- 1Xiangya School of Medicine, Central South University, Changsha, China
- 2School of Life Sciences, Central South University, Changsha, China
- 3Hunan Key Laboratory of Animal Models for Human Diseases, Central South University, Changsha, China
As microtubule-based structures, primary cilia are typically present on the cells during the G0 or G1-S/G2 phase of the cell cycle and are closely related to the development of the central nervous system. The presence or absence of this special organelle may regulate the central nervous system tumorigenesis (e.g., glioblastoma) and several degenerative diseases. Additionally, the development of primary cilia can be regulated by several pathways. Conversely, primary cilia are able to regulate a few signaling transduction pathways. Therefore, development of the central nervous system tumors in conjunction with abnormal cilia can be regulated by up- or downregulation of the pathways related to cilia and ciliogenesis. Here, we review some pathways related to ciliogenesis and tumorigenesis, aiming to provide a potential target for developing new therapies at genetic and molecular levels.
Introduction
Glioblastoma
GBM is the most common and aggressive primary brain tumor in adults, accounting for 45.6% of primary malignant brain tumors, despite its low annual incidence of 3.1 per 100,000 compared with that of cancers deriving from other organs such as the breast and lung, with increased incidence and decreased survival rate with age (1). Majority of GBM patients suffer recurrence within 32–36 months, and median survival is approximately 10 months (2). Necrosis and endothelial proliferation are the defining histopathologic features of primary GBM, qualifying it as the highest grade of brain tumors in the World Health Organization (WHO) classification, i.e., grade IV. Secondary GBM refers to a minority of GBMs evolving from WHO grade III or IV gliomas (3). Clinical course of tissue destruction, edema, and epilepsy contribute to the clinical symptoms including personality changes, mood disorders, sensorimotor deficits, aphasia, and epilepsy (4, 5). Previous studies have suggested that adult GBM may be derived from neural stem and progenitor cells, which are located in the subventricular zone, subcortical white matter, and dentate gyrus of the hippocampus (6, 7). Standard treatment of GBM includes surgery, radiotherapy, antiangiogenic therapy, and alkylating chemotherapy with drugs such as temozolomide (TMZ). However, therapeutic efficacy is still limited despite rapid progress in cancer therapy. Consequently, there is a need for new therapeutic strategies for GBM. Thus, finding molecules and signaling pathways associated with proliferation, malignant development, and therapeutic resistance is significantly necessary to identify new targets for developing novel approaches for GBM treatment.
O6-methylguanyl DNA methyltransferase (MGMT), a DNA repair enzyme that counteracts the toxicity of TMZ, plays an important role in the proliferation and therapeutic resistance of GBM and remains the most significant biomarker in clinical decision making, especially in GBM recurrence (8, 9). As a downstream gene of the sonic hedgehog (SHH) pathway, whose activation depends on the presence of primary cilia, the expression level of MGMT is closely correlated with primary cilia and related pathways, accounting for the close relationship between GBM and primary cilia. Additionally, aberrant cilia have been found in six of seven biopsies of GBM patients, providing further evidence that primary cilia play an important role in the genesis and development of GBM (10).
Primary Cilia
Cilia are microtubule-based organelles extending from the surface of cells, and they can be classified into motile and non-motile (primary) cilia (11). The latter are confirmed as highly conserved sensorial antenna that may detect several types of stimulations from the environment, growth factors, neurotransmitters, and even light (e.g., photoreceptor cells). This organelle is 1~10 μm long and 0.2~0.3 μm wide, and its core is an axoneme characterized as 9 + 0 ring structure composed of nine pairs of microtubule doublets surrounded by a bilayer lipid membrane in continuity with the plasma membrane (12). The cilia possess specific proteins and lipids that are required to maintain their bioactivity. The continuous region sinks in the cell body to form a structure called “ciliary pit” or “ciliary pocket,” which is closely related to endocytosis and formation of endosomes (13). Primary cilia usually exist in quiescent cells as basal bodies, which are modified centrioles, to initiate ciliogenesis, and they may be utilized to organize the mitotic spindle in cell division (11, 14). When the mitosis is complete, the mother centriole tends to transform into two distinct centrioles, but only one mature centriole is capable of nucleating a primary cilium at its distal end, and it carries fibrous appendages and satellites (15). To organize and maintain the organized cilia, the intraflagellar transport (IFT) is developed to transport the components used to form the axoneme to the ciliary tip via a bidirectional microtubule-based transportation system between the ciliary tip and base, which is operated by kinesin and dynein (16). Additionally, the primary cilia regulate the proliferation of other ciliated cells living in the microenvironment adjacent to the ciliated cell via ciliary tip excision and release of ciliary vesicles (10).
