
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 27 July 2021
Sec. Cancer Genetics
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.709796
This article is part of the Research Topic Emerging Role and Potential Application of Non-coding RNAs in Gastrointestinal Cancers View all 7 articles
 Zhuoyuan Xin1,2,3
Zhuoyuan Xin1,2,3 Luping Zhang1
Luping Zhang1 Mingqing Liu1
Mingqing Liu1 Yachen Wang1
Yachen Wang1 Yingli Zhang1
Yingli Zhang1 Weidan Zhao1
Weidan Zhao1 Yongxiao Sun1
Yongxiao Sun1 Lei Shi1
Lei Shi1 Na Xu1
Na Xu1 Nan Zhang1*
Nan Zhang1* Hong Xu1*
Hong Xu1*Background: Helicobacter pylori (H. pylori) is a type I biological carcinogen, which may cause about 75% of the total incidence of gastric cancer worldwide. H. pylori infection can induce and activate the cancer-promoting signaling pathway and affect the occurrence and outcome of gastric cancer through controlling the regulatory functions of long non-coding RNAs (lncRNAs). However, we have no understanding of the prognostic worth of lncRNAs for gastric cancer patients infected with H. pylori.
Method: We screened differentially expressed lncRNAs using DESeq2 method among TCGA database. And we built the H. pylori infection-related lncRNAs regulatory patterns. Then, we constructed H. pylori infection-based lncRNAs prognostic signatures for gastric cancer patients together with H. pylori infection, via uni-variable and multi-variable COX regression analyses. Based on receiver operator characteristic curve (ROC) analysis, we evaluated the prediction effectiveness for this model.
Results: We identified 115 H. pylori infection–related genes were differentially expressed among H. pylori–infected gastric cancer tissues versus gastric cancer tissues. Functional enrichment analysis implies that H. pylori infection might interfere with the immune-related pathways among gastric cancer tissues. Then, we built H. pylori infection–related dys-regulated lncRNA regulatory networks. We also identified 13 differentially expressed lncRNAs were associated with prognosis for gastric cancer patients together with H. pylori infection. Kaplan-Meier analysis demonstrated that the lncRNA signatures were correlated with the poor prognosis. What is more, the AUC of the lncRNA signatures was 0.712. Also, this prognostic prediction model was superior to the traditional clinical characters.
Conclusion: We successfully constructed a H. pylori–related lncRNA risk signature and nomogram associated with H. pylori–infected gastric cancer patients prognosis, and the signature and nomogram can predict the prognosis of these patients.
Gastric cancer (GC) is one of the most common cancers of gastrointestinal system. According to the world cancer report provided by World Health Organization (WHO), there are more than 1 million new cases of gastric cancer at 2018, together with 783,000 attributed deaths, which makes it the third leading cause of cancer-related mortality worldwide (1). Helicobacter pylori (H. pylori) is the type I biological carcinogens, which would cause nearly 75% of the total incidence of gastric cancer worldwide (2). Infection of H. pylori would induce or regulate the oncogenic metabolic pathways, and then affect the occurrences and outcomes of gastric cancer (3). So, H. pylori infection is considered to be one of the most serious risk factor and would affect the prognosis of gastric cancer patients. The identification of prognostic indicators complicated with the effects of H. pylori infection for GC patients is urgently needed.
Long noncoding RNAs (lncRNAs) are new kinds of linear functional RNAs, categorized by long sequence sizes (over 200 nucleotides), which are widely known to be involved in physiological and pathological regulations, such as cell cycle regulation, epigenetic regulation, as well as cell differentiations, and so on (4). Recently, lncRNAs are newly identified as gastric cancer regulators, which would control the activation or inhibition of cancer-related metabolic pathways caused by H. pylori infection (5). However, we have no understanding of the prognostic worth of lncRNAs for gastric cancer patients complicated with H. pylori infection.
Hence, in this study, we first collected H. pylori infection–related genes and identified the expression status, based on the RNA-Sequencing data provided by The Cancer Genome Atlas (TCGA) database. Then, we constructed the aberrant lncRNAs regulatory networks caused by H. pylori infection and extracted the epithelial cell signaling in Helicobacter pylori infection lncRNA regulatory pattern. And based on the lncRNAs profiles and clinical features, we built a multi-lncRNA prognostic signature for GC patients complicated with H. pylori infection that are correlated with poor prognosis. Also, this prognostic prediction model is superior to traditional clinical characters.
