
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 27 October 2021
Sec. Molecular and Cellular Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.758653
This article is part of the Research TopicAltered Expression of Proteins in Cancer: Function and Potential Therapeutic TargetsView all 40 articles
The long non-coding RNA metastasis-associated lung adenocarcinoma transcript-1 (MALAT1) was initially found to be overexpressed in early non-small cell lung cancer (NSCLC). Accumulating studies have shown that MALAT1 is overexpressed in the tissue or serum of NSCLC and plays a key role in its occurrence and development. In addition, the expression level of MALAT1 is significantly related to the tumor size, stage, metastasis, and distant invasion of NSCLC. Therefore, MALAT1 could be used as a biomarker for the early diagnosis, severity assessment, or prognosis evaluation of NSCLC patients. This review describes the basic properties and biological functions of MALAT1, focuses on the specific molecular mechanism of MALAT1 as a microRNA sponge in the occurrence and development of NSCLC in recent years, and emphasizes the application and potential prospect of MALAT1 in molecular biological markers and targeted therapy of NSCLC.
Non-small cell lung cancer (NSCLC) is one of the leading causes of cancer-related deaths worldwide (1). Although great advances have been made in surgery, chemotherapy, and immunotherapy, the 5-year survival rate of patients with NSCLC is still only about 15% due to the high rate of distant metastasis and recurrence (2, 3). Therefore, the invasion and the metastasis of cancer cells are serious challenges in the treatment of NSCLC. In-depth understanding of the potential mechanisms of the occurrence and development of NSCLC is of great significance in order to improve the effect of clinical treatment.
Long non-coding RNA (lncRNA) is a transcript consisting of more than 200 nucleotides in length (4). It is well known that lncRNA can regulate the expressions of many genes and participate in the development of tumors (5). Metastasis-associated lung adenocarcinoma transcript-1 (MALAT1) was initially found to be overexpressed in early NSCLC, which is a type of non-coding ribonucleic acid (6). Although there have been many studies on MALAT1 in the past, the specific molecular mechanism of MALAT1 regulation of NSCLC is still not very clear (7). In the past decade, more and more studies have found that MALAT1 can regulate its downstream target molecules by directly binding to microRNA (miRNA), thus playing an important role in the cell proliferation, metastasis, invasion, and treatment of drug resistance in NSCLC (8–11). In this review, we first briefly introduce the basic properties and biological functions of MALAT1, focus on the molecular mechanism of MALAT1 as an miRNA sponge in the occurrence and the development of NSCLC, and highlight the application and potential prospect of MALAT1 in molecular biological markers and targeted therapy in NSCLC.
MALAT1 is also termed nuclear enriched abundant transcript 2 (NEAT2) (12). The structure and biogenesis of its genes are located in human chromosome 11q13 and mouse chromosome 19qA (13, 14). The MALAT1 transcript is about 7 kb in humans and 6.7 kb in mice (12, 15). Previously, MALAT1 was named because of its clinical significance in predicting the metastasis and survival of early NSCLC, but a subsequent study showed that MALAT1 is widely expressed in normal tissues and is extremely abundant and widely conserved in 33 species of mammals (6, 16), which indicates that MALAT1 may have potentially important biological functions (17).
Different from the typical mechanism of cleavage and polyadenylation, the MALAT1 3′ end lacks the structure of poly(A) tail (18). With the cleavage of ribonuclease (RNase P), the primary transcript of MALAT1 forms a mature transcript of 7 kb and a small transcript fragment at the 3′ end (Figure 1) (18). The mature transcript is mainly located in nuclear bodies known as nuclear speckles, which are subnuclear structures enriched with RNA processing factors and poly(A)+ RNAs and involved in posttranscriptional regulation of gene expression (19, 20). Its 3′ end is highly conserved and forms a unique triple-helix structure that can protect it from the damage of 3′–5′ exonucleases, which is beneficial to the stability of MALAT1 (21, 22). The small transcript fragment is bound by ribonuclease Z (RNase Z) and further cleaved and modified by the CCA-adding enzyme to produce a 61-nt-long lncRNA called MALAT1-associated small cytoplasmic RNA (mascRNA), then folds into the transfer RNA (tRNA) cloverleaf structure and is exported to the cytoplasm (Figure 1) (18). MALAT1 located in nuclear speckles can regulate other physiological and pathological processes such as embryonic development, tumor progression, cardiovascular remodeling, and tissue inflammation mainly by affecting gene transcription, interfering with messenger RNA (mRNA) cleavage, regulating epigenetic changes, or acting as a competitive endogenous RNA (23–28). There are few reports on the role of mascRNA, which may participate in cardiovascular innate immunity by affecting fas ligand (FASLG), tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), etc. (29) It may also be part of the molecular mechanism of function in cancer to regulate the glutaminyl-tRNA synthetase (QARS) protein levels and promote global protein translation and cell proliferation (30).
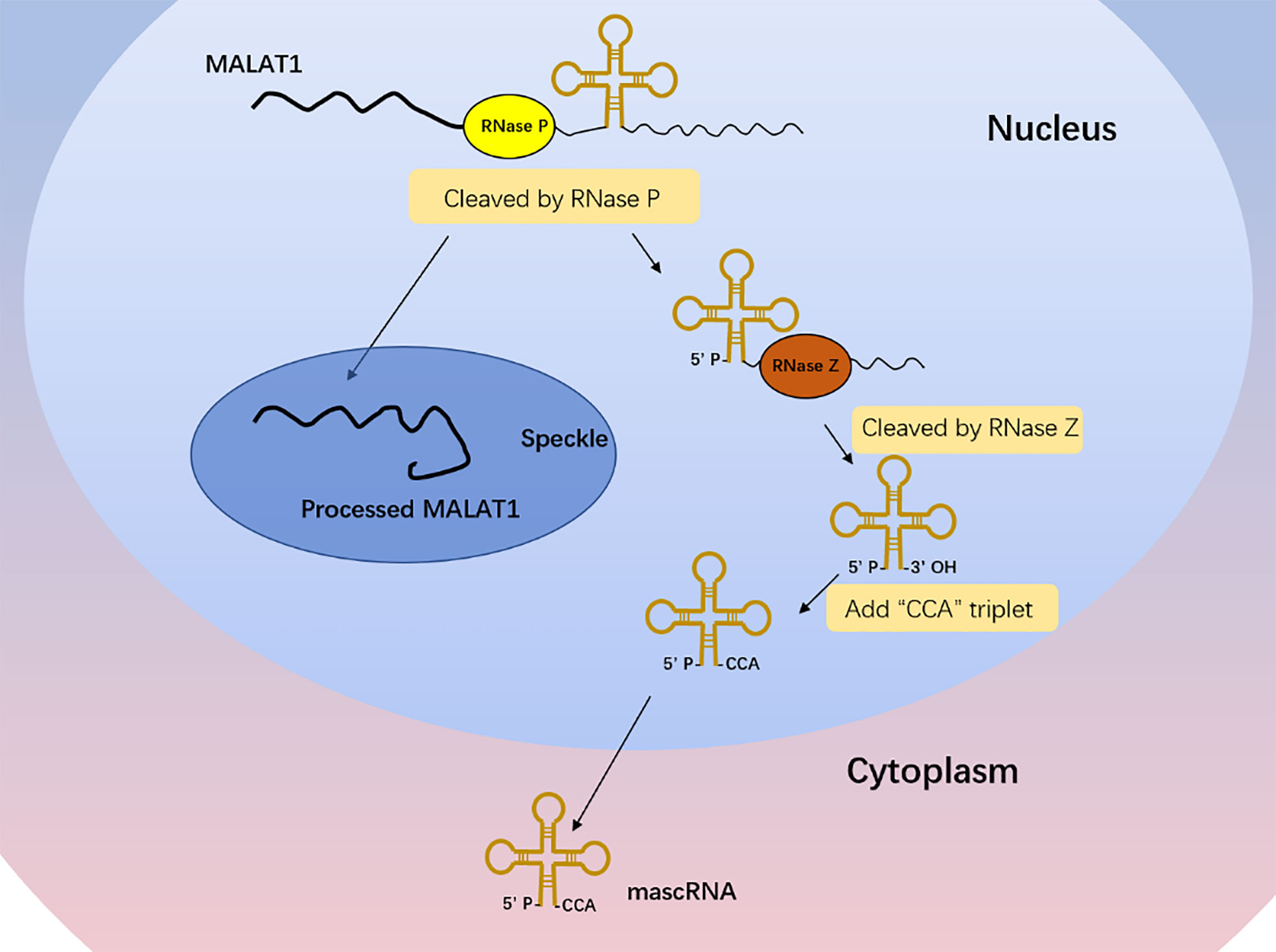
Figure 1 MALAT1 biogenesis. The primary transcript of MALAT1 forms a mature transcript of 7 kb and a small transcript fragment at the 3′ end with the cleavage of RNase P. The mature transcript is mainly located in nuclear speckles, and its 3′ end is highly conserved and forms a unique triple-helix structure that can protect it from the damage of 3′–5′ exonucleases, which is beneficial to the stability of MALAT1. The small transcript fragment is bound by RNase Z and further cleaved and modified by the CCA-adding enzyme to produce a 61-nt-long lncRNA called MALAT1-associated small cytoplasmic RNA (mascRNA), then folds into the tRNA cloverleaf structure and is exported to the cytoplasm.
