- 1Department of Urology, The Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University, Changzhou, China
- 2Department of Graduate School, Dalian Medical University, Dalian, China
Objective: As a result of the inconsistency between reports, a meta-analysis was designed to appraise the clinical implications of long non-coding RNAs (lncRNAs) in exosomes for the diagnosis of bladder cancer.
Methods: The PubMed, EMBASE, and Cochrane library databases were searched to identify the relevant literature on lncRNAs in exosomes for bladder cancer diagnosis from database inception to May 2021. The literature was screened according to the inclusion and exclusion criteria, and the Quality Assessment of Diagnostic Accuracy Studies-2 entry tool was applied to evaluate the quality of the literature, and the sources of heterogeneity were explored using meta-regression and subgroup analysis. Stata 14.0 and RevMan 5.3 software were used for statistical analysis.
Results: A total of 23 studies described in 10 articles were included, with a total of 1883 patients with bladder cancer and 1721 patients in the non-cancerous control group. The exosome-derived lncRNAs performed better in the diagnosis of bladder cancer with a pooled sensitivity of 0.74 (95% CI, 0.69-0.77), specificity of 0.76 (95% CI, 0.72-0.80), and area under the curve of 0.83. The heterogeneity between studies was partly as a result of differences in specimen type, number of lncRNAs, lncRNA expression form, and reference gene type. Subgroup analysis showed that the detection efficacy based on the combination of multiple lncRNAs (0.86, 95% CI, 0.82-0.88) was higher than that based on a single lncRNA (0.81, 95% CI, 0.78-0.85), and exosomal lncRNAs with blood as the detection sample had a high diagnostic efficacy (0.86, 95% CI, 0.82-0.86).
Conclusions: Exosome-derived lncRNAs hold great promise as non-invasive diagnostic biomarkers of bladder cancer. However, their clinical value needs to be examined in further comprehensive prospective studies.
Introduction
Bladder cancer is a common malignancy in urology worldwide and ranks 11th among the most common malignancies throughout the body (1). Approximately 80% of patients with bladder cancer have non-muscle invasive bladder cancer at initial onset with a better prognosis. But they are at risk of recurrence after transurethral resection of the bladder tumor. The remaining patients have muscle-invasive bladder cancer, which is prone to distant metastasis and has a high risk of death (2). A good prognosis for bladder cancer relies on early detection. Cystoscopy and tissue biopsy are the gold standard for the diagnosis of bladder cancer. However, the invasive nature of the procedure and high cost limit its application. Non-invasive and low-cost urine exfoliation cytology is not indicated for low-grade bladder cancer (3). Other currently known bladder cancer tumor biomarkers lack diagnostic sensitivity and specificity, despite their high detection rates. Therefore, it is important to find non-invasive humoral tumor markers with high sensitivity and specificity to establish a safe and effective detection method for the early diagnosis of bladder cancer.
Exosomes are found in many types of body fluids with high abundance. They contain multiple components such as proteins and RNAs and can participate in material and information exchange between cells. Many studies have suggested that exosomes can be used as a new approach for the screening and early diagnosis of tumors. Long non-coding RNAs (lncRNAs) are longer than 200 nucleotides encoding no protein, which play key roles in gene expression, differentiation. In recent years, increasing studies have highlighted the role of lncRNAs in the carcinogenesis of bladder cancer and suggested that lncRNAs might be used as biomarkers in cancer (4, 5). An increasing number of studies have suggested that lncRNAs in exosomes may serve as potential markers for bladder cancer diagnosis. However, we found that several recent studies had discrepancies in their diagnostic accuracy when assessing the diagnostic value of lncRNAs in exosomes in bladder cancer. Chen et al. had a diagnostic specificity of 85% for bladder cancer, while Maryam et al. had a diagnostic sensitivity of 46.67% for bladder cancer (6, 7). Therefore, we performed a meta-analysis to evaluate the comprehensive diagnostic efficacy of lncRNAs in exosomes for patients with bladder cancer.
Materials and Methods
Search Strategy
The present meta-analysis was performed in accordance with the guidelines for diagnostic meta-analysis (8). The PubMed, EMBASE, and Cochrane library databases were searched to identify published studies related to the diagnosis of bladder cancer using the detection of lncRNA in exosomes from database inception to May 2021. Search terms included urinary bladder neoplasms, RNA, long non-coding, lncRNA, RNA, long non-translated, long non-coding RNA, long non-protein coding RNA, lincRNA, and exosome. A combination of subject headings and free words was used for searching, and supplementary searches of references from retrieved articles were conducted to ensure comprehensiveness.
Inclusion and Exclusion Criteria
The inclusion criteria were as follows: (1) published clinical diagnostic studies employing detection of lncRNAs in exosomes for the diagnosis of bladder cancer; (2) study subjects were patients with clinically already pathologically confirmed bladder cancer; and (3) studies provided sufficient data in the full text to calculate sensitivity and specificity. For studies with overlapping data, the study with the superior results was included. The exclusion criteria were as follows: (1) valid data (true positives, false positives, false negatives and true negatives) could not be extracted for merging; (2) non-clinical studies; and (3) reviews, case reports, and meeting abstracts.
Data Extraction
Data extraction was performed individually by two investigators, with disagreements resolved with the assistance of a third researcher. The extracted contents included: (1) basic characteristics of the study, including the first author, publication year, country of the study, sample size, specimen type, tumor type, tumor stage, tumor grade, lncRNA species, detection methods, and internal reference genes; and (2) diagnostic implications including TP, FP, FN, TN, sensitivity, and specificity.
Quality Assessment
The Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) was used to evaluate the quality of the included accuracy trials. The QUADAS-2 tool consists of four evaluation domains: selection of cases, diagnostic tests to be evaluated, gold standard tests, study flow, and progress. Each evaluation area had three or four landmark issues, which were used to evaluate the risk of bias in each field. The first three parts were also used to evaluate the applicability of the documents included. A total score for the seven items of more than four points means that the quality of literature research is high (9).
Statistical Methods
The QUADAS-2 evaluation function in RevMan 5.3 was used to evaluate the quality of the included studies for diagnostic accuracy studies, and the pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR) were calculated using Stata 14.0. The size of between-study heterogeneity was estimated using I2; a fixed effects model was used if I2 < 50%, which was considered to indicate small study heterogeneity, and a random effects model was used if I2 > 50%, which was considered to indicate substantial heterogeneity (10). The cumulative receiver operating characteristic curve (SROC) was plotted, and the area under the curve (AUC) was calculated to evaluate the diagnostic experimental value. The post-diagnostic effect after the pooled analysis was assessed using Fagan’s nomogram. Meta-regression, subgroup analysis, and sensitivity analysis were used to analyze the sources of heterogeneity. Publication bias was assessed using Deek’s funnel plot asymmetry test, with P < 0.10 indicating significant bias (11).