The RNA-sequencing data for 163 patients with H. pylori infection information were extracted from The Cancer Genome Atlas (TCGA) database, including lncRNA, miRNA, and mRNA profiles. We set up the gastric cancer tissues without H. pylori infection as a control group. Meanwhile, the clinical information of these patients was also obtained (Table 1). H. pylori infection–related genes were obtained from Gene Set Enrichment Analysis (https://www.gsea-msigdb.org/gsea/index.jsp) (Figure S1). Immunohistochemical images from the Human Protein Atlas (HPA) (https://www.proteinatlas.org) were used to identify the protein expression levels of five differentially expressed genes.
Based on R version 4.0.3, “DESeq2” method was used to screen the differentially expressed lncRNAs, miRNAs, and mRNAs. Then, we used The Encyclopedia of RNA Interactomes (ENCORI) database (6) to construct H. pylori infection–related lncRNAs regulatory networks. We also used the STRING database to construct the PPI networks, and the networks were visualized by Cytoscape version 3.8.2.AB.
Based on R version 4.0.3, we used “clusterProfiler” package to perform gene ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was also used. Then, we used “pheatmap”, “enrichplot”, “pathview”, and “ggplot2” packages to visualize the results.
Lasso-penalized COX regression, univariate COX regression, and multivariate COX regression analyses were used to identify the H. pylori infection–related lncRNA signatures. Multivariate Cox regression analysis is based on the results of univariate regression analysis. The risk scores were calculated for each gastric cancer patients. And based on the risk scores, the patients were classified into high-risk (over median number) and low-risk (no more than median number) groups.
We used “rms” and “regplot” packages to construct the hybrid nomogram, based on R version 4.0.3. A nomogram was constructed integrating the prognostic signatures, for predicting 1-, 2-, and 3‐year OS of gastric cancer patients.
The significance of differences between comparative two groups was evaluated by Student’s t-test and the Wilcoxon test, respectively. Differences with P value less than 0.05 were considered significant. The data were analyzed using R version 4.0.3.
The work flowchart for this study is shown in Figure 1. First, we have collected 402 H. pylori infection–related genes from Gene Set Enrichment Analysis (https://www.gsea-msigdb.org/gsea/index.jsp) (Figure S1) and identified 115 genes, which were differentially expressed among H. pylori–infected gastric cancer tissues (among which 77 were up-regulated and 38 were down-regulated; Figure S2). We selected five genes that are significantly differentially expressed and searched their protein expression levels from the Human Protein Atlas (HPA). Immunohistochemical images from HPA indicated high levels of GBP5, ATP6V1G2 protein in gastric cancer tissues and GBP5, ATP6V1G2 protein were not detected in normal stomach tissues (Figures S2A–D). However, the expression levels of CXCL8 are opposite (Figures S2E, F). In addition, HPA indicated low levels of DEPDC1 protein in normal stomach tissues and moderate expression level in gastric cancer tissues (Figures S2G, H); however, the expression levels of MET are the opposite (Figures S2I, J). GO Biological Process annotation identified that these genes participated in “GO:0071219 Cellular response to molecule of bacterial origin,” “GO:0002685 Regulation of leukocyte migration,” as well as “GO:0007159 Leukocyte cell-cell adhesion” (Figure 2A and Table S3). Following which, KEGG enrichment analysis has shown that these aberrantly expressed genes were significantly involved in “hsa05120: Epithelial cell signaling in H. pylori infection” and “hsa04061: Viral protein interaction with cytokine and cytokine receptor” (Figure 2B and Table S4). Furthermore, H. pylori infection–related oncogenic signaling pathways, including “hsa04064: NF-kappa B signaling pathway” and “hsa04010: MAPK signaling pathway,” were also significantly enriched (Figures 2C, D and S1). So, these items imply that the H. pylori infection can induce and activate the cancer-promoting signaling pathway and affect the occurrence and outcome of gastric cancer.
