
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Oncol. , 23 November 2021
Sec. Molecular and Cellular Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.755578
 Tian Fang1†
Tian Fang1† Tingting Liang1†
Tingting Liang1† Yizhuo Wang1
Yizhuo Wang1 Haitao Wu1
Haitao Wu1 Shuhan Liu1
Shuhan Liu1 Linying Xie2
Linying Xie2 Zhihao Zhang3
Zhihao Zhang3 Jiaying Liang1
Jiaying Liang1 Cheng Yao1
Cheng Yao1 Yehui Tan1*
Yehui Tan1* Chang Wang1*
Chang Wang1*Mutations in KRAS (codon 12/13), NRAS, BRAFV600E, and amplification of ERBB2 and MET account for 70–80% of anti-epidermal growth factor receptor (EGFR) monoclonal antibody primary resistance. However, the list of anti-EGFR monoclonal antibody primary resistance biomarkers is still incomplete. Herein, we report a case of wild-type RAS/BRAF metastatic colorectal cancer (CRC) with resistance to anti-EGFR monoclonal antibody and chemotherapy. Initially, mutation detection in postoperative tumor tissue by using amplification-refractory mutation system polymerase chain reaction indicated wild-type RAS/BRAF without point mutations, insertion deletions, or fusion mutations. Therefore, we recommended combined therapy of cetuximab and FOLFIRI after failure of platinum-based adjuvant chemotherapy, but the disease continued to progress. Next generation sequencing analysis of the postoperative tumor tissue revealed that KRAS copy number was increased and detected SMAD4, RNF43, and PREX2 mutations. This is the first case of advanced CRC with increased copy numbers of KRAS resistant to cetuximab and chemotherapy, which results in poor patient survival, and other mutated genes may be associated with the outcomes. Our findings indicate KRAS copy number alterations should also be examined, especially with anti-EGFR monoclonal antibody therapy in CRC, since it may be related with the primary resistance to these drugs.
Colorectal cancer (CRC) is one of the most common causes of cancer-related deaths worldwide (1). The mean age of patients with CRC ranges from 49 to 60 years. Early onset of CRC generally refers to the onset in patients younger than 50 years of age at the time of diagnosis. It is characterized by a more advanced stage at diagnosis, poorer cell differentiation, higher prevalence of signet ring cell histology, and a primary tumor located on the left colon or rectum (2).
The combination of biological monoclonal antibodies and chemotherapeutic cytotoxic drugs provides clinical benefits to patients with advanced or metastatic CRC (mCRC) (3). Bevacizumab is an anti-vascular endothelial growth factor monoclonal antibody (MoAb) used as a first-line treatment in RAS- or BRAF-mutated mCRC (4). Cetuximab and panitumumab can promote survival in patients with wild-type RAS and BRAF tumors, which target the epidermal growth factor receptor (EGFR) extracellular domain and subsequently inhibit the mitogen-activated protein kinase signaling pathway. The principal downstream effectors of EGFR activation are the RAS/RAF/MEK/ERK, PI3K/AKT/mTOR, and PLCγ/PKC pathways (5), which is a key regulator of cell proliferation, differentiation, division, and survival, and the metastatic potential of tumor cells (6). Mutations in any of the upstream genes may be transmitted to the protein through transcription or translation, resulting in abnormal activation of the signaling pathway (7). Alterations in RAS, BRAF, PIK3CA, EGFR, PTEN, and HER2 are key determinants of resistance to anti-EGFR MoAb therapies (8–10). However, little is known regarding patients harboring KRAS amplification, and data on the response to anti-EGFR treatment are lacking.
Here, we present a patient with an initial diagnosis of KRAS-amplified locally advanced rectal adenocarcinoma who demonstrated no clinical response to anti-EGFR MoAb and chemotherapy, and the disease progressed rapidly. This case report indicates the potential of increased KRAS gene copy numbers (GCN) in primary resistance to anti-EGFR MoAb in patients with CRC.
A 40-year-old man was admitted in our hospital with complaints of diarrhea for one month. Physical examination was normal, but electronic colonoscopy revealed a large polypoid mass in the rectum, situated approximately 9–15 cm from the anal verge. The lesion almost completely occluded the rectal lumen, allowing only a small endoscope to pass through. Pathology revealed an adenocarcinoma. Enhanced computed tomography (CT) of the abdomen revealed occupying lesions in the middle and upper rectum. Considering the possibility of rectal cancer, CT findings were consistent with T4aN1M0 (Figure 1). Blood tests showed a slight increase in tumor markers (CA724, 7.79 U/mol; carcinoembryonic antigen, 9.16 ng/mol), creatinine (102.8 μmol/L), direct bilirubin (7.2 μmol/L), and C-reactive protein (13 mg/L). The patient underwent total resection of the rectal cancer and ileostomy on March 24, 2021. After surgery, all blood test results returned to normal.
