- 1Department of Biochemistry and Molecular Biology, Mudanjiang Medical University, Mudanjiang, China
- 2Department of Dermatological, Hongqi Hospital Affiliated to Mudanjiang Medical University, Mudanjiang, China
- 3Department of Nursing Youth League Committee, Mudanjiang Medical University, Mudanjiang, China
Purpose: Breast cancer type 1 susceptibility (BRCA) mutations not only increase breast cancer (BC) risk but also result in poor survival and prognosis for BC patients. This study will analyze the effect and safety of therapeutic regimens for the treatment of BC patients with germline BRCA (gBRCA) mutations by network meta-analysis.
Methods: Public databases were searched from inception to 29 April 2021. Frequentist network meta-analysis was conducted to analyze the benefit of chemotherapy and targeted drug-related strategies.
Results: Seventeen articles were included in the analysis. For progression-free survival (PFS), olaparib (hazard ratio (HR): 0.58; 95% confidence interval (CI): 0.43 – 0.79), platinum (HR: 0.45; 95% CI: 0.22 – 0.89), and talazoparib (HR: 0.54; 95% CI: 0.41 – 0.71) were significantly better than platinum-free chemotherapy (Chemo). The results based on indirect comparisons showed that veliparib (Vel) + platinum + Chemo was also significantly better than Chemo (HR: 0.37; 95% CI: 0.20 – 0.69). For overall survival (OS), olaparib was significantly better than Chemo only in the population who did not receive prior chemotherapy. For pathologic complete response (pCR), bevacizumab+Chemo had a significant advantage over platinum agents (OR: 3.64; 95% CI: 1.07 - 12.39). Olaparib and talazoparib both showed significantly higher objective response rates (ORRs) than Chemo.
Conclusion: The PFS results suggested that olaparib, talazoparib, and Vel+platinum agent+Chemo were ideal regimens for overall, TNBC, and advanced BC patients with gBRCA mutations. Whether PARPis are suitable for patients with gBRCA mutations who have received prior platinum therapy still needs to be clarified.
Introduction
Breast cancer (BC) is one of the three most common cancers in the world (1) and is also the most common cancer and the leading cause of cancer-related death in women globally (2). Approximately 2.1 million women were newly diagnosed with BC in 2018, with BC accounting for nearly a quarter of female cancer cases worldwide (3).
Breast cancer type 1 susceptibility (BRCA), a tumor suppressor gene, encodes a protein that repairs DNA double-strand breaks by homologous recombination repair (HRR) and can inhibit the occurrence of malignant tumors (4). Germline BRCA1/2 mutation reduces gene expression and has been considered an important risk factor for the onset of BC; in addition, mutant BRCA1/2 allele carriers have a lifetime risk of BC as high as 80%-85% (5). Approximately 75% of BC patients with germline BRCA1 (gBRCA) mutations are classified as having triple-negative breast cancer (TNBC), and the TNBC rate among patients with gBRCA2 mutations is reported to be 20%-25% (6, 7), which often has a high nuclear grade and a larger tumor burden (8). In addition to the risk of BC, BRCA mutations are also considered predive of prognosis, and BC patients with gBRCA mutations have worse survival outcomes than those without gBRCA mutations (9, 10). Therefore, screening for gBRCA mutation carriers may help formulate therapeutic strategies to improve survival outcomes.
Among TNBC patients, the incidence of gBRCA mutation is 11.2% (11). Both gBRCA mutation-associated BC and sporadic TNBC are characterized by abnormal DNA repair ability and extensive genome instability, which support the application of DNA-damaging agents, such as platinum agents (12). Platinum agents can cause DNA strand breaks and lead to cancer cell apoptosis, which makes them more effective in BC cancer cells with DNA repair defects caused by BRCA mutations (13, 14). However, the clinical results are still controversial (15–17).
In addition, for BRCA-mutated human epidermal growth factor receptor 2 (HER2)-negative metastatic or advanced BC, the US Food and Drug Administration (FDA) has approved treatment with the poly (ADP-ribose) polymerase inhibitors (PARPis) talazoparib and olaparib (18). A network meta-analysis showed that these two PARPis have similar effects, safety and acceptability in the treatment of BRCA-mutated HER2-negative metastatic or advanced BC (19).
Because BC patients with gBRCA mutations have defects in homologous recombination repair (HRR), which weakens the ability of cancer cells to repair DNA damage, they are likely to be sensitive to both PARPis and platinum agents (20). In previous meta-analyses, the effectiveness of platinum agents (17, 21) and PARPis (3) was analyzed, but the analysis of combination regimens, patient characteristics, and survival outcomes was inadequate. In particular, no ideal combination regimen containing platinum agents, PARPis and other agents has been identified for BC patients with gBRCA mutations. This study will analyze the effectiveness and safety of various drug regimens for the treatment of BC patients with gBRCA mutations by network meta-analysis.
Methods
Search Strategy
The literature search was conducted in PubMed, Embase, Scopus, EBSCOhost and the Cochrane Central Register of Controlled Trials from database inception to 29 April 2021, without language restriction. Keywords included (BRCA*, breast cancer susceptibility gene), (breast, mammary), (malignant, neoplasms, cancer, tumor, carcinoma, adenocarcinoma) and (random*, randomized, randomised). The references of important reviews were also screened to avoid omissions.
Study Selection and Eligibility Criteria
We included randomized controlled trials (RCTs) of parallel design for BC patients with gBRCA mutations. Eligibility criteria included the following: 1, studies with an RCT design; 2, studies including BC patients with gBRCA mutations or reporting a gBRCA-mutated BC population as a subgroup; 3, studies in which the intervention regimen was chemotherapy or targeted drug-related strategies; and 4, studies reporting survival outcomes in terms of progression-free survival (PFS) or overall survival (OS) based on Cox regression analysis, pathologic complete response (pCR), or objective response rate (ORR). The exclusion criteria included the following: 1, studies without an RCT design; 2, studies that included patients with gBRCA mutations with other types of cancer, such as ovarian cancer, endometrial cancer, or colon cancer; 3, studies that grouped patients by BRCA expression level or methylation instead of by gBRCA mutation; and 4, studies that grouped patients not only by gBRCA mutation but also by other HRR mutations.
Data Extraction and Quality Assessment
The extracted data included the first author’s name, publication date, research location, study abbreviation, registration ID, patient types, interventions, controls, and follow-up period. Primary outcomes included survival results; secondary outcomes included pCR, ORR, and serious adverse events (SAEs). The Cochrane tool, which includes consideration of random sequence generation, allocation concealment, blinding of investigator and patients, blinding of assessor, missing outcome data, selective reporting and other factors, was used to assess the risk of bias in RCTs (22). The GRADE approach was used to assess the quality of the evidence for direct, indirect and network comparisons. Evidence quality rating factors for comparisons included considerations of risk of bias, inconsistency, indirectness, imprecision and publication bias. If direct and indirect comparison results provided similar results, the higher quality level of the direct and indirect comparisons was used for the network comparison quality level, and if it provided different results, the network comparison quality level was downgraded because of inconsistency. The quality of evidence was divided into high, moderate, low and very low (23).
Statistical Analysis
Odds ratios (ORs) with corresponding 95% confidence intervals (CIs) were used for dichotomous outcomes, and hazard ratios (HRs) with 95% CIs were used for Cox regression model results of survival analyses. Frequentist network meta-analysis was conducted using random-effects models (24). We assessed agreement between direct and indirect comparisons in every closed loop of evidence using node splitting approaches and for the entire network using a design-by-treatment interaction model. The P-score metric, which measures the extent of certainty that an intervention is better than others, was used to compare the effectiveness and safety of all analyzed therapeutic interventions (25). Study bias was evaluated at the outcome level with Egger’s regression test. Subgroup analysis was performed according to patient mutation type and molecular subtype and receipt of prior chemotherapy or platinum agents. All pairwise and network meta-analyses were performed using RevMan software and R software with the “netmeta” and “meta” packages.
