- 1Division of Hemato-Oncology, Department of Internal Medicine, Kangnam Sacred-Heart Hospital, Hallym University Medical Center, College of Medicine, Hallym University, Seoul, South Korea
- 2Department of Obstetrics and Gynecology, Kangnam Sacred-Heart Hospital, Hallym University Medical Center, College of Medicine, Hallym University, Seoul, South Korea
- 3Division of Gastroenterology, Department of Internal Medicine, Dongtan Sacred-Heart Hospital, Hallym University Medical Center, College of Medicine, Hallym University, Hwasung, South Korea
The fibroblast growth factor-4 receptor (FGFR4) is a member of receptor tyrosine kinase. The FGFR4 Gly388Arg polymorphism in the transmembrane domain of the receptor has been shown to increase genetic susceptibility to cancers. However, its prognostic impact in cancer patients still remains controversial. Herein, we performed this meta-analysis to evaluate the clinicopathological and prognostic impacts of the FGFR4 Gly388Arg polymorphism in patients with cancer. We carried out a computerized extensive search using PubMed, Medline, and Ovid Medline databases up to July 2021. From 44 studies, 11,574 patients were included in the current meta-analysis. Regardless of the genetic models, there was no significant correlation of the FGFR4 Gly388Arg polymorphism with disease stage 3/4. In the homozygous model (Arg/Arg vs. Gly/Gly), the Arg/Arg genotype tended to show higher rate of lymph node metastasis compared with the Gly/Gly genotype (odds ratio = 1.21, 95% confidence interval (CI): 0.99-1.49, p = 0.06). Compared to patients with the Arg/Gly or Arg/Arg genotype, those with the Gly/Gly genotype had significantly better overall survival (hazard ratios (HR) = 1.19, 95% CI: 1.05-1.35, p = 0.006) and disease-free survival (HR = 1.25, 95% CI: 1.03-1.53, p = 0.02). In conclusion, this meta-analysis showed that the FGFR4 Gly388Arg polymorphism was significantly associated with worse prognosis in cancer patients. Our results suggest that this polymorphism may be a valuable genetic marker to identify patients at higher risk of recurrence or mortality.
Introduction
The fibroblast growth factor receptors (FGFRs), a subfamily of transmembrane receptor tyrosine kinase (RTK), composed of four related members (FGFR1-4) (1, 2). The activation of FGFR pathway by binding of various ligands to FGFRs triggers several downstream cascades and then activates multiple signal transduction pathways, including the STAT, PI3K/Akt, Ras/Raf/MapK, and phospholipase Cγ (1, 3–5). These signaling pathways regulate a variety of cellular functions such as cell survival, proliferation, migration, differentiation, angiogenesis, and epithelial-mesenchymal transition, and can thereby involve in carcinogenesis (4–6). Actually, numerous studies have demonstrated the aberrant activation of FGFR signaling in carcinogenesis. A recent analysis of 4,853 tumors by the next-generation sequencing has revealed that 7% of cancers carry the FGFR aberrations, including gene amplifications (66%), mutations (26%), and rearrangements (8%) (7). The cancer types most commonly affected were urothelial cancer, breast cancer (BC), endometrial cancer, squamous cell lung cancer, ovarian cancer, carcinoma of unknown primary, glioma and cholangiocarcinoma.
FGFR4 is frequently overexpressed in various types of cancer. It is a highly versatile protein with more than 20 known ligands (8). Specific functions of FGFR4 in carcinogenesis still remain unknown. Whereas Falvella et al. reported downregulation of FGFR4 expression in lung adenocarcinomas (ADC) (9), Sahadevan et al. indicated that FGFR4 expression was upregulated in prostate cancer (PC) (10). The FGFR4 gene is mapped to chromosome 5 (5q35.1) and is highly polymorphic (11). Single-nucleotide polymorphisms (SNP) are the most common genetic variation, representing 90% of sequence differences with an overall frequency of 1 per 1,000 bases. A common nonsynonymous SNP at codon 388 (rs351855 G>A) located in exon 9, which resulted in the substitution of arginine for glycine (Gly388Arg), was identified in the transmembrane domain of FGFR4 receptor (12). Many researchers have reported that the FGFR4 rs351855 G>A polymorphism is involved with the development of various types of cancer, including BC (12–14), PC (15, 16), colorectal cancer (CRC) (12, 13, 17), lung cancer (LC) (18–20), hepatocellular carcinoma (HCC) (21, 22), gastric cancer (GC) (23), head and neck squamous cell carcinoma (HNSCC) (24–26), and cervical cancer (27, 28). In the last decade, several meta-analyses were conducted to find that the FGFR4 Gly388Arg polymorphism was associated with increased risk of some cancers (29–33). In addition, the FGFR4 Arg388 allele has been linked to advanced stage and more frequent lymph node (LN) metastases than the wild-type homozygote (Gly/Gly) in the cohorts of CRC (12), BC (12, 34), PC (15), and LC (18). It has also been implicated in reducing disease-free survival or overall survival in patients with BC (12, 35), HNSCC (24, 25, 36), GC (37, 38), PC (39), CRC (40), and LC (9, 41). Interestingly, Serra et al. reported that the FGFR4 Arg388 allele significantly reduced the response to everolimus, an immunosuppressant medication in patients with pancreatic neuroendocrine tumors (panNET) (42). In addition, Thussbas et al. reported that FGFR4 Arg388 was significantly associated with shorter disease-free survival or overall survival among BC patients receiving adjuvant systemic therapy (35). However, other researchers have reported contrasting results in several cancer types (43–46). There have also been many studies that failed to observe any significant contribution of the FGFR4 Gly388Arg SNP to the clinicopathological parameters or prognosis in patients with cancer (13, 14, 16, 17, 21, 23, 26, 27, 35, 47–65). Therefore, the prognostic impact of the FGFR4 Gly388Arg polymorphism in cancer patients still remains controversial.
In 2010, Frullanti et al. conducted a meta-analysis of 21 studies to evaluate the role of the FGFR4 Gly388Arg polymorphism as a prognostic factor in cancers. They found a statistically significant association of the FGFR4 Arg388 allele and overall survival (hazard ratio (HR) = 1.21, 95% confidence interval (CI) 1.05 – 1.40) and LN involvement (odds ratio (OR) = 1.33, 95% CI 1.01-1.74) (66). Because of a limited number of eligible articles, however, they only included three or less studies in the subgroup analysis according to the primary tumor type. Given the amount of accumulated data thereafter, an updated quantitative synthesis has been deemed worthy. Herein, we performed this meta-analysis to evaluate the clinicopathological and prognostic impacts of the FGFR4 Gly388Arg polymorphism in patients with cancer.
Methods
Publication Search Strategy
This meta-analysis was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (67). We considered all studies that examined the clincopathological or prognostic value of the FGFR4 Gly388Arg polymorphism in any types of cancer. We carried out a computerized extensive search using PubMed, Medline, and Ovid Medline databases up to July 2021. The search used the following keywords variably combined: ‘fibroblast growth factor receptor 4 or FGFR4’, ‘polymorphism or SNP’, ‘Gly388Arg or rs351855’ ‘prognosis or survival’ and ‘cancer or carcinoma or tumor.’ The ‘snowball’ method was adopted to identify additional relevant articles and the reference lists of identified articles were hand-searched (68). In case of duplicate publication, the recent paper was selected.
Eligible Criteria
Studies should meet the following eligible criteria: (i) prospective or retrospective cohort study; (ii) study investigating the association of the FGFR4 Gly388Arg polymorphism with clinicopathological features (LN metastasis or disease stage) or survival outcomes (disease-free survival or overall survival); (iii) the use of adequate method to assess the FGFR4 Gly388Arg SNP; (iv) adequate data to estimate OR with 95% confidence interval (CI) for pathological parameters and/or HR with 95% CIs for survival; (v) study published only in peer-reviewed journal; and (vi) article written in English.
Data Extraction
Two investigators (STP and SYJ), working independently and in parallel, screened the literature and extracted from the eligible articles according to the inclusion criteria. The following data were collected from each article and recorded using the predesigned data-collection form: first author, year of publication, country, ethnicity, inclusion period, sample size, cancer type, genotyping method, genotype counts for the FGFR4 Gly388Arg polymorphism, LN status, disease stage, survival outcomes (disease-free survival or overall survival), and ORs with 95% CIs for pathological parameters and HRs with their 95% CIs for survival outcomes. When both univariate and multivariate analysis were performed for survival times, the HR with 95% CI from multivariate analysis was selected. Any conflicts were resolved by discussion, with input from the other investigators (JHK and HSK).
Statistical Analyses
The strength of the association between the FGFR4 Gly388Arg polymorphism and pathological findings was estimated by the ORs with their 95% CIs in the four genetic models: homozygous [Arg/Arg (AA) vs. Gly/Gly (GG)]; heterozygous (Arg/Gly (AG) vs. GG)); recessive (AA vs. AG+GG); and dominant (AG+AA vs. GG). For the survival analyses, HRs with 95% CIs according to the FGFR4 Gly388Arg polymorphism status were combined. Statistical values were directly obtained from the original articles. If HRs with their 95% CIs were not reported, the Engauge Digitizer software was utilized to calculate them from the corresponding data and Kaplan-Meier curves. The RevMan version 5.4 software was used to combine the data. The heterogeneity across studies was assessed by the Q statistic and I2 inconsistency test. If significant heterogeneity was detected (p < 0.1 or I2 > 50%), the random-effects model was selected. Otherwise (p ≥ 0.1 and I2 ≤ 50%), the fixed-effects model was used. Statistical significance of the pooled HR or OR was determined by Z test. The combined OR or HR > 1.0 implies that cancers harboring the FGFR4 Gly388Arg polymorphism had worse clinicopathological features or survival, respectively.
