- 1Department of Orthopedics, Affiliated Hospital of Chifeng University, Chifeng, China
- 2Department of Pathology, Harbin Medical University, Harbin, China
- 3Department of Urology, Hainan General Hospital, Hainan, China
The RNA-binding motif (RBM) proteins are a class of RNA-binding proteins named, containing RNA-recognition motifs (RRMs), RNA-binding domains, and ribonucleoprotein motifs. RBM proteins are involved in RNA metabolism, including splicing, transport, translation, and stability. Many studies have found that aberrant expression and dysregulated function of RBM proteins family members are closely related to the occurrence and development of cancers. This review summarizes the role of RBM proteins family genes in cancers, including their roles in cancer occurrence and cell proliferation, migration, and apoptosis. It is essential to understand the mechanisms of these proteins in tumorigenesis and development, and to identify new therapeutic targets and prognostic markers.
Introduction
RNA-binding proteins (RBPs) are a kind of crucial intracellular protein, which can be widely involved in a variety of post-transcriptional regulation processes, such as RNA splicing, transport, localization, and translation. RBPs are divided into many kinds according to different functions, including Hu-antigen R (HuR), heterogeneous nuclear ribonucleoprotein family (hnRNP), the arginine/serine-rich splicing factor protein family (SRSF), and RNA-binding motif (RBM) proteins family, etc. (1). RBM proteins family is a subgroup of RBPs, which has the same domain characteristics as RBPs, including RNA-recognition motifs (RRMs), RNA-binding domains (RBM), ribonucleoprotein (RNP), cold-shock domain (CSD), and zinc finger (ZnF), etc. (1). RRM is a central structural motif of the RBM proteins family; usually, RBM protein has one or more RRMs, such as RBM3, which includes one RRM, and RBM19 contains up to six RRMs. The member of the RBM proteins family is named sequentially after confirming that they contain RRM. Up to now, more than 50 RBM proteins have been identified (Table S1). It is worth noting that not all the RRM-containing RNA-binding proteins are designated as RBM proteins. Once the exact functions of the RBM protein are determined, the RBM protein will be renamed according to its function, and the ‘‘RBM’’ designation can be removed (2).
Like RBPs, the RBM proteins family are involved in multiple biological activities, such as RNA metabolism, including pre-mRNA splicing, RNA stability, mRNA translation, etc. (Figure 1) (3–7). RBM proteins can regulate alternative splicing by binding to the exon/intron region near the splice site of mRNA. For example, RBM10 can bind to the intron region near the splice site on mRNA, thus interfering with the recognition of splice site, while RBM5 and RBM6 can bind to the exon region near the splice site and recruit splicing components (8). And RBM4 has been reported can regulate the selection of 5’ splice sites or exons in vitro and antagonize the effect of SRSF protein on the selection of 5’ splice sites (9). In addition, RBM proteins can regulate the stability of RNA by directly binding its target mRNAs, such as RBMS1, RBM38, and RBM3 (10–13). Among them, RBM3 also participates in the translation regulation of cyclooxygenase-2 (COX-2) mRNA by recognizing and binding COX-2 AU-rich elements (ARE) sequence. Overexpression of RBM3 can improve the mRNA translation of COX-2 in HCT116 cells (13). Over the past few decades, different effects of the RBM proteins family have been gradually found in various cancer-related studies. In this review, we focused on the role of the RBM proteins family in cancer and summarized the effects of the RBM proteins family members on the occurrence, progression, and treatment of cancer.
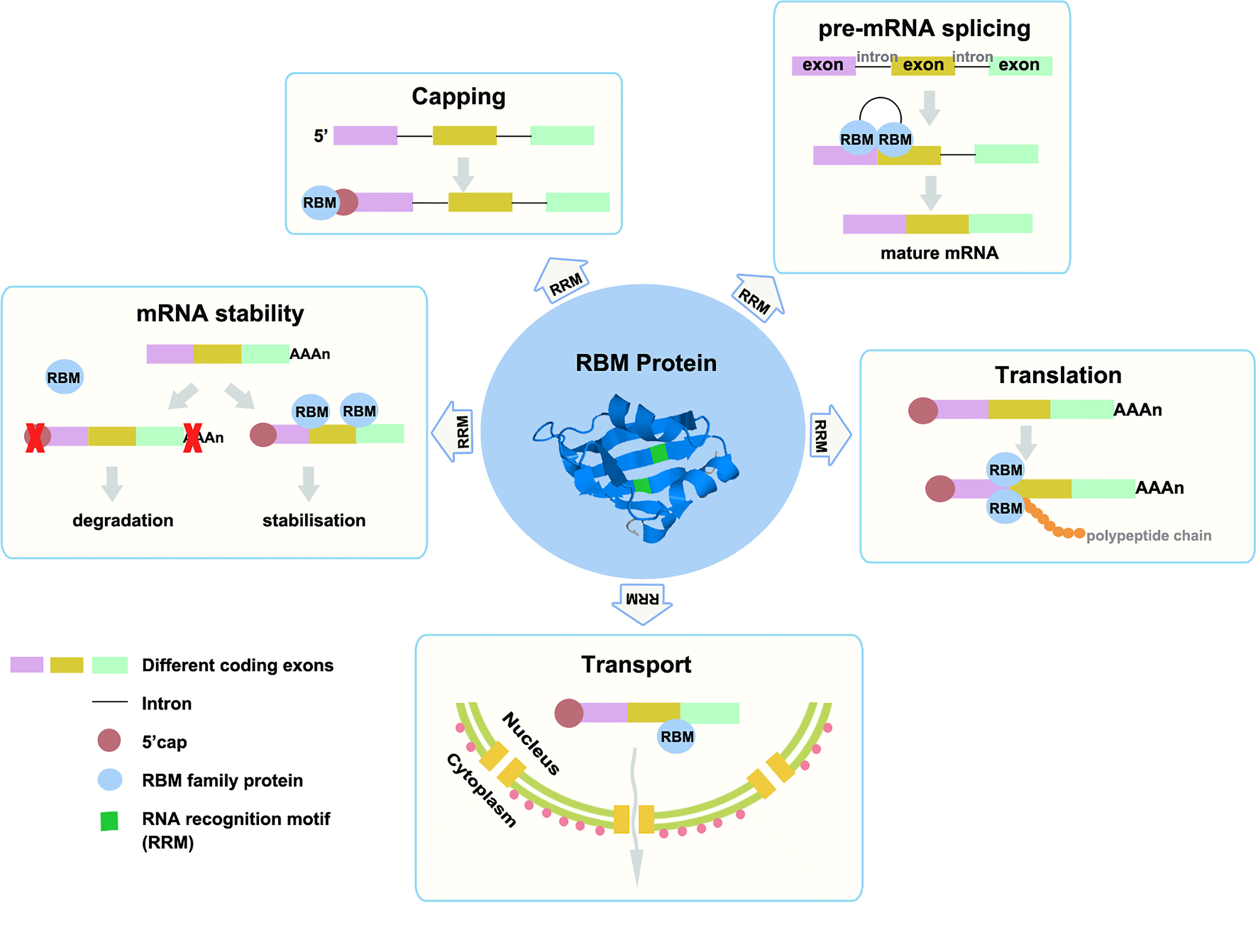
Figure 1 The RBM proteins family can affect gene expression and function by intervening mRNA transport, translation, capping, splicing, and stability. The three colored rectangles represent different exons, black lines represent introns, reddish brown circles represent 5 ‘cap, light blue circles represent RBM family proteins, and green rectangles represent the RRM domain. The central circle is the structure of RBM protein, usually, it has one or more RRMs.