Results
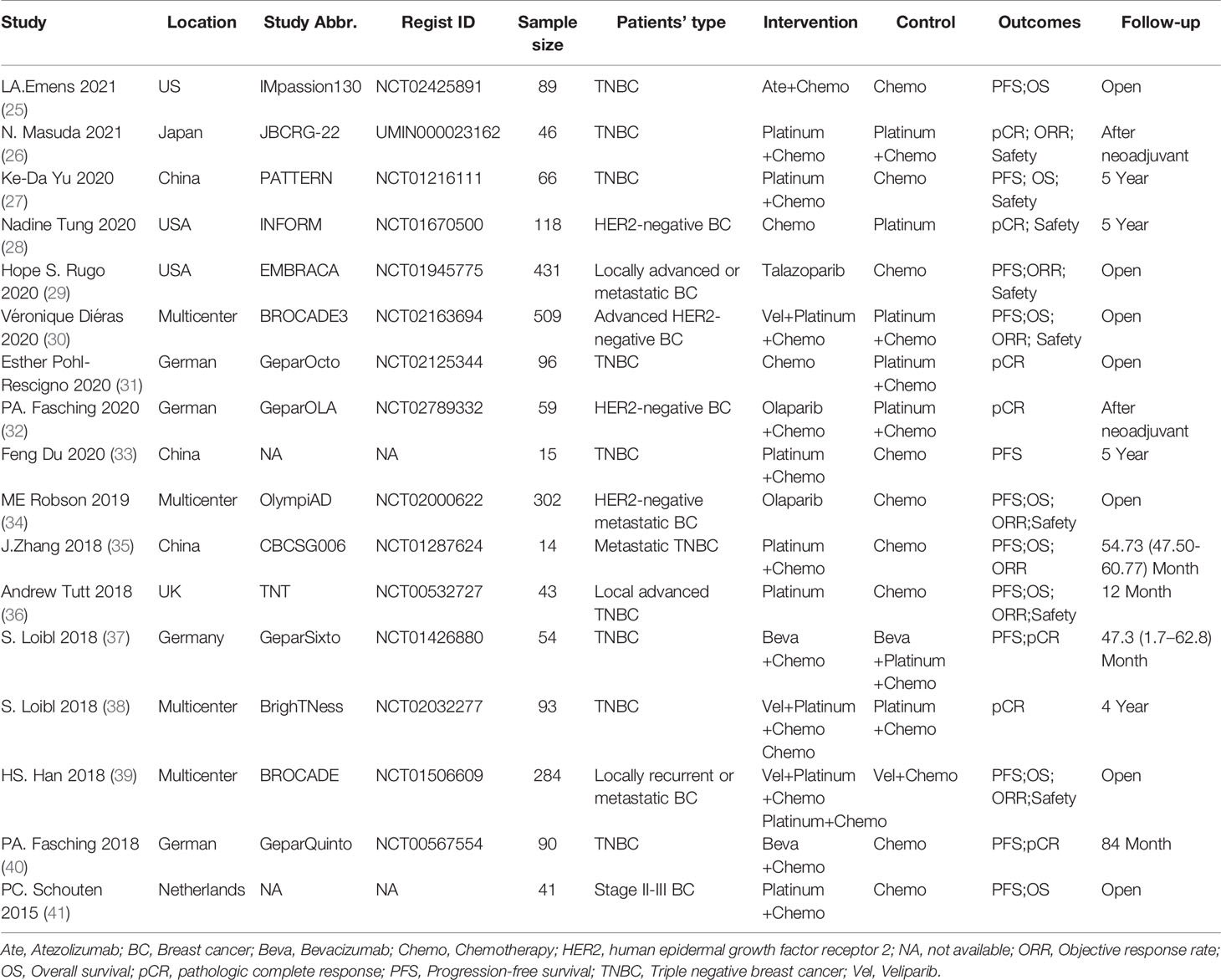
After searching, a total of 2,803 items were obtained, and 2,003 items remained after removing duplications. After screening the titles and abstracts, 142 articles were obtained and underwent full-text review. In addition, 125 documents were excluded for the following reasons: duplicate reports (64), studies with no inclusion of a population with gBRCA mutation in the entire population or subgroups (30), studies with no report of the above outcomes or use of the above protocols (16), reviews or comments (9), studies with non-RCT designs (4), studies reporting gBRCA mutation-related BC risk (1), and studies researching populations with mutations in other HRR-related genes (1). Because conference abstracts were included in the search results, many duplicate reports existed. The study abbreviation and registration ID were mainly used for identifying duplicate reports and adopting the latest reported results. Ultimately, 17 articles were included in the analysis (25–41) (Figure 1 and Table 1).
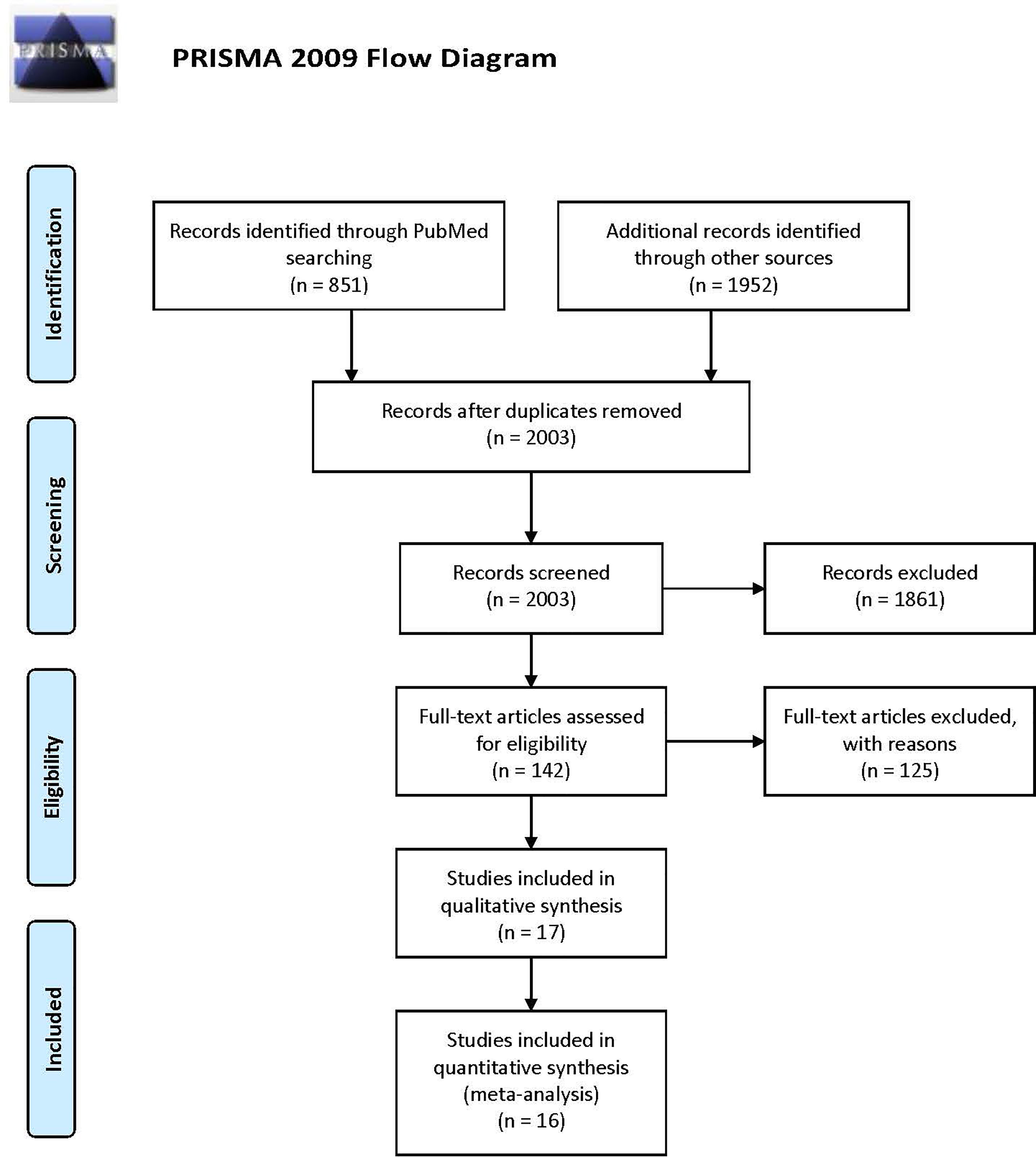
Figure 1 PRISMA flowchart describing the study identification and selection process, which was adopted to document the number of items identified from the databases, the number of duplicates removed, the number of full-text articles screened, and the number of studies included in the final analysis.
A total of 2,350 BC patients with gBRCA mutations were included. With a publication time cutoff from 2015 to 2021, all publications were published in the past 6 years. This is mainly because early-phase studies did not consider the presence of gBRCA mutations in patients. Four studies reported BC patients with gBRCA mutations as a subgroup (27, 35, 36, 38), and others considered BC patients with gBRCA mutations overall. The targeted agents analyzed included atezolizumab (Ate), talazoparib, olaparib, veliparib (Vel), bevacizumab (Beva), and platinum agents, including carboplatin and cisplatin (Supplementary Table 1). The subgroup meta-analysis of PFS results showed that PARPi (HR: 0.64; 95% CI: 0.56, 0.75; p<0.001) and platinum (HR: 0.51; 95% CI: 0.33, 0.78; p=0.002) were significantly better than the control (Figure 2). There was a study not included in the quantitative synthesis (26) because both the intervention and control were platinum agents + chemo according to our regimen classification method. Its pCR and ORR results showed no significant difference between paclitaxel plus carboplatin and eribulin plus carboplatin regimens for BRCA mutation TNBC patients (26). The included studies were all RCTs, and most of the studies have been publicly registered. Although there were deficiencies in the blinding method design, the overall design quality was good (Figure 2).