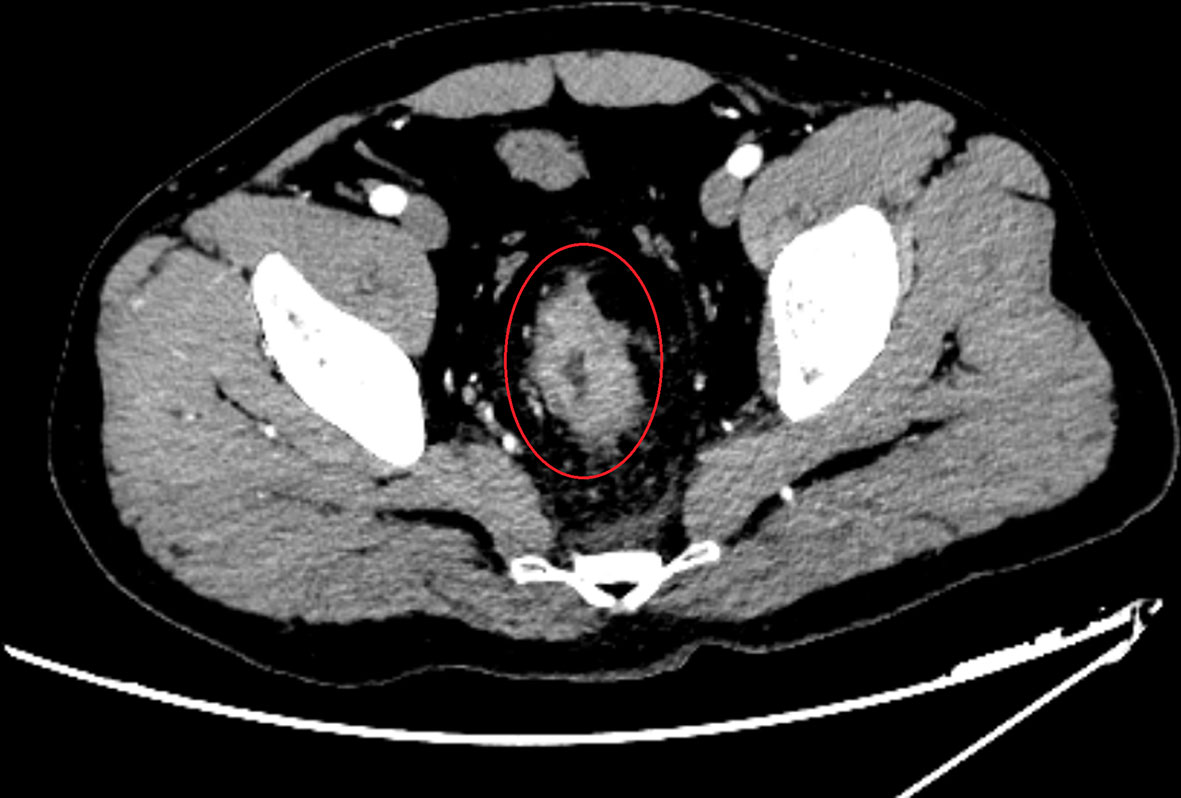
Figure 1 Imaging examinations performed before surgery. Enhanced CT scans on March 21, 2020 of abdomen revealed that occupying lesions in the middle and upper rectum, the intestinal lumen was narrowed, and the serosal layer was hairy. After enhancement, the lesion was uneven and enhanced, and the length of the lesion was about 5.7 cm, considering that was rectal cancer (T4aN1M0).
Surgical pathology findings showed a moderate-to-poorly differentiated adenocarcinoma in the rectum (pT3N2bMx). The tumor volume was approximately 4 cm × 3 cm × 1.2 cm. Cancer infiltration was observed in vessels and nerves. Residual tumors can be detected on the edges of the surgical specimens. Meanwhile, 20 mesenteric lymph nodes were observed, and cancer embolus was distinguished in the local submucosal vessels. Immunohistochemistry results were as follows: pMMR, Ki-67 (+70%); CKpan (+), P53 (−), CgA (−), and Syn (−) (Figure 2).