RBM Proteins Family Is Frequently Related to the Occurrence of Cancer
Several studies showed that the RBM proteins family is closely related to the occurrence of cancer. RBM3, a cold-induced RNA-binding protein, was found to be upregulated in several types of cancers (14–17). However, Zeng et al. found that the overexpression of RBM3 in PC3 cells (a human prostate cancer cell line) weakened the stem cell-like characteristics of these cells (18). They indicated that RBM3 hindered the occurrence of prostate cancer because the tumor formation rate of PC3 cells overexpressed with RBM3 in nude mice was significantly lower than that in the control group (18). p53 is the most common mutant gene in human cancer and mutant p53 has been reported to promote tumor metastasis. RBM38, also known as RNPC1, is a target gene of the p53 family; it can inhibit p53 translation by interacting with eIF4E on p53 mRNA (11, 19, 20). Zhang et al. found that RBM38 can jointly regulate mutant p53 and PTEN, a key regulator of T cell development, to affect the occurrence of T cell lymphomas. They showed that the deletion of RBM38 enhanced the expression of mutant p53, and decreased the expression of tumor suppressor PTEN, which promoted the occurrence of lymphoma (21). And Zhang et al. found that mice who deleted RBM38 were more prone to aging and spontaneous tumors (20). These researches indicated that RBM38 could interact with p53 to form a negative feedback regulatory loop to involve tumorgenesis. RNA-binding motif single-stranded interacting protein 3 (RBMS3), another member of the RBM proteins family, has been reported could suppress the morphogenesis of non-small cell lung cancer (NSCLC) (22). Based on these studies, some RBM proteins, such as RBM38, RBM3, and RBMS3, play an inhibitory role in tumorigenesis. However, whether other RBM proteins family have the same effect in tumorigenesis has not been reported. Therefore, the role of the RBM proteins family in tumorigenesis and related molecular mechanisms still needs to be further explored.
RBM Proteins Family Can Promote Tumor Cell Proliferation
Studies have found that RBM proteins family can promote the proliferation of tumor cells. The mechanism of the RBM proteins family promoting proliferation is complex and usually involves the following aspects.
RBM Proteins Family Can Affect Tumor Cell Proliferation by Regulating Cancer-Related Genes and Signaling Pathways
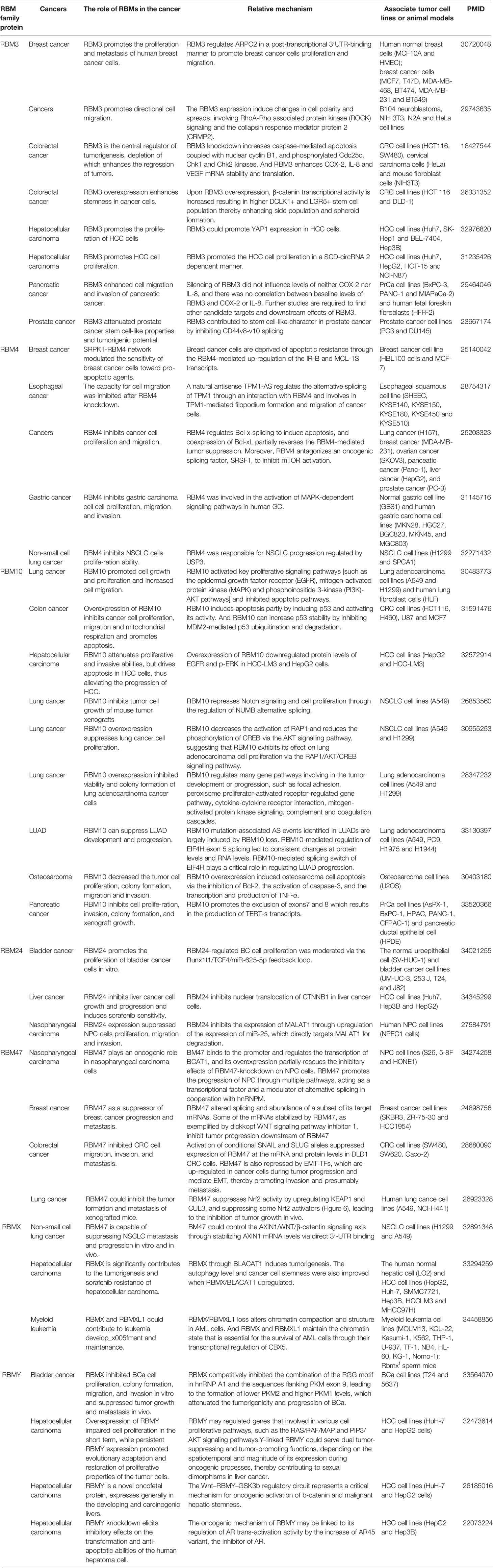
SM Sureban et al. reported that RBM3 could promote the proliferation of colon cancer cells by enhancing the stability and translation ability of COX-2, IL-8 and VEGF mRNA (13). Hypoxic and other adverse conditions that are detrimental to cell growth, RBM3 participates in the survival of colon cancer cells mainly through a COX-2 signal transduction mechanism (23). Furthermore, RBM3 could promote the growth and proliferation of hepatocellular carcinoma (HCC) cells in the stearoyl-CoA desaturase (SCD)-circRNA-2-dependent manner by control SCD-circRNA-2 formation (24). Lin et al. found that RBM4 inhibits the apoptosis of breast cancer cells by upregulating the expression of transcripts IR-B and MCL-1S (25). In the U251 cell line, RBM17 decreased the expression of apoptosis related factors caspase 3, caspase 9 and PARP, and promoted the proliferation of glioma cells (26).
RBM Proteins Family Can Promote Cell Proliferation by Participating in the Regulation of Cell Cycle
HAN et al. found that inhibiting RBM17 expression can significantly reduce the proliferation of hypopharyngeal carcinoma cells, promote their apoptosis, and block their cell cycle progression at the G2/M phase (27). RBM17 plays a similar role in HCC and glioma. Li et al. showed that inhibiting RBM17 expression can decrease the proliferation of HCC cells, arrest cells at the G2/M phase, and significantly increase the apoptosis rate (28). In breast cancer cells, knocking down the RBM7 gene also inhibits cell proliferation, and induces G1 cell cycle arrest. Whereas overexpressing RBM7 promotes the proliferation of breast cancer cells by binding to AU-rich elements of cyclin-dependent kinase1 (CDK1) 3’-UTR and then stabilizing CDK1 mRNA (29).
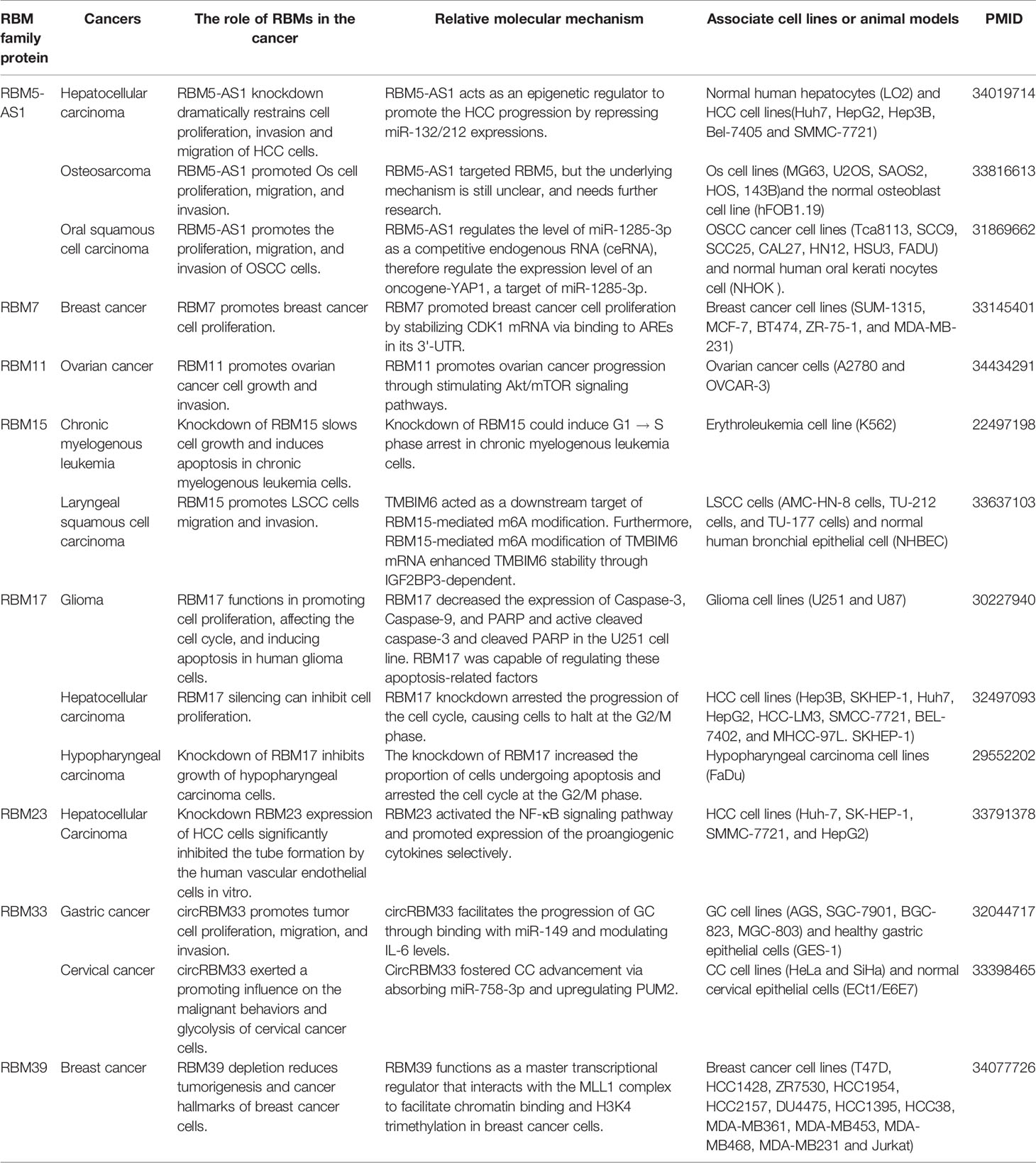
Other RBM proteins family members also participate in promoting proliferation in various cancers, such as RBM5-AS1, RBM11, RBM15, RBM23, RBM33, etc. (Table 1). The proliferative effects of the RBM protein family members, as mentioned above that on tumor cells, may contribute to tumor progression. Nevertheless, the RBM proteins family can also play anti-tumor effects in cancers.
