
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 30 September 2021
Sec. Cancer Molecular Targets and Therapeutics
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.711642
This article is part of the Research TopicRadiochemotherapy in Multimodal Cancer TherapyView all 20 articles
 Qiyu Luo1,2†
Qiyu Luo1,2† Wenwu He3†
Wenwu He3† Tianqin Mao1
Tianqin Mao1 Xuefeng Leng3
Xuefeng Leng3 Hong Wu4
Hong Wu4 Wen Li4
Wen Li4 Xuyang Deng3
Xuyang Deng3 Tingci Zhao5
Tingci Zhao5 Ming Shi6
Ming Shi6 Chuan Xu4*
Chuan Xu4* Yongtao Han3*
Yongtao Han3*Long-term survival in oesophageal squamous cell carcinoma (ESCC) is related with pathological response after neoadjuvant chemoradiotherapy (NCRT) followed by surgery. However, effective biomarkers to predict the pathologic response are still lacking. Therefore, a systematic analysis focusing on genes associated with the efficacy of chemoradiotherapy in ESCC will provide valuable insights into the regulation of molecular processes. By screening publications deposited in PubMed, we collected genes associated with the efficacy of chemoradiotherapy. A specific subnetwork was constructed using the Steiner minimum tree algorithm. Survival analysis in Kaplan-Meier Plotter online resources was performed to explore the relationship between gene mRNA expression and the prognosis of patients with ESCC. Quantitative real-time polymerase chain reaction (qRT-PCR), Western blotting, and immunohistochemical staining (IHC) were used to evaluate the expression of key genes in cell lines and human samples. The areas under the receiver operating characteristic (ROC) curves (AUCs) were used to describe performance and accuracy. Transwell assays assessed cell migration, and cell viability was detected using the Cytotoxicity Assay. Finally, we identified 101 genes associated with efficacy of chemoradiotherapy. Additionally, specific molecular networks included some potential related genes, such as CUL3, MUC13, MMS22L, MME, UBC, VAPA, CYP1B1, and UGDH. The MMS22L mRNA expression level showed the most significant association with the ESCC patient outcome (p < 0.01). Furthermore, MMS22L was downregulated at both the mRNA (p < 0.001) and protein levels in tumour tissues compared with that in normal tissues. Lymph node metastasis was significantly associated with low MMS22L expression (p < 0.01). MMS22L levels were inversely correlated with the NCRT response in ESCC (p < 0.01). The resulting area under the ROC curve was 0.847 (95% CI: 0.7232 to 0.9703; p < 0.01). In conclusion, low expression of MMS22L is associated with poor response to NCRT, worse survival, lymph node metastasis, and enhanced migration of tumour cells in ESCC.
According to the 2018 Global Cancer Statistics report, oesophageal cancer is among the 10 most frequent carcinomas globally (1). Squamous cell carcinoma (SCC) and adenocarcinoma are the major histologic types of oesophageal carcinoma, and SCC is the main histological type in China (1, 2). Despite advances in surgery, radiotherapy, and chemotherapy, the 5-year survival rate for patients with oesophageal squamous cell carcinoma (ESCC) remains markedly poor because of an advanced stage at diagnosis, the presence of tumour heterogeneity, and insufficient tumour prognostic factors (3, 4). Regarding ESCC at moderate-to-advanced stages, both effective preoperative chemotherapy and chemoradiotherapy have been widely used to shrink tumour size, repress tumour growth or metastasis, increase the R0 resection rate, and reduce the local recurrence rate of ESCC, to improve overall survival compared with surgery alone (5–8). No significant benefits were found in patients who did not respond to the conventional therapy because of toxicity from neoadjuvant chemoradiation therapy (NCRT); they might miss the best timing of treatment and obtain worse prognosis (9–11).
Tumour heterogeneity presents a challenge to successfully treat cancer using chemoradiotherapy, and it is a major factor in chemoradiotherapy failure (12, 13). To enable individualised treatment, screening out response cases and avoiding overtreatment of patients who would not benefit from the inclusion of chemotherapy with sensitive biomarkers are critical. In recent years, although some studies have attempted to reveal the biomarkers that predict responses to chemoradiotherapy, no reliable biomarkers have been identified to assess the efficacy of NCRT in ESCC. Therefore, large amount of studies are needed to further refine the biomarkers for easy use and validate them as biomarkers for future clinical trials.
Identifying new biomarkers is warranted to assist in screening patients who can benefit from chemotherapy and chemoradiotherapy based on bioinformatics analysis, offering consolidated validation for the individual candidate genes. In this study, we collected genes potentially associated with efficacy of chemoradiotherapy to infer specific molecular networks associated with the efficacy of chemoradiotherapy, and some potential related genes were identified. Additionally, the primary aim of the study was to measure potential related gene expression levels in ESCC tissues and evaluate their value as potential predictive biomarkers for the NCRT response in ESCC.
As reported previously (4), the efficacy of chemoradiotherapy in patients with ESCC is associated with multiple genes. In the present study, all the genes were obtained by systematic analysis of the human genetic association studies deposited in PubMed (https://www.ncbi.nlm.nih.gov/pubmed). Briefly, with reference to published studies, the search terms were: “(chemotherapy OR radiotherapy OR chemoradiotherapy) AND (cancer OR carcinoma OR neoplasm OR tumour) AND (esophagus OR gastroesophageal) AND (genetic polymorphism OR genes)”, and the date of the last search was September 15, 2019. In total, 217 abstracts were identified by the final electronic searches in PubMed. We included abstracts with sufficient evaluation data, including the methodology, the definition of outcomes, and an appropriate evaluation matrix. Studies without any kind of validation (external validation or internal validation) were excluded. We excluded reviews, editorials, nonhuman studies, and letters without sufficient data. In total, 138 studies were excluded because they failed to meet the above criteria, and 79 articles met the prespecified inclusion criteria. The publications used and the discarded publications can be found in the supplementary documentation (Supplementary Table S1). Two reviewers (QL and MT) independently screened the full text and extracted the following information from each study: patient race, number of positive cases, interventions, histological type, the origin of specimens, and genes. Finally, we assembled the purpose genes associated with the response to chemoradiotherapy in ESCC histological type or cell lines.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analyses were performed using the R package “cluster Profiler” (14). For the GO and pathway enrichment analysis results, the p-value and q-values were calculated using Fisher’s exact test and the R package (p-value <0.05 and q-value <0.05).
In the context of a human protein-protein interaction (PPI) data set obtained from Protein Interaction Network Analysis platform (PINA) (15), we applied the Steiner minimum tree algorithm implemented in our software framework GenRev to construct a specific subnetwork by using the candidate gene set as seeds (16). A PPI network was built using the Search Tool for the Retrieval of Interacting Genes (STRING) database (version 11.0; https://string-db.org/) (17) and visualised by Cytoscape (an open-source software platform) (18).
The prognostic value of the expression of new gene mRNAs was evaluated using publicly available miRNA expression datasets (Pan-cancer RNA-seq) in Kaplan-Meier plotter (http://kmplot.com/analysis/), an online database including gene expression data and clinical data (19, 20). To assess the prognostic value of a specific gene, the patient samples were divided into two cohorts (high and low expression groups) according to the median mRNA expression of the gene. We analysed the overall survival (OS) of ESCC patients by using a Kaplan-Meier survival plot. Briefly, eight genes (CUL3, MUC13, MMS22L, MME, UBC, VAPA, CYP1B1, and UGDH) were uploaded into the database respectively to obtain the Kaplan-Meier survival plots, in which the number-at-risk is shown below the main plot. Log rank p-value and hazard ratio (HR) with 95% confidence intervals were calculated and displayed on the picture. A p-value <0.05 was considered statistically significant.
Sixty-one ESCC tissues were obtained from the Department of Thoracic Surgery of Sichuan Cancer Hospital (Chengdu, China) from Jan 2018 to Sept 2019 in this study, and all the samples were histologically confirmed to be ESCC tissues by a postoperative pathologist. Twenty-three paired human ESCC cancer tissues and matched adjacent normal tissues (located at least 5 cm from the tumour border, Supplementary Figure S1) from 23 patients with surgery alone were snap frozen after they were taken from surgery and then stored in liquid nitrogen for quantitative real-time PCR and Western blotting. Another 38 ESCC biopsy specimens from the gastroscopies were paraffin embedded for immunohistochemical staining analysis before patients received NCRT. The samples used in the study were approved by the Ethical Committee of the Sichuan Cancer Hospital (No. SCCHEC-02-2017-043), and the patients provided written informed consent to participants.
