- Department of Laboratory Medicine, The Affiliated Hospital of Southwest Medical University, Sichuan, China
The E2F family of transcription factors (E2Fs) consist of eight genes in mammals. These genes encode ten proteins that are usually classified as transcriptional activators or transcriptional repressors. E2Fs are important for many cellular processes, from their canonical role in cell cycle regulation to other roles in angiogenesis, the DNA damage response and apoptosis. A growing body of evidence demonstrates that cancer stem cells (CSCs) are key players in tumor development, metastasis, drug resistance and recurrence. This review focuses on the role of E2Fs in CSCs and notes that many signals can regulate the activities of E2Fs, which in turn can transcriptionally regulate many different targets to contribute to various biological characteristics of CSCs, such as proliferation, self-renewal, metastasis, and drug resistance. Therefore, E2Fs may be promising biomarkers and therapeutic targets associated with CSCs pathologies. Finally, exploring therapeutic strategies for E2Fs may result in disruption of CSCs, which may prevent tumor growth, metastasis, and drug resistance.
Introduction
The study of the E2F family began in 1987 and E2Fs were initially identified as activators of the adenovirus gene E2 promoter that can induce host cell proliferation (1, 2). The E2F family of transcription factors (E2Fs) have been recognized for nearly 30 years. A total of 8 genes have been found, and 10 protein products encoded by these genes form a core transcription axis crucial for regulating cell cycle progression, apoptosis, differentiation, DNA damage repair, metabolism, and angiogenesis (3–7). According to initial reporter gene detection and evaluation of their expression patterns during the cell cycle, E2Fs have been classified as transcriptional activators (E2F1-3) or transcriptional repressors (E2F4-8) (Figure 1), and are thus predicted to play a dual role in human cancers (8, 9). E2F activators are predicted to be oncogenic, while E2F repressors are predicted to have tumor suppressor functions. However, due to the complexity of their structure and function, it is still quite challenging to study the role of E2Fs in cancer.
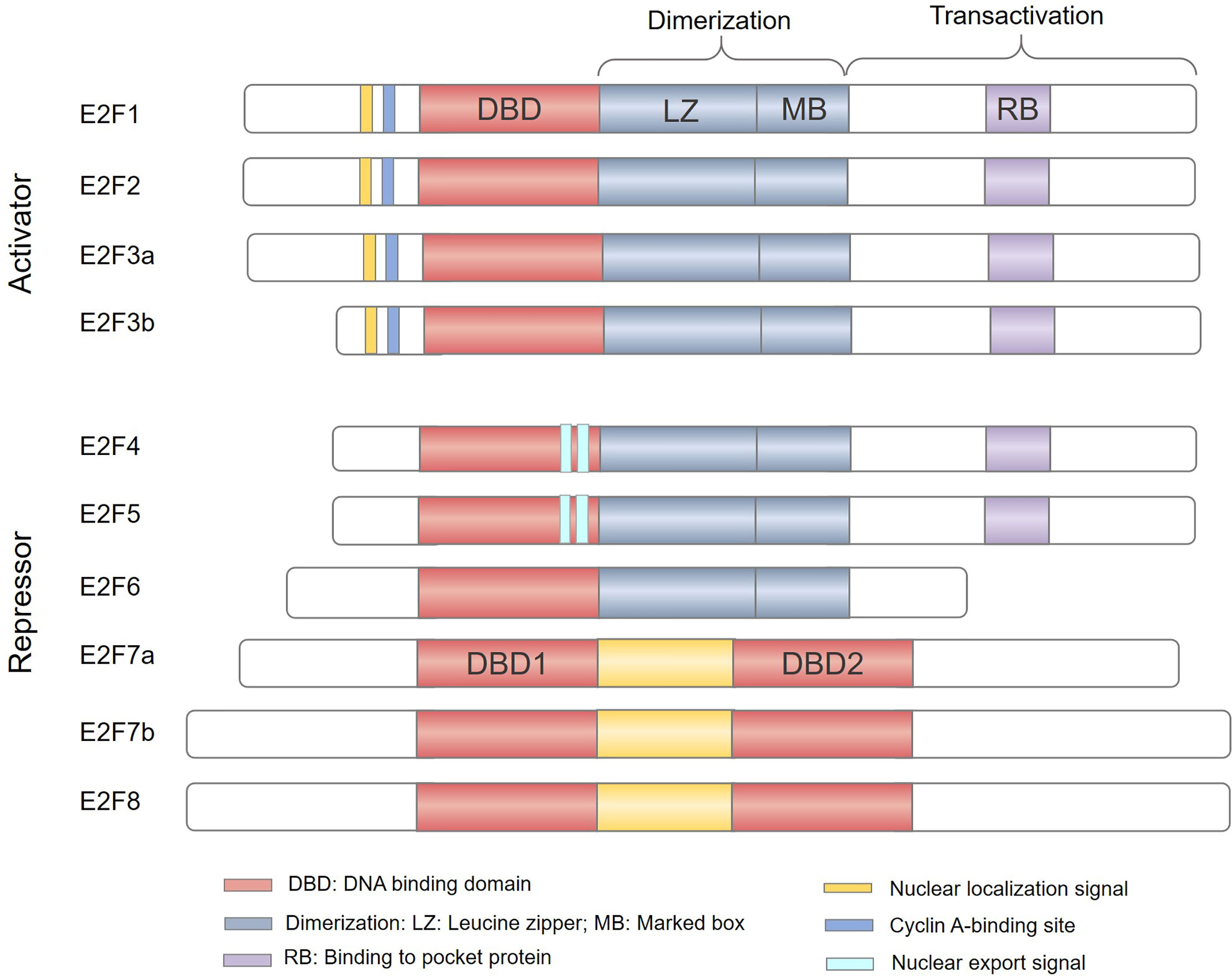
Figure 1 The E2F family of transcription factors. Based predominantly on the results of in vitro studies, E2Fs are generally divided into transcriptional activators (E2F1-3) and transcriptional repressors (E2F4-8). E2F family members E2F1–E2F6 contain a distinctive winged-helix DNA binding domain (DBD) and a dimerization partner (DP) binding domain composed of a leucine zipper (LZ) and a marker box (MB) domain to bind DNA. In addition, E2F1-5 has a pocket protein (RB, p107 and p130)-binding domain in the transactivation domain; the minimum site is shown in light purple (RB). E2F1-3 also have a nuclear localization sequence (NLS) and a Cyclin A-binding site, while E2F4-5 have a nuclear export signal (NES). E2F7-8 have two tandem DBDs but lack dimerization and transactivation domains and do not bind to DP or pocket proteins.
Cancer stem cells (CSCs, also called tumor-initiating cells) are a small subpopulation of cancer cells that possess self-renewal capacity and lead to the heterogeneous lineages of cancer cells that constitute the tumor, and they are largely responsible for tumor growth and progression (10). CSCs were first identified in acute myeloid leukemia (AML) and have since been purified from diverse types of solid malignancies, such as breast cancer (11), liver cancer (12), lung cancer (13), colon cancer (14), prostate cancer (15), ovarian cancer (16) and brain cancer (17). The CSC model provides a satisfactory explanation for the origin of complex tumors and intratumoral heterogeneity. Only a few CSCs with self-renewal and differentiation potential can initiate tumor formation and produce intratumoral heterogeneity (18). The CSC model has been well established for cancer research, and accumulated evidence has demonstrated that a high recurrence rate and high mortality rate in cancers are intimately related to the biological properties of CSCs (19, 20).
In cancer literature, the role of E2Fs in CSCs is widely regarded as an activator gene. Up-regulation of E2Fs is reported to be involved in proliferation promotion (21), maintenance and acquisition of self-renewal (22, 23), invasion and metastatic progression (24) and resistance to chemotherapy and radiotherapy (25) in many CSCs. However, the unexpected positive effect of repressive E2Fs on stem cell attributes of CSCs could potentially be a consequence of repression of negative regulators such as miRNAs (26). The discrepancies between these findings and those on E2Fs as a CSCs promoter or suppressor have not been thoroughly investigated. In addition, whether E2Fs may regulate the CSCs as it does in normal stem cells has not been clearly elucidated (27). In summary, this review has provided new evidence demonstrating the biological roles of E2Fs in CSCs and its underlying mechanisms, which opens up a new perspective for biomarkers or therapeutic targets for cancer.
E2Fs: Structure And Functions
Currently, there are 8 E2F genes (called E2F1-8) that encode 10 proteins in mammalian cells (Figure 1). All proteins encoded by E2Fs contain one or more highly conserved DNA binding domains (DBDs), which regulate promoter expression by targeting transcription (28). Among E2F genes, the E2F3 and E2F7 loci can undergo two alternative splicing events to encode four protein isoforms: E2F3a, E2F3b, E2F7a, and E2F7b (29–31). The E2F family can be broadly divided into two categories, typical E2Fs (E2F1-6) and atypical E2Fs (E2F7-8), based on their unique structural characteristics (32, 33). Canonical E2F1-6 members possess a DBD upstream of a dimerization partner (DP) binding domain composed of a leucine zipper (LZ) and a marker box (MB) domain (33–35). Unlike E2F6, E2F1-5 have a transactivation domain in the C-terminus and contain a binding region for pocket proteins. Therefore, E2F1-5 are widely regulated by the pocket proteins (also called “retinoblastoma family proteins”) RB, p107 and p130 (36). In addition, E2F1-3 have a nuclear localization signal (NLS) and a cyclin A binding site in the N-terminus to ensure their translocation to the nucleus to regulate cell cycle activity (37, 38). E2F4-5 have bipartite nuclear export signals (NESs) that mediate their cytoplasmic translocation (39, 40). However, atypical E2F7 and E2F8 have two distinct DBDs but lack DP binding domains, pocket protein binding regions, and transcriptional activation domains (41, 42). Consequently, atypical E2F7-8 bind to DNA as homodimers or heterodimers via the DBD to regulate gene transcription (43). Owing to their structural differences (Figure 1), E2Fs are destined to perform different functions in the cells.