RBM Proteins Family Inhibits Tumor Cell Proliferation
To date, studies on the anti-tumor proliferation effects of the RBM proteins family on cancer have mainly focused on the following aspect.
RBM Protein Inhibits Cell Proliferation by Targeting the Expression of Proto-Oncogene or Anti-Oncogene
RBM38 can suppress c-Myc protein expression to suppressed cell proliferation by directly binding to target AU-rich elements in the 3’-UTR of c-Myc mRNA. Conversely, c-Myc negatively regulates RBM38 expression by binding to the E-box in the promoter region of the RBM38 gene in breast cancer (30). RBM38 can also increase the expression of phosphatase and tensin homolog gene on chromosome 10 (PTEN) by binding to the 3’-UTR of PTEN transcript, thereby inhibiting the cell proliferation of breast cancer (31). Zhang et al. reported that RBM38 is phosphorylated at Ser195 by glycogen synthase kinase 3 (GSK3), promoting the translation of p53 mRNA and inhibiting tumor cell growth and proliferation (32).
RBM Proteins Family Can Inhibit Cell Proliferation by Regulating the Cell Cycle in Cancer
P21 protein is a cyclin-dependent kinase inhibitor that can arrest the cell cycle and prevent cell proliferation. RBMS2 positively regulates the stability of P21 mRNA by binding to its 3’ -UTR and therefore inhibits the proliferation of breast cancer cells (33). RBM43 is another tumor suppressor gene in the RBM proteins family. It is significantly downregulated in tumors, and its low expression is associated with a poor prognosis (34). Overexpression of RBM43 can inhibit the cell cycle progression by directly binding to the 3’ -UTR of CyclinB1 and then reducing CyclinB1 expression in HCC cells (34).
RBM Protein Can Inhibit Tumor Proliferation by Regulating Signal Pathway
Yong et al. found that RBM4 can inhibit the proliferation of gastric cancer cells in vitro and in vivo. RBM4 inhibits the activity of MAPK dependent signal pathway by inhibiting the expression of MAPK pathway protein, so it plays a role in inhibiting the proliferation of gastric cancer cells (35). RBM5 is a tumor suppressor gene in lung cancer and breast cancer, but its role in the pathogenesis of medulloblastoma (MB) remains unclear. Yu et al. Found that RBM5 knockdown induced Daoy and ons-76 cells proliferation, and the β-Catenin protein expression level was up-regulated in Daoy cells, Therefore, RBM5 may regulate Wnt/β- Catenin signal transduction plays a tumor suppressive role in MB (36). Jiang et al. Found similar results. In human glioma, RBM5 inhibits Wnt/β- Catenin signal transduction to play a role in tumor inhibition (37). Rbm10 can inhibit Notch signal transduction and cell proliferation by regulating the variable splicing of numb. RBM10, a splicing factor, inhibits cell proliferation by switching hTERT transcripts to generate a function-less isoform and suppressing the telomerase activity in pancreatic cancer (38). RBM10 is also an alternative splicing regulator of the Notch regulator gene NUMB. Jordi Hernandez et al. found that RBM10 can inhibit cell proliferation by promoting exon 9 skipping of NUMB in lung adenocarcinoma (LUAD) (39). The inhibitory effect of RBM10 on cell proliferation can be obtained through inactivating RAP1/Akt/CREB signaling pathway in LUAD cells (40).
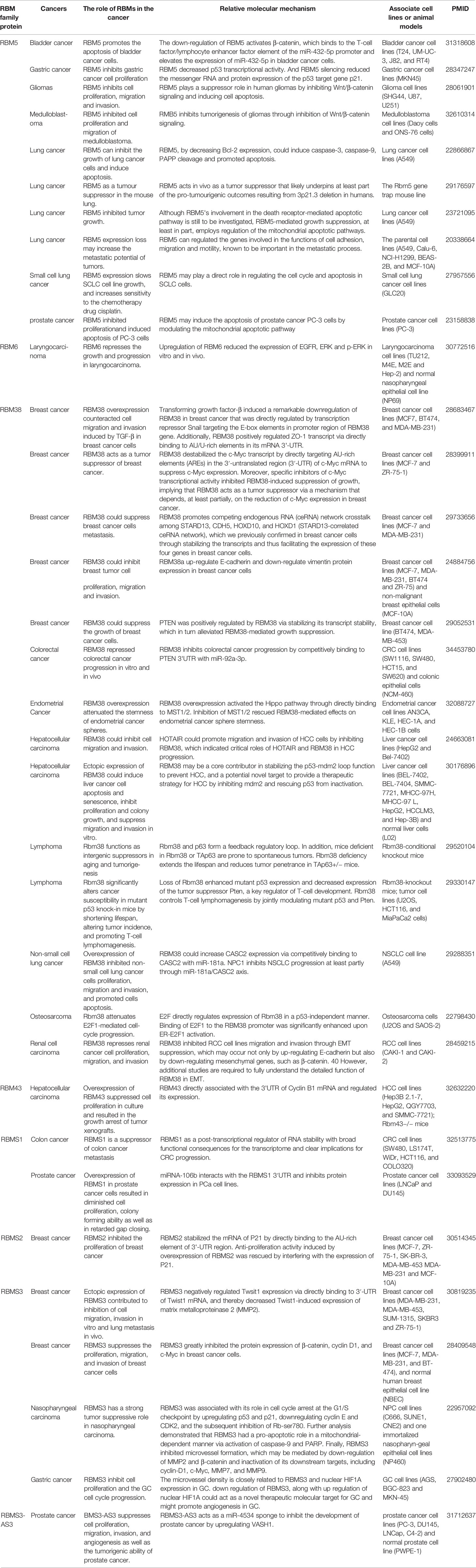
Other RBM proteins family members also participate in suppressing proliferation in different cancers, such as RBM6, RBMS1, RBMS2, RBMS3, etc. (Table 2). In fact, many RBM proteins family members have dual effects on tumor cells, namely promoting proliferation and inhibiting proliferation, including RBM3, RBM4, RBM10, RBMX, etc. The mechanism of these genes’ dual effect in different tumors is not precise. It may be related to the characteristics of tumors and the location of gene expression, still need more research.
