
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 09 December 2021
Sec. Cancer Genetics
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.746577
 Zhongqi Li1†
Zhongqi Li1† Fang Yu2†
Fang Yu2† Wenle Ye3†
Wenle Ye3† Liping Mao3,4,5,6
Liping Mao3,4,5,6 Jiansong Huang3,4,5,6
Jiansong Huang3,4,5,6 Yang Shao7
Yang Shao7 Junrong Yan7
Junrong Yan7 Wenjuan Yu3,4,5,6
Wenjuan Yu3,4,5,6 Jie Jin3,4,5,6*
Jie Jin3,4,5,6* Jinghan Wang3,4,5,6*
Jinghan Wang3,4,5,6*Diffuse large B-cell lymphoma (DLBCL) is a heterogeneous group of large lymphoid B cell malignancy with distinct clinical and genetic features. Recently, NOTCH1 mutations were identified in DLBCL cases by Next-generation sequencing (NGS), but the clinical features and prognostic impact were not systematically studied. Here, NOTCH1 genes in 161 DLBCL samples were sequenced by NGS. The prognostic value of NOTCH1 mutations was assessed in the context of clinical and laboratory factors, such as international prognostic index (IPI), cell-of-origin classification, double expression of BCL2 and c-MYC. The combined data from three Western cohorts were used to validate these results. As a result, NOTCH1 mutations were found in 17(10.6%) patients, and three patients had a hotspot mutation of c.7541_7542delCT. The presence of NOTCH1 mutations was significantly associated with poor complete response and progression free survival(PFS), which was independent of established clinical and laboratory parameters. In addition, 30 (1.92%) of 1562 patients treated with R-CHOP regimen in those combined Western cohorts had NOTCH1 mutations. Meta-analysis of the Western cohorts confirmed that NOTCH1 mutations were also associated with poor PFS and OS. In conclusion, DLBCL patients with the NOTCH1 mutations have worse PFS and OS, and the NOTCH1 mutations can be used as an independent predictor for patients with DLBCL.
The NOTCH pathway is a highly conserved signaling pathway, which is widely involved in cellular proliferation, differentiation, and apoptosis (1). There are four types of Notch receptors in mammals, such as NOTCH1, NOTCH2, NOTCH3, and NOTCH4 proteins. NOTCH1 and NOTCH2 receptors are highly expressed in many tissues, while NOTCH3 is mainly seen in vascular smooth muscles, and NOTCH4 is usually observed in endothelium (2). Notably, most of the genetic changes of the Notch receptors were observed in the NOTCH1 gene (3–6). This receptor consists of an extracellular component, followed by a transmembrane domain and an intracellular region (NICD). There are at least two forms to activate the Notch-1 mediated signals: ligand-dependent and ligand-independent activation pathways, respectively (7). When the extracellular domain binds to its ligand, the ligand-dependent NOTCH 1 signaling is activated (8). While, gain-of-function mutations in NOTCH1 gene often lead to the ligand-independent activation in pathological conditions (8). After activation, NICD is cleaved from the intracellular domain and then translocated into the nucleus, leading to the transcription of Notch target genes, including the MYC oncogene (5, 8).
The first report about NOTCH1 receptors in malignancies was the observation of a constitutive activation of NOTCH1 signals in T-cell acute lymphoblastic leukemia with a t(7;9)(q34;q34.3) chromosome translocation (9). Subsequently, more and more studies discovered activation of NOTCH1 receptor occurs in several solid tumors such as colorectal cancer (8), head and neck cancer (10), lung cancer (11), and melanoma (12), and other hematologic malignancies such as chronic lymphocytic leukemia (13, 14), mantle cell lymphoma (15), and Hodgkin’s lymphoma (5). Biologically, defects in the Notch signaling pathway would contribute to the development of congenital disorders, viral infections, and cancer (1–3). Clinically, patients with NOTCH1 mutations are often associated with poorer clinical outcomes and a higher risk of disease progression (13, 16). In contrast, tumor suppressive effect was also reported in some studies. For example, NOTCH1 deficiency in skin can lead to the development of skin tumors (17). Thus, the multifaceted role of NOTCH1 signaling in cancer inhibition or promotion depends on the influence of cellular microenvironment (18).
Diffuse large B cell lymphoma (DLBCL) is the most common type of non-Hodgkin lymphoma and has received extensive attention in terms of genetic findings and clinical outcomes (19). Using whole genome/exome sequencing, plenty of mutations were found in DBLCL (6). However, the biological significance and clinical associations of each mutated gene still need further investigation. It has been reported that the NOTCH1 mutations are associated with reduced benefit of anti-CD20 chemoimmunotherapy regimens in chronic lymphocytic leukemia (20), and its clinical significance in DLBCL is unclear (6, 21, 22). In this study, we enrolled a relatively large cohort of DLBCL patients to investigate the clinical and biological characteristics of NOTCH1 mutations in DLBCL patients.