Publication biases were evaluated by the Begg’s funnel plot (69) and the Egger’s linear regression test (70). All the statistics were two-sided, with p-value < 0.05 considered significant.
Results
Results of Search
The flow diagram of search process is shown in Figure 1. Except for duplicates, a total of 190 relevant articles were initially retrieved, but 103 of them were excluded after careful screening of the titles and abstracts. Of the remaining 87 potentially eligible studies, 43 which did not meet the eligible criteria were further excluded. Finally, 44 studies were selected for the qualitative synthesis (9, 12–18, 21, 23–27, 34–45, 47–53, 55–65).
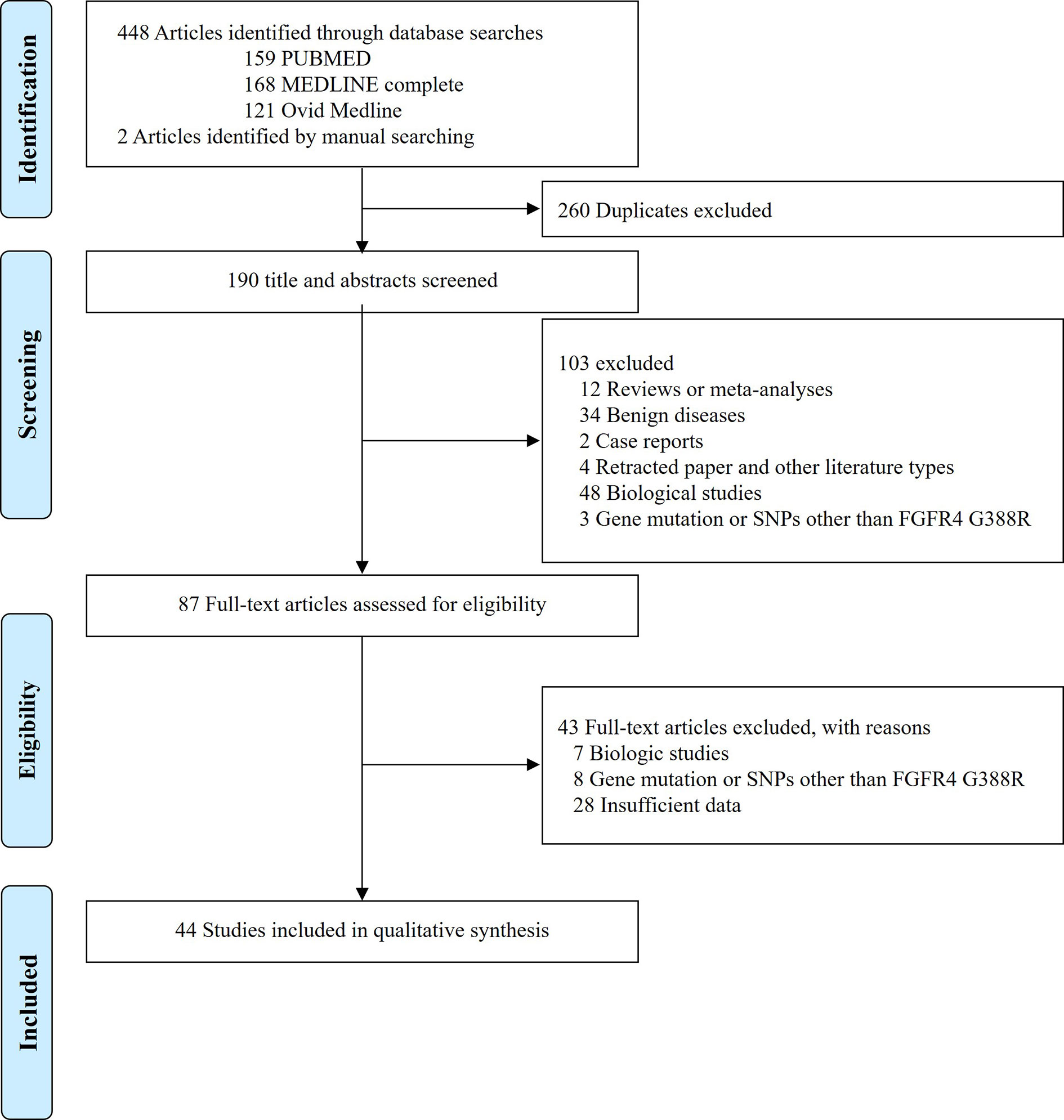
Figure 1 Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) flow diagram showing the selection process of the included studies.
Characteristics of the Included Studies
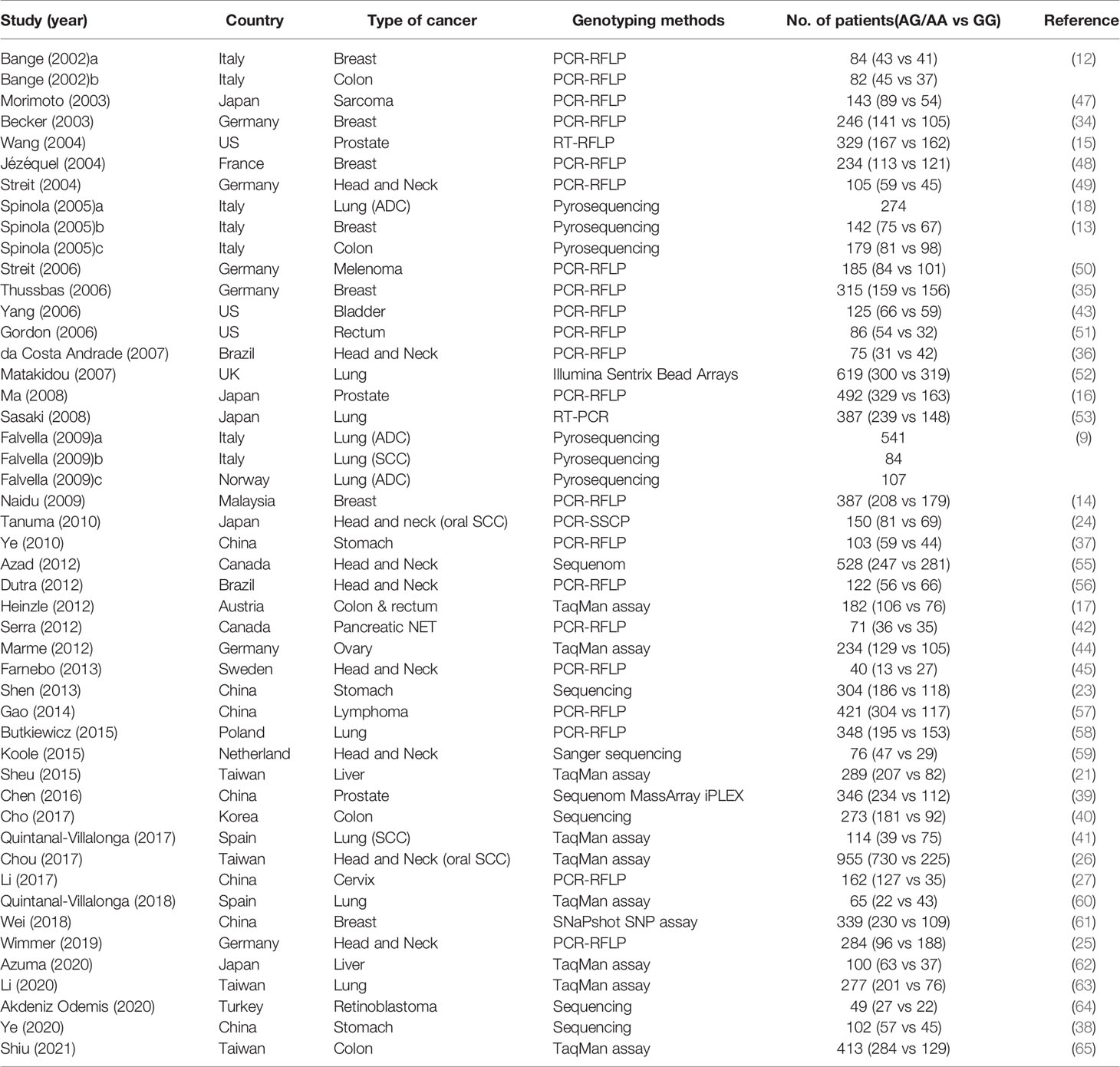
The main characteristics and clinicopathological findings of the included studies are summarized in Table 1 and its supplement. In total, data were obtained on 11,574 subjects from the 44 included studies. Studies were more commonly conducted in Western countries (26 studies). Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method was most frequently used to assess the FGFR4 Gly388Arg polymorphism status. Most studies used formalin-fixed and paraffin-embedded tissue to extract DNA, except for some utilizing fresh frozen tumor tissue or peripheral blood. The impact of the FGFR4 Gly388Arg SNP was most often studied in HNSCC (9 studies), followed by LC (8 studies), BC (7 studies), and CRC (6 studies). Other investigated tumor types were GC, PC, HCC, retinoblastoma, sarcoma, melanoma, lymphoma, bladder cancer, ovarian cancer, cervical cancer, and panNET. Twenty-four studies analyzed the pathological or prognostic parameters according to the four genetic models. Seven studies had a small sample size with less than 100 subjects in total (36, 42, 45, 51, 59, 60, 64), and most studies used univariate statistical method to compare survival times, except for seven studies (37, 39, 40, 44, 56, 58, 61).