The human ESCC cell lines TE-1, Kyse150, and Eca109 and the human normal oesophageal cell line HEEC were provided by the Shanghai Institute of Cell Biology, Chinese Academy of Sciences. All the cell lines were maintained in RPMI-1640 (HyClone, USA) supplemented with 100 U/ml of penicillin-streptomycin (HyClone, USA) and 10% foetal bovine serum (FBS; Gibco, USA) at 37°C with 5% CO2 in a humidified atmosphere.
The mRNA expression of MMS22L was measured by quantitative real-time polymerase chain reaction (qRT-PCR). The above cell clines and 23 paired human ESCC cancer tissues and matched adjacent normal tissues were individually homogenised in liquid nitrogen, and total RNA was extracted from cells and tissues using TRIzol reagent (Invitrogen, Waltham, MA, USA). The total RNA products were immediately transcribed into cDNA using a cDNA synthesis kit (Takara, Kyoto, Japan) following the manufacturer’s instructions. Complementary DNA was amplified using TB Green™ Advantage® qPCR Premix (Takara, Kyoto, Japan) on the CFX-Connect Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The MMS22L primers were as follows: forward 5′-CAGAGAATGTCACAGGTAGTGCC-3′; reverse 5′-TCTGATGGAGCTGTGCTTGGCA-3′. The conditions for qRT-PCR were as follows: initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 5 s and 55°C–57°C for 30 s, and melting curves were generated by heating from 55°C to 95°C with 0.5°C increments each cycle. The results were normalised to β-actin using the 2−ΔΔCt method (21), the forward primer: 5′-CTTAGTTGCGTTACACCCTTTCTTG-3′ and reverse primer 3′-ACTGCTGTCACCTTCACCGTTC-5′. All the experiments were performed in triplicate.
The Total proteins of tissues and cells were extracted using RIPA buffer with protease inhibitors (Biyuntian, China). The protein concentrations were measured using the bicinchoninic acid (BCA) Protein Assay Kit (Beijing Suolabao Biotech, China). The MMS22L protein levels in cancer tissue and adjacent tissue were evaluated by Western blotting (WB). Briefly, equal amounts of total protein extract were first separated in an 8% SDS-PAGE gel and transferred onto PVDF membranes (Millipore, Billerica, MA, USA). Next, the membranes were blocked for 2 h with PBST and 5% milk at room temperature, incubated with primary antibody overnight at 4°C and then with secondary antibody for 2 h at room temperature. Protein expression was detected using an anti-MMS22L (C6orf167) antibody (ab181047, 1:1,000, Abcam, Cambridge, MA, USA) and a β-actin polyclonal antibody (1:1,000 CST, Danvers, MA, USA). Protein bands were detected using an Immobilon® Western system (Millipore; #WBKLS0100) and imaged on the Minichemi machine (Sage Creation, Beijing, China).
Tissues from paraffin-embedded blocks were sectioned at 5-μm thickness. Immunohistochemical staining (IHC) was performed using DAB kit (Zhongshan Golden Bridge, Beijing, China) following the manufacturer’s protocol. The anti-MMS22L antibody (bs17689R) used for IHC was purchased from Bioss (Beijing, China). To quantify MMS22L staining, five randomly chosen fields per section were evaluated at ×200 magnification for each sample. Image-Pro Plus 6.0 was used to determine integrated optical density (IOD) values, and the IOD per unit area (mean density) represented the relative MMS22L expression level.
Two target small hairpin RNA (shRNA) lentiviruses of the MMS22L gene, sh147 and sh148 and a negative control shRNA lentivirus con077 were designed and synthesised by Shanghai Ji Kai Gene Technology Co., Ltd. (Shanghai, China). TE-1 cells were infected with lentivirus at an MOI of 10 PFU per cell. Stable transformants were selected with 2 μg/ml of puromycin. The knockdown efficiency was detected by qRT-PCR and Western blotting.
Transwell migration assays were performed using plates (Corning, Corning, NY, USA) with 8‐μm‐pore size membranes. Briefly, 2 × 104 cells were suspended in 200 μl of FBS‐free RPMI‐1640 medium and added to the upper chambers of Transwell plates. RPMI‐1640 medium (500 μl) supplemented with 5% FBS was seeded into the lower chamber. After a 24‐h incubation period at 37°C and 5% CO2, the migrated cells were fixed with 4% paraformaldehyde (in 1 × PBS) for 20 min at room temperature and then were stained with crystal violet.
Cytotoxicity was assayed using cell counting kit (CCK8; Hanheng, Shanghai, China) following the manufacturer’s instructions. Briefly, cells were seeded on 96-well plates at a density of 2.0 × 103 per well. After cells were treated with 5-FU (Selleck, Houston, TX, USA) at concentrations of 0, 0.1, 1, 5, 10, and 100 μM for 72 h, the medium containing 5-FU was exchanged for 100 μl of RPMI-1640, and 10 μl of CCK8 reagent was added. Two hours later, absorbance at 450 nm was measured using a microplate reader. Each group had three repeats, and the experiment was repeated three times.
All the data were obtained from at least three independent measurements and were shown as means ± SEM. Student’s t-test, Mann-Whitney tests, and receiver and IC50 operating characteristic (ROC) curves were performed using GraphPad Prism 7 software (GraphPad Software Inc., San Diego, CA, USA). One-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test was used for data analysis among three or more groups. Statistical analysis was conducted, and statistical significance was set at p < 0.05.
As described in the Materials and Methods section, 101 candidate genes associated with the efficacy of chemoradiotherapy in patients with ESCC were assembled after removing duplicates. The GO annotations were classified as biological process, cellular component, and molecular function (p < 0.05), and the top 10 GO terms are shown in Figures 1A–C, respectively. The top biological process GO enrichment terms were cotranslational protein targeting the membrane, targeting the ER, and establishment of protein localisation to endoplasmic reticulum. Ribosome component was the most enriched cellular component GO term. Additionally, we found several enriched molecular functions such as structural constituent of ribosome, damaged DNA binding, antigen binding, single-stranded DNA binding, and ubiquitin protein ligase binding. Furthermore, the KEGG pathways in which 101 candidate genes were mostly enriched were ribosome, followed by human T-cell leukaemia virus 1 infection, platinum drug resistance, human cytomegalovirus infection, proteoglycans in cancer, etc. (Figure 1D).