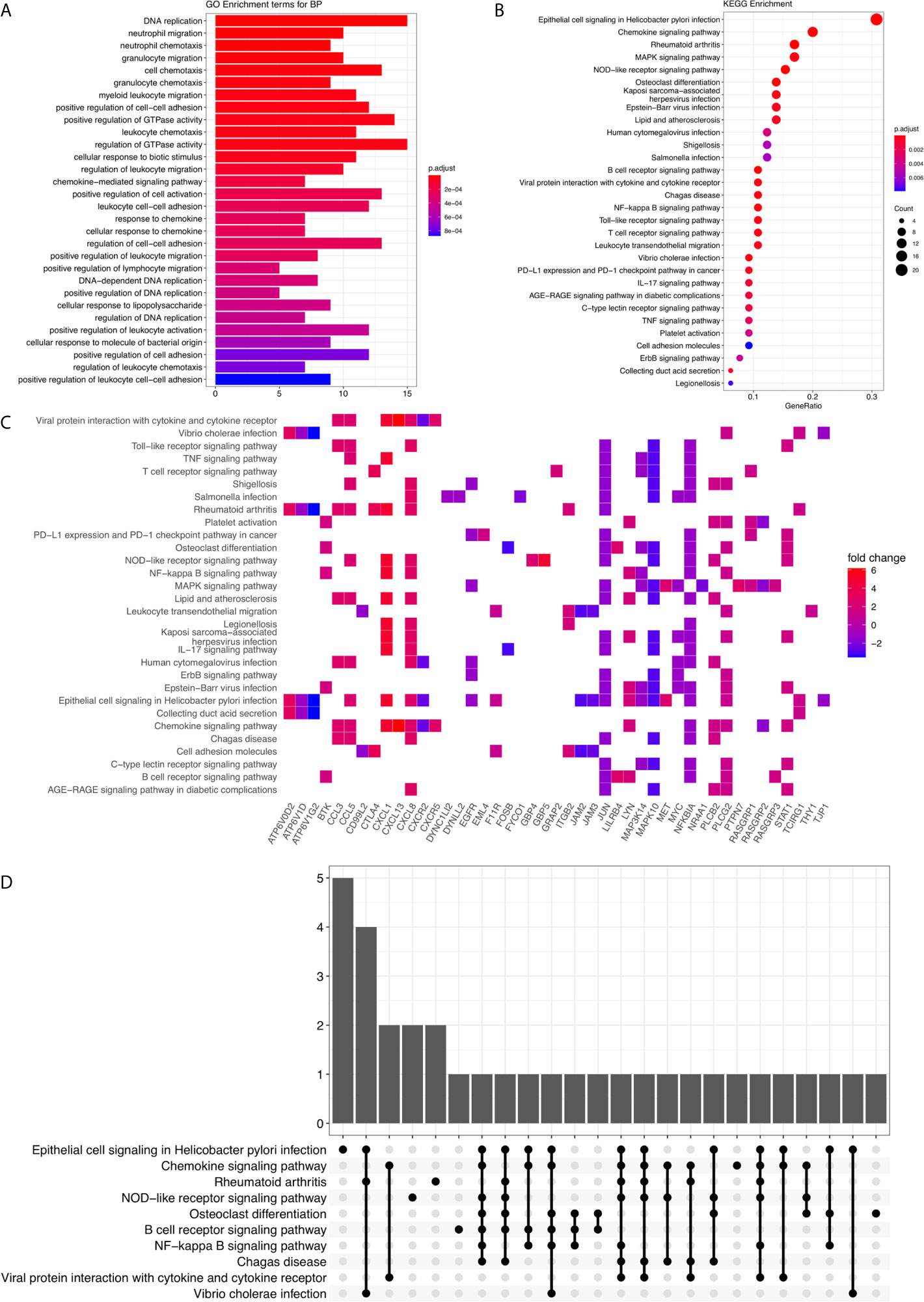
Figure 2 GO annotation and KEGG enrichment analysis. (A) The barplot for GO Biological Process terms. The x-axis represents the enriched gene counts. The bar colors represent the adjusted P-value. (B) The bubble diagram for KEGG terms. The x-axis represents the Gene Ratio. The dot colors represent the adjusted P-value. And the dot size represents the enriched gene counts. (C) The KEGG enrichment heatmap. (D) The KEGG enrichment upset plot.