Previous studies have found that MALAT1 can participate in the regulation of biological function through the following main mechanisms (Figure 2): 1) affecting the gene transcription. MALAT1 can recruit Sp1, a transcription factor, in multiple myeloma. Sp1 can activate and promote the secretion of growth factor TGF-β by binding to the prompter of latent transforming growth factor beta binding protein 3 (LTBP3) (31). MALAT1 can promote the transcription of telomeric repeat-binding factor 2 (TRF2) by recruiting RNApol II, P300, and CRUPT to bind to the promoter region of TRF2, which promotes the growth of liver cancer stem cells (32). 2) Affecting the alternative splicing of pre-mRNAs. MALAT1 is identified as a nuclear-retained regulatory RNA that can interact with the serine- and arginine-rich (SR) protein splicing factors such as SRSF1, SRSF2, and SRSF3, affect the distribution of splicing factors in nuclear speckle domains, and regulate alternative splicing of pre-mRNAs (33). Additionally, MALAT1 can promote ovarian cancer progression by regulating the splicing factor RBFOX2-mediated alternative splicing (34). Furthermore, MALAT1 can induct the oncogenic splicing factor SRSF1 and modulate the alterative splicing of SK61 in hepatocellular carcinoma (35). 3) Regulating protein activity. MALAT1 can competitively bind to SFPQ leading to PTBP2 release from the SFPQ/PTBP2 complex, which enhances the function of PTBP2 in promoting tumor cell proliferation and migration (36). 4) Mediating epigenetic changes. Malat1 can cause the trimethylation of histone 3 lysine 9 (H3K9me3) by recruiting the suppressor of variegation 3–9 homolog 1 (Suv39h1) to MyoD-binding loci. This trimethylation suppresses the transcriptional activity of MyoD, which represses myoblast differentiation (37). In addition, the overexpression of MALAT1 could increase the expression of acetyl-H4 histone in the IQ motif-containing GTPase-activating protein 1 (IQGAP1) promoter, which may promote the proliferation and invasion of thyroid cancer cells (38). 5) Promoting the nuclear and cytoplasmic translocation of cellular proteins. MALAT1 retains the serine/arginine-rich proteins SF2/ASF from the cytoplasm to the nucleus, thus promoting the development of gastric cancer cells (39). MALAT1 can bind to an abundant nuclear factor heterogeneous nuclear ribonucleoprotein C (hnRNPC) protein, which could transfer from the nucleus to the cytoplasm during cell division, to assist its translocation (40). (6) Acting as an endogenous miRNA sponge. MiRNAs play an important role in cell proliferation, differentiation, apoptosis, and development. Recent evidence suggests that other RNAs such as lncRNA can also compete with mRNAs by sponging miRNAs (41). Among these lncRNAs, MALAT1 is one of the most studied RNAs involved in various molecular processes such as endogenous miRNA sponging (42). Here, we will focus on the potential function of MALAT1 as a miRNA sponge in NSCLC (Table 1).
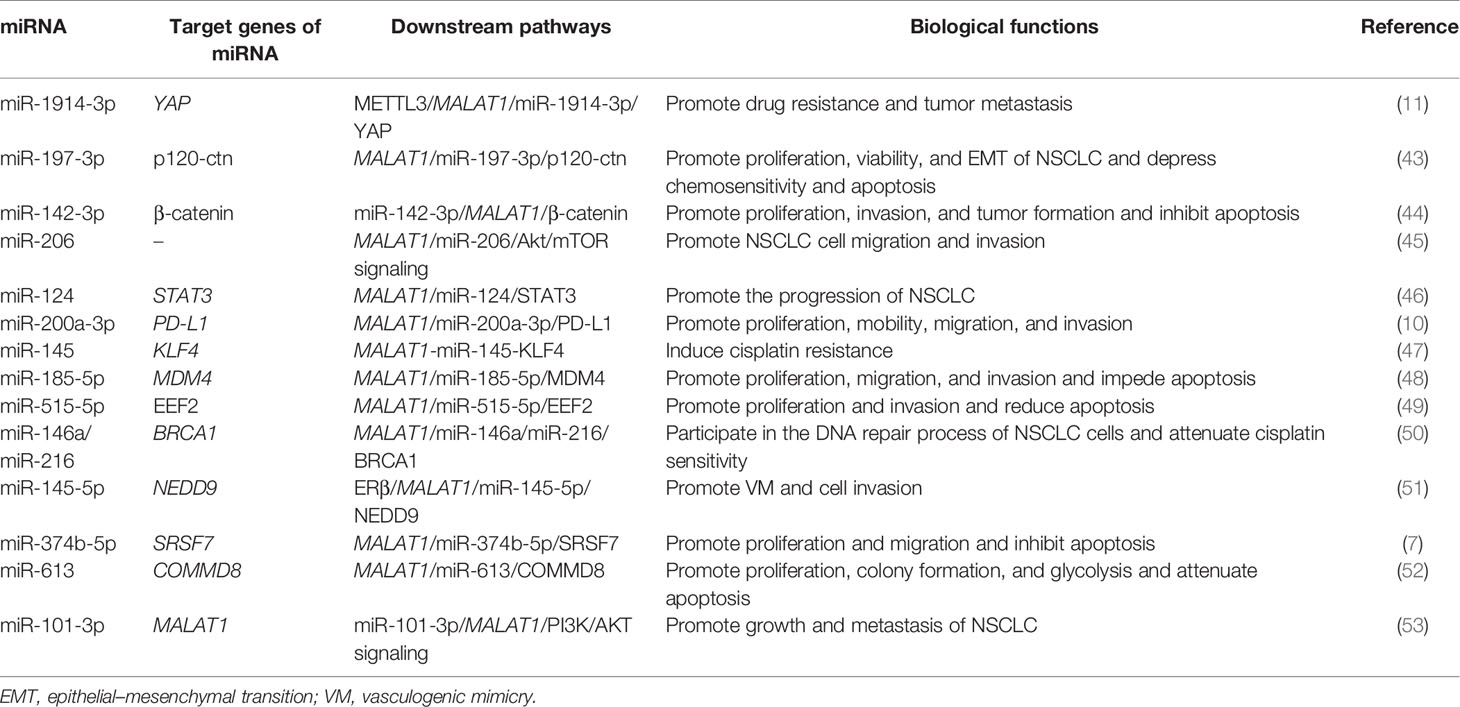
Table 1 Mechanism and roles of the metastasis-associated lung adenocarcinoma transcript-1 (MALAT1) in non-small cell lung cancer (NSCLC) progression.