Clinicopathological Impact of the FGFR4 Gly388Arg Polymorphism
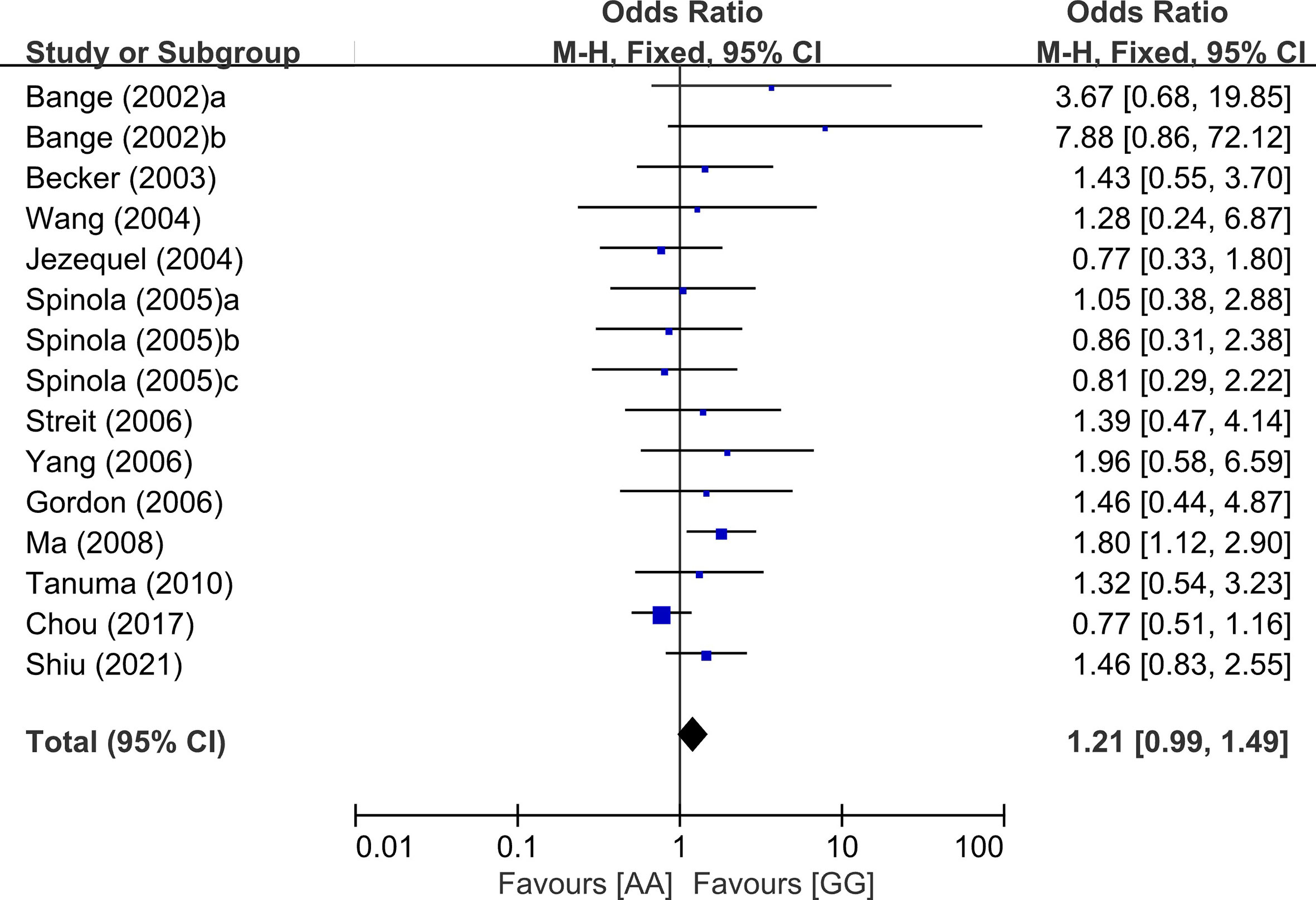
From 24 studies, 6,157 patients were included in the meta-analysis of ORs with 95% CIs for LN metastasis. The odds of LN metastasis at the time of diagnosis were not different between patients with the GG genotype and those with the AG or AA genotype (OR = 1.08, 95% CI: 0.91-1.29, p = 0.39, random-effects, Table 2, forest plot not shown). In the homozygous model (amino acid: AA vs. GG), the AA genotype tended to show higher rate of LN metastasis compared with the GG genotype. (OR = 1.21, 95% CI: 0.99-1.49, p = 0.06, fixed-effects) (Figure 2 and Table 2).
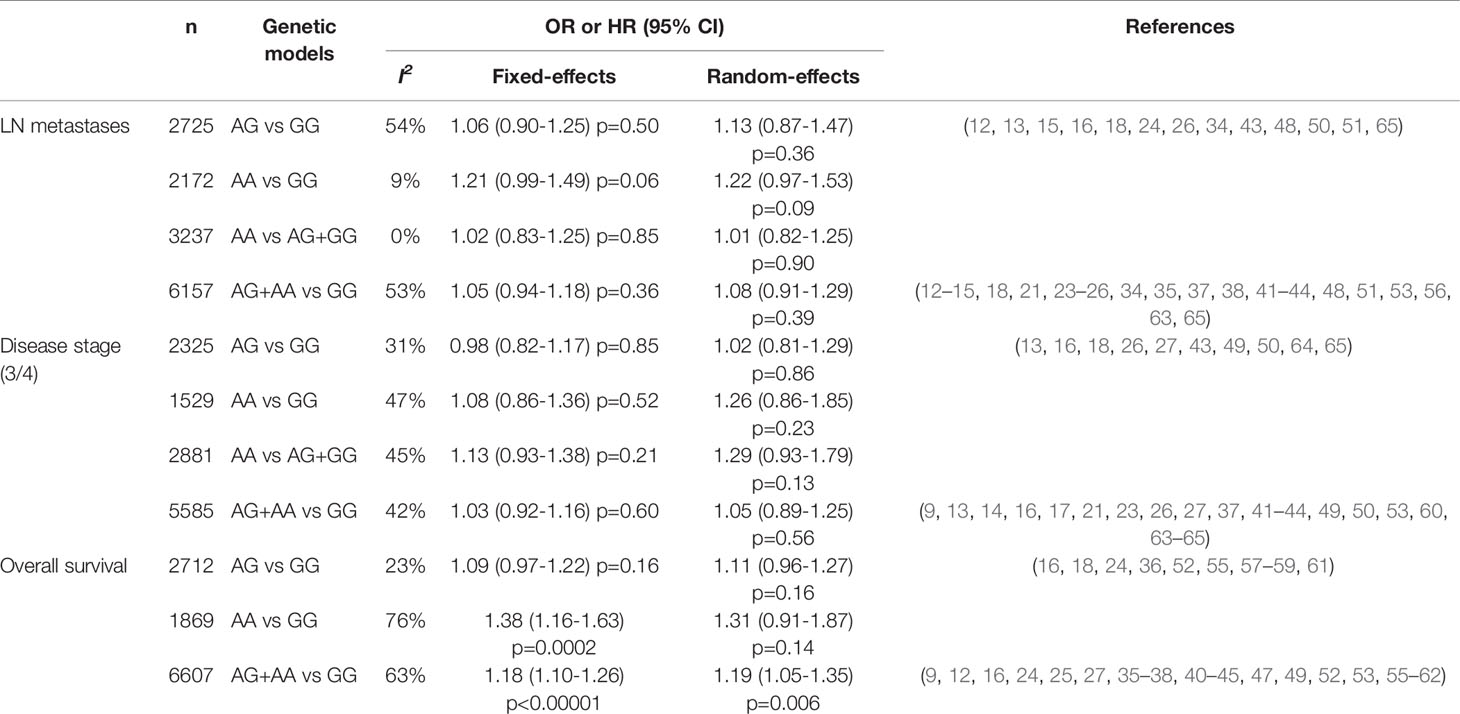
Table 2 The meta-analyses of clinicopathological and prognostic parameters among the genetic models.
From 21 studies, 5,585 patients were pooled to assess the effect of the FGFR4 Gly388Arg SNP on disease stage. There was no significant difference of advanced disease (stage 3/4) between patients with the GG genotype and those with the AG or AA genetic type (OR = 1.05, 95% CI: 0.89-1.25, p = 0.56, random-effects, forest plot not shown). The association of the FGFR4 Gly388Arg SNP with disease stage was not significant in the other genetic models, either (Table 2).
Prognostic Significance of the FGFR4 Gly388Arg Polymorphism
From 28 studies, 6,607 patients were included in the meta-analysis of HRs for overall survival. Patients with the GG genotype tended to show better overall survival, compared to those with the AG (HR = 1.09, 95% CI: 0.97-1.22, p = 0.16, fixed-effects) or those with the AA genotype (HR = 1.31, 95% CI: 0.91-1.87, p = 0.14, random-effects) (Table 2). Compared to patients with the AG or AA genotypes, those with the wild-type homozygote (GG) had significantly longer overall survival (HR = 1.19, 95% CI: 1.05-1.35, p = 0.006, random-effects) (Figure 3A).
