with a shorter disease-free period (Table 1; Supplementary Figure S4). By using the GEO dataset, we further performed the RFS analyses. As shown in Supplementary Figure S5 and Table 1, upregulated MMP2, MMP11, MMP14, MMP17, MMP19, MMP24, and MMP28 were significantly associated with a shorter relapse-free time, while the downregulated MMP8, MMP13, MMP16, MMP20, and MMP27 was significantly associated with a shorter relapse-free survival time.
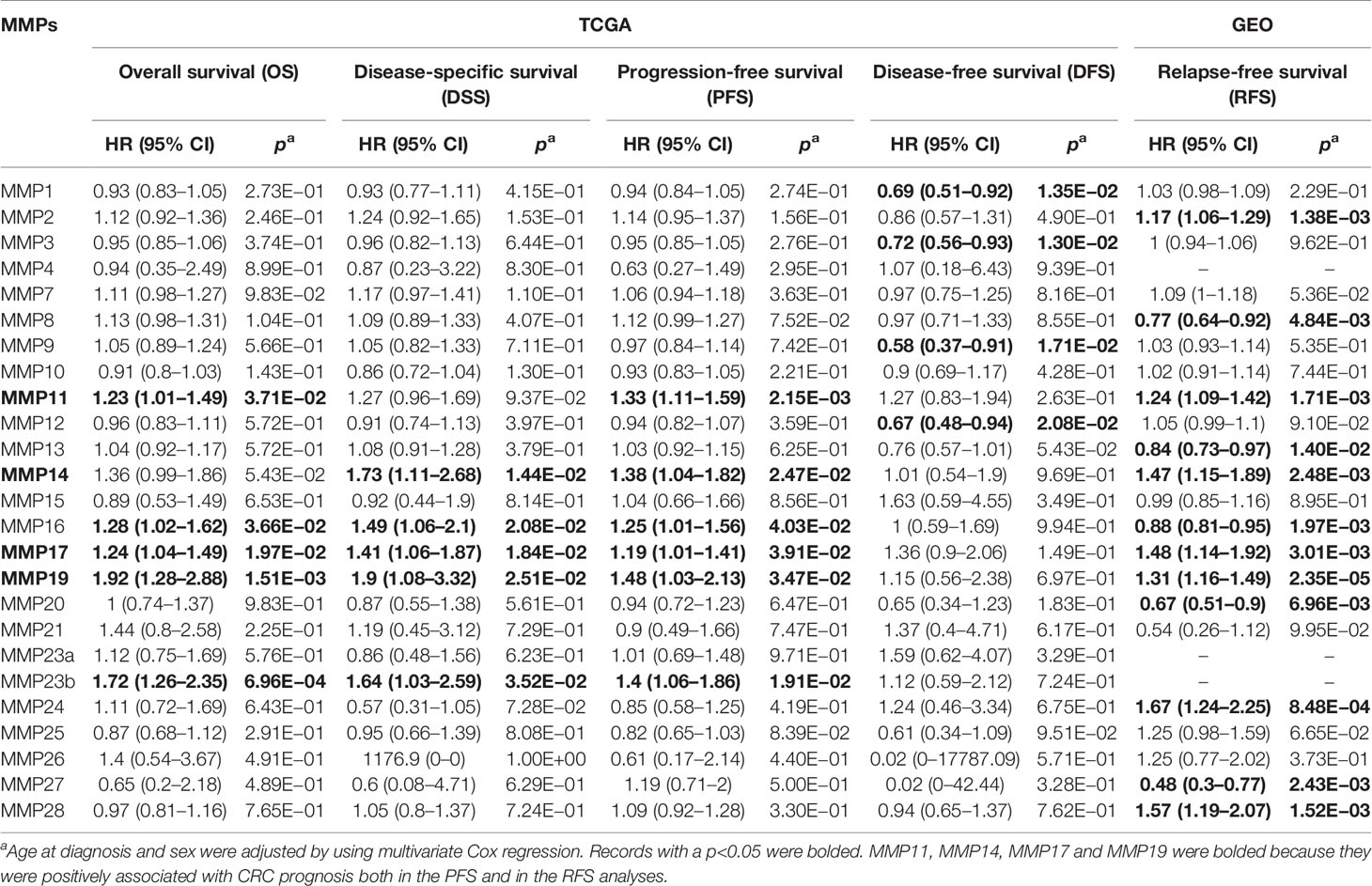
Table 1 Association of the mRNA expression of MMPs with the prognosis of patients with colorectal cancer.