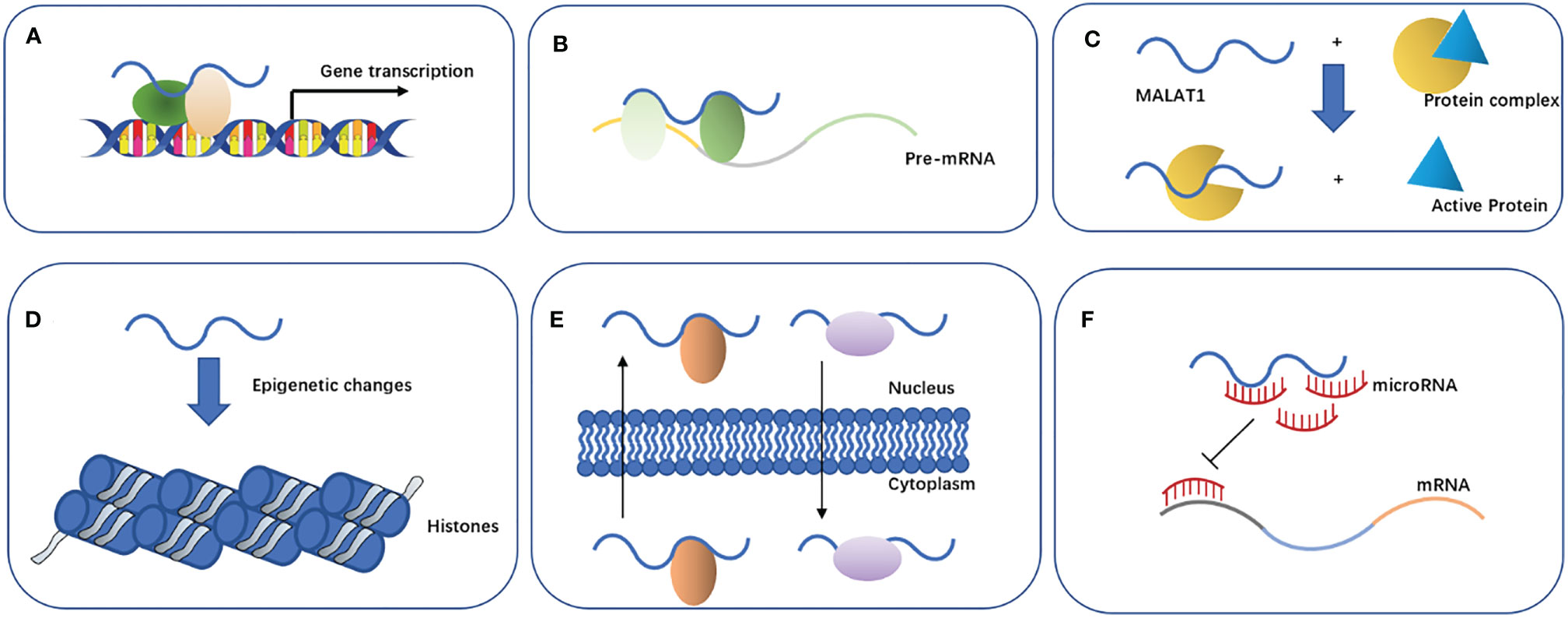
Figure 2 Properties and biological functions of MALAT1. (A) Affects gene transcription. (B) Affects the alternative splicing of pre-mRNAs. (C) Regulates protein activity. (D) Mediates epigenetic changes. (E) Promotes nuclear and cytoplasmic translocation of cellular proteins. (F) Acts as an endogenous miRNA sponge.
N6-methyladenosine (m6A) mRNA methylation initiated by methyltransferase-like 3 (METTL3) promotes the translation of YAP mRNA by recruiting YTHDF1/3 and eIF3b into the translation initiation complex, so the expression of METTL3 is positively correlated with the level of YAP protein (54). On the other hand, METTL3 improved the m6A modification level of the lncRNA MALAT1 and increased its stability. MALAT1 sponging miR-1914-3p weakens the ability of miR-1914-3p to target and inhibit YAP, thus increasing the expression of YAP in NSCLC (54). The increased expression and activity of YAP lead to cisplatin (DDP) resistance and metastasis of NSCLC (54). Therefore, the increased activity of the METTL3/MALAT1/miR-1914-3p/YAP axis promotes the metastasis and drug resistance of NSCLC.
The high expressions of MALAT1 and miR-197-3p were closely related to the survival and growth of NSCLC (43). Luciferase activity assay showed that MALAT1 was complementary to miR-197-3p at certain sites. P120 catenin (p120-ctn) regulates the proliferation of cancer cells by regulating cell adhesion and the cell cycle (55). Yang et al. found that p120-ctn was confirmed to be a targeted downstream molecule of MALAT1 and miR-197-3p (43). Reducing the expression of p120-ctn can repress the epithelial–mesenchymal transition (EMT) and the survival and proliferation ability of NSCLC, while it enhances the apoptosis rate of cancer cells. Moreover, p120-ctn can mediate the role of MALAT1 and miR-197-3p in promoting the progression and chemotherapy resistance of NSCLC cells (43). The results of in vivo experiments using NSCLC mouse models showed that a low expression of MALAT1, miR-197-3p, or p120-ctn can decrease the tumor volume and weight compared with the control group (43). Consequently, the MALAT1/miR-197-3p/p120-ctn axis may play a potential role in the regulation of NSCLC, which will provide a direction for improving the prognosis of NSCLC patients after chemotherapy.
The expression of miR-142-3p decreased, while β-catenin and MALAT1 increased in NSCLC tissues. RT-PCR and luciferase reporter assays showed that miR-142-3p negatively inhibited the level of MALAT1 by directly binding to the 3′-UTR of MALAT1 mRNA (44). On the one hand, upregulation of miR-142-3p mimic transfection can significantly reduce the proliferation and migration of NSCLC H1299 cells while inducing G0/G1 phase arrest and reducing that of the S phase; on the other hand, the overexpression of miR-142-3p can downregulate the expression of β-catenin in H1299 cells (44). In vivo experiments showed that the upregulation of miR-142-3p and the downregulation of β-catenin or MALAT1 could significantly reduce the tumorigenicity of NSCLC cells (44). To sum up, miR-142-3p can play a tumor-suppressing role in the progression of NSCLC by inhibiting the MALAT1/β-catenin signaling pathway.
Tang et al. detected the expression of MALAT1 in tumor tissues and adjacent normal tissues in 36 cases of NSCLC using real-time quantitative PCR (qRT-PCR) and found that the expression of MALAT1 was significantly upregulated in NSCLC tissues (45). In addition, MALAT1 promoted EMT, cell migration, and invasion by activating the Akt/mTOR signals in A549 and H1299 cells. MiR-206 is the direct downstream target of MALAT1 in NSCLC, and there was a negative correlation between the expressions of MALAT1 and miR-206 in NSCLC (45). MALAT1 promoted cell migration and invasion in NSCLC cells by sponging miR-206. In addition, miR-206 could also inhibit the activation of the Akt/mTOR signal mediated by MALAT1 in A549 and H1299 cells (45). Taken together, MALAT1 can promote the migration and invasion of NSCLC by targeting miR-206 and activating the Akt/mTOR signaling pathway, which provides a molecular basis for the metastasis of MALAT1 in NSCLC.
It was found that the level of miR-124 in A549, H23, H522, H1299, and H460 NSCLC cells was significantly downregulated (46). Luciferase reporter assays showed that miR-124 is the direct target of MALAT1, and there was a potential negative correlation between miR-124 and MALAT1. shMALAT1 can suppress the proliferation, colony formation, and apoptosis of NSCLC cells, while miR-124 inhibitors can reverse this effect. In addition, it was also found that STAT3 is a new mRNA target of miR-124 (46). The downregulation of MALAT1 can inhibit the development of NSCLC by enhancing the expression of miR-124 and reducing the expression of STAT3 (46). In summary, it is speculated that MALAT1 may participate in the occurrence and development of NSCLC as an endogenous miRNA sponge through the MALAT1/miR-124/STAT3 signaling axis.
The targeting relationship between MALAT1 and miR-200a-3p and programmed death-ligand 1 (PD-L1) was further verified by qRT-PCR and dual-luciferase reporter gene detection (10). The researchers found that MALAT1 sponged miR-200a-3p, and PD-L1 was identified as the target of miR-200a-3p and indirectly regulated by MALAT1. Moreover, the level of MALAT1 was negatively correlated with the expression of miR-200a-3p in NSCLC, but positively correlated with the expression of PD-L1 (10). Furthermore, MALAT1 promoted the proliferation, migration, and invasion of NSCLC cells through sponging miR-200a-3p (10). Overall, MALAT1 promotes the progress of NSCLC by regulating the miR-200a-3p/PD-L1 axis, which is of positive significance to the selection of new targeted drugs and the enrichment of therapeutic methods in the future.
Kruppel-like factor 4 (KLF4) has been shown to be associated with DDP resistance in some cancers (56, 57). KLF4 is negatively regulated by miR-145 and positively regulated by MALAT1 at the mRNA and protein levels in NSCLC A549 cells. Luciferase reporter assay, qRT-RCR, and Western blotting confirmed that MALAT1 indirectly regulated KLF4 by directly sponging miR-145, suggesting that MALAT1 may be involved in DDP resistance by regulating the level of KLF4 (47). In addition, MALAT1 knockout reversed the resistance of A549rCDDP cells to DDP. Collectively, the MALAT1/miR-145/KLF4 axis is an important inducer of DDP resistance in NSCLC (47). Therefore, MALAT1 may serve as a promising predictor and therapeutic target of DDP in patients with NSCLC.