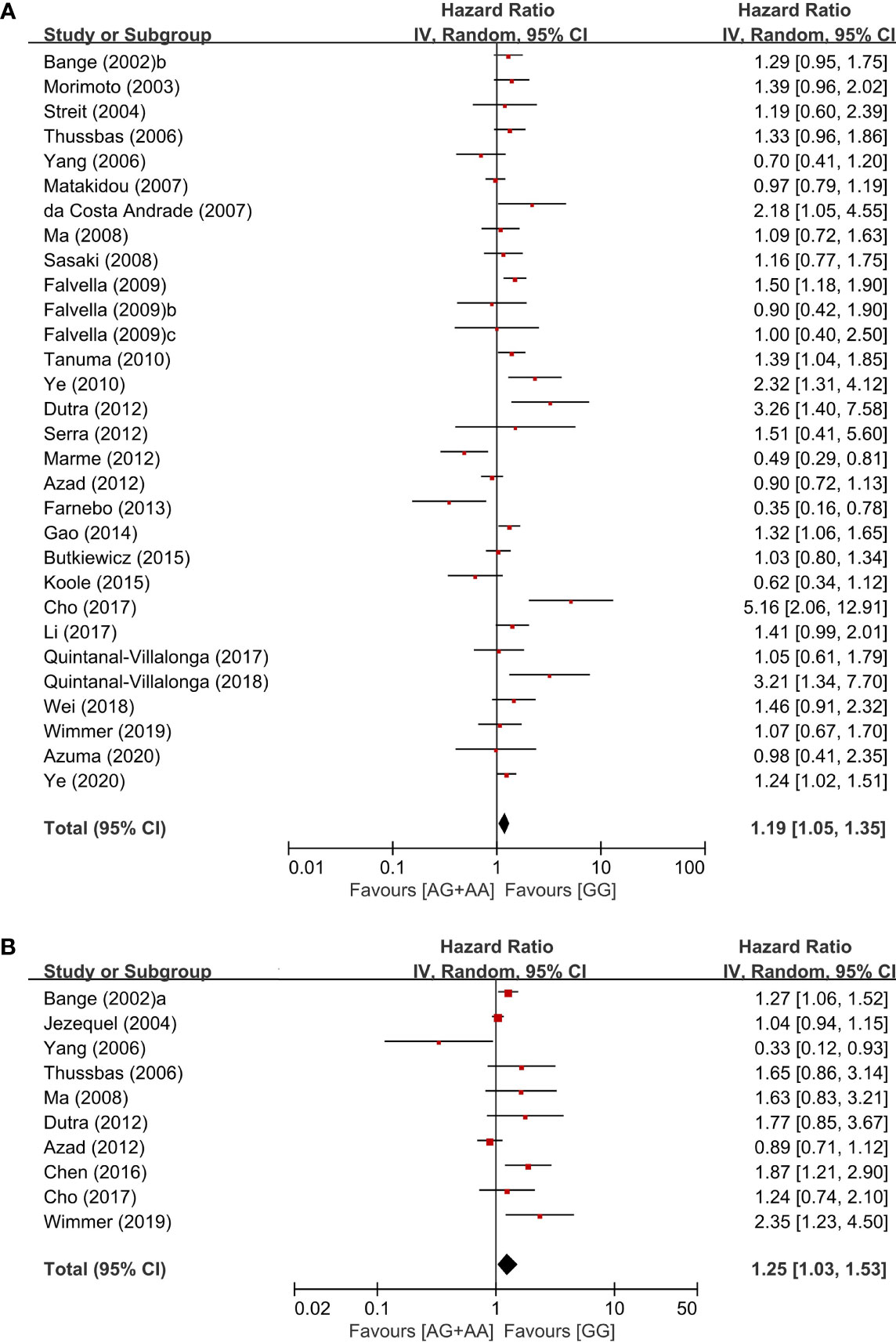
Figure 3 Forest plots for overall survival (A) and disease-free survival (B) in the dominant model (AG+AA vs. GG).
From 10 studies, 2,803 patients were included in the pooled analysis to evaluate the correlation between the FGFR4 Gly388Arg SNP and disease-free survival. Patients with the GG genotype showed significantly longer disease-free survival than those with the AG or AA genotype (HR = 1.25, 95% CI: 1.03-1.53, p = 0.02, random-effects) (Figure 3B).
Subgroup Analysis According to the Primary Site and Ethnicity
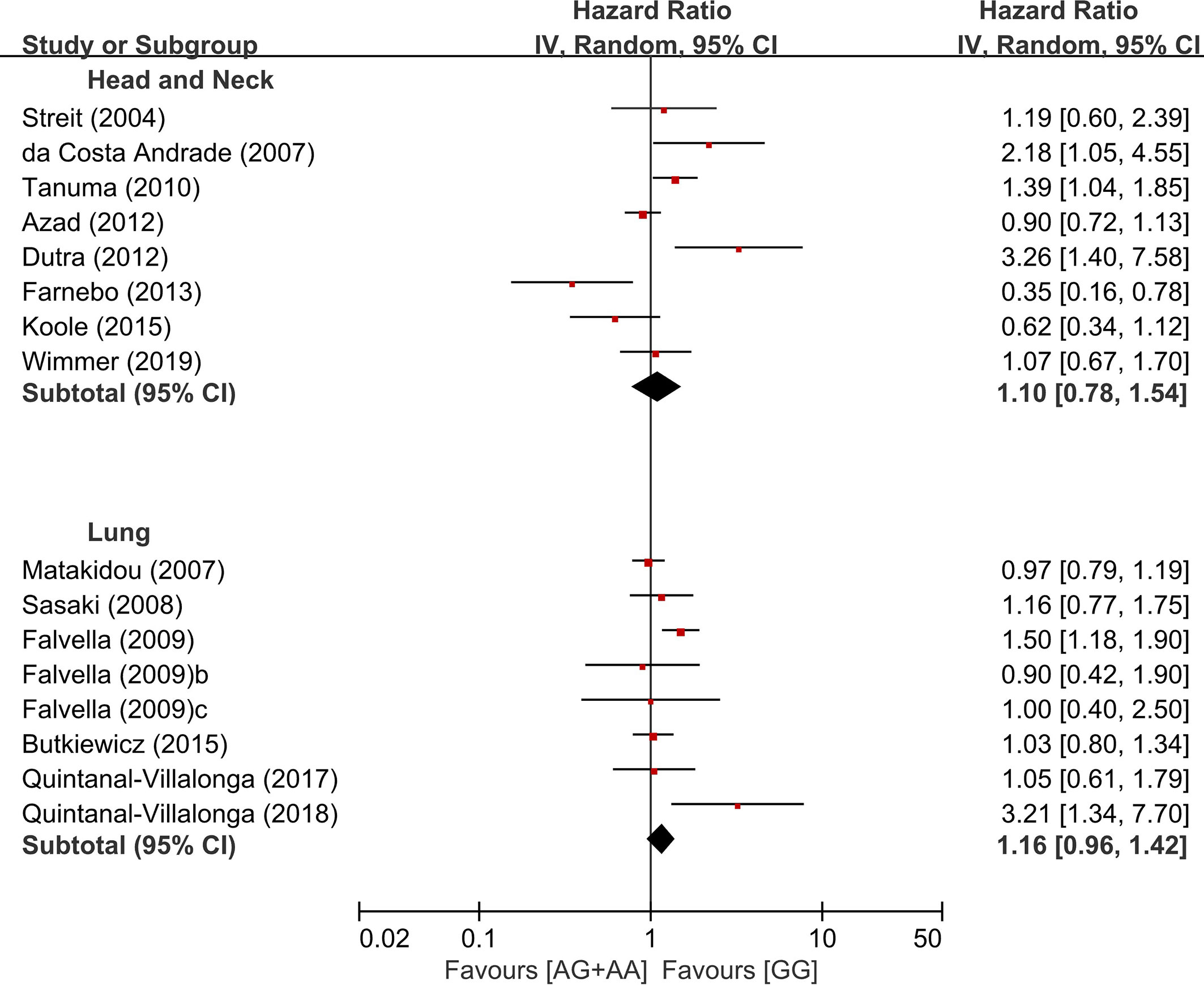
In the subgroup analysis according to the primary sites, there was no significant overall survival differences between the genotypes of the dominant model (AG+AA vs. GG) in patients with HNSCC (HR = 1.10, 95% CI: 0.78-1.54, p=0.59, random-effects) or LC (HR = 1.16, 95% CI: 0.96-1.42, p = 0.13, random-effects) (Figure 4). We did not perform the subgroup analyses for other types of cancer in which only two or less studies were included.