We analyzed the MMP alterations and networks by using the cBioPortal online tool for colorectal cancer. As shown in Figure 6, of these 220 colorectal cancer patients, MMPs were altered in more than 30% of them (Figure 6A). The top 5 altered genes were MMP24 (10%), MMP9 (9%), and MMP16 (5%) (Figure 6B). As shown in Supplementary Figure S6, we also calculated the correlation of MMPs with each other and several cancer-associated genes, including MYC, TP53, cyclin-D, as well as CDK4/6. We found that multiple MMPs including MMP1, MMP3, MMP4, MMP7, MMP8, and MMP10–MMP14 were positively correlated with the expression of MYC, CCND1, and CDK4/6. We then constructed the network for MMPs and the 80 most frequently altered neighbor genes (Figure 6C). The results showed that collagen-related genes (for example, COL1A1) and metalloproteinase inhibitor-related genes (for example, TIMP2) were closely associated with MMP alterations. The functions of MMPs and the genes significantly associated with MMP alterations were predicted by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) in the Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/summary.jsp).
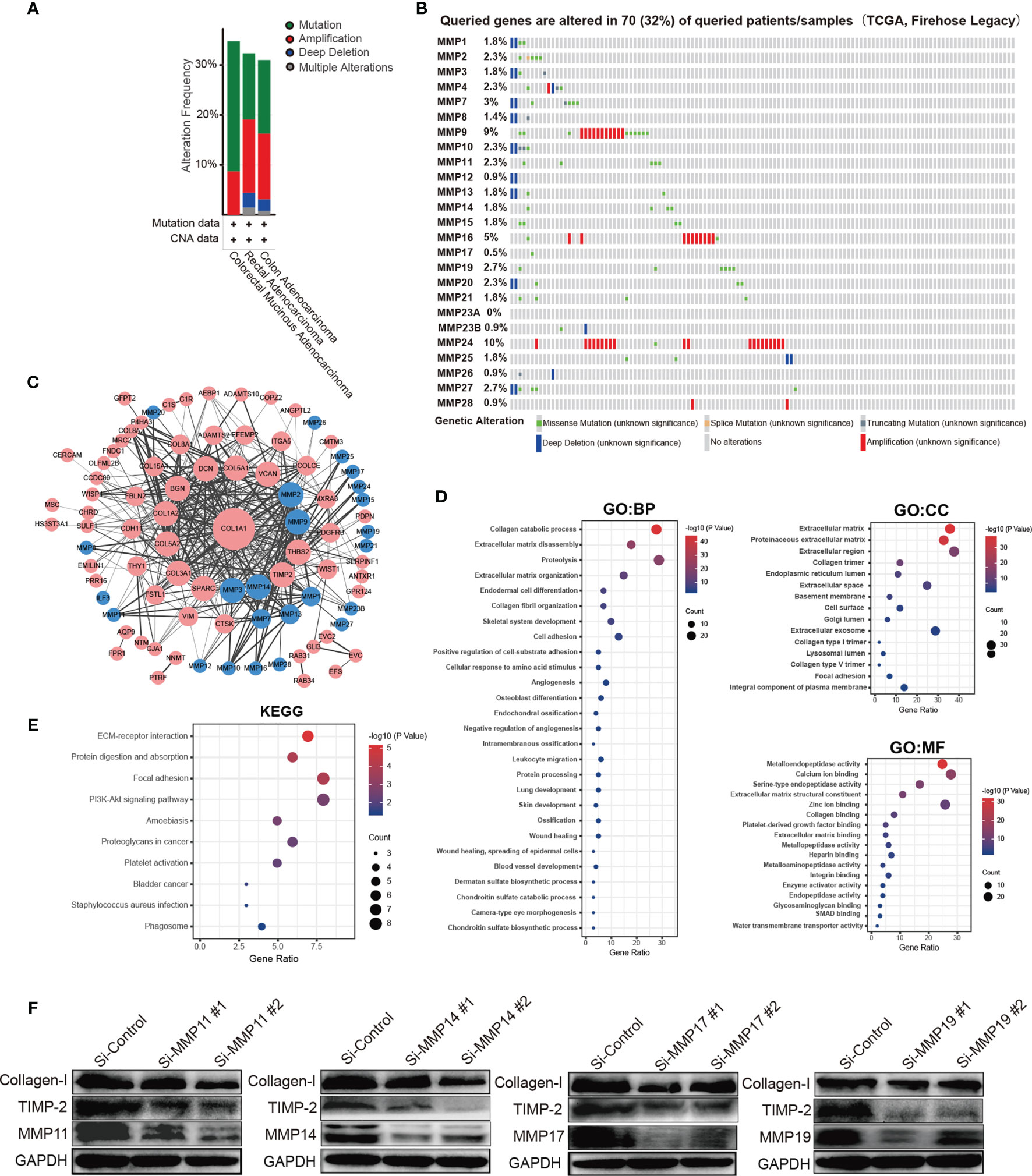
Figure 6 Prediction function and pathways of the changes in MMPs and their frequently altered neighbor genes in patients with colorectal cancer. (A) Overview of mutation and copy number validation of MMPs in different types of colorectal cancer, (B) detailed alteration proportion and types of each MMP gene, (C) network analyses for MMPs and their 50 most frequently altered neighbor genes. (D) GO pathway analyses for MMPs and the genes significantly associated with MMP alterations. (E) KEGG pathway analyses for MMPs and the genes significantly associated with MMP alterations. (F) the expression levels of collagen-I (COL1A1) and TIMP2 after knocking down of MMP11, MMP14, MMP17, and MMP19 in HCT116 cell line by using siRNAs.
GO enrichment analyses predicted the functional roles of target host genes on the basis of three aspects, including biological processes, cellular components, and molecular functions. For biological processes, the top 3 pathways were collagen catabolic process, extracellular matrix disassembly, and proteolysis, respectively. For cellular components, the top 3 pathways were extracellular matrix, proteinaceous