Wang et al. found that the expressions of MALAT1 and MDM4 were significantly high in 30 cases of NSCLC, and MALAT1 could positively regulate the expression of MDM4 in NSCLCs cells (48). The deletion of MALAT1 and MDM4 could significantly decrease the proliferation and metastasis of NSCLC cells and promote apoptosis. In addition, the binding sites of miR-185-5p and MALAT1 or MDM4 were predicted using a database, and their relationship was further confirmed by dual-luciferase report assays. The results showed that miR-185-5p can be a target of MALAT1 and could also directly regulate MDM4, and its overexpression can obviously suppress NSCLC cells (48). It was further confirmed that MALAT1 can promote the proliferation, migration, invasion, and apoptosis of NSCLC cells by regulating the expression of MDM4 mediated by miR-185-5p (48). These results may provide not only a new regulatory mechanism but also a new potential therapeutic target for the treatment of NSCLC.
It has been reported that MALAT1 is involved in the repair pathway of DNA double-strand breaks, and targeting MALAT1 can induce apoptosis in myeloma cells (58). BRCA1 is a multifunctional protein that plays a key role in the homologous recombination DNA repair pathway (59). Through the MALAT1 pull-down assay, the researchers found that miR-146a and miR-216 directly interact with MALAT1 in A549 and H1299 cells and that they can specifically inhibit the expression of BRCA1 (50). By inhibiting MALAT1, miR-146a and miR-216 can be released to further inhibit the expression of BRCA1 and induce DNA damage. Therefore, MALAT1 can participate in the DNA repair process of NSCLC cells by regulating the miR-146a/miR-216/BRCA1 pathway. In addition, targeting MALAT1 can also increase the sensitivity of NSCLC cells to DDP (50). In summary, MALAT1 may become a new target for the treatment of NSCLC.
Estrogen receptor beta (ERβ) may affect the progression of NSCLC (51). Yu et al. found that ERβ can increase the expression of MALAT1 by directly binding to the estrogen response elements (EREs) located on the MALAT1 promoter, thus inhibiting miR-145-5p. Because miR-145-5p directly targets the 3′-UTR of the neural precursor cell expressed, developmentally downregulated 9 (NEDD9) mRNA, increasing the expression of MALAT1 can indirectly upregulate the protein expression of NEDD9. Further experiments showed that ERβ could promote the vasculogenic mimicry (VM) formation and cell invasion of NSCLC by the ERβ/MALAT1/miR-145-5p/NEDD9 signaling pathway (51). This may help in providing new strategies to better inhibit the metastasis of NSCLC in the future.
The expressions of MALAT1 and serine/arginine-rich splicing factor 7 (SRSF7) were upregulated and the expression of miR-374b-5p was downregulated in NSCLC (7). The expression of MALAT1 was negatively correlated with the expression of miR374b-5p and positively correlated with the expression of SRSF7. MiR-374b-5p is the target of MALAT1. Knockout of MALAT1 and miR-374b-5p overexpression can inhibit the proliferation, migration, and invasion of NSCLC cells and induce apoptosis. In vivo experiments showed that the overexpression of MALAT1 promoted the tumor growth of NSCLC (7). SRSF7 is the downstream target molecule of miR-374b-5p. The overexpression of SRSF7 reverses the effects of MALAT1 gene knockout on the proliferation, apoptosis, migration, and invasion of NSCLC cells (7). Therefore, it was concluded that MALAT1 participates in the progress of NSCLC through the MALAT1/miR-374b-5p/SRSF7 axis. This study may provide a theoretical basis for the diagnosis and treatment of NSCLC.
The expressions of MALAT1 and COMMD8 were abnormally increased in NSCLC tissues and cells (52). We found that miR-613 is the target of MALAT1 and that it can bind to the 3′-UTR of COMMD8. MALAT1 upregulated the level of COMMD8 by competitively targeting miR-613, thus playing a carcinogenic role in NSCLC (52). MALAT1 or COMMD8 gene knockout inhibited cell proliferation, clone formation, and glycolysis, but promoted cell apoptosis. In vivo experiments have shown that MALAT1 gene knockout reduced the tumor growth. In addition, researchers also found that extracellular MALAT1 was released by packaging into exosomes (52). These pieces of evidence provide new insights into the treatment of NSCLC, and the MALAT1/miR-613/COMMD8 axis will be a promising approach for future treatment options.
The relative expression of miR-101-3p in NSCLC cells decreased significantly, while the relative expression of MALAT1 increased significantly (53). MiR-101-3p can significantly inactivate the PI3K/AKT pathway; inhibit the expression of Bcl-2 and MMP-9; and suppress the proliferation, migration, and invasion of NSCLC cells by directly binding to MALAT1 (53). On the contrary, the overexpression of MALAT1 reversed the inhibitory effect of miR-101-3p on the activation of the PI3K/AKT signaling pathway and the expressions of Bcl-2 and MMP-9 in NSCLC. These results suggest that miR-101-3p blocks the PI3K/AKT signaling pathway by targeting the inhibition of MALAT1, thus inhibiting the growth and metastasis of NSCLC (53). Therefore, miR-101-3p is expected to become an effective target for the prevention and treatment of NSCLC.
Although there are many methods for the diagnosis of NSCLC, these may not fully meet the needs of early diagnosis of the cancer. MALAT1 is a relatively stable RNA transcript with a half-life of 9–12 h, which may be due to its triple-helix structure at the 3′-end (21, 22, 60). This characteristic of having a long half-life makes MALAT1 easy to detect in tumor tissues and body fluids. Research has shown that MALAT1 can be used as a biomarker for the diagnosis of many kinds of malignant tumors (61–63). Especially in NSCLC, the high expression of MALAT1 was significantly correlated with tumor node metastasis (TNM) stage, vascular invasion, pathological differentiation, and recurrence (64). Further studies have shown that the overexpression of MALAT1 was significantly related to the prognosis of lung squamous cell carcinoma, which is one type of NSCLC (65). Moreover, different expression levels of MALAT1 in peripheral blood were observed between cancer patients and healthy controls (66).
Rong et al. found that the levels of MALAT1 in serum exosomes were higher in patients with NSCLC, suggesting that exosome-derived MALAT1 may also reflect the biological changes of NSCLC cells (49). Zhang et al. found that the expression of MALAT1 in serum exosomes of NSCLC patients was upregulated and that the level of exosomal MALAT1 was positively correlated with tumor stage and lymph node metastasis (67). The above data suggest that MALAT1 in exosomes may also be used as a serum-based tumor biomarker to diagnose and predict NSCLC. Liquid biopsy provides the opportunity of detecting and monitoring cancer in various body fluids by detecting free circulating tumor cells, circulating tumor DNA fragments, circulating RNA, and exosomes (68). Its advantage lies in that it can reduce the harm of biopsy through noninvasive sampling and has important significance for the early diagnosis of cancer, but the low expression level of MALAT1 in blood makes sensitive analysis difficult (66). Although some progress has been made in the detection of MALAT1 in blood with traditional RT-PCR, the procedure is complicated, the amount of serum required is large, and the equipment is expensive. A recent study by Chen et al. showed that the detection of the levels of MALAT1 in blood was more rapid, sensitive, and inexpensive when using a novel ultrasensitive screen-printed carbon electrode (SPCE)-based electrochemical biosensor that uses a Au nanocluster (NC)/multi-walled carbon nanotube (MWCNT)–NH2 nanostructure (69). This new methodology for the detection of MALAT1 will increase its applicability to clinical diagnosis of NSCLC.
In addition, the expression level of MALAT1 can also be used as a biomarker of chemosensitivity in different cancers (43, 70–72). Resistance to multiple drugs is the main cause of chemotherapy failure in patients with lung cancer (73). Studies have shown that MALAT1 is also involved in the drug resistance of NSCLC. For example, Fang et al. found that the expression of MALAT1 was upregulated in DDP-resistant A549 cells. MALAT1 upregulated MRP1 and MDR1 by activating STAT3, thus reducing the sensitivity to DDP in vitro and in vivo (74). NSCLC patients carrying epidermal growth factor receptor (EGFR) mutations initially respond to EGFR tyrosine kinase inhibitors (EGFR-TKIs) such as gefitinib, but gradually developed acquired drug resistance (75, 76). It was found that the overexpression of MALAT1 could eliminate not only the inhibitory effect of polyphyllin I (PPI) on the activity of gefitinib-resistant NSCLC cells but also the apoptosis induced by PPI, while MALAT1 gene knockout could enhance the inhibition and apoptosis induced by PPI (77). These data suggest that MALAT1 may represent a candidate biomarker and therapeutic target for chemotherapy drug resistance.