Based on their functional properties, E2Fs are generally subdivided into two categories: transcriptional activators (E2F1-3) and repressors (E2F4-8) (29). The transcriptional activity of E2F1-3a is dependent on cell cycle regulatory and their binding partners, which include DPs and pocket proteins. Pocket proteins (RB/p107/p130) exhibit varying degrees of binding specificity for E2F subunits. For example, E2F1-3a preferentially bind to RB (28, 44). Generally, when RB is hypophosphorylated, it can hinder the transcriptional activity of the E2F-DP heterodimer by masking the transcription activation domains of E2F1-3a (3, 35). During cell cycle progression from G1 to S phase, RB is phosphorylated by activated cyclin-dependent kinases (CDKs), resulting in its dissociation from the RB-E2F complex and the unmasking of the E2F1-3a transcriptional activation domain (45, 46). E2F3b acts as both a transcriptional activator and repressor protein due to its unique transcriptional program and expression patterns (47). E2F3b remains constitutively expressed throughout the cell cycle, similar to the E2F4-5 repressors (35, 46, 48). During cell cycle progression in G0 phase, E2F3b-5 interact with one of the three RB/p107/p130 proteins, subsequently recruiting corepressor complexes to alter the local chromatin structure of E2F target genes and inducing transcriptional repression (46, 49, 50). E2F6-8 are considered repressors that are independent of pocket proteins, and their primary function is to modulate cell cycle progression from S to G2 phase. E2F6 can suppress transcription by recruiting chromatin remodeling complexes (51), while E2F7-8 can directly modulate gene transcription (33, 52). E2Fs are critical regulators of the cell cycle, and they regulate every phase of the cell cycle by controlling the transcription of numerous target genes involved in DNA replication and cell cycle progression.
CSCs: Biological Characteristics
Tumor initiation, development, metastasis, recurrence and acquisition of therapeutic resistance in numerous different human cancers has been attributed to the properties of CSCs, which include proliferation potential, self-renewal capacity, differentiation potential, high metastatic capacity, and drug resistance (20, 53). Compared with normal stem cells, CSCs, with their abnormal expression of cell cycle-related regulatory factors and dysregulation of negative feedback mechanisms, are often able to proliferate extensively, potentially indefinitely (54, 55).
A prominent feature of CSCs is their extraordinary self-renewal ability, a unique stem-cell associated cell division event maintaining the undifferentiated state and long-term proliferation potential of at least one daughter cell, which is the direct cause of tumorigenesis (10). CSCs can divide symmetrically producing two CSCs that are undifferentiated (amplification of renewing CSCs) or asymmetrically producing one undifferentiated CSC and one lineage-restricted and partially differentiated daughter cell excessively increases cell growth and eventually leads to together driving heterogeneous tumor formation (19, 56). Regardless of the degree of differentiation of a given tumor, the undifferentiated and self-renewing CSC subset provides for the long term proliferative potential driving tumor development, tumor maintenance and metastasis, and are thus widely considered the key link to tumorigenesis (57). Therefore, a further understanding the regulatory mechanism of CSC self-renewal is vital to preventing tumorigenesis, and it can also provide clear targets for cancer treatment.
Along with their self-renewal ability, CSCs also have differentiation potential. CSCs can differentiate into a series of distinct cell types present within the tumor, which constitute the bulk of the tumor (58). It should be noted, however, that CSC differentiation into non-stem tumor cells (non-CSCs) is not a one-way pathway but can be reversible or plastic (59). For example, tumor cells can also dedifferentiate and acquire stem cell properties in response to specific stimuli (56, 60). In summary, CSC populations are dynamic populations with high cellular plasticity. In heterogeneous tumor cell populations, cells can undergo phenotypic switching between CSCs and tumor cells phenotype, a phenomenon that is essential for tumor progression and recurrence (61, 62).
A high metastatic potential is another key trait of CSCs. In vivo, CSC populations in tumors preferentially metastasize, and single-cell analysis has shown that early metastatic cells have unique stemness gene expression patterns (63, 64). Epithelial‐to‐mesenchymal transition (EMT) is the basis for cell invasion and metastasis. It is currently clear that EMT signaling, which enhances the metastatic potential of CSCs, and CSC phenotypes are tightly connected (65, 66). These studies have shown that the metastatic potential of CSCs is far higher than that of ordinary tumor cells and plays a crucial role in tumor metastasis and development.
Furthermore, drug resistance is regarded as an important feature of CSCs. CSCs display high resistance to chemotherapy and radiotherapy. Many mechanisms have been proposed for CSC resistance, such as expression of multidrug resistance proteins, enhancement of the DNA repair capacity, inhibition of cell death-related pathways, apoptosis evasion, cell cycle promotion and metabolic alteration (67). Additionally, the hypoxia microenvironment is also a key component of CSC maintenance and acquisition of drug resistance, especially in the enhancement of drug resistance mechanisms (68, 69). Because of their therapeutic resistance, CSCs are considered to be the root of treatment failure and tumor recurrence.
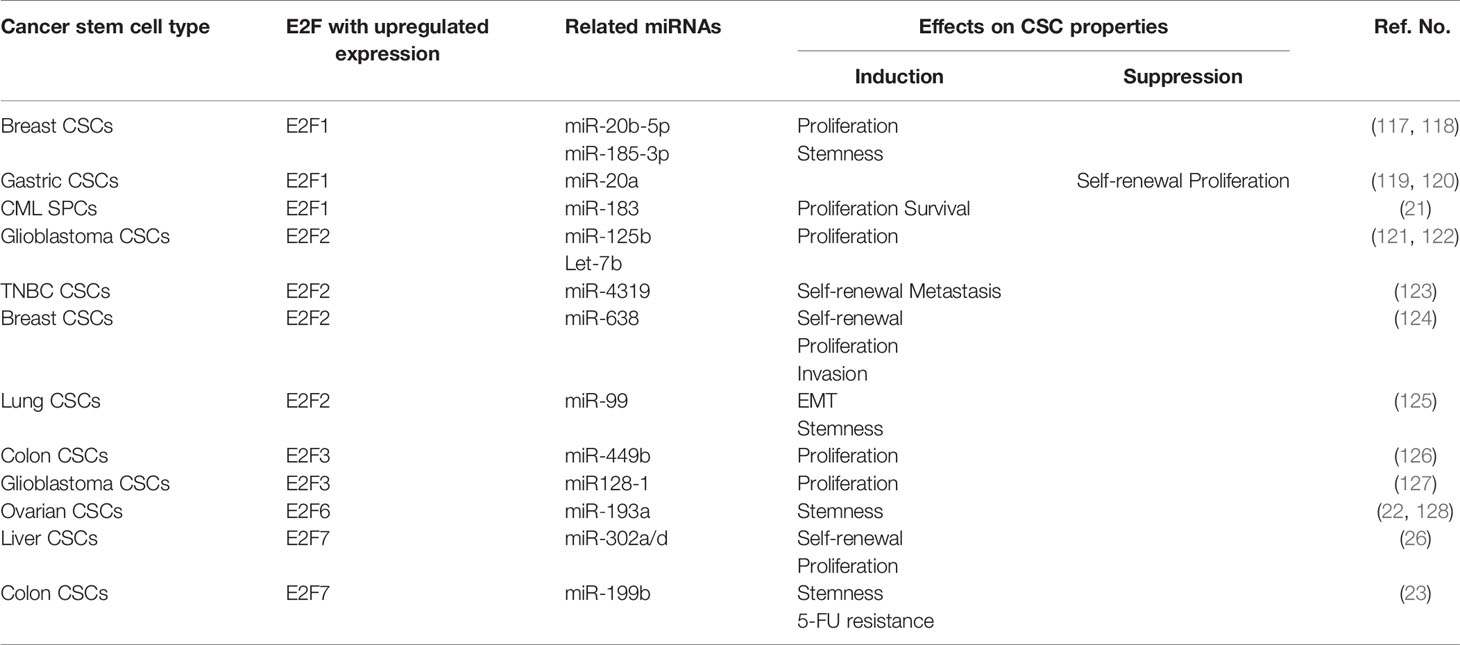
A continually increasing number of studies have researched E2Fs in CSCs and have found that E2Fs are widely involved in the regulation of the biological characteristics of CSCs, such as their proliferation, self-renewal, metastasis, and drug resistance (Figure 2).
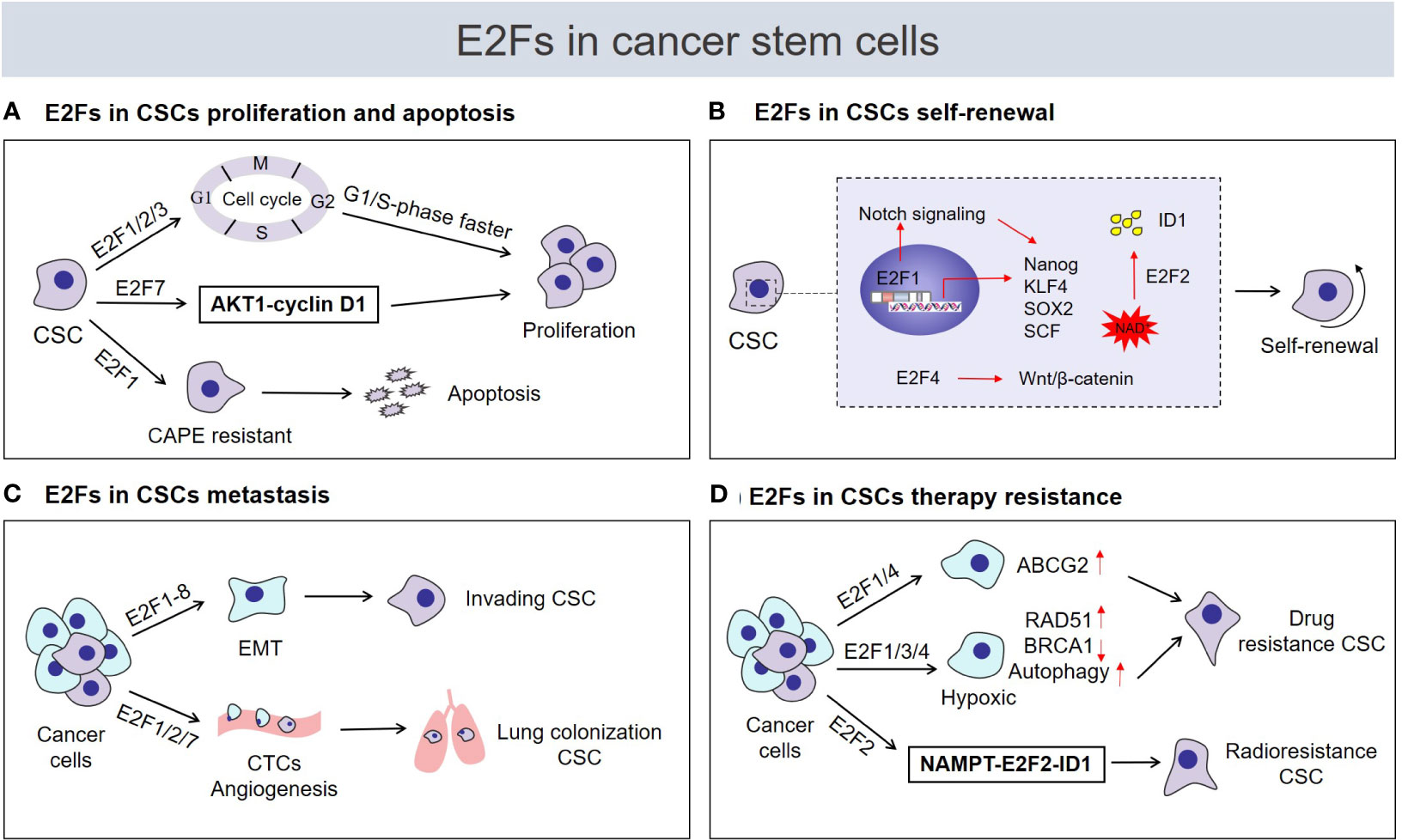
Figure 2 Roles of E2Fs in cancer stem cell (CSC) biological characteristics. Tumor cells are heterogeneous, including immortal cancer stem cells. CSCs usually express high levels of E2F transcription factors and are characterized by (A) enhanced proliferation or induction of apoptosis, (B) acquisition and maintenance of self‐renewal capability, (C) enhanced invasion and metastasis potential, (D) regulation of drug or radiation resistance. CAPE, caffeic acid phenethyl ester; ID1, inhibitor of differentiation 1; NAD+, nicotinamide adenine dinucleotide; CTCs, circulating tumor cells; NAMPT, nicotinamide phosphoribosyl transferase.