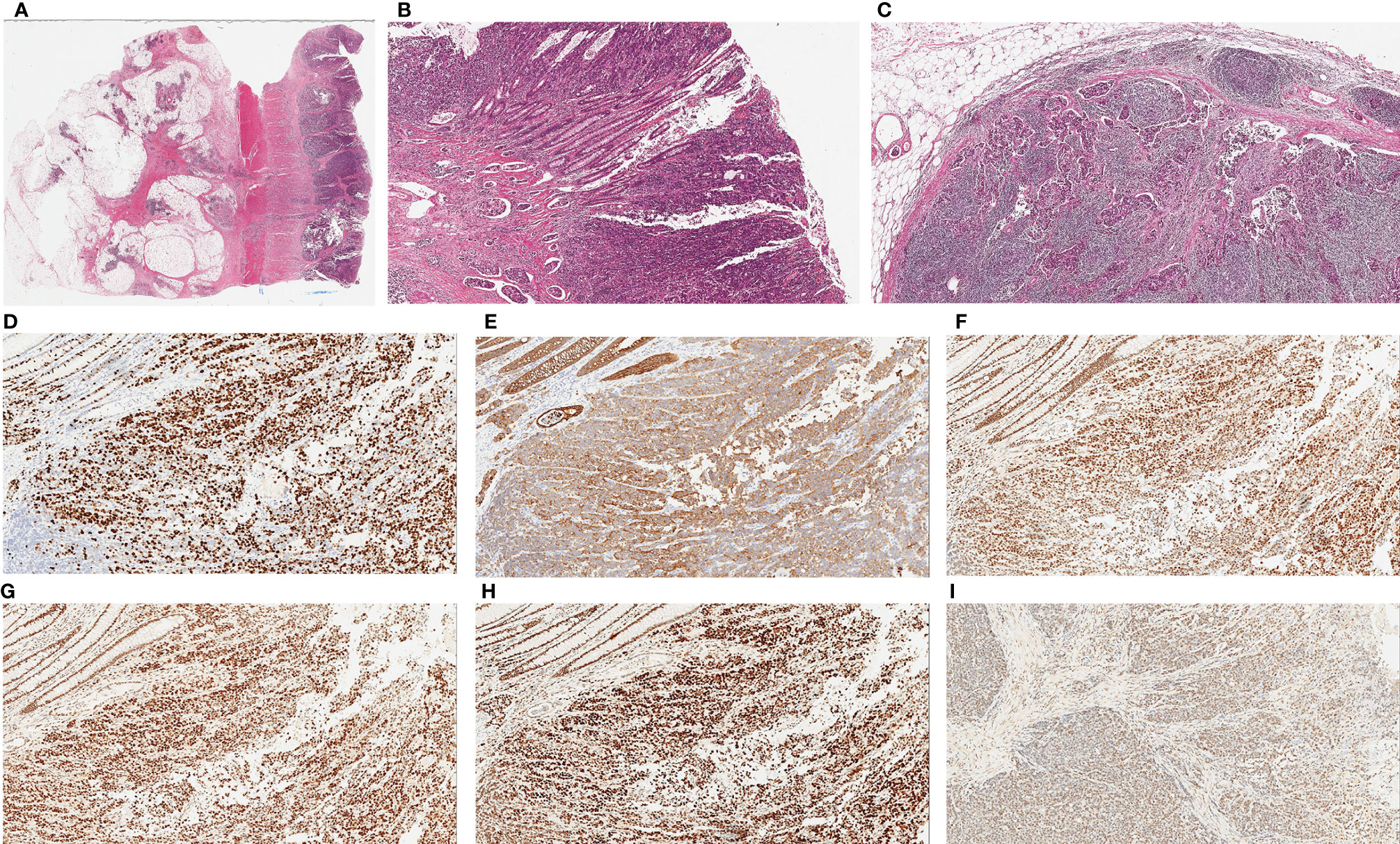
Figure 2 Histopathological examination of tumor. (A) Hematoxylin and eosin (H&E) staining of colonic primary tumor shows glandular differentiation and invasion into the entire intestinal wall (×20). (B) Poorly differentiated adenocarcinoma on the surface of mucosa with multiple intravascular thrombus in submucosa, (×40). (C) Metastatic tumors can be seen in lymph node, (×40). Immunohistochemical staining of tumor cells. Ki-67 partial expression in tumor (+70%) (D, ×100), CKpan expression in tumor (E, ×100.), Immunostaining for MLH1 (+70%) (F, ×100), MSH2 (+70%) (G, ×100), MSH6 (+80%) (H, ×100)and PMS2 (+50%) (I, ×100) confirmed the tumor with proficient MMR.
Enhanced pelvic Magnetic Resonance Imaging (MRI) was performed 21 days after the surgery and revealed multiple lymph node shadows with visible enhancement beside bilateral iliac vessels with a short diameter of approximately 0.6–0.7 cm, but postoperative inflammatory changes were not excluded. Therefore, five cycles of adjuvant chemotherapy (XELOX: oxaliplatin, 130 mg/m2 on day 1; capecitabine, 1,000 mg/m2 twice daily on days 1–14, orally) strategy was recommended from April 14 to July 28, 2020. The CT and MRI evaluations were progressive disease (PD) after chemotherapy, detecting significantly enlarged lymph nodes around the abdominal aorta (0.6-1.5cm) on August 12, 2020 (Figure 3A).