In this study, 161 newly diagnosed DLBCL patients were enrolled from 2013 to 2020 in the hematological department of our hospital. The pathological diagnoses of DLBCL was based on the World Health Organization Classification (23). We included DLBCL patients with fresh frozen tumor tissues, older than 18 years, and received R-CHOP(rituximab 375 mg/m2 on Day 0, cyclophosphamide 750 mg/m2, doxorubicin 50 mg/m2, and vincristine 1.4 mg/m2 on Day 1, and prednisone 50 mg/m2 orally on Days 1-5) chemotherapy. Patients with HIV infection, pregnancy, another cancer, and double and/or triple hits lymphoma were excluded in this study. Clinical and laboratory information were retrospectively collected from the medical records at the time of DLBCL diagnosis. The computed tomography (CT) scans and/or positron emission tomography-CT, and bone marrow biopsy were used to assess the treatment response, and disease progression. All of the subjects were well-informed about the study and provided written informed consent to participate in the study. The study was approved by the Institutional Review Board of our hospital (No : IIT20210369A).
Formalin-fixed paraffin-embedded tissue sections were used for IHC and FISH analyses. Automated IHC for CD20, CD10, BCL2, BCL6, MUM1, c-MYC, Ki-67 were performed on 4-μm-think tissue sections using an automated slide stainer, the VentanaBenchmark XT (Ventana Medical Systems). Cases with more than 40% positive cells of MYC and 50% of BCL2 were identified as double expressor lymphoma(DEL). Bcl-2, Bcl-6 and c-Myc fracture probes were applied to the sections, and details of FISH methods were previously described (24). COO classification was determined by Hans’s algorithm (25).
NOTCH1 mutations were performed by the targeted NGS tests. Genomic DNA was extracted from the formalin-fixed paraffin-embedded tissue sections. The detailed methods were reported in supplementary methods. Mutation analyses of NOTCH1 were carried out as described previously (26). The primers were depicted in Table S1.
Our major aim was to evaluate the prognostic significance of NOTCH1 mutations on progression free survival (PFS) in DLBCL patients after RCHOP chemotherapy. PFS was defined as the time from disease diagnosis until the time of progression, relapse or death from any cause. Overall survival (OS) was defined as time from the date of diagnosis until death due to any cause or the last follow-up. Complete response (CR) was defined according to the Revised Response Criteria for Malignant Lymphoma (27). The log-rank test in the Kaplan-Meier survival model was used to evaluate the prognostic impact of categorical variables. Univariate and multivariate analyses with Cox proportional hazards models were performed to assess significant predictors. The proportional-hazards assumption was checked for each variable before fitting Cox models. The survival meta-analyses were conducted by the “meta” package (28), the detailed information about the mutation sites was illustrated by the “trackViewer” package (29). The median, interquartile range and frequency counts were used to summarize the distribution of clinical data. Fisher’s exact test and nonparameter T-test were used to test the categorical and continuous variables, respectively. All statistical analyses were conducted with R statistic packages, version 3.6.1 (www.r-project.org). The two-sided level of significance was set at p-value < 0.05.
As illustrated in Figure 1, NOTCH1 mutations were detected in 17 of 161 DLBCL patients (10.6%), specifically, including one splice mutation, two non-sense mutations, five frame shift mutations, and eleven missense mutations (Figure 1A and Table S2). We conducted the Sanger Sequencing to examine 8 out of 16 mutated sites of NOTCH1, and validated 2 mutated sites in extracellular regions such as c.2537 A>C (p.Q846P) and c.2542G>A (p.E848K), and four sites in intracellular domains like c.6392G>T (p.G2131V), c.6598G>A (p.V2200M), c.7541_7542delCT(p.P2514Rfs) and c.7216C>T (p.Q2406*). Two sites (R207C and P837L) in the EGF-like repeats regions were not validated by the Sanger Sequencing probably due to the relatively low tumor alleles. The details of Sanger sequencing were depicted in the Figure S1 and Table S2. Generally, splice, framing and non-sense mutations often lead to large-scale changes in proteins. However, missense mutations lead to the substitution of different amino acids, which in turn have different effects on the protein’s function. Therefore, we further estimated the effects of missense mutations on protein function by using the PANTHER cSNP tool (30). There results showed that all of the missense mutations may impair the function of NOTCH1 protein, among which the highest score were G2131V, E334K, and V2200M missense mutations, implying the more likely deleterious effect on proteins (Figure S2). NOTCH1 missense mutations and the recurrent c.7541_7542delCT (validated by Sanger sequencing in Figure S1) are supposed to affect the NOTCH activity. However, the non-sense mutations and the frameshift deletion in the initial region of NOTCH1 gene probably lead to the absence of protein expression. Thus, we named mutations potentially affecting the NOCH1 activity as type 1 group and mutations probably leading to the absence of protein expression as type 2 group. In this study, there were no differences in their relationship with the clinical parameters (Table S3).