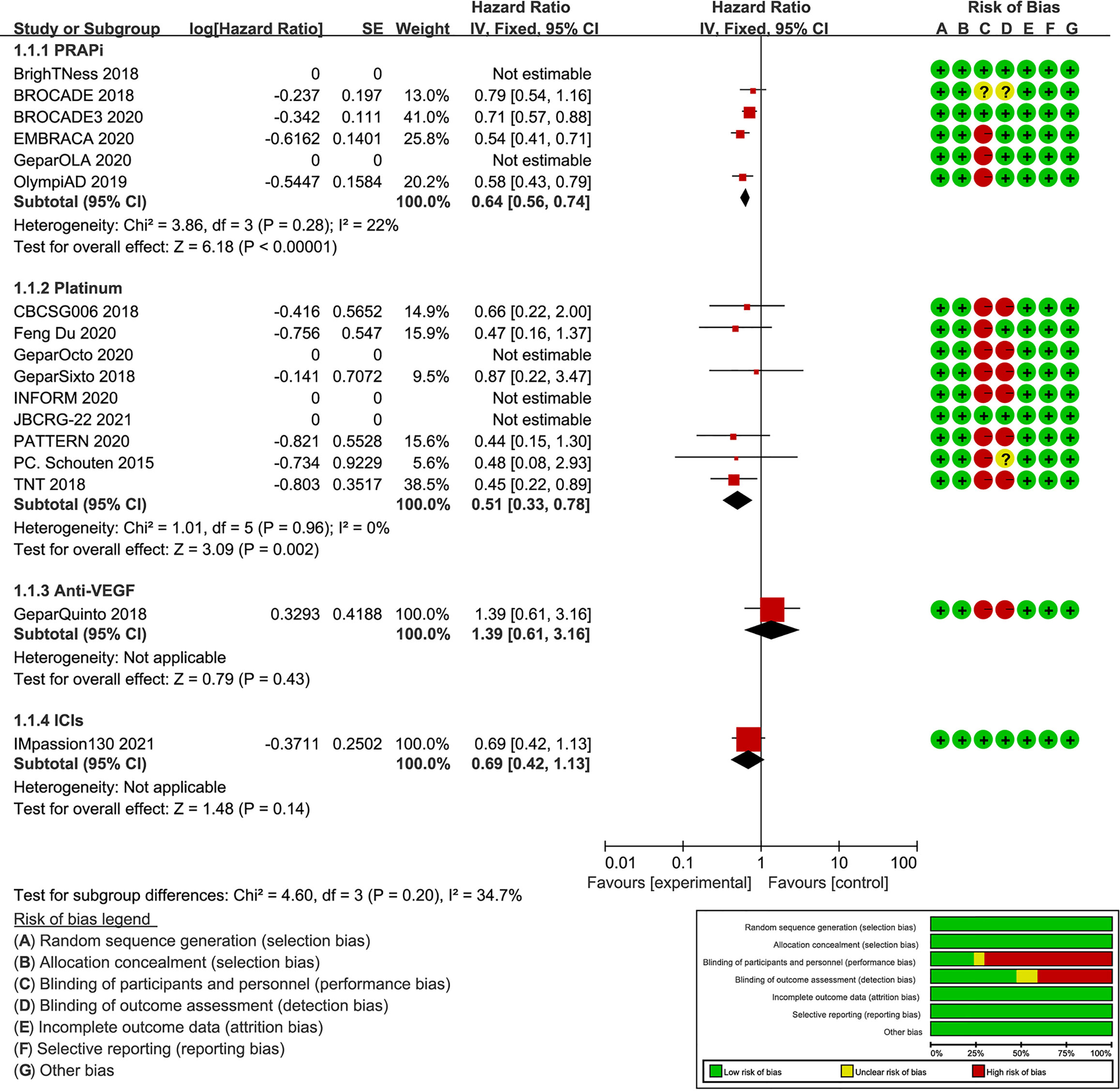
Figure 2 Forest plot of PFS results and risk of bias graph for each included study. On the left, the authors’ name, publication years, and the logHR results of individual studies are listed. Subgroup analysis is conducted according to different intervention drugs, including PARPi, platinum, anti-VEGF, and ICIs. The forest plot in the middle shows the PFS results of each study, and the center of diamonds represent the pooled HR. The center is left to vertical line, favoring the experimental groups. The horizontal tips represent the 95%CIs. The right side of the figure shows the risk of bias assessment for each study according to the Cochrane tool.
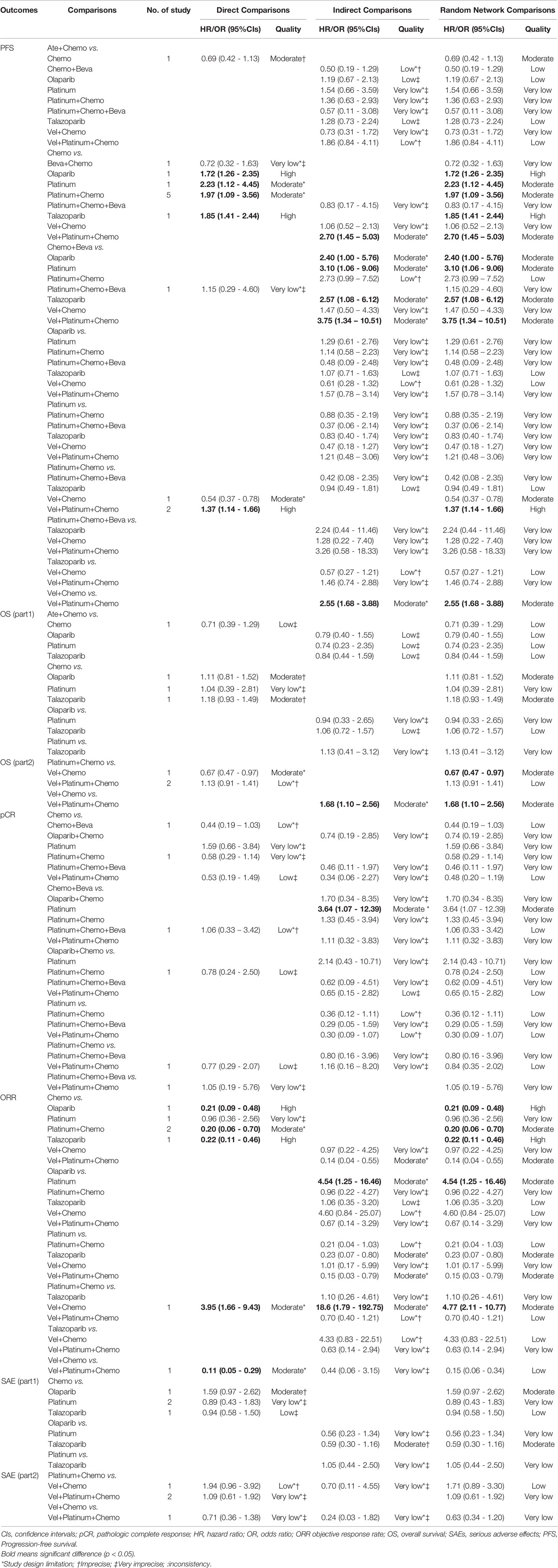
For the PFS analysis, ten regimens were included (Figure 3A). In the pairwise comparisons based on the network random effects model, olaparib (HR: 0.58; 95% CI: 0.43 – 0.79), platinum agents (HR: 0.45; 95% CI: 0.22 – 0.89), and talazoparib (HR: 0.54; 95% CI: 0.41 – 0.71) were significantly better than platinum-free chemotherapy (Chemo). The quality of evidence supporting the superiority of olaparib and talazoparib compared to Chemo was high (Table 2). In the results based on indirect comparisons, Vel + platinum agents + Chemo was also significantly better than Chemo (HR: 0.37; 95% CI: 0.20 – 0.69), with the highest p-score ranking. In the subgroup analysis, olaparib, talazoparib, and platinum agents were ideal regimens for advanced BC patients. The rank results were similar between the TNBC subgroup and the overall population. The non-TNBC patient population was divided into two network parts due to major differences. Talazoparib and Vel+platinum agent+Chemo remained ideal regimens. In non-TNBC patients and patients with BRCA2 mutations, talazoparib may have a relative advantage over olaparib. Olaparib and talazoparib were superior to Chemo regardless of whether the patient received prior chemotherapy. In addition, for patients who received prior platinum therapy, talazoparib and olaparib were not significantly better or worse than Chemo (Figure 4A). The study bias analysis did not show any publication bias (Figure 5A).