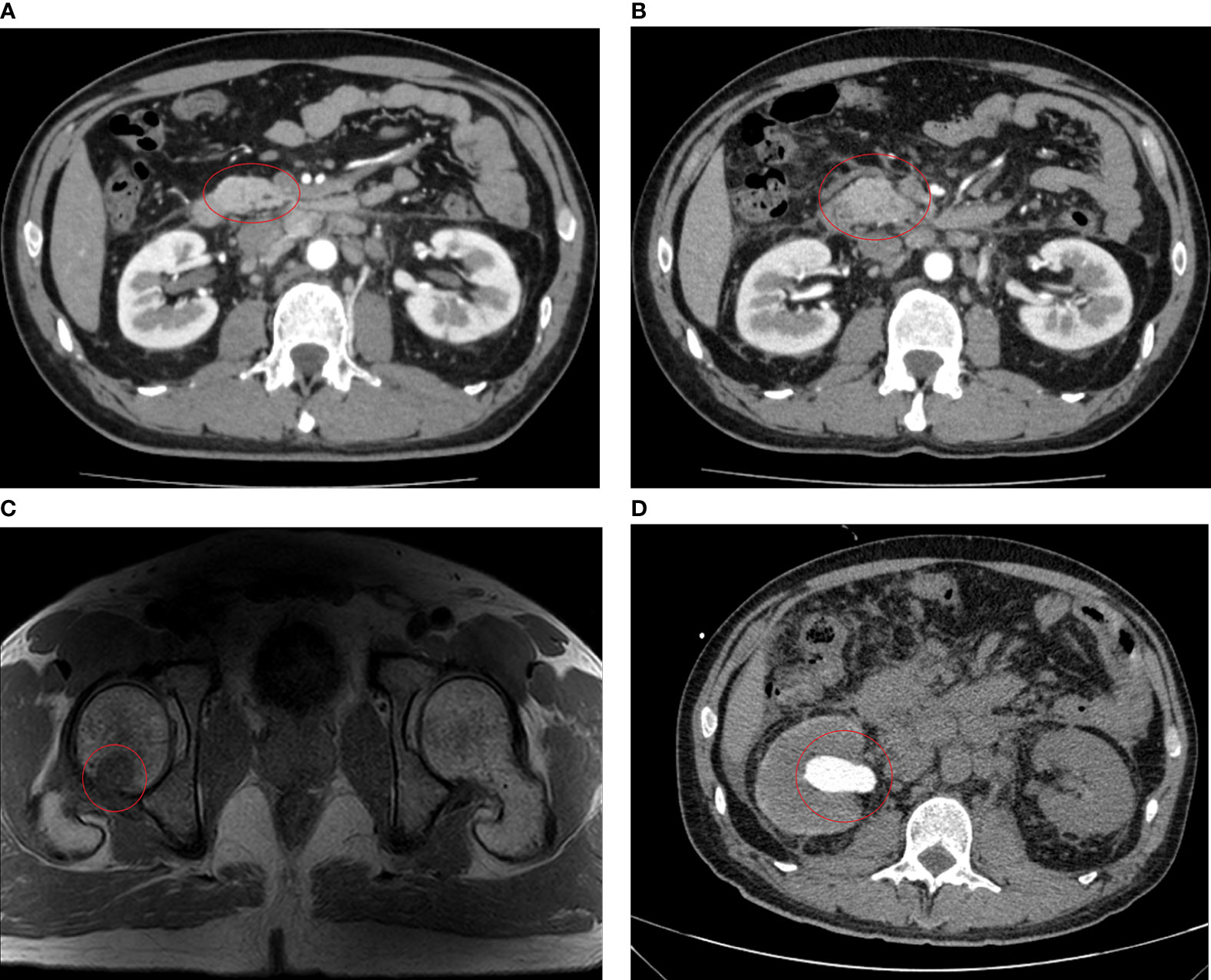
Figure 3 Imaging examinations performed after chemotherapy. (A) After five cycles of XELOX, CT scans on August 12, 2020 revealed enlargement of the abdominal para-aortic and bilateral para-iliac lymph nodes, about 0.5–1.5 cm, with slightly enhanced. (B) After six cycles of FOLFIRI plus cetuximab treatment, CT scans on November 28, 2020 revealed significant enlargement of lymph nodes in the hilar area, adjacent to the abdominal aorta, and bilateral iliac vessels, with a maximum of about 2.3 cm and slightly enhancement. (C) MRI scans on November 29, 2020 show patchy shadows are seen on the right femoral head and greater trochanter, enhanced scans are seen to be enhanced, consider metastasis. (D) CT scans on December 22, 2020 showed hydronephrodilation of the right renal pelvis and space-occupying lesion about 16.2 mm × 9.3 mm in the initial segment of the right ureter.