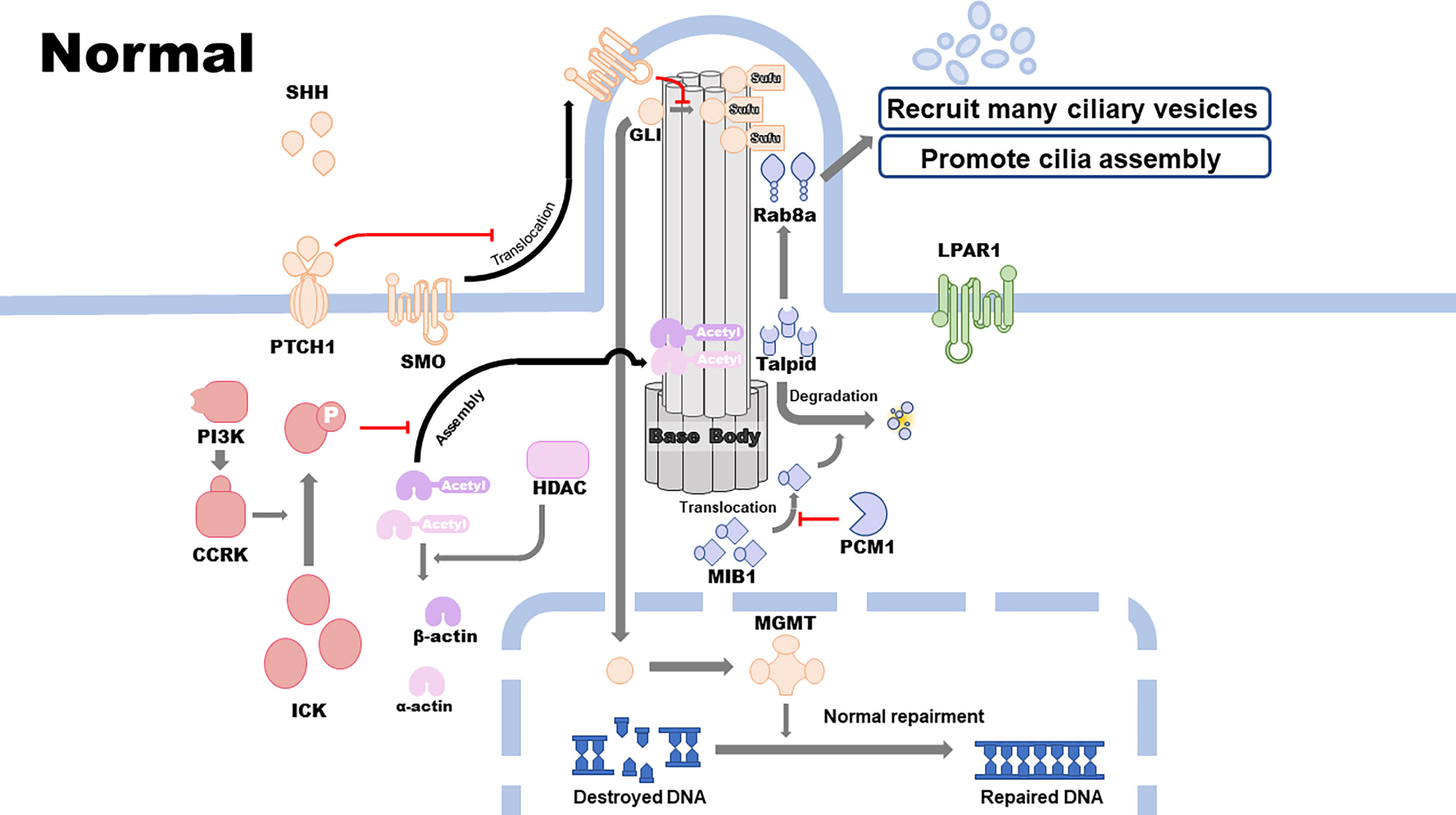
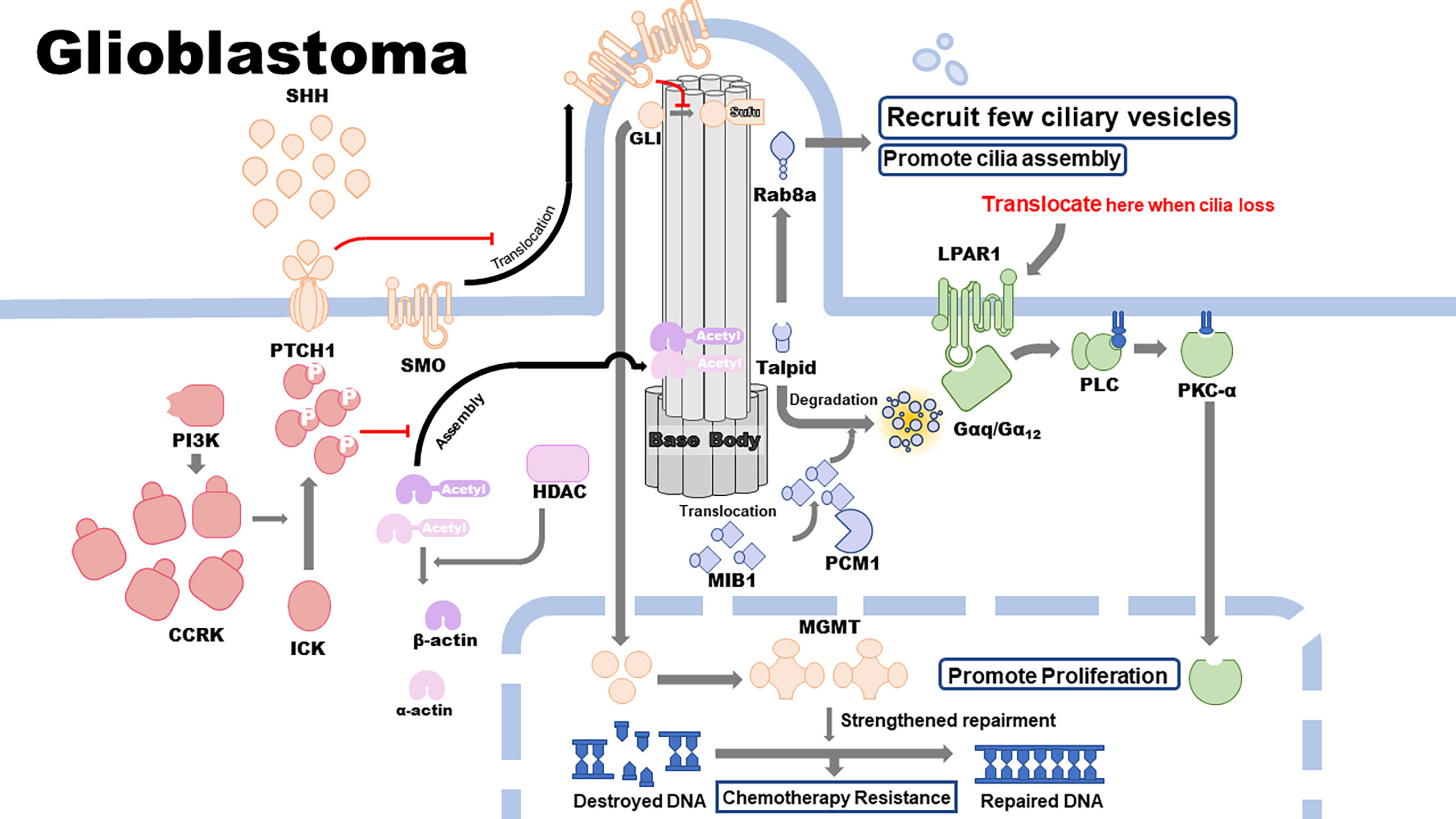
Primary cilia are highly associated with GBM development and therapeutic resistance. Cilia-related signaling pathways, including the SHH, cell cycle–related kinase (CCRK), and histone deacetylase 6 (HDAC6) pathways, are closely correlated with the proliferation, malignant development, and therapeutic resistance of GBM. Other cilium-related pathways, including the lysophosphatidic acid receptor 1 (LPAR1) and pericentriolar material 1 (PCM1) pathways, inhibit the proliferation and development of GBM cell lines. Epidermal growth factor receptor (EGFR), alpha-type platelet-derived growth factor receptor (PDGFRɑ), MGMT, and isocitrate dehydrogenase 1 (IDH1) pathways promote GBM therapeutic resistance, which is associated with or modulated by the assembly and disassembly of primary cilia (Figures 1 and 2).