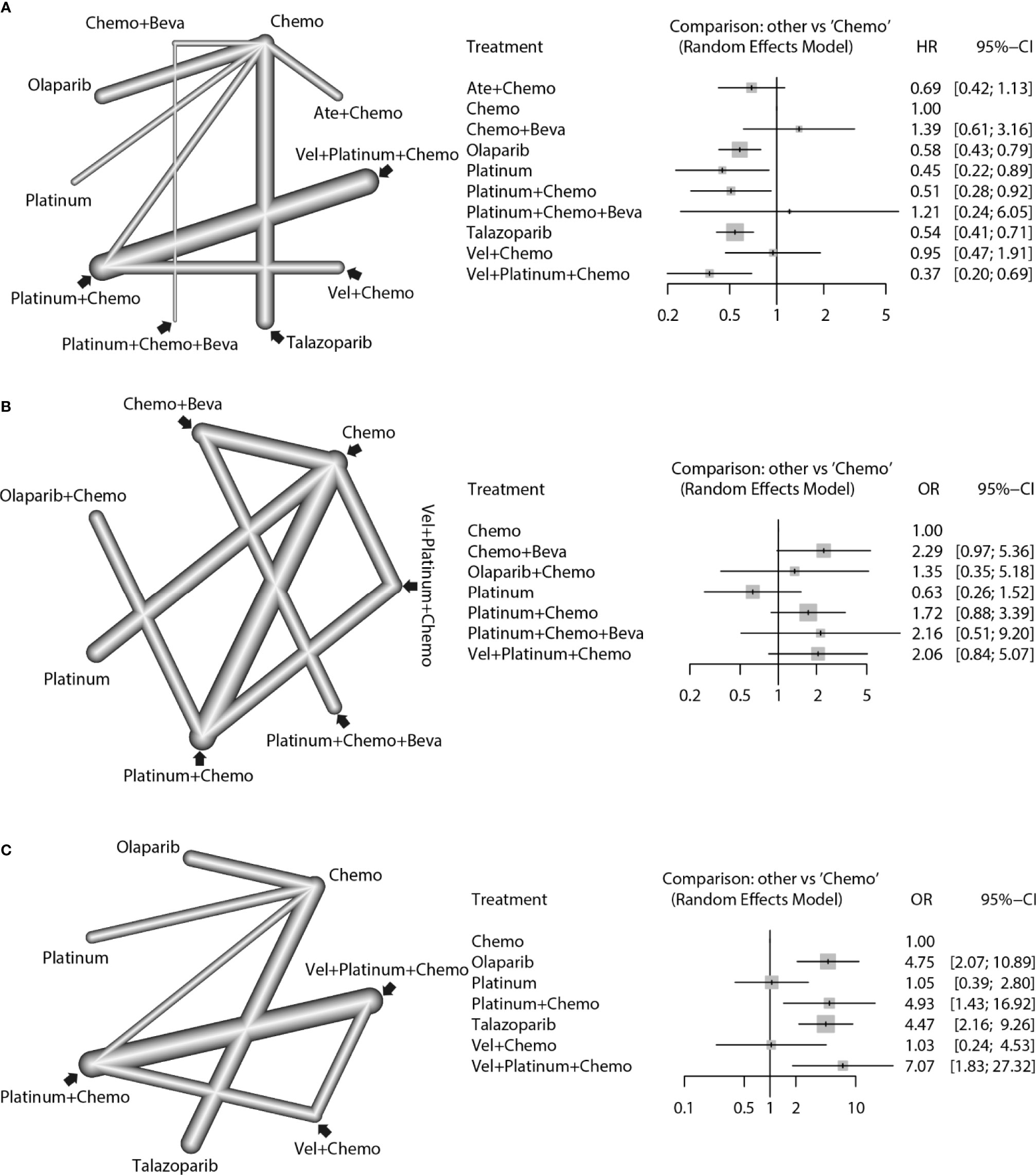
Figure 3 Network comparisons of PFS (A), pCR (B), and ORR (C) results for the strategies included in the analyses. The left side of the figure shows the network diagrams. Each dot indicates an intervention, and the edge between the two dots indicates that there is a direct comparison from the RCT. The thickness of the edge represents the precision of the comparisons. On the right, forest plots show the network meta-analysis results for all comparisons between Chemo and other interventions.
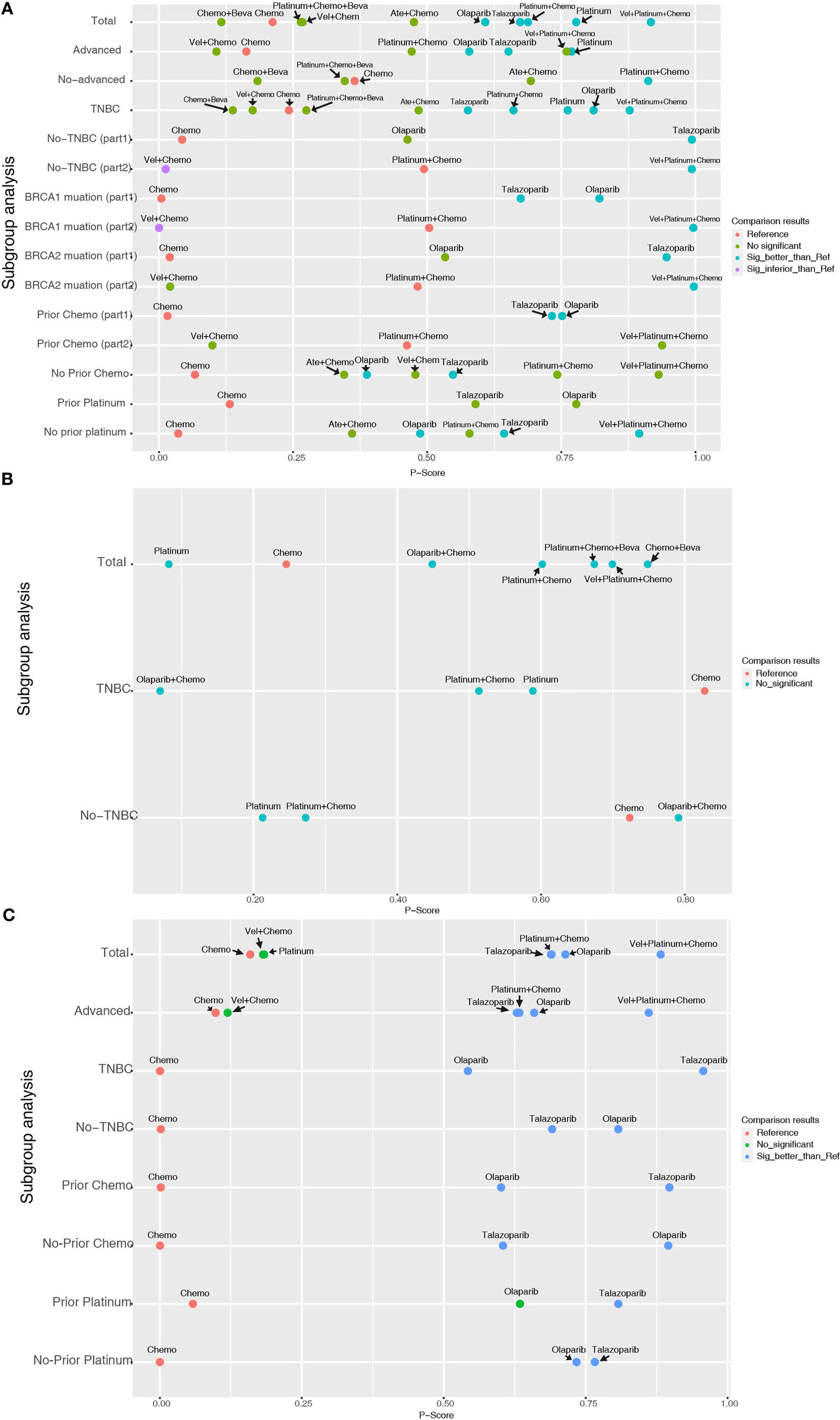
Figure 4 Entire and subgroup analyses of PFS (A), pCR (B), and ORR (C) results by network meta-analysis according to the P-score. Each dot represents an intervention, its ordinate represents the subgroup to which it belonged, and its abscissa represents the p-score results from the network meta-analysis. The various colors of the dots indicate whether there is a statistical difference compared with the reference (commonly Chemo).