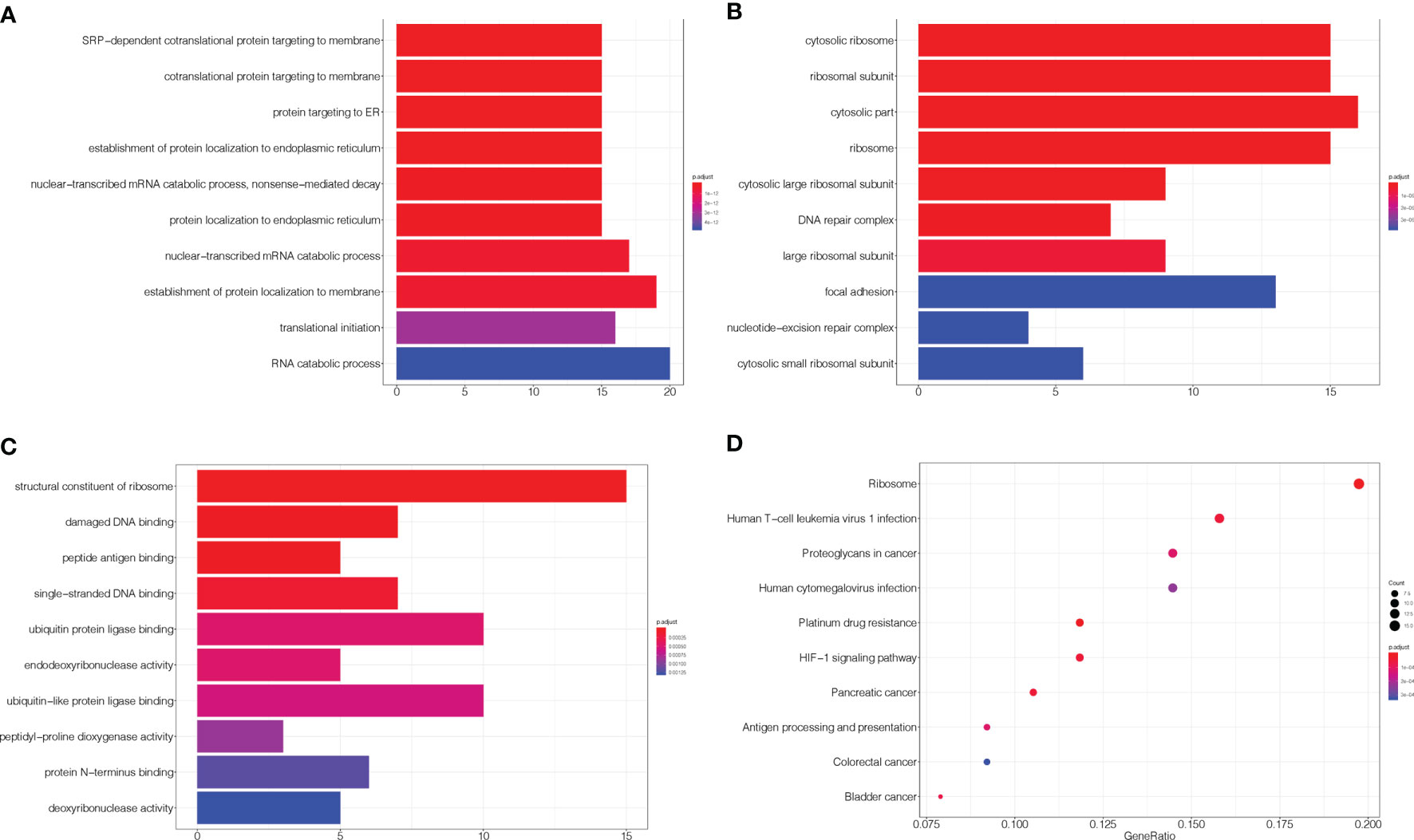
Figure 1 GO and KEGG enrichment of related genes. (A–C) The top 10 GO terms in biological process, cellular component, and molecular function, respectively. (D) KEGG pathways were analysed, and the top 10 pathways are shown. The x-axis shows the GeneRatio, whereby a higher value indicates more genes enriched in the pathway; the y-axis shows the enriched pathways, and the redder the dot is, the more significant is the pathway.
The protein subnetwork comprised 105 nodes and 379 edges (interactions), and 97 of 101 candidate genes were included in the subnetwork, accounting for 92.38% of 105 genes in the network and 96% of 101 candidate genes, and demonstrating a high coverage of the candidate genes set in the subnetwork (Figure 2). Additionally, eight genes in the subnetwork outside of the candidate gene set were obtained (Table 1).
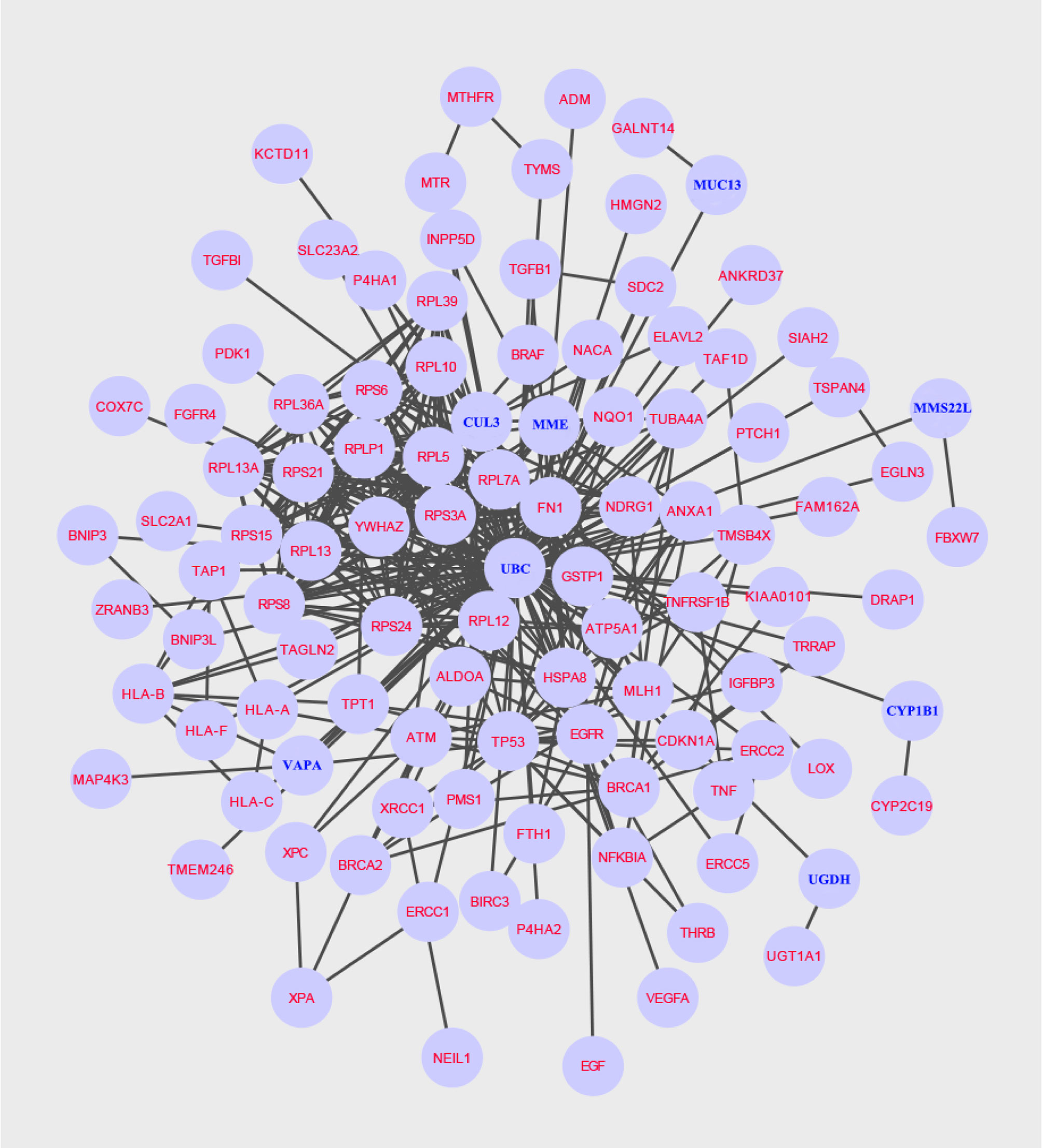
Figure 2 A specific protein network associated with NCRT efficacy is built using the Steiner minimal tree algorithm, including 105 nodes and 379 edges. Additionally, eight new genes are labelled in blue.
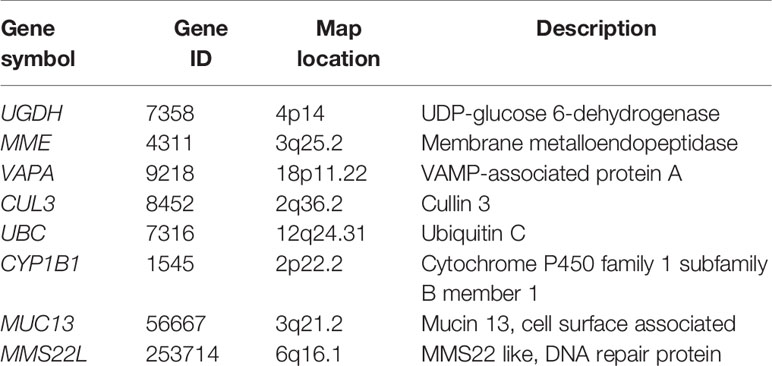
Table 1 Identification of eight potentially related genes associated with chemoradiotherapy efficacy: UGDH, VAPA, MME, CUL3, UBC, CYP1B1, MUC13, and MMS22L.