extracellular matrix, and extracellular region, respectively, and for the molecular functions, the top 3 pathways were metalloendopeptidase activity, calcium ion binding, and serine-type endopeptidase activity, respectively (Figure 6D). In the KEGG enrichment analyses, the top 3 pathways were ECM-receptor interaction pathway, protein digestion and absorption pathway, and focal adhesion pathway, respectively (Figure 6E). Finally, by knocking down the expression of MMP11, MMP14, MMP17, and MMP19, we found that the expression of TIMP2 were significantly downregulated (Figure 6F). Similar trends were found for collagen-I (COL1A1) but not so obvious as TIMP2.
MMPs were reported to be associated with the progression of colorectal cancer; however, a comprehensive bioinformatic analysis for all MMPs has yet to be performed. In this study, we systematically explored the mRNA expression level of all 24 MMPs and their prognosis value in colorectal cancer. We found that, the transcriptional level of MMP1, MMP3, MMP7, MMP9–MMP12, and MMP14 in tumor were significantly upregulated, both in public database and in our samples. Also, in the clinicopathological and prognosis analyses, upregulated MMP11, MMP14, MMP17, and MMP19 were significantly associated with a higher tumor stage and a worse prognosis.
In this study, five survival endpoints were used in the survival analyses. OS is an important endpoint and is easy to define (the patient is either alive or dead). However, using OS as an endpoint may weaken a clinical study as deaths because of noncancer causes that do not necessarily reflect tumor biology. DSS can overcome the shortage of OS as DSS only considers the people who have not died from a specific disease in a defined period of time. However, both OS and DSS demand longer follow-up times; thus, in many clinical trials, DFS or PFS are preferred. PFS is defined as the time to disease progression or death from any cause. Whereas, DFS is used to describe the period after a successful treatment during which there are no signs and symptoms of the disease that was treated. The above four endpoints of TCGA dataset were standardized by Liu et al. in 2018 (29). Another survival endpoint, RFS which was used by the GEO database (GSE39582), was defined as the time from surgery to the first relapse and was censored at 5 years (30).