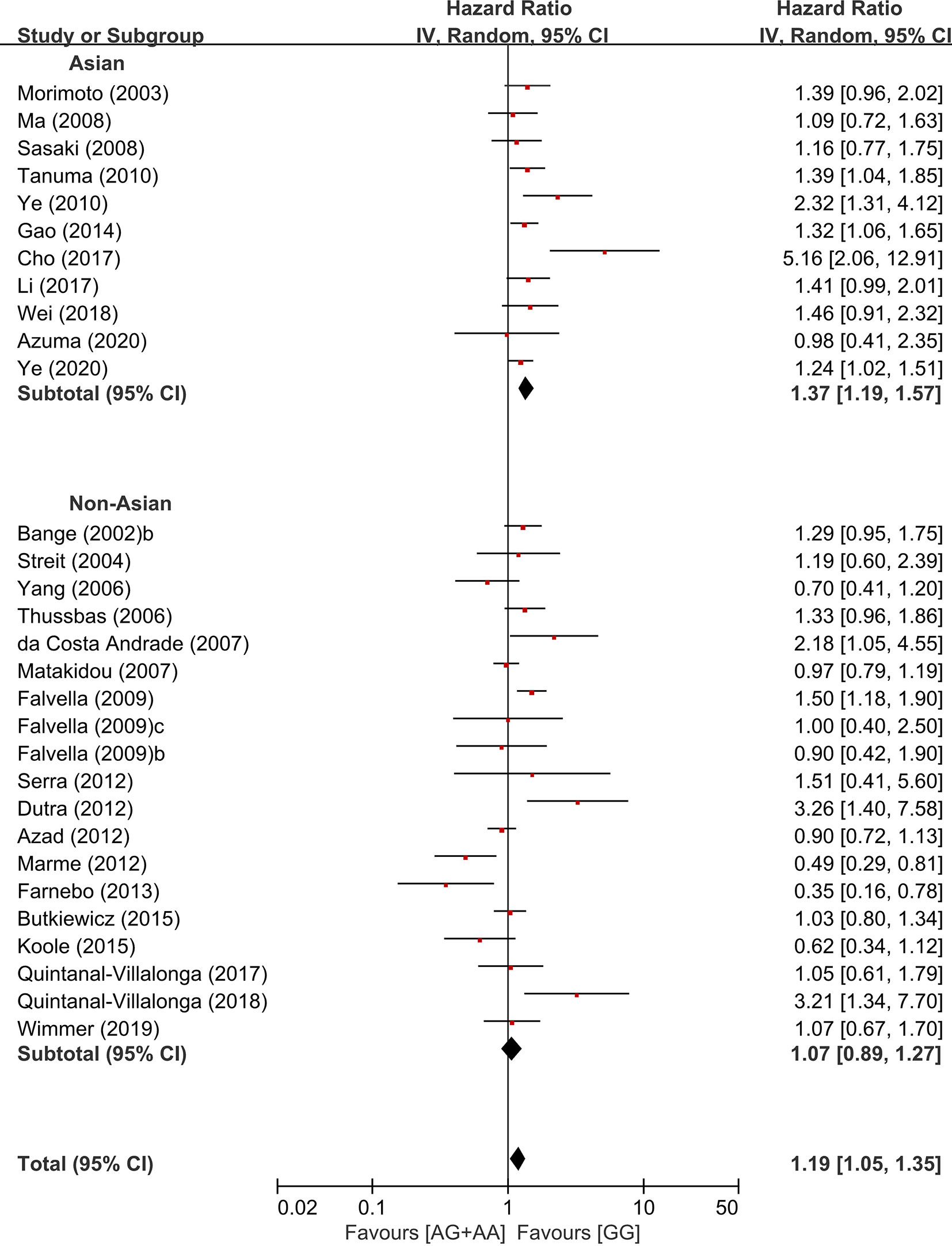
In the stratification by the ethnicity, there was a significant association of the common homozygous genotype (GG) with better overall survival in the Asian population (HR = 1.37, 95% CI: 1.19-1.57, p < 0.00001, random-effects), but not in non-Asians (HR = 1.07, 95% CI: 0.89-1.27, p = 0.47, random-effects) (Figure 5).
Publication Bias
The funnel plots of LN metastasis (Figure 6A), disease stage (Figure 6B), overall survival (Figure 6C), and disease-free survival (Figure 6D) were graphically symmetric. The Egger’s test indicated no evidence of substantial publication bias for LN metastasis (p = 0.134), stage 3/4 (p = 0.078), overall survival (p = 0.410) and disease-free survival (p = 0.696).
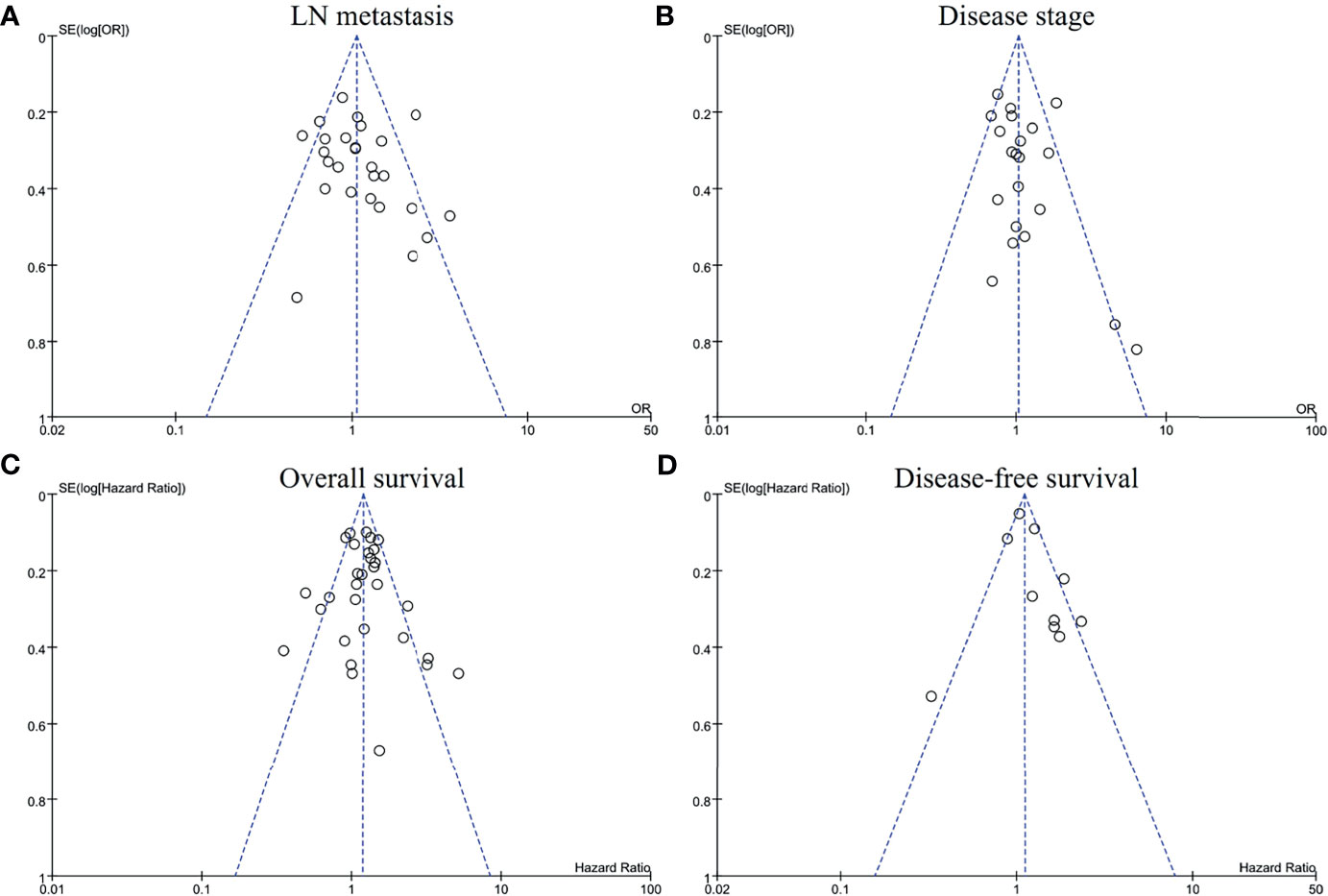
Figure 6 Funnel plots for publication bias regarding lymph node metastasis (A), disease stage (B), overall survival (C), and disease-free survival (D).
Discussions
The FGFR4 Gly388Arg polymorphism results in the replacement of the glycine residue with a charged arginine residue in the transmembrane domain of the receptor, which consequently exposes proximal STAT3 binding site and then enhances STAT3 signal to stimulate cell proliferation (71). Thus, the FGFR4 Arg388 variants may promote tumorigenesis by enhancing cell motility, invasiveness, and proliferation. The FGFR4 gene rs351855 G>A polymorphism has been known to confer increased genetic susceptibility to cancers (29–33). Many researchers also examined the relationship between the FGFR4 gene SNP and its pathological or prognostic roles among diverse cancer types. However, the results were inconsistent. In the current meta-analysis, we evaluated the clinicopathological and prognostic significance of the FGFR4 Gly388Arg polymorphism in cancers. The study included 11,574 patients with various types of cancer from the 44 eligible articles.
In the current study, the FGFR4 Gly388Arg SNP failed to show a significant correlation with disease stage 3/4 at the time of diagnosis, regardless of the genetic models. In the homozygous model (AA vs. GG), patients with the AA genotype showed a tendency of higher rate of LN metastases than those with the GG genotype (OR = 1.21, 95% CI: 0.99-1.49, p = 0.06). In terms of survival times, patients with the GG genotype tended to show longer overall survival, compared to those with the AG (HR = 1.09, 95% CI: 0.97-1.22, p = 0.16) or those with the AA genotype (HR = 1.31, 95% CI: 0.91-1.87, p = 0.14). In the dominant model (AG/AA. vs GG), moreover, the Arg388 allele carriers showed a significantly increased hazard of worse overall survival (HR = 1.19, 95% CI: 1.05-1.35, p = 0.006) and disease-free survival (HR = 1.25, 9% CI: 1.03-1.53, p = 0.02). The previous meta-analysis in 2011 by Frullanti et al. reported that there was a significant association between the AA genotype and LN involvement (OR = 1.33, 95% CI: 1.01-1.74, p = 0.04) (66). They also found that the Arg388 allele carriers showed worse prognosis compared to the homozygous carriers of the common Gly388 allele (HR = 1.21, 95% CI: 1.05-1.40, p = 0.01) (66). These results indicate that the FGFR4 Gly388Arg polymorphism is a potential genetic marker associated with worse prognosis in cancer patients.