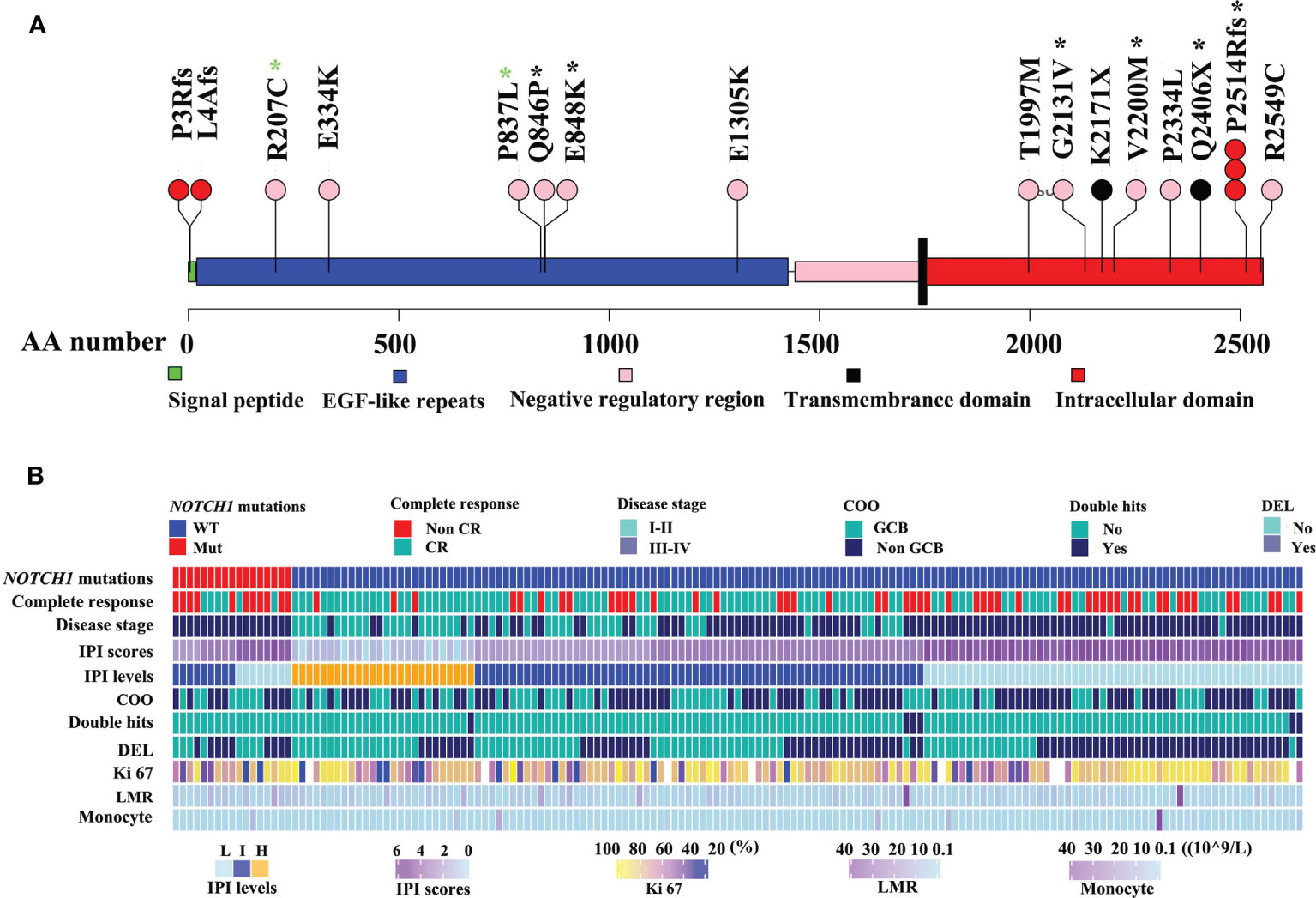
Figure 1 Mutation maps of the NOTCH1 protein (A). The x-axis reports the amino acid(AA) number. The circles are colored with respect to the corresponding mutation types: “black” representing Non-sense mutations, “red” equaling to frameshift mutations, “pink” representing missense mutations. Black stars representing mutations identified by Sanger sequencing, while green stars representing no mutations identified by Sanger sequencing. The detailed clinical information of DLBCL was illustrated (B). IPI, International Prognostic Index; non-GCB, non-germinal center B-cell-like lymphoma; DEL, double expressor lymphoma; HB, hemoglobin; LMR, lymphoma-to-monocyte ratio.