MMP11 also named stromelysin-3 is a member of the stromelysin subgroup belonging to the MMP superfamily. In this study, MMP11 was significantly upregulated in tumor, both in public database (Oncomine and GEPIA) and in our samples. The protein levels of MMP11 in our 12 pair samples were upregulated in tumor tissue for patients 2, 3, 6, 9, and 10 but not for other patients. Also, in the clinicopathological and survival analyses, upregulated MMP11 was significantly associated with a higher tumor stage (p = 0.003), a shorter OS (HR = 1.23, p = 3.71 × 10−2), a shorter PFS time (HR = 1.33, p = 2.15 × 10−3), and a shorter RFS time (HR = 1.24, p = 1.71 × 10−3). In the DSS and DFS analyses, although the association did not reach a significant level, a similar trend was found (HR >1). In a previous study, Li et al. measured the serum levels of MMP11 in 92 colon cancer patients and 92 healthy individuals using ELISA. They found that the serum levels of MMP11 were substantially higher in colon cancer patients than in healthy controls and was an independent predictor of the OS and DFS of colon cancer (32). MMP11 also played an important role in the tumorigenesis, proliferation, and invasion process of other cancers (33, 34). The mechanism behind it, may by inhibiting apoptosis as well as enhancing migration and invasion of cancer cells (35).
MMP14 plays an important role in extracellular matrix remodeling during aging. It has been reported to interact with TIMP2 (36). In our network analyses (Figure 6C), TIMP2 was indeed the closest gene of MMP14. Thus, by knocking down MMP14, the expression of TIMP2 was significantly downregulated (Figure 6F). In the transcriptional level, MMP14 was significantly upregulated in tumor tissue both in the public database and our own subjects. In the protein level, MMP14 was significantly upregulated in tumor tissue both in the UALCAN database and in patients 2, 3, 7, 9, 10, 11, and 12 of our own subjects. Furthermore, in the clinicopathological and prognosis analyses, upregulated MMP14 was significantly associated with a higher tumor stage (p = 0.007), a shorter DSS survival time (HR = 1.73, p = 0.01), a shorter PFS time (HR = 1.38, p = 0.02), and a significantly shorter RFS time (HR = 1.47, p = 2.48 × 10−3). The association of MMP14 with the prognosis of colorectal cancer was also reported by Cui et al. in 2019. In addition, Cui et al. found that patient with upregulated MMP14 was significantly associated with a lower 5-year DFS and OS (37). Recently, Ragusa and coworkers found that upregulated MMP14 levels correlated with blood vessel dysfunction and a lack of cytotoxic T cells (38).
MMP17 and MMP19 were another two MMPs. In this study, we found that upregulated MMP17 and MMP19 were significantly associated with a higher tumor stage (p = 4 × 10−4 and p = 2 × 10−3 for MMP17 and MMP19, respectively), a shorter OS time (HR = 1.24, p = 0.02 for MMP17 and HR = 1.92, p = 1.51 × 10−3 for MMP19), a shorter DSS time (HR = 1.41, p = 0.02 for MMP17 and HR = 1.9, p = 0.03 for MMP19), a shorter PFS (HR = 1.19, p = 0.04 for MMP17 and HR = 1.48, p = 0.03 for MMP19), and a shorter RFS (HR = 1.48, p = 3.01 × 10−3 for MMP17 and HR = 1.31, p = 2.35 × 10−5 for MMP19). However, in the transcriptional analyses, MMP17 and MMP19 were significantly upregulated in Oncomine and our 12 samples but not in the GEPIA. In the protein analyses of our samples, MMP17 was upregulated in tumor tissue for patients 4, 6, and 10. Recently, by detecting MMP19 mRNA expression in 198 CRC cancer tissues and paired normal controls, Chen et al. found that MMP19 expression was significantly upregulated in cancer tissues than in normal controls. In addition, by using immunohistochemistry to detect the expression of MMP19 protein in 42 patients, they further found that MMP19 mRNA expression is highly correlated with their protein levels. In their prognosis analyses, significant association between upregulated MMP19 expression and worse prognosis was also found (39). The transcriptional level of MMP17 and MMP19 in colorectal cancer tissue and normal tissue may need to be further confirmed.
Recently, a Pan-cancer analysis for MMPs in TCGA was implemented by Emily et al. (40) Different from our study, they focus on the overall performance of MMPs in several cancers. In their study, they only used the colon cancer patients (COPD) of TCGA (without rectal cancer) and there is no validation dataset. In addition, the only survival endpoint used in their analyses was OS. As we described above, due to shorter follow-up time in TCGA, the accuracy of OS may not be as good as PFS. Finally, without adjusting the effect of age at diagnosis and sex, the log-rank test used in their study may bias the final result.
In summary, our study was among the first study to systematically evaluate the performance of MMPs in colorectal cancer. This study will deepen our understanding of the prognosis mechanism of colorectal cancer. Also, MMP11, MMP14, MMP17, and MMP19 are potential targets of precision therapy for patients with colorectal cancer.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the Academic Committee of Sun Yat-Sen University. The patients/participants provided their written informed consent to participate in this study.