Role Of E2Fs In The Biological Behaviors Of Cancer Stem Cells
E2Fs Roles in Proliferation and Apoptosis of Cancer Stem Cells
E2Fs are critical regulators of genes required for cell cycle progression and play an integral role in the control of cell proliferation. Previous studies have suggested that E2Fs are required to control cell proliferation differently in carcinogenic environments than in normal cells (29). As shown in recent studies, the proliferative potential of CSCs seems to be strongly correlated with cell cycle regulation by E2Fs. The transcriptional activators E2F1-3 regulate cell proliferation by activating genes essential for G1/S-phase progression in CSCs (Figure 2A). ENPP1 (ectonucleotide pyrophosphatase/phosphodiesterase 1) has been proved to induce the stemness features of cancer cells (70, 71). Bageritz et al (72) also discovered that E-NPP1 could facilitate glioblastoma CSC proliferation by controlling cell cycle progression. And E-NPP1-deficient glioblastoma CSCs led to a decrease in the transcriptional function of E2F1, and classical E2F1 target genes facilitating G1/S transition were downregulated, resulting in the accumulation of cells in G1 phase and inhibition of their proliferation. Similarly, E2F1 plays a pivotal role in regulating the proliferation status of chronic myeloid leukemia (CML) stem/progenitor cells (SPCs). In terms of cell cycle regulation, the E2F1 signaling pathway was found to be deregulated in CML SPCs, and inhibition of E2F1 led to cell cycle arrest and induced blockade of proliferation (21). Moreover, the dissociation of E2F1 from the RB protein was found to restore its transcriptional activity, which is required for cell cycle progression from G1 to S phase (73). In glioblastoma CSCs, the bromodomain inhibitor JQ1 was found to cause cell cycle arrest and suppress CSC proliferation by preventing the release of E2F1 from the RB/E2F1 complex (74). Additionally, glioblastoma CSCs display the dependence of HELLS (helicase, lymphoid-specific) to maintain proliferation. Both E2F3a and E2F3b were found to interact with HELLS, thereby increasing the expression of cell cycle progression-related genes to maintain glioblastoma CSC proliferation, while proliferation was impaired after E2F3 gene knockdown (75). Remarkably, although the function of E2F repressors is generally opposite that of E2F activators in the regulation of the cell cycle, the effects of E2F repressors are inconsistent with the predicted inhibitory effect on proliferation. In fact, current experimental evidence supports the role of E2F repressors in promoting cell cycle progression and proliferation of CSCs. For example, high levels of E2F7 detected in liver CSC populations were found to be vital for the maintenance of CSC proliferation via E2F7 activation of AKT1-cyclin D1 signaling and downstream cell cycle mediators to promote cell cycle progression and proliferation (26) (Figure 2A). The unexpected positive effect of repressive E2Fs on stem cell attributes of CSCs, such as proliferation by modulation of the cell cycle could potentially be a consequence of repression of negative regulators such as miRNAs (see below). However, E2F1 is also an effective activator of apoptosis (34), which may lead to the opposite effect of E2F1 on proliferation of CSCs. As demonstrated by El-Khattouti et al (76), the apoptosis of CD133 (+) melanoma subpopulation cells induced by the anticancer drug caffeic acid phenethyl ester (CAPE) requires the participation of E2F1. Functional analysis of E2F1 in the CD133(+) melanoma subpopulation demonstrated that overexpression of E2F1 induced both degradation of the antiapoptotic protein Mcl-1 and activation of the ASK1/JNK and p38 pathways, which are involved in regulating the localization of Bax to mitochondria to trigger apoptosis (76) (Figure 2A). Overall, E2Fs mainly regulate the proliferation of CSCs in a cell cycle-dependent manner, and E2Fs are seem to necessary factors in the control of CSC proliferation. But E2F1 can promote the apoptosis of CSCs, whether this is attributed to the CSC-type specific or other mechanisms is worth further study.
E2Fs in the Maintenance and Acquisition of Cancer Stem Cell Self-Renewal
Self-renewal is the process by which cells divide while maintaining an undifferentiated state. This process requires maintenance of stem cell transcription factor expression, activation of self-renewal signaling pathways and strict control of cell metabolism (77–79). Accumulating evidence suggests the substantial engagement of distinct E2F members in each of the abovementioned self‐renewal programs and the contributions of E2Fs to stemness acquisition and maintenance of the self-renewal ability of CSCs. Several studies have confirmed that E2F1 can directly transcriptionally activate the stem cell transcription factors NANOG, KLF4, SOX2 and SCF to act as a regulator of CSC self‐renewal (Figure 2B). For example, using liver tumor-initiating stem-like cells (TICs), Chen et al (80) demonstrated that E2F1 transcriptionally activated NANOG expression, which in turn inhibited oxidative phosphorylation (OXPHOS) and activated fatty acid oxidation (FAO) through metabolic reprogramming, maintaining the TIC self-renewal ability. In ovarian CSCs, E2F1 acts as an upstream transcription factor to activate KLF4 expression and participates in EIF5A2-mediated modulation of CSC self-renewal properties (81). In addition, YAP1 is known to be a major mediator of Hippo signaling pathway and plays an important role in CSC self-renewal (82). Schaal et al (83) proved that E2F1 transcriptionally activates the Sox2 promoter via YAP1 mediated in response to the signaling events caused by the binding of nicotine to α7 nAChR, which enhances the self-renewal of lung CSCs. In the same context, E2F1-3a was also found to induce SCF expression at the transcriptional level under nicotine exposure. Research on the underlying mechanism has mainly involved E2F1. When the binding of nicotine to nAChRs initiates a signaling cascade, it results in the induction of E2F1-mediated SCF promoter activation and stimulates c-Kit expressed on stem-like cells, facilitating the self-renewal of lung cancer side population cells (84). Additionally, the NOTCH and WNT/β-catenin signaling pathways are essential for the self-renewal of CSCs (57). Limited studies have demonstrated regulation of the NOTCH and β‐catenin pathways by E2Fs to promote the acquisition of self-renewal capability. In prostate cancer cells, this involvement of MUC1-C (mucin 1 C-terminal transmembrane subunit) in lineage plasticity is associated with induction the CSCs state and self-renewal capacity (85, 86). A recent study confirmed that MUC1-C could directly bind to E2F1 activated the BAF remodeling complex, induced NOTCH1 signaling pathway and promoted NANOG expression, which in turn led to the promotion of CSCs stemness (86). An additional study reported that TREX1 (three prime repair exonuclease 1) negatively regulates the self-renewal of osteosarcoma CSCs. With respect to underlying mechanisms, TREX1 suppression allowed the activation of β-catenin signaling in the dependence of E2F4, thus possibly enhancing the self-renewal ability of osteosarcoma CSCs by upregulating OCT4 (87) (Figure 2B). Intriguingly, a recent finding suggests crosstalk among E2F2, the NAD+ biosynthesis pathway and CSC self‐renewal. Gujar et al (25) found that NAD+ metabolism governs glioblastoma CSC self-renewal via the NAMPT-E2F2-ID axis. NAMPT is the rate-limiting enzyme in the NAD salvage pathway, and E2F2 acts downstream of NAMPT and controls ID1 gene transcription to drive glioblastoma CSC self-renewal. E2F2 is required for the link NAD+ metabolism and the self-renewal transcriptional program in glioblastoma CSCs (Figure 2B).
Recent studies have highlighted that E2Fs promote CSC self‐renewal by enhancing the activity of stem cell transcription factors, regulating self‐renewal signaling pathways, and modulating cell metabolism. However, to unequivocally define E2Fs as regulators of CSC self‐renewal, further studies are needed to clarify that E2Fs are required for the regulation of CSC self‐renewal.
The Roles of E2Fs in Cancer Stem Cell Metastasis
Metastasis is the process by which malignant tumor cells travel from the primary site through lymphatic vessels, blood vessels, or body cavities and spread to other sites (88). In metastasis, CSCs show more invasive and metastatic phenotypes than non-stem tumor cells and play the lead role (89). Because CSCs have the ability to initiate tumors and a high metastatic capacity, they are considered to be the cells responsible for repopulating metastatic tumor (89, 90). In recent years, emerging research evidence has documented that some circulating tumor cells (CTCs) are CSC-like cells with strong metastatic potential that can drive macroscopic tumor growth in distant tissues (91–95). Interestingly, in a mouse model of metastatic breast cancer, the lung colonization capacity of CTCs and the numbers of metastatic lesions in the lung were strikingly reduced in mice on both the E2F1 and E2F2 knockout backgrounds (24, 96) (Figure 2C). Moreover, E2F1 loss also caused loss of VEGFA expression and tumor angiogenesis defects (24). CSCs are mostly located around blood vessels and rely on the microvasculature for metastasis (97, 98). These findings raise the possibility that cell invasiveness and tumor angiogenesis regulated by the activity of E2F1/2 might be effectively involved in the metastasis of CSC-like cells, thus facilitating tumor metastasis. Moreover, a recent study by Teng et al. (99) found that upregulation of E2F7 resulted in increased activation of the VEGFR-2 signaling pathway and promoted metastasis, invasion, and angiogenesis in hepatocellular carcinoma (HCC). Concomitantly, high expression of E2F7 in HCC stem cells (26) provides the potential for E2F7 to enhance CSC metastasis.
