- 1Department of Neonatology, First Teaching Hospital of Tianjin University of Traditional Chinese Medicine, Tianjin, China
- 2Department of Neonatology, National Clinical Research Center for Chinese Medicine Acupuncture and Moxibustion, Tianjin, China
- 3Department of Urology, Tianjin Medical University General Hospital, Tianjin, China
Background: Although the effect of the LEP G19A (rs2167270) polymorphism on cancers is assumed, the results of its influence have been contradictory. A meta-analysis was conducted to precisely verify the relationships between LEP G19A and the development of digestion-related cancers.
Methods: Investigators systematically searched the literature in PubMed, Embase, and Web of Science and used STATA software 14.0 for the meta−analysis. The odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to evaluate the associations. Subgroup analyses stratified by ethnicity, cancer type, and cancer system were further conducted to assess the relationship between the LEP G19A polymorphism and digestion-related cancers.
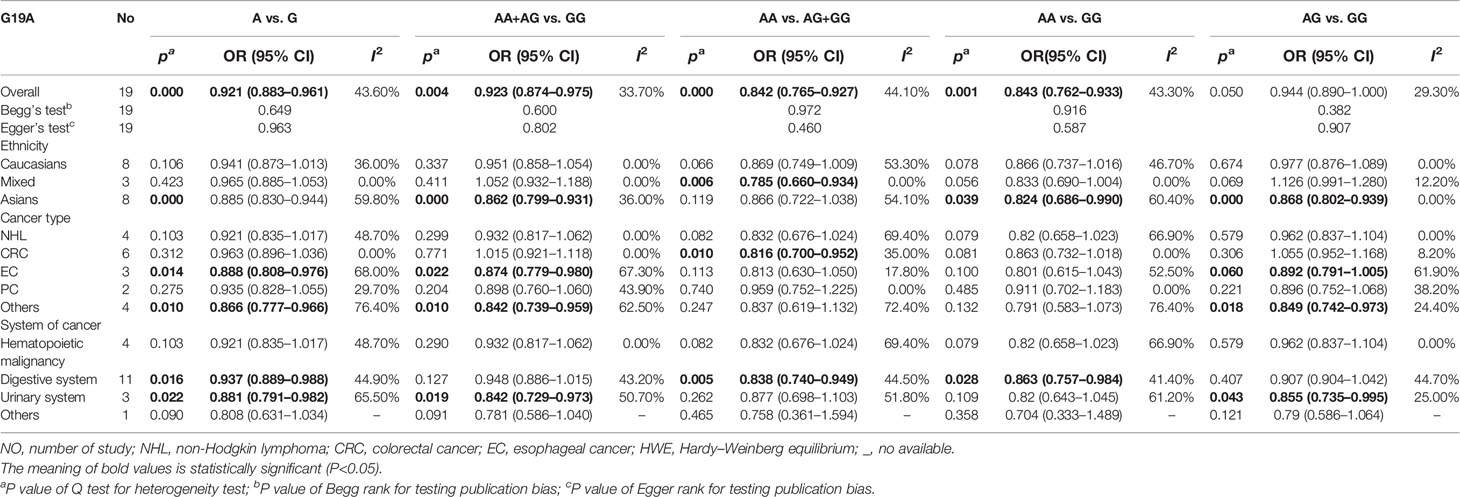
Results: In the overall population, we found a significant relationship with overall cancer (allele comparison: OR = 0.921, p = 0.000; dominant comparison: OR = 0.923, p = 0.004; recessive comparison: OR = 0.842, p = 0.000; homozygote model: OR = 0.0843, p = 0.001). In a subgroup analysis conducted by ethnicity, we obtained significant results in Asians (Asian allele comparison: OR = 0.885, p = 0.000; dominant comparison: OR = 0.862, p = 0.000; homozygote model: OR = 0.824, p = 0.039; and heterozygote comparison: OR = 0.868, p = 0.000) but not in Caucasians. In a subgroup analysis conducted by cancer type and cancer system, we obtained significant results that the LEP G19A polymorphism may decrease the risk of colorectal cancer, esophageal cancer, digestive system cancer, and urinary system cancer.
Conclusions: This meta-analysis revealed that the LEP G19A polymorphism may decrease the risk of cancer.
Introduction
It is well known that cancer is one of major causes of death with over 6.1 million projected to die each year, and morbidity rates have increased gradually over the past decade (1, 2), so it has been a public health burden worldwide. The reason for cancer is complicated and the etiology and mechanism of carcinogenesis are not clearly elucidated to date. It was widely accepted that the interplay between environmental factors, genetics, and lifestyle plays an important role in the carcinogenesis according to epidemiology. There is mounting evidence indicating that many metabolic diseases such as obesity and diabetes may significantly increase the risk of cancer (3–5). The polymorphism of obesity and diabetes gene may be associated with genetic susceptibility of cancer.
Leptin (LEP), a 16-kDa hormone of energy expenditure, is a balancing mediator of homeostasis by regulating acquisition and consumption of energy, which was a basic pathophysiological process in normal cells and cancer cells. Many epidemiological studies have revealed the link between LEP and the development of many kinds of cancers (6–8). Among the pathophysiological mechanisms of cancer, LEP seems relevant to the proliferation of cancer stem cells (9). Some studies also revealed that LEP through its signal pathways regulating energy intake and expenditure [MAPK, PI3K, mTOR, and JAK/STAT (10, 11)] produced an effect in angiogenesis processes that were critical in the genesis and development of cancer (12). Pathophysiological mechanisms of cancer such as inflammation, invasion, and metastasis are also favored by LEP (13–15). So, LEP may be involved in various pathological processes of carcinogenesis.
Single-nucleotide polymorphism can change the functions of genes and the expression of protein. LEP G19A polymorphism, positioning at the 5′-untranslated region of gene, may impact mRNA translation and change the serum level of LEP. With the development of molecular epidemiology, various studies have demonstrated that LEP G19A polymorphism is related to cancer risk (16–19). However, results between G19A polymorphism with cancers have been inconsistent or inconclusive. Therefore, we performed a metaanalysis to verify the correlation between the G19A mutation of the LEP gene and susceptibility to cancers.
In this study, we conducted a meta-analysis to verify whether the G19A polymorphism of the LEP gene affects the risk of cancer.
Methods
Literature Search
A comprehensive literature search of PubMed, Embase, and Web of Science was performed to search all potential studies that involved the relevance between the G19A polymorphism and cancers prior to June 2021. Our study contained the following terms: (“leptin” OR “LEP” OR “G19A” OR “rs2167270”) AND (“polymorphism” OR “variant” OR “mutation”) AND (“malignancy” OR “cancer” OR “carcinoma” OR “neoplasm”).