The Effect of RBM Proteins Family on Tumor Cell Apoptosis
The studies mentioned above indicate that the RBM proteins family members play essential roles in tumor proliferation. In addition, many studies have found that the RBM proteins family also involve in the regulation of apoptosis in cancer, mainly in two aspects.
RBM Proteins Family Can Regulate Tumor Cell Apoptosis by Induced Pro-Apoptotic Genes and Apoptosis Regulatory Proteins, Including Bax and p53
Garabito et al. found that the expression of RBM genes (RBMX, RBM3, and RBM10) on the X chromosome is remarkably associated with the pro-apoptotic gene Bax in breast cancer cells (41). RBM10, a vital member of the RBM genes on the X chromosome, can also promote cell apoptosis by enhancing the expression of TNF-α and regulating the alternative splicing of related genes, including FAS and BCL-X (42, 43). Rbm10 can also increase the stability of p53 by inhibiting MDM2 mediated ubiquitination and degradation of p53 and prolong the half-life of p53 to induce apoptosis to inhibit cancer cell proliferation and induce apoptosis.
RBM Proteins Family Can Affect Apoptosis by Enhanced Mitochondrial Apoptotic Activity and Upregulated the Expression of Autophagy-Related Proteins
Zhao et al. found that RBM5 protein expression significantly decreased in prostate cancer tissues than in normal tissues. Mitochondrial apoptotic activity is significantly increased when RBM5 is overexpressed in prostate cancer cells (44). The upregulation of RBM5 can induce cell apoptosis and increased cell sensitivity to certain apoptotic stimuli by altering the apoptosis regulatory proteins (44, 45). Loiselle et al. reported that RBM5 could directly regulate the cell cycle and apoptosis in small-cell lung cancer (SCLC) (46). RBM5 upregulates the level of autophagy-related proteins, such as LC3, Beclin1, and LAMP1, which further induce cell autophagy in LUAD (47). Similarly, down-regulation of RBM5 in bladder cancer cells leads to inhibition of apoptosis by increasing the expression of β-catenin-mediated mir-432-5p (48). The pro-apoptotic effect of RBM protein may contribute to inhibit tumor progression. Hence, it can accelerate tumor cell death by inducing the expression of RBM proteins family members. And this mechanism may benefit targeted therapy of tumors in the future.
The RBM Proteins Family Affect the Invasion and Migration of Tumor Cells
In addition to playing a role in tumor cell proliferation and apoptosis, the RBM proteins family can also affect the migration and invasion of tumor cells. Most RBM proteins family effect as an inhibitor in the invasion and metastasis of cancer, the primary biology mechanism as the following.
RBM Proteins Family Protein Can Inhibit Tumor Cell Invasion and Metastasis by Targeted Gene Expression
Zonula occludens-1 (ZO-1) is a member of the membrane-associated guanylate kinase (MAGUK) family of proteins, which can control endothelial cell-cell tension, cell migration, and barrier formation (49). RBM38 can positively regulate the ZO-1 gene by directly binding to AU/U-rich elements in the ZO-1 mRNA 3’-UTR. Therefore, overexpression of RBM38 can reverse the invasion and migration of breast cancer cells caused by the knockdown of ZO-1 (50). RBMS3 negatively regulates the expression of Twsit1 and reduces the level of Matrix metalloproteinase 2 (MMP2) induced by Twist1, thus inhibiting the invasion and metastasis of breast cancer cells (51). In human prostate cancer, RBM25 binding directly to an Amotl1-derived circRNA, circAMOTL1L, resulted in the relief of the miR-193a-5p repression of the Pcdha gene cluster, whereas p53 regulates EMT via directly activating the RBM25 gene (52).
RBM Proteins Family Could Inhibit Cancer Invasion and Metastasis by Involving Signaling Pathways, and Regulation mRNA Stability, etc
RBM47 could inhibit the metastasis of NSCLC by increasing the stability of AXIN1 mRNA and then inhibiting Wnt/β-catenin signal transduction (53). In breast cancer, RBM47 also plays a similar role. Dickkopf-1(DKK-1), as a WNT signaling pathway inhibitor 1, was bound with RBM47 to inhibit the activation of the WNT pathway and exert a tumor suppressor effect (54). As a post-transcriptional regulator of RNA stability, RBMS1 has clear significance for the progression of colon cancer. In a mouse model of xenotransplantation, silencing RBMS1 increased the metastatic ability of colon cancer cells while restoring RBMS1 weakened the metastatic capacity of colon cancer cells (10). Some studies have also shown that RBM5 could inhibit the metastasis and invasion of lung cancer (55, 56). Moreover, RBM3 downregulation is related to the distant metastasis of esophageal squamous cell carcinoma (57).
RBM Proteins Family Also Can Promote Cancer Cell Invasion and Metastasis
However, Huang et al. reported that RBM4 promotes the migration and invasion of esophageal cancer. They found that RBM family protein could promote tumor invasion and metastasis by participating in the alternative splicing of some genes, such as tropomyosin I (TPM1). Knockout of the RBM4 gene resulted in specific down-regulation of TPM1 variants V2 and V7, which might inhibit migration and filamentous group formation in esophageal cancer cells (58). RBM5-AS1 can be used as an oncogenic factor in multiple cancers, such as hepatocellular carcinoma, osteosarcoma, and oral squamous cell carcinoma. Mu et al. showed that RBM5-AS1 could decrease miR-132/212 by recruit PRC2 complex, and facilitate HCC cell migration and invasion (59). Fu et al. found that RBM11 was highly expressed in ovarian cancer and could promote tumor cell invasion and metastasis by activating Akt/mTOR signaling (60). circRBM33 was generated from the RBM33 and could promote gastric cancer cells migration and invasion through the circRBM33/miR-149/IL-6 axis (61).
In conclusion, RBM proteins family protein has a dual role in different cancers. Most RBM proteins family effect as an inhibitor in the migration and invasion of cancer cells, while a small group of RBM proteins family members could facilitate tumor cell migration and invasion. It is necessary to explore further why RBM proteins family play different roles in tumor cells. And the relative molecular mechanism of the RBM proteins family may provide a theoretical basis for future research and clinical application.
RBM Proteins Family Can Use As a Predictor for a Prognosis of Cancer
Recently, it was reported that in various tumors, such as liver cancer, gastric cancer, lung cancer, and breast cancer, the expression level of RBM proteins is related to the tumor size, invasion depth, lymph node metastasis, and prognosis. Yong et al. found that RBM4 expression in gastric cancer tissues was significantly lower than that in adjacent normal tissues. The downregulation of RBM4 was significantly associated with poor differentiation, lymph node metastasis, distant metastasis, and advanced Tumor Node Metastasis (TNM) stage in gastric cancer (62). They found that compared with the RBM4 high-expression group, the RBM4 low expression group had worse overall survival (OS) and disease-free survival (DFS) (62). Gao et al. showed that the overexpression of RBM3 in patients with colorectal cancer, gastric cancer, or melanoma predicted a good prognosis (63). Besides, RBM15 was identified as high-confidence interactors with Wilms tumor-associated protein (WTAP) in proteomic analysis. WTAP binds METTL3, the methyltransferase that mediates methylation of m6A in mRNA16, and is recruited to RNAs via an unknown adaptor protein to trigger m6A formation. Patil et al. found RBM15 is part of the WTAP-METTL3 N6-methyladenosine (m6A) methyltransferase complex and participates in M6A modification (64). Studies on RNA methylation regulators in papillary thyroid carcinoma (PTC) and gastric cancer have shown that RBM15 was significantly positively correlated with a better prognosis (65, 66). But in LUAD, the high expression level of RBM15 is related to a poor prognosis (67, 68).
And we further explored the relationship between the expression level of RBM proteins family members and prognosis in different cancers by TCGA. The results showed that members of the RBM proteins family were significantly correlated with tumor prognosis, and the expression levels of many RBM members could predict the prognosis of tumor patients (Figure 2). For example, a high expression of the RBM proteins was associated with shorter survival in ACC. Conversely, low expression of the RBM proteins in kidney renal clear cell carcinoma (KIRC) was associated with a worse prognosis.