JY: acquisition of data, statistical analysis and technical interpretation of data, drafting of the manuscript, and critical revision of the manuscript for important intellectual content. ZH: acquisition of data and material, technical, and administrative support. XH, ZL, and LL: acquisition of data or material support. PL: study concept and design, acquisition of data, material support, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, and administrative support. HC: study concept and design, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, and administrative support. All authors contributed to the article and approved the submitted version.
This study was supported by the National Key R&D Program of China (PL, 2017YFC1308800); National Natural Science Foundation of China (PL, 81970452; ZH, 81902938); Science and Technology Program of Shenzhen, China (PL, JCYJ20190807161807867); Natural Science Foundation of Guangdong Province, China (ZH, 2020A1515011248); Fundamental Research Funds for the Central Universities (ZH, 19ykzd02); Guangdong Science and Technology Department (HC and BW, 2020B1212060018); the National Natural Science Foundation of China (HC, 81802833); and the 100 Top Talent Programs of Sun Yat-Sen University (HC, 58000-18841290).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.771099/full#supplementary-material
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Hankey BF, Ries LA, Edwards BK. The Surveillance, Epidemiology, and End Results Program: A National Resource. Cancer Epidemiol Biomarkers Prev (1999) 8(12):1117–21 https://cebp.aacrjournals.org/content/8/12/1117.long.
3. Lech G, Slotwinski R, Slodkowski M, Krasnodebski IW. Colorectal Cancer Tumour Markers and Biomarkers: Recent Therapeutic Advances. World J Gastroenterol (2016) 22(5):1745–55. doi: 10.3748/wjg.v22.i5.1745
4. Vandenbroucke RE, Libert C. Is There New Hope for Therapeutic Matrix Metalloproteinase Inhibition? Nat Rev Drug Discov (2014) 13(12):904–27. doi: 10.1038/nrd4390
5. Gialeli C, Theocharis AD, Karamanos NK. Roles of Matrix Metalloproteinases in Cancer Progression and Their Pharmacological Targeting. FEBS J (2011) 278(1):16–27. doi: 10.1111/j.1742-4658.2010.07919.x
6. Coussens LM, Fingleton B, Matrisian LM. Matrix Metalloproteinase Inhibitors and Cancer: Trials and Tribulations. Science (2002) 295(5564):2387–92. doi: 10.1126/science.1067100
7. Bramhall SR, Schulz J, Nemunaitis J, Brown PD, Baillet M, Buckels JA. A Double-Blind Placebo-Controlled, Randomised Study Comparing Gemcitabine and Marimastat With Gemcitabine and Placebo as First Line Therapy in Patients With Advanced Pancreatic Cancer. Br J Cancer (2002) 87(2):161–7. doi: 10.1038/sj.bjc.6600446
8. Sparano JA, Bernardo P, Stephenson P, Gradishar WJ, Ingle JN, Zucker S, et al. Randomized Phase III Trial of Marimastat Versus Placebo in Patients With Metastatic Breast Cancer Who Have Responding or Stable Disease After First-Line Chemotherapy: Eastern Cooperative Oncology Group Trial E2196. J Clin Oncol (2004) 22(23):4683–90. doi: 10.1200/JCO.2004.08.054
9. Hirte H, Vergote IB, Jeffrey JR, Grimshaw RN, Coppieters S, Schwartz B, et al. A Phase III Randomized Trial of BAY 12-9566 (Tanomastat) as Maintenance Therapy in Patients With Advanced Ovarian Cancer Responsive to Primary Surgery and Paclitaxel/Platinum Containing Chemotherapy: A National Cancer Institute of Canada Clinical Trials Group Study. Gynecol Oncol (2006) 102(2):300–8. doi: 10.1016/j.ygyno.2005.12.020