Inclusion and Exclusion Criteria
The inclusion criteria were as follows: (1) investigate the association between the LEP G19A (rs2167270) mutation and cancers; (2) meet cohort design or case–control design; (3) abundant data should behave to estimate an odds ratio (OR) and 95% confidence interval; (4) results were reported in English; and (5) include human subjects. We adopted the following exclusion criteria: (1) duplicated studies; (2) studies in which subjects were not human; and (3) studies in which we could not obtain sufficient raw data.
Data Extraction
Investigators extracted genotype data independently, and every data point reached a consensus. The extracted data contained the (1) name of the first author; (2) year of publication; (3) ethnicity of cases and controls; (4) cancer type of studies; and (5) frequency of LEP G19A in genes.
Statistical Analysis
We computed ORs and their 95% CIs to estimate the association between the LEP G19A (rs2167270) mutation and cancers. The pooled ORs and their 95% CIs were computed for genes using the following five models: dominant model (AA + AG vs. GG), recessive model (AA vs. GG + AG), allele model (A vs. G), homozygous model (AA vs. GG), and heterozygote model (AG vs. GG).
The Q test was used to estimate heterogeneity between different studies, and p < 0.05 was considered significant for heterogeneity. In addition, inconsistency was quantified by the I2 statistic. Twenty-five percent and 50% of the I2 values indicated low and high levels of heterogeneity, respectively. An I2 < 50% suggested that no heterogeneity existed. When heterogeneity existed, the fixed effects model (FEM) was utilized; otherwise, the random-effects model (REM) was utilized for calculation.
To evaluate the specific effects of ethnicity, cancer type, and cancer system, investigators performed subgroup analyses by ethnicity, cancer type, and cancer system.