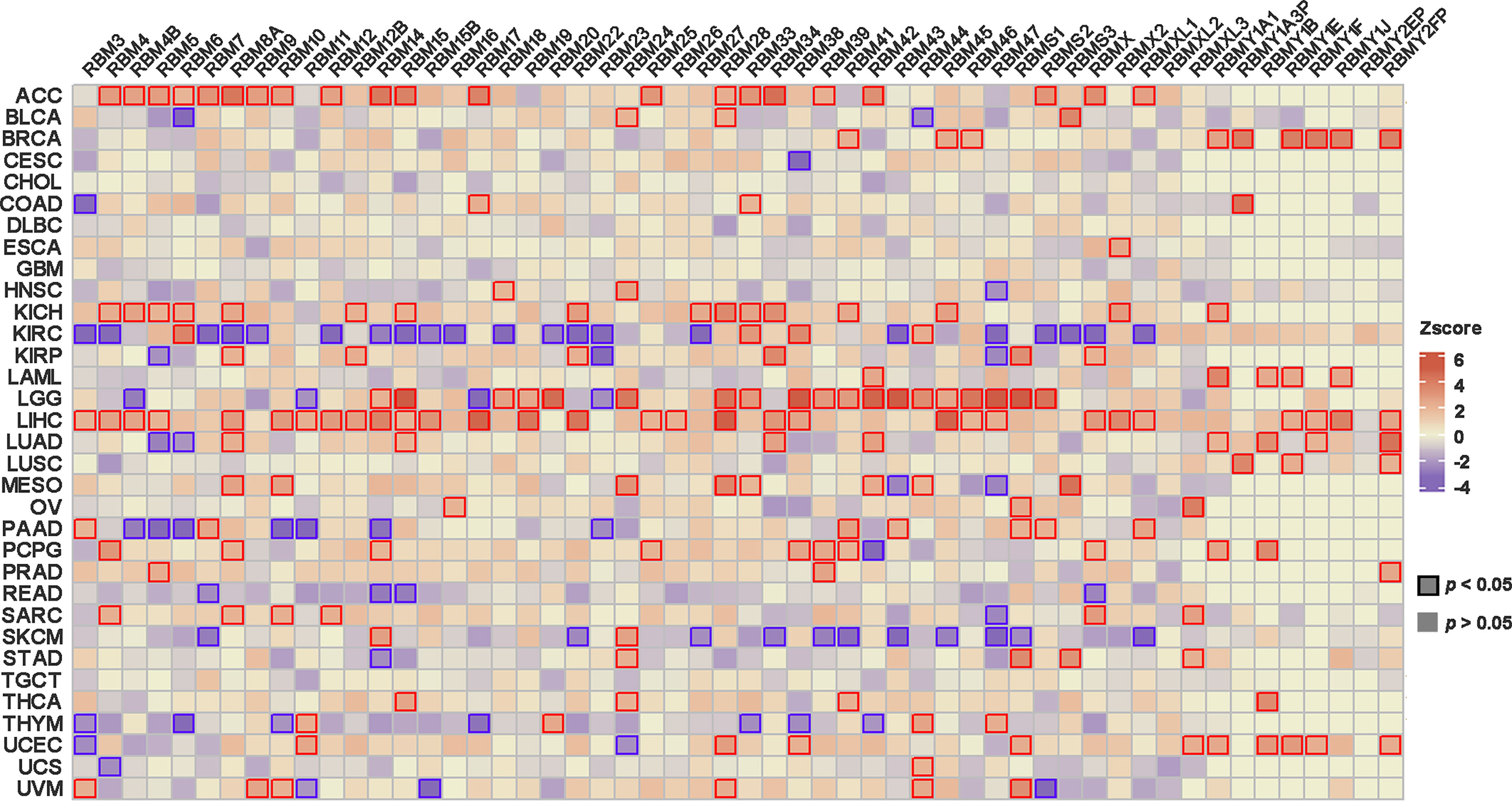
Figure 2 The correlation analysis between RBM proteins family expression and the patient’s prognosis in the TCGA. p < 0.05 was considered statistically significant.
RBM Can Be Used as a Potential Therapeutic Target in Cancers
As mentioned above, there are significant differences in the expression levels of RBM proteins in lung cancer, breast cancer, liver cancer, colon cancer, and other human cancers, and their expression levels are significantly correlated with prognosis. Therefore, targeting RBM protein may be a new therapeutic strategy for the treatment of human cancer. In fact, previous studies have shown that RBMX is highly expressed in HCC tissues and cell lines, resulting in increased drug resistance of HCC cells (69, 70). And targeting RBMX can be used as a new strategy for HCC treatment. RBM39 can bind to c-Jun and stimulate its transcriptional activity, promoting its involvement in many aspects of cancer development. Studies have found that RBM39 is highly expressed in breast cancer tissues and can promote tumor cell proliferation. Considering the role of RBM39 in breast cancer, Shannon D Chilewski et al. developed an RBM39 peptide to treat triple-negative breast cancer (71). A bioinformatics analysis also showed that RBM39, a target gene of miR-494, can be used as a biomarker to predict trastuzumab resistance in breast cancer (72). Additionally, Wu et al. found that loss of RBMS3 might increase the chemical resistance of epithelial ovarian cancer (73). Downregulation of mir-383 induced RBM24 mediated NF-κB signal activation. Therefore, RBM24 can become a potential therapeutic target to reverse the chemoresistance of lung adenocarcinoma cells (74). In patients treated with oxaliplatin, a first-line chemotherapy drug, high expression of RBM3 is an independent predictor of prolonged survival in patients with metastatic colorectal cancer (75). In epithelial ovarian cancer cell lines, RBM3 expression silencing resulted in decreased sensitivity to cisplatin. It was suggested that RBM3 might be a useful therapeutic predictor in epithelial ovarian cancer (76).
Although the role of other members of the RBM proteins family in tumor treatment and prognosis is not clear, existing studies have shown that some members of RBM proteins family, such as RBM3, RBM4, and RBM39, play an essential role in tumor treatment and prognostic markers. Therefore, RBM proteins may be a potential target for tumor treatment and prognosis.
Conclusion
In recent years, increasing attention has been paid to the RBM proteins family, and their various roles in multiple cancers have been continuously revealed. It was shown that some members of the RBM proteins family play a tumor-suppressive role in cancers, inhibiting tumorigenesis and cell proliferation, promoting tumor cell apoptosis, and limiting cell migration and invasion, such as RBM6 and RBM38. While some members play the opposite role, promoting cell proliferation and the invasion of cancer, including RBM7, RBM11, and RBM15. Besides, another part of RBM proteins plays a dual function of cancers (Table 3). For instance, RBM3 plays a cancer-promoting role in breast and colorectal cancer, while it inhibits tumorigenesis in prostate cancer. And RBM5 and RBM5-AS1 play opposite effects in tumor cells. RBM5 can inhibit tumor cell growth in gastric cancer and lung cancer. RBM5-AS1 promotes the migration and invasion of osteosarcoma tumor cells. It was not clarified that why RBM proteins family members play a dual role in tumors. Kido et al. found that the dual role of RBM proteins family genes may be related to time and space (77). They discovered that RBMY acts as a suppressor in the early stages of the tumor and shows a cancer-promoting effect in the long-term progression of tumors. However, the specific mechanism still needs more in-depth exploration. The dual function of RBMs may provide a novel idea for the treatment and research of tumors, as some tumor-promoting factors may also be turned into tumor suppressor factors under some conditions. Besides, it can also amplify the tumor suppressor role of RBM protein that may be used as a new target for clinical treatment of tumors. In future studies, further exploration of the dual role of RBM proteins family in tumors and elucidating the related molecular mechanisms may contribute to the development of new therapeutic targets.
In summary, based on the current research, the influence of the RBM proteins family on cancer is diverse, and these proteins are involved in various aspects of tumorigenesis and development. And RBM protein can be used as novel tumor markers for clinical application in early diagnosis and prognosis evaluation of multiple cancers.
Author Contributions
ZL, QG and ZF searched the pubmed and literatures. JZ and QG performed tables and figures. QG, JZ, and ZF wrote the first draft of the manuscript. YW and ZL wrote sections of the manuscript. JT and TW review and editing manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.757135/full#supplementary-material.