10. Winer A, Adams S, Mignatti P. Matrix Metalloproteinase Inhibitors in Cancer Therapy: Turning Past Failures Into Future Successes. Mol Cancer Ther (2018) 17(6):1147–55. doi: 10.1158/1535-7163.MCT-17-0646
11. Shah MA, Starodub A, Sharma S, Berlin J, Patel M, Wainberg ZA, et al. Andecaliximab/GS-5745 Alone and Combined With Mfolfox6 in Advanced Gastric and Gastroesophageal Junction Adenocarcinoma: Results From a Phase I Study. Clin Cancer Res (2018) 24(16):3829–37. doi: 10.1158/1078-0432.CCR-17-2469
12. Shiozawa J, Ito M, Nakayama T, Nakashima M, Kohno S, Sekine I. Expression of Matrix Metalloproteinase-1 in Human Colorectal Carcinoma. Mod Pathol (2000) 13(9):925–33. doi: 10.1038/modpathol.3880169
13. Sunami E, Tsuno N, Osada T, Saito S, Kitayama J, Tomozawa S, et al. MMP-1 Is a Prognostic Marker for Hematogenous Metastasis of Colorectal Cancer. Oncologist (2000) 5(2):108–14. doi: 10.1634/theoncologist.5-2-108
14. Bendardaf R, Buhmeida A, Ristamaki R, Syrjanen K, Pyrhonen S. MMP-1 (Collagenase-1) Expression in Primary Colorectal Cancer and Its Metastases. Scand J Gastroenterol (2007) 42(12):1473–8. doi: 10.1080/00365520701485449
15. Langenskiold M, Holmdahl L, Falk P, Ivarsson ML. Increased Plasma MMP-2 Protein Expression in Lymph Node-Positive Patients With Colorectal Cancer. Int J Colorectal Dis (2005) 20(3):245–52. doi: 10.1007/s00384-004-0667-4
16. Adachi Y, Yamamoto H, Itoh F, Hinoda Y, Okada Y, Imai K. Contribution of Matrilysin (MMP-7) to the Metastatic Pathway of Human Colorectal Cancers. Gut (1999) 45(2):252–8. doi: 10.1136/gut.45.2.252
17. Ii M, Yamamoto H, Adachi Y, Maruyama Y, Shinomura Y. Role of Matrix Metalloproteinase-7 (Matrilysin) in Human Cancer Invasion, Apoptosis, Growth, and Angiogenesis. Exp Biol Med (Maywood) (2006) 231(1):20–7. doi: 10.1177/153537020623100103
18. Loesch M, Zhi HY, Hou SW, Qi XM, Li RS, Basir Z, et al. P38gamma MAPK Cooperates With C-Jun in Trans-Activating Matrix Metalloproteinase 9. J Biol Chem (2010) 285(20):15149–58. doi: 10.1074/jbc.M110.105429
19. Zhang B, Halder SK, Kashikar ND, Cho YJ, Datta A, Gorden DL, et al. Antimetastatic Role of Smad4 Signaling in Colorectal Cancer. Gastroenterology (2010) 138(3):969–80.e1-3. doi: 10.1053/j.gastro.2009.11.004
20. Papageorgis P, Cheng K, Ozturk S, Gong Y, Lambert AW, Abdolmaleky HM, et al. Smad4 Inactivation Promotes Malignancy and Drug Resistance of Colon Cancer. Cancer Res (2011) 71(3):998–1008. doi: 10.1158/0008-5472.CAN-09-3269
21. Garg P, Sarma D, Jeppsson S, Patel NR, Gewirtz AT, Merlin D, et al. Matrix Metalloproteinase-9 Functions as a Tumor Suppressor in Colitis-Associated Cancer. Cancer Res (2010) 70(2):792–801. doi: 10.1158/0008-5472.CAN-09-3166
22. Yang W, Arii S, Gorrin-Rivas MJ, Mori A, Onodera H, Imamura M. Human Macrophage Metalloelastase Gene Expression in Colorectal Carcinoma and Its Clinicopathologic Significance. Cancer (2001) 91(7):1277–83. doi: 10.1002/1097-0142(20010401)91:7<1277::AID-CNCR1129>3.0.CO;2-H
23. Huang MY, Chang HJ, Chung FY, Yang MJ, Yang YH, Wang JY, et al. MMP13 Is a Potential Prognostic Marker for Colorectal Cancer. Oncol Rep (2010) 24(5):1241–7. doi: 10.3892/or_00000978
24. Porte H, Chastre E, Prevot S, Nordlinger B, Empereur S, Basset P, et al. Neoplastic Progression of Human Colorectal Cancer Is Associated With Overexpression of the Stromelysin-3 and BM-40/SPARC Genes. Int J Cancer (1995) 64(1):70–5. doi: 10.1002/ijc.2910640114
25. Baker EA, Bergin FG, Leaper DJ. Matrix Metalloproteinases, Their Tissue Inhibitors and Colorectal Cancer Staging. Br J Surg (2000) 87(9):1215–21. doi: 10.1046/j.1365-2168.2000.01531.x