Using the Kaplan-Meier plotter database, the prognostic value of the eight novel genes was evaluated in ESCC patients. We found that higher and lower expression of five biomarkers was significantly associated with overall survival. ESCC patients with higher mRNA levels of MMS22L (HR = 0.33; CI: 0.14−0.75; logrank p = 0.0052) and MUC13 (HR = 0.37; CI: 0.14−0.96; logrank p = 0.033) had higher OS (Figures 3A, B), and ESCC patients with lower mRNA levels of VAPA (HR = 2.32; CI: 1.04−5.17; logrank p = 0.035), CYP1B1 (HR = 2.67; CI: 1.02−6.97; logrank p = 0.039), and UBC (HR = 2.26; CI: 1.03−4.96; logrank p = 0.038) had higher OS (Figures 3C–E), while the mRNA expression of UGDH, MME and CUL13 were not associated with ESCC patient survival (Figures 3F–H).
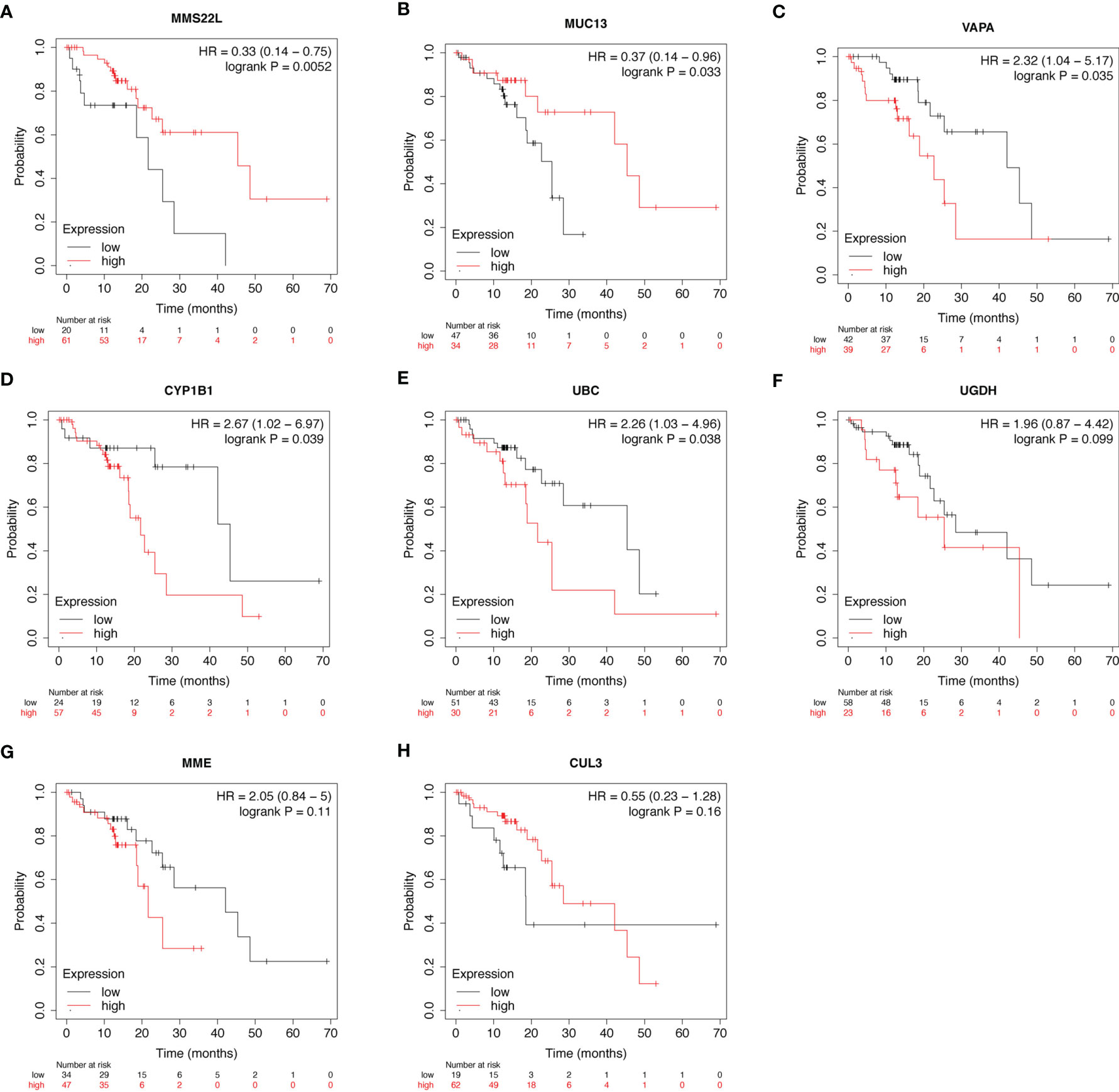
Figure 3 Prognostic value of mRNA expression of eight genes in patients with ESCC. (A–H) Overall Survival curves of MMS22L (A), MUC13 (B), VAPA (C), CYP1B1 (D), UBC (E), UGDH (F), MME (G), and CUL13 (H) were plotted in Kaplan-Meier plotter. “Probability” on the y-axis represents the survival rates, the red line represents the patients with mRNA expression above the median, and the black line represents the patients with mRNA expression below the median. ESCC, Oesophageal squamous cell carcinoma; HR, hazard ratio.
MMS22L is not a well-studied protein, and information about this gene product is very limited. As described above, the expression of MMS22L had the most significant association with ESCC patient outcome. Cellular components associated with the function of MMS22L were significantly enriched, such as damaged DNA binding and ubiquitin protein ligase binding. Therefore, we further detected the expression level of the MMS22L gene in ESCC tissues and adjacent normal tissues using quantitative real-time PCR and WB. MMS22L mRNA expression was significantly decreased in ESCC tissue compared with that in adjacent normal tissues (p < 0.05) (Figure 4A), and WB results were consistent with the RT-qPCR (Figure 4B). MMS22L protein expression was significantly reduced in seven tumour tissues compared with that in normal adjacent tissues.
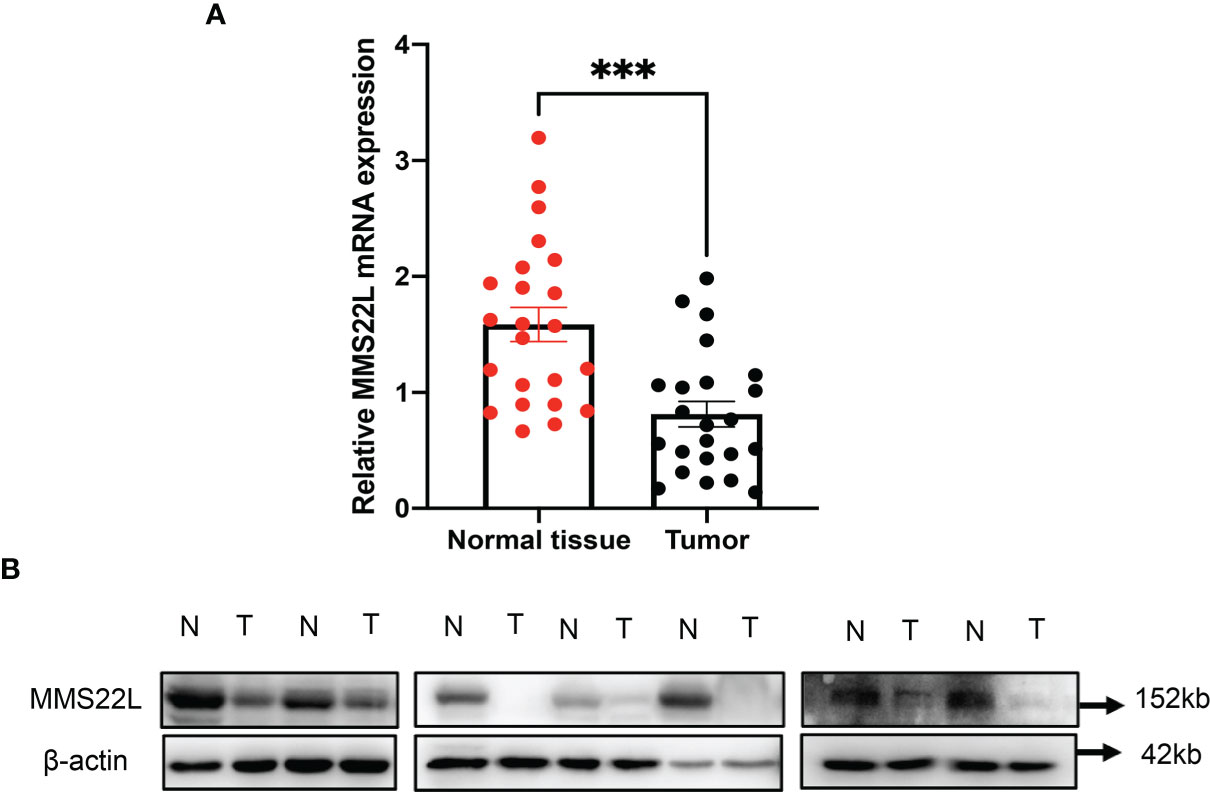
Figure 4 The MMS22L mRNA and protein levels are significantly lower in cancerous tissues than in normal adjacent tissues. (A) MMS22L mRNA was detected by qRT-PCR, and relative quantification analysis was normalised to β-actin mRNA (***p < 0.001). (B) MMS22L protein expression was detected in randomly selected seven ESCC tissues and adjacent normal tissues using Western blotting. T, tumour tissues; N, normal adjacent tissues.