Due to the enrichment and high expression of MALAT1 in the nucleus, its effect on traditional shRNAs or siRNAs may not be ideal and prone to off-target effects (78). The application of antisense oligonucleotides (ASOs) is a valuable method to antagonize MALAT1. ASOs, which are small RNA/DNA-based oligonucleotides capable of crossing cell membranes and binding to the target RNA in the nucleus and cytoplasm, are divided into two main categories: mixmeRs and gapmeRs (79, 80). Gutschner et al. found that MALAT1 could be targeted with second-generation ASOs, thus leading to the drastic reduction of lung cancer metastasis in a pulmonary metastatic model in vivo (78). Moreover, the same investigators achieved functional knockout of MALAT1 through zinc finger nuclease (ZFN)-mediated site-specific integration of RNA destabilizing elements into the human genome, which showed efficient silencing of the highly abundant MALAT1 in human lung cancer cells (78).
As an important and highly conserved lncRNA, MALAT1 has been widely studied, especially its role in tumorigenesis, metastasis, drug resistance, and clinical prognosis (81–83). However, the specific role of MALAT1 in the occurrence and development of NSCLC has not been fully elucidated. Based on the basic biological properties of MALAT1, more and more studies have shown that it can be used as a bait for miRNA to share miRNA response elements (MREs) with mRNAs, which indirectly affects the expression of some specific downstream genes, thus promoting the proliferation, invasion, apoptosis, drug resistance, and tumor growth of NSCLC. In general, MALAT1 is mostly known to be enriched in nuclear speckles, and we also agree that cytoplasmic P-bodies are the localizing site of the RNA-induced silencing complex (RISC) effector proteins Ago1–4 and the functional site of miRNA-mediated gene silencing (84). The vast majority of researchers used to apply bioinformatics program such as ChipBase, LncRNAdb, and StarBase to predict the interaction between MALAT1 and miRNA in previous research on MALAT1 as a miRNA sponge in NSCLC. Subsequently, they verified the direct interaction using luciferase reporter, RNA immunoprecipitation (RIP), and MALAT1 pull-down assays. However, there was little focus on the sites (cytoplasm or nucleus) where these interactions occur. On the contrary, Jin et al. demonstrated that MALAT1 and miR-1914-3p are abundant and stable in the cellular cytoplasm using RNA fluorescence in situ hybridization assay and confirmed that MALAT1 directly binds miR-1914-3p using luciferase reporter assay, RIP for argonaute 2 (Ago2) in A549 cells, and RNA pull-down assay (54). Additionally, Leucci et al. showed that miR-9 targets MALAT1 for degradation in the nucleus by directly binding to two miRNA binding sites (85). Furthermore, Wu et al. found that Ago2 was expressed both in the nucleus and cytoplasm of sw480 cells (86). Moreover, Gagnon et al. reported that 75% of the miRNAs in the cytoplasm could shuttle into the nucleus and then bind to nuclear Ago2 (87). These studies showed that the distribution of MALAT1 or miRNA is not limited to the nucleus or cytoplasm. Hence, we wondered whether MALAT1 or miRNA might be involved in some cases with nucleoplasmic translocation. Additionally, the locations of MALAT1 and various miRNA interactions in NSCLC cells need to be further verified and explored.
Taken together, based on the literature, some miRNAs such as miR-142-3p and miR-101-3p can target MALAT1 for degradation, thereby negatively inhibiting the lever of MALAT1 in NSCLC (44, 53). On the contrary, MALAT1 can also act as a miRNA sponge by sequestering the target miRNAs and affecting downstream gene expression, and the expression level of MALAT1 was negatively correlated with the expressions of miRNAs in NSCLC (53). Whether miRNA is degraded or recycled remains to be investigated. It also has been reported that MALAT1 and some miRNAs were more abundant in the Ago2 pellet than in the immunoglobulin G (IgG) pellet by conducting an RIP assay, which suggested that MALAT1 might be a target of miRNA through an Ago2-dependent manner.
Intriguingly, there is an exosome-derived MALAT1 in the serum of NSCLC patients, and the expression of MALAT1 in exosomes is highly correlated with the TNM stage and lymphatic metastasis of NSCLC. However, at present, the mechanism of MALAT1 in the exosomes of NSCLC patients remains in the preliminary research stage and needs to be further clarified. It is interesting to note that, due to the enrichment and high expression of MALAT1 in the nucleus, the specific mechanism of MALAT1 packing into exosomes that are rarely reported remains to be explored in the future, although it is common for lncRNA as a cargo to be loaded into exosomes. Moreover, MALAT1 may be a key actor in the hallmark of resisting cell death as it can decrease the levels of cleaved CASP3 in NSCLCs, which leads to escaping apoptosis (77, 88). On the contrary, whether MALAT1 detection in serum due to cell death may involve complex mechanisms needs to be further studied.
In addition, MALAT1 knockout mice did not cause obvious phenotype in development, gene expression, and physiological function, which is not consistent with MALAT1 being involved in the occurrence and development of NSCLC in vitro, so this also needs to be further explored (89). In-depth understanding of the function and regulatory mechanism of MALAT1 in NSCLC may provide a new breakthrough for the diagnosis and targeted therapy of NSCLC in the future.
QZ wrote the manuscript and designed the figures. DC constructed the topic. LL, JZ, and YC provided scientific suggestions and participated in manuscript preparation. DX and YY provided guidance and revised this manuscript. All authors contributed to the article and approved the submitted version.
The study was supported by grants provided by The National Natural Science Foundation of China (81871709), the Natural Science Foundation of Suzhou (KJXW2017063), Natural Science Foundation of Zhangjiagang (ZKS2022).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the American Journal Experts (AJE) for English language editing.
NSCLC, non-small-cell lung cancer; LncRNA, long non-coding RNA; MALAT1, metastasis-associated lung adenocarcinoma transcript-1; NEAT2, nuclear enriched abundant transcript 2; RNase, ribonuclease; FASLG, fas ligand; TNF-α, tumor necrosis factor-α; IL-6, interleukin-6; QARS, glutaminyl-tRNA synthetase; LTBP3, latent transforming growth factor beta binding protein 3; TRF2, transcription of telomeric repeat-binding factor 2; H3K9me3, trimethylation of histone 3 lysine 9; Suv39h1, suppressor of variegation 3–9 homolog 1; IQGAP1, IQ motif-containing GTPase-activating protein 1; hnRNPC, heterogeneous nuclear ribonucleoprotein C; m6A, N6-methyladenosine; METTL3, methyltransferase-like 3; DDP, cisplatin; p120-ctn, P120 catenin; EMT, epithelial–mesenchymal transition; qRT-PCR, real-time quantitative PCR; PD-L1, programmed death-ligand 1; KLF4, Kruppel-like factor 4; ERβ, estrogen receptor β; EREs, estrogen response elements; VM, vasculogenic mimicry; SRSF7, serine/arginine-rich splicing factor 7; EGFR-TKI, epidermal growth factor receptor tyrosine kinase inhibitor; PPI, polyphyllin I; ASOs, antisense oligonucleotides; ZFN, zinc finger nuclease; MRE, miRNA response element; RIP, RNA immunoprecipitation.