Pathways Related to Primary Cilia and GBM
The SHH Pathway
It is widely known that the SHH pathway is implicated in several physiologic and pathologic processes such as angiogenesis, development of the central nervous system (CNS), tumor proliferation, therapeutic drug resistance, and progression of some of the CNS tumors including GBM. Shh is one of the putative signaling molecules, implicated in the regulation of the CNS polarity and neural patterning (17). Shh is initially synthesized as a 45 kDa precursor protein, which is automatically cleaved into two secreted peptides, namely, 19 kDa Shh-N (amino terminus) and 26 kDa Shh-C (carboxy terminus). Shh-N has been shown to be the key location of the signal-mediating function of Shh, while Shh-C mediates its auto-proteolysis reaction. Auto-proteolysis results in the addition of a cholesterol molecule at the C-terminus of Shh-N and a palmitoyl group at the N-terminus of Shh, resulting in the formation of a dual lipid modified Shh-N, an activated form of Shh with increased inductive potency and better secretive regulation, transportation, and distribution ability, thereby enabling long-range SHH signaling (18–20).
The genes that regulate the presence or absence of cilia affect the activation of the SHH pathway, as the transduction and activation of this pathway require primary cilia as the structural base. It has been reported that Shh levels are much higher in the microenvironment of GBMs than in other tissues and that the activation of SHH signaling pathway requires the binding of Shh to the Patched (PTCH) mediated Smoothened (SMO) (PTCH-SMO) receptor complex (21), consequently resulting in a more activated SHH pathway in GBM. PTCH contains a sterol-sensing domain, which interacts with the cholesterol portion of the dual-lipid-modified Shh-N (21). PTCH is believed to suppress SMO activity directly and indirectly; however, when it binds to Shh, SMO is activated and stabilized, initiating the downstream signaling cascade by encoding membrane proteins, which are similar to G-protein-coupled receptors (22). While primary cilia exist, the signal transduction process tends to be initiated by the binding of Shh protein in the extracellular matrix to PTCH1 located on the membrane surrounding the axoneme of the primary cilia. The binding between these two molecules triggers PTCH1 removal from the primary cilia, followed by translocation of SMO from the adjacent membrane to the cilia, where it inhibits the transformation of the active form of Gli transcription factor family (GLIA) into the repressor form, consequently leading to the accumulation of GLIA. GLI, Cos2, Fu, and suppressor of fused (Sufu) form a tetrameric complex for the downstream signaling process. This complex results in the transcription activity of GLI. Cos2 is a kinesin-like protein, which is mainly a motor domain that binds to ATP and microtubules (23). Fu is a segment polarity protein, which binds to Cos2 via its carboxy-terminal and is phosphorylated in response to hedgehog signaling (24). Sufu is a negative regulator of the SHH signaling pathway. When Shh ligand is not present, Sufu directly binds to GLI and inhibits its translocation to the nucleus, resulting in the cleavage of GLI by proteasomes into transcriptional repressors, thereby preventing the downstream pathway activation. Various molecules in the Gli transcription factor family display different functions, such as regulation of the transcription of several oncogenes and tumor-suppressor genes, modulation of tumor progression, and development of resistance to some therapeutic drugs (25). For instance, MGMT, a DNA repair enzyme that can repair damage in cancer cell DNA induced by some alkylating agents (e.g., TMZ), contributes significantly to the development of resistance to therapeutic drugs specific to target cell DNA (e.g., TMZ) (26). Moreover, MGMT is considered a downstream gene whose transcript level can be regulated by GLI1 (26, 27), a member of Gli transcription factor family. When the expression of genes upstream of the SHH pathway is upregulated, the active form of GLI1 leads to its accumulation and translocation to the nucleus, and then, it upregulates the expression of MGMT at the transcription level. The increased expression of MGMT results in enhanced repair of destroyed DNA, which in turn leads to development of resistance to TMZ. A recent study revealed that acquired kinase inhibitor resistance is associated with upregulation of ciliogenesis, failure of control of cilia length, and increased activation of the SHH pathway. Manipulating cilia length and integrity via Kif7 knockdown can resensitize GBM cells and confer drug resistance (28). Collectively, the SHH pathway, the activation of which depends on primary cilia, can regulate the development of malignant phenotype and resistance towards therapeutic drugs via downstream transcription factors, which can regulate several oncogenes and tumor-suppressor genes.
The LPAR1 Signaling Pathway
Lysophosphatidic acid (LPA), which exists widely in the human body, is a small bioactive glycophospholipid extracted from membrane phospholipids with many different biological functions associated with GBM progression, such as stimulation of cell proliferation, cytoskeleton recombination, cell survival, cell differentiation, and DNA synthesis and ion transport, via binding to a variety of G-protein-coupled receptors (9). LPAR1 acts as the central signaling hub for extracellular-intracellular and intracellular-extracellular signal transduction, mediating various molecular and biological functions, including cell survival, cell migration, cell adhesion, cytoskeletal changes, cell proliferation, immune function, Ca2+ mobilization, and myelination (29, 30).
LPAR1 is associated with GBM, and it has been confirmed that the primary cilia can moderate GBM development and cell proliferation in vivo and in vitro via the LPAR1 signaling pathway (31). A recent study clearly demonstrated the interaction between cilia and LPAR1 signaling pathway in GBM cells (31). When cilia are present, LPAR1 is sequestered in the primary cilia, and compartmentalization of LPAR1 and its downstream effectors, Gɑ12 and Gɑq, which are located on the cell membrane, prevents the interaction between LPAR1 and G-protein-coupled receptor subunits, restricting its cell proliferative signaling. In contrast, loss of the primary cilia eliminates spatial barriers and results in redistribution of LPAR1 to the plasma membrane, where it binds to Gɑ12 and Gɑq and facilitates the activation of downstream pathways, promoting the proliferative function of LPA and unlocking the potential for unlimited proliferation of GBM cells, consequently maintaining a highly proliferative phenotype. LPAR1 inhibitor Ki16425 reduces cell growth only in deciliated astrocytes, thereby validating the abovementioned findings. Furthermore, Gɑq activates phospholipase C, which produces diacylglycerol (DAG) from phosphatidylinositol 4,5-bisphosphate. DAG activates classic and novel protein kinase C (PKC) (32). Valdés-Rives et al. conducted in vitro experiments and found that LPAR1 induces PKC ɑ isozyme (PKCɑ) translocation from cell membrane to the nucleus in GBM cells via G-protein-coupled receptors, promoting GBM cell growth (33). Taken together, the loss of cilia can induce LPAR1 relocation and activate downstream G-protein-coupled receptor signaling pathways, enhancing the proliferation of GBM cells. This is suggestive of a potential target site of GBM prevention and treatment.