Results
Search Results and Included Literature
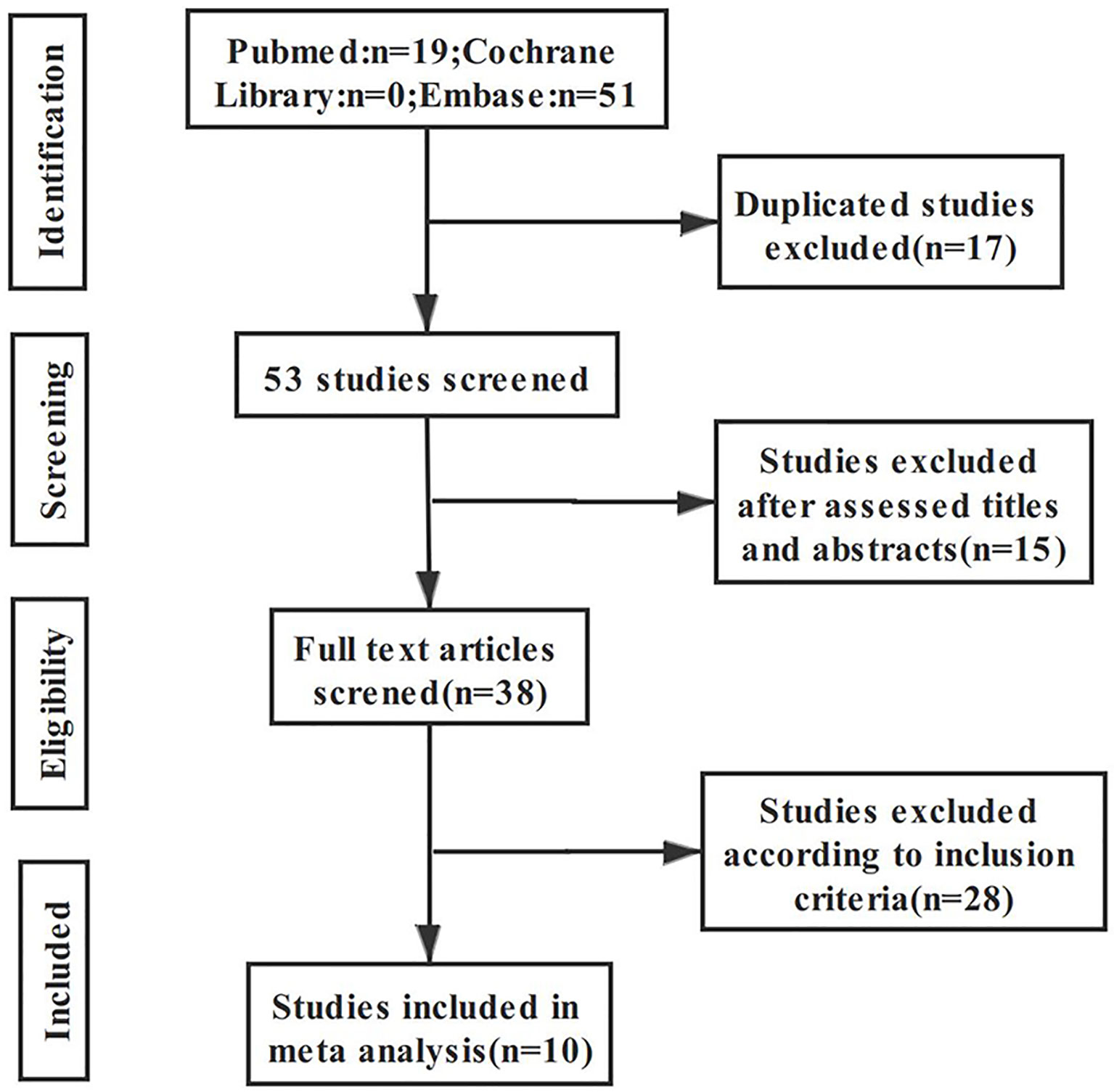
A total of 70 articles were retrieved through the search strategy; after excluding 17 duplicate articles and 15 articles after primary screening of the title, 38 articles remained for further assessment. The remaining articles were subjected to full-text assessment; 28 articles were further excluded due to ineligibility, resulting in a total of 10 articles being included in the final analysis. The literature screening process and results are shown in Figure 1.
Literature Characteristics and Quality Assessment
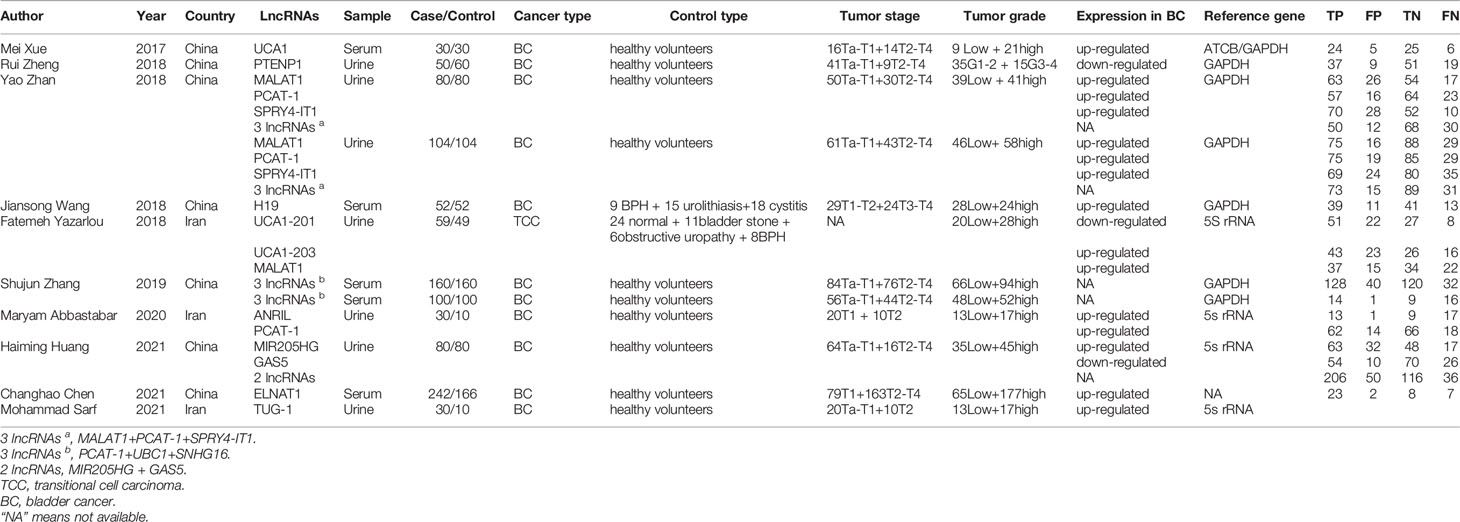
Twenty-three studies from 10 articles published between 2017 and 2021 were included in this meta-analysis (6, 7, 12–19). The included population comprised 1883 patients with bladder cancer and 1721 control patients. In terms of sample selection, 18 studies extracted exosomes from urine, and five studies extracted exosomes from blood. Only five studies utilized multiple lncRNAs to detect bladder cancer in combination, and 18 employed a single lncRNA for detection. In terms of patient tumor stage, 20 studies included both patients with NMIBC and patients with MIBC, and all studies involved both low- and high-grade bladder cancer. In terms of lncRNA detection, 22 studies used real-time reverse transcription polymerase chain reaction (qRT-PCR) to detect the expression levels of lncRNAs and convert the lncRNA concentrations to the expression levels of internal reference genes. The basic information of the included studies is presented in Table 1.