Then, we have identified 800 differentially expressed lncRNA among H. pylori–infected gastric cancer tissues (among which 443 were up-regulated and 357 were down-regulated, Table S5). Using The Encyclopedia of RNA Interactomes (ENCORI) database (6), we constructed the H. pylori infection–related lncRNAs regulatory network (Figure 3A and Table S6), based on the 115 aberrant expressed H. pylori infection–related genes we just identified. This network contains 370 lncRNAs, 92 miRNAs, and 100 genes. Using STRING database version 11.0, the protein-protein interaction (PPI) relationships for the 100 target genes were constructed (Figure 3B and Table S7). Next, based on the KEGG enrichment items, we mapped the genes on hsa04064 KEGG graph (Figure 3C) and constructed the “Epithelial cell signaling in H. pylori infection lncRNA regulatory pattern” (Figure 3D and Table S8). Furthermore, we also identified the PPI relationships for the genes involved in this lncRNA regulatory pattern (Figure 3E and Table S9).
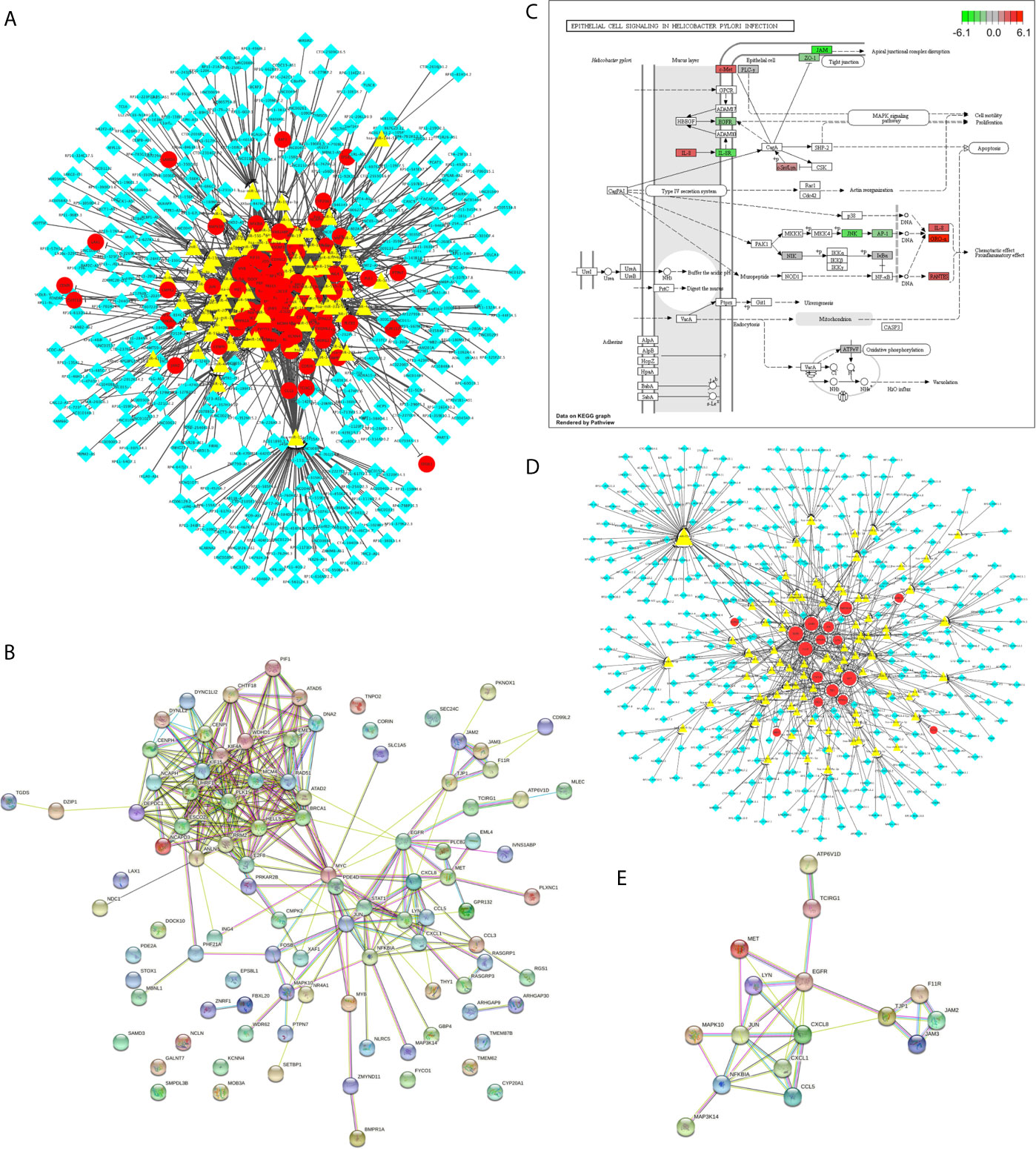