Clinical features of DLBCL patients with NOTCH1 mutations are summarized in Table S4. Patients with NOTCH1 mutations were predominated in stage III-IV(P=0.003, Figure 1B). NOTCH1 mutations were significantly associated with lower blood monocyte counts (P=0.031) but higher lymphoma/monocyte ratio (P=0.02) and higher hemoglobin levels (P=0.006). Notably, patients with the NOTCH1 mutations had a lower complete response rate (P=0.028) than those without NOTCH1 mutations. There was no statistically significant correlation between NOTCH1 mutations and gender, age, international prognostic index (IPI), cell-of-origin (COO) classification, double expressor lymphoma (DEL), white blood cell count (WBC), platelet count, neutrophil counts and other variables (Table S4).
At the median follow-up of 43.3 months, 3-year progression-free survival (PFS) and overall survival (OS) rates for DLBCL patients were 28% and 60%, respectively. In this study, we also evaluated the influence of recognized prognostic factors such as IPI, COO and DEL classifications on prognosis (Figures S3–S5 and Table S5). Consistent with other studies, higher levels of IPI, non-GCB and DEL predicted shorter PFS and OS, respectively. Additionally, hemoglobin (HB) and lymphoma/monocyte ratio(LMR) also have some prognostic values for PFS or OS. Notably, there was a significant difference in PFS and OS between patients with and without NOTCH1 mutations in our DLBCL patients (Figures 2A, B). In multivariate analyses, the effect of NOTCH1 mutations on poor PFS[HR(95% CI), 2.373(1.296,4.344); P=0.005] and OS [HR(95% CI), 5.025(2.001,12.62); P<0.001] persisted, and its prognostic impact was independent of GCB subtypes and/or non-DEL in DLPCL patients (Table 1). In addition, we performed a multivariate analysis of NOTCH1 mutations and treatment response. Similarly, NOTCH1 mutations were inversely and independently associated with complete remission after chemotherapy (Figure S6).
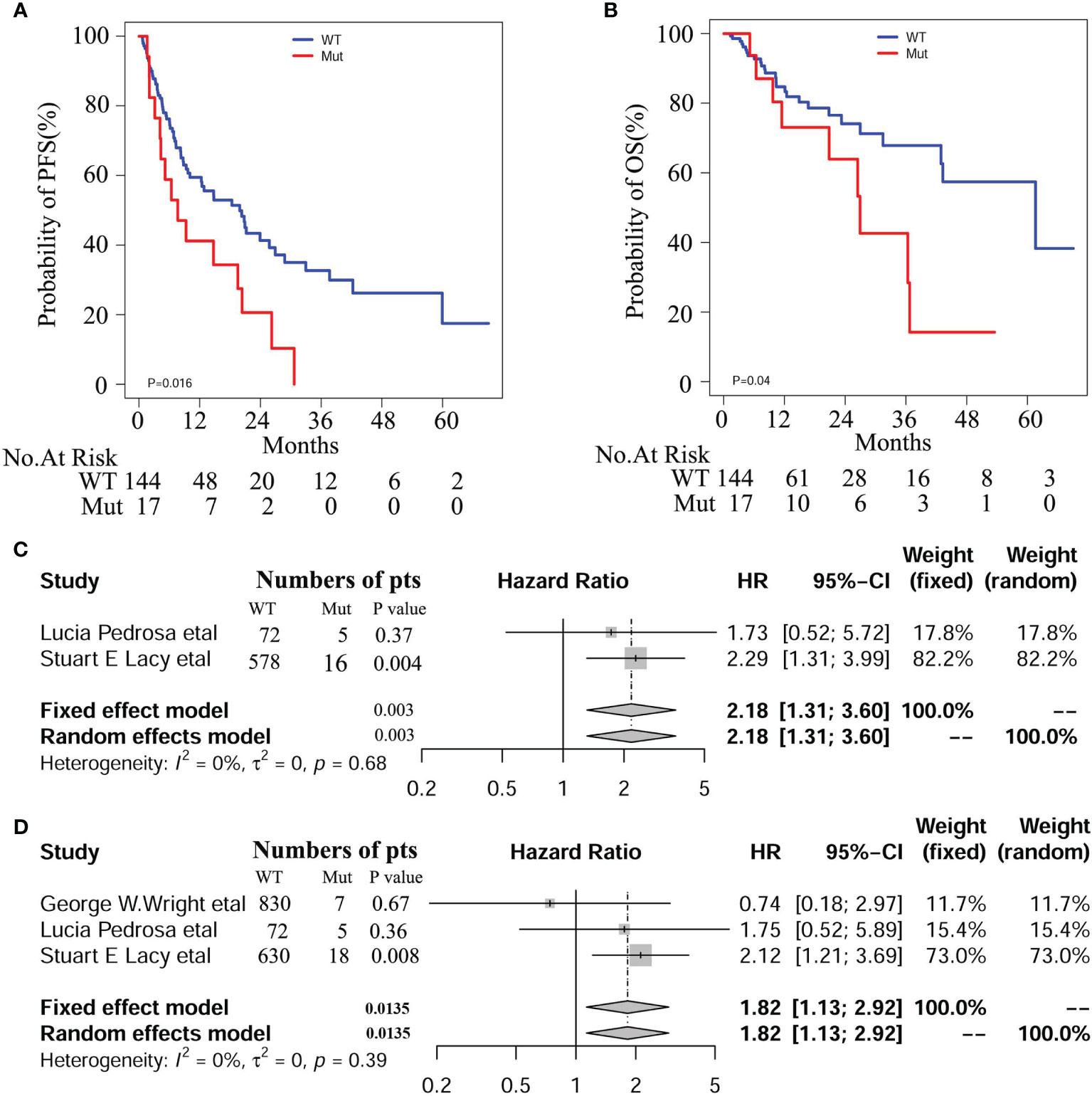
Figure 2 Survival curves of PFS (A) and OS (B) in our DLBCL patients with and with NOTCH1 mutations. Meta-analyses of PFS (C) and OS (D) in the Western cohorts of DLBCL patients.