The results of the evaluation of the quality of articles performed using QUADAS-2 (Figure 2) showed that all 23 included studies reached medium to high quality level, but there was some bias in case selection and diagnostic tests to be evaluated. This may be due to the fact that the time of testing was not defined for some of the included samples, and most studies did not explicitly state whether the reviewers interpreted the results of the trials to be evaluated without knowing the results of the gold standard assessment.
Diagnostic Performance
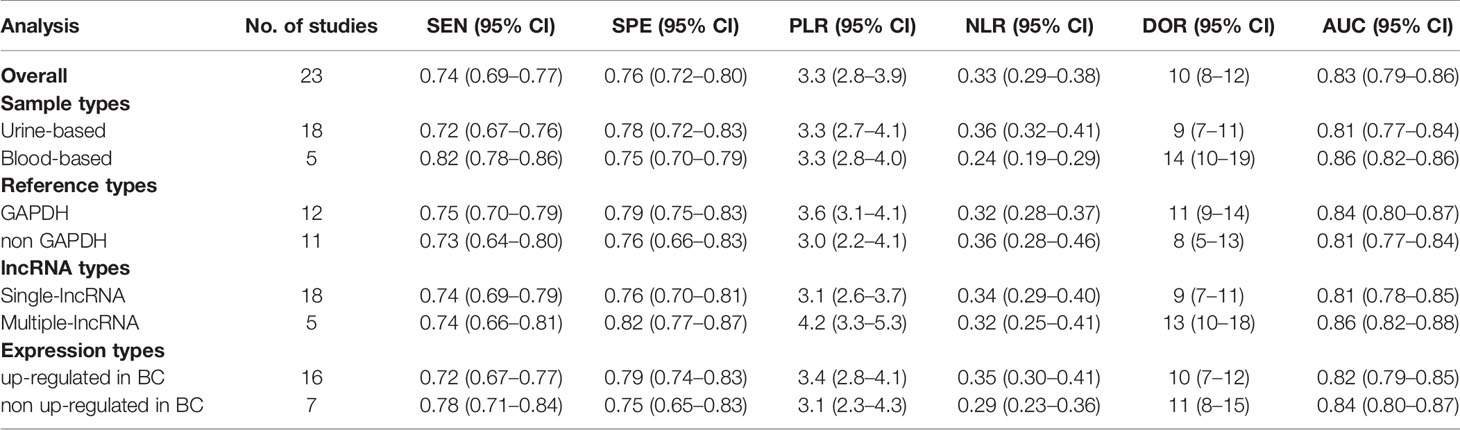
Because of significant heterogeneity (P < 0.01) among the studies in terms of sensitivity (I2 = 74.00%) and specificity (I2 = 72.11%), a random effects model was chosen to combine effect sizes. The specific results are shown in Figure 3. The pooled results across all 23 studies were: sensitivity, 0.74 (0.69–0.77) and specificity, 0.76 (0.72–0.80). The PLR was 3.3 (2.8–3.9), and the NLR was 0.33 (0.29–0.38), indicating that bladder cancer patients had 3.8 times more positive test results than healthy individuals. The DOR was 10 (8–12), indicating that lncRNAs in exosomes can be used to distinguish bladder cancer patients from controls (Table 2). The AUC was 0.83 (0.79–0.86), indicating that lncRNAs in exosomes could be a better diagnostic indicator for bladder cancer, and the results are shown in Figure 4A. According to the Fagan plot (Figure 4B), the pre-test probability was 52%, the post-test probability of a positive bladder cancer test was 78%, and the post-test probability of a negative bladder cancer test was 27%, which illustrated that the post-test probability and likelihood ratio were moderate.