According to the RECIST 1.1 criteria (22), the response to NCRT was defined as complete response (CR), partial response (PR), stable disease, (SD), and progressive disease (PD), and the 38 patients receiving NCRT were divided into the responding group (CR plus PR, 29 patients) and nonresponding group (SD, nine patients) according to the response to NCRT in this study. MMS22L IHC in representative tumour tissues are shown in Figure 5A. The MMS22L protein in the responding group was significantly higher than that in the nonresponding groups (p < 0.01, Figure 5B). This result indicated that the MMS22L expression significantly predicted a response to NACRT. ROC curve analyses revealed that MMS22L was a valuable biomarker for differentiating responding from nonresponding for NCRT. MMS22L yielded an AUC of 0.847 (95% CI: 0.7232 to 0.9703; p < 0.01; Figure 5C) with 100% sensitivity and 65.52% specificity in predicting the efficacy of NCRT in ESCC. Additionally, among the various clinicopathological characteristics evaluated, no significant associations were found between the MMS22L expression level and age, sex, differentiation, location, depth of invasion, or TNM clinical stage, but a low expression level of MMS22L was markedly associated with lymph node metastasis (p < 0.01; Table 2).
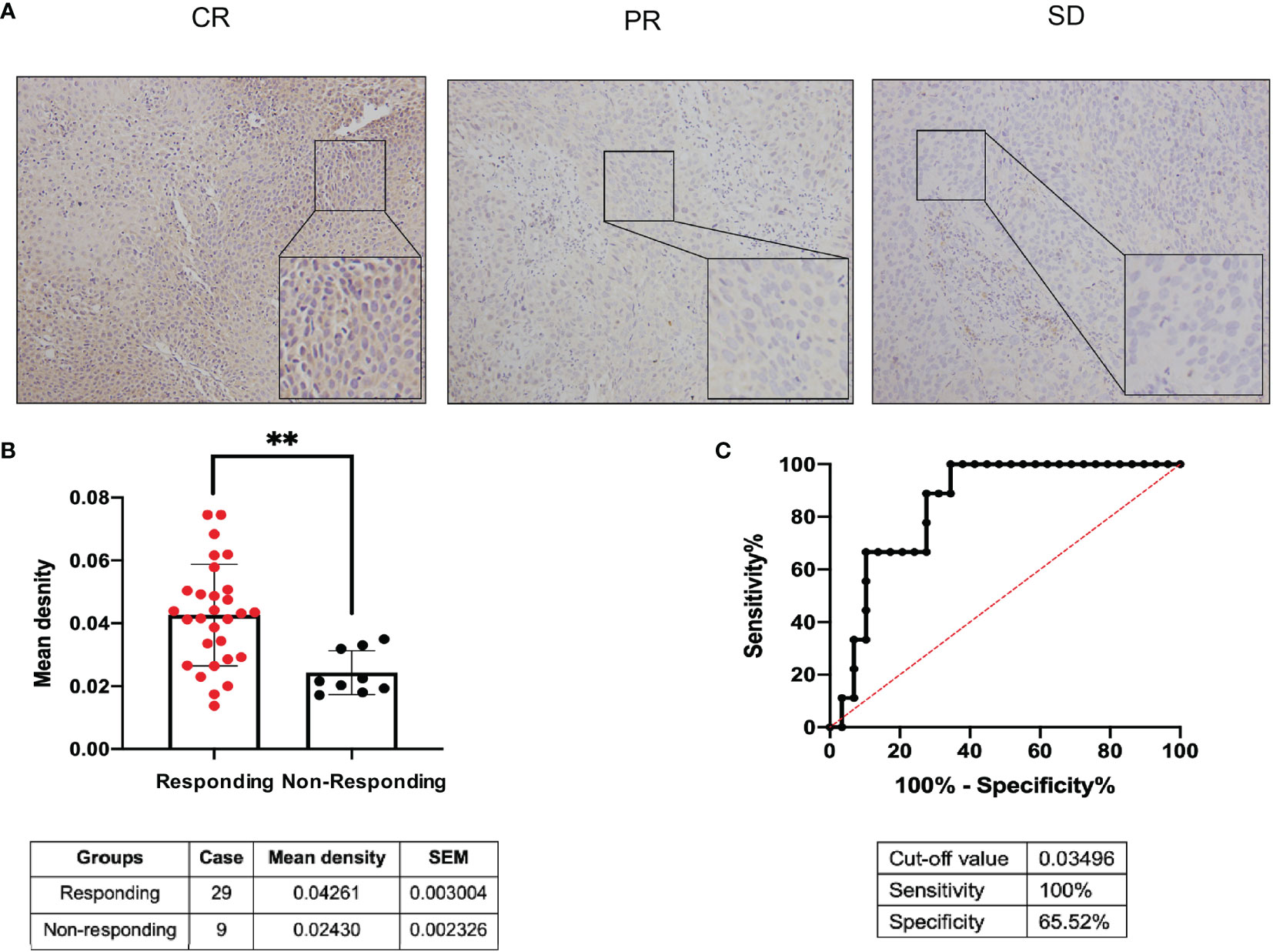
Figure 5 Predictive value of MMS22L for different responses to NCRT in patients with ESCC. (A) MMS22L IHC in representative tumour tissues. (B) The mean density of the responding group and nonresponding group from all 38 cases who received NCRT is summarised in the table below. (C) ROC curve for evaluating the predictive value of MMS22L for ESCC patients with NCRT. The AUC was 0.847, the sensitivity was 100%, the specificity was 65.52%, and the cutoff value was 0.03495 (**p < 0.01). NCRT, neoadjuvant chemoradiotherapy; AUC, area under the ROC curve; ROC, receiver operating characteristic.
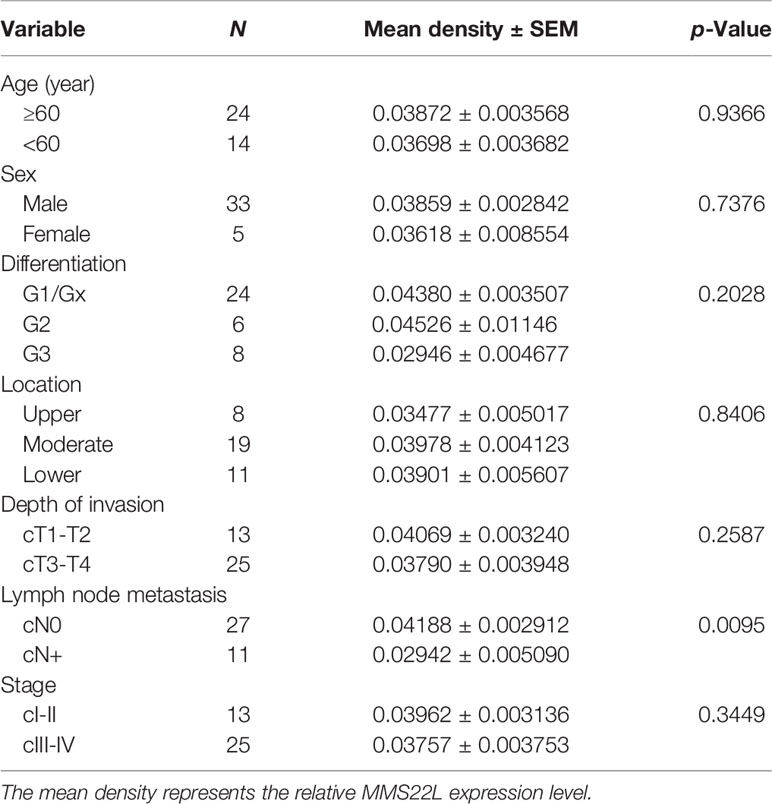
Table 2 Association of MMS22L expression in the biopsy specimens with clinicopathological parameters.