26. Kikuchi R, Noguchi T, Takeno S, Kubo N, Uchida Y. Immunohistochemical Detection of Membrane-Type-1-Matrix Metalloproteinase in Colorectal Carcinoma. Br J Cancer (2000) 83(2):215–8. doi: 10.1054/bjoc.2000.1195
27. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res (2017) 45(W1):W98–W102. doi: 10.1093/nar/gkx247
28. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (2017) 19(8):649–58. doi: 10.1016/j.neo.2017.05.002
29. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell (2018) 173(2):400–16.e11. doi: 10.1016/j.cell.2018.02.052
30. Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene Expression Classification of Colon Cancer Into Molecular Subtypes: Characterization, Validation, and Prognostic Value. PloS Med (2013) 10(5):e1001453. doi: 10.1371/journal.pmed.1001453
31. Cancer Genome Atlas N. Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature (2012) 487(7407):330–7. doi: 10.1038/nature11252
32. Pang L, Wang DW, Zhang N, Xu DH, Meng XW. Elevated Serum Levels of MMP-11 Correlate With Poor Prognosis in Colon Cancer Patients. Cancer Biomark (2016) 16(4):599–607. doi: 10.3233/CBM-160601
33. Deng H, Guo RF, Li WM, Zhao M, Lu YY. Matrix Metalloproteinase 11 Depletion Inhibits Cell Proliferation in Gastric Cancer Cells. Biochem Biophys Res Commun (2005) 326(2):274–81. doi: 10.1016/j.bbrc.2004.11.027
34. Chen C, Liu X, Jiang J, Li S, Wang G, Ju L, et al. Matrix Metalloproteinase 11 Is a Potential Biomarker in Bladder Cancer Diagnosis and Prognosis. Onco Targets Ther (2020) 13:9059–69. doi: 10.2147/OTT.S243452
35. Zhang X, Huang S, Guo J, Zhou L, You L, Zhang T, et al. Insights Into the Distinct Roles of MMP-11 in Tumor Biology and Future Therapeutics (Review). Int J Oncol (2016) 48(5):1783–93. doi: 10.3892/ijo.2016.3400
36. Zucker S, Drews M, Conner C, Foda HD, DeClerck YA, Langley KE, et al. Tissue Inhibitor of Metalloproteinase-2 (TIMP-2) Binds to the Catalytic Domain of the Cell Surface Receptor, Membrane Type 1-Matrix Metalloproteinase 1 (MT1-MMP). J Biol Chem (1998) 273(2):1216–22. doi: 10.1074/jbc.273.2.1216
37. Cui G, Cai F, Ding Z, Gao L. MMP14 Predicts a Poor Prognosis in Patients With Colorectal Cancer. Hum Pathol (2019) 83:36–42. doi: 10.1016/j.humpath.2018.03.030
38. Ragusa S, Prat-Luri B, Gonzalez-Loyola A, Nassiri S, Squadrito ML, Guichard A, et al. Antiangiogenic Immunotherapy Suppresses Desmoplastic and Chemoresistant Intestinal Tumors in Mice. J Clin Invest (2020) 130(3):1199–216. doi: 10.1172/JCI129558
39. Chen Z, Wu G, Ye F, Chen G, Fan Q, Dong H, et al. High Expression of MMP19 Is Associated With Poor Prognosis in Patients With Colorectal Cancer. BMC Cancer (2019) 19(1):448. doi: 10.1186/s12885-019-5673-6
Keywords: colorectal cancer, MMPs, prognosis, expression, tumor stage
Citation: Yu J, He Z, He X, Luo Z, Lian L, Wu B, Lan P and Chen H (2021) Comprehensive Analysis of the Expression and Prognosis for MMPs in Human Colorectal Cancer. Front. Oncol. 11:771099. doi: 10.3389/fonc.2021.771099
Received: 05 September 2021; Accepted: 13 October 2021;
Published: 05 November 2021.
Edited by:
Walter Hernán Pavicic, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Weifeng Ding, Nantong University, ChinaCopyright © 2021 Yu, He, He, Luo, Lian, Wu, Lan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haitao Chen, Y2hlbmh0NTZAbWFpbC5zeXN1LmVkdS5jbg==; Ping Lan, bGFucGluZ0BtYWlsLnN5c3UuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.