Sensitivity analyses were performed to evaluate the stability of the results. A funnel plot of Egger’s or Begg’s test was conducted to reveal possible publication bias. We used the Newcastle-Ottawa Scale to assess the including literature quality. All meta-analyses were performed using STATA software (Version 12.0, College Station, TX).
Results
Study Characteristics
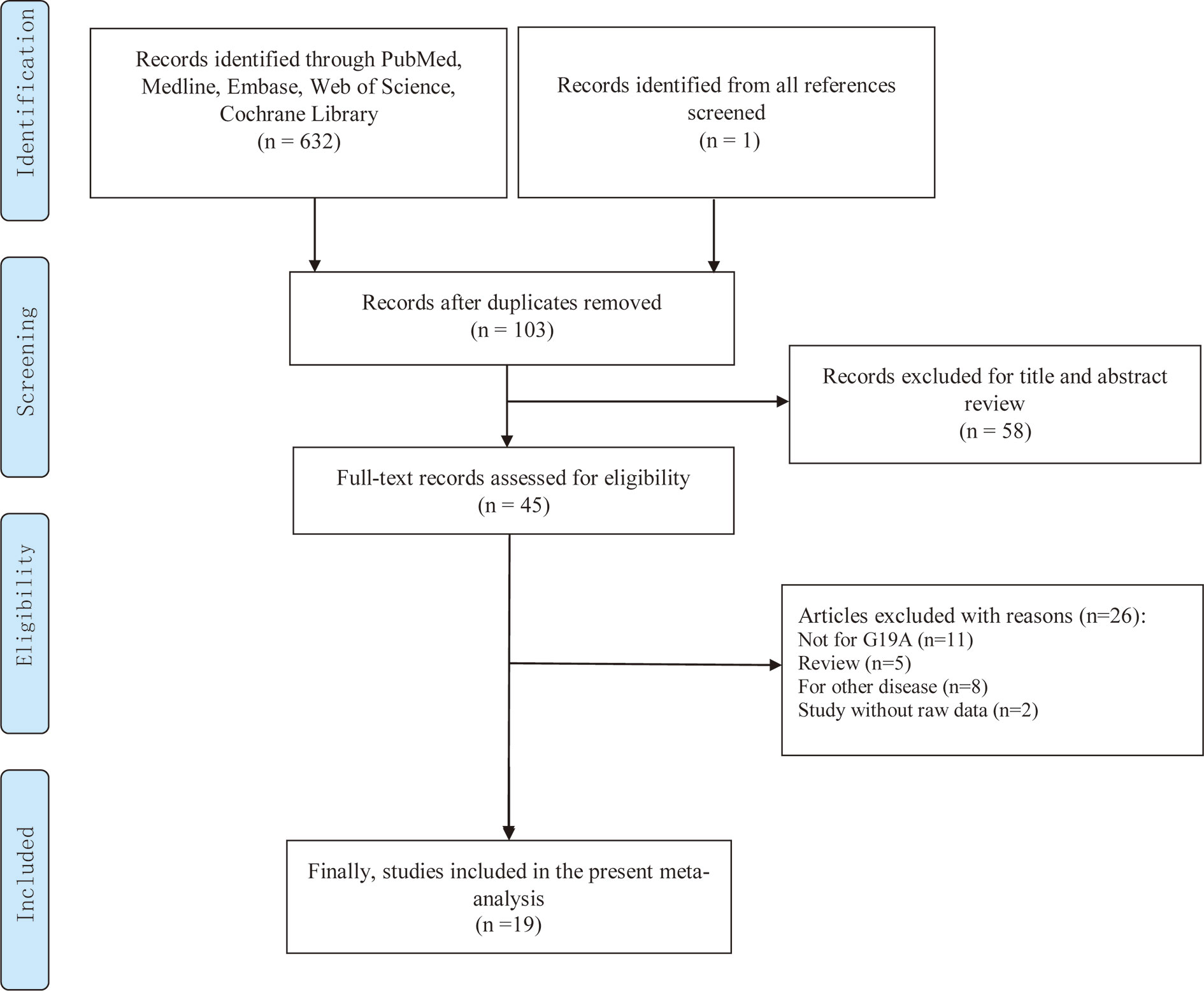
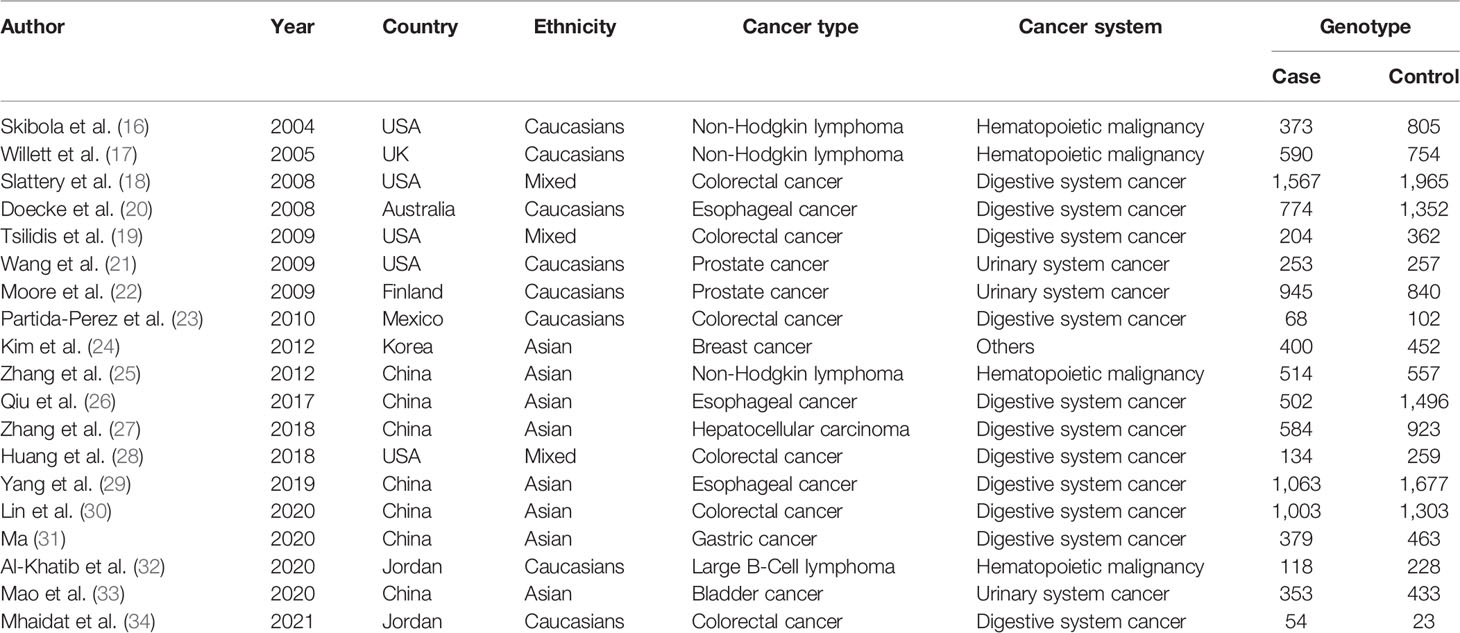
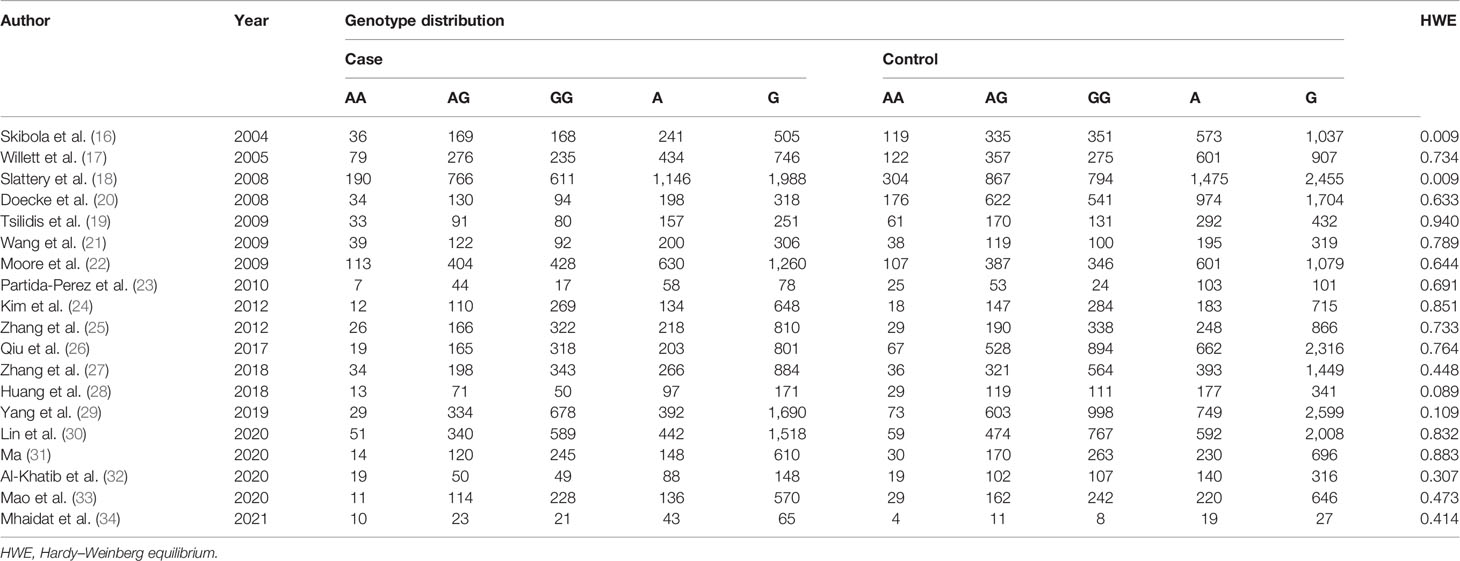
Depending on the search strategy, 633 articles were retrieved (Figure 1). Among them, 103 articles were eligible after excluding repeated publications. By reviewing the titles and study abstracts, 58 articles were excluded. Of the remaining 45 studies, 26 articles were excluded, including 11 studies that were not focused on the LEP G19A genetic mutation. Five studies were meta-analyses. Eight studies were on other disorders that were not cancer. Two articles did not provide raw data. Finally, 19 studies conformed to our meta-analyses, and Tables 1 and 2 summarize the extracted data (16–34).
Effect of the LEP G19A Polymorphism on Cancers
We investigated the effect of the LEP G19A mutation on cancer susceptibility in five genetic models. In all models, if the heterogeneity was less than 50%, the authors applied fixed models, whereas if the heterogeneity was greater than 50%, random models were used.
In the overall population, we found a significant relationship with cancer in four models (allele comparison: OR = 0.921, p = 0.000; dominant comparison: OR = 0.923, p = 0.004; recessive comparison: OR = 0.842, p = 0.000; homozygote model: OR = 0.0843, p = 0.001), and no relevance was observed in the heterozygote model (OR = 0.944, p = 0.05) (Table 3 and Figure 2).