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2015. CA Cancer J Clin (2015) 65:5–29. doi: 10.3322/caac.21254
2. Trinidad Lopez C, Souto Bayarri M, Oca Pernas R, Delgado Sanchez-Gracian C, Gonzalez Vazquez M, Vaamonde Liste A, et al. Characteristics of Computed Tomography Perfusion Parameters in Non-Small-Cell-Lung-Cancer and Its Relationship to Histology, Size, Stage an Treatment Response. Clin Imaging (2018) 50:5–12. doi: 10.1016/j.clinimag.2017.12.001
3. Molinier O, Goupil F, Debieuvre D, Auliac JB, Jeandeau S, Lacroix S, et al. Five-Year Survival and Prognostic Factors According to Histology in 6101 Non-Small-Cell Lung Cancer Patients. Respir Med Res (2020) 77:46–54. doi: 10.1016/j.resmer.2019.10.001
4. Thum T. Noncoding RNAs and Myocardial Fibrosis. Nat Rev Cardiol (2014) 11:655–63. doi: 10.1038/nrcardio.2014.125
5. Kim C, Kang D, Lee EK, Lee JS. Long Noncoding RNAs and RNA-Binding Proteins in Oxidative Stress, Cellular Senescence, and Age-Related Diseases. Oxid Med Cell Longev (2017) 2017:2062384. doi: 10.1155/2017/2062384
6. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a Novel Noncoding RNA, and Thymosin Beta4 Predict Metastasis and Survival in Early-Stage Non-Small Cell Lung Cancer. Oncogene (2003) 22:8031–41. doi: 10.1038/sj.onc.1206928
7. Song J, Su ZZ, Shen QM. Long Non-Coding RNA MALAT1 Regulates Proliferation, Apoptosis, Migration and Invasion via miR-374b-5p/SRSF7 Axis in Non-Small Cell Lung Cancer. Eur Rev Med Pharmacol Sci (2020) 24:1853–62. doi: 10.26355/eurrev_202002_20363
8. Chen R, Liu Y, Zhuang H, Yang B, Hei K, Xiao M, et al. Quantitative Proteomics Reveals That Long Non-Coding RNA MALAT1 Interacts With DBC1 to Regulate P53 Acetylation. Nucleic Acids Res (2017) 45:9947–59. doi: 10.1093/nar/gkx600
9. Yu Q, Xiang L, Chen Z, Liu X, Ou H, Zhou J, et al. MALAT1 Functions as a Competing Endogenous RNA to Regulate SMAD5 Expression by Acting as a Sponge for miR-142-3p in Hepatocellular Carcinoma. Cell Bioscience (2019) 9:39. doi: 10.1186/s13578-019-0299-6
10. Wei S, Wang K, Huang X, Zhao Z, Zhao Z. LncRNA MALAT1 Contributes to Non-Small Cell Lung Cancer Progression via Modulating miR-200a-3p/Programmed Death-Ligand 1 Axis. Int J Immunopathol Pharmacol (2019) 33:2058738419859699. doi: 10.1177/2058738419859699
11. Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. M(6)A mRNA Methylation Initiated by METTL3 Directly Promotes YAP Translation and Increases YAP Activity by Regulating the MALAT1-miR-1914-3p-YAP Axis to Induce NSCLC Drug Resistance and Metastasis. J Hematol Oncol (2019) 12:135. doi: 10.1186/s13045-019-0830-6
12. Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A Screen for Nuclear Transcripts Identifies Two Linked Noncoding RNAs Associated With SC35 Splicing Domains. BMC Genomics (2007) 8:39. doi: 10.1186/1471-2164-8-39
13. Wilusz JE. Long Noncoding RNAs: Re-Writing Dogmas of RNA Processing and Stability. Biochim Biophys Acta (2016) 1859:128–38. doi: 10.1016/j.bbagrm.2015.06.003
14. Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, et al. The lncRNA Malat1 Is Dispensable for Mouse Development But Its Transcription Plays a Cis-Regulatory Role in the Adult. Cell Rep (2012) 2:111–23. doi: 10.1016/j.celrep.2012.06.003
15. Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, et al. A Long Nuclear-Retained Non-Coding RNA Regulates Synaptogenesis by Modulating Gene Expression. EMBO J (2010) 29:3082–93. doi: 10.1038/emboj.2010.199
16. Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, et al. Loss of the Abundant Nuclear Non-Coding RNA MALAT1 Is Compatible With Life and Development. RNA Biol (2012) 9:1076–87. doi: 10.4161/rna.21089
17. Zhao M, Wang S, Li Q, Ji Q, Guo P, Liu X. MALAT1: A Long Non-Coding RNA Highly Associated With Human Cancers. Oncol Lett (2018) 16:19–26. doi: 10.3892/ol.2018.8613
18. Wilusz JE, Freier SM, Spector DL. 3’ End Processing of a Long Nuclear-Retained Noncoding RNA Yields a tRNA-Like Cytoplasmic RNA. Cell (2008) 135:919–32. doi: 10.1016/j.cell.2008.10.012
19. Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An Architectural Role for a Nuclear Noncoding RNA: NEAT1 RNA Is Essential for the Structure of Paraspeckles. Mol Cell (2009) 33:717–26. doi: 10.1016/j.molcel.2009.01.026
20. Kurogi Y, Matsuo Y, Mihara Y, Yagi H, Shigaki-Miyamoto K, Toyota S, et al. Identification of a Chemical Inhibitor for Nuclear Speckle Formation: Implications for the Function of Nuclear Speckles in Regulation of Alternative pre-mRNA Splicing. Biochem Biophys Res Commun (2014) 446:119–24. doi: 10.1016/j.bbrc.2014.02.060
21. Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A Triple Helix Stabilizes the 3’ Ends of Long Noncoding RNAs That Lack Poly(A) Tails. Genes Dev (2012) 26:2392–407. doi: 10.1101/gad.204438.112
22. Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of Triple-Helical Structures by the 3’-End Sequences of MALAT1 and MENbeta Noncoding RNAs. Proc Natl Acad Sci USA (2012) 109:19202–7. doi: 10.1073/pnas.1217338109
23. Huang Q, Chi Y, Deng J, Liu Y, Lu Y, Chen J, et al. Fine Particulate Matter 2.5 Exerted Its Toxicological Effect by Regulating a New Layer, Long Non-Coding RNA. Sci Rep (2017) 7:9392. doi: 10.1038/s41598-017-09818-6
24. Leti F, Legendre C, Still CD, Chu X, Petrick A, Gerhard GS, et al. Altered Expression of MALAT1 lncRNA in Nonalcoholic Steatohepatitis Fibrosis Regulates CXCL5 in Hepatic Stellate Cells. Transl Res (2017) 190:25–39.e21. doi: 10.1016/j.trsl.2017.09.001
25. Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, et al. Pathogenic Role of lncRNA-MALAT1 in Endothelial Cell Dysfunction in Diabetes Mellitus. Cell Death Dis (2014) 5:e1506. doi: 10.1038/cddis.2014.466
26. Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, et al. Long Noncoding RNA MALAT1 Regulates Endothelial Cell Function and Vessel Growth. Circ Res (2014) 114:1389–97. doi: 10.1161/CIRCRESAHA.114.303265
27. Shen S, Jiang H, Bei Y, Xiao J, Li X. Long Non-Coding RNAs in Cardiac Remodeling. Cell Physiol Biochem (2017) 41:1830–7. doi: 10.1159/000471913
28. Zhang M, Gu H, Chen J, Zhou X. Involvement of Long Noncoding RNA MALAT1 in the Pathogenesis of Diabetic Cardiomyopathy. Int J Cardiol (2016) 202:753–5. doi: 10.1016/j.ijcard.2015.10.019
29. Gast M, Schroen B, Voigt A, Haas J, Kuehl U, Lassner D, et al. Long Noncoding RNA MALAT1-Derived mascRNA Is Involved in Cardiovascular Innate Immunity. J Mol Cell Biol (2016) 8:178–81. doi: 10.1093/jmcb/mjw003
30. Lu X, Huang J, Wu S, Zheng Q, Liu P, Feng H, et al. The tRNA-Like Small Noncoding RNA mascRNA Promotes Global Protein Translation. EMBO Rep (2020) 21:e49684. doi: 10.15252/embr.201949684