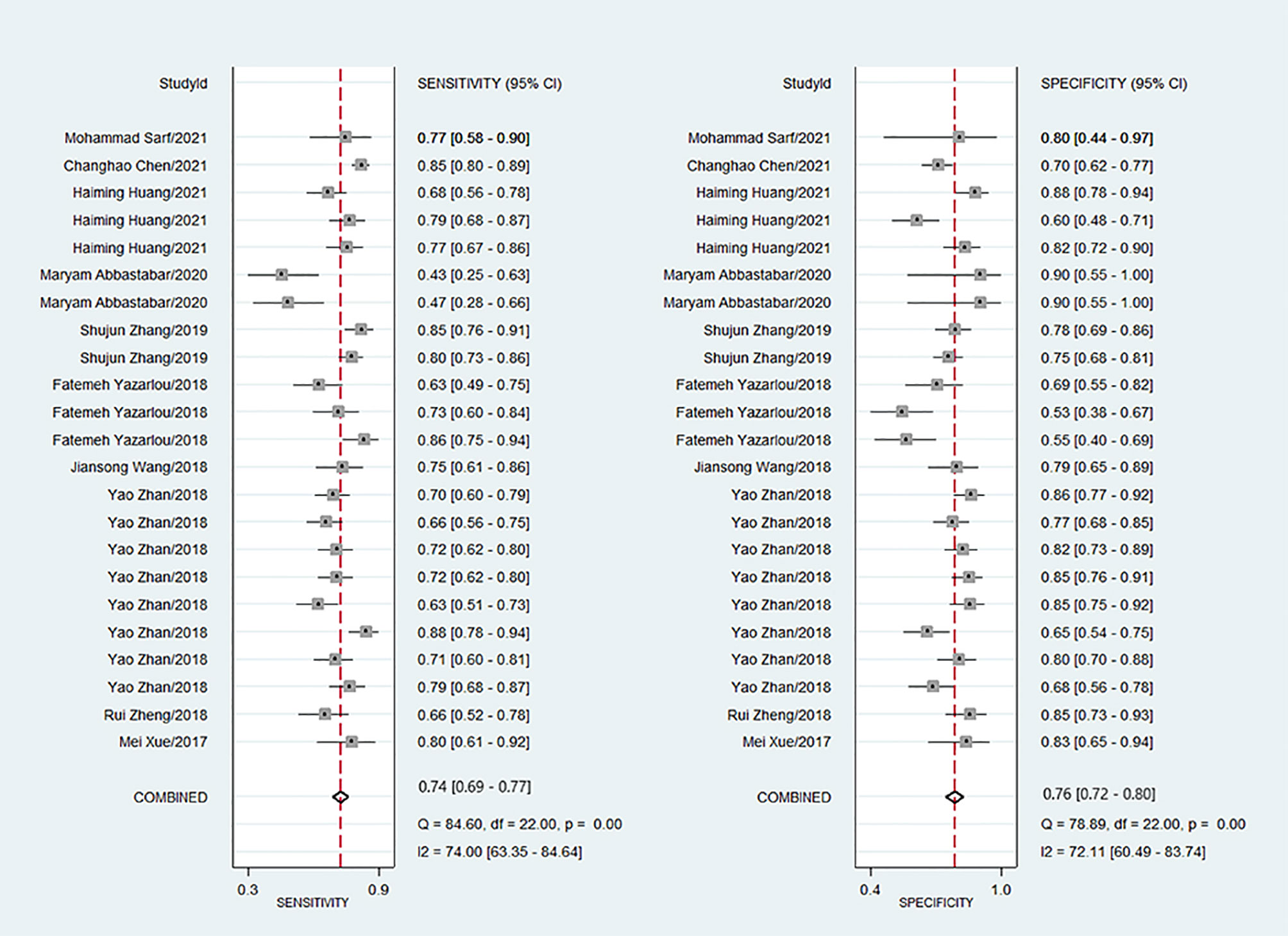
Figure 3 Forest plots of sensitivity and specificity for exosome derived lncRNAs in the diagnosis of bladder cancer.
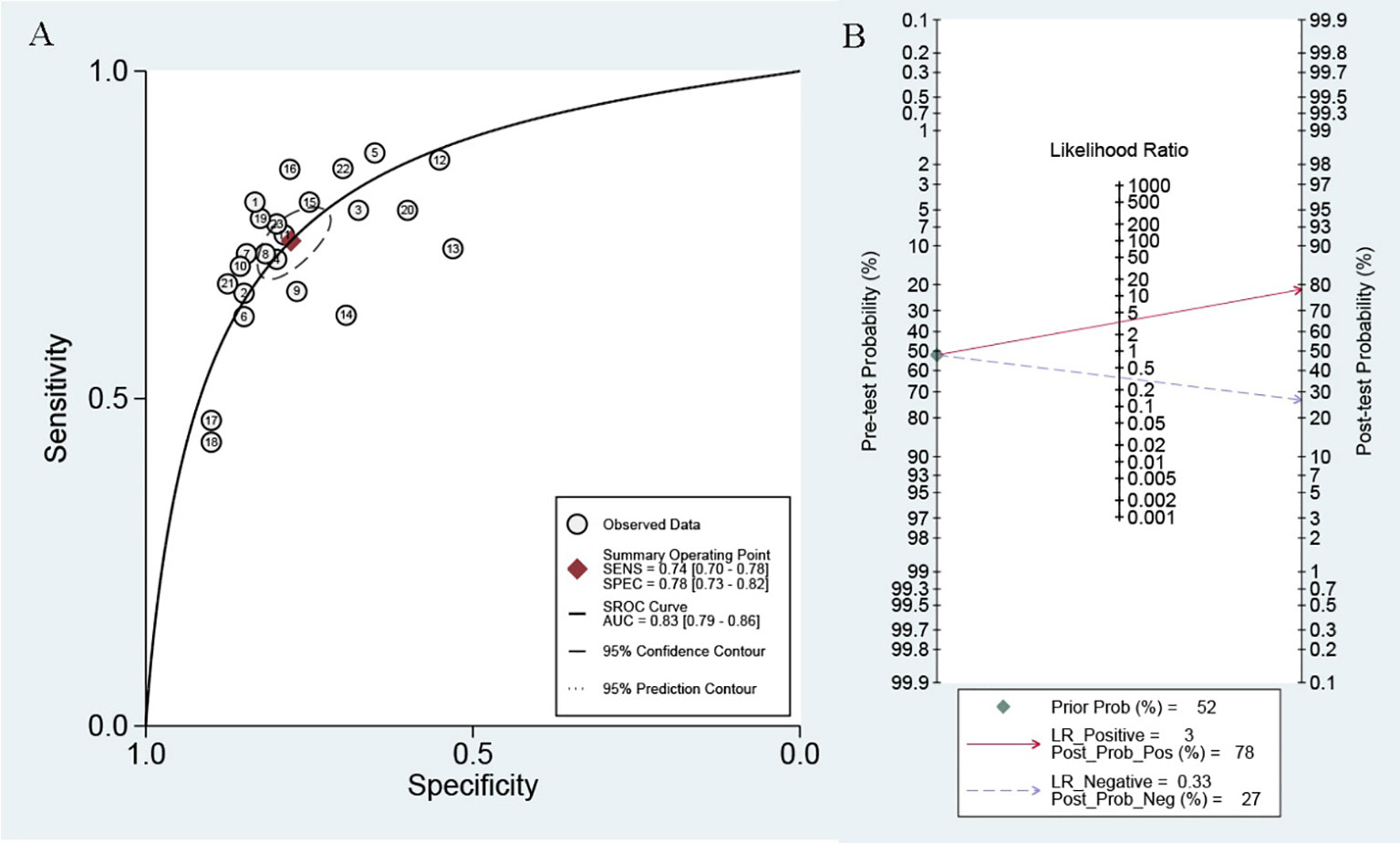
Figure 4 (A) SROC curve with pooled estimates of sensitivity, specificity and AUC of overall studies. (B) Fagan’s nomogram for evaluation of post-test probabilities based on pooled estimates of PLR and NLR of overall studies.