The association of the FGFR4 Arg388 polymorphism with the susceptibility to cancer has mainly been described in PC and BC (15, 16, 29–31, 33). In the meta-analysis of 27 studies comprising 8,682 cases by Xiong et al., interestingly, the FGFR4 rs351855 G>A polymorphism increased the risk of PC and BC, but decreased the susceptibility of LC (33). This finding indicates that the FGFR4 Arg388 SNP might has opposite effects on different types of cancer, suggesting that this polymorphism may modify cancer susceptibility in a tissue specific manner. Therefore, the prognostic role of the FGFR4 Arg388 polymorphism may also be different among different types of cancer. In our subgroup analyses using two most common primary sites, however, there was no significant overall survival difference between the GG and AG/AA genotypes in both HNSCC (HR = 1.10, 95% CI: 0.78-1.54, p=0.65) and LC (HR = 1.16, 95% CI: 0.96-1.42, p = 0.13). In HNSCC, various tumor locations among studies may explain the negative result on the prognostic value of the FGFR4 Arg388 SNP, since different anatomical locations show different clinical and molecular characteristics (72). In terms of LC, the prognostic impact of the FGFR4 Arg388 variant may be different among histologic subtypes. Indeed, the prognostic role of this SNP was more frequently observed in patients with ADC (9, 18). In the current study, unfortunately, we could not perform the subgroup analysis according to the tumor location in HNSCC and histologic subtype in LC due to a limited number of relevant articles.
The previous studies have reported that there is significantly different in the prevalence of the FGFR4 Arg388 allele between Asians (37.2-40.1%) and Caucasians (29.5–30.4%) (29, 33). The higher frequency of the Arg388 allele in Asian populations might lead to a higher statistical power of studies in Asians than in non-Asians. However, the meta-analysis by Xu at al. reported that the association between Arg variant genotype and increase risk of cancer was significant only in Asians, not in Caucacians or Africans (33). In another meta-analysis by Liwei et al., the significant association between Arg variant genotype and susceptibility of PC was observed among European ethic descents, not among African-Americans (31). When we stratified by the ethnicity in the current study, the FGFR4 Arg388 allele was associated with worse overall survival in Asian population (HR = 1.37, 95% CI: 1.19-1.57, p < 0.00001), but not in non-Asians (HR = 1.07, 95% CI: 0.89-1.27, p = 0.47). These findings suggest that the genetic effect of the FGFR4 Gly388Arg SNP may be different among the ethnicities.
Beside the FGFR4 rs351855 G>A polymorphism, there are also other SNPs or mutations of FGFR4 which may affect the risk of cancer development or prognosis of cancer patients. The recent meta-analysis by Moazeni-Roodi et al. revealed that the FGFR4 rs1966265 C>T polymorphism significantly reduced the risk of cancer in the recessive model (TT vs CT+CC) and the rs7708357 G>A variant was significantly associated with increased cancer development in the dominant model (AG+AA vs GG) (32). The Y367C FGFR4 mutation in the extracellular juxtamembrane domain promotes the FGFR4 dimerization on the cell surface and thereby leads to ligand-independent activation of downstream signaling pathways (73). In addition, the mutations in FGFR4 kinase domain such as N535K and V550E cause receptor autophospholyation and then activate the STAT3 signal pathway (73). Some FGFR4 mutations (N535K, V548M and V550L) were reported to be relatively resistant to tyrosine kinase inhibitors (74). However, the clinical and pathological significance of these genetic variations involving FGFR4 are still needed to be investigated in further studies.
There were some inherent limitations of this meta-analysis. First, there might be a selection bias since we were only able to acquire data from published articles written in English. Second, most studies were performed retrospectively and therefore, may carry the biases of the retrospective design. Third, considerable number of included studies had a relatively small sample size, and most studies utilized univariate statistical method to compare survival outcomes. Forth, because of a paucity of relevant articles, we could not perform the subgroup analyses in other types of cancers than HNSCC and LC. Finally, there was a substantial heterogeneity in the pooled outcomes, which might weaken reliability of the meta-analysis although the random-effects model was adopted.
In conclusion, this meta-analysis elucidated that FGFR4 Gly388Arg polymorphism was associated with worse prognosis in cancer patients. Our results suggest that this SNP may be a valuable genetic marker to identify patients at higher risk of recurrence or mortality. Considering the limitations of the current study, however, large prospective researches with genotyping of the whole FGFR4 locus are warranted to reveal the clinicopathological and prognostic roles of the FGFR4 Gly388Arg SNP among various types of cancer, histology, and ethnicity.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
JHK and HSK conceived and designed the study. STP and SYJ searched the literatures and extracted the data. HSK and STP carried out the statistical analyses and data interpretation. JHK and HJJ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) and funded by the Korean government, Ministry of Science and ICT (MSIT) (NRF- 2020R1G1A1005483).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.762528/full#supplementary-material
References
1. Eswarakumar VP, Lax I, Schlessinger J. Cellular Signaling by Fibroblast Growth Factor Receptors. Cytokine Growth Factor Rev (2005) 16(2):139–49. doi: 10.1016/j.cytogfr.2005.01.001
2. Burke D, Wilkes D, Blundell TL, Malcolm S. Fibroblast Growth Factor Receptors: Lessons From the Genes. Trends Biochem Sci (1998) 23(2):59–62. doi: 10.1016/s0968-0004(97)01170-5
3. Haugsten EM, Wiedlocha A, Olsnes S, Wesche J. Roles of Fibroblast Growth Factor Receptors in Carcinogenesis. Mol Cancer Res (2010) 8(11):1439–52. doi: 10.1158/1541-7786.MCR-10-0168
4. Dailey L, Ambrosetti D, Mansukhani A, Basilico C. Mechanisms Underlying Differential Responses to FGF Signaling. Cytokine Growth Factor Rev (2005) 16(2):233–47. doi: 10.1016/j.cytogfr.2005.01.007
5. Turner N, Grose R. Fibroblast Growth Factor Signalling: From Development to Cancer. Nat Rev Cancer (2010) 10(2):116–29. doi: 10.1038/nrc2780
6. Grose R, Dickson C. Fibroblast Growth Factor Signaling in Tumorigenesis. Cytokine Growth Factor Rev (2005) 16(2):179–86. doi: 10.1016/j.cytogfr.2005.01.003
7. Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR Landscape in Cancer: Analysis of 4,853 Tumors by Next-Generation Sequencing. Clin Cancer Res (2016) 22(1):259–67. doi: 10.1158/1078-0432.CCR-14-3212
8. Powers CJ, McLeskey SW, Wellstein A. Fibroblast Growth Factors, Their Receptors and Signaling. Endocr Relat Cancer (2000) 7(3):165–97. doi: 10.1677/erc.0.0070165
9. Falvella FS, Frullanti E, Galvan A, Spinola M, Noci S, De Cecco L, et al. FGFR4 Gly388Arg Polymorphism may Affect the Clinical Stage of Patients With Lung Cancer by Modulating the Transcriptional Profile of Normal Lung. Int J Cancer (2009) 124(12):2880–5. doi: 10.1002/ijc.24302
10. Sahadevan K, Darby S, Leung HY, Mathers ME, Robson CN, Gnanapragasam VJ. Selective Over-Expression of Fibroblast Growth Factor Receptors 1 and 4 in Clinical Prostate Cancer. J Pathol (2007) 213(1):82–90. doi: 10.1002/path.2205
11. Lang L, Teng Y. Fibroblast Growth Factor Receptor 4 Targeting in Cancer: New Insights Into Mechanisms and Therapeutic Strategies. Cells (2019) 8(1):31–43. doi: 10.3390/cells8010031
12. Bange J, Prechtl D, Cheburkin Y, Specht K, Harbeck N, Schmitt M, et al. Cancer Progression and Tumor Cell Motility Are Associated With the FGFR4 Arg(388) Allele. Cancer Res (2002) 62(3):840–7.