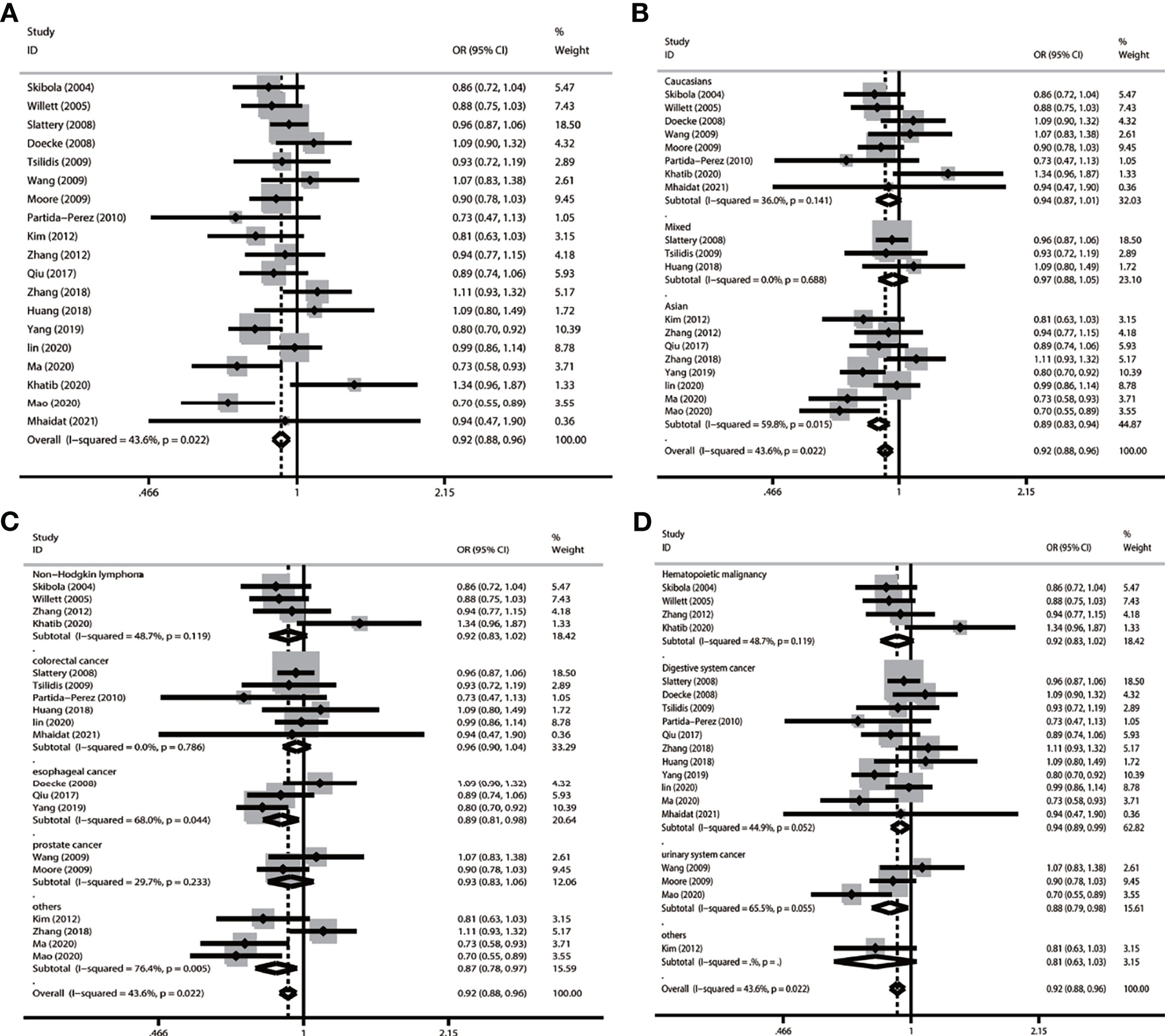
Figure 2 Forest plot of subgroup analysis of LEP G19A and cancer risk in the allele model (A vs. G) (A) LEP G19A polymorphism and overall cancer risk; (B) LEP G19A polymorphism and cancer risk on ethnicity; (C) LEP G19A polymorphism and risk of cancer type; (D) LEP G19A polymorphism and the risk of cancer system).
In a subgroup analysis conducted by ethnicity, we obtained significant results in Asians in four models (allele comparison: OR = 0.885, p = 0.000; dominant comparison: OR = 0.862, p = 0.000; homozygote model: OR = 0.824, p = 0.039; and heterozygote comparison: OR = 0.868, p = 0.000); we also obtained significant results in the mixed recessive model: OR = 0.785, p = 0.1006. We obtained no significant results in the Caucasian population in five models (Table 3 and Figure 2).
In a subgroup analysis conducted by cancer type, we obtained significant results that the LEP G19A polymorphism decreased the risk of colorectal cancer in one model (recessive model: OR = 0.816, p = 0.010); decreased the risk of esophageal cancer in two models (allele model: OR = 0.888, p = 0.014; dominant comparison: OR = 0.874, p = 0.022); and decreased the risk of other types of cancer in three models (allele comparison: OR = 0.866, p = 0.010; dominant comparison: OR = 0.842, p = 0.010; heterozygote comparison: OR = 0.849, p = 0.018) (Table 3 and Figure 2).
In a subgroup analysis conducted by cancer system, we obtained significant results that the LEP G19A polymorphism decreased the risk of digestive system cancer in three models (allele comparison: OR = 0.937, p = 0.016; recessive comparison: OR = 0.838, p = 0.005; homozygote comparison: OR = 0.863, p = 0.028); we also obtained significant results that the LEP G19A polymorphism decreased the risk of urinary system cancer in three models (allele comparison: OR = 0.881, p = 0.022; dominant comparison: OR = 0.842, p = 0.019; heterozygote comparison: OR = 0.855, p = 0.043) (Table 3 and Figure 2).