EMT is closely related to the migration and invasion of CSCs. Of note, EMT combines two of the most important attributes to facilitate metastasis: invasiveness and stemness (89, 100). To the best of our knowledge, E2F1-8 extensively activate the EMT program in a variety of cancers, leading to the acquisition of a very invasive and metastatic phenotype (101–106), which may further result in the acquisition of a metastatic CSC phenotype (Figure 2C).
To date, the exact mechanism of several E2Fs in the regulation of CSC metastasis is still unclear, although their involvement seems essential. Further research is necessary to understand how E2F-dependent regulation of EMT and angiogenesis are interconnected in CSC metastasis.
E2Fs Regulate the Drug Resistance of Cancer Stem Cells
CSCs are the main culprit of resistance to chemotherapy and radiotherapy. The mechanisms underlying drug resistance involves in efficiently inducing drug efflux through ATP-binding cassette (ABC) transporters (including MDR1, MRP1, and ABCG2), activating DNA repair programs, enhancing autophagy, and/or inhibiting apoptotic signals to avoid death (67, 107). In a variety of human cancers, the resistance mechanism of ABCG2 protein overexpression in CSCs has been used as an effective functional mechanism to identify and isolate CSCs (19). E2F1 was further reported to drive chemotherapeutic drug resistance in lung and breast cancer cells via activation of ABCG2 expression (108). This observation demonstrates new scenarios that E2F1 may participate in ABCG2-mediated regulation of CSC resistance. Additionally, an increasing amount of evidence shows that tumor cells under hypoxic conditions are induced to transform into CSCs phenotype and mediate tumor radiotherapy or chemotherapy resistance (68). Meanwhile, many cancer literatures have investigated the close link between E2Fs and hypoxia. For example, E2F1 can transactivate the RAD51 promoter under hypoxia to facilitate homologous recombination repair and drug resistance of prostate cancer cells (109). E2F4 was reported to repress BRCA1 expression with hypoxia-induced (110). Concomitantly, DNA repair gene BRCA1 is a critical tumor suppressor that helps CSCs acquire drug resistance under hypoxic conditions (111). And E2F1 and E2F3 can mediate autophagy activation of cancer cells with hypoxia (112, 113). Moreover, autophagy caused by hypoxia commonly results in enhancing CSCs chemoresistance (114, 115). Thus, this set of observations indicate the possibilities that E2Fs could induce the DNA repair and autophagy enhancement through hypoxia regulation, thereby increasing CSC drug resistance. Other studies have also suggested that E2F2 mediates the radioresistance of glioblastoma CSCs (25). The NAMPT-E2F2-ID1 pathway was found to be upregulated in glioblastoma CSCs after radiation and represented a protective response to radiation (Figure 2D).
In summary, these studies, together with the limited research focus on E2Fs and CSC resistance, accentuate the necessity to redirect our attention to the study of E2Fs, because these proteins perform unique transcriptional functions to mediate resistance mechanisms in CSCs.
E2F-Related miRNAs in Regulation of Cancer Stem Cells
MicroRNAs (miRNAs) are small noncoding RNAs that function as posttranscriptional regulators. Previous works have described relationships between the expression of miRNAs and E2Fs. E2F activity can be regulated by miRNAs; in contrast, miRNAs themselves are targets of E2F proteins (116). Subsequent studies found that this feedback relationship plays a pivotal role in CSCs (Table 1). Regarding E2F1, oncogenic miR-20b-5p was found to increase the proliferation of breast CSCs by upregulating E2F1 protein expression (117). Concomitantly, E2F1 was found to act as a downstream target of miR-185-3p and promote CSC stemness properties through the LINC00511/miR-185-3p/E2F1/Nanog axis in breast cancer (118). Additionally, regulation of the E2F1 pathway by miRNAs is a novel strategy for combating CSCs. For example, hsa-mir183/EGR1–mediated upregulation of E2F1 is required for CML SPC survival and proliferation. Downstream of this event, inhibition of E2F1 was found to reduce the proliferation of CML SPCs and induce p53-mediated apoptosis, but healthy SPCs were not affected by inhibition of the E2F1 pathway (21). The researches above indicated that E2F1 seems to be a positive regulator of CSC-traits. However, E2F1 can also negatively regulate the characteristics of CSC. In gastric CSCs, upregulated miRNA-20a was found to contribute to the self-renewal and proliferation of CSCs by targeting the inhibition of E2F1 protein expression and subsequently activating Wnt/β-catenin signaling (119, 120). Taken together, E2F1-mediated regulation promotes the properties of breast cancer and CML CSCs, but has an inhibitory effect on the characteristics of gastric CSCs. And targeting blockage of E2F1 anticipated is expected to be a potential therapeutic applicable to breast cancer and CML. However, whether the promotion and inhibition of E2F1-mediated CSCs regulatory activities are tumor-type specific or miRNAs regulation, which requires further evidences to predict outcome.
A variety of miRNAs regulate the biological characteristics of CSCs by directly targeting E2F2 (Table 1). For instance, miR‐125b was found to suppress the proliferation of CD133-positive glioblastoma CSCs by direct downregulation of E2F2 (121). Elevated Let-7b also repressed E2F2 expression in glioblastoma CSCs, resulting in reductions in tumorsphere growth and CSC populations (122). In triple-negative breast cancer (TNBC), miR-4319 was found to negatively regulate the self-renewal and metastasis of CSCs through targeted inhibition of E2F2 (123). Concomitantly, overexpression of miR-638 also inhibited the self-renewal, proliferation, and invasion abilities of breast CSCs by suppressing E2F2 (124). Moreover, E2F2 expression was shown to inversely correlate with miR-99 expression and directly with elevated level of vimentin in lung cancer biopsies. By inhibiting E2F2, miR-99a repressed the EMT process, accompanied by suppression of stemness features, consequently decreasing the CSC population (125). Collectively, targeting E2F2 is expected to become a therapeutic target for a variety of CSCs.
Fewer studies have been devoted to studying the role of miRNAs related to the remaining E2Fs (E2F3-E2F8) in CSCs (Table 1). For example, miR-449b was found to inhibit the proliferation of SW1116 colon CSCs through downregulation of E2F3 expression (126). miR128-1 was found to suppress the proliferation and tumorigenicity of glioblastoma CSCs by targeting E2F3 (127). Furthermore, E2F7 was found to be the direct target of miRNA-302a/d. miRNA-302a/d negatively regulates the self-renewal ability and cell cycle entry pathway of liver CSCs through direct repression of the target gene E2F7 and its downstream AKT/β-catenin/CCND1 signaling pathway (26).
On the other hand, E2Fs play a role in CSCs by regulating the transcription and maturation of various miRNAs (Table 1). Recent research findings have suggested that E2F6 functions as a competing endogenous RNA (ceRNA) for miR‐193a. Upregulation of E2F6 mRNA expression, in turn, was found to upregulate the stemness marker c-Kit through E2F6-mediated silencing of miR‐193a and to promote ovarian cancer stemness and tumorigenesis (22, 128). In colon CSCs, E2F7 was found to transcriptionally inhibit miR-199b expression to promote USP47 expression, thereby increasing colon CSC stemness and accelerating the occurrence of colon cancer (23). In addition, E2F7 silencing was shown to decrease the production of ALDH1+ cells and repress antagonistic effects of ALDH1+ cells on 5-fluorouracil (5-FU) treatment.
The above findings show that E2Fs and miRNAs regulate one another and play important roles in the proliferation, self-renewal, metastasis, and drug resistance of CSCs. However, there are still some E2Fs (E2F4, E2F5 and E2F8) that are relatively infrequently studied in CSCs, and the related miRNAs have not been studied, which may be the direction of future research.
E2Fs Are Novel Biomarkers And Therapeutic Targets Associated With CSCs
In recent studies, according to published bioinformatics papers, E2Fs have been found to be dysregulated in most tumors and can be considered prognostic and diagnostic biomarkers for cancer as well as potential therapeutic targets (129–136). As demonstrated previously, E2Fs are widely involved in the regulation of the biological characteristics of CSCs and play an important role in the occurrence, progression, metastasis and drug resistance of cancer, suggesting that they might be novel cancer biomarkers and therapeutic targets for CSC-associated. Indeed, using the logic model of core circuits, Khan et al (102) identified several EMT receptor proteins that, in combination with E2F1 upregulation, could be regarded as more reliable biomarkers for predicting the malignant progression of bladder cancer and breast cancer. In invasive bladder cancer and breast cancer, the survival times of patients with low expression of E2F1 and EMT receptor protein signatures were found to be approximately twice as long as those of patients with high expression (102). Similarly, Lee et al. (137) found that high expression of the E2F1-EZH2-SUZ12 signature reflected the invasion and CSC-like characteristics of bladder cancer and predicted poor prognosis of patients. In CD133(+) cells isolated from human astrocytomas, the expression of E2F2 was found to be upregulated and associated with the transformation of human astrocytes. Therefore, it was suggested that E2F2 be used as a therapeutic target for astrocytoma eradication (138). In high-grade muscle-invasive bladder cancer (MIBC), E2F3 is often highly expressed in the 6p22 amplification region, similar to the stem-like signature. E2F3 is epigenetically regulated and might be a potential therapeutic target (139, 140). In addition, E2F7 is highly expressed in HCC and colon cancer and can promote stemness in these cancers, suggesting that E2F7 may be a novel therapeutic target for HCC and colon cancer (23, 26). There are few studies on other E2F family members (E2F4, E2F5, E2F6, E2F8) in CSCs, but these E2Fs also have diagnostic and prognostic value and are therapeutic targets in different cancers. The E2F4 activity level can be used as an indicator of the survival and prognosis of patients with breast cancer and prostate cancer. Patients with negative E2F4 activity scores tend to survive, while upregulation of E2F4 activity is related to poor prognosis (141, 142). The E2F5 status significantly improves the diagnostic accuracy in epithelial ovarian cancer (OEC), and the presence of CA125 or E2F5 increases the sensitivity of OEC detection to 97.9% and the specificity to 72.5% (143). Additionally, E2F5 was identified as an independent prognostic factor in esophageal squamous cell carcinoma; the 5-year survival rate of the E2F5-positive group was 39.3%, which was significantly lower than that of the E2F5-negative group (83.8%) (144). In glioblastoma, research by Huang et al (145) showed that E2F6 is a potential therapeutic target for combating temozolomide (TMZ) resistance and that the progression-free survival (PFS) times of TMZ-treated patients with high levels of E2F6 were significantly shorter. Finally, E2F8 was identified as a novel therapeutic target for controlling the progression of lung cancer and liver cancer (43, 146). High E2F8 expression was found to be associated with poor RFS in patients with ER+ breast cancer (147). The above studies showed that E2Fs, either alone or in combination with other proteins or markers, can be used as diagnostic and prognostic biomarkers or therapeutic targets for cancer in a tumor-type specific manner.