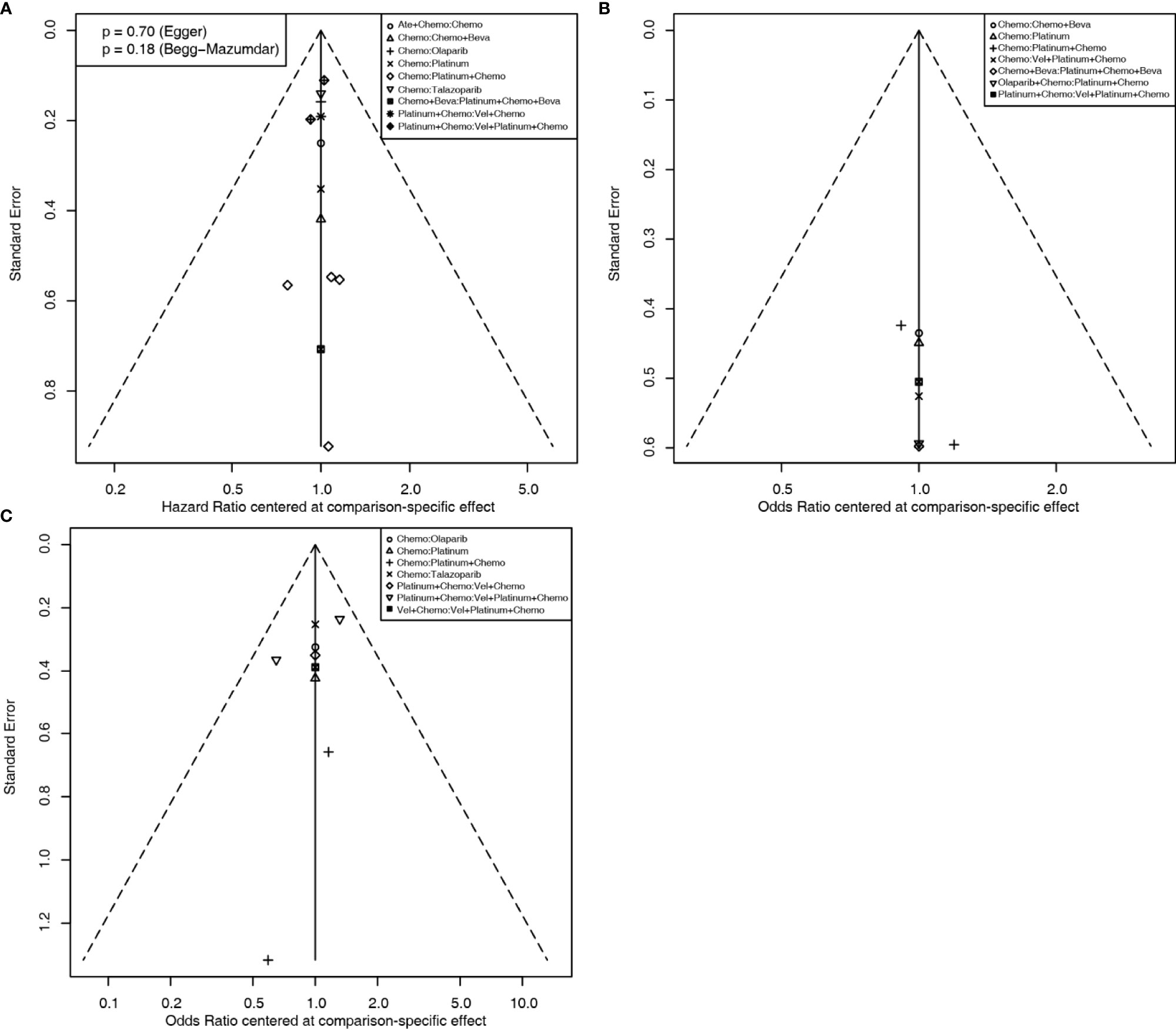
Figure 5 Comparison-adjusted funnel plots for assessing PFS (A), pCR (B), and ORR (C) results. Potential publication bias is assessed by whether the data point distribution is symmetrical, and the Egger’s and Begg’s test results are calculated if the number of data point is more than ten.
In the OS results, due to the large differences between subgroups, the analysis of regimens was divided into two parts (Supplementary Figures 1A, B). The first analysis included the Ate+Chemo, Chemo, olaparib, platinum agent, and talazoparib regimens, but no significant difference was identified by pairwise comparisons. The second analysis included platinum agent+Chemo, Vel+Chemo, and Vel+platinum agent+Chemo. Among them, Vel+Chemo was inferior to platinum agent+Chemo (HR: 1.48; 95% CI: 1.03 - 2.13), and Vel+platinum agent+Chemo was significantly better than Vel+Chemo (HR: 1.68; 95% CI: 1.10-2.56) (Table 2). In the subgroup analysis, olaparib was found to be significantly better than Chemo only in the population who did not receive prior chemotherapy (Supplementary Figure 2A).
In the pCR analysis, only Chemo+Beva had a significant advantage (OR: 3.64; 95% CI: 1.07-12.39) over the platinum agent (Figure 3B and Table 2). The application of platinum agents alone was not ideal for the improvement of pCR. In addition, the small sample size affected the credibility of the subgroup results (Figure 4B). The study bias analysis did not show any publication bias (Figure 5B).
In the ORR analysis, Chemo was significantly inferior to olaparib (OR: 4.75; 95% CI: 2.07 - 10.89), platinum agent+Chemo (OR: 4.93; 95% CI: 1.43-16.92), and talazoparib (OR: 4.47; 95% CI: 2.16 – 9.26) (Figure 3C). Olaparib was significantly better than platinum agents (OR: 4.54; 95% CI: 1.25 - 16.46). Platinum agent+Chemo was also significantly better than Vel+Chemo (OR: 4.77; 95% CI: 2.11 - 10.77) (Table 2). In the subgroup analysis, the rank results of advanced patients were similar to those of the overall patients. Olaparib and talazoparib both showed significant advantages over Chemo, except for in patients who received prior platinum therapy (Figure 4C). The study bias analysis did not show any publication bias (Figure 5C). In the SAE analysis, there were no significant differences in SAEs between the two subgroups (Supplementary Figures 1C, D and Supplementary Figure 2B).
K-means cluster analysis was performed on the P-score results of PFS, pCR, and ORR. The results showed that Vel + platinum agents + Chemo exhibited high effectiveness. Ate+ Chemo, olaparib, talazoparib, platinum agent + chemo, and platinum agent alone had moderate effectiveness. Others had relative low effectiveness (Figure 6). However, there were still missing values in the cluster analysis. More research is needed to correct the cluster results.
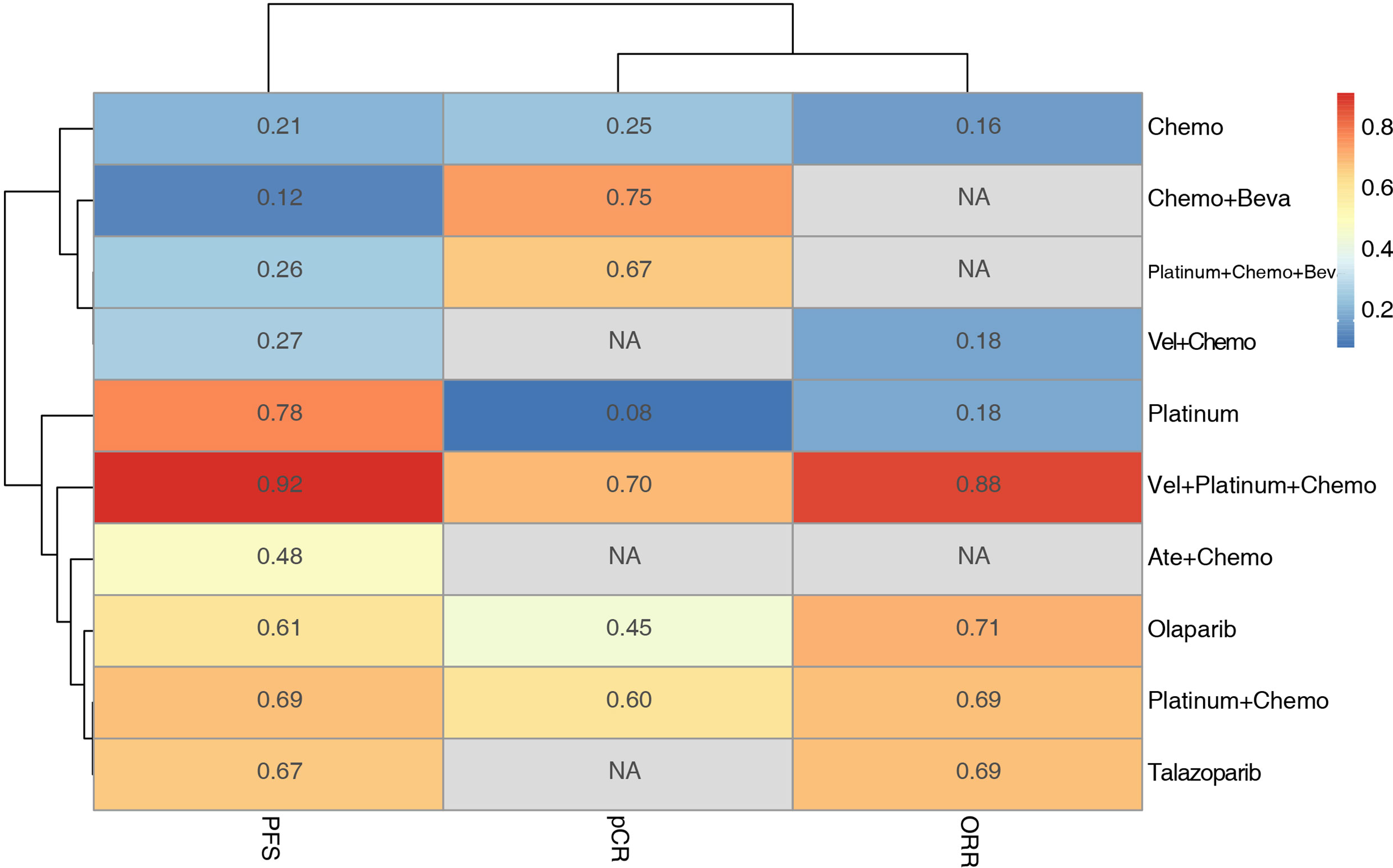
Figure 6 Hierarchical clustering of PFS pCR and ORR results based on the P-score. The numbers and colors in the grid represent the P-score value of each intervention method for each outcome, and the lines on the left and upper side represent the similarities among the elements by K-means cluster analysis.