Sensitivity Analysis and Publication Bias
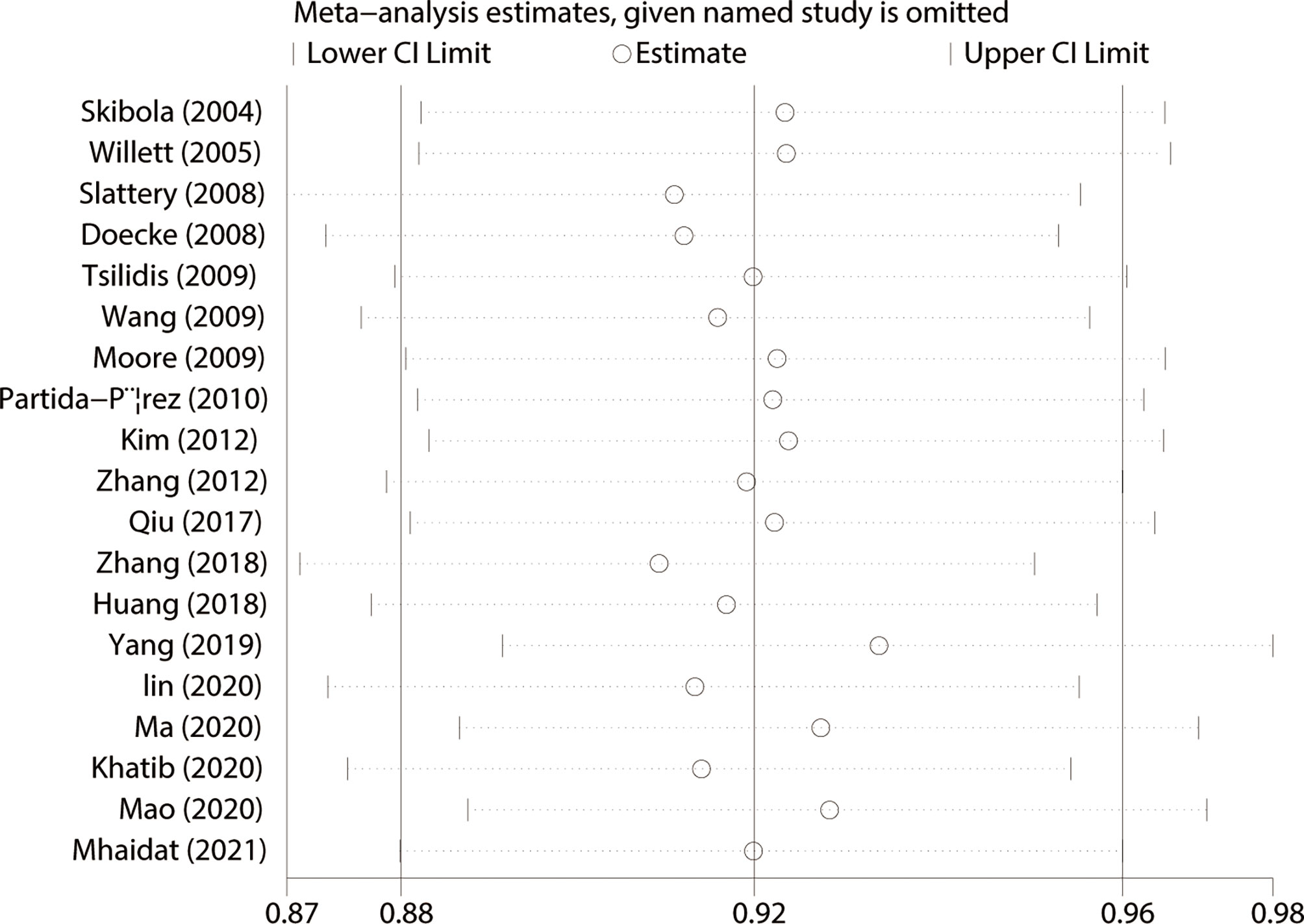
We used Begg’s and Egger’s tests to evaluate publication bias in all models. All results of Begg’s and Egger’s tests were >0.05 in all models and funnel plots, revealing that publication bias may not exist among our studies (Table 2, Figures 3 and 4). We conducted a sensitivity analysis, and pooled ORs and the corresponding 95% CIs were computed. The results did not show a significant change even though one study was deleted each time, which suggested that the results were statistically stable (Figure 5).
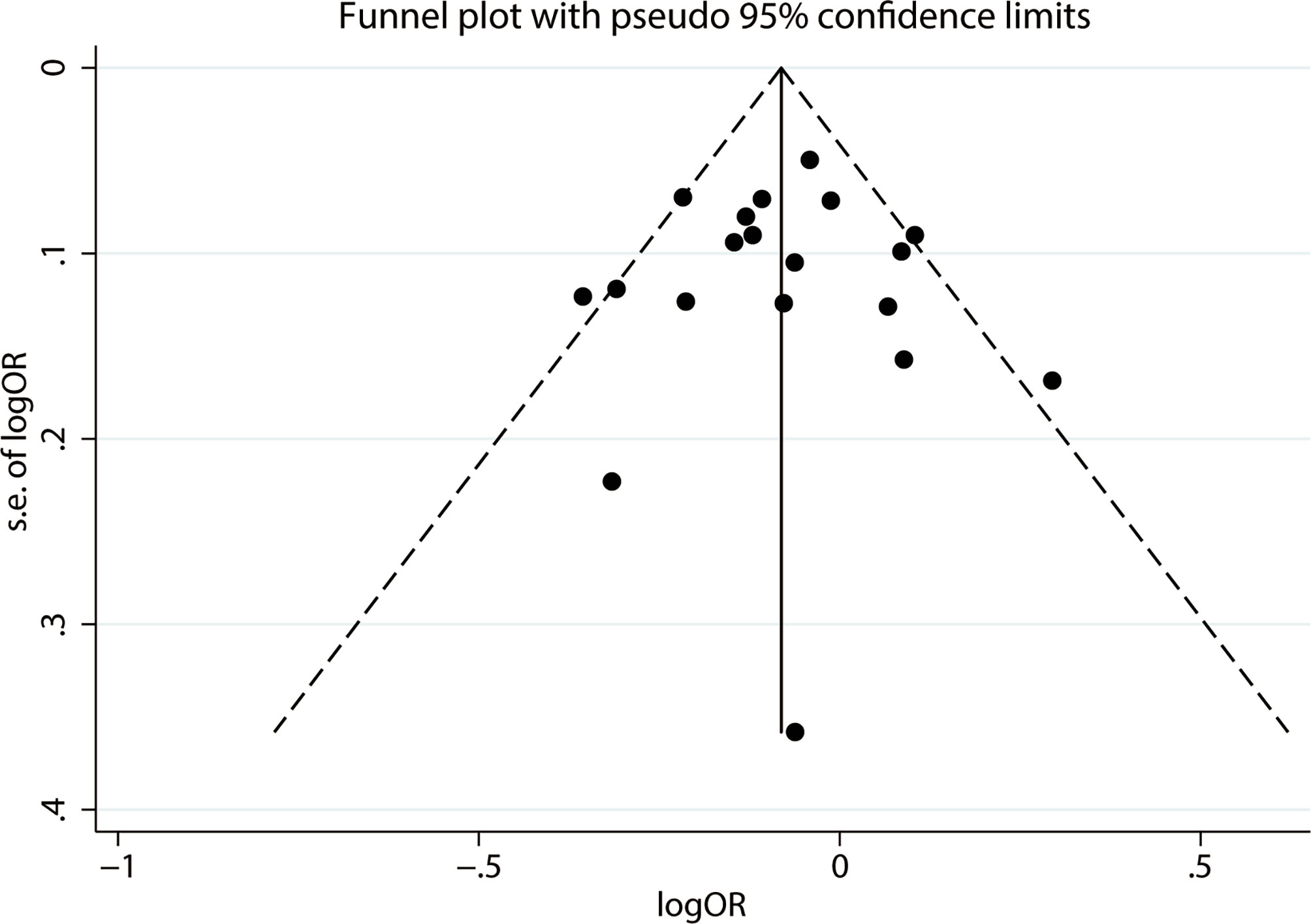
Figure 3 Funnel plot of publication bias on the relationship between LEP G19A polymorphism and the risk of digestion-related cancer in allele model (A vs. G).