Conclusions And Future Perspectives
In summary, E2Fs are major players in regulating the biological characteristics of CSCs. E2Fs regulate the proliferation, self-renewal, metastasis, and drug resistance of CSCs via distinct mechanisms and can be considered stemness regulators in tumors. Therefore, E2Fs play integral roles in tumor growth, progression, metastasis and anticancer drug resistance. Notably, E2Fs are highly expressed in most solid tumors and are closely related to malignant progression and poor prognosis. A large number of studies have suggested that E2Fs can be used as new diagnostic and prognostic biomarkers and are potential therapeutic targets for cancer. However, there are still some problems that need to be solved. For example, do E2F transcriptional repressors promote the proliferation of CSCs? What are the regulation roles of E2Fs involved in CSCs apoptosis? How do E2Fs regulate EMT and tumor angiogenesis to affect CSC metastasis? Most importantly, there is no specific drug targeting E2Fs, a limitation that needs further exploration. Future research focused on answering the above questions may help us to better understand the roles of E2Fs in CSCs and may be a key step in combating CSCs.
Author Contributions
DX and QP contributed to drafting and editing the manuscript, shared the first authorship. JL and XW contributed to the literature search. TY participated in the design, revision and finalization of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Youth Fund, China (Grant No. 82003138), the Sichuan Science and Technology Program for key Research and Development, China (Grant No. 2021YFS0226), Doctoral Research Initiation Fund of Affiliated Hospital of Southwest Medical University, China (Grant No. 19077) and University’s scientific research project of Southwest Medical University, China (Grant No. 2020ZRQNA019).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the members in the research group who are not listed in the authors.
References
1. Yee AS, Reichel R, Kovesdi I, Nevins JR. Promoter Interaction of the E1A-Inducible Factor E2F and its Potential Role in the Formation of a Multi-Component Complex. EMBO J (1987) 6:2061–8. doi: 10.1002/j.1460-2075.1987.tb02471.x
2. Kovesdi I, Reichel R, Nevins JR. Role of an Adenovirus E2 Promoter Binding Factor in E1A-Mediated Coordinate Gene Control. Proc Natl Acad Sci USA (1987) 84:2180–4. doi: 10.1073/pnas.84.8.2180
3. DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med (2006) 6:739–48. doi: 10.2174/1566524010606070739
4. Biswas AK, Johnson DG. Transcriptional and Nontranscriptional Functions of E2F1 in Response to DNA Damage. Cancer Res (2012) 72:13–7. doi: 10.1158/0008-5472.Can-11-2196
5. Blanchet E, Annicotte JS, Lagarrigue S, Aguilar V, Clapé C, Chavey C, et al. E2F Transcription Factor-1 Regulates Oxidative Metabolism. Nat Cell Biol (2011) 13:1146–52. doi: 10.1038/ncb2309
6. Weijts BG, Bakker WJ, Cornelissen PW, Liang KH, Schaftenaar FH, Westendorp B, et al. E2F7 and E2F8 Promote Angiogenesis Through Transcriptional Activation of VEGFA in Cooperation With HIF1. EMBO J (2012) 31:3871–84. doi: 10.1038/emboj.2012.231
7. Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, et al. E2F Integrates Cell Cycle Progression With DNA Repair, Replication, and G(2)/M Checkpoints. Genes Dev (2002) 16:245–56. doi: 10.1101/gad.949802
8. Dimova DK, Stevaux O, Frolov MV, Dyson NJ. Cell Cycle-Dependent and Cell Cycle-Independent Control of Transcription by the Drosophila E2F/RB Pathway. Genes Dev (2003) 17:2308–20. doi: 10.1101/gad.1116703
9. Takahashi Y, Rayman JB, Dynlacht BD. Analysis of Promoter Binding by the E2F and pRB Families In Vivo: Distinct E2F Proteins Mediate Activation and Repression. Genes Dev (2000) 14:804–16.
10. Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer Stem Cells–Perspectives on Current Status and Future Directions: AACR Workshop on Cancer Stem Cells. Cancer Res (2006) 66:9339–44. doi: 10.1158/0008-5472.Can-06-3126
11. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective Identification of Tumorigenic Breast Cancer Cells. Proc Natl Acad Sci USA (2003) 100:3983–8. doi: 10.1073/pnas.0530291100
12. Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, et al. Identification and Characterization of Tumorigenic Liver Cancer Stem/Progenitor Cells. Gastroenterology (2007) 132:2542–56. doi: 10.1053/j.gastro.2007.04.025
13. Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, et al. Identification and Expansion of the Tumorigenic Lung Cancer Stem Cell Population. Cell Death Differ (2008) 15:504–14. doi: 10.1038/sj.cdd.4402283
14. Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and Expansion of Human Colon-Cancer-Initiating Cells. Nature (2007) 445:111–5. doi: 10.1038/nature05384
15. Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective Identification of Tumorigenic Prostate Cancer Stem Cells. Cancer Res (2005) 65:10946–51. doi: 10.1158/0008-5472.Can-05-2018
16. Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, et al. Identification and Characterization of Ovarian Cancer-Initiating Cells From Primary Human Tumors. Cancer Res (2008) 68:4311–20. doi: 10.1158/0008-5472.Can-08-0364
17. Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of Human Brain Tumour Initiating Cells. Nature (2004) 432:396–401. doi: 10.1038/nature03128
18. Lobo NA, Shimono Y, Qian D, Clarke MF. The Biology of Cancer Stem Cells. Annu Rev Cell Dev Biol (2007) 23:675–99. doi: 10.1146/annurev.cellbio.22.010305.104154
19. Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, et al. Targeting Cancer Stem Cell Pathways for Cancer Therapy. Signal Transduct Target Ther (2020) 5:8. doi: 10.1038/s41392-020-0110-5
20. Nassar D, Blanpain C. Cancer Stem Cells: Basic Concepts and Therapeutic Implications. Annu Rev Pathol (2016) 11:47–76. doi: 10.1146/annurev-pathol-012615-044438
21. Pellicano F, Park L, Hopcroft LEM, Shah MM, Jackson L, Scott MT, et al. Hsa-Mir183/EGR1-Mediated Regulation of E2F1 Is Required for CML Stem/Progenitor Cell Survival. Blood (2018) 131:1532–44. doi: 10.1182/blood-2017-05-783845
22. Cheng FHC, Lin HY, Hwang TW, Chen YC, Huang RL, Chang CB, et al. E2F6 Functions as a Competing Endogenous RNA, and Transcriptional Repressor, to Promote Ovarian Cancer Stemness. Cancer Sci (2019) 110:1085–95. doi: 10.1111/cas.13920
23. Guo X, Liu L, Zhang Q, Yang W, Zhang Y. E2F7 Transcriptionally Inhibits MicroRNA-199b Expression to Promote USP47, Thereby Enhancing Colon Cancer Tumor Stem Cell Activity and Promoting the Occurrence of Colon Cancer. Front Oncol (2020) 10:565449. doi: 10.3389/fonc.2020.565449
24. Hollern DP, Honeysett J, Cardiff RD, Andrechek ER. The E2F Transcription Factors Regulate Tumor Development and Metastasis in a Mouse Model of Metastatic Breast Cancer. Mol Cell Biol (2014) 34:3229–43. doi: 10.1128/mcb.00737-14
25. Gujar AD, Le S, Mao DD, Dadey DY, Turski A, Sasaki Y, et al. An NAD+-Dependent Transcriptional Program Governs Self-Renewal and Radiation Resistance in Glioblastoma. Proc Natl Acad Sci USA (2016) 113:E8247–e8256. doi: 10.1073/pnas.1610921114
26. Ma YS, Lv ZW, Yu F, Chang ZY, Cong XL, Zhong XM, et al. MicroRNA-302a/D Inhibits the Self-Renewal Capability and Cell Cycle Entry of Liver Cancer Stem Cells by Targeting the E2F7/AKT Axis. J Exp Clin Cancer Res (2018) 37:252. doi: 10.1186/s13046-018-0927-8
27. Hsu J, Arand J, Chaikovsky A, Mooney NA, Demeter J, Brison CM, et al. E2F4 Regulates Transcriptional Activation in Mouse Embryonic Stem Cells Independently of the RB Family. Nat Commun (2019) 10:2939. doi: 10.1038/s41467-019-10901-x
28. Attwooll C, Lazzerini Denchi E, Helin K. The E2F Family: Specific Functions and Overlapping Interests. EMBO J (2004) 23:4709–16. doi: 10.1038/sj.emboj.7600481
29. Chen HZ, Tsai SY, Leone G. Emerging Roles of E2Fs in Cancer: An Exit From Cell Cycle Control. Nat Rev Cancer (2009) 9:785–97. doi: 10.1038/nrc2696
30. Di Stefano L, Jensen MR, Helin K. E2F7, a Novel E2F Featuring DP-Independent Repression of a Subset of E2F-Regulated Genes. EMBO J (2003) 22:6289–98. doi: 10.1093/emboj/cdg613