Figure 3 The H. pylori infection–related lncRNAs regulatory patterns. (A) The aberrant lncRNA regulatory networks among H. pylori–infected gastric cancer tissues. The red circles represent the differentially expressed H. pylori infection–related genes. The yellow triangles represent the miRNAs. And the blue diamonds represent the lncRNAs. (B) PPI network for H. pylori infection–related genes involved in A. (C) KEGG graph for “hsa05120: Epithelial cell signaling in Helicobacter pylori infection”. The colored molecules were enriched differentially expressed H. pylori infection–related genes. The color represents the log2 (Fold Change) values for each molecule. Red represents the up regulation. Green represents the down regulation. (D) The Epithelial cell signaling in H. pylori infection lncRNA regulatory pattern. The red circles represent the differentially expressed H. pylori infection–related genes. The yellow triangles represent the miRNAs. And the blue diamonds represent the lncRNAs. (E) PPI network for target genes involved in Epithelial cell signaling in H. pylori infection lncRNA regulatory pattern.
To analyze the correlation between H. pylori infection–related lncRNAs and prognosis, we used univariate COX analysis and identified 43 significant H. pylori infection (Table 2), which would be used in multivariate COX analysis (Table 3). In summary, we found that 13 aberrantly expressed lncRNAs may be independent prognosis prediction factors for H. pylori–infected gastric cancer (including AC007319.1, AP000593.7, LINC00304, LINC01526, RP11-197P3.5, RP11-284F21.7, RP11-319E16.2, RP11-334C17.5, RP11-617F23.2, RP11-649G15.2, RP11-706O15.3, RP11-867G23.12, as well as RP4-791M13.3, Table 3). Also, the risk scores were also calculated to construct a prognostic signature for H. pylori–infected gastric cancer (Table S10).
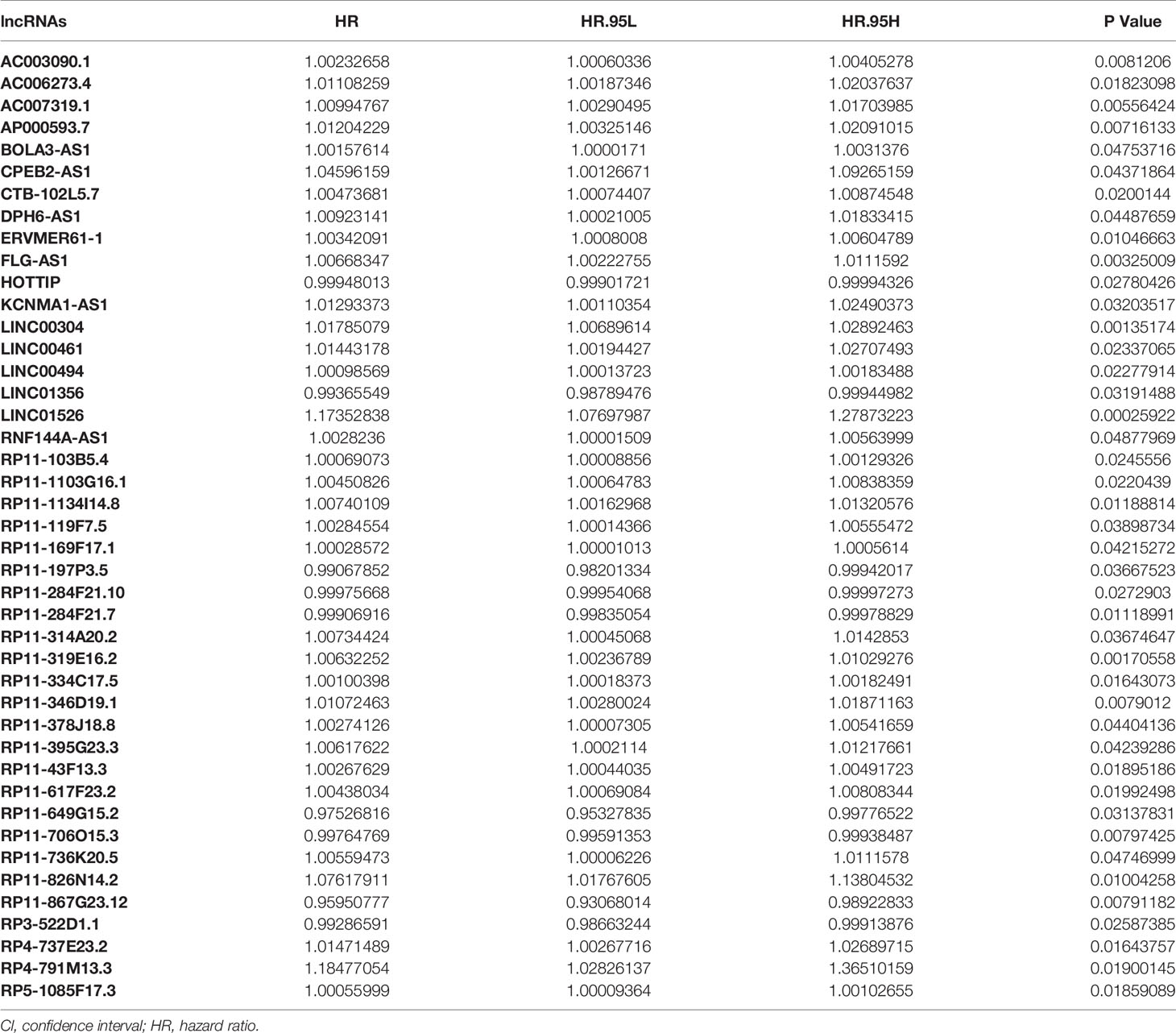
Table 2 Univariate COX regression analysis with the Lasso method for the H. pylori infection–related lncRNA prognostic signature.
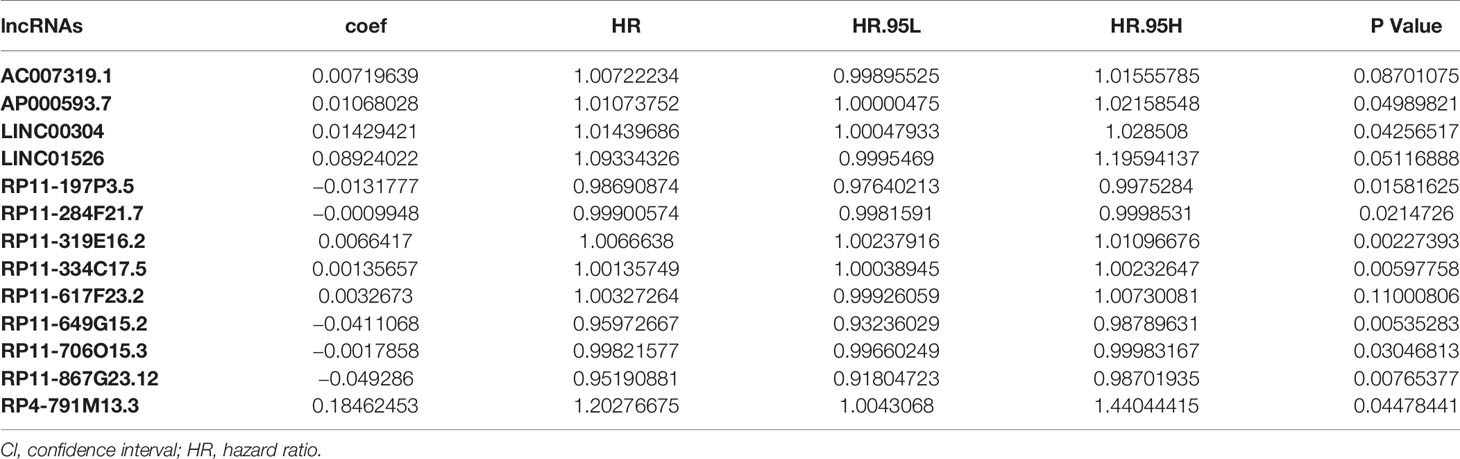
Table 3 Multivariate