Discussion
It has been confirmed that the occurrence of cancer is a complex, multistep, and multifactorial event that contains various genetic, environmental, and lifestyle factors, such as smoking, drinking, obesity, and genetic factor. Multiple studies have revealed that metabolic-related factors are associated with the risk of cancer (35–37). The LEP, metabolic-related factors regulating balancing by regulating acquisition and energy consumption, was confirmed relevant to cancer (38–40). The LEP G19A polymorphism may alter the transcription of mRNA and the level of LEP was confirmed to be associated with any kind of cancer (21, 22, 24–26). However, the conclusions of those studies were inconsistent. Two meta-analyses were researched by Liu et al. (41), including 10 studies, and Yang et al. (29), including 13 studies, generating conflicting results in subgroup analysis and lacking subgroup analysis of the cancer system. Meanwhile, an expanding body of literature on the relationship between LEP G19A polymorphism and cancer risk has been published. Therefore, we conducted this meta-analysis to address this relevance between the LEP G19A polymorphism and cancer risk.
Our current meta-analysis contained 19 studies of cancers containing 9,878 patients, and 14,251 controls were pooled, which contained more participants and cancer types than the previous meta-analysis. Overall, we found a significant correlation between the LEP G19A mutation and susceptibility to cancers under four models (allele model, dominant model, recessive model, and homozygote model), which means that this mutation may decrease the risk of overall cancer. This result was confirmed in a meta-analysis conducted by Liu et al. (41) and Yang et al. (29). Studies (42, 43) confirmed that the LEP G19A mutation might reduce mRNA translation with a lower serum level of LEP, which may attenuate the cancer risk as a protective factor.
Obesity was defined as an imbalance between caloric consumption and energy expenditure. Meanwhile, the LEP is a metabolic-related factor regulating balancing by regulating acquisition and consumption of energy. So it seems that obesity has a positive correlation with LEP polymorphism. However, some studies showed that there was no association between LEP polymorphism and obesity (44, 45). Mizuta et al. (46) study showed that LEP G19A was not associated with obesity. The study by Nesrine et al. (47) even showed that different polymorphisms of the LEP gene have distinct correlations with obesity. Our study showed that LEP G19A polymorphism decreases cancer risk, but the exact mechanism is unknown and mounting evidence indicates that obesity may greatly increase the risk of cancer (3–5). This provides us with a hint that LEP G19A polymorphism may not lead to cancer by gaining weight. Further studies are needed to elucidate the mechanism of action of LEP G19A polymorphism and cancer.
When stratified by ethnicity, we found a significant correlation between this mutation and Asians and no significant in Caucasians, which means that this mutation may decrease the risk of Asian people not Caucasians. This difference might be caused by a discrepancy in the interplay between genes and the environment. Moreover, the frequency of the A allele in Caucasians (68%) and Asians (44%) might be the reason for contributing to the discrepancy in the non-significant results in Caucasians. When stratified by cancer type and cancer system, it was first to describe the association between LEP G19A mutation and the cancer system. We found a significant correlation between this mutation and colorectal cancer, esophageal cancer, digestive system cancer, and urinary system cancer, which means that this mutation may decrease the risk of colorectal cancer, esophageal cancer, digestive system cancer, and urinary system cancer, but we found no correlation between this mutation and the other cancer system; the reason for this difference in risk with different tumors is as yet unknown, possibly due to LEP and its receptors playing various roles in the mediation of physiological reactions and carcinogenesis in different pathological types of cancer.
Heterogeneity may exist in our meta-analysis of cancer in the overall analysis. Stratified analyses indicated that heterogeneity was significant in some subgroups (e.g., Asians, esophageal cancer, and urinary system cancer). These factors may cause heterogeneity in our study. We checked the stability of our pooled results by sensitivity analyses. The trend of relevance was not significantly changed in the sensitivity analyses, which meant that the pooled results in our meta-analysis were statistically stable. We used Begg’s and Egger’s tests to evaluate publication bias. Begg’s and Egger’s tests’ p-values > 0.05 in all models, so that publication bias may exist in this meta-analysis.
The following limitations should be mentioned: (1) The number of studies focused on the relationship between LEP G19A and cancer was relatively small, so little information about stratified analyses of ethnicity, cancer type, and cancer system was available; therefore, further studies are required to determine the actual relationship in all populations. (2) Our study had no access to other potential factors influencing the results, such as other lifestyles, environments, and ages.
Conclusion
In conclusion, this meta-analysis suggests that the LEP G19A mutation may decrease the risk of overall cancer, colorectal cancer, esophageal cancer, digestive system cancer, and urinary system cancer. In the future, more comprehensive objects containing genetic environmental interaction are warranted to discover the correlation between LEP G19A mutation and the risk of cancer.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author Contributions