13. Spinola M, Leoni VP, Tanuma J, Pettinicchio A, Frattini M, Signoroni S, et al. FGFR4 Gly388Arg Polymorphism and Prognosis of Breast and Colorectal Cancer. Oncol Rep (2005) 14(2):415–9. doi: 10.3892/or.14.2.415
14. Naidu R, Har YC, Taib NA. Polymorphism of FGFR4 Gly388Arg Does Not Confer an Increased Risk to Breast Cancer Development. Oncol Res (2009) 18(2–3):65–71. doi: 10.3727/096504009789954609
15. Wang J, Stockton DW, Ittmann M. The Fibroblast Growth Factor Receptor-4 Arg388 Allele Is Associated With Prostate Cancer Initiation and Progression. Clin Cancer Res (2004) 10(18 Pt 1):6169–78. doi: 10.1158/1078-0432.CCR-04-0408
16. Ma Z, Tsuchiya N, Yuasa T, Inoue T, Kumazawa T, Narita S, et al. Polymorphisms of Fibroblast Growth Factor Receptor 4 Have Association With the Development of Prostate Cancer and Benign Prostatic Hyperplasia and the Progression of Prostate Cancer in a Japanese Population. Int J Cancer (2008) 123(11):2574–9. doi: 10.1002/ijc.23578
17. Heinzle C, Gsur A, Hunjadi M, Erdem Z, Gauglhofer C, Stattner S, et al. Differential Effects of Polymorphic Alleles of FGF Receptor 4 on Colon Cancer Growth and Metastasis. Cancer Res (2012) 72(22):5767–77. doi: 10.1158/0008-5472.CAN-11-3654
18. Spinola M, Leoni V, Pignatiello C, Conti B, Ravagnani F, Pastorino U, et al. Functional FGFR4 Gly388Arg Polymorphism Predicts Prognosis in Lung Adenocarcinoma Patients. J Clin Oncol (2005) 23(29):7307–11. doi: 10.1200/JCO.2005.17.350
19. Fang HM, Tian G, Zhou LJ, Zhou HY, Fang YZ. FGFR4 Genetic Polymorphisms Determine the Chemotherapy Response of Chinese Patients With Non-Small Cell Lung Cancer. Acta Pharmacol Sin (2013) 34(4):549–54. doi: 10.1038/aps.2012.206
20. Ture M, Yakut T, Deligonul A, Karkucak M, Sag SO, Hartavi M, et al. Investigation of FGFR4 (Gly388Arg) Gene Polymorphism in Primary Lung Cancer Patients. Int J Hum Genet (2015) 15(1):7–12. doi: 10.1080/09723757.2015.11886245
21. Sheu MJ, Hsieh MJ, Chiang WL, Yang SF, Lee HL, Lee LM, et al. Fibroblast Growth Factor Receptor 4 Polymorphism Is Associated With Liver Cirrhosis in Hepatocarcinoma. PloS One (2015) 10(4):e0122961. doi: 10.1371/journal.pone.0122961
22. Yang Y, Zhou Y, Lu M, An Y, Li R, Chen Y, et al. Association Between Fibroblast Growth Factor Receptor 4 Polymorphisms and Risk of Hepatocellular Carcinoma. Mol Carcinog (2012) 51(7):515–21. doi: 10.1002/mc.20805
23. Shen YY, Lu YC, Shen DP, Liu YJ, Su XY, Zhu GS, et al. Fibroblast Growth Factor Receptor 4 Gly388Arg Polymorphism in Chinese Gastric Cancer Patients. World J Gastroenterol (2013) 19(28):4568–75. doi: 10.3748/wjg.v19.i28.4568
24. Tanuma J, Izumo T, Hirano M, Oyazato Y, Hori F, Umemura E, et al. FGFR4 Polymorphism, TP53 Mutation, and Their Combinations Are Prognostic Factors for Oral Squamous Cell Carcinoma. Oncol Rep (2010) 23(3):739–44. doi: 10.3892/or_00000692
25. Wimmer E, Ihrler S, Gires O, Streit S, Issing W, Bergmann C. Fibroblast Growth Factor Receptor 4 Single Nucleotide Polymorphism Gly388Arg in Head and Neck Carcinomas. World J Clin Oncol (2019) 10(3):136–48. doi: 10.5306/wjco.v10.i3.136
26. Chou CH, Hsieh MJ, Chuang CY, Lin JT, Yeh CM, Tseng PY, et al. Functional FGFR4 Gly388Arg Polymorphism Contributes to Oral Squamous Cell Carcinoma Susceptibility. Oncotarget (2017) 8(56):96225–38. doi: 10.18632/oncotarget.21958
27. Li YP, Zhang L, Zou YL, Yu Y. Association Between FGFR4 Gene Polymorphism and High-Risk HPV Infection Cervical Cancer. Asian Pac J Trop Med (2017) 10(7):680–4. doi: 10.1016/j.apjtm.2017.07.008
28. Chen TH, Yang SF, Liu YF, Lin WL, Han CP, Wang PH. Association of Fibroblast Growth Factor Receptor 4 Genetic Polymorphisms With the Development of Uterine Cervical Cancer and Patient Prognosis. Reprod Sci (2018) 25(1):86–93. doi: 10.1177/1933719117702250
29. Xu W, Li Y, Wang X, Chen B, Wang Y, Liu S, et al. FGFR4 Transmembrane Domain Polymorphism and Cancer Risk: A Meta-Analysis Including 8555 Subjects. Eur J Cancer (2010) 46(18):3332–8. doi: 10.1016/j.ejca.2010.06.017
30. Xu B, Tong N, Chen SQ, Hua LX, Wang ZJ, Zhang ZD, et al. FGFR4 Gly388Arg Polymorphism Contributes to Prostate Cancer Development and Progression: A Meta-Analysis of 2618 Cases and 2305 Controls. BMC Cancer (2011) 11:84. doi: 10.1186/1471-2407-11-84
31. Liwei L, Chunyu L, Jie L, Ruifa H. Association Between Fibroblast Growth Factor Receptor-4 Gene Polymorphism and Risk of Prostate Cancer: A Meta-Analysis. Urol Int (2011) 87(2):159–64. doi: 10.1159/000329069
32. Moazeni-Roodi A, Sarabandi S, Karami S, Hashemi M, Ghavami S. An Updated Meta-Analysis of the Association Between Fibroblast Growth Factor Receptor 4 Polymorphisms and Susceptibility to Cancer. Biosci Rep (2020) 40(10). doi: 10.1042/BSR20192051
33. Xiong SW, Ma J, Feng F, Fu W, Shu SR, Ma T, et al. Functional FGFR4 Gly388Arg Polymorphism Contributes to Cancer Susceptibility: Evidence From Meta-Analysis. Oncotarget (2017) 8(15):25300–9. doi: 10.18632/oncotarget.15811
34. Becker N, Nieters A, Chang-Claude J. The Fibroblast Growth Factor Receptor Gene Arg388 Allele Is Not Associated With Early Lymph Node Metastasis of Breast Cancer. Cancer Epidemiol Biomarkers Prev (2003) 12(6):582–3.
35. Thussbas C, Nahrig J, Streit S, Bange J, Kriner M, Kates R, et al. FGFR4 Arg388 Allele Is Associated With Resistance to Adjuvant Therapy in Primary Breast Cancer. J Clin Oncol (2006) 24(23):3747–55. doi: 10.1200/JCO.2005.04.8587
36. da Costa Andrade VC, Parise O Jr, Hors CP, de Melo Martins PC, Silva AP, Garicochea B. The Fibroblast Growth Factor Receptor 4 (FGFR4) Arg388 Allele Correlates With Survival in Head and Neck Squamous Cell Carcinoma. Exp Mol Pathol (2007) 82(1):53–7. doi: 10.1016/j.yexmp.2006.05.003
37. Ye Y, Shi Y, Zhou Y, Du C, Wang C, Zhan H, et al. The Fibroblast Growth Factor Receptor-4 Arg388 Allele Is Associated With Gastric Cancer Progression. Ann Surg Oncol (2010) 17(12):3354–61. doi: 10.1245/s10434-010-1323-6
38. Ye Y, Li J, Jiang D, Li J, Xiao C, Li Y, et al. FGFR4 Gly388Arg Polymorphism Affects the Progression of Gastric Cancer by Activating STAT3 Pathway to Induce Epithelial to Mesenchymal Transition. Cancer Res Treat (2020) 52(4):1162–77. doi: 10.4143/crt.2020.138
39. Chen L, Lei Z, Ma X, Huang Q, Zhang X, Zhang Y, et al. Prognostic Significance of Fibroblast Growth Factor Receptor 4 Polymorphisms on Biochemical Recurrence After Radical Prostatectomy in a Chinese Population. Sci Rep (2016) 6:33604. doi: 10.1038/srep33604
40. Cho SH, Hong CS, Kim HN, Shin MH, Kim KR, Shim HJ, et al. FGFR4 Arg388 Is Correlated With Poor Survival in Resected Colon Cancer Promoting Epithelial to Mesenchymal Transition. Cancer Res Treat (2017) 49(3):766–77. doi: 10.4143/crt.2016.457
41. Quintanal-Villalonga A, Carranza-Carranza A, Melendez R, Ferrer I, Molina-Pinelo S, Paz-Ares L. Prognostic Role of the FGFR4-388arg Variant in Lung Squamous-Cell Carcinoma Patients With Lymph Node Involvement. Clin Lung Cancer (2017) 18(6):667–74.e1. doi: 10.1016/j.cllc.2017.05.008
42. Serra S, Zheng L, Hassan M, Phan AT, Woodhouse LJ, Yao JC, et al. The FGFR4-G388R Single-Nucleotide Polymorphism Alters Pancreatic Neuroendocrine Tumor Progression and Response to mTOR Inhibition Therapy. Cancer Res (2012) 72(22):5683–91. doi: 10.1158/0008-5472.CAN-12-2102
43. Yang YC, Lu ML, Rao JY, Wallerand H, Cai L, Cao W, et al. Joint Association of Polymorphism of the FGFR4 Gene and Mutation TP53 Gene With Bladder Cancer Prognosis. Br J Cancer (2006) 95(11):1455–8. doi: 10.1038/sj.bjc.6603456