To assess MMS22L expression in human ESCC cells, we detected the mRNA and protein expression of MMS22L in HEEC cells and three human ESCC cell lines (TE-1, Kyse150, and Eca109) by qRT-PCR and WB, respectively. The expression of MMS22L was lower in TE-1, Kyse150, and Eca109 cells than in HEEC cells (Figure 6A). TE-1 cells showed the highest MMS22L expression in three human ESCC cell lines. Thus, TE-1 cells were selected for subsequent assays. After targeting MMS22L with two different shRNAs, the knockdown efficiency was detected (Figure 6B). We performed Transwell migration assays to analyse whether MMS22L inhibits the migration ability of TE-1 cells. MMS22L inhibited the migration of TE-1 cells (Figure 6C). Different TE-1 cells were determined by CCK8 assays with different doses of 5-FU to calculate the IC50 and were performed in triplicate. The IC50 value decreased significantly after MMS22L knockout (Figure 6D). These results from in vitro experiments indicated that the knockdown of MMS22L in the TE-1 cell line is associated with enhanced migration and resistance to 5-FU.
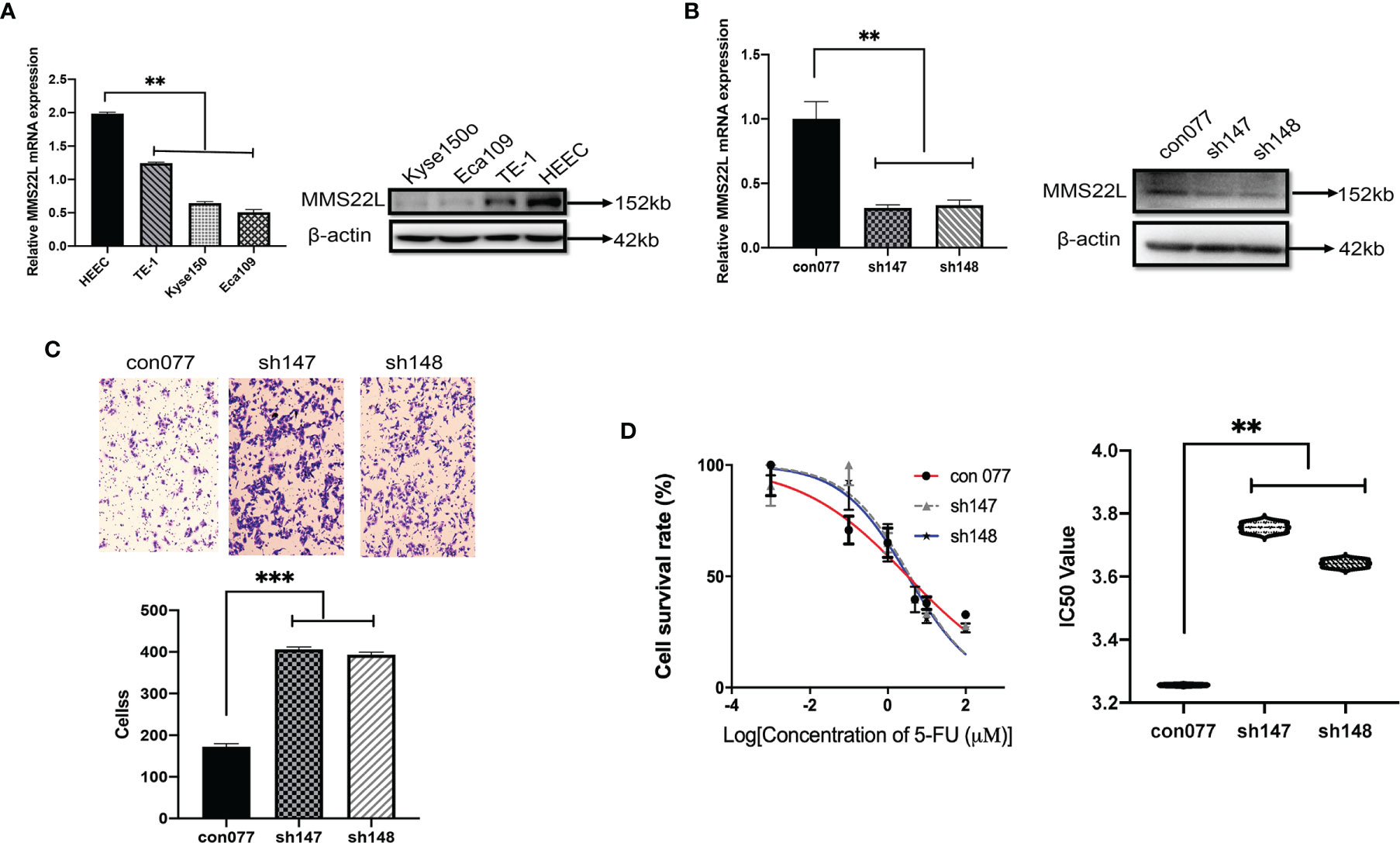
Figure 6 Lower expression of MMS22L increases the migration capabilities and the 5-FU resistance of TE-1 cells. (A) MMS22L expression in a human normal oesophageal cell line (HEEC) and human ESCC cell lines. (B) The transfection efficiency was measured after 48 h. (C) In the shRNA groups, the migration capabilities increased and the number of migrating cells increased compared with those in the con077 group. (D) IC50 values of TE-1 cells to 5-FU were detected via the CCK8 assay after transfection (**p < 0.01; ***p < 0.001). CCK8, cell counting kit; IC50, half-maximal inhibitory concentration; 5-FU, 5-fluorouracil.
According to published statistics, approximately 572,034 new oesophageal cancer cases (3.2%) and an estimated 508,585 related deaths (5.3%) occurred (1). With the development of comprehensive treatment strategies, patients with ESCC benefit from NCRT (5). However, the postoperative pathological complete response rate was only 43.2% according to a phase III, multicentre, randomised open-label clinical trial (NEOCRTEC5010) (23). Therefore, further understanding of NCRT resistance-related genes as novel prognostic biomarkers in ESCC is necessary. Recently, an increasing number of potential genes have been found to be associated with chemoradiotherapy efficacy and prognosis in patients with cancer. However, a few of these predicted genes have been identified in ESCC, and a few biomarkers are available for clinical monitoring.
In our study, we applied the Steiner minimum tree algorithm to explore novel biomarkers in the context of a human PPI background. First, a list of 101 candidate genes associated with the efficacy of chemoradiotherapy in patients with ESCC was assembled in 79 relevant articles. Simultaneously, to better understand the function of these genes in ESCC, we performed GO and KEGG analyses. Notably, KEGG analysis revealed that the pathways in which these genes are mainly enriched are ribosome, human T-cell leukaemia virus 1 infection, platinum drug resistance, human cytomegalovirus infection, and proteoglycans in cancer. Second, we obtained eight novel genes associated with linking genes potentially related to chemoradiotherapy efficacy in ESCC outside the candidate gene set. Referring to published studies, the findings of our study validate previous reported outcomes because different functions of these genes have been identified to be associated with ESCC. MUC13 is a potential biomarker to predict the efficacy of neoadjuvant chemotherapy in ESCC patients (24, 25). Moghadam et al. suggest a relationship between the CYP1B1-rs1056836 genetic polymorphism and clinical features of ESCC (26). Therefore, the results of the present study are considered to be reliable.