AZ, SW, FZ, WL, QL, and XL conceived the study. FZ, WL, and QL contributed to data acquisition, data interpretation, and statistical analysis. AZ, SW, and XL contributed to the study design, statistical analysis, writing, and revising of the manuscript critically. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Funds of China (82171594) and Zhao Yi-Cheng Medical Science Foundation (ZYYFY2018031).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all authors whose studies were included in our meta-analysis.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71:7–33. doi: 10.3322/caac.21654
2. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2020. CA Cancer J Clin (2020) 70:7–30. doi: 10.3322/caac.21590
3. Kolb R, Sutterwala FS, Zhang W. Obesity and Cancer: Inflammation Bridges the Two. Curr Opin Pharmacol (2016) 29:77–89. doi: 10.1016/j.coph.2016.07.005
4. Dalamaga M, Christodoulatos GS, Mantzoros CS. The Role of Extracellular and Intracellular Nicotinamide Phosphoribosyl-Transferase In Cancer: Diagnostic and Therapeutic Perspectives and Challenges. Metabolism (2018) 82:72–87. doi: 10.1016/j.metabol.2018.01.001
5. Garg SK, Maurer H, Reed K, Selagamsetty R. Diabetes and Cancer: Two Diseases With Obesity as a Common Risk Factor. Diabetes Obes Metab (2014) 16:97–110. doi: 10.1111/dom.12124
6. Bieńkiewicz J, Romanowicz H, Wilczyński M, Jabłoński G, Stepowicz A, Obłękowska A, et al. Association of Single Nucleotide Polymorphism LEP-R C.668A>G (P.Gln223Arg, Rs1137101) of Leptin Receptor Gene With Endometrial Cancer. BMC Cancer (2021) 21:925. doi: 10.1186/s12885-021-08620-y
7. Hołysz H, Paszel-Jaworska A, Romaniuk-Drapała A, Grodecka-Gazdecka S, Rubiś B. LEP (-2548g>A LEP) and LEPR (223gln>Arg, 109lys>Arg) Polymorphisms as Breast Cancer Risk Factors in the Polish Female Population. Mol Biol Rep (2021) 48:3237–44. doi: 10.1007/s11033-021-06328-7
8. Tayel SI, Alhanafy AM, Ahmed SM, Eltorgoman AA, Elsayed IE. Biochemical Study on Modifying Role of Variants of Leptin Gene and its Receptor on Serum Leptin Levels in Breast Cancer. Mol Biol Rep (2020) 47:3807–20. doi: 10.1007/s11033-020-05436-0
9. Delort L, Bougaret L, Cholet J, Vermerie M, Billard H, Decombat C, et al. Hormonal Therapy Resistance and Breast Cancer: Involvement of Adipocytes and Leptin. Nutrients (2019) 11(12):2839–55. doi: 10.3390/nu11122839
10. Ghasemi A, Saeidi J, Azimi-Nejad M, Hashemy SI. Leptin-Induced Signaling Pathways in Cancer Cell Migration and Invasion. Cell Oncol (Dordr) (2019) 42:243–60. doi: 10.1007/s13402-019-00428-0
11. Olea-Flores M, Juárez-Cruz JC, Mendoza-Catalán MA, Padilla-Benavides T, Navarro-Tito N. Signaling Pathways Induced by Leptin During Epithelial–Mesenchymal Transition in Breast Cancer. Int J Mol Sci (2018) 19(11):3493–513. doi: 10.3390/ijms19113493
12. Llanos A, Yao S, Singh A, Aremu JB, Khiabanian H, Lin Y, et al. Gene Expression of Adipokines and Adipokine Receptors in the Tumor Microenvironment: Associations of Lower Expression With More Aggressive Breast Tumor Features. Breast Cancer Res Treat (2021) 185:785–98. doi: 10.1007/s10549-020-05972-0
13. Nwadozi E, Ng A, Strömberg A, Liu HY, Olsson K, Gustafsson T, et al. Leptin is a Physiological Regulator of Skeletal Muscle Angiogenesis and is Locally Produced by PDGFRα and PDGFRβ Expressing Perivascular Cells. Angiogenesis (2019) 22:103–15. doi: 10.1007/s10456-018-9641-6
14. Clements VK, Long T, Long R, Figley C, Smith D, Ostrand-Rosenberg S. Frontline Science: High Fat Diet and Leptin Promote Tumor Progression by Inducing Myeloid-Derived Suppressor Cells. J Leukoc Biol (2018) 103:395–407. doi: 10.1002/JLB.4HI0517-210R
15. Ray A, Cleary MP. The Potential Role of Leptin in Tumor Invasion and Metastasis. Cytokine Growth Factor Rev (2017) 38:80–97. doi: 10.1016/j.cytogfr.2017.11.002
16. Skibola CF, Holly EA, Forrest MS, Hubbard A, Bracci PM, Skibola DR, et al. Body Mass Index, Leptin and Leptin Receptor Polymorphisms, and non-Hodgkin Lymphoma. Cancer Epidemiol Biomarkers Prev (2004) 13:779–86.
17. Willett EV, Skibola CF, Adamson P, Skibola DR, Morgan GJ, Smith MT, et al. Non-Hodgkin's Lymphoma, Obesity and Energy Homeostasis Polymorphisms. Br J Cancer (2005) 93:811–6. doi: 10.1038/sj.bjc.6602762
18. Slattery ML, Wolff RK, Herrick J, Caan BJ, Potter JD. Leptin and Leptin Receptor Genotypes and Colon Cancer: Gene-Gene and Gene-Lifestyle Interactions. Int J Cancer (2008) 122:1611–7. doi: 10.1002/ijc.23135
19. Tsilidis KK, Helzlsouer KJ, Smith MW, Grinberg V, Hoffman-Bolton J, Clipp SL, et al. Association of Common Polymorphisms in IL10, and in Other Genes Related to Inflammatory Response and Obesity With Colorectal Cancer. Cancer Causes Control (2009) 20:1739–51. doi: 10.1007/s10552-009-9427-7
20. Doecke JD, Zhao ZZ, Stark MS, Green AC, Hayward NK, Montgomery GW, et al. Single Nucleotide Polymorphisms in Obesity-Related Genes and the Risk of Esophageal Cancers. Cancer Epidemiol Biomarkers Prev (2008) 17:1007–12. doi: 10.1158/1055-9965.EPI-08-0023
21. Wang MH, Helzlsouer KJ, Smith MW, Hoffman-Bolton JA, Clipp SL, Grinberg V, et al. Association of IL10 and Other Immune Response- and Obesity-Related Genes With Prostate Cancer in CLUE II. Prostate (2009) 69:874–85. doi: 10.1002/pros.20933
22. Moore SC, Leitzmann MF, Albanes D, Weinstein SJ, Snyder K, Virtamo J, et al. Adipokine Genes and Prostate Cancer Risk. Int J Cancer (2009) 124:869–76. doi: 10.1002/ijc.24043
23. Partida-Pérez M, de la Luz AM, Peregrina-Sandoval J, Macías-Gómez N, Moreno-Ortiz J, Leal-Ugarte E, et al. Association of LEP and ADIPOQ Common Variants With Colorectal Cancer in Mexican Patients. Cancer Biomark (2010) 7:117–21. doi: 10.3233/CBM-2010-0154
24. Kim KZ, Shin A, Lee YS, Kim SY, Kim Y, Lee ES. Polymorphisms in Adiposity-Related Genes are Associated With Age at Menarche and Menopause in Breast Cancer Patients and Healthy Women. Hum Reprod (2012) 27:2193–200. doi: 10.1093/humrep/des147