44. Marme F, Hielscher T, Hug S, Bondong S, Zeillinger R, Castillo-Tong DC, et al. Fibroblast Growth Factor Receptor 4 Gene (FGFR4) 388Arg Allele Predicts Prolonged Survival and Platinum Sensitivity in Advanced Ovarian Cancer. Int J Cancer (2012) 131(4):E586–91. doi: 10.1002/ijc.27329
45. Farnebo L, Tiefenbock K, Ansell A, Thunell LK, Garvin S, Roberg K. Strong Expression of Survivin Is Associated With Positive Response to Radiotherapy and Improved Overall Survival in Head and Neck Squamous Cell Carcinoma Patients. Int J Cancer (2013) 133(8):1994–2003. doi: 10.1002/ijc.28200
46. FitzGerald LM, Karlins E, Karyadi DM, Kwon EM, Koopmeiners JS, Stanford JL, et al. Association of FGFR4 Genetic Polymorphisms With Prostate Cancer Risk and Prognosis. Prostate Cancer Prostatic Dis (2009) 12(2):192–7. doi: 10.1038/pcan.2008.46
47. Morimoto Y, Ozaki T, Ouchida M, Umehara N, Ohata N, Yoshida A, et al. Single Nucleotide Polymorphism in Fibroblast Growth Factor Receptor 4 at Codon 388 Is Associated With Prognosis in High-Grade Soft Tissue Sarcoma. Cancer (2003) 98(10):2245–50. doi: 10.1002/cncr.11778
48. Jezequel P, Campion L, Joalland MP, Millour M, Dravet F, Classe JM, et al. G388R Mutation of the FGFR4 Gene Is Not Relevant to Breast Cancer Prognosis. Br J Cancer (2004) 90(1):189–93. doi: 10.1038/sj.bjc.6601450
49. Streit S, Bange J, Fichtner A, Ihrler S, Issing W, Ullrich A. Involvement of the FGFR4 Arg388 Allele in Head and Neck Squamous Cell Carcinoma. Int J Cancer (2004) 111(2):213–7. doi: 10.1002/ijc.20204
50. Streit S, Mestel DS, Schmidt M, Ullrich A, Berking C. FGFR4 Arg388 Allele Correlates With Tumour Thickness and FGFR4 Protein Expression With Survival of Melanoma Patients. Br J Cancer (2006) 94(12):1879–86. doi: 10.1038/sj.bjc.6603181
51. Gordon MA, Gil J, Lu B, Zhang W, Yang D, Yun J, et al. Genomic Profiling Associated With Recurrence in Patients With Rectal Cancer Treated With Chemoradiation. Pharmacogenomics (2006) 7(1):67–88. doi: 10.2217/14622416.7.1.67
52. Matakidou A, El Galta R, Rudd MF, Webb EL, Bridle H, Eisen T, et al. Further Observations on the Relationship Between the FGFR4 Gly388Arg Polymorphism and Lung Cancer Prognosis. Br J Cancer (2007) 96(12):1904–7. doi: 10.1038/sj.bjc.6603816
53. Sasaki H, Okuda K, Kawano O, Yukiue H, Yano M, Fujii Y. Fibroblast Growth Factor Receptor 4 Mutation and Polymorphism in Japanese Lung Cancer. Oncol Rep (2008) 20(5):1125–30. doi: 10.3892/or_00000119
54. Choi KY, Rho YS, Kwon KH, Chung EJ, Kim JH, Park IS, et al. ECRG1 and FGFR4 Single Nucleotide Polymorphism as Predictive Factors for Nodal Metastasis in Oral Squamous Cell Carcinoma. Cancer Biomark (2012) 12(3):115–24. doi: 10.3233/CBM-130299
55. Azad AK, Bairati I, Samson E, Cheng D, Mirshams M, Qiu X, et al. Validation of Genetic Sequence Variants as Prognostic Factors in Early-Stage Head and Neck Squamous Cell Cancer Survival. Clin Cancer Res (2012) 18(1):196–206. doi: 10.1158/1078-0432.CCR-11-1759
56. Dutra RL, de Carvalho MB, Dos Santos M, Mercante AM, Gazito D, de Cicco R, et al. FGFR4 Profile as a Prognostic Marker in Squamous Cell Carcinoma of the Mouth and Oropharynx. PloS One (2012) 7(11):e50747. doi: 10.1371/journal.pone.0050747
57. Gao L, Feng Z, Li Q, Li L, Chen L, Xiao T. Fibroblast Growth Factor Receptor 4 Polymorphism Is Associated With Increased Risk and Poor Prognosis of Non-Hodgkin's Lymphoma. Tumour Biol (2014) 35(4):2997–3002. doi: 10.1007/s13277-013-1386-7
58. Butkiewicz D, Krzesniak M, Drosik A, Giglok M, Gdowicz-Klosok A, Kosarewicz A, et al. The VEGFR2, COX-2 and MMP-2 Polymorphisms Are Associated With Clinical Outcome of Patients With Inoperable Non-Small Cell Lung Cancer. Int J Cancer (2015) 137(10):2332–42. doi: 10.1002/ijc.29605
59. Koole K, van Kempen PM, van Bockel LW, Smets T, van der Klooster Z, Dutman AC, et al. FGFR4 Is a Potential Predictive Biomarker in Oral and Oropharyngeal Squamous Cell Carcinoma. Pathobiology (2015) 82(6):280–9. doi: 10.1159/000439536
60. Quintanal-Villalonga A, Ojeda-Marquez L, Marrugal A, Yague P, Ponce-Aix S, Salinas A, et al. The FGFR4-388arg Variant Promotes Lung Cancer Progression by N-Cadherin Induction. Sci Rep (2018) 8(1):2394. doi: 10.1038/s41598-018-20570-3
61. Wei W, You Z, Sun S, Wang Y, Zhang X, Pang D, et al. Prognostic Implications of Fibroblast Growth Factor Receptor 4 Polymorphisms in Primary Breast Cancer. Mol Carcinog (2018) 57(8):988–96. doi: 10.1002/mc.22819
62. Azuma S, Uojima H, Chuma M, Shao X, Hidaka H, Nakazawa T, et al. Influence of NOS3 Rs2070744 Genotypes on Hepatocellular Carcinoma Patients Treated With Lenvatinib. Sci Rep (2020) 10(1):17054. doi: 10.1038/s41598-020-73930-3
63. Li JP, Huang HC, Yang PJ, Chang CY, Chao YH, Tsao TC, et al. FGFR4 Gene Polymorphism Reduces the Risk of Distant Metastasis in Lung Adenocarcinoma in Taiwan. Int J Environ Res Public Health (2020) 17(16):5694–704. doi: 10.3390/ijerph17165694
64. Akdeniz Odemis D, Tuncer SB, Adamnejad Ghafour A, Jabbarli K, Gider Y, Celik B, et al. FGFR4 C.1162G > A (P.Gly388Arg) Polymorphism Analysis in Turkish Patients With Retinoblastoma. J Oncol (2020) 2020:9401038. doi: 10.1155/2020/9401038
65. Shiu BH, Hsieh MH, Ting WC, Chou MC, Chang LC, Huang CC, et al. Impact of FGFR4 Gene Polymorphism on the Progression of Colorectal Cancer. Diagnostics (Basel) (2021) 11(6):978–87. doi: 10.3390/diagnostics11060978
66. Frullanti E, Berking C, Harbeck N, Jezequel P, Haugen A, Mawrin C, et al. Meta and Pooled Analyses of FGFR4 Gly388Arg Polymorphism as a Cancer Prognostic Factor. Eur J Cancer Prev (2011) 20(4):340–7. doi: 10.1097/CEJ.0b013e3283457274
67. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. BMJ (2009) 339:b2535. doi: 10.1136/bmj.b2535
68. Greenhalgh T, Peacock R. Effectiveness and Efficiency of Search Methods in Systematic Reviews of Complex Evidence: Audit of Primary Sources. BMJ (2005) 331(7524):1064–5. doi: 10.1136/bmj.38636.593461.68
69. Sterne JA, Sutton AJ, Ioannidis JP, Terrin N, Jones DR, Lau J, et al. Recommendations for Examining and Interpreting Funnel Plot Asymmetry in Meta-Analyses of Randomised Controlled Trials. BMJ (2011) 343:d4002. doi: 10.1136/bmj.d4002
70. Egger M, Davey Smith G, Schneider M, Minder C. Bias in Meta-Analysis Detected by a Simple, Graphical Test. BMJ (1997) 315(7109):629–34. doi: 10.1136/bmj.315.7109.629
71. Ulaganathan VK, Ullrich A. Membrane-Proximal Binding of STAT3 Revealed by Cancer-Associated Receptor Variants. Mol Cell Oncol (2016) 3(3):e1145176. doi: 10.1080/23723556.2016.1145176
72. Ipenburg NA, Koole K, Liem KS, van Kempen PM, Koole R, van Diest PJ, et al. Fibroblast Growth Factor Receptor Family Members as Prognostic Biomarkers in Head and Neck Squamous Cell Carcinoma: A Systematic Review. Target Oncol (2016) 11(1):17–27. doi: 10.1007/s11523-015-0374-9
73. Tang S, Hao Y, Yuan Y, Liu R, Chen Q. Role of Fibroblast Growth Factor Receptor 4 in Cancer. Cancer Sci (2018) 109(10):3024–31. doi: 10.1111/cas.13759
Keywords: FGFR4, Gly388Arg, polymorphism, prognosis, meta-analysis
Citation: Kim JH, Jeong SY, Jang HJ, Park ST and Kim HS (2021) FGFR4 Gly388Arg Polymorphism Reveals a Poor Prognosis, Especially in Asian Cancer Patients: A Meta-Analysis. Front. Oncol. 11:762528. doi: 10.3389/fonc.2021.762528
Received: 23 August 2021; Accepted: 04 October 2021;
Published: 19 October 2021.
Edited by:
Alexandre Harlé, Institut de Cancérologie de Lorraine, FranceReviewed by:
Michal Linial, Hebrew University of Jerusalem, IsraelZhilian Jia, City of Hope National Medical Center, United States
Copyright © 2021 Kim, Jeong, Jang, Park and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sung Taek Park, cGFya3N0OTZAbmF2ZXIuY29t; Hyeong Su Kim, bmVwMm5AaGFsbHltLm9yLmty
 Jung Han Kim1
Jung Han Kim1 Hyeong Su Kim
Hyeong Su Kim