We enrolled 1562 patients with DLBCL treated with R-CHOP regimen, including 837 from George W. Wright et al. (22), 77 from Lucía Pedrosa et al. (31), and 648 from Stuart E. Lacy and colleagues (32), respectively. Among these patients, 30 (1.92%) cases were identified as NOTCH1 mutations. Detailed mutation information is illustrated in Figure S7, including frame shift mutation, non-sense mutation, splicing site mutation, and missense mutation. We conducted a meta-analysis on these three cohorts and found a significant correlation between NOTCH1 mutations and PFS (HR 95%(CI), 2.18 [1.31; 3.60]; P=0.0025), and OS (HR 95%(CI), 1.82 [1.13; 2.92]; P=0.014, Figure 2). Besides, we combined the individual data of the three cohorts to obtain similar results. As shown in the Figure S8, there was a significant correlation between NOTCH1 mutations and PFS (P=0.005 and OS(P=0.02), respectively.
Whole genome and exome sequencings have revealed numerous somatic mutations that occur repeatedly in DLBCL. A systematic and in-depth study of these mutant genes can help us better screen out high-risk cases and predict new therapeutic targets for DLBCL. In this study, we included a relatively large cohort of DLBCL patients, analyzed NOTCH1 gene mutations by NGS sequencing, and evaluated the prognostic value of NOTCH1 mutations and other recognized clinical and laboratory risk stratification factors. Finally, we performed a meta-analysis on three published Western cohorts to verify our findings.
As a result, NOTCH1 mutations were found in 17(10.6%) patients, and three patients had a hotspot mutation of c.7541_7542delCT. In comparison, the frequency of NOTCH1 mutations in 1562 Western patients treated with R-CHOP was just 1.92%. NOTCH1 mutations are more common in the extracellular regions in the Chinese patients. Additionally, most mutated sites in the intracellular domains are different between the Chinese and Western patients. In order to confirm these new mutations in the Chinese patients, we conducted the Sanger Sequencing. In this study, we found 2 mutated sites in extracellular regions such as c.2537 A>C and c.2542G>A, and four mutated sites like c.6392G>T, c.6598G>A, c.7541_7542delCT, and c.7216C>T in the intracellular domains. Due to no high quality samples and PCR failure, we cannot validate the other mutation sites by Sanger sequencing, particularly for two mutations in the signal peptide. Different technology platforms, analysis pipelines and statistical methods may be one of the main reasons for the differences. For example, Noel F. C. C. de Miranda et al. used Sanger sequencing to detect only hotspots mutations(p.1500-1800 and p.2300-2555), 6% of DLBCL samples were identified (33). Another possible reason may be the difference in target populations. DLBCL gene expression profiles in different ethnic groups have been confirmed to differ between Western and Asian DLBCL patients (33).