25. Zhang Y, Wang MY, He J, Wang JC, Yang YJ, Jin L, et al. Tumor Necrosis Factor-α Induced Protein 8 Polymorphism and Risk of non-Hodgkin's Lymphoma in a Chinese Population: A Case-Control Study. PloS One (2012) 7:e37846. doi: 10.1371/journal.pone.0037846
26. Qiu H, Lin X, Tang W, Liu C, Chen Y, Ding H, et al. Investigation of TCF7L2, LEP and LEPR Polymorphisms With Esophageal Squamous Cell Carcinomas. Oncotarget (2017) 8:109107–19. doi: 10.18632/oncotarget.22619
27. Zhang S, Jiang J, Chen Z, Wang Y, Tang W, Liu C, et al. Investigation of LEP and LEPR Polymorphisms With the Risk of Hepatocellular Carcinoma: A Case-Control Study in Eastern Chinese Han Population. Onco Targets Ther (2018) 11:2083–9. doi: 10.2147/OTT.S153931
28. Huang BZ, Tsilidis KK, Smith MW, Hoffman-Bolton J, Visvanathan K, Platz EA, et al. Polymorphisms in Genes Related to Inflammation and Obesity and Colorectal Adenoma Risk. Mol Carcinog (2018) 57:1278–88. doi: 10.1002/mc.22842
29. Yang J, Zhong Z, Tang W, Chen J. Leptin Rs2167270 G > A (G19A) Polymorphism may Decrease the Risk of Cancer: A Case-Control Study and Meta-Analysis Involving 19 989 Subjects. J Cell Biochem (2019) 120:10998–1007. doi: 10.1002/jcb.28378
30. Lin J, Xie Z, Lan B, Guo Z, Tang WF, Liu C, et al. Investigation of Leptin and its Receptor (LEPR) for Single Nucleotide Polymorphisms In Colorectal Cancer: A Case-Control Study Involving 2,306 Subjects. Am J Transl Res (2020) 12:3613–28. doi: 10.1200/JCO.2020.38.15_suppl.e16100
31. Ma R, He Q. A Variant of Leptin Gene Decreases the Risk of Gastric Cancer in Chinese Individuals: Evidence From a Case-Control Study. Pharmgenomics Pers Med (2020) 13:397–404. doi: 10.2147/PGPM.S258672
32. Al-Khatib SM, Abdo N, Al-Eitan LN, Al-Mistarehi AW, Zahran DJ, Kewan TZ. LTA, LEP, and TNF-A Gene Polymorphisms are Associated With Susceptibility and Overall Survival of Diffuse Large B-Cell Lymphoma in an Arab Population: A Case-Control Study. Asian Pac J Cancer Prev (2020) 21:2783–91. doi: 10.31557/APJCP.2020.21.9.2783
33. Mao F, Niu XB, Gu S, Ji L, Wei BJ, Wang HB. Investigation of Leptin G19A Polymorphism With Bladder Cancer Risk: A Case-Control Study. J Clin Lab Anal (2020) 34:e23351. doi: 10.1002/jcla.23351
34. Mhaidat NM, Alzoubi KH, Kubas MA, Banihani MN, Hamdan N, Al-Jaberi TM. High Levels of Leptin and non-High Molecular Weight-Adiponectin in Patients With Colorectal Cancer: Association With Chemotherapy and Common Genetic Polymorphisms. BioMed Rep (2021) 14:13. doi: 10.3892/br.2020.1389
35. Vander HM, DeBerardinis RJ. Understanding the Intersections Between Metabolism and Cancer Biology. Cell (2017) 168:657–69. doi: 10.1016/j.cell.2016.12.039
36. Xu H, Zhou S, Tang Q, Xia H, Bi F. Cholesterol Metabolism: New Functions and Therapeutic Approaches in Cancer. Biochim Biophys Acta Rev Cancer (2020) 1874:188394. doi: 10.1016/j.bbcan.2020.188394
37. Li Z, Zhang H. Reprogramming of Glucose, Fatty Acid and Amino Acid Metabolism for Cancer Progression. Cell Mol Life Sci (2016) 73:377–92. doi: 10.1007/s00018-015-2070-4
38. Lee KN, Choi HS, Yang SY, Park HK, Lee YY, Lee OY, et al. The Role of Leptin in Gastric Cancer: Clinicopathologic Features and Molecular Mechanisms. Biochem Biophys Res Commun (2014) 446:822–9. doi: 10.1016/j.bbrc.2014.02.072
39. Dong Z, Fu S, Xu X, Yang Y, Du L, Li W, et al. Leptin-Mediated Regulation of ICAM-1 is Rho/ROCK Dependent and Enhances Gastric Cancer Cell Migration. Br J Cancer (2014) 110:1801–10. doi: 10.1038/bjc.2014.70
40. Aleksandrova K, Boeing H, Jenab M, Bueno-de-Mesquita HB, Jansen E, van Duijnhoven FJ, et al. Leptin and Soluble Leptin Receptor in Risk of Colorectal Cancer in the European Prospective Investigation Into Cancer and Nutrition Cohort. Cancer Res (2012) 72:5328–37. doi: 10.1158/0008-5472.CAN-12-0465
41. Liu P, Shi H, Huang C, Shu H, Liu R, Yang Y, et al. Association of LEP A19G Polymorphism With Cancer Risk: A Systematic Review and Pooled Analysis. Tumour Biol (2014) 35:8133–41. doi: 10.1007/s13277-014-2088-5
42. Pan H, Deng LL, Cui JQ, Shi L, Yang YC, Luo JH, et al. Association Between Serum Leptin Levels and Breast Cancer Risk: An Updated Systematic Review and Meta-Analysis. Medicine (Baltimore) (2018) 97:e11345. doi: 10.1097/MD.0000000000011345
43. Savino F, Rossi L, Di Stasio L, Galliano I, Montanari P, Bergallo M. Mismatch Amplification Mutation Assay Real-Time PCR Analysis of the Leptin Gene G2548A and A19G Polymorphisms and Serum Leptin in Infancy: A Preliminary Investigation. Horm Res Paediatr (2016) 85:318–24. doi: 10.1159/000444484
44. Fairbrother U, Kidd E, Malagamuwa T, Walley A. Genetics of Severe Obesity. Curr Diabetes Rep (2018) 18:85. doi: 10.1007/s11892-018-1053-x
45. Zhang L, Yuan LH, Xiao Y, Lu MY, Zhang LJ, Wang Y. Association of Leptin Gene -2548 G/A Polymorphism With Obesity: A Meta-Analysis. Ann Nutr Metab (2014) 64:127–36. doi: 10.1159/000363392
46. Mizuta E, Kokubo Y, Yamanaka I, Miyamoto Y, Okayama A, Yoshimasa Y, et al. Leptin Gene and Leptin Receptor Gene Polymorphisms are Associated With Sweet Preference and Obesity. Hypertens Res (2008) 31:1069–77. doi: 10.1291/hypres.31.1069
Keywords: leptin (LEP), cancer, polymorphism, A19G, rs2167270
Citation: Zhang A, Wang S, Zhang F, Li W, Li Q and Liu X (2021) The Prognosis of Leptin rs2167270 G > A (G19A) Polymorphism in the Risk of Cancer: A Meta-Analysis. Front. Oncol. 11:754162. doi: 10.3389/fonc.2021.754162
Received: 06 August 2021; Accepted: 29 October 2021;
Published: 18 November 2021.
Edited by:
Walter Hernán Pavicic, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Manuel Pires Bicho, University of Lisbon, PortugalRajeev K. Singla, Sichuan University, China
Copyright © 2021 Zhang, Wang, Zhang, Li, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoqiang Liu, MTI5MTMxMzYzOEBxcS5jb20=
†These authors have contributed equally to this work
 Aiqiao Zhang
Aiqiao Zhang Shangren Wang3†
Shangren Wang3†