31. Li B, Chen P, Qu J, Shi L, Zhuang W, Fu J, et al. Activation of LTBP3 Gene by a Long Noncoding RNA (lncRNA) MALAT1 Transcript in Mesenchymal Stem Cells From Multiple Myeloma. J Biol Chem (2014) 289:29365–75. doi: 10.1074/jbc.M114.572693
32. Wu M, Lin Z, Li X, Xin X, An J, Zheng Q, et al. HULC Cooperates With MALAT1 to Aggravate Liver Cancer Stem Cells Growth Through Telomere Repeat-Binding Factor 2. Sci Rep (2016) 6:36045. doi: 10.1038/srep36045
33. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol Cell (2010) 39:925–38. doi: 10.1016/j.molcel.2010.08.011
34. Gordon MA, Babbs B, Cochrane DR, Bitler BG, Richer JK. The Long Non-Coding RNA MALAT1 Promotes Ovarian Cancer Progression by Regulating RBFOX2-Mediated Alternative Splicing. Mol Carcinog (2019) 58:196–205. doi: 10.1002/mc.22919
35. Malakar P, Shilo A, Mogilevsky A, Stein I, Pikarsky E, Nevo Y, et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res (2017) 77:1155–67. doi: 10.1158/0008-5472.CAN-16-1508
36. Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, et al. Long Non-Coding RNA MALAT1 Promotes Tumour Growth and Metastasis in Colorectal Cancer Through Binding to SFPQ and Releasing Oncogene PTBP2 From SFPQ/PTBP2 Complex. Br J Cancer (2014) 111:736–48. doi: 10.1038/bjc.2014.383
37. Chen X, He L, Zhao Y, Li Y, Zhang S, Sun K, et al. Malat1 Regulates Myogenic Differentiation and Muscle Regeneration Through Modulating MyoD Transcriptional Activity. Cell Discovery (2017) 3:17002. doi: 10.1038/celldisc.2017.2
38. Huang JK, Ma L, Song WH, Lu BY, Huang YB, Dong HM, et al. MALAT1 Promotes the Proliferation and Invasion of Thyroid Cancer Cells via Regulating the Expression of IQGAP1. BioMed Pharmacother (2016) 83:1–7. doi: 10.1016/j.biopha.2016.05.039
39. Wang J, Su L, Chen X, Li P, Cai Q, Yu B, et al. MALAT1 Promotes Cell Proliferation in Gastric Cancer by Recruiting SF2/ASF. BioMed Pharmacother (2014) 68:557–64. doi: 10.1016/j.biopha.2014.04.007
40. Yang F, Yi F, Han X, Du Q, Liang Z. MALAT-1 Interacts With hnRNP C in Cell Cycle Regulation. FEBS Lett (2013) 587:3175–81. doi: 10.1016/j.febslet.2013.07.048
41. Tay Y, Rinn J, Pandolfi PP. The Multilayered Complexity of ceRNA Crosstalk and Competition. Nature (2014) 505:344–52. doi: 10.1038/nature12986
42. Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G, Zhu YS. MALAT1: A Potential Biomarker in Cancer. Cancer Manage Res (2018) 10:6757–68. doi: 10.2147/CMAR.S169406
43. Yang T, Li H, Chen T, Ren H, Shi P, Chen M. LncRNA MALAT1 Depressed Chemo-Sensitivity of NSCLC Cells Through Directly Functioning on miR-197-3p/P120 Catenin Axis. Molecules Cells (2019) 42:270–83. doi: 10.14348/molcells.2019.2364
44. Liu J, Tian W, Zhang W, Jia Y, Yang X, Wang Y, et al. MicroRNA-142-3p/MALAT1 Inhibits Lung Cancer Progression Through Repressing β-Catenin Expression. BioMed Pharmacother (2019) 114:108847. doi: 10.1016/j.biopha.2019.108847
45. Tang Y, Xiao G, Chen Y, Deng Y. LncRNA MALAT1 Promotes Migration and Invasion of Non-Small-Cell Lung Cancer by Targeting miR-206 and Activating Akt/mTOR Signaling. Anticancer Drugs (2018) 29:725–35. doi: 10.1097/CAD.0000000000000650
46. Li S, Mei Z, Hu HB, Zhang X. The lncRNA MALAT1 Contributes to Non-Small Cell Lung Cancer Development via Modulating miR-124/STAT3 Axis. J Cell Physiol (2018) 233:6679–88. doi: 10.1002/jcp.26325
47. Cui Y, Li G, Zhang X, Dai F, Zhang R. Increased MALAT1 Expression Contributes to Cisplatin Resistance in Non-Small Cell Lung Cancer. Oncol Lett (2018) 16:4821–8. doi: 10.3892/ol.2018.9293
48. Wang D, Zhang S, Zhao M, Chen F. LncRNA MALAT1 Accelerates Non-Small Cell Lung Cancer Progression via Regulating miR-185-5p/MDM4 Axis. Cancer Med (2020) 9:9138–49. doi: 10.1002/cam4.3570
49. Rong F, Liu L, Zou C, Zeng J, Xu Y. MALAT1 Promotes Cell Tumorigenicity Through Regulating miR-515-5p/EEF2 Axis in Non-Small Cell Lung Cancer. Cancer Manage Res (2020) 12:7691–701. doi: 10.2147/CMAR.S242425
50. Huang J, Lin C, Dong H, Piao Z, Jin C, Han H, et al. Targeting MALAT1 Induces DNA Damage and Sensitize Non-Small Cell Lung Cancer Cells to Cisplatin by Repressing BRCA1. Cancer Chemother Pharmacol (2020) 86:663–72. doi: 10.1007/s00280-020-04152-7
51. Yu W, Ding J, He M, Chen Y, Wang R, Han Z, et al. Estrogen Receptor β Promotes the Vasculogenic Mimicry (VM) and Cell Invasion via Altering the lncRNA-MALAT1/miR-145-5p/NEDD9 Signals in Lung Cancer. Oncogene (2019) 38:1225–38. doi: 10.1038/s41388-018-0463-1
52. Wang S, Wang T, Liu D, Kong H. LncRNA MALAT1 Aggravates the Progression of Non-Small Cell Lung Cancer by Stimulating the Expression of COMMD8 via Targeting miR-613. Cancer Manage Res (2020) 12:10735–47. doi: 10.2147/CMAR.S263538
53. Zhang X, He X, Liu Y, Zhang H, Chen H, Guo S, et al. MiR-101-3p Inhibits the Growth and Metastasis of Non-Small Cell Lung Cancer Through Blocking PI3K/AKT Signal Pathway by Targeting MALAT-1. BioMed Pharmacother (2017) 93:1065–73. doi: 10.1016/j.biopha.2017.07.005
54. Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. M(6)A mRNA Methylation Initiated by METTL3 Directly Promotes YAP Translation and Increases YAP Activity by Regulating the MALAT1-miR-1914-3p-YAP Axis to Induce NSCLC Drug Resistance and Metastasis. J Hematol Oncol (2021) 14:32. doi: 10.1186/s13045-021-01048-8
55. Cheung LW, Leung PC, Wong AS. Cadherin Switching and Activation of P120 Catenin Signaling Are Mediators of Gonadotropin-Releasing Hormone to Promote Tumor Cell Migration and Invasion in Ovarian Cancer. Oncogene (2010) 29:2427–40. doi: 10.1038/onc.2009.523
56. Huang J, Liu K, Song D, Ding M, Wang J, Jin Q, et al. Krüppel-Like Factor 4 Promotes High-Mobility Group Box 1-Induced Chemotherapy Resistance in Osteosarcoma Cells. Cancer Sci (2016) 107:242–9. doi: 10.1111/cas.12864
57. Jia Y, Zhang W, Liu H, Peng L, Yang Z, Lou J. Inhibition of Glutathione Synthesis Reverses Krüppel-Like Factor 4-Mediated Cisplatin Resistance. Cancer Chemotherapy Pharmacol (2012) 69:377–85. doi: 10.1007/s00280-011-1708-7
58. Hu Y, Lin J, Fang H, Fang J, Li C, Chen W, et al. Targeting the MALAT1/PARP1/LIG3 Complex Induces DNA Damage and Apoptosis in Multiple Myeloma. Leukemia (2018) 32:2250–62. doi: 10.1038/s41375-018-0104-2
59. Anantha RW, Simhadri S, Foo TK, Miao S, Liu J, Shen Z, et al. Functional and Mutational Landscapes of BRCA1 for Homology-Directed Repair and Therapy Resistance. Elife (2017) 6. doi: 10.7554/eLife.21350
60. Tani H, Nakamura Y, Ijiri K, Akimitsu N. Stability of MALAT-1, a Nuclear Long Non-Coding RNA in Mammalian Cells, Varies in Various Cancer Cells. Drug Discov Ther (2010) 4:235–9.