The CCRK-ICK/MAK Pathway
CCRK overexpression is found in conjunction with loss of cilia among various CNS tumors, including GBM, and the high expression level is typically in concert with the high mortality and poor prognosis of patients with CNS tumors. Additionally, homologs of CCRK, such as LF2 in Chlamydomonas and NIH3T3 in zebrafish, can regulate the formation and length of primary cilia (34, 35). Collectively, these findings suggest that CCRK may play a key role in the human primary cilia formation, subsequently contributing to the occurrence and development of several tumors. Yang et al. indicated that overexpression of CCRK, whose upstream regulator is phosphoinositide 3-kinase, can induce loss of cilia through its substrate ICK/MAK, consequently promoting the proliferation of GBM cells (36). Overexpression of CCRK leads to a loss of cilia induced by serum starvation via phosphorylation activation of ICK, resulting in a reduced ability to inhibit cell cycle re-entry of cilia and deregulation of the cell cycle progression, as determined via EdU incorporation (37). The majority of cilia loss is induced by the activation of ICK, which is caused by the kinase activity of CCRK, and the rest is caused by the structure of non-phosphorylated ICK. Therefore, CCRK is considered as an oncogene owing to its ability to induce cell cycle progression and increase the proliferative capacity of GBM cells. Accordingly, depletion of CCRK and ICK can inhibit the proliferation of cancer cells by stabilizing the structure and function of the primary cilia in a KIF3a-dependent manner (35). In addition, the primary cilia–dependent SHH pathway is also regulated by CCRK, as CCRK possesses the ability to inhibit ciliogenesis. As an early event in the activation of the SHH pathway, translocation of Gli3 is significantly upregulated following depletion of CCRK (36).
The HDAC6-Related Pathway
As an epigenetic modification, acetylation induced by histone acetyltransferase (HAT) can reduce the expression level of target genes. HDAC6 was initially found to be a deacetylase that can remove an acetyl group from the histone and, subsequently, restore the expression level of genes downregulated by acetylation (38). Therefore, the balance between HAT and HDAC6 levels plays an important role in the maintenance of normal expression of genes encoding structural or non-structural proteins, including some oncogenes and tumor-suppressor genes, and consequently, this balance maintains the homeostasis in the human body and prevents the occurrence and development of some tumors. When the balance between HAT and HDAC6 levels is dysregulated, the probability of tumor occurrence significantly increases due to an imbalance in the expression of oncogenes and tumor-suppressor genes. Apart from histone deacetylation, HDAC6 can also deacetylate some proteins located in the plasma or on the cell membrane, such as acetylated α-tubulin (27). As the assembly of the primary cilia requires acetylated α-tubulin, HDAC6 may deacetylate and destroy the protein component needed to form the axoneme of the primary cilia. Therefore, high expression of HDAC6 tends to appear in concert with the loss of the primary cilia in GBM biopsies and cultured cell lines. Taken together, HDAC6 has been gradually considered as an oncogene as its overexpression may promote malignancy and enhanced proliferation via inhibition of assembly of the primary cilia. Thus, depletion of HDAC6 levels, which results in promotion of ciliogenensis as well as cell apoptosis induced by TMZ and reversal of malignancy, is becoming a potential strategy in developing new therapies for GBM. In addition to the reversion of malignancy induced by HDAC6 depletion, alteration in the sensitivity of GBM cells to TMZ is triggered by not only HDAC6 depletion but also deactivation of the SHH pathway due to the loss of cilia, leading to a reduction in the MGMT level and DNA repairing ability (39, 40).
The PCM1-Regulated Pathway
PCM1 is an essential pericentriolar protein that plays a significant role in cell division and survival, especially in centrosome stability, microtubule and actin organization, and centriolar duplication prior to mitosis and ciliogenesis, in normal and malignant cells, such as GBM cells (41). Centriolar satellites, majorly composed of PCM1, are non-membrane cytoplasmic granules that are located around the centrosome and play crucial roles in transporting centrosome proteins from the cytoplasm to the centrosome during centrosome assembly or ciliogenesis. Therefore, diseases, such as ciliopathies, may possibly occur when centriolar satellites become deficient (42). In addition, PCM1 has been shown to bind to several centrosome proteins to ensure that they are accurately located (43, 44). Thus, PCM1, centriolar satellites, and the centrosome are significantly implicated, closely correlated, and function together in cell cycle dynamics and ciliogenesis in GBM cells.
As a key component of centriolar satellites and the functional activity of the centrosome, PCM1 plays a vital role in GBM cell mitosis, cell cycle, cell proliferation, and sensitivity to standard-of-care therapeutic agent TMZ via regulation of ciliogenesis (41). PCM1 restricts Mindbomb 1, an E3 ligase associated with Notch signaling, to satellites via tethering, and thus, prevents its translocation to centrioles, which would otherwise promote Talpid3 destabilization through polyubiquitination, failure to recruit ciliary vesicles associated with Talpid3-bianding protein Rab8a, or inhibition of cilium assembly (45, 46). PCM1-associated cilia formation thus inhibits GBM progression and cell proliferation. Furthermore, studies have found that sorting nexin 17, a P-selectin binding protein, can recruit and stabilize ubiquitin-specific protease 9X, a deubiquitinating enzyme that is able to regulate the integrity of centriolar satellites in mitotic cells, antagonize Mindbomb 1-induced ubiquitination and degradation of PCM1 via binding to PCM1, and inhibit PCM1 ubiquitination during ciliogenesis (47, 48). In addition, Plk4, a conserved protein kinase that regulates the centriole duplication cycle, is required for PCM1 phosphorylation, followed by adequate spatial distribution and organization of centriolar satellites, which facilitate ciliogenesis (49, 50). By sequestering proteins in the centriolar satellite compartment, PCM1 promotes ciliogenesis, thereby inhibiting GBM progression.
Pathways Related to Cilia and Therapy Resistance of GBM Subtypes
Based on genomic and transcriptomic data, the Cancer Genome Atlas Research Network (2008) has provided a detailed view of the genomic alterations and affected signaling pathways and stratified GBM into four subtypes: classical, mesenchymal, proneural, and neural GBM (51). Each subtype displays different genomic features that affect the resistance mechanisms associated with or modulated by the primary cilia (Table 1).