Since the first-line platinum-based regiment was unsuccessful, chemotherapy (FOLFIRI: 5-fluorouracil 400 mg/m2, IV bolus, folinic acid 400 mg/m2, and irinotecan 180 mg/m2 on day 1, followed by a continuous 46-h infusion of fluorouracil 2,400 mg/m2) in combination with cetuximab (500 mg/m² biweekly on day 1) was administered for six cycles from August 17 to November 12, 2020, considering that the patient harbored no RAS or BRAF alterations according to the mutation profile. After three cycles of chemotherapy and targeted therapy, the patient developed a grade 2 cutaneous toxicity, a single skin impetigo with a diameter greater than 10 mm, and the curative effect was evaluated as stable disease (SD), so we continued the therapy for three more cycles. On November 28, 2020, CT and MRI scans indicated PD, detecting a lymph node around the abdominal aorta enlarged to 2.3 cm (Figure 3B), tumor metastases to the right femoral head and greater trochanter (Figure 3C), and left cervical lymph nodes enlarged to 11 mm × 8 mm.
Likewise, abdominal CT scans indicated PD again, observing a space-occupying lesion (1.6 cm × 0.9 cm) in the initial segment of the right ureter on December 22, 2020 (Figure 3D). Renal function tests suggested that uric acid and creatinine levels continuously increased to 470 µmol/L (normal: 210–430 µmol/L) and 226 µmol/L (normal: 57–97 µmol/L), respectively. The patient decided to stop the treatment and never returned to the hospital after four days of palliative radiotherapy.
Wild-type KRAS, NRAS, and BRAF were amplified using amplification refractory mutation system polymerase chain reaction (ARMS-PCR) performed on March 20, 2020. Results revealed that the patient had wild-type RAS and BRAF. However, the tumor failed to respond to both treatment options and continued to progress.
Molecular characterization of the blood and postoperative tumor DNA of the patient was performed using next-generation sequencing (NGS) on December 1, 2020. Sequence analyses are shown in Table 1. We detected that the patient had mutations in three genes (SMAD4, RNF43, and PREX2), and GCN variations of KRAS increased to 7.8 gene copies before chemotherapy. In addition, the mutation abundance of the three mutant genes was observed using blood-based circulating tumor DNA (ctDNA) analysis: SMAD4 increased from 38.69 to 58.0%, RNF43 increased from 41.25 to 48.03%, and PREX2 increased from 39.22 to 52.99%, after the failure of second-line treatment. Likewise, the GCN of KRAS also increased to 8.31 gene copies, along with low abundance mutations in the other nine newly confirmed genes—CTNNB1, EPHA5, ETV1, FANCA, JAK2, MYC, PALB2, PIK3CA, and SPEN.
We analyzed MSI through surgically resected tumor tissues, and the results were reported as microsatellite stability (MSS)/MSI-low. The TMB detected in the tumor tissues was described as low level (2.95 mutations/MB). Furthermore, the TMB detected in ctDNA was reported as medium level (11.82 mutations/MB) after the failure of second-line therapy.
Here, we present a patient with an early onset CRC with an initial diagnosis of moderate-to-poorly differentiated locally advanced rectal adenocarcinoma. Unlike the traditional RAS/BRAF mutant CRCs with poor prognosis, the patient had RAS/BRAF wild-type CRC accompanied with KRAS amplification and other less-reported gene mutations. Moreover, the patient’s prognosis was extremely poor, the disease progressed rapidly even with the use cetuximab in combination with chemotherapy. Therefore, we hypothesized that the disease in our patient with early onset CRC was more aggressive due to gene alterations, especially the increase in copy number of KRAS. Therefore, we report this case and hope to bring clinical benefits to clinicians.
After the rapid progress of second-line treatment in our case, blood tests and postoperative tumor tissues were performed and analyzed using NGS and retrospective NGS, respectively. Results suggested an increase in KRAS copy number before treatment, accompanied by SMAD4, RNF43, and PREX2 mutations (mutation abundance was much more than 5%). After treatment failure, blood analysis indicated that the copy number of KRAS and clonal mutation abundance of the three genes continued to increase, indicating that the tumor developed from the original clone.
KRAS amplification in CRC is a rare event, with an overall prevalence of 0.67–2% (11, 12). Previous studies have identified somatic mutations in KRAS as biomarkers for inherent resistance to EGFR-targeted drugs in patients with CRC (13), with a positive tissue mutation rate of 32–52.1% (7). However, alterations are responsive to the anti-EGFR inhibitors cetuximab or panitumumab (11, 14). GCN amplification of KRAS leads to the activation of RAS-RAF-ERK or PIK-AKT-mTOR pathways, even without additional activating mutations in the genes (15). In the presence of KRAS amplification, cetuximab can partially eliminate the phosphorylation of MEK and ERK but cannot induce growth arrest (13). A retrospective clinical study conducted by Valtorta et al. (14) detected that all four KRAS-amplified cases were found among the 53 patients who were resistant to anti-EGFR antibodies, while none of the 44 responders had tumors carrying this molecular alteration. Similarly, tumor biopsies of 10 patients who developed resistance to anti-EGFR showed the emergence of KRAS amplification in one case and acquisition of secondary KRAS mutations in six cases (13). Another retrospective study by Favazza et al. (11) reported that all eight patients with KRAS-amplified mCRC showed disease progression at the time of anti-EGFR therapy, and they concluded that KRAS amplification is responsible for the primary resistance to EGFR inhibitors. Meanwhile, RAS amplifications (involving KRAS, NRAS, and HRAS) were correlated with a younger median patient age at initial diagnosis and a history of inflammatory bowel disease (11). In the present case, KRAS amplification was present before chemotherapy, resulting in resistance to cetuximab. Therefore, patients with advanced mCRC should be monitored not only based on the RAS/BRAF mutation status but also RAS amplification, and patients with CRC that do not respond to anti-EGFR MoAbs should be excluded. Moreover, we suggest that genetic testing using NGS should be performed in younger patients to guide decision-making and prolong overall survival, because KRAS amplification is more likely to occur in younger patients.