To further identify effective biomarkers with diagnostic and prognostic value, we evaluated the effects of the eight novel genes on the survival of ESCC patients using the online Kaplan-Meier plotter. ESCC patients with higher mRNA levels of MMS22L and MUC13 had higher OS. Additionally, ESCC patients with lower mRNA levels of VAPA, CYP1B1, and UBC had higher OS. As shown in the Kaplan-Meier plotter, MMS22L expression was the most likely candidate gene among many novel genes. The MMS22L gene is mapped to chromosome 6 open-reading frame 167, also known as C6orf167. In our study, MMS22L expression in 23 ESCC tissues was notably lower than that in their para-carcinoma tissues, and this trend was consistently observed across the ESCC cell lines and the human normal oesophageal cell line. Thus, MMS22L may be a tumour suppressor gene in ESCC. In contrast to our findings, a previous study found that MMS22L expression was upregulated in lung and oesophageal cancer tissues compared with that in adjacent normal lung and oesophageal tissues, and MMS22L was identified as an oncogene (27). This discrepancy may arise from the inadequate sample size and unknown oesophageal cancer histologic type in their study. Many genes play both tumour suppressor or oncogenic roles in different tissues, tumour types, or cellular contexts (28).
Previous studies have shown that MMS22L plays critical roles in the DNA damage response and repair, affecting the response of tumour cells to DNA-damaging agents, such as camptothecin (CPT), ionising radiation (IR), hydroxyurea (HU), and methyl methanesulfonate (MMS) (29–32). According to previous reports, knocking down MMS22L promoted apoptosis by activating caspase-3 and downregulating Bcl-XL (29), an inhibitor of apoptosis. Importantly, Duro et al. found that tumour cells lacking MMS22L were resistant to HU, cisplatin, and IR. However, this finding is inconsistent with that reported by O’Connell et al. who found that Hela cells lacking MMS22L were sensitive to IR, CPT, and MMS (30). Additionally, downregulation of MMS22L is associated the bone metastasis in breast cancer (33). Previous controversial results have generated considerable interest regarding the function of MMS22L in ESCC. In the present study, we found that low MMS22L expression, immunohistochemically stained and analysed semiquantitatively in ESCC samples, was a useful predictor of worse responses to NCRT and lymph node metastasis, and enhanced migration and resistance to 5-FU in the TE-1 cell line. These findings were corroborated by in vitro studies showing that the knockdown of MMS22L in the TE-1 cell line was associated with enhanced migration and resistance to 5-FU.
Proteins associated with each other in the String Protein-Protein Interaction Network have related functions, and FBXW7 is most closely related to MMS22L. FBXW7 is an F-box protein that binds to key regulators of cell division and growth, including cyclin E, MYC, JUN, and Notch (34). FBXW7 is a critical tumour suppressor involved in the ubiquitin-proteasome system in human cancer, and loss of FBXW7 function leads to chromosomal instability (34, 35). FBXW7 expression is downregulated in ESCC tissues and correlates with the TNM stage, the degree of differentiation, the invasion depth, the lymph node metastasis, and a worse prognosis in ESCC (36, 37). Mutations in FBXW7 are associated with metastasis and correlates with increased expression of T-cell proliferation and antigen presentation functions (38). These results may suggest that an indirect or direct regulatory function between MMS22L and FBXW7 will contribute to tumour progression and metastasis through immunological effects or the tumour microenvironment. However, this interpretation of these data remains speculative at this point and will require further cell biological studies.
In conclusion, network analysis based on molecular aspects associated with the efficacy of chemoradiotherapy in ESCC may facilitate identification of novel biomarkers and a deeper understanding of the mechanisms. Our approach presents interesting approaches for future studies. Additionally, our data indicate that low MMS22L expression status in biopsy specimens is a predictive factor for the unfavourable efficacy of NACRT in ESCC. Therefore, patients with ESCC receiving NCRT are likely to fail when MMS22L downregulation is observed on a biopsy specimen. However, a larger sample size will be needed to validate and possibly extend these findings.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the Ethical Committee of the Sichuan Cancer Hospital (No. SCCHEC-02-2017-043). The patients/participants and legal guardian/next of kin provided written informed consent to participate in this study.
Conception and design: YH and CX. Acquisition of data: QL and WH. Analysis and interpretation of data: TM and XL. Drafting of the manuscript: QL and WH. Critical revision of the manuscript for important intellectual content: YH and CX. Harvesting of the specimen: XD and MS. Administrative, technical, or material support: HW, WL, and TZ. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Key R&D Program of China (2016YFC0901401), Bethune Charitable Foundation (HZB-20190528-19) and the Applied Basic Research Programs of Science and Technology Commission Foundation of Sichuan Province, China (2020YJ0174).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.711642/full#supplementary-material
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Liang H, Fan JH, Qiao YL. Epidemiology, Etiology, and Prevention of Esophageal Squamous Cell Carcinoma in China. Cancer Biol Med (2017) 14(1):33–41. doi: 10.20892/j.issn.2095-3941.2016.0093
3. Liu M, He Z, Guo C, Xu R, Li F, Ning T, et al. Effectiveness of Intensive Endoscopic Screening for Esophageal Cancer in China: A Community-Based Study. Am J Epidemiol (2019) 188(4):776–84. doi: 10.1093/aje/kwy291
4. Gusella M, Pezzolo E, Modena Y, Barile C, Menon D, Crepaldi G, et al. Predictive Genetic Markers in Neoadjuvant Chemoradiotherapy for Locally Advanced Esophageal Cancer: A Long Way to Go. Review of the Literature. Pharmacogenomics J (2018) 18(1):14–22. doi: 10.1038/tpj.2017.25
5. Shapiro J, van Lanschot JJB, Hulshof M, van Hagen P, van Berge Henegouwen MI, Wijnhoven BPL, et al. Neoadjuvant Chemoradiotherapy Plus Surgery Versus Surgery Alone for Oesophageal or Junctional Cancer (CROSS): Long-Term Results of a Randomised Controlled Trial. Lancet Oncol (2015) 16(9):1090–8. doi: 10.1016/s1470-2045(15)00040-6
6. van Hagen P, Hulshof MC, van Lanschot JJ, Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, et al. Preoperative Chemoradiotherapy for Esophageal or Junctional Cancer. New Engl J Med (2012) 366(22):2074–84. doi: 10.1056/NEJMoa1112088
7. Meredith KL, Weber JM, Turaga KK, Siegel EM, McLoughlin J, Hoffe S, et al. Pathologic Response After Neoadjuvant Therapy is the Major Determinant of Survival in Patients With Esophageal Cancer. Ann Surg Oncol (2010) 17(4):1159–67. doi: 10.1245/s10434-009-0862-1
8. Oppedijk V, van der Gaast A, van Lanschot JJ, van Hagen P, van Os R, van Rij CM, et al. Patterns of Recurrence After Surgery Alone Versus Preoperative Chemoradiotherapy and Surgery in the CROSS Trials. J Clin oncology: Off J Am Soc Clin Oncol (2014) 32(5):385–91. doi: 10.1200/jco.2013.51.2186