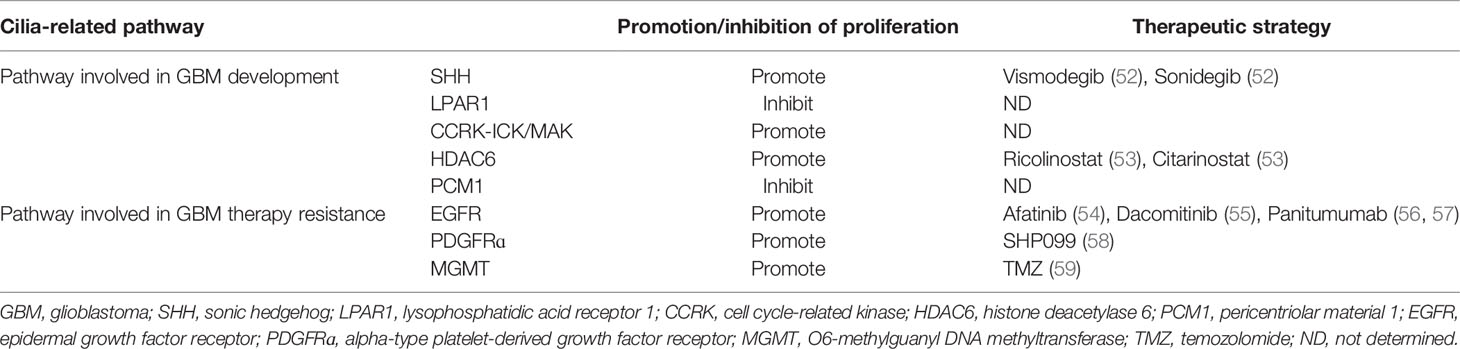
Table 1 Therapeutic strategy and cilia-related pathways associated with glioblastoma development and therapy resistance.
Classical GBM
Classical GBM is defined by EGFR amplification and CDKN2 depletion with a distinct lack of additional abnormalities in TP53, NF1, PDGFRɑ, or IDH1 genes (60). EGFR extensively regulates cellular processes during cell development, and it is critically associated with tumorigenesis and tumor cell invasion (61). EGFR has been found to be localized on the primary cilia in astrocytes and neuroblasts (62). EGFR activates phosphatidylinositol (3,4,5)-trisphosphate, partly through phosphoinositide 3-kinase-induced phosphorylation of phosphatidylinositol 4,5-bisphosphate into phosphatidylinositol (3,4,5)-trisphosphate and the hydrolysis of phosphatidylinositol 4,5-bisphosphate by activation of phospholipase C-gamma 2 (63). Following this, phosphatidylinositol (3,4,5)-trisphosphate activates the Akt signaling pathway and regulates the cell cycle (64). Amplified EGFR expression in classical GBM can sustain EGFR signaling, Akt activity, and DNA repair mechanisms via the primary cilia and, thus, promote resistance towards EGFR-targeted therapy (65). In addition, classical GBM has the highest MGMT methylation rate among the four subtypes, which confers a strong ability of repairing destroyed DNA and, hence, resistance to TMZ therapy (66).
Mesenchymal GBM
Mesenchymal GBM is characterized by a high frequency of NF1 mutation/deletion, low NF1 mRNA expression, and PTEN mutation (60). Studies have shown that a loss of NF1 expression results in resistance to endocrine therapy, through both estrogen receptor-dependent and -independent mechanisms in advanced breast cancer (67). However, associations between NF1 activity and cilia-related therapeutic resistance in mesenchymal GBM remain unclear.
Proneural GBM
Proneural GBM is characterized by amplification of PDGFRɑ expression and point mutations in IDH1 and TP53 genes (60). PDGFRα localizes to the primary cilia in astrocytes and neuroblasts. PDGFRα receptor in the primary cilium leads to the onset of a number of downstream signaling pathways within the cilium, including the Akt and Mek1/2-Erk1/2 pathways, to regulate the cell cycle and directional cell migration (64). IDH1 mutations have previously been associated with secondary GBM (68). Hence, amplification of PDGFRα expression can sustain PDGFRα signaling via the primary cilia and enhance resistance of proneural GBM to therapy.
Neural GBM
Neural GBM is genetically associated with neural, astrocytic, and oligodendrocytic gene signatures. Their expression patterns are similar to those in the normal brain tissue, which can be differentiated based on the morphology using light microscopy (60).
Conclusion
GBM is the most common and malignant brain tumor in adults with poor prognoses, short recurrence period, and low survival rate. Common cancer therapies, such as surgical resection, radiotherapy, and chemotherapy, are widely applied for treating GBM, but the therapeutic effect remains to be limited. Therefore, there is a need to explore new therapeutic strategies to combat GBM. Studies have shown that the primary cilia may play dual roles in GBM progression, which should be highly considered for devising potential methods of GBM treatment. Thus, cilia-elated signaling pathways could be novel targets for GBM treatment.
In this review, we summarized five primary cilium-related signaling pathways moderating the development and therapeutic resistance of GBM. The SHH pathway promotes DNA repair and TMZ resistance. Drugs targeting the SHH pathway have been developed, of which vismodegib and sonidegib have been approved by the Food and Drug Administration for treating basal cell carcinoma (52). Thus, it is necessary to develop SHH inhibitors for GBM treatment, which is possible. The CCRK-ICK/MAK pathway induces cilium loss and promotes GBM development. Drugs targeting CCRK are being developed, but no CCRK-specific inhibitor is publicly available due to lacking information on the 3D structure of the CCRK protein (69). Accordingly, research on the 3D structure of the CCRK protein is essential to develop CCRK inhibitors for GBM therapy. HDAC6 inhibits cilia formation, and HDAC6 inhibitors ricolinostat and citarinostat are currently being tested for cancer treatment (53). Therefore, it is necessary to test the efficacy of these HDAC6 inhibitors and conduct clinical trials for use in GBM treatment. The PCM1 pathway facilitates cilia formation and inhibits GBM progression, while the LPAR1 pathway promotes GBM cell proliferation. Considering that no drugs targeting these two pathways have been reported, it is possible and suggestive to develop new GBM therapies targeting the PCM1 and LPAR1 pathways.
In conclusion, further research is required to obtain a deeper understanding of the potential involvement of and the signaling pathways related to the primary cilia in regulating cell proliferation and the impact on GBM development and its resistance to therapeutic drugs. We expect major progress in the following years, which may allow for designing cilia-based and molecular targeted therapies to offer new treatment strategies against GBM.