The mutation frequency and abundance of clonal mutations of SMAD4, RNF43, and PREX2 indicated that the tumor was progressing from the original clone. SMAD4, PREX2, and RNF43 are involved in the TGF-β, PI3K, and Wnt signaling pathways, respectively. These pathways are involved in cell death resistance, growth suppression, and sustained proliferative signaling, respectively. Several studies have reported chemoresistance and anti-EGFR resistance in patients with SMAD4-mutated mCRC (16–18), leading to poor prognosis. Furthermore, SMAD4 and RNF43 mutations were more commonly observed in early onset mCRC (≤55 years) (19).
NGS could be considered as the best choice to analyze genetic tests for anti-EGFR therapy because it is superior to ARMS-PCR in copy number detection. Moreover, it saves the amount of samples, is cost- and time-efficient, and has great potential for clinical application to expand testing to include mutations in RAS and other less reported CRC-related genes (20). NGS analysis must be carried out in early onset CRCs, family history-related CRCs, and CRC-related cancers to identify cause-effective mutations, elucidate the clinical diagnosis, guide decision-making, and prevent the development of the disease in other family members.
Furthermore, blood-based NGS testing suggested low-abundance mutations (<5%) of nine newly emerged genes, indicating the emergence of tumor subcloning and heterogeneity. These genes have been previously reported to be associated with poor prognosis (21–27), but few reports on the efficacy of chemotherapy or target drugs are available. This suggests that tumors with polygenic mutations tend to be more aggressive in terms of biological behavior, leading to poor survival rates. NGS was partially proven to have a higher sensitivity and specificity for detecting mutations with low abundance than ARMS-PCR. Blood-based NGS testing—ctDNA analysis—offers a convenient way to monitor tumor progression and treatment response. Since tumor mutational profiles are highly variable from person to person, a fixed content panel may be insufficient to track the treatment response in all patients. Compared to tumor tissue biopsies, blood-based ctDNA analyses are minimally invasive and accessible for the regular follow-up of cancer patients. The amount of ctDNA can be quantified, and genetic changes can be identified (28). The ctDNA analysis improves both the specificity and sensitivity of monitoring treatment response across several tumor types. It can identify tumor recurrence potentially earlier than imaging-based diagnosis. When augmented with tumor hotspot genes, it can track acquired drug-related mutations in patients (29).
In conclusion, the present case report identified a rare early onset CRC accompanied by KRAS GCN amplification. The disease progressed rapidly, and the effects of chemotherapy and anti-EGFR therapy were poor. This suggests that KRAS amplification might be responsible for the primary resistance to anti-EGFR treatment in a small proportion of patients, and targeted drugs should not only be based on RAS and BRAF mutations in CRCs. Furthermore, NGS has the advantage of discovering genes that affect the efficacy of anti-EGFR therapies, and blood-based serial ctDNA analysis provides a convenient way to monitor the efficacy and resistance mechanism of anti-EGFR MoAbs. With improvements and developments in NGS technology, the identification of copy number variations may provide more implications for the diagnosis and treatment of CRC. Further validation in a larger population is needed to establish a predictive biomarker for resistance to anti-EGFR therapy.
All data generated or analysed during this study are included in this published article. Additional data and materials related to the genetic tests, pathologic reports, treatment information, and images are available for review upon reasonable request.
Written informed consent was obtained from the patient for the publication of any potentially identifiable images or data included in this manuscript.
TF and TL contributed equally to this work. All authors were involved in the drafting of the manuscript. TF, TL, and YW designed the clinical treatment for the patient. ZZ, HW, LX, JL, and CY performed the clinical treatment for the patients. CW and YT provided comments and edited the manuscript to become the final version for submission. All authors contributed to the article and approved the submitted version.
This study was supported by Health commission of Jilin Province (NO. 2017J063) and Department of Finance of Jilin Province (NO. JLSWSRCZ2020-0048).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Editage (www.editage.cn) for English language editing.