Supplementary Table 1 | The member of the RBM protein family.
References
1. Qin H, Ni H, Liu Y, Yuan Y, Xi T, Li X, et al. RNA-Binding Proteins in Tumor Progression. J Hematol Oncol (2020) 1(13):90. doi: 10.1186/s13045-020-00927-w
2. Sutherland LC, Rintala-Maki ND, White RD, Morin CD. RNA Binding Motif (RBM) Proteins: A Novel Family of Apoptosis Modulators? J Cell Biochem (2005) 1(94):5–24. doi: 10.1002/jcb.20204
3. Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-Binding Proteins and the Messages They Carry. Nat Rev Mol Cell Biol (2002) 3(3):195–205. doi: 10.1038/nrm760
4. Lee MS, Henry M, Silver PA. A Protein That Shuttles Between the Nucleus and the Cytoplasm is an Important Mediator of RNA Export. Genes Dev (1996) 10(10):1233–46. doi: 10.1101/gad.10.10.1233
5. Jonas K, Calin GA, Pichler M. RNA-Binding Proteins as Important Regulators of Long Non-Coding RNAs in Cancer. Int J Mol Sci (2020) 21(8):2969. doi: 10.3390/ijms21082969
6. Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, Xiao R, et al. A Large-Scale Binding and Functional Map of Human RNA-Binding Proteins. Nature (2020) 7818(583):711–19. doi: 10.1038/s41586-020-2077-3
7. Maris C, Dominguez C, Allain FH. The RNA Recognition Motif, a Plastic RNA-Binding Platform to Regulate Post-Transcriptional Gene Expression. FEBS J (2005) 9(272):2118–31. doi: 10.1111/j.1742-4658.2005.04653.x
8. Bechara EG, Sebestyen E, Bernardis I, Eyras E, Valcarcel J. RBM5, 6, and 10 Differentially Regulate NUMB Alternative Splicing to Control Cancer Cell Proliferation. Mol Cell (2013) 5(52):720–33. doi: 10.1016/j.molcel.2013.11.010
9. Lai MC, Kuo HW, Chang WC, Tarn WY. A Novel Splicing Regulator Shares a Nuclear Import Pathway With SR Proteins. EMBO J (2003) 6(22):1359–69. doi: 10.1093/emboj/cdg126
10. Yu J, Navickas A, Asgharian H, Culbertson B, Fish L, Garcia K, et al. RBMS1 Suppresses Colon Cancer Metastasis Through Targeted Stabilization of Its mRNA Regulon. Cancer Discov (2020) 9(10):1410–23. doi: 10.1158/2159-8290.CD-19-1375
11. Shu L, Yan W, Chen X. RNPC1, an RNA-Binding Protein and a Target of the P53 Family, Is Required for Maintaining the Stability of the Basal and Stress-Induced P21 Transcript. Genes Dev (2006) 21(20):2961–72. doi: 10.1101/gad.1463306
12. Zhang J, Jun Cho S, Chen X. RNPC1, an RNA-Binding Protein and a Target of the P53 Family, Regulates P63 Expression Through mRNA Stability. Proc Natl Acad Sci USA (2010) 21(107):9614–9. doi: 10.1073/pnas.0912594107
13. Sureban SM, Ramalingam S, Natarajan G, May R, Subramaniam D, Bishnupuri KS, et al. Translation Regulatory Factor RBM3 Is a Proto-Oncogene That Prevents Mitotic Catastrophe. Oncogene (2008) 33(27):4544–56. doi: 10.1038/onc.2008.97
14. Jogi A, Brennan DJ, Ryden L, Magnusson K, Ferno M, Stal O, et al. Nuclear Expression of the RNA-Binding Protein RBM3 Is Associated With an Improved Clinical Outcome in Breast Cancer. Mod Pathol (2009) 12(22):1564–74. doi: 10.1038/modpathol.2009.124
15. Salomonsson A, Micke P, Mattsson JSM, La Fleur L, Isaksson J, Jonsson M, et al. Comprehensive Analysis of RNA Binding Motif Protein 3 (RBM3) in Non-Small Cell Lung Cancer. Cancer Med (2020) 15(9):5609–19. doi: 10.1002/cam4.3149
16. Jonsson L, Gaber A, Ulmert D, Uhlen M, Bjartell A, Jirstrom K. High RBM3 Expression in Prostate Cancer Independently Predicts a Reduced Risk of Biochemical Recurrence and Disease Progression. Diagn Pathol (2011) 6):91. doi: 10.1186/1746-1596-6-91
17. Grupp K, Wilking J, Prien K, Hube-Magg C, Sirma H, Simon R, et al. High RNA-Binding Motif Protein 3 Expression Is an Independent Prognostic Marker in Operated Prostate Cancer and Tightly Linked to ERG Activation and PTEN Deletions. Eur J Cancer (2014) 4(50):852–61. doi: 10.1016/j.ejca.2013.12.003
18. Zeng Y, Wodzenski D, Gao D, Shiraishi T, Terada N, Li Y, et al. Stress-Response Protein RBM3 Attenuates the Stem-Like Properties of Prostate Cancer Cells by Interfering With CD44 Variant Splicing. Cancer Res (2013) 13(73):4123–33. doi: 10.1158/0008-5472.CAN-12-1343
19. Zhang J, Cho SJ, Shu L, Yan W, Guerrero T, Kent M, et al. Translational Repression of P53 by RNPC1, a P53 Target Overexpressed in Lymphomas. Genes Dev (2011) 14(25):1528–43. doi: 10.1101/gad.2069311
20. Zhang J, Xu E, Ren C, Yan W, Zhang M, Chen M, et al. Mice Deficient in Rbm38, a Target of the P53 Family, are Susceptible to Accelerated Aging and Spontaneous Tumors. Proc Natl Acad Sci USA (2014) 52(111):18637–42. doi: 10.1073/pnas.1415607112
21. Zhang J, Xu E, Ren C, Yang HJ, Zhang Y, Sun W, et al. Genetic Ablation of Rbm38 Promotes Lymphomagenesis in the Context of Mutant P53 by Downregulating PTEN. Cancer Res (2018) 6(78):1511–21. doi: 10.1158/0008-5472.CAN-17-2457
22. Huang J, Li Y, Lu Z, Che Y, Sun S, Mao S, et al. Analysis of Functional Hub Genes Identifies CDC45 as an Oncogene in Non-Small Cell Lung Cancer - a Short Report. Cell Oncol (Dordr) (2019) 4(42):571–78. doi: 10.1007/s13402-019-00438-y
23. Wellmann S, Truss M, Bruder E, Tornillo L, Zelmer A, Seeger K, et al. The RNA-Binding Protein RBM3 Is Required for Cell Proliferation and Protects Against Serum Deprivation-Induced Cell Death. Pediatr Res (2010) 1(67):35–41. doi: 10.1203/PDR.0b013e3181c13326
24. Dong W, Dai ZH, Liu FC, Guo XG, Ge CM, Ding J, et al. The RNA-Binding Protein RBM3 Promotes Cell Proliferation in Hepatocellular Carcinoma by Regulating Circular RNA SCD-circRNA 2 Production. EBioMedicine (2019) 45:155–67. doi: 10.1016/j.ebiom.2019.06.030
25. Lin JC, Lin CY, Tarn WY, Li FY. Elevated SRPK1 Lessens Apoptosis in Breast Cancer Cells Through RBM4-Regulated Splicing Events. RNA (2014) 10(20):1621–31. doi: 10.1261/rna.045583.114
26. Lu J, Li Q, Cai L, Zhu Z, Guan J, Wang C, et al. RBM17 Controls Apoptosis and Proliferation to Promote Glioma Progression. Biochem Biophys Res Commun (2018) 1(505):20–8. doi: 10.1016/j.bbrc.2018.09.056
27. Han Y, Zhang M, Chen D, Li H, Wang X, Ma S. Downregulation of RNA Binding Motif Protein 17 Expression Inhibits Proliferation of Hypopharyngeal Carcinoma FaDu Cells. Oncol Lett (2018) 4(15):5680–84. doi: 10.3892/ol.2018.8012