Discussion
In this study, the effect and safety of therapeutic regimens in BC patients with BRCA mutations were analyzed by network meta-analysis. The PFS analysis suggested that olaparib, talazoparib, and Vel+platinum agent+Chemo were ideal regimens for overall, TNBC, and advanced BC patients with gBRCA mutations. It was also shown that talazoparib and olaparib had no obvious advantages over Chemo in patients who received prior platinum treatment. The OS analysis showed that olaparib was significantly better than Chemo only in the population who did not receive prior chemotherapy. In the pCR analysis, only Chemo+Beva was significantly better than the platinum agent. In the ORR analysis, olaparib and talazoparib had significant advantages over Chemo. However, olaparib had a relatively weak effect on patients who received prior platinum treatment. In addition, there were no significant differences in SAEs.
Mutations in the tumor suppressor gene BRCA1/2 cause HRR dysfunction, and in tumors with such mutations, PARP inhibition more effectively kills tumor cells, resulting in synthetic lethality (42). PARP1, which is mainly involved in single-strand DNA break repair, is the main target of PARPis. For tumor cells without HRR dysfunction, DNA damage caused by these agents will be repaired through HRR pathways. However, when HRR dysfunction occurs, such as HRR dysfunction caused by gBRCA mutation, DNA damage caused by PARPis can produce effective cytotoxicity and induce apoptosis. The results of this work prove that olaparib and talazoparib can prolong the PFS of patients with BRCA-mutated BC, and these agents are also suitable for TNBC and advanced BC patients. PARPis did not increase the SAE risk, suggesting that they specifically affected cancer cells and not normal cells. However, in the OS analysis, olaparib was beneficial compared with Chemo only in the TNBC subgroup. No other regimens were found to provide survival benefits to BC patients with gBRCA mutations.
Vel is a selective PARP1/2 inhibitor with relatively weak affinity, while olaparib and talazoparib have relatively strong affinity. These agents cannot cause the same scale of synthetic lethality (43). However, Vel mainly selectively inhibits the activity of PARP without holding the PARP protein to DNA damage repair intermediates, and combination regimens of Vel and platinum agents have been tested (44). PARP-1 has also been suggested to be a marker of the response to DNA-damaging platinum agents, which also supports the combination of Vel and platinum (45). This study showed the significant therapeutic advantage of Vel+platinum agent+Chemo but also showed that for non-TNBC patients, patients with BRCA1 mutations, and patients who do not receive prior chemotherapy, Vel+platinum agent will even be significantly inferior to platinum agent+Chemo. In patients who received prior platinum therapy, the Vel+Chemo regimen achieved significantly inferior OS compared to the platinum agent+Chemo regimen.
There is evidence that patients with gBRCA mutations show overexpression of vascular endothelial growth factor, angiopoietin-1, and angiopoietin-2 (46, 47). This finding supports the application of Beva to inhibit angiogenesis and cause hypoxia-related DNA damage and synthetic lethal reactions with few adverse events. Our analysis showed that Beva can significantly improve the pCR rate, but in the P-score ranking results of PFS, Beva-Chemo was even inferior to Chemo alone, although the difference was not statistically significant. The pCR and PFS were also different. In general, Beva combined with Chemo should not be recommended based on the survival outcomes.
However, whether PARPis are suitable for patients with gBRCA mutations who have received prior platinum agents still needs to be clarified. In the PFS subgroup analysis, olaparib and talazoparib were not significantly superior to Chemo in patients who had received prior platinum agents. The abovementioned differences between pCR and PFS and the poor effect of PARPis in patients who received prior platinum agents indicate that patients with gBRCA mutations who have received prior platinum therapy can regain BRCA function via additional mutations or the activation of other HRR mechanisms (48). Cross-resistance between platinum agents and PARPis has also been suggested. Therefore, the combined application of PARPis and platinum drugs can be used to maximize the therapeutic benefits in patients with gBRCA mutations. However, based on concerns about SAEs, especially hematological toxicity, only the combination of veliparib and platinum-related chemotherapy has been tested in RCTs, and this combination showed benefits that were similar to those of olaparib and talazoparib in patients with gBRCA mutations. However, for patients who received prior chemotherapy, Vel+platinum agent+Chemo also did not achieve significant PFS benefits compared to platinum agent+Chemo. Therefore, for this population, olaparib and talazoparib are still the best strategies.
One study (MEDIOLA) was excluded because it used a non-RCT design, and this study showed that the combination of olaparib and durvalumab has promising antitumor activity and safety that are similar to those of olaparib and durvalumab monotherapies (49). There are also some important ongoing RCTs. In one study, the PARPi niraparib showed superior survival results compared to the physician’s choice Chemo, but the final results were awaited (NCT01905592). In another study, the novel PARPi fluzoparib was combined with apatinib for the treatment of patients with gBRCA-mutated HER2-negative metastatic BC (NCT04296370). Olaparib combined with the PD-1 inhibitor pembrolizumab (NCT04191135), the PD-L1 antibody atezolizumab (NCT02849496), or durvalumab (NCT03167619) is being tested for the treatment of patients with gBRCA1/2-mutated BC. In addition, olaparib combined with ceralasertib or adavosertib is being used for TNBC with the gBRCA mutation (NCT03330847). With the completion of these RCTs, more therapeutic regimens and more accurate results will be provided to help uncover the ideal regimens for patients with gBRCA-mutated BC.
There are still several limitations in this work. First, due to the differences in groups, some outcomes and subgroup results were divided into two parts. Second, this work only analyzed patients with gBRCA mutations; patients with BRCA epigenetic modifications were not analyzed because epigenetic modification and expression levels might change with disease development and treatment. Patients who have loss of BRCA function due to causes besides mutation have more sensitivity to platinum therapy than those who have gBRCA mutation (50). Third, a variety of cytotoxic agents in platinum-free Chemo regimens (anthracyclines and alkylating agents) still have DNA-damaging effects. This diversity of cytotoxic agents in the Chemo arm increased the heterogeneity of the results, which was difficult to eliminate in this study.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
YJ and X-YM: Conceptualization, Methodology, Writing - Original draft preparation. CM: Methodology. N-ND and L-HL: Data curation, Software. ZH and X-YW: Software, Visualization. Z-YS: Software, Validation. R-JC: Supervision, Writing - Reviewing and Editing. YJ and X-YM contribute equally to this work. All authors contributed to the article and approved the submitted version.
Funding
Youth Innovative Talents Training Program in General Undergraduate Colleges and Universities in Heilongjiang Province (No.2020-QC23).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.718761/full#supplementary-material
Supplementary Figure 1 | Network comparisons of OS (A, B) and SAE (C, D) results for the strategies included in the analyses. The left side of the figure shows the network diagrams; each dot indicates an intervention, and the edge between the two dots indicates that there is a direct comparison from the RCT. The thickness of the edge represents the precision of the comparisons. On the right side, forest plots show the network meta-analysis results for all comparisons between Chemo (or Platinum+Chemo) and other interventions.
Supplementary Figure 2 | Entire and subgroup analyses of OS (A) and SAE (B) results by network meta-analysis according to the P-score. Each dot represents an intervention, its ordinate represents the subgroup to which it belonged, and its abscissa represents the p-score results from the network meta-analysis. The various colors of the dots indicate whether there is a statistical difference compared with the reference (commonly Chemo).