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2020. CA Cancer J Clin (2020) 70(1):7–30. doi: 10.3322/caac.21590
2. Mauri G, Sartore-Bianchi A, Russo AG, Marsoni S, Bardelli A, Siena S. Early-Onset Colorectal Cancer in Young Individuals. Mol Oncol (2019) 13(2):109–31. doi: 10.1002/1878-0261.12417
3. Venook AP, Niedzwiecki D, Lenz HJ, Innocenti F, Fruth B, Meyerhardt JA, et al. Effect of First-Line Chemotherapy Combined With Cetuximab or Bevacizumab on Overall Survival in Patients With KRAS Wild-Type Advanced or Metastatic Colorectal Cancer: A Randomized Clinical Trial. JAMA (2017) 317(23):2392–401. doi: 10.1001/jama.2017.7105
4. Mody K, Baldeo C, Bekaii-Saab T. Antiangiogenic Therapy in Colorectal Cancer. Cancer J (2018) 24(4):165–70. doi: 10.1097/PPO.0000000000000328
5. Ciardiello F, Tortora G. EGFR Antagonists in Cancer Treatment. N Engl J Med (2008) 358(11):1160–74. doi: 10.1056/NEJMra0707704
6. Sabbah DA, Hajjo R, Sweidan K. Review on Epidermal Growth Factor Receptor (EGFR) Structure, Signaling Pathways, Interactions, and Recent Updates of EGFR Inhibitors. Curr Top Med Chem (2020) 20(10):815–34. doi: 10.2174/1568026620666200303123102
7. Wan XB, Wang AQ, Cao J, Dong ZC, Li N, Yang S, et al. Relationships Among KRAS Mutation Status, Expression of RAS Pathway Signaling Molecules, and Clinicopathological Features and Prognosis of Patients With Colorectal Cancer. World J Gastroenterol (2019) 25(7):808–23. doi: 10.3748/wjg.v25.i7.808
8. Miyamoto Y, Suyama K, Baba H. Recent Advances in Targeting the EGFR Signaling Pathway for the Treatment of Metastatic Colorectal Cancer. Int J Mol Sci (2017) 18(4):752. doi: 10.3390/ijms18040752
9. Montagut C, Dalmases A, Bellosillo B, Crespo M, Pairet S, Iglesias M, et al. Identification of a Mutation in the Extracellular Domain of the Epidermal Growth Factor Receptor Conferring Cetuximab Resistance in Colorectal Cancer. Nat Med (2012) 18(2):221–3. doi: 10.1038/nm.2609
10. Morelli MP, Overman MJ, Dasari A, Kazmi SMA, Mazard T, Vilar E, et al. Characterizing the Patterns of Clonal Selection in Circulating Tumor DNA From Patients With Colorectal Cancer Refractory to Anti-EGFR Treatment. Ann Oncol (2015) 26(4):731–6. doi: 10.1093/annonc/mdv005
11. Favazza LA, Parseghian CM, Kaya C, Nikiforova MN, Roy S, Wald AI, et al. KRAS Amplification in Metastatic Colon Cancer is Associated With a History of Inflammatory Bowel Disease and may Confer Resistance to Anti-EGFR Therapy. Mod Pathol (2020) 33(9):1832–43. doi: 10.1038/s41379-020-0560-x
12. Ye J, Lin M, Zhang C, Zhu X, Li S, Liu H, et al. Tissue Gene Mutation Profiles in Patients With Colorectal Cancer and Their Clinical Implications. BioMed Rep (2020) 13(1):43–8. doi: 10.3892/br.2020.1303
13. Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS Mutations and Acquired Resistance to Anti-EGFR Therapy in Colorectal Cancer. Nature (2012) 486(7404):532–6. doi: 10.1038/nature11156
14. Valtorta E, Misale S, Sartore-Bianchi A, Nagtegaal ID, Paraf F, Lauricella C, et al. KRAS Gene Amplification in Colorectal Cancer and Impact on Response to EGFR-Targeted Therapy. Int J Cancer (2013) 133(5):1259–65. doi: 10.1002/ijc.28106
15. Essakly A, Loeser H, Kraemer M, Alakus H, Chon SH, Zander T, et al. PIK3CA and KRAS Amplification in Esophageal Adenocarcinoma and Their Impact on the Inflammatory Tumor Microenvironment and Prognosis. Transl Oncol (2020) 13(2):157–64. doi: 10.1016/j.tranon.2019.10.013
16. Fang T, Liang T, Wang Y, Wu H, Liu S, Xie L, et al. Prognostic Role and Clinicopathological Features of SMAD4 Gene Mutation in Colorectal Cancer: A Systematic Review and Meta-Analysis. BMC Gastroenterol (2021) 21(1):297. doi: 10.1186/s12876-021-01864-9