Author Contributions
ML wrote the manuscript and took charge of the figures. JZ researched the subject and wrote the manuscript. HZ was responsible for the figures. Embellishing the article was conducted by RX. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Science and Technology Major Project of the Ministry of Science and Technology of China (2017ZX10103005-006), the National Natural Science Foundation of China (81970403).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Dr. Liangliang Fan (School of life sciences, Central South University; Hunan Key Laboratory of Animal Models for Human Diseases).
References
1. Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2007-2011. Neuro Oncol (2014) 16 Suppl 4(Suppl 4):iv1–63. doi: 10.1093/neuonc/nou223
2. Loskutov Y, Pugacheva EN. Targeting Primary Cilia - Associated Signaling in Glioblastoma: Guided Approach for Drug Development. Oncoscience (2019) 6(1-2):289–90. doi: 10.18632/oncoscience.475
3. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol (2007) 114(2):97–109. doi: 10.1007/s00401-007-0243-4
4. Yuile P, Dent O, Cook R, Biggs M, Little N. Survival of Glioblastoma Patients Related to Presenting Symptoms, Brain Site and Treatment Variables. J Clin Neurosci (2006) 13(7):747–51. doi: 10.1016/j.jocn.2005.10.011
5. Chaichana KL, Parker SL, Olivi A, Quiñones-Hinojosa A. Long-Term Seizure Outcomes in Adult Patients Undergoing Primary Resection of Malignant Brain Astrocytomas. Clinical Article. J Neurosurg (2009) 111(2):282–92. doi: 10.3171/2009.2.Jns081132
6. Sanai N, Nguyen T, Ihrie RA, Mirzadeh Z, Tsai HH, Wong M, et al. Corridors of Migrating Neurons in the Human Brain and Their Decline During Infancy. Nature (2011) 478(7369):382–6. doi: 10.1038/nature10487
7. Spalding KL, Bergmann O, Alkass K, Bernard S, Salehpour M, Huttner HB, et al. Dynamics of Hippocampal Neurogenesis in Adult Humans. Cell (2013) 153(6):1219–27. doi: 10.1016/j.cell.2013.05.002
8. Weller M, van den Bent M, Hopkins K, Tonn JC, Stupp R, Falini A, et al. EANO Guideline for the Diagnosis and Treatment of Anaplastic Gliomas and Glioblastoma. Lancet Oncol (2014) 15(9):e395–403. doi: 10.1016/s1470-2045(14)70011-7
9. Weller M, Tabatabai G, Kästner B, Felsberg J, Steinbach JP, Wick A, et al. MGMT Promoter Methylation Is a Strong Prognostic Biomarker for Benefit From Dose-Intensified Temozolomide Rechallenge in Progressive Glioblastoma: The DIRECTOR Trial. Clin Cancer Res (2015) 21(9):2057–64. doi: 10.1158/1078-0432.Ccr-14-2737
10. Moser JJ, Fritzler MJ, Rattner JB. Ultrastructural Characterization of Primary Cilia in Pathologically Characterized Human Glioblastoma Multiforme (GBM) Tumors. BMC Clin Pathol (2014) 14:40. doi: 10.1186/1472-6890-14-40
11. Youn YH, Han YG. Primary Cilia in Brain Development and Diseases. Am J Pathol (2018) 188(1):11–22. doi: 10.1016/j.ajpath.2017.08.031
12. Álvarez-Satta M, Matheu A. Primary Cilium and Glioblastoma. Ther Adv Med Oncol (2018) 10:1758835918801169. doi: 10.1177/1758835918801169
13. Moser JJ, Fritzler MJ, Rattner JB. Primary Ciliogenesis Defects Are Associated With Human Astrocytoma/Glioblastoma Cells. BMC Cancer (2009) 9:448. doi: 10.1186/1471-2407-9-448
14. Hoang-Minh LB, Dutra-Clarke M, Breunig JJ, Sarkisian MR. Glioma Cell Proliferation is Enhanced in the Presence of Tumor-Derived Cilia Vesicles. Cilia (2018) 7:6. doi: 10.1186/s13630-018-0060-5
15. Lange BM, Gull K. A Molecular Marker for Centriole Maturation in the Mammalian Cell Cycle. J Cell Biol (1995) 130(4):919–27. doi: 10.1083/jcb.130.4.919
16. Álvarez-Satta M, Moreno-Cugnon L, Matheu A. Primary Cilium and Brain Aging: Role in Neural Stem Cells, Neurodegenerative Diseases and Glioblastoma. Ageing Res Rev (2019) 52:53–63. doi: 10.1016/j.arr.2019.04.004
17. Machold R, Fishell G. Hedgehog Patterns Midbrain ARChitecture. Trends Neurosci (2002) 25(1):10–1. doi: 10.1016/s0166-2236(00)01982-2
18. Chamoun Z, Mann RK, Nellen D, von Kessler DP, Bellotto M, Beachy PA, et al. Skinny Hedgehog, an Acyltransferase Required for Palmitoylation and Activity of the Hedgehog Signal. Science (2001) 293(5537):2080–4. doi: 10.1126/science.1064437
19. Lewis PM, Dunn MP, McMahon JA, Logan M, Martin JF, St-Jacques B, et al. Cholesterol Modification of Sonic Hedgehog is Required for Long-Range Signaling Activity and Effective Modulation of Signaling by Ptc1. Cell (2001) 105(5):599–612. doi: 10.1016/s0092-8674(01)00369-5
20. Zeng X, Goetz JA, Suber LM, Scott WJ Jr., Schreiner CM, Robbins DJ. A Freely Diffusible Form of Sonic Hedgehog Mediates Long-Range Signalling. Nature (2001) 411(6838):716–20. doi: 10.1038/35079648
21. Choudhry Z, Rikani AA, Choudhry AM, Tariq S, Zakaria F, Asghar MW, et al. Sonic Hedgehog Signalling Pathway: A Complex Network. Ann Neurosci (2014) 21(1):28–31. doi: 10.5214/ans.0972.7531.210109
22. Taipale J, Cooper MK, Maiti T, Beachy PA. Patched Acts Catalytically to Suppress the Activity of Smoothened. Nature (2002) 418(6900):892–7. doi: 10.1038/nature00989
23. Kamal A, Goldstein LS. Principles of Cargo Attachment to Cytoplasmic Motor Proteins. Curr Opin Cell Biol (2002) 14(1):63–8. doi: 10.1016/s0955-0674(01)00295-2