31. He Y, Armanious MK, Thomas MJ, Cress WD. Identification of E2F-3B, an Alternative Form of E2F-3 Lacking a Conserved N-Terminal Region. Oncogene (2000) 19:3422–33. doi: 10.1038/sj.onc.1203682
32. Dimova DK, Dyson NJ. The E2F Transcriptional Network: Old Acquaintances With New Faces. Oncogene (2005) 24:2810–26. doi: 10.1038/sj.onc.1208612
33. Lammens T, Li J, Leone G, De Veylder L. Atypical E2Fs: New Players in the E2F Transcription Factor Family. Trends Cell Biol (2009) 19:111–8. doi: 10.1016/j.tcb.2009.01.002
34. Iaquinta PJ, Lees JA. Life and Death Decisions by the E2F Transcription Factors. Curr Opin Cell Biol (2007) 19:649–57. doi: 10.1016/j.ceb.2007.10.006
35. Xanthoulis A, Tiniakos DG. E2F Transcription Factors and Digestive System Malignancies: How Much do We Know? World J Gastroenterol (2013) 19:3189–98. doi: 10.3748/wjg.v19.i21.3189
36. Swiss VA, Casaccia P. Cell-Context Specific Role of the E2F/Rb Pathway in Development and Disease. Glia (2010) 58:377–90. doi: 10.1002/glia.20933
37. Magae J, Wu CL, Illenye S, Harlow E, Heintz NH. Nuclear Localization of DP and E2F Transcription Factors by Heterodimeric Partners and Retinoblastoma Protein Family Members. J Cell Sci (1996) 109( Pt 7):1717–26. doi: 10.1242/jcs.109.7.1717
38. Müller H, Moroni MC, Vigo E, Petersen BO, Bartek J, Helin K. Induction of S-Phase Entry by E2F Transcription Factors Depends on Their Nuclear Localization. Mol Cell Biol (1997) 17:5508–20. doi: 10.1128/mcb.17.9.5508
39. Verona R, Moberg K, Estes S, Starz M, Vernon JP, Lees JA. E2F Activity Is Regulated by Cell Cycle-Dependent Changes in Subcellular Localization. Mol Cell Biol (1997) 17:7268–82. doi: 10.1128/mcb.17.12.7268
40. Gaubatz S, Lees JA, Lindeman GJ, Livingston DM. E2F4 Is Exported From the Nucleus in a CRM1-Dependent Manner. Mol Cell Biol (2001) 21:1384–92. doi: 10.1128/mcb.21.4.1384-1392.2001
41. Logan N, Delavaine L, Graham A, Reilly C, Wilson J, Brummelkamp TR, et al. E2F-7: A Distinctive E2F Family Member With an Unusual Organization of DNA-Binding Domains. Oncogene (2004) 23:5138–50. doi: 10.1038/sj.onc.1207649
42. Logan N, Graham A, Zhao X, Fisher R, Maiti B, Leone G, et al. E2F-8: An E2F Family Member With a Similar Organization of DNA-Binding Domains to E2F-7. Oncogene (2005) 24:5000–4. doi: 10.1038/sj.onc.1208703
43. Lv Y, Xiao J, Liu J, Xing F. E2F8 Is a Potential Therapeutic Target for Hepatocellular Carcinoma. J Cancer (2017) 8:1205–13. doi: 10.7150/jca.18255
44. Poppy Roworth A, Ghari F, La Thangue NB. To Live or Let Die - Complexity Within the E2F1 Pathway. Mol Cell Oncol (2015) 2:e970480. doi: 10.4161/23723548.2014.970480
45. Cam H, Dynlacht BD. Emerging Roles for E2F: Beyond the G1/S Transition and DNA Replication. Cancer Cell (2003) 3:311–6. doi: 10.1016/s1535-6108(03)00080-1
46. Kent LN, Leone G. The Broken Cycle: E2F Dysfunction in Cancer. Nat Rev Cancer (2019) 19:326–38. doi: 10.1038/s41568-019-0143-7
47. Kent LN, Bae S, Tsai SY, Tang X, Srivastava A, Koivisto C, et al. Dosage-Dependent Copy Number Gains in E2f1 and E2f3 Drive Hepatocellular Carcinoma. J Clin Invest (2017) 127:830–42. doi: 10.1172/jci87583
48. Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, et al. Identification of a Novel E2F3 Product Suggests a Mechanism for Determining Specificity of Repression by Rb Proteins. Mol Cell Biol (2000) 20:3626–32. doi: 10.1128/mcb.20.10.3626-3632.2000
49. Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A Complex With Chromatin Modifiers That Occupies E2F- and Myc-Responsive Genes in G0 Cells. Science (2002) 296:1132–6. doi: 10.1126/science.1069861
50. Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, et al. E2F Mediates Cell Cycle-Dependent Transcriptional Repression In Vivo by Recruitment of an HDAC1/mSin3B Corepressor Complex. Genes Dev (2002) 16:933–47. doi: 10.1101/gad.969202
51. Trimarchi JM, Fairchild B, Wen J, Lees JA. The E2F6 Transcription Factor Is a Component of the Mammalian Bmi1-Containing Polycomb Complex. Proc Natl Acad Sci USA (2001) 98:1519–24. doi: 10.1073/pnas.041597698
52. Moon NS, Dyson N. E2F7 and E2F8 Keep the E2F Family in Balance. Dev Cell (2008) 14:1–3. doi: 10.1016/j.devcel.2007.12.017
53. Batlle E, Clevers H. Cancer Stem Cells Revisited. Nat Med (2017) 23:1124–34. doi: 10.1038/nm.4409
54. Santoro A, Vlachou T, Carminati M, Pelicci PG, Mapelli M. Molecular Mechanisms of Asymmetric Divisions in Mammary Stem Cells. EMBO Rep (2016) 17:1700–20. doi: 10.15252/embr.201643021
55. Pardee AB, Li CJ. Two Controls of Cell Proliferation Underlie Cancer Relapse. J Cell Physiol (2018) 233:8437–40. doi: 10.1002/jcp.26597
56. Prasetyanti PR, Medema JP. Intra-Tumor Heterogeneity From a Cancer Stem Cell Perspective. Mol Cancer (2017) 16:41. doi: 10.1186/s12943-017-0600-4
57. Yang Y, Li X, Wang T, Guo Q, Xi T, Zheng L. Emerging Agents That Target Signaling Pathways in Cancer Stem Cells. J Hematol Oncol (2020) 13:60. doi: 10.1186/s13045-020-00901-6
58. Vermeulen L, de Sousa e Melo F, Richel DJ, Medema JP. The Developing Cancer Stem-Cell Model: Clinical Challenges and Opportunities. Lancet Oncol (2012) 13:e83–9. doi: 10.1016/s1470-2045(11)70257-1
59. Thankamony AP, Saxena K, Murali R, Jolly MK, Nair R. Cancer Stem Cell Plasticity - A Deadly Deal. Front Mol Biosci (2020) 7:79. doi: 10.3389/fmolb.2020.00079
60. Xu J, Liao K, Zhou W. Exosomes Regulate the Transformation of Cancer Cells in Cancer Stem Cell Homeostasis. Stem Cells Int (2018) 2018:4837370. doi: 10.1155/2018/4837370
61. Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, et al. Normal and Neoplastic Nonstem Cells can Spontaneously Convert to a Stem-Like State. Proc Natl Acad Sci USA (2011) 108:7950–5. doi: 10.1073/pnas.1102454108
62. Tsui YM, Chan LK, Ng IO. Cancer Stemness in Hepatocellular Carcinoma: Mechanisms and Translational Potential. Br J Cancer (2020) 122:1428–40. doi: 10.1038/s41416-020-0823-9
63. Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, et al. Cancer Stem Cells From Human Breast Tumors Are Involved in Spontaneous Metastases in Orthotopic Mouse Models. Proc Natl Acad Sci USA (2010) 107:18115–20. doi: 10.1073/pnas.1006732107
64. Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, et al. Single-Cell Analysis Reveals a Stem-Cell Program in Human Metastatic Breast Cancer Cells. Nature (2015) 526:131–5. doi: 10.1038/nature15260
65. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The Epithelial-Mesenchymal Transition Generates Cells With Properties of Stem Cells. Cell (2008) 133:704–15. doi: 10.1016/j.cell.2008.03.027
66. Shibue T, Weinberg RA. EMT, CSCs, and Drug Resistance: The Mechanistic Link and Clinical Implications. Nat Rev Clin Oncol (2017) 14:611–29. doi: 10.1038/nrclinonc.2017.44
67. Garcia-Mayea Y, Mir C, Masson F, Paciucci R, ME LL. Insights Into New Mechanisms and Models of Cancer Stem Cell Multidrug Resistance. Semin Cancer Biol (2020) 60:166–80. doi: 10.1016/j.semcancer.2019.07.022
68. Sun X, Lv X, Yan Y, Zhao Y, Ma R, He M, et al. Hypoxia-Mediated Cancer Stem Cell Resistance and Targeted Therapy. BioMed Pharmacother (2020) 130:110623. doi: 10.1016/j.biopha.2020.110623
69. Najafi M, Farhood B, Mortezaee K, Kharazinejad E, Majidpoor J, Ahadi R. Hypoxia in Solid Tumors: A Key Promoter of Cancer Stem Cell (CSC) Resistance. J Cancer Res Clin Oncol (2020) 146:19–31. doi: 10.1007/s00432-019-03080-1
70. Hu M, Guo W, Liao Y, Xu D, Sun B, Song H, et al. Dysregulated ENPP1 Increases the Malignancy of Human Lung Cancer by Inducing Epithelial-Mesenchymal Transition Phenotypes and Stem Cell Features. Am J Cancer Res (2019) 9:134–44.
71. Takahashi RU, Miyazaki H, Takeshita F, Yamamoto Y, Minoura K, Ono M, et al. Loss of microRNA-27b Contributes to Breast Cancer Stem Cell Generation by Activating ENPP1. Nat Commun (2015) 6:7318. doi: 10.1038/ncomms8318
72. Bageritz J, Puccio L, Piro RM, Hovestadt V, Phillips E, Pankert T, et al. Stem Cell Characteristics in Glioblastoma Are Maintained by the Ecto-Nucleotidase E-Npp1. Cell Death Differ (2014) 21:929–40. doi: 10.1038/cdd.2014.12
73. Fang Z, Lin M, Li C, Liu H, Gong C. A Comprehensive Review of the Roles of E2F1 in Colon Cancer. Am J Cancer Res (2020) 10:757–68.