17. Wasserman I, Lee LH, Ogino S, Marco MR, Wu C, Chen X, et al. SMAD4 Loss in Colorectal Cancer Patients Correlates With Recurrence, Loss of Immune Infiltrate, and Chemoresistance. Clin Cancer Res (2019) 25(6):1948–56. doi: 10.1158/1078-0432.CCR-18-1726
18. Lupini L, Bassi C, Mlcochova J, Musa G, Russo M, Vychytilova-Faltejskova P, et al. Prediction of Response to Anti-EGFR Antibody-Based Therapies by Multigene Sequencing in Colorectal Cancer Patients. BMC Cancer (2015) 15:808. doi: 10.1186/s12885-015-1752-5
19. Xu T, Zhang Y, Zhang J, Qi C, Liu D, Wang Z, et al. Germline Profiling and Molecular Characterization of Early Onset Metastatic Colorectal Cancer. Front Oncol (2020) 10:568911. doi: 10.3389/fonc.2020.568911
20. Gao J, Wu H, Wang L, Zhang H, Duan H, Lu J, et al. Validation of Targeted Next-Generation Sequencing for RAS Mutation Detection in FFPE Colorectal Cancer Tissues: Comparison With Sanger Sequencing and ARMS-Scorpion Real-Time PCR. BMJ Open (2016) 6(1):e009532. doi: 10.1136/bmjopen-2015-009532
21. Zhang W, Meyfeldt J, Wang H, Kulkarni S, Lu J, Mandel JA, et al. Beta-Catenin Mutations as Determinants of Hepatoblastoma Phenotypes in Mice. J Biol Chem (2019) 294(46):17524–42. doi: 10.1074/jbc.RA119.009979
22. Lee KS, Kwak Y, Nam KH, Kim DW, Kang SB, Choe G, et al. C-MYC Copy-Number Gain Is an Independent Prognostic Factor in Patients With Colorectal Cancer. PloS One (2015) 10(10):e0139727. doi: 10.1371/journal.pone.0139727
23. Oh S, Song H, Freeman WM, Shin S, Janknecht R. Cooperation Between ETS Transcription Factor ETV1 and Histone Demethylase JMJD1A in Colorectal Cancer. Int J Oncol (2020) 57(6):1319–32. doi: 10.3892/ijo.2020.5133
24. Huang W, Lin A, Luo P, Liu Y, Xu W, Zhu W, et al. EPHA5 Mutation Predicts the Durable Clinical Benefit of Immune Checkpoint Inhibitors in Patients With Lung Adenocarcinoma. Cancer Gene Ther (2021) 28(7–8):864–74. doi: 10.1038/s41417-020-0207-6. 2020.10.1038/s41417-020-0207-6.
25. Du W, Hong J, Wang YC, Zhang YJ, Wang P, Su WY, et al. Inhibition of JAK2/STAT3 Signalling Induces Colorectal Cancer Cell Apoptosis via Mitochondrial Pathway. J Cell Mol Med (2012) 16(8):1878–88. doi: 10.1111/j.1582-4934.2011.01483.x
26. Bougatef K, Ouerhani S, Moussa A, Kourda N, Coulet F, Colas C, et al. Prevalence of Mutations in APC, CTNNB1, and BRAF in Tunisian Patients With Sporadic Colorectal Cancer. Cancer Genet Cytogenet (2008) 187(1):12–8. doi: 10.1016/j.cancergencyto.2008.06.016
27. Nepomuceno TC, De Gregoriis G, de Oliveira FMB, Suarez-Kurtz G, Monteiro AN, Carvalho MA. The Role of PALB2 in the DNA Damage Response and Cancer Predisposition. Int J Mol Sci (2017) 18(9):1886. doi: 10.3390/ijms18091886
28. Pessoa LS, Heringer M, Ferrer VP. ctDNA as a Cancer Biomarker: A Broad Overview. Crit Rev Oncol Hematol (2020) 155:103109. doi: 10.1016/j.critrevonc.2020.103109
Keywords: KRAS, gene copy number, colorectal cancer, cetuximab, case report, anti-EGFR monoclonal antibody, gene amplification
Citation: Fang T, Liang T, Wang Y, Wu H, Liu S, Xie L, Zhang Z, Liang J, Yao C, Tan Y and Wang C (2021) An Early-Onset Advanced Rectal Cancer Patient With Increased KRAS Gene Copy Number Showed A Primary Resistance to Cetuximab in Combination With Chemotherapy: A Case Report. Front. Oncol. 11:755578. doi: 10.3389/fonc.2021.755578
Received: 10 August 2021; Accepted: 28 October 2021;
Published: 23 November 2021.
Edited by:
Todd M. Pitts, University of Colorado, United StatesReviewed by:
Anna Capasso, University of Texas at Austin, United StatesCopyright © 2021 Fang, Liang, Wang, Wu, Liu, Xie, Zhang, Liang, Yao, Tan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang Wang, d2FuZ2NoYW5nQGpsdS5lZHUuY24=; Yehui Tan, eWh0YW5Aamx1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.