24. Nybakken K, Perrimon N. Hedgehog Signal Transduction: Recent Findings. Curr Opin Genet Dev (2002) 12(5):503–11. doi: 10.1016/s0959-437x(02)00333-7
25. Qiu ZK, Shen D, Chen YS, Yang QY, Guo CC, Feng BH, et al. Enhanced MGMT Expression Contributes to Temozolomide Resistance in Glioma Stem-Like Cells. Chin J Cancer (2014) 33(2):115–22. doi: 10.5732/cjc.012.10236
26. Yu W, Zhang L, Wei Q, Shao A. O(6)-Methylguanine-DNA Methyltransferase (MGMT): Challenges and New Opportunities in Glioma Chemotherapy. Front Oncol (2019) 9:1547. doi: 10.3389/fonc.2019.01547
27. Urdiciain A, Erausquin E, Meléndez B, Rey JA, Idoate MA, Castresana JS. Tubastatin A, an Inhibitor of HDAC6, Enhances Temozolomide−Induced Apoptosis and Reverses the Malignant Phenotype of Glioblastoma Cells. Int J Oncol (2019) 54(5):1797–808. doi: 10.3892/ijo.2019.4739
28. Jenks AD, Vyse S, Wong JP, Kostaras E, Keller D, Burgoyne T, et al. Primary Cilia Mediate Diverse Kinase Inhibitor Resistance Mechanisms in Cancer. Cell Rep (2018) 23(10):3042–55. doi: 10.1016/j.celrep.2018.05.016
29. Kihara Y, Maceyka M, Spiegel S, Chun J. Lysophospholipid Receptor Nomenclature Review: IUPHAR Review 8. Br J Pharmacol (2014) 171(15):3575–94. doi: 10.1111/bph.12678
30. Chrencik JE, Roth CB, Terakado M, Kurata H, Omi R, Kihara Y, et al. Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1. Cell (2015) 161(7):1633–43. doi: 10.1016/j.cell.2015.06.002
31. Loskutov YV, Griffin CL, Marinak KM, Bobko A, Margaryan NV, Geldenhuys WJ, et al. LPA Signaling Is Regulated Through the Primary Cilium: A Novel Target in Glioblastoma. Oncogene (2018) 37(11):1457–71. doi: 10.1038/s41388-017-0049-3
32. Steinberg SF. Structural Basis of Protein Kinase C Isoform Function. Physiol Rev (2008) 88(4):1341–78. doi: 10.1152/physrev.00034.2007
33. Valdés-Rives SA, de la Fuente-Granada M, Velasco-Velázquez MA, González-Flores O, González-Arenas A. LPA(1) Receptor Activation Induces Pkcα Nuclear Translocation in Glioblastoma Cells. Int J Biochem Cell Biol (2019) 110:91–102. doi: 10.1016/j.biocel.2019.03.003
34. Tam LW, Wilson NF, Lefebvre PA. A CDK-Related Kinase Regulates the Length and Assembly of Flagella in Chlamydomonas. J Cell Biol (2007) 176(6):819–29. doi: 10.1083/jcb.200610022
35. Ko HW, Norman RX, Tran J, Fuller KP, Fukuda M, Eggenschwiler JT. Broad-Minded Links Cell Cycle-Related Kinase to Cilia Assembly and Hedgehog Signal Transduction. Dev Cell (2010) 18(2):237–47. doi: 10.1016/j.devcel.2009.12.014
36. Yang Y, Roine N, Mäkelä TP. CCRK Depletion Inhibits Glioblastoma Cell Proliferation in a Cilium-Dependent Manner. EMBO Rep (2013) 14(8):741–7. doi: 10.1038/embor.2013.80
37. Wang LY, Kung HJ. Male Germ Cell-Associated Kinase is Overexpressed in Prostate Cancer Cells and Causes Mitotic Defects via Deregulation of APC/Ccdh1. Oncogene (2012) 31(24):2907–18. doi: 10.1038/onc.2011.464
38. Guo P, Chen W, Li H, Li M, Li L. The Histone Acetylation Modifications of Breast Cancer and Their Therapeutic Implications. Pathol Oncol Res (2018) 24(4):807–13. doi: 10.1007/s12253-018-0433-5
39. Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-Repair Gene MGMT and the Clinical Response of Gliomas to Alkylating Agents. N Engl J Med (2000) 343(19):1350–4. doi: 10.1056/nejm200011093431901
40. Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT Gene Silencing and Benefit From Temozolomide in Glioblastoma. N Engl J Med (2005) 352(10):997–1003. doi: 10.1056/NEJMoa043331
41. Hoang-Minh LB, Deleyrolle LP, Nakamura NS, Parker AK, Martuscello RT, Reynolds BA, et al. PCM1 Depletion Inhibits Glioblastoma Cell Ciliogenesis and Increases Cell Death and Sensitivity to Temozolomide. Transl Oncol (2016) 9(5):392–402. doi: 10.1016/j.tranon.2016.08.006
42. Hori A, Toda T. Regulation of Centriolar Satellite Integrity and Its Physiology. Cell Mol Life Sci (2017) 74(2):213–29. doi: 10.1007/s00018-016-2315-x
43. Dammermann A, Merdes A. Assembly of Centrosomal Proteins and Microtubule Organization Depends on PCM-1. J Cell Biol (2002) 159(2):255–66. doi: 10.1083/jcb.200204023
44. Staples CJ, Myers KN, Beveridge RD, Patil AA, Lee AJ, Swanton C, et al. The Centriolar Satellite Protein Cep131 is Important for Genome Stability. J Cell Sci (2012) 125(Pt 20):4770–9. doi: 10.1242/jcs.104059
45. Wang L, Lee K, Malonis R, Sanchez I, Dynlacht BD. Tethering of an E3 Ligase by PCM1 Regulates the Abundance of Centrosomal KIAA0586/Talpid3 and Promotes Ciliogenesis. Elife (2016) 5:e12950. doi: 10.7554/eLife.12950
46. Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, et al. Mind Bomb is a Ubiquitin Ligase That is Essential for Efficient Activation of Notch Signaling by Delta. Dev Cell (2003) 4(1):67–82. doi: 10.1016/s1534-5807(02)00409-4
47. Wang P, Xia J, Zhang L, Zhao S, Li S, Wang H, et al. SNX17 Recruits USP9X to Antagonize MIB1-Mediated Ubiquitination and Degradation of PCM1 During Serum-Starvation-Induced Ciliogenesis. Cells (2019) 8(11):1335. doi: 10.3390/cells8111335
48. Han KJ, Wu Z, Pearson CG, Peng J, Song K, Liu CW. Deubiquitylase USP9X Maintains Centriolar Satellite Integrity by Stabilizing Pericentriolar Material 1 Protein. J Cell Sci (2019) 132(2):jcs221663. doi: 10.1242/jcs.221663