Meta-Regression and Subgroup Analysis
Potential sources of heterogeneity were explored using a meta-regression analysis. As shown in Figure 5, specimen type, number of lncRNAs, lncRNA expression form, and reference gene type were the main sources of heterogeneity in exosomal lncRNA detection in bladder cancer. Subgroup analysis was further performed according to the above factors, and the pooled results of diagnostic value in different subgroups are shown in Table 2. Among them, exosomal lncRNAs in blood samples showed higher sensitivity, DOR, and AUC (sensitivity = 0.82, DOR = 14, AUC = 0.86). The combination of multiple lncRNAs had better predictive ability than a single lncRNA (DOR = 13, AUC = 0.86).
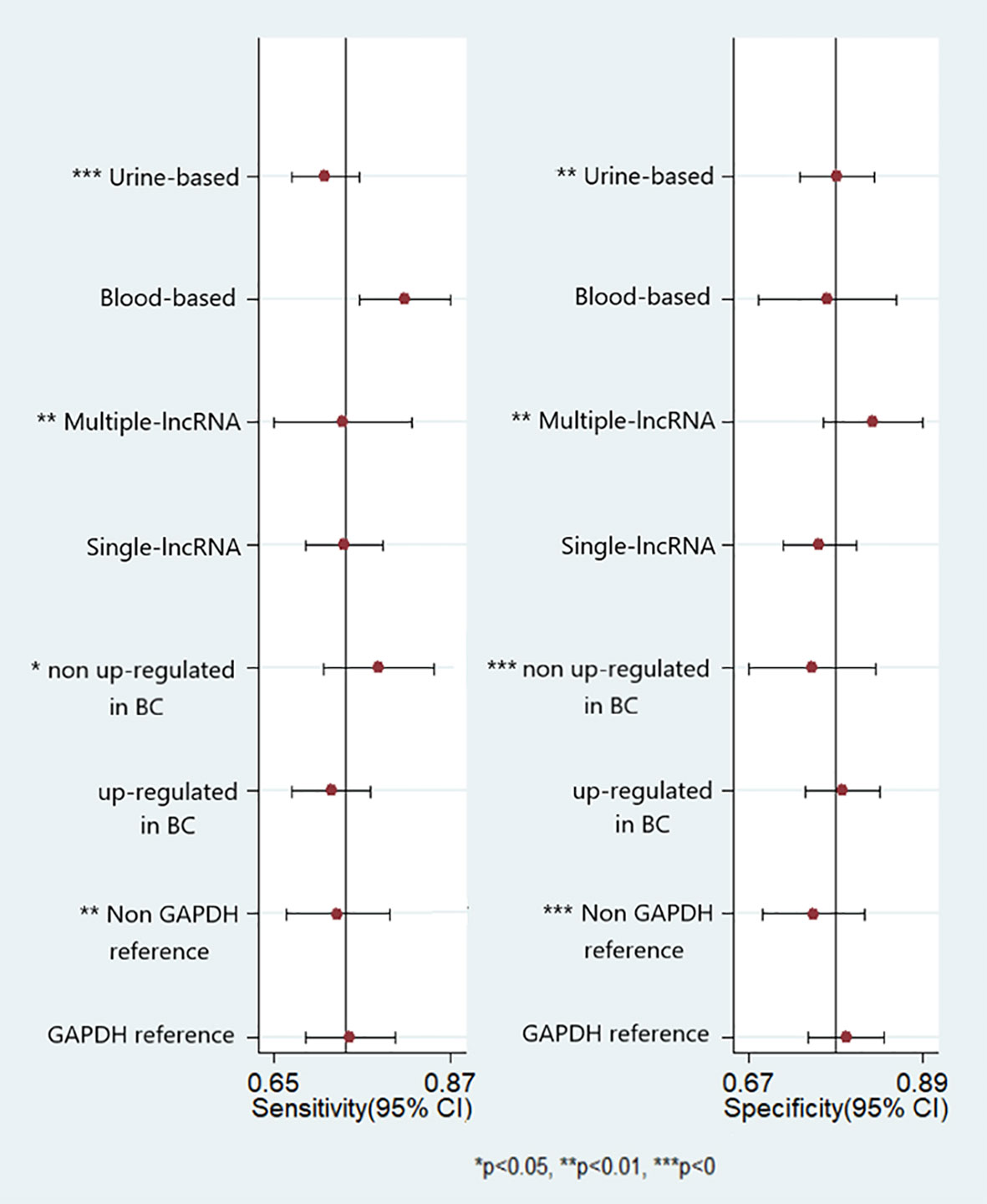
Figure 5 Forest plots of multivariable meta-regression analyses for sensitivity and specificity (vertical lines signify pooled estimates of sensitivity and specificity respectively).