61. Xie ZC, Dang YW, Wei DM, Chen P, Tang RX, Huang Q, et al. Clinical Significance and Prospective Molecular Mechanism of MALAT1 in Pancreatic Cancer Exploration: A Comprehensive Study Based on the GeneChip, GEO, Oncomine, and TCGA Databases. OncoTargets Ther (2017) 10:3991–4005. doi: 10.2147/OTT.S136878
62. Zidan HE, Karam RA, El-Seifi OS, Abd Elrahman TM. Circulating Long Non-Coding RNA MALAT1 Expression as Molecular Biomarker in Egyptian Patients With Breast Cancer. Cancer Genet (2018) 220:32–7. doi: 10.1016/j.cancergen.2017.11.005
63. Pan J, Bian Y, Cao Z, Lei L, Pan J, Huang J, et al. Long Noncoding RNA MALAT1 as a Candidate Serological Biomarker for the Diagnosis of Non-Small Cell Lung Cancer: A Meta-Analysis. Thorac Cancer (2020) 11:329–35. doi: 10.1111/1759-7714.13265
64. Lin L, Li H, Zhu Y, He S, Ge H. Expression of Metastasis-Associated Lung Adenocarcinoma Transcript 1 Long Non-Coding RNA In Vitro and in Patients With Non-Small Cell Lung Cancer. Oncol Lett (2018) 15:9443–9. doi: 10.3892/ol.2018.8531
65. Schmidt LH, Spieker T, Koschmieder S, Schäffers S, Humberg J, Jungen D, et al. The Long Noncoding MALAT-1 RNA Indicates a Poor Prognosis in Non-Small Cell Lung Cancer and Induces Migration and Tumor Growth. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer (2011) 6:1984–92. doi: 10.1097/JTO.0b013e3182307eac
66. Weber DG, Johnen G, Casjens S, Bryk O, Pesch B, Jockel KH, et al. Evaluation of Long Noncoding RNA MALAT1 as a Candidate Blood-Based Biomarker for the Diagnosis of Non-Small Cell Lung Cancer. BMC Res Notes (2013) 6:518. doi: 10.1186/1756-0500-6-518
67. Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, et al. Serum Long Non Coding RNA MALAT-1 Protected by Exosomes Is Up-Regulated and Promotes Cell Proliferation and Migration in Non-Small Cell Lung Cancer. Biochem Biophys Res Commun (2017) 490:406–14. doi: 10.1016/j.bbrc.2017.06.055
68. Poulet G, Massias J, Taly V. Liquid Biopsy: General Concepts. Acta Cytol (2019) 63:449–55. doi: 10.1159/000499337
69. Chen M, Wu D, Tu S, Yang C, Chen D, Xu Y. A Novel Biosensor for the Ultrasensitive Detection of the lncRNA Biomarker MALAT1 in Non-Small Cell Lung Cancer. Sci Rep (2021) 11:3666. doi: 10.1038/s41598-021-83244-7
70. Hu N, Chen L, Wang C, Zhao H. MALAT1 Knockdown Inhibits Proliferation and Enhances Cytarabine Chemosensitivity by Upregulating miR-96 in Acute Myeloid Leukemia Cells. BioMed Pharmacother (2019) 112:108720. doi: 10.1016/j.biopha.2019.108720
71. Voce DJ, Bernal GM, Wu L, Crawley CD, Zhang W, Mansour NM, et al. Temozolomide Treatment Induces lncRNA MALAT1 in an NF-κb and P53 Codependent Manner in Glioblastoma. Cancer Res (2019) 79:2536–48. doi: 10.1158/0008-5472.CAN-18-2170
72. Bai L, Wang A, Zhang Y, Xu X, Zhang X. Knockdown of MALAT1 Enhances Chemosensitivity of Ovarian Cancer Cells to Cisplatin Through Inhibiting the Notch1 Signaling Pathway. Exp Cell Res (2018) 366:161–71. doi: 10.1016/j.yexcr.2018.03.014
73. Costea T, Vlad OC, Miclea LC, Ganea C, Szöllősi J, Mocanu MM. Alleviation of Multidrug Resistance by Flavonoid and Non-Flavonoid Compounds in Breast, Lung, Colorectal and Prostate Cancer. Int J Mol Sci (2020) 21. doi: 10.3390/ijms21020401
74. Fang Z, Chen W, Yuan Z, Liu X, Jiang H. LncRNA-MALAT1 Contributes to the Cisplatin-Resistance of Lung Cancer by Upregulating MRP1 and MDR1 via STAT3 Activation. BioMed Pharmacother (2018) 101:536–42. doi: 10.1016/j.biopha.2018.02.130
75. Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Screening for Epidermal Growth Factor Receptor Mutations in Lung Cancer. New Engl J Med (2009) 361:958–67. doi: 10.1056/NEJMoa0904554
76. Kim HR, Lee JC, Kim YC, Kim KS, Oh IJ, Lee SY, et al. Clinical Characteristics of Non-Small Cell Lung Cancer Patients Who Experienced Acquired Resistance During Gefitinib Treatment. Lung Cancer (Amsterdam Netherlands) (2014) 83:252–8. doi: 10.1016/j.lungcan.2013.11.008
77. Yang Q, Chen W, Xu Y, Lv X, Zhang M, Jiang H. Polyphyllin I Modulates MALAT1/STAT3 Signaling to Induce Apoptosis in Gefitinib-Resistant Non-Small Cell Lung Cancer. Toxicol Appl Pharmacol (2018) 356:1–7. doi: 10.1016/j.taap.2018.07.031
78. Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The Noncoding RNA MALAT1 Is a Critical Regulator of the Metastasis Phenotype of Lung Cancer Cells. Cancer Res (2013) 73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850
79. Amodio N, Raimondi L, Juli G, Stamato MA, Caracciolo D, Tagliaferri P, et al. MALAT1: A Druggable Long Non-Coding RNA for Targeted Anti-Cancer Approaches. J Hematol Oncol (2018) 11:63. doi: 10.1186/s13045-018-0606-4
80. Chery J. RNA Therapeutics: RNAi and Antisense Mechanisms and Clinical Applications. Postdoc J (2016) 4:35–50. doi: 10.14304/SURYA.JPR.V4N7.5
81. Liu N, Feng S, Li H, Chen X, Bai S, Liu Y. Long Non-Coding RNA MALAT1 Facilitates the Tumorigenesis, Invasion and Glycolysis of Multiple Myeloma via miR-1271-5p/SOX13 Axis. J Cancer Res Clin Oncol (2020) 146:367–79. doi: 10.1007/s00432-020-03127-8
82. Abbastabar M, Sarfi M, Golestani A, Khalili E. lncRNA Involvement in Hepatocellular Carcinoma Metastasis and Prognosis. EXCLI J (2018) 17:900–13. doi: 10.17179/excli2018-1541
83. Liu P, Li X, Cui Y, Chen J, Li C, Li Q, et al. LncRNA-MALAT1 Mediates Cisplatin Resistance via miR-101-3p/VEGF-C Pathway in Bladder Cancer. Acta Biochim Biophys Sin (2019) 51:1148–57. doi: 10.1093/abbs/gmz112
84. Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics (2010) 11:537–61. doi: 10.2174/138920210793175895
85. Leucci E, Patella F, Waage J, Holmstrom K, Lindow M, Porse B, et al. microRNA-9 Targets the Long Non-Coding RNA MALAT1 for Degradation in the Nucleus. Sci Rep (2013) 3:2535. doi: 10.1038/srep02535
86. Wu S, Sun H, Wang Y, Yang X, Meng Q, Yang H, et al. MALAT1 Rs664589 Polymorphism Inhibits Binding to miR-194-5p, Contributing to Colorectal Cancer Risk, Growth, and Metastasis. Cancer Res (2019) 79:5432–41. doi: 10.1158/0008-5472.CAN-19-0773
87. Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi Factors Are Present and Active in Human Cell Nuclei. Cell Rep (2014) 6:211–21. doi: 10.1016/j.celrep.2013.12.013
88. Poulet C, Njock MS, Moermans C, Louis E, Louis R, Malaise M, et al. Exosomal Long Non-Coding RNAs in Lung Diseases. Int J Mol Sci (2020) 21. doi: 10.3390/ijms21103580
Keywords: long non-coding RNA, metastasis-associated lung adenocarcinoma transcript-1, non-small cell lung cancer, metastasis, invasion, microRNA
Citation: Zhou Q, Liu L, Zhou J, Chen Y, Xie D, Yao Y and Cui D (2021) Novel Insights Into MALAT1 Function as a MicroRNA Sponge in NSCLC. Front. Oncol. 11:758653. doi: 10.3389/fonc.2021.758653
Received: 14 August 2021; Accepted: 06 October 2021;
Published: 27 October 2021.
Edited by:
Marta Martins, Universidade de Lisboa, PortugalReviewed by:
Nicola Amodio, University of Catanzaro, ItalyCopyright © 2021 Zhou, Liu, Zhou, Chen, Xie, Yao and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dawei Cui, ZGF3ZWljdWlAemp1LmVkdS5jbg==; Dacheng Xie, ZGFjaGVuZ3hpZUAxNjMuY29t; Yinan Yao, eWFveWluYW5Aemp1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.