28. Li C, Ge S, Zhou J, Peng J, Chen J, Dong S, et al. Exploration of the Effects of the CYCLOPS Gene RBM17 in Hepatocellular Carcinoma. PLoS One (2020) 6(15):e0234062. doi: 10.1371/journal.pone.0234062
29. Xi PW, Zhang X, Zhu L, Dai XY, Cheng L, Hu Y, et al. Oncogenic Action of the Exosome Cofactor RBM7 by Stabilization of CDK1 mRNA in Breast Cancer. NPJ Breast Cancer (2020) 6:58. doi: 10.1038/s41523-020-00200-w
30. Li XX, Shi L, Zhou XJ, Wu J, Xia TS, Zhou WB, et al. The Role of C-Myc-RBM38 Loop in the Growth Suppression in Breast Cancer. J Exp Clin Cancer Res (2017) 1(36):49. doi: 10.1186/s13046-017-0521-5
31. Zhou XJ, Wu J, Shi L, Li XX, Zhu L, Sun X, et al. PTEN Expression Is Upregulated by a RNA-Binding Protein RBM38 via Enhancing Its mRNA Stability in Breast Cancer. J Exp Clin Cancer Res (2017) 1(36):149. doi: 10.1186/s13046-017-0620-3
32. Zhang M, Zhang J, Chen X, Cho SJ, Chen X. Glycogen Synthase Kinase 3 Promotes P53 mRNA Translation via Phosphorylation of RNPC1. Genes Dev (2013) 20(27):2246–58. doi: 10.1101/gad.221739.113
33. Sun X, Hu Y, Wu J, Shi L, Zhu L, Xi PW, et al. RBMS2 Inhibits the Proliferation by Stabilizing P21 mRNA in Breast Cancer. J Exp Clin Cancer Res (2018) 1(37):298. doi: 10.1186/s13046-018-0968-z
34. Feng H, Liu J, Qiu Y, Liu Y, Saiyin H, Liang X, et al. RNA-Binding Motif Protein 43 (RBM43) Suppresses Hepatocellular Carcinoma Progression Through Modulation of Cyclin B1 Expression. Oncogene (2020) 33(39):5495–506. doi: 10.1038/s41388-020-1380-7
35. Yong H, Zhao W, Zhou X, Liu Z, Tang Q, Shi H, et al. RNA-Binding Motif 4 (RBM4) Suppresses Tumor Growth and Metastasis in Human Gastric Cancer. Med Sci Monit (2019) 25:4025–34. doi: 10.12659/MSM.914513
36. Yu J, Ji G, Shi W, Zhao R, Shen W, Zheng J, et al. RBM5 Acts as Tumor Suppressor in Medulloblastoma Through Regulating Wnt/beta-Catenin Signaling. Eur Neurol (2020) 3(83):242–50. doi: 10.1159/000507759
37. Jiang Y, Sheng H, Meng L, Yue H, Li B, Zhang A, et al. RBM5 Inhibits Tumorigenesis of Gliomas Through Inhibition of Wnt/beta-Catenin Signaling and Induction of Apoptosis. World J Surg Oncol (2017) 1(15):9. doi: 10.1186/s12957-016-1084-1
38. Xiao W, Chen X, Li X, Deng K, Liu H, Ma J, et al. RBM10 Regulates Human TERT Gene Splicing and Inhibits Pancreatic Cancer Progression. Am J Cancer Res (2021) 1(11):157–70.
39. Hernandez J, Bechara E, Schlesinger D, Delgado J, Serrano L, Valcarcel J. Tumor Suppressor Properties of the Splicing Regulatory Factor RBM10. RNA Biol (2016) 4(13):466–72. doi: 10.1080/15476286.2016.1144004
40. Jin X, Di X, Wang R, Ma H, Tian C, Zhao M, et al. RBM10 Inhibits Cell Proliferation of Lung Adenocarcinoma via RAP1/AKT/CREB Signalling Pathway. J Cell Mol Med (2019) 6(23):3897–904. doi: 10.1111/jcmm.14263
41. Martin-Garabato E, Martinez-Arribas F, Pollan M, Lucas AR, Sanchez J, Schneider J. The Small Variant of the Apoptosis-Associated X-Chromosome RBM10 Gene is Co-Expressed With Caspase-3 in Breast Cancer. Cancer Genomics Proteomics (2008) 3-4(5):169–73.
42. Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the Hallmarks of Lung Adenocarcinoma With Massively Parallel Sequencing. Cell (2012) 6(150):1107–20. doi: 10.1016/j.cell.2012.08.029
43. Serrano P, Hammond JA, Geralt M, Wuthrich K. Splicing Site Recognition by Synergy of Three Domains in Splicing Factor Rbm10. Biochemistry (2018) 10(57):1563–67. doi: 10.1021/acs.biochem.7b01242
44. Zhao L, Li R, Shao C, Li P, Liu J, Wang K. 3p21.3 Tumor Suppressor Gene RBM5 Inhibits Growth of Human Prostate Cancer PC-3 Cells Through Apoptosis. World J Surg Oncol (2012) 10:247. doi: 10.1186/1477-7819-10-247
45. Shao C, Zhao L, Wang K, Xu W, Zhang J, Yang B. The Tumor Suppressor Gene RBM5 Inhibits Lung Adenocarcinoma Cell Growth and Induces Apoptosis. World J Surg Oncol (2012) 10:160. doi: 10.1186/1477-7819-10-160
46. Loiselle JJ, Roy JG, Sutherland LC. RBM5 Reduces Small Cell Lung Cancer Growth, Increases Cisplatin Sensitivity and Regulates Key Transformation-Associated Pathways. Heliyon (2016) 11(2):e00204. doi: 10.1016/j.heliyon.2016.e00204
47. Su Z, Wang K, Li R, Yin J, Hao Y, Lv X, et al. Overexpression of RBM5 Induces Autophagy in Human Lung Adenocarcinoma Cells. World J Surg Oncol (2016) 14:57. doi: 10.1186/s12957-016-0815-7
48. Zhang YP, Liu KL, Wang YX, Yang Z, Han ZW, Lu BS, et al. Down-Regulated RBM5 Inhibits Bladder Cancer Cell Apoptosis by Initiating an miR-432-5p/Beta-Catenin Feedback Loop. FASEB J (2019) 10(33):10973–85. doi: 10.1096/fj.201900537R
49. Fanning AS, Anderson JM. Zonula Occludens-1 and -2 are Cytosolic Scaffolds That Regulate the Assembly of Cellular Junctions. Ann N Y Acad Sci (2009) 1165:113–20. doi: 10.1111/j.1749-6632.2009.04440.x
50. Wu J, Zhou XJ, Sun X, Xia TS, Li XX, Shi L, et al. RBM38 is Involved in TGF-Beta-Induced Epithelial-to-Mesenchymal Transition by Stabilising Zonula Occludens-1 mRNA in Breast Cancer. Br J Cancer (2017) 5(117):675–84. doi: 10.1038/bjc.2017.204
51. Zhu L, Xi PW, Li XX, Sun X, Zhou WB, Xia TS, et al. The RNA Binding Protein RBMS3 Inhibits the Metastasis of Breast Cancer by Regulating Twist1 Expression. J Exp Clin Cancer Res (2019) 1(38):105. doi: 10.1186/s13046-019-1111-5
52. Yang Z, Qu CB, Zhang Y, Zhang WF, Wang DD, Gao CC, et al. Dysregulation of P53-RBM25-Mediated Circamotl1l Biogenesis Contributes to Prostate Cancer Progression Through the Circamotl1l-miR-193a-5p-Pcdha Pathway. Oncogene (2019) 14(38):2516–32. doi: 10.1038/s41388-018-0602-8
53. Shen DJ, Jiang YH, Li JQ, Xu LW, Tao KY. The RNA-Binding Protein RBM47 Inhibits non-Small Cell Lung Carcinoma Metastasis Through Modulation of AXIN1 mRNA Stability and Wnt/beta-Catentin Signaling. Surg Oncol (2020) 34):31–9. doi: 10.1016/j.suronc.2020.02.011
54. Vanharanta S, Marney CB, Shu W, Valiente M, Zou Y, Mele A, et al. Loss of the Multifunctional RNA-Binding Protein RBM47 as a Source of Selectable Metastatic Traits in Breast Cancer. Elife (2014) 3:e02734. doi: 10.7554/eLife.02734