49. Hori A, Barnouin K, Snijders AP, Toda T. A Non-Canonical Function of Plk4 in Centriolar Satellite Integrity and Ciliogenesis Through PCM1 Phosphorylation. EMBO Rep (2016) 17(3):326–37. doi: 10.15252/embr.201541432
50. Nigg EA, Stearns T. The Centrosome Cycle: Centriole Biogenesis, Duplication and Inherent Asymmetries. Nat Cell Biol (2011) 13(10):1154–60. doi: 10.1038/ncb2345
51. Cancer Genome Atlas Research Network. Comprehensive Genomic Characterization Defines Human Glioblastoma Genes and Core Pathways. Nature (2008) 455(7216):1061–8. doi: 10.1038/nature07385
52. Skoda AM, Simovic D, Karin V, Kardum V, Vranic S, Serman L. The Role of the Hedgehog Signaling Pathway in Cancer: A Comprehensive Review. Bosn J Basic Med Sci (2018) 18(1):8–20. doi: 10.17305/bjbms.2018.2756
53. Pulya S, Amin SA, Adhikari N, Biswas S, Jha T, Ghosh B. HDAC6 as Privileged Target in Drug Discovery: A Perspective. Pharmacol Res (2021) 163:105274. doi: 10.1016/j.phrs.2020.105274
54. Reardon DA, Nabors LB, Mason WP, Perry JR, Shapiro W, Kavan P, et al. Phase I/randomized Phase II Study of Afatinib, an Irreversible ErbB Family Blocker, With or Without Protracted Temozolomide in Adults With Recurrent Glioblastoma. Neuro Oncol (2015) 17(3):430–9. doi: 10.1093/neuonc/nou160
55. Zahonero C, Aguilera P, Ramírez-Castillejo C, Pajares M, Bolós MV, Cantero D, et al. Preclinical Test of Dacomitinib, an Irreversible EGFR Inhibitor, Confirms Its Effectiveness for Glioblastoma. Mol Cancer Ther (2015) 14(7):1548–58. doi: 10.1158/1535-7163.Mct-14-0736
56. Gan HK, Burgess AW, Clayton AH, Scott AM. Targeting of a Conformationally Exposed, Tumor-Specific Epitope of EGFR as a Strategy for Cancer Therapy. Cancer Res (2012) 72(12):2924–30. doi: 10.1158/0008-5472.Can-11-3898
57. An Z, Aksoy O, Zheng T, Fan QW, Weiss WA. Epidermal Growth Factor Receptor and EGFRvIII in Glioblastoma: Signaling Pathways and Targeted Therapies. Oncogene (2018) 37(12):1561–75. doi: 10.1038/s41388-017-0045-7
58. Sang Y, Hou Y, Cheng R, Zheng L, Alvarez AA, Hu B, et al. Targeting Pdgfrα-Activated Glioblastoma Through Specific Inhibition of SHP-2-Mediated Signaling. Neuro Oncol (2019) 21(11):1423–35. doi: 10.1093/neuonc/noz107
59. Jiapaer S, Furuta T, Tanaka S, Kitabayashi T, Nakada M. Potential Strategies Overcoming the Temozolomide Resistance for Glioblastoma. Neurol Med Chir (Tokyo) (2018) 58(10):405–21. doi: 10.2176/nmc.ra.2018-0141
60. Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell (2010) 17(1):98–110. doi: 10.1016/j.ccr.2009.12.020
61. Takeuchi K, Ito F. EGF Receptor in Relation to Tumor Development: Molecular Basis of Responsiveness of Cancer Cells to EGFR-Targeting Tyrosine Kinase Inhibitors. FEBS J (2010) 277(2):316–26. doi: 10.1111/j.1742-4658.2009.07450.x
62. Danilov AI, Gomes-Leal W, Ahlenius H, Kokaia Z, Carlemalm E, Lindvall O. Ultrastructural and Antigenic Properties of Neural Stem Cells and Their Progeny in Adult Rat Subventricular Zone. Glia (2009) 57(2):136–52. doi: 10.1002/glia.20741
63. Ma R, Li WP, Rundle D, Kong J, Akbarali HI, Tsiokas L. PKD2 Functions as an Epidermal Growth Factor-Activated Plasma Membrane Channel. Mol Cell Biol (2005) 25(18):8285–98. doi: 10.1128/mcb.25.18.8285-8298.2005
64. Christensen ST, Clement CA, Satir P, Pedersen LB. Primary Cilia and Coordination of Receptor Tyrosine Kinase (RTK) Signalling. J Pathol (2012) 226(2):172–84. doi: 10.1002/path.3004
65. Jutten B, Rouschop KM. EGFR Signaling and Autophagy Dependence for Growth, Survival, and Therapy Resistance. Cell Cycle (2014) 13(1):42–51. doi: 10.4161/cc.27518
66. Lee E, Yong RL, Paddison P, Zhu J. Comparison of Glioblastoma (GBM) Molecular Classification Methods. Semin Cancer Biol (2018) 53:201–11. doi: 10.1016/j.semcancer.2018.07.006
67. Pearson A, Proszek P, Pascual J, Fribbens C, Shamsher MK, Kingston B, et al. Inactivating NF1 Mutations Are Enriched in Advanced Breast Cancer and Contribute to Endocrine Therapy Resistance. Clin Cancer Res (2020) 26(3):608–22. doi: 10.1158/1078-0432.Ccr-18-4044
68. Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 Mutations in Gliomas. N Engl J Med (2009) 360(8):765–73. doi: 10.1056/NEJMoa0808710
Keywords: glioblastoma, primary cilia, CCRK, LPAR1, HDAC6, PCM1, sonic hedgehog
Citation: Li M, Zhang J, Zhou H and Xiang R (2021) Primary Cilia–Related Pathways Moderate the Development and Therapy Resistance of Glioblastoma. Front. Oncol. 11:718995. doi: 10.3389/fonc.2021.718995
Received: 01 June 2021; Accepted: 04 August 2021;
Published: 26 August 2021.
Edited by:
Cecilia Ana Suarez, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Brunella Franco, Telethon Institute of Genetics and Medicine (TIGEM), ItalyWon-Jing Wang, National Yang Ming Chiao Tung University, Taiwan
Vasileios Toulis, University College London, United Kingdom
Copyright © 2021 Li, Zhang, Zhou and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong Xiang, c2hpcmxlc21pbGVAY3N1LmVkdS5jbg==
 Minghao Li
Minghao Li Jiaxun Zhang
Jiaxun Zhang Haonan Zhou
Haonan Zhou Rong Xiang
Rong Xiang