The human NOTCH1 gene is located in the neoplasia-associated region of position 34 of the long arm of chromosome 9 (34). The produced protein may have multiple functions: either an oncogene or a tumor suppressor gene. In this study, we found that patients with the NOTCH1 mutations had poor PFS and OS, implying an oncogenic role in DLBCL progression. Furthermore, we found NOTCH1 mutations were negatively associated with complete remission after 6-8 cycles of immunochemotherapy, implying the NOTCH1 mutation may have predictive potential in the clinical response of DLBCL patients treated with RCHOP chemotherapy. In fact, previous study has reported that NOTCH1 mutations were associated with lack of benefit of CD20 antibody therapies in chronic lymphocytic leukemia (35). Due to the relatively low mutation frequency, the prognostic value of NOTCH1 mutations for DLBCL has not been systematically studied previously. In this study, we recruited 161 DLBCL cases in our hospital, among whom patients with the NOTCH1 mutation had a lower complete response rate than patients without the NOTCH1 mutation. Similarly, we also enrolled 1562 DLBCL patients treated with R-CHOP from the published DLBCL database to perform meta-analysis, and found a significant association between NOTCH1 mutations and short PFS and OS, respectively. In addition, we combined personal data from three databases and obtained same results. Thus, NOTCH1 is conformed to be a potential predictor for DLBCL patients.
DLBCL is a highly heterogeneous tumor type. COO classification and diphenotypic lymphoma (high expression of Bcl2 and c-Myc protein) are commonly known prognostic indicators for clinicians. Non-GCB type DLBCL and double-expression DLBCL both predict poor prognosis. In this study, NOTCH1 mutation was found to be an independent risk factor for prognosis in our DLBCL patients. This systematic analysis of NOTCH1 mutation in DLBCL provides data, which support for application of NOTCH1 mutation detection in clinical diagnosis and treatment, and also provides ideas for finding new therapeutic targets for DLBCL.
However, how the NOTCH1 mutations affect prognosis and the efficacy of chemotherapeutic drugs remains unclear. It was reported that tumor-infiltrating macrophages (TIMs) are involved in microenvironmental interactions in NOTCH1-mutated patients (36). Monocytes are innate immune cells of the host mononuclear phagocyte system, and its distribution and the transition with macrophages are disrupted in cancer and can affect patient prognosis (37). In fact, peripheral blood monocyte count could reflect the number of local TIMs (38). Our results showed that the NOTCH1 mutations were significantly associated with low blood monocyte count and high lymphoma/monocyte ratio (LMR). LMR is regarded as a prognostic factor for DLBCL patients (39). The above result supported the fact tumor proliferation promoted by NOTCH1 signals outweighs immune clearance by the host immune system (40). This hypothesis is needed to study in the future.
In conclusion, NOTCH1 mutations predict a poor progression free survival in DLBCL patients. Targeting of NOTCH1 mutations could be a potentially effective approach to improve survival of patients.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
All of the subjects were well-informed about the study and provided written informed consent to participate in the study. The study was approved by the Institutional Review Board of our hospital (No: IIT20210369A).
JW, ZL, FY, and JJ designed the research and/or analyzed the data. WLY, YS, JS, and JY carried out the molecular genetic studies, LM and WJY provided clinical data. JW and FY wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work is supported by Zhejiang Provincial Natural Science Foundation of China (LY19H080009). The funders had no role in study design, data collection, data analysis, interpretation, writing of this report.
Authors YS and JY were employed by Nanjing Geneseeq Technology Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the patients for donating specimens.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.746577/full#supplementary-material
1. Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science (1999) 284(5415):770–6. doi: 10.1126/science.284.5415.770
2. Aster JC, Pear WS, Blacklow SC. The Varied Roles of Notch in Cancer. Annu Rev Pathol (2017) 12:245–75. doi: 10.1146/annurev-pathol-052016-100127