74. Wen N, Guo B, Zheng H, Xu L, Liang H, Wang Q, et al. Bromodomain Inhibitor Jq1 Induces Cell Cycle Arrest and Apoptosis of Glioma Stem Cells Through the VEGF/PI3K/AKT Signaling Pathway. Int J Oncol (2019) 55:879–95. doi: 10.3892/ijo.2019.4863
75. Zhang G, Dong Z, Prager BC, Kim LJ, Wu Q, Gimple RC, et al. Chromatin Remodeler HELLS Maintains Glioma Stem Cells Through E2F3 and MYC. JCI Insight (2019) 4:e126140. doi: 10.1172/jci.insight.126140
76. El-Khattouti A, Sheehan NT, Monico J, Drummond HA, Haikel Y, Brodell RT, et al. CD133⁺ Melanoma Subpopulation Acquired Resistance to Caffeic Acid Phenethyl Ester-Induced Apoptosis Is Attributed to the Elevated Expression of ABCB5: Significance for Melanoma Treatment. Cancer Lett (2015) 357:83–104. doi: 10.1016/j.canlet.2014.10.043
77. Jaworska AM, Wlodarczyk NA, Mackiewicz A, Czerwinska P. The Role of TRIM Family Proteins in the Regulation of Cancer Stem Cell Self-Renewal. Stem Cells (2020) 38:165–73. doi: 10.1002/stem.3109
78. Sancho P, Barneda D, Heeschen C. Hallmarks of Cancer Stem Cell Metabolism. Br J Cancer (2016) 114:1305–12. doi: 10.1038/bjc.2016.152
79. Hadjimichael C, Chanoumidou K, Papadopoulou N, Arampatzi P, Papamatheakis J, Kretsovali A. Common Stemness Regulators of Embryonic and Cancer Stem Cells. World J Stem Cells (2015) 7:1150–84. doi: 10.4252/wjsc.v7.i9.1150
80. Chen CL, Uthaya Kumar DB, Punj V, Xu J, Sher L, Tahara SM, et al. NANOG Metabolically Reprograms Tumor-Initiating Stem-Like Cells Through Tumorigenic Changes in Oxidative Phosphorylation and Fatty Acid Metabolism. Cell Metab (2016) 23:206–19. doi: 10.1016/j.cmet.2015.12.004
81. Wang K, Wang Y, Wang Y, Liu S, Wang C, Zhang S, et al. EIF5A2 Enhances Stemness of Epithelial Ovarian Cancer Cells via a E2F1/KLF4 Axis. Stem Cell Res Ther (2021) 12:186. doi: 10.1186/s13287-021-02256-2
82. Park JH, Shin JE, Park HW. The Role of Hippo Pathway in Cancer Stem Cell Biology. Mol Cells (2018) 41:83–92. doi: 10.14348/molcells.2018.2242
83. Schaal CM, Bora-Singhal N, Kumar DM, Chellappan SP. Regulation of Sox2 and Stemness by Nicotine and Electronic-Cigarettes in non-Small Cell Lung Cancer. Mol Cancer (2018) 17:149. doi: 10.1186/s12943-018-0901-2
84. Perumal D, Pillai S, Nguyen J, Schaal C, Coppola D, Chellappan SP. Nicotinic Acetylcholine Receptors Induce C-Kit Ligand/Stem Cell Factor and Promote Stemness in an ARRB1/ β-Arrestin-1 Dependent Manner in NSCLC. Oncotarget (2014) 5:10486–502. doi: 10.18632/oncotarget.2395
85. Yasumizu Y, Rajabi H, Jin C, Hata T, Pitroda S, Long MD, et al. MUC1-C Regulates Lineage Plasticity Driving Progression to Neuroendocrine Prostate Cancer. Nat Commun (2020) 11:338. doi: 10.1038/s41467-019-14219-6
86. Hagiwara M, Yasumizu Y, Yamashita N, Rajabi H, Fushimi A, Long MD, et al. MUC1-C Activates the BAF (mSWI/SNF) Complex in Prostate Cancer Stem Cells. Cancer Res (2021) 81:1111–22. doi: 10.1158/0008-5472.Can-20-2588
87. Feng J, Lan R, Cai G, Lin J. TREX1 Suppression Imparts Cancer-Stem-Cell-Like Characteristics to CD133(-) Osteosarcoma Cells Through the Activation of E2F4 Signaling. Int J Clin Exp Pathol (2019) 12:1134–53.
88. Chaffer CL, Weinberg RA. A Perspective on Cancer Cell Metastasis. Science (2011) 331:1559–64. doi: 10.1126/science.1203543
89. Celià-Terrassa T, Jolly MK. Cancer Stem Cells and Epithelial-To-Mesenchymal Transition in Cancer Metastasis. Cold Spring Harb Perspect Med (2020) 10:a036905. doi: 10.1101/cshperspect.a036905
90. Steinbichler TB, Savic D, Dudás J, Kvitsaridze I, Skvortsov S, Riechelmann H, et al. Cancer Stem Cells and Their Unique Role in Metastatic Spread. Semin Cancer Biol (2020) 60:148–56. doi: 10.1016/j.semcancer.2019.09.007
91. Grillet F, Bayet E, Villeronce O, Zappia L, Lagerqvist EL, Lunke S, et al. Circulating Tumour Cells From Patients With Colorectal Cancer Have Cancer Stem Cell Hallmarks in Ex Vivo Culture. Gut (2017) 66:1802–10. doi: 10.1136/gutjnl-2016-311447
92. Schölch S, García SA, Iwata N, Niemietz T, Betzler AM, Nanduri LK, et al. Circulating Tumor Cells Exhibit Stem Cell Characteristics in an Orthotopic Mouse Model of Colorectal Cancer. Oncotarget (2016) 7:27232–42. doi: 10.18632/oncotarget.8373
93. Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, et al. Identification of a Population of Blood Circulating Tumor Cells From Breast Cancer Patients That Initiates Metastasis in a Xenograft Assay. Nat Biotechnol (2013) 31:539–44. doi: 10.1038/nbt.2576
94. Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell (2019) 176:98–112.e14. doi: 10.1016/j.cell.2018.11.046
95. Papadaki MA, Stoupis G, Theodoropoulos PA, Mavroudis D, Georgoulias V, Agelaki S. Circulating Tumor Cells With Stemness and Epithelial-To-Mesenchymal Transition Features Are Chemoresistant and Predictive of Poor Outcome in Metastatic Breast Cancer. Mol Cancer Ther (2019) 18:437–47. doi: 10.1158/1535-7163.Mct-18-0584
96. Andrechek ER. HER2/Neu Tumorigenesis and Metastasis Is Regulated by E2F Activator Transcription Factors. Oncogene (2015) 34:217–25. doi: 10.1038/onc.2013.540
97. Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, et al. A Perivascular Niche for Brain Tumor Stem Cells. Cancer Cell (2007) 11:69–82. doi: 10.1016/j.ccr.2006.11.020
98. Ping YF, Zhang X, Bian XW. Cancer Stem Cells and Their Vascular Niche: Do They Benefit From Each Other? Cancer Lett (2016) 380:561–7. doi: 10.1016/j.canlet.2015.05.010
99. Teng F, Zhang JX, Chang QM, Wu XB, Tang WG, Wang JF, et al. LncRNA MYLK-AS1 Facilitates Tumor Progression and Angiogenesis by Targeting miR-424-5p/E2F7 Axis and Activating VEGFR-2 Signaling Pathway in Hepatocellular Carcinoma. J Exp Clin Cancer Res (2020) 39:235. doi: 10.1186/s13046-020-01739-z
100. Morel AP, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of Breast Cancer Stem Cells Through Epithelial-Mesenchymal Transition. PloS One (2008) 3:e2888. doi: 10.1371/journal.pone.0002888
101. Wang T, Chen X, Qiao W, Kong L, Sun D, Li Z. Transcription Factor E2F1 Promotes EMT by Regulating ZEB2 in Small Cell Lung Cancer. BMC Cancer (2017) 17:719. doi: 10.1186/s12885-017-3701-y
102. Khan FM, Marquardt S, Gupta SK, Knoll S, Schmitz U, Spitschak A, et al. Unraveling a Tumor Type-Specific Regulatory Core Underlying E2F1-Mediated Epithelial-Mesenchymal Transition to Predict Receptor Protein Signatures. Nat Commun (2017) 8:198. doi: 10.1038/s41467-017-00268-2
103. Kim LK, Park SA, Eoh KJ, Heo TH, Kim YT, Kim HJ. E2F8 Regulates the Proliferation and Invasion Through Epithelial-Mesenchymal Transition in Cervical Cancer. Int J Biol Sci (2020) 16:320–9. doi: 10.7150/ijbs.37686
104. Ye YY, Mei JW, Xiang SS, Li HF, Ma Q, Song XL, et al. MicroRNA-30a-5p Inhibits Gallbladder Cancer Cell Proliferation, Migration and Metastasis by Targeting E2F7. Cell Death Dis (2018) 9:410. doi: 10.1038/s41419-018-0444-x
105. Lu Z, Nian Z, Jingjing Z, Tao L, Quan L. MicroRNA-424/E2F6 Feedback Loop Modulates Cell Invasion, Migration and EMT in Endometrial Carcinoma. Oncotarget (2017) 8:114281–91. doi: 10.18632/oncotarget.23218
106. Jusino S, Saavedra HI. Role of E2Fs and Mitotic Regulators Controlled by E2Fs in the Epithelial to Mesenchymal Transition. Exp Biol Med (Maywood) (2019) 244:1419–29. doi: 10.1177/1535370219881360
107. Nazio F, Bordi M, Cianfanelli V, Locatelli F, Cecconi F. Autophagy and Cancer Stem Cells: Molecular Mechanisms and Therapeutic Applications. Cell Death Differ (2019) 26:690–702. doi: 10.1038/s41418-019-0292-y
108. Rosenfeldt MT, Bell LA, Long JS, O’Prey J, Nixon C, Roberts F, et al. E2F1 Drives Chemotherapeutic Drug Resistance via ABCG2. Oncogene (2014) 33:4164–72. doi: 10.1038/onc.2013.470
109. Wu M, Wang X, McGregor N, Pienta KJ, Zhang J. Dynamic Regulation of Rad51 by E2F1 and P53 in Prostate Cancer Cells Upon Drug-Induced DNA Damage Under Hypoxia. Mol Pharmacol (2014) 85:866–76. doi: 10.1124/mol.113.090688
110. Bindra RS, Gibson SL, Meng A, Westermark U, Jasin M, Pierce AJ, et al. Hypoxia-Induced Down-Regulation of BRCA1 Expression by E2Fs. Cancer Res (2005) 65:11597–604. doi: 10.1158/0008-5472.Can-05-2119
111. Kim H, Lin Q, Yun Z. BRCA1 Regulates the Cancer Stem Cell Fate of Breast Cancer Cells in the Context of Hypoxia and Histone Deacetylase Inhibitors. Sci Rep (2019) 9:9702. doi: 10.1038/s41598-019-46210-y
112. Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 Is an RB/E2F Target Gene Required for Hypoxia-Induced Autophagy. Mol Cell Biol (2007) 27:6229–42. doi: 10.1128/mcb.02246-06