9. Dittrick GW, Weber JM, Shridhar R, Hoffe S, Melis M, Almhanna K, et al. Pathologic Nonresponders After Neoadjuvant Chemoradiation for Esophageal Cancer Demonstrate No Survival Benefit Compared With Patients Treated With Primary Esophagectomy. Ann Surg Oncol (2012) 19(5):1678–84. doi: 10.1245/s10434-011-2078-4
10. Donahue JM, Nichols FC, Li Z, Schomas DA, Allen MS, Cassivi SD, et al. Complete Pathologic Response After Neoadjuvant Chemoradiotherapy for Esophageal Cancer Is Associated With Enhanced Survival. Ann Thorac Surg (2009) 87(2):392–8; discussion 8-9. doi: 10.1016/j.athoracsur.2008.11.001
11. Pasini F, de Manzoni G, Zanoni A, Grandinetti A, Capirci C, Pavarana M, et al. Neoadjuvant Therapy With Weekly Docetaxel and Cisplatin, 5-Fluorouracil Continuous Infusion, and Concurrent Radiotherapy in Patients With Locally Advanced Esophageal Cancer Produced a High Percentage of Long-Lasting Pathological Complete Response: A Phase 2 Study. Cancer (2013) 119(5):939–45. doi: 10.1002/cncr.27822
12. Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. New Engl J Med (2012) 366(10):883–92. doi: 10.1056/NEJMoa1113205
13. Wu D, Wang DC, Cheng Y, Qian M, Zhang M, Shen Q, et al. Roles of Tumor Heterogeneity in the Development of Drug Resistance: A Call for Precision Therapy. Semin Cancer Biol (2017) 42:13–9. doi: 10.1016/j.semcancer.2016.11.006
14. Yu G, Wang LG, Han Y, He QY. Clusterprofiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics (2012) 16(5):284–7. doi: 10.1089/omi.2011.0118
15. Wu J, Vallenius T, Ovaska K, Westermarck J, Mäkelä TP, Hautaniemi S. Integrated Network Analysis Platform for Protein-Protein Interactions. Nat Methods (2009) 6(1):75–7. doi: 10.1038/nmeth.1282
16. Zheng S, Zhao Z. Genrev: Exploring Functional Relevance of Genes in Molecular Networks. Genomics (2012) 99(3):183–8. doi: 10.1016/j.ygeno.2011.12.005
17. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING V11: Protein-Protein Association Networks With Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res (2019) 47(D1):D607–d13. doi: 10.1093/nar/gky1131
18. Wiredja D, Bebek G. Identifying Gene Interaction Networks. Methods Mol Biol (2017) 1666:539–56. doi: 10.1007/978-1-4939-7274-6_27
19. Madamsetty VS, Pal K, Dutta SK, Wang E, Mukhopadhyay D. Targeted Dual Intervention-Oriented Drug-Encapsulated (DIODE) Nanoformulations for Improved Treatment of Pancreatic Cancer. Cancers (Basel) (2020) 12(5):1189–208. doi: 10.3390/cancers12051189
20. Quiñones-Díaz BI, Reyes-González JM, Sánchez-Guzmán V, Conde-Del Moral I, Valiyeva F, Santiago-Sánchez GS, et al. MicroRNA-18a-5p Suppresses Tumor Growth via Targeting Matrix Metalloproteinase-3 in Cisplatin-Resistant Ovarian Cancer. Front Oncol (2020) 10:602670. doi: 10.3389/fonc.2020.602670
21. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
22. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1). Eur J Cancer (2009) 45(2):228–47. doi: 10.1016/j.ejca.2008.10.026
23. Yang H, Liu H, Chen Y, Zhu C, Fang W, Yu Z, et al. Neoadjuvant Chemoradiotherapy Followed by Surgery Versus Surgery Alone for Locally Advanced Squamous Cell Carcinoma of the Esophagus (NEOCRTEC5010): A Phase III Multicenter, Randomized, Open-Label Clinical Trial. J Clin Oncol (2018) 36(27):2796–803. doi: 10.1200/jco.2018.79.1483
24. Shen LY, Wang H, Dong B, Yan WP, Lin Y, Shi Q, et al. Possible Prediction of the Response of Esophageal Squamous Cell Carcinoma to Neoadjuvant Chemotherapy Based on Gene Expression Profiling. Oncotarget (2016) 7(4):4531–41. doi: 10.18632/oncotarget.6554
25. Wang H, Shen L, Lin Y, Shi Q, Yang Y, Chen K. The Expression and Prognostic Significance of Mucin 13 and Mucin 20 in Esophageal Squamous Cell Carcinoma. J Cancer Res Ther (2015) 11 Suppl 1:C74–9. doi: 10.4103/0973-1482.163846
26. Moghadam AR, Mehramiz M, Entezari M, Aboutalebi H, Kohansal F, Dadjoo P, et al. A Genetic Polymorphism in the CYP1B1 Gene in Patients With Squamous Cell Carcinoma of the Esophagus: An Iranian Mashhad Cohort Study Recruited Over 10 Years. Pharmacogenomics (2018) 19(6):539–46. doi: 10.2217/pgs-2018-0197
27. Nguyen MH, Ueda K, Nakamura Y, Daigo Y. Identification of a Novel Oncogene, MMS22L, Involved in Lung and Esophageal Carcinogenesis. Int J Oncol (2012) 41(4):1285–96. doi: 10.3892/ijo.2012.1589
28. Stepanenko AA, Vassetzky YS, Kavsan VM. Antagonistic Functional Duality of Cancer Genes. Gene (2013) 529(2):199–207. doi: 10.1016/j.gene.2013.07.047
29. Duro E, Lundin C, Ask K, Sanchez-Pulido L, MacArtney TJ, Toth R, et al. Identification of the MMS22L-TONSL Complex That Promotes Homologous Recombination. Mol Cell (2010) 40(4):632–44. doi: 10.1016/j.molcel.2010.10.023
30. O’Connell BC, Adamson B, Lydeard JR, Sowa ME, Ciccia A, Bredemeyer AL, et al. A Genome-Wide Camptothecin Sensitivity Screen Identifies a Mammalian MMS22L-NFKBIL2 Complex Required for Genomic Stability. Mol Cell (2010) 40(4):645–57. doi: 10.1016/j.molcel.2010.10.022
31. O’Donnell L, Panier S, Wildenhain J, Tkach JM, Al-Hakim A, Landry MC, et al. The MMS22L-TONSL Complex Mediates Recovery From Replication Stress and Homologous Recombination. Mol Cell (2010) 40(4):619–31. doi: 10.1016/j.molcel.2010.10.024
32. Piwko W, Olma MH, Held M, Bianco JN, Pedrioli PG, Hofmann K, et al. Rnai-Based Screening Identifies the Mms22L-Nfkbil2 Complex as a Novel Regulator of DNA Replication in Human Cells. EMBO J (2010) 29(24):4210–22. doi: 10.1038/emboj.2010.304
33. Savci-Heijink CD, Halfwerk H, Koster J, van de Vijver MJ. A Novel Gene Expression Signature for Bone Metastasis in Breast Carcinomas. Breast Cancer Res Treat (2016) 156(2):249–59. doi: 10.1007/s10549-016-3741-z
34. Welcker M, Clurman BE. FBW7 Ubiquitin Ligase: A Tumour Suppressor at the Crossroads of Cell Division, Growth and Differentiation. Nat Rev Cancer (2008) 8(2):83–93. doi: 10.1038/nrc2290
35. Lin PC, Yeh YM, Lin BW, Lin SC, Chan RH, Chen PC, et al. Intratumor Heterogeneity of MYO18A and FBXW7 Variants Impact the Clinical Outcome of Stage III Colorectal Cancer. Front Oncol (2020) 10:588557. doi: 10.3389/fonc.2020.588557
36. Yu H, Ling T, Shi R, Shu Q, Li Y, Tan Z. [Expression of FBXW7 in Esophageal Squamous Cell Carcinoma and Its Clinical Significance]. Zhonghua Zhong Liu Za Zhi (2015) 37(5):347–51.
37. Yokobori T, Mimori K, Iwatsuki M, Ishii H, Tanaka F, Sato T, et al. Copy Number Loss of FBXW7 is Related to Gene Expression and Poor Prognosis in Esophageal Squamous Cell Carcinoma. Int J Oncol (2012) 41(1):253–9. doi: 10.3892/ijo.2012.1436
Keywords: oesophageal squamous cell carcinoma, MMS22L, bioinformatics analysis, neoadjuvant chemoradiotherapy, lymph node metastasis, migration
Citation: Luo Q, He W, Mao T, Leng X, Wu H, Li W, Deng X, Zhao T, Shi M, Xu C and Han Y (2021) MMS22L Expression as a Predictive Biomarker for the Efficacy of Neoadjuvant Chemoradiotherapy in Oesophageal Squamous Cell Carcinoma. Front. Oncol. 11:711642. doi: 10.3389/fonc.2021.711642
Received: 18 May 2021; Accepted: 31 August 2021;
Published: 30 September 2021.
Edited by:
Anne Vehlow, Technical University of Dresden, GermanyReviewed by:
Stephanie Hehlgans, Goethe University Frankfurt, GermanyCopyright © 2021 Luo, He, Mao, Leng, Wu, Li, Deng, Zhao, Shi, Xu and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongtao Han, eW9uZ3Rhb19oYW5AMTI2LmNvbQ==; Chuan Xu, eHVjaHVhbjEwMEB1ZXN0Yy5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.