Sensitivity Analysis and Publication Bias
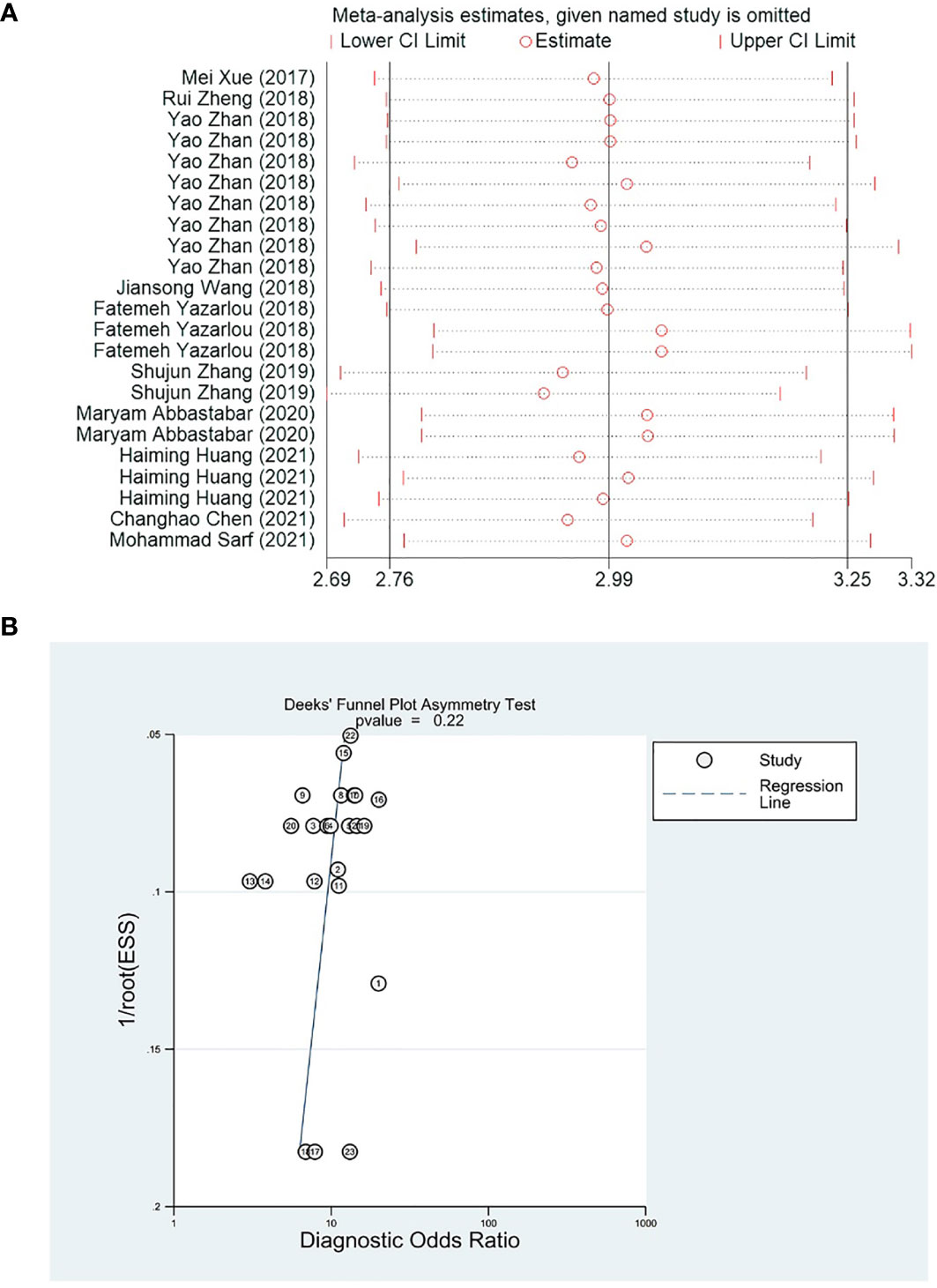
Removing single studies one by one, the difference between the pooled effect size of the remaining studies and the total effect size was observed for sensitivity analysis. The results showed that the outcome measures did not change significantly after removal, which indicated that the present meta-analysis was robust (Figure 6A). Deek’s funnel plot was used to assess the potential publication bias of the included studies, and suggested that there was no publication bias in the included studies (p = 0.22) (Figure 6B).
Discussion
Bladder cancer is one of the most common malignancies of the urinary system. Recently, with increased understanding of the molecular mechanisms of tumorigenesis, many studies of bladder cancer diagnosis, recurrence, and progression prediction have been conducted, mainly focusing on the abnormalities at the gene, protein, and molecular levels. Several studies have shown that nuclear matrix protein 22, telomerase, vascular endothelial growth factor, hyaluronidase, and proliferation-associated nuclear antigen Ki‐67 have some value in the diagnosis and prognosis of bladder cancer. However, all of the above markers lack sufficient sensitivity or specificity to change the current situation regarding bladder cancer diagnosis based on cystoscopy (20–22). Unfortunately, cystoscopy is an invasive examination with high cost and low patient acceptance, which makes it difficult to be widely used in clinical practice. Therefore, searching for non-invasive bladder cancer tumor markers with high sensitivity and high specificity has become the focus of current clinical attention.
Liquid biopsy is non-invasive, cost-effective, and convenient compared with traditional bladder cancer detection methods, and its sensitivity for tumor diagnosis is higher (23, 24). Exosomes are spherical or ellipsoid shaped secretory vesicles containing bioactive substances, such as nucleic acids and proteins, which are actively produced and secreted by tissue cells. There are some protein molecules on their surface that can act as ligands to specifically bind to the receptor molecules on the surface of target cells. After exosomes fuse with the target cell membrane, the bioactive substances they carry are transferred to the target cell to activate or inhibit the related signaling pathways within the target cell, ultimately affecting the biological function of the target cell (25, 26). Since Trams et al. first found that a subset of tumor cell lines and normal cell lines are able to release exosomes into culture medium, tumor-derived circulating exosomes have attracted increasing attention as a promising alternative to liquid biopsy in non-invasive cancer diagnosis and monitoring treatment response (27).
LncRNAs are non-coding RNAs longer than 200 nucleotides and have limited potential to encode proteins. Relevant studies have reported that lncRNAs account for 3.36% of the total RNA content of exosomes (28). LncRNAs derived from exosomes can mediate intercellular communication signaling molecules to regulate tumor cell proliferation, invasion, and migration, and promote cancer development. Many studies have found that lncRNAs in exosomes play an important role in the early diagnosis of tumors. Zhang et al. found that exosomal Mala T-1 expression was significantly higher in patients with NSCLC than in healthy controls, and the increased exosomal Mala T-1 expression significantly correlated with TNM stage and lymphatic metastasis (29). A study by Mustafa et al. found that lncRNA p21 in the serum exosomes could be used to discriminate between patients with prostate cancer and prostatitis (30). Therefore, an increasing number of researchers believe that lncRNAs of exosomal origin are promising novel diagnostic biomarkers for bladder cancer. We performed a meta-analysis to explore the diagnostic performance of exosome-derived lncRNAs as non-invasive diagnostic biomarkers for bladder cancer.
To the best of our knowledge, this is one of the few evidence-based meta-analyses focused on elucidating the diagnostic significance of exosome-derived lncRNAs in bladder cancer. Our report validated the potential diagnostic performance of exosome-derived lncRNAs as non-invasive biomarkers for discriminating bladder cancer from controls, with a pooled sensitivity of 74% and pooled specificity of 76%. In addition, a DOR value of 10 (8–12) indicated that patients with a positive test for lncRNA of exosomal origin had 10-fold higher odds of developing bladder cancer than controls. Because of the threshold effect between studies, the SROC curve was a better method to evaluate the pooled diagnostic accuracy for discrimination between cases and controls. The AUC of 0.83 suggested high diagnostic accuracy.