References
1. Harbeck N, Gnant M. Breast Cancer. Lancet (2017) 389(10074):1134–50. doi: 10.1016/S0140-6736(16)31891-8
2. Winters S, Martin C, Murphy D, Shokar NK. Breast Cancer Epidemiology, Prevention, and Screening. Prog Mol Biol Transl Sci (2017) 151:1–32. doi: 10.1016/bs.pmbts.2017.07.002
3. Wang J, Zhang Y, Yuan L, Ren L, Zhang Y, Qi X. Comparative Efficacy, Safety, and Acceptability of Single-Agent Poly (ADP-Ribose) Polymerase (PARP) Inhibitors in BRCA-Mutated HER2-Negative Metastatic or Advanced Breast Cancer: A Network Meta-Analysis. Aging (Albany NY) (2020) 13(1):450–9. doi: 10.6004/jnccn.2019.7423
4. Tarsounas M, Sung P. The Antitumorigenic Roles of BRCA1-BARD1 in DNA Repair and Replication. Nat Rev Mol Cell Biol (2020) 21(5):284–99. doi: 10.1038/s41580-020-0218-z
5. Cullinane CM, Creavin B, O’Connell EP, Kelly L, O’Sullivan MJ, Corrigan MA, et al. Risk of Colorectal Cancer Associated With BRCA1 and/or BRCA2 Mutation Carriers: Systematic Review and Meta-Analysis. Br J Surg (2020) 107(8):951–9. doi: 10.1002/bjs.11603
6. Mahfoudh W, Bettaieb I, Ghedira R, Snoussi K, Bouzid N, Klayech Z, et al. Contribution of BRCA1 5382insc Mutation in Triple Negative Breast Cancer in Tunisia. J Transl Med (2019) 17(1):123. doi: 10.1186/s12967-019-1873-8
7. Wang N, Li K, Huang W, Kong W, Liu X, Shi W, et al. Efficacy of Platinum in Advanced Triple-Negative Breast Cancer With Germline BRCA Mutation Determined by Next Generation Sequencing. Chin J Cancer Res (2020) 32(2):149–62. doi: 10.21147/j.issn.1000-9604.2020.02.03
8. Chen H, Wu J, Zhang Z, Tang Y, Li X, Liu S, et al. Association Between BRCA Status and Triple-Negative Breast Cancer: A Meta-Analysis. Front Pharmacol (2018) 9:909. doi: 10.3389/fphar.2018.00909
9. Zhong Q, Peng HL, Zhao X, Zhang L, Hwang WT. Effects of BRCA1- and BRCA2-Related Mutations on Ovarian and Breast Cancer Survival: A Meta-Analysis. Clin Cancer Res (2015) 21(1):211–20. doi: 10.1158/1078-0432.CCR-14-1816
10. Liu M, Xie F, Liu M, Zhang Y, Wang S. Association Between BRCA Mutational Status and Survival in Patients With Breast Cancer: A Systematic Review and Meta-Analysis. Breast Cancer Res Treat (2021) 186(3):591–605. doi: 10.1007/s10549-021-06104-y
11. Couch FJ, Hart SN, Sharma P, Toland AE, Wang X, Miron P, et al. Inherited Mutations in 17 Breast Cancer Susceptibility Genes Among a Large Triple-Negative Breast Cancer Cohort Unselected for Family History of Breast Cancer. J Clin Oncol (2015) 33(4):304–11. doi: 10.1200/JCO.2014.57.1414
12. Foulkes WD, Smith IE, Reis-Filho JS. Triple-Negative Breast Cancer. N Engl J Med (2010) 363(20):1938–48. doi: 10.1056/NEJMra1001389
13. Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC, et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients With Triple-Negative Breast Cancer. Clin Cancer Res (2016) 22(15):3764–73. doi: 10.1158/1078-0432.CCR-15-2477
14. Lin PH, Chen M, Tsai LW, Lo C, Yen TC, Huang TY, et al. Using Next-Generation Sequencing to Redefine Brcaness in Triple-Negative Breast Cancer. Cancer Sci (2020) 111(4):1375–84. doi: 10.1111/cas.14313
15. Poggio F, Bruzzone M, Ceppi M, Ponde NF, La Valle G, Del Mastro L, et al. Platinum-Based Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer: A Systematic Review and Meta-Analysis. Ann Oncol (2018) 29(7):1497–508. doi: 10.1093/annonc/mdy127
16. Pandy JGP, Balolong-Garcia JC, Cruz-Ordinario MVB, Que FVF. Triple Negative Breast Cancer and Platinum-Based Systemic Treatment: A Meta-Analysis and Systematic Review. BMC Cancer (2019) 19(1):1065. doi: 10.1186/s12885-019-6253-5
17. Wang CJ, Xu Y, Lin Y, Zhu HJ, Zhou YD, Mao F, et al. Platinum-Based Neoadjuvant Chemotherapy for Breast Cancer With BRCA Mutations: A Meta-Analysis. Front Oncol (2020) 10:592998. doi: 10.3389/fonc.2020.592998
18. Zhao Y, Zhang LX, Jiang T, Long J, Ma ZY, Lu AP, et al. The Ups and Downs of Poly(ADP-Ribose) Polymerase-1 Inhibitors in Cancer Therapy-Current Progress and Future Direction. Eur J Med Chem (2020) 203:112570. doi: 10.1016/j.ejmech.2020.112570
19. Poggio F, Bruzzone M, Ceppi M, Conte B, Martel S, Maurer C, et al. Single-Agent PARP Inhibitors for the Treatment of Patients With BRCA-Mutated HER2-Negative Metastatic Breast Cancer: A Systematic Review and Meta-Analysis. ESMO Open (2018) 3(4):e000361. doi: 10.1136/esmoopen-2018-000361
20. Faraoni I, Graziani G. Role of BRCA Mutations in Cancer Treatment With Poly(ADP-Ribose) Polymerase (PARP) Inhibitors. Cancers (Basel) (2018) 10(12):487. doi: 10.3390/cancers10120487
21. Caramelo O, Silva C, Caramelo F, Frutuoso C, Almeida-Santos T. The Effect of Neoadjuvant Platinum-Based Chemotherapy in BRCA Mutated Triple Negative Breast Cancers -Systematic Review and Meta-Analysis. Hered Cancer Clin Pract (2019) 17:11. doi: 10.1186/s13053-019-0111-y
22. Zeng X, Zhang Y, Kwong JS, Zhang C, Li S, Sun F, et al. The Methodological Quality Assessment Tools for Preclinical and Clinical Studies, Systematic Review and Meta-Analysis, and Clinical Practice Guideline: A Systematic Review. J Evid Based Med (2015) 8(1):2–10. doi: 10.1111/jebm.12141
23. Busse JW, Sadeghirad B, Oparin Y, Chen E, Goshua A, May C, et al. Management of Acute Pain From Non-Low Back, Musculoskeletal Injuries: A Systematic Review and Network Meta-Analysis of Randomized Trials. Ann Intern Med (2020) 173(9):730–8. doi: 10.7326/M19-3601
24. Palmer SC, Tendal B, Mustafa RA, Vandvik PO, Li S, Hao Q, et al. Sodium-Glucose Cotransporter Protein-2 (SGLT-2) Inhibitors and Glucagon-Like Peptide-1 (GLP-1) Receptor Agonists for Type 2 Diabetes: Systematic Review and Network Meta-Analysis of Randomised Controlled Trials. BMJ (2021) 372:m4573. doi: 10.1136/bmj.m4573
25. Emens LA, Molinero L, Loi S, Rugo HS, Schneeweiss A, Dieras V, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer: Biomarker Evaluation of the Impassion130 Study. J Natl Cancer Inst (2021) 113(8):1005–16. doi: 10.1093/jnci/djab004
26. Masuda N, Bando H, Yamanaka T, Kadoya T, Takahashi M, Nagai SE, et al. Eribulin-Based Neoadjuvant Chemotherapy for Triple-Negative Breast Cancer Patients Stratified by Homologous Recombination Deficiency Status: A Multicenter Randomized Phase II Clinical Trial. Breast Cancer Res Treat (2021) 118(1):117–31. doi: 10.1007/s10549-021-06184-w
27. Yu KD, Ye FG, He M, Fan L, Ma D, Mo M, et al. Effect of Adjuvant Paclitaxel and Carboplatin on Survival in Women With Triple-Negative Breast Cancer: A Phase 3 Randomized Clinical Trial. JAMA Oncol (2020) 6(9):1390–6. doi: 10.1001/jamaoncol.2020.2965
28. Tung N, Arun B, Hacker MR, Hofstatter E, Toppmeyer DL, Isakoff SJ, et al. TBCRC 031: Randomized Phase II Study of Neoadjuvant Cisplatin Versus Doxorubicin-Cyclophosphamide in Germline BRCA Carriers With HER2-Negative Breast Cancer (the INFORM Trial). J Clin Oncol (2020) 38(14):1539–48. doi: 10.1200/JCO.19.03292