3. Mao L. NOTCH Mutations: Multiple Faces in Human Malignancies. Cancer Prev Res (Phila) (2015) 8(4):259–61. doi: 10.1158/1940-6207.CAPR-15-0063
4. Mutvei AP, Fredlund E, Lendahl U. Frequency and Distribution of Notch Mutations in Tumor Cell Lines. BMC Cancer (2015) 15:311. doi: 10.1186/s12885-015-1278-x
5. Arruga F, Vaisitti T, Deaglio S. The NOTCH Pathway and Its Mutations in Mature B Cell Malignancies. Front Oncol (2018) 8:550. doi: 10.3389/fonc.2018.00550
6. Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N Engl J Med (2018) 378(15):1396–407. doi: 10.1056/NEJMoa1801445
7. Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, et al. Leukemia-Associated Mutations Within the NOTCH1 Heterodimerization Domain Fall Into at Least Two Distinct Mechanistic Classes. Mol Cell Biol (2006) 26(12):4642–51. doi: 10.1128/MCB.01655-05
8. Tyagi A, Sharma AK, Damodaran C. A Review on Notch Signaling and Colorectal Cancer. Cells (2020) 9(6):1549. doi: 10.3390/cells9061549
9. Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, et al. TAN-1, the Human Homolog of the Drosophila Notch Gene, Is Broken by Chromosomal Translocations in T Lymphoblastic Neoplasms. Cell (1991) 66(4):649–61. doi: 10.1016/0092-8674(91)90111-b
10. Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science (2011) 333(6046):1157–60. doi: 10.1126/science.1208130
11. Licciulli S, Avila JL, Hanlon L, Troutman S, Cesaroni M, Kota S, et al. Notch1 is Required for Kras-Induced Lung Adenocarcinoma and Controls Tumor Cell Survival via P53. Cancer Res (2013) 73(19):5974–84. doi: 10.1158/0008-5472.CAN-13-1384
12. Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, et al. Activation of Notch1 Signaling Is Required for Beta-Catenin-Mediated Human Primary Melanoma Progression. J Clin Invest (2005) 115(11):3166–76. doi: 10.1172/JCI25001
13. Jain N, Keating MJ. Richter Transformation of CLL. Expert Rev Hematol (2016) 9(8):793–801. doi: 10.1080/17474086.2016.1199948
14. Nadeu F, Delgado J, Royo C, Baumann T, Stankovic T, Pinyol M, et al. Clinical Impact of Clonal and Subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM Mutations in Chronic Lymphocytic Leukemia. Blood (2016) 127(17):2122–30. doi: 10.1182/blood-2015-07-659144
15. Pararajalingam P, Coyle KM, Arthur SE, Thomas N, Alcaide M, Meissner B, et al. Coding and Noncoding Drivers of Mantle Cell Lymphoma Identified Through Exome and Genome Sequencing. Blood (2020) 136(5):572–84. doi: 10.1182/blood.2019002385
16. Aref S, Rizk R, El Agder M, Fakhry W, El Zafarany M, Sabry M. NOTCH-1 Gene Mutations Influence Survival in Acute Myeloid Leukemia Patients. Asian Pac J Cancer Prev (2020) 21(7):1987–92. doi: 10.31557/APJCP.2020.21.7.1987
17. Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, et al. Notch1 Functions as a Tumor Suppressor in Mouse Skin. Nat Genet (2003) 33(3):416–21. doi: 10.1038/ng1099
18. Lobry C, Oh P, Aifantis I. Oncogenic and Tumor Suppressor Functions of Notch in Cancer: It’s NOTCH What You Think. J Exp Med (2011) 208(10):1931–5. doi: 10.1084/jem.20111855
19. Li S, Young KH, Medeiros LJ. Diffuse Large B-Cell Lymphoma. Pathology (2018) 50(1):74–87. doi: 10.1016/j.pathol.2017.09.006
20. Pozzo F, Bittolo T, Arruga F, Bulian P, Macor P, Tissino E, et al. NOTCH1 Mutations Associate With Low CD20 Level in Chronic Lymphocytic Leukemia: Evidence for a NOTCH1 Mutation-Driven Epigenetic Dysregulation. Leukemia (2016) 30(1):182–9. doi: 10.1038/leu.2015.182
21. Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular Subtypes of Diffuse Large B Cell Lymphoma Are Associated With Distinct Pathogenic Mechanisms and Outcomes. Nat Med (2018) 24(5):679–90. doi: 10.1038/s41591-018-0016-8
22. Wright GW, Huang DW, Phelan JD, Coulibaly ZA, Roulland S, Young RM, et al. A Probabilistic Classification Tool for Genetic Subtypes of Diffuse Large B Cell Lymphoma With Therapeutic Implications. Cancer Cell (2020) 37(4):551–568 e514. doi: 10.1016/j.ccell.2020.03.015
23. Sukswai N, Lyapichev K, Khoury JD, Medeiros LJ. Diffuse Large B-Cell Lymphoma Variants: An Update. Pathology (2020) 52(1):53–67. doi: 10.1016/j.pathol.2019.08.013
24. Chen Y, Chen H, Chen L, Zheng X, Yang X, Zheng Z, et al. Immunohistochemical Overexpression of BCL-2 Protein Predicts an Inferior Survival in Patients With Primary Central Nervous System Diffuse Large B-Cell Lymphoma. Medicine (Baltimore) (2019) 98(45):e17827. doi: 10.1097/MD.0000000000017827
25. Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the Molecular Classification of Diffuse Large B-Cell Lymphoma by Immunohistochemistry Using a Tissue Microarray. Blood (2004) 103(1):275–82. doi: 10.1182/blood-2003-05-1545
26. Mi JQ, Wang X, Yao Y, Lu HJ, Jiang XX, Zhou JF, et al. Newly Diagnosed Acute Lymphoblastic Leukemia in China (II): Prognosis Related to Genetic Abnormalities in a Series of 1091 Cases. Leukemia (2012) 26(7):1507–16. doi: 10.1038/leu.2012.23
27. Cheson BD, Pfistner B, Juweid ME, Gascoyne RD, Specht L, Horning SJ, et al. Revised Response Criteria for Malignant Lymphoma. J Clin Oncol (2007) 25(5):579–86. doi: 10.1200/JCO.2006.09.2403
28. Balduzzi S, Rucker G, Schwarzer G. How to Perform a Meta-Analysis With R: A Practical Tutorial. Evid Based Ment Health (2019) 22(4):153–60. doi: 10.1136/ebmental-2019-300117
29. Ou J, Zhu LJ. Trackviewer: A Bioconductor Package for Interactive and Integrative Visualization of Multi-Omics Data. Nat Methods (2019) 16(6):453–4. doi: 10.1038/s41592-019-0430-y
30. Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, et al. Applications for Protein Sequence-Function Evolution Data: mRNA/Protein Expression Analysis and Coding SNP Scoring Tools. Nucleic Acids Res (2006) 34(Web Server issue):W645–50. doi: 10.1093/nar/gkl229
31. Pedrosa L, Fernandez-Miranda I, Perez-Callejo D, Quero C, Rodriguez M, Martin-Acosta P, et al. Proposal and Validation of a Method to Classify Genetic Subtypes of Diffuse Large B Cell Lymphoma. Sci Rep (2021) 11(1):1886. doi: 10.1038/s41598-020-80376-0
32. Lacy SE, Barrans SL, Beer PA, Painter D, Smith AG, Roman E, et al. Targeted Sequencing in DLBCL, Molecular Subtypes, and Outcomes: A Haematological Malignancy Research Network Report. Blood (2020) 135(20):1759–71. doi: 10.1182/blood.2019003535
33. de Miranda NF, Georgiou K, Chen L, Wu C, Gao Z, Zaravinos A, et al. Exome Sequencing Reveals Novel Mutation Targets in Diffuse Large B-Cell Lymphomas Derived From Chinese Patients. Blood (2014) 124(16):2544–53. doi: 10.1182/blood-2013-12-546309
34. Larsson C, Lardelli M, White I, Lendahl U. The Human NOTCH1, 2, and 3 Genes are Located at Chromosome Positions 9q34, 1p13-P11, and 19p13.2-P13.1 in Regions of Neoplasia-Associated Translocation. Genomics (1994) 24(2):253–8. doi: 10.1006/geno.1994.1613
35. Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi M, Dohner K, et al. Gene Mutations and Treatment Outcome in Chronic Lymphocytic Leukemia: Results From the CLL8 Trial. Blood (2014) 123(21):3247–54. doi: 10.1182/blood-2014-01-546150
36. Arruga F, Gizdic B, Serra S, Vaisitti T, Ciardullo C, Coscia M, et al. Functional Impact of NOTCH1 Mutations in Chronic Lymphocytic Leukemia. Leukemia (2014) 28(5):1060–70. doi: 10.1038/leu.2013.319
37. Huang YH, Cai K, Xu PP, Wang L, Huang CX, Fang Y, et al. CREBBP/EP300 Mutations Promoted Tumor Progression in Diffuse Large B-Cell Lymphoma Through Altering Tumor-Associated Macrophage Polarization via FBXW7-NOTCH-CCL2/CSF1 Axis. Signal Transduct Target Ther (2021) 6(1):10. doi: 10.1038/s41392-020-00437-8
38. Hayashi T, Fujita K, Nojima S, Hayashi Y, Nakano K, Ishizuya Y, et al. Peripheral Blood Monocyte Count Reflecting Tumor-Infiltrating Macrophages is a Predictive Factor of Adverse Pathology in Radical Prostatectomy Specimens. Prostate (2017) 77(14):1383–8. doi: 10.1002/pros.23398
39. Stefaniuk P, Szymczyk A, Podhorecka M. The Neutrophil to Lymphocyte and Lymphocyte to Monocyte Ratios as New Prognostic Factors in Hematological Malignancies - A Narrative Review. Cancer Manag Res (2020) 12:2961–77. doi: 10.2147/CMAR.S245928
Keywords: next generation sequencing (NGS), diffuse large B-cell lymphoma, clinical decision making, NOTCH1 mutations, clinical outcome
Citation: Li Z, Yu F, Ye W, Mao L, Huang J, Shao Y, Yan J, Yu W, Jin J and Wang J (2021) Clinical Features and Prognostic Significance of NOTCH1 Mutations in Diffuse Large B-Cell Lymphoma. Front. Oncol. 11:746577. doi: 10.3389/fonc.2021.746577
Received: 24 July 2021; Accepted: 12 November 2021;
Published: 09 December 2021.
Edited by:
Ricardo Ribeiro, Universidade do Porto, PortugalReviewed by:
Margarita Sánchez-Beato, Hospital Universitario Puerta de Hierro Majadahonda, SpainCopyright © 2021 Li, Yu, Ye, Mao, Huang, Shao, Yan, Yu, Jin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinghan Wang, MTUxMzA4NEB6anUuZWR1LmNu; Jie Jin, amllajA1MDNAemp1LmVkdS5jbg==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.