113. Araki K, Kawauchi K, Sugimoto W, Tsuda D, Oda H, Yoshida R, et al. Mitochondrial Protein E2F3d, a Distinctive E2F3 Product, Mediates Hypoxia-Induced Mitophagy in Cancer Cells. Commun Biol (2019) 2:3. doi: 10.1038/s42003-018-0246-9
114. Dykstra KM, Hanekamp DW, Johnson M, Guzman ML, Wang ES. Abstract 2864: Inhibition of the Late Stages of Autophagy Overcomes Hypoxia-Induced Chemoresistance and Targets Leukemic Stem Cells in Acute Myeloid Leukemia. Cancer Res (2018) 78:2864. doi: 10.1158/1538-7445.AM2018-2864
115. Dykstra KM, Fay HRS, Massey AC, Yang N, Johnson M, Portwood S, et al. Inhibiting Autophagy Targets Human Leukemic Stem Cells and Hypoxic AML Blasts by Disrupting Mitochondrial Homeostasis. Blood Adv (2021) 5:2087–100. doi: 10.1182/bloodadvances.2020002666
116. Emmrich S, Pützer BM. Checks and Balances: E2F-microRNA Crosstalk in Cancer Control. Cell Cycle (2010) 9:2555–67. doi: 10.4161/cc.9.13.12061
117. Xia L, Li F, Qiu J, Feng Z, Xu Z, Chen Z, et al. Oncogenic miR-20b-5p Contributes to Malignant Behaviors of Breast Cancer Stem Cells by Bidirectionally Regulating CCND1 and E2F1. BMC Cancer (2020) 20:949. doi: 10.1186/s12885-020-07395-y
118. Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q, et al. Long Noncoding RNA LINC00511 Contributes to Breast Cancer Tumourigenesis and Stemness by Inducing the miR-185-3p/E2F1/Nanog Axis. J Exp Clin Cancer Res (2018) 37:289. doi: 10.1186/s13046-018-0945-6
119. Wu Q, Yang Z, Wang F, Hu S, Yang L, Shi Y, et al. MiR-19b/20a/92a Regulates the Self-Renewal and Proliferation of Gastric Cancer Stem Cells. J Cell Sci (2013) 126:4220–9. doi: 10.1242/jcs.127944
120. Shao Q, Xu J, Guan X, Zhou B, Wei W, Deng R, et al. In Vitro and In Vivo Effects of miRNA-19b/20a/92a on Gastric Cancer Stem Cells and the Related Mechanism. Int J Med Sci (2018) 15:86–94. doi: 10.7150/ijms.21164
121. Wu N, Xiao L, Zhao X, Zhao J, Wang J, Wang F, et al. miR-125b Regulates the Proliferation of Glioblastoma Stem Cells by Targeting E2F2. FEBS Lett (2012) 586:3831–9. doi: 10.1016/j.febslet.2012.08.023
122. Song H, Zhang Y, Liu N, Zhang D, Wan C, Zhao S, et al. Let-7b Inhibits the Malignant Behavior of Glioma Cells and Glioma Stem-Like Cells via Downregulation of E2F2. J Physiol Biochem (2016) 72:733–44. doi: 10.1007/s13105-016-0512-6
123. Chu J, Li Y, Fan X, Ma J, Li J, Lu G, et al. MiR-4319 Suppress the Malignancy of Triple-Negative Breast Cancer by Regulating Self-Renewal and Tumorigenesis of Stem Cells. Cell Physiol Biochem (2018) 48:593–604. doi: 10.1159/000491888
124. Lin QY, Wang JQ, Wu LL, Zheng WE, Chen PR. miR-638 Represses the Stem Cell Characteristics of Breast Cancer Cells by Targeting E2F2. Breast Cancer (2020) 27:147–58. doi: 10.1007/s12282-019-01002-0
125. Feliciano A, Garcia-Mayea Y, Jubierre L, Mir C, Hummel M, Castellvi J, et al. miR-99a Reveals Two Novel Oncogenic Proteins E2F2 and EMR2 and Represses Stemness in Lung Cancer. Cell Death Dis (2017) 8:e3141. doi: 10.1038/cddis.2017.544
126. Fang Y, Gu X, Li Z, Xiang J, Chen Z. miR-449b Inhibits the Proliferation of SW1116 Colon Cancer Stem Cells Through Downregulation of CCND1 and E2F3 Expression. Oncol Rep (2013) 30:399–406. doi: 10.3892/or.2013.2465
127. Shan ZN, Tian R, Zhang M, Gui ZH, Wu J, Ding M, et al. Mir128-1 Inhibits the Growth of Glioblastoma Multiforme and Glioma Stem-Like Cells via Targeting BMI1 and E2F3. Oncotarget (2016) 7:78813–26. doi: 10.18632/oncotarget.12385
128. Cheng FH, Aguda BD, Tsai JC, Kochańczyk M, Lin JM, Chen GC, et al. A Mathematical Model of Bimodal Epigenetic Control of miR-193a in Ovarian Cancer Stem Cells. PloS One (2014) 9:e116050. doi: 10.1371/journal.pone.0116050
129. Li Y, Huang J, Yang D, Xiang S, Sun J, Li H, et al. Expression Patterns of E2F Transcription Factors and Their Potential Prognostic Roles in Breast Cancer. Oncol Lett (2018) 15:9216–30. doi: 10.3892/ol.2018.8514
130. Liu ZL, Bi XW, Liu PP, Lei DX, Wang Y, Li ZM, et al. Expressions and Prognostic Values of the E2F Transcription Factors in Human Breast Carcinoma. Cancer Manag Res (2018) 10:3521–32. doi: 10.2147/cmar.S172332
131. Liu X, Hu C. Novel Potential Therapeutic Target for E2F1 and Prognostic Factors of E2F1/2/3/5/7/8 in Human Gastric Cancer. Mol Ther Methods Clin Dev (2020) 18:824–38. doi: 10.1016/j.omtm.2020.07.017
132. Yao H, Lu F, Shao Y. The E2F Family as Potential Biomarkers and Therapeutic Targets in Colon Cancer. PeerJ (2020) 8:e8562. doi: 10.7717/peerj.8562
133. Huang YL, Ning G, Chen LB, Lian YF, Gu YR, Wang JL, et al. Promising Diagnostic and Prognostic Value of E2Fs in Human Hepatocellular Carcinoma. Cancer Manag Res (2019) 11:1725–40. doi: 10.2147/cmar.S182001
134. Sun CC, Zhou Q, Hu W, Li SJ, Zhang F, Chen ZL, et al. Transcriptional E2F1/2/5/8 as Potential Targets and Transcriptional E2F3/6/7 as New Biomarkers for the Prognosis of Human Lung Carcinoma. Aging (Albany NY) (2018) 10:973–87. doi: 10.18632/aging.101441
135. Yu H, Li Z, Wang M. Expression and Prognostic Role of E2F Transcription Factors in High-Grade Glioma. CNS Neurosci Ther (2020) 26:741–53. doi: 10.1111/cns.13295
136. Zhang Y, Wang Z, Ma J, Huo J, Li Y, Wang Y, et al. Bioinformatics Identification of the Expression and Clinical Significance of E2F Family in Endometrial Cancer. Front Genet (2020) 11:557188. doi: 10.3389/fgene.2020.557188
137. Lee SR, Roh YG, Kim SK, Lee JS, Seol SY, Lee HH, et al. Activation of EZH2 and SUZ12 Regulated by E2F1 Predicts the Disease Progression and Aggressive Characteristics of Bladder Cancer. Clin Cancer Res (2015) 21:5391–403. doi: 10.1158/1078-0432.Ccr-14-2680
138. Okamoto OK, Oba-Shinjo SM, Lopes L, Nagahashi Marie SK. Expression of HOXC9 and E2F2 Are Up-Regulated in CD133(+) Cells Isolated From Human Astrocytomas and Associate With Transformation of Human Astrocytes. Biochim Biophys Acta (2007) 1769:437–42. doi: 10.1016/j.bbaexp.2007.05.002
139. Bellmunt J. Stem-Like Signature Predicting Disease Progression in Early Stage Bladder Cancer. The Role of E2F3 and SOX4. Biomedicines (2018) 6:85. doi: 10.3390/biomedicines6030085
140. Shen H, Morrison CD, Zhang J, Underwood W 3rd, Yang N, Frangou C, et al. 6p22.3 Amplification as a Biomarker and Potential Therapeutic Target of Advanced Stage Bladder Cancer. Oncotarget (2013) 4:2124–34. doi: 10.18632/oncotarget.1485
141. Khaleel SS, Andrews EH, Ung M, DiRenzo J, Cheng C. E2F4 Regulatory Program Predicts Patient Survival Prognosis in Breast Cancer. Breast Cancer Res (2014) 16:486. doi: 10.1186/s13058-014-0486-7
142. Cheng C, Varn FS, Marsit CJ. E2F4 Program Is Predictive of Progression and Intravesical Immunotherapy Efficacy in Bladder Cancer. Mol Cancer Res (2015) 13:1316–24. doi: 10.1158/1541-7786.Mcr-15-0120
143. Kothandaraman N, Bajic VB, Brendan PN, Huak CY, Keow PB, Razvi K, et al. E2F5 Status Significantly Improves Malignancy Diagnosis of Epithelial Ovarian Cancer. BMC Cancer (2010) 10:64. doi: 10.1186/1471-2407-10-64
144. Ishimoto T, Shiozaki A, Ichikawa D, Fujiwara H, Konishi H, Komatsu S, et al. E2F5 as an Independent Prognostic Factor in Esophageal Squamous Cell Carcinoma. Anticancer Res (2013) 33:5415–20.
145. Huang K, Liu X, Li Y, Wang Q, Zhou J, Wang Y, et al. Genome-Wide CRISPR-Cas9 Screening Identifies NF-κb/E2F6 Responsible for EGFRvIII-Associated Temozolomide Resistance in Glioblastoma. Adv Sci (Weinh) (2019) 6:1900782. doi: 10.1002/advs.201900782
146. Park SA, Platt J, Lee JW, López-Giráldez F, Herbst RS, Koo JS. E2F8 as a Novel Therapeutic Target for Lung Cancer. J Natl Cancer Inst (2015) 107:djv151. doi: 10.1093/jnci/djv151
Keywords: E2Fs, cancer stem cells, cancer, biomarkers, therapeutic targets
Citation: Xie D, Pei Q, Li J, Wan X and Ye T (2021) Emerging Role of E2F Family in Cancer Stem Cells. Front. Oncol. 11:723137. doi: 10.3389/fonc.2021.723137
Received: 10 June 2021; Accepted: 27 July 2021;
Published: 12 August 2021.
Edited by:
Matiullah Khan, AIMST University, MalaysiaReviewed by:
Collene Jeter, University of Texas MD Anderson Cancer Center, United StatesBikul Das, Indian Institute of Technology Guwahati, India
Copyright © 2021 Xie, Pei, Li, Wan and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting Ye, eWV0aW5nMTEwM0AxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Dan Xie†
Dan Xie† Qin Pei
Qin Pei Jingyuan Li
Jingyuan Li Xue Wan
Xue Wan Ting Ye
Ting Ye