55. Liang H, Zhang J, Shao C, Zhao L, Xu W, Sutherland LC, et al. Differential Expression of RBM5, EGFR and KRAS mRNA and Protein in non-Small Cell Lung Cancer Tissues. J Exp Clin Cancer Res (2012) 31):36. doi: 10.1186/1756-9966-31-36
56. Jamsai D, Watkins DN, O’Connor AE, Merriner DJ, Gursoy S, Bird AD, et al. In Vivo Evidence That RBM5 is a Tumour Suppressor in the Lung. Sci Rep (2017) 1(7):16323. doi: 10.1038/s41598-017-15874-9
57. Grupp K, Hofmann B, Kutup A, Bachmann K, Bogoevski D, Melling N, et al. Reduced RBM3 Expression Is Associated With Aggressive Tumor Features in Esophageal Cancer But Not Significantly Linked to Patient Outcome. BMC Cancer (2018) 1(18):1106. doi: 10.1186/s12885-018-5032-z
58. Huang GW, Zhang YL, Liao LD, Li EM, Xu LY. Natural Antisense Transcript TPM1-AS Regulates the Alternative Splicing of Tropomyosin I Through an Interaction With RNA-Binding Motif Protein 4. Int J Biochem Cell Biol (2017) 90:59–67. doi: 10.1016/j.biocel.2017.07.017
59. Mu JY, Tian JX, Chen YJ. lncRNA RBM5-AS1 Promotes Cell Proliferation and Invasion by Epigenetically Silencing miR-132/212 in Hepatocellular Carcinoma Cells. Cell Biol Int (2021) 45(11):2201–10. doi: 10.1002/cbin.11649
60. Fu C, Yuan M, Sun J, Liu G, Zhao X, Chang W, et al. RNA-Binding Motif Protein 11 (RBM11) Serves as a Prognostic Biomarker and Promotes Ovarian Cancer Progression. Dis Markers (2021) 2021):3037337. doi: 10.1155/2021/3037337
61. Wang N, Lu K, Qu H, Wang H, Chen Y, Shan T, et al. CircRBM33 Regulates IL-6 to Promote Gastric Cancer Progression Through Targeting miR-149. BioMed Pharmacother (2020) 125):109876. doi: 10.1016/j.biopha.2020.109876
62. Yong H, Zhu H, Zhang S, Zhao W, Wang W, Chen C, et al. Prognostic Value of Decreased Expression of RBM4 in Human Gastric Cancer. Sci Rep (2016) 6:28222. doi: 10.1038/srep28222
63. Gao G, Shi X, Long Y, Yao Z, Shen J, Shen L. The Prognostic and Clinicopathological Significance of RBM3 in the Survival of Patients With Tumor: A Prisma-Compliant Meta-Analysis. Med (Baltimore) (2020) 19(99):e20002. doi: 10.1097/MD.0000000000020002
64. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. M(6)A RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature (2016) 7620(537):369–73. doi: 10.1038/nature19342
65. Hou J, Shan H, Zhang Y, Fan Y, Wu B. M(6)A RNA Methylation Regulators Have Prognostic Value in Papillary Thyroid Carcinoma. Am J Otolaryngol (2020) 4(41):102547. doi: 10.1016/j.amjoto.2020.102547
66. Zhang J, Piao HY, Wang Y, Meng XY, Yang D, Zhao Y, et al. To Develop and Validate the Combination of RNA Methylation Regulators for the Prognosis of Patients With Gastric Cancer. Onco Targets Ther (2020) 13:10785–95. doi: 10.2147/OTT.S276239
67. Zhang Y, Liu X, Liu L, Li J, Hu Q, Sun R. Expression and Prognostic Significance of M6a-Related Genes in Lung Adenocarcinoma. Med Sci Monit (2020) 26:e919644. doi: 10.12659/MSM.919644
68. Wang H, Zhao X, Lu Z. M(6)A RNA Methylation Regulators Act as Potential Prognostic Biomarkers in Lung Adenocarcinoma. Front Genet (2021) 12:622233. doi: 10.3389/fgene.2021.622233
69. Song Y, He S, Ma X, Zhang M, Zhuang J, Wang G, et al. RBMX Contributes to Hepatocellular Carcinoma Progression and Sorafenib Resistance by Specifically Binding and Stabilizing BLACAT1. Am J Cancer Res (2020) 11(10):3644–65.
70. Antonello ZA, Hsu N, Bhasin M, Roti G, Joshi M, Van Hummelen P, et al. Vemurafenib-Resistance via De Novo RBM Genes Mutations and Chromosome 5 Aberrations Is Overcome by Combined Therapy With Palbociclib in Thyroid Carcinoma With BRAF(V600E). Oncotarget (2017) 49(8):84743–60. doi: 10.18632/oncotarget.21262
71. Chilewski SD, Bhosale D, Dees S, Hutchinson I, Trimble R, Pontiggia L, et al. Development of CAPER Peptides for the Treatment of Triple Negative Breast Cancer. Cell Cycle (2020) 4(19):432–47. doi: 10.1080/15384101.2020.1711579
72. Hermawan A, Putri H. Integrative Bioinformatics Analysis Reveals miR-494 and Its Target Genes as Predictive Biomarkers of Trastuzumab-Resistant Breast Cancer. J Egypt Natl Canc Inst (2020) 1(32):16. doi: 10.1186/s43046-020-00028-2
73. Wu G, Cao L, Zhu J, Tan Z, Tang M, Li Z, et al. Loss of RBMS3 Confers Platinum Resistance in Epithelial Ovarian Cancer via Activation of miR-126-5p/Beta-Catenin/CBP Signaling. Clin Cancer Res (2019) 3(25):1022–35. doi: 10.1158/1078-0432.CCR-18-2554
74. He B, Wu C, Sun W, Qiu Y, Li J, Liu Z, et al. Mir383 Increases the Cisplatin Sensitivity of Lung Adenocarcinoma Cells Through Inhibition of the RBM24mediated NFkappaB Signaling Pathway. Int J Oncol (2021) 59(5):87. doi: 10.3892/ijo.2021.5267
75. Siesing C, Sorbye H, Dragomir A, Pfeiffer P, Qvortrup C, Ponten F, et al. High RBM3 Expression is Associated With an Improved Survival and Oxaliplatin Response in Patients With Metastatic Colorectal Cancer. PLoS One (2017) 8(12):e0182512. doi: 10.1371/journal.pone.0182512
76. Ehlen A, Brennan DJ, Nodin B, O’Connor DP, Eberhard J, Alvarado-Kristensson M, et al. Expression of the RNA-Binding Protein RBM3 Is Associated With a Favourable Prognosis and Cisplatin Sensitivity in Epithelial Ovarian Cancer. J Transl Med (2010) 8:78. doi: 10.1186/1479-5876-8-78
Keywords: RNA-binding motif protein, cancer, prognosis, tumorigenesis, RNA binding protein
Citation: Li Z, Guo Q, Zhang J, Fu Z, Wang Y, Wang T and Tang J (2021) The RNA-Binding Motif Protein Family in Cancer: Friend or Foe? Front. Oncol. 11:757135. doi: 10.3389/fonc.2021.757135
Received: 11 August 2021; Accepted: 19 October 2021;
Published: 04 November 2021.
Edited by:
Carlos Pérez-Plasencia, National Autonomous University of Mexico, MexicoReviewed by:
Irene Diaz-Moreno, Sevilla University, SpainYing Hu, Harbin Institute of Technology, China
Copyright © 2021 Li, Guo, Zhang, Fu, Wang, Wang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianzhen Wang, d3R6cGF0aEAxNjMuY29t; Jing Tang, amluZ2htdUAxNjMuY29t
†These authors have contributed equally to this work
 Zhigang Li1†
Zhigang Li1† Tianzhen Wang
Tianzhen Wang Jing Tang
Jing Tang