29. Rugo HS, Ettl J, Hurvitz SA, Goncalves A, Lee KH, Fehrenbacher L, et al. Outcomes in Clinically Relevant Patient Subgroups From the EMBRACA Study: Talazoparib vs Physician’s Choice Standard-of-Care Chemotherapy. JNCI Cancer Spectr (2020) 4(1):pkz085. doi: 10.1093/jncics/pkz085
30. Dieras V, Han HS, Kaufman B, Wildiers H, Friedlander M, Ayoub JP, et al. Veliparib With Carboplatin and Paclitaxel in BRCA-Mutated Advanced Breast Cancer (BROCADE3): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet Oncol (2020) 21(10):1269–82. doi: 10.1016/S1470-2045(20)30447-2
31. Pohl-Rescigno E, Hauke J, Loibl S, Mobus V, Denkert C, Fasching PA, et al. Association of Germline Variant Status With Therapy Response in High-Risk Early-Stage Breast Cancer: A Secondary Analysis of the Geparocto Randomized Clinical Trial. JAMA Oncol (2020) 6(5):744–8. doi: 10.1001/jamaoncol.2020.0007
32. Fasching PA, Link T, Hauke J, Seither F, Jackisch C, Klare P, et al. Neoadjuvant Paclitaxel/Olaparib in Comparison to Paclitaxel/Carboplatinum in Patients With HER2-Negative Breast Cancer and Homologous Recombination Deficiency (Geparola Study). Ann Oncol (2021) 32(1):49–57. doi: 10.1016/j.annonc.2020.10.471
33. Du F, Wang W, Wang Y, Li M, Zhu A, Wang J, et al. Carboplatin Plus Taxanes Are Non-Inferior to Epirubicin Plus Cyclophosphamide Followed by Taxanes as Adjuvant Chemotherapy for Early Triple-Negative Breast Cancer. Breast Cancer Res Treat (2020) 182(1):67–77. doi: 10.1007/s10549-020-05648-9
34. Robson ME, Tung N, Conte P, Im SA, Senkus E, Xu B, et al. Olympiad Final Overall Survival and Tolerability Results: Olaparib Versus Chemotherapy Treatment of Physician’s Choice in Patients With a Germline BRCA Mutation and HER2-Negative Metastatic Breast Cancer. Ann Oncol (2019) 30(4):558–66. doi: 10.1093/annonc/mdz012
35. Zhang J, Lin Y, Sun XJ, Wang BY, Wang ZH, Luo JF, et al. Biomarker Assessment of the CBCSG006 Trial: A Randomized Phase III Trial of Cisplatin Plus Gemcitabine Compared With Paclitaxel Plus Gemcitabine as First-Line Therapy for Patients With Metastatic Triple-Negative Breast Cancer. Ann Oncol (2018) 29(8):1741–7. doi: 10.1093/annonc/mdy209
36. Tutt A, Tovey H, Cheang MCU, Kernaghan S, Kilburn L, Gazinska P, et al. Carboplatin in BRCA1/2-Mutated and Triple-Negative Breast Cancer Brcaness Subgroups: The TNT Trial. Nat Med (2018) 24(5):628–37. doi: 10.1038/s41591-018-0009-7
37. Loibl S, Weber KE, Timms KM, Elkin EP, Hahnen E, Fasching PA, et al. Survival Analysis of Carboplatin Added to an Anthracycline/Taxane-Based Neoadjuvant Chemotherapy and HRD Score as Predictor of Response-Final Results From Geparsixto. Ann Oncol (2018) 29(12):2341–7. doi: 10.1093/annonc/mdy460
38. Loibl S, O’Shaughnessy J, Untch M, Sikov WM, Rugo HS, McKee MD, et al. Addition of the PARP Inhibitor Veliparib Plus Carboplatin or Carboplatin Alone to Standard Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer (Brightness): A Randomised, Phase 3 Trial. Lancet Oncol (2018) 19(4):497–509. doi: 10.1016/S1470-2045(18)30111-6
39. Ignatiadis M, Litiere S, Rothe F, Riethdorf S, Proudhon C, Fehm T, et al. Trastuzumab Versus Observation for HER2 Nonamplified Early Breast Cancer With Circulating Tumor Cells (EORTC 90091-10093, BIG 1-12, Treat CTC): A Randomized Phase II Trial. Ann Oncol (2018) 29(8):1777–83. doi: 10.1093/annonc/mdy211
40. Fasching PA, Loibl S, Hu C, Hart SN, Shimelis H, Moore R, et al. BRCA1/2 Mutations and Bevacizumab in the Neoadjuvant Treatment of Breast Cancer: Response and Prognosis Results in Patients With Triple-Negative Breast Cancer From the Geparquinto Study. J Clin Oncol (2018) 36(22):2281–7. doi: 10.1200/JCO.2017.77.2285
41. Schouten PC, Vollebergh MA, Opdam M, Jonkers M, Loden M, Wesseling J, et al. High XIST and Low 53BP1 Expression Predict Poor Outcome After High-Dose Alkylating Chemotherapy in Patients With a BRCA1-Like Breast Cancer. Mol Cancer Ther (2016) 15(1):190–8. doi: 10.1158/1535-7163.MCT-15-0470
42. Mateo J, Lord CJ, Serra V, Tutt A, Balmana J, Castroviejo-Bermejo M, et al. A Decade of Clinical Development of PARP Inhibitors in Perspective. Ann Oncol (2019) 30(9):1437–47. doi: 10.1093/annonc/mdz192
43. Shen Y, Rehman FL, Feng Y, Boshuizen J, Bajrami I, Elliott R, et al. BMN 673, a Novel and Highly Potent PARP1/2 Inhibitor for the Treatment of Human Cancers With DNA Repair Deficiency. Clin Cancer Res (2013) 19(18):5003–15. doi: 10.1158/1078-0432.CCR-13-1391
44. Wagner LM. Profile of Veliparib and Its Potential in the Treatment of Solid Tumors. Onco Targets Ther (2015) 8:1931–9. doi: 10.2147/OTT.S69935
45. Zhu G, Chang P, Lippard SJ. Recognition of Platinum-DNA Damage by Poly(ADP-Ribose) Polymerase-1. Biochemistry (2010) 49(29):6177–83. doi: 10.1021/bi100775t
46. Danza K, Pilato B, Lacalamita R, Addati T, Giotta F, Bruno A, et al. Angiogenetic Axis Angiopoietins/Tie2 and VEGF in Familial Breast Cancer. Eur J Hum Genet (2013) 21(8):824–30. doi: 10.1038/ejhg.2012.273
47. Sharma M, Piplani S, Madan M, Manjari M, Garg S, Kaur K. Hypoxia-Induced Factor-1 Alpha, Vascular Endothelial Growth Factor Expression in BRCA1-Related Breast Cancer: A Prospective Study in Tertiary Care Hospital. Indian J Pathol Microbiol (2017) 60(4):469–74. doi: 10.4103/IJPM.IJPM_524_16
48. Tobalina L, Armenia J, Irving E, O’Connor MJ, Forment JV. A Meta-Analysis of Reversion Mutations in BRCA Genes Identifies Signatures of DNA End-Joining Repair Mechanisms Driving Therapy Resistance. Ann Oncol (2021) 32(1):103–12. doi: 10.1016/j.annonc.2020.10.470
49. Domchek SM, Postel-Vinay S, Im SA, Park YH, Delord JP, Italiano A, et al. Olaparib and Durvalumab in Patients With Germline BRCA-Mutated Metastatic Breast Cancer (MEDIOLA): An Open-Label, Multicentre, Phase 1/2, Basket Study. Lancet Oncol (2020) 21(9):1155–64. doi: 10.1016/S1470-2045(20)30324-7
Keywords: BRCA mutation, chemotherapy, targeted therapy, meta-analysis, breast cancer
Citation: Jiang Y, Meng X-Y, Deng N-N, Meng C, Li L-H, He Z-K, Wang X-Y, Song Z-Y and Cui R-J (2021) Effect and Safety of Therapeutic Regimens for Patients With Germline BRCA Mutation-Associated Breast Cancer: A Network Meta-Analysis. Front. Oncol. 11:718761. doi: 10.3389/fonc.2021.718761
Received: 01 June 2021; Accepted: 05 August 2021;
Published: 20 August 2021.
Edited by:
Tomoharu Sugie, Kansai Medical University Hospital, JapanReviewed by:
Camila O. Dos Santos, Cold Spring Harbor Laboratory, United StatesAkira Yamauchi, Kitano Hospital, Japan
Copyright © 2021 Jiang, Meng, Deng, Meng, Li, He, Wang, Song and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong-Jun Cui, Y3Vpcm9uZ2p1bjA0M0AxMjYuY29t
†These authors have contributed equally to this work
 Ying Jiang
Ying Jiang Xiang-Yu Meng1†
Xiang-Yu Meng1† Zi-Kang He
Zi-Kang He Rong-Jun Cui
Rong-Jun Cui