Heterogeneity is a potential concern, which affects the interpretation of pooled effects and meta-analysis results. Although we developed strict inclusion and exclusion criteria to identify eligible studies, heterogeneity still existed due to the presence of potential confounding factors. We performed meta-analysis and subgroup analyses to explore potential sources of heterogeneity. Our results showed that factors such as specimen type, number of lncRNAs, lncRNA expression form, and reference gene type all influenced pooled sensitivity and specificity, suggesting that the above factors may be major sources of heterogeneity in lncRNA detection in bladder cancer. The specimen type appeared to contribute to the heterogeneity of circulating lncRNA detection, with a pooled AUC for the diagnostic performance of blood-derived exosomal lncRNA of 0.86. This may be due to the fact that urine contains more deposits, and the heterogeneous cellular components may affect its use as a reliable biomarker. A diagnostic model consisting of multiple lncRNAs appeared to have better diagnostic performance, which may be because the development of bladder cancer was itself the result of a complex multistage process of genomic and epigenetic abnormalities, which should also be affected by multiple lncRNAs. Reference gene type appeared to be another possible source of heterogeneity, with a pooled AUC of 0.84 for GAPDH as the reference gene and 0.81 for a non-GAPDH reference gene. This may be attributed to the different sample situations and test conditions between studies. As reference gene type differences and mechanisms are not well understood, extensive investigations are warranted to further confirm whether such differences are truly present. In terms of lncRNA expression, up-regulated lncRNAs were less predictive than those which non up-regulated. It is worth mentioning that UCA1 has been cloned from human bladder TCC cell line. Xue et al. found that the expression levels of UCA1 in the exosomes of bladder cancer patients were higher. Yazarlou et al. further investigated the expression of two splicing variants of UCA1, UCA1-201 and UCA1-203, in tumor patients. The results showed up-regulation of UCA1-203 while down-regulation of UCA1-201 in TCC samples compared with normal subjects. Such different pattern of expression of these two variants might imply specific roles for them which should be assessed in future studies.
Wang et al. performed an impressive meta-analysis of the diagnostic value of lncRNAs in bladder cancer (31). They reported a pooled sensitivity of 0.76 (0.72 - 0.80) and pooled specificity of 0.77 (0.73 - 0.81) for lncRNAs of exosomal origin in their subgroup analysis. These results support our conclusions. However, in terms of heterogeneity analysis, they did not perform subgroup analysis of reference gene categories and expression forms (up - or downregulated) of lncRNAs in bladder cancer patients. In addition, we found that lncRNAs were stably present in exosomes from serum or urine in each study. The mechanism may be that the membrane structure of exosomes could act as a protective membrane to protect these molecules from degradation. Given the stability of lncRNAs in exosomes and the simplicity and repeatability of detection of serum and urine samples, we deduced that exosome-derived lncRNAs may also be used as clinical biomarkers for other diseases and might be a desirable material for basic research and clinical testing.
The present meta-analysis has several limitations that need to be addressed. First, the significant statistical heterogeneity in our analysis with respect to specimen type, number of lncRNAs, lncRNA expression form, and reference gene type will have an inevitable influence on the results. Second, a good biological marker needs to be able to distinguish cancer from other diseases with similar symptoms. However, most studies included in this meta-analysis simply tried to distinguish bladder cancer patients from healthy populations, without involving bladder cancer patients with similar symptoms. In addition, the populations included in this meta-analysis were mostly Asian with a smaller Caucasian population, with no relevant data on African populations. Finally, although all studies provided information on tumor stage and grade, most did not provide cut-off values; further comprehensive studies are needed to address this issue.
Conclusion
Our comprehensive analysis confirmed that exosome-derived lncRNAs might serve as potential clinical biomarkers for bladder cancer diagnosis with high AUC values. However, their clinical value still needs to be tested in further comprehensive prospective studies.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author Contributions
All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from Changzhou Sci & Tech program (CJ20190100), the Young Scientists Foundation of Changzhou No. 2 People’s Hospital (YJRC202039; 2019K008), the Innovation team funding (XK201803), the Young Talent Development Plan of Changzhou Health Commission (No. CZQM2020065), the Faculty-level subject funding (YJXK202013), the Top Talent Project (RC201620), and the National Natural Science Foundation (No. 81902565).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Witjes JA, Bruins HM, Cathomas R, Comperat EM, Cowan NC, Gakis G, et al. European Association of Urology Guidelines on Muscle-Invasive and Metastatic Bladder Cancer: Summary of the 2020 Guidelines. Eur Urol (2021) 79(1):82–104. doi: 10.1016/j.eururo.2020.03.055
2. Karl A, Grimm T, Jokisch F, Gaisa NT, Stief CG. [Non-Muscle Invasive Bladder Cancer: Current Aspects of Diagnostics, Local Therapy Options and the Update of the 2016 WHO Classification]. Urologe A (2016) 55(9):1247–58. doi: 10.1007/s00120-016-0192-4
3. Kamat AM, Hegarty PK, Gee JR, Clark PE, Svatek RS, Hegarty N, et al. ICUD-EAU International Consultation on Bladder Cancer 2012: Screening, Diagnosis, and Molecular Markers. Eur Urol (2013) 63(1):4–15. doi: 10.1016/j.eururo.2012.09.057
4. Sarko DK, McKinney CE. Exosomes: Origins and Therapeutic Potential for Neurodegenerative Disease. Front Neurosci (2017) 11:82. doi: 10.3389/fnins.2017.00082
5. Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res (2017) 77(15):3965–81. doi: 10.1158/0008-5472.CAN-16-2634
6. Chen C, Zheng H, Luo Y, Kong Y, An M, Li Y, et al. Sumoylation Promotes Extracellular Vesicle-Mediated Transmission of LncRNA ELNAT1 and Lymph Node Metastasis in Bladder Cancer. J Clin Invest (2021) 131(8). doi: 10.1172/JCI146431
7. Abbastabar M, Sarfi M, Golestani A, Karimi A, Pourmand G, Khalili E. Tumor-Derived Urinary Exosomal Long Non-Coding RNAs as Diagnostic Biomarkers for Bladder Cancer. EXCLI J (2020) 19:301–10. doi: 10.17179/excli2019-1683
8. Leeflang MM. Systematic Reviews and Meta-Analyses of Diagnostic Test Accuracy. Clin Microbiol Infect (2014) 20(2):105–13. doi: 10.1111/1469-0691.12474
9. Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann Intern Med (2011) 155(8):529–36. doi: 10.7326/0003-4819-155-8-201110180-00009
10. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring Inconsistency in Meta-Analyses. BMJ (2003) 327(7414):557–60. doi: 10.1136/bmj.327.7414.557
11. Deeks JJ, Macaskill P, Irwig L. The Performance of Tests of Publication Bias and Other Sample Size Effects in Systematic Reviews of Diagnostic Test Accuracy was Assessed. J Clin Epidemiol (2005) 58(9):882–93. doi: 10.1016/j.jclinepi.2005.01.016
12. Xue M, Chen W, Xiang A, Wang R, Chen H, Pan J, et al. Hypoxic Exosomes Facilitate Bladder Tumor Growth and Development Through Transferring Long Non-Coding RNA-UCA1. Mol Cancer (2017) 16(1). doi: 10.1186/s12943-017-0714-8
13. Zheng R, Du M, Wang X, Xu W, Liang J, Wang W, et al. Exosome-Transmitted Long Non-Coding RNA PTENP1 Suppresses Bladder Cancer Progression. Mol Cancer (2018) 17(1). doi: 10.1186/s12943-018-0880-3
14. Zhan Y, Du L, Wang L, Jiang X, Zhang S, Li J, et al. Expression Signatures of Exosomal Long Non-Coding RNAs in Urine Serve as Novel Non-Invasive Biomarkers for Diagnosis and Recurrence Prediction of Bladder Cancer 11 Medical and Health Sciences 1112 Oncology and Carcinogenesis. Mol Cancer (2018) 17(1). doi: 10.1186/s12943-018-0893-y
15. Wang J, Yang K, Yuan W, Gao Z. Determination of Serum Exosomal H19 as a Noninvasive Biomarker for Bladder Cancer Diagnosis and Prognosis. Med Sci Monitor (2018) 24:9307–16. doi: 10.12659/MSM.912018
16. Yazarlou F, Modarressi MH, Mowla SJ, Oskooei VK, Motevaseli E, Tooli LF, et al. Urinary Exosomal Expression of Long Non-Coding RNAs as Diagnostic Marker in Bladder Cancer. Cancer Manage Res (2018) 10:6357–65. doi: 10.2147/CMAR.S186108
17. Zhang S, Du L, Wang L, Jiang X, Zhan Y, Li J, et al. Evaluation of Serum Exosomal LncRNA-Based Biomarker Panel for Diagnosis and Recurrence Prediction of Bladder Cancer. J Cell Mol Med (2019) 23(2):1396–405. doi: 10.1111/jcmm.14042
18. Huang H, Du J, Jin B, Pang L, Duan N, Huang C, et al. Combination of Urine Exosomal mRNAs and LncRNAs as Novel Diagnostic Biomarkers for Bladder Cancer. Front Oncol (2021) 11:667212. doi: 10.3389/fonc.2021.667212
19. Sarfi M, Abbastabar M, Khalili E. Increased Expression of Urinary Exosomal LncRNA TUG-1 in Early Bladder Cancer. Gene Rep (2021) 22. doi: 10.1016/j.genrep.2020.101010
20. Noel N, Couteau J, Maillet G, Gobet F, D’Aloisio F, Minier C, et al. TP53 and FGFR3 Gene Mutation Assessment in Urine: Pilot Study for Bladder Cancer Diagnosis. Anticancer Res (2015) 35(9):4915–21.
21. Onal B, Han U, Yilmaz S, Koybasioglu F, Altug U. The Use of Urinary Nuclear Matrix Protein 22 (NMP22) as a Diagnostic Adjunct to Urine Cytology for Monitoring of Recurrent Bladder Cancer–Institutional Experience and Review. Diagn Cytopathol (2015) 43(4):307–14. doi: 10.1002/dc.23239
22. Puntoni M, Petrera M, Campora S, Garrone E, Defferrari C, Torrisi R, et al. Prognostic Significance of VEGF After Twenty-Year Follow-Up in a Randomized Trial of Fenretinide in Non-Muscle-Invasive Bladder Cancer. Cancer Prev Res (Phila) (2016) 9(6):437–44. doi: 10.1158/1940-6207.CAPR-15-0345
23. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid Biopsy: Monitoring Cancer-Genetics in the Blood. Nat Rev Clin Oncol (2013) 10(8):472–84. doi: 10.1038/nrclinonc.2013.110
24. Di Meo A, Bartlett J, Cheng Y, Pasic MD, Yousef GM. Liquid Biopsy: A Step Forward Towards Precision Medicine in Urologic Malignancies. Mol Cancer (2017) 16(1). doi: 10.1186/s12943-017-0644-5
25. Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon T, Kim BC, et al. Exosome RNA Unshielding Couples Stromal Activation to Pattern Recognition Receptor Signaling in Cancer. Cell (2017) 170(2):352–66.e13. doi: 10.1016/j.cell.2017.06.031
26. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-Mediated Transfer of Mrnas and MicroRNAs is a Novel Mechanism of Genetic Exchange Between Cells. Nat Cell Biol (2007) 9(6):654–9. doi: 10.1038/ncb1596
27. Thery C. Exosomes: Secreted Vesicles and Intercellular Communications. F1000 Biol Rep (2011) 3:15. doi: 10.3410/B3-15
28. Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, et al. Characterization of Human Plasma-Derived Exosomal RNAs by Deep Sequencing. BMC Genomics (2013) 14:319. doi: 10.1186/1471-2164-14-319
29. Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, et al. Serum Long non Coding RNA MALAT-1 Protected by Exosomes is Up-Regulated and Promotes Cell Proliferation and Migration in Non-Small Cell Lung Cancer. Biochem Biophys Res Commun (2017) 490(2):406–14. doi: 10.1016/j.bbrc.2017.06.055
30. Isin M, Uysaler E, Ozgur E, Koseoglu H, Sanli O, Yucel OB, et al. Exosomal LncRNA-P21 Levels may Help to Distinguish Prostate Cancer From Benign Disease. Front Genet (2015) 6:168. doi: 10.3389/fgene.2015.00168
Keywords: exosomes, lncRNAs, bladder cancer, noninvasive tests, pooled analysis
Citation: Su Q, Wu H, Zhang Z, Lu C, Zhang L and Zuo L (2021) Exosome-Derived Long Non-Coding RNAs as Non-Invasive Biomarkers of Bladder Cancer. Front. Oncol. 11:719863. doi: 10.3389/fonc.2021.719863
Received: 07 June 2021; Accepted: 30 July 2021;
Published: 19 August 2021.
Edited by:
Francesca Sanguedolce, University of Foggia, ItalyReviewed by:
Chunlin Ou, Central South University, ChinaMahdieh Razmi, Iran University of Medical Sciences, Iran
Copyright © 2021 Su, Wu, Zhang, Lu, Zhang and Zuo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Zuo, enVvbGkxOTc4QGhvdG1haWwuY29t; Lifeng Zhang, bmotbGlra3lAMTYzLmNvbQ==
 Quanxin Su1,2
Quanxin Su1,2 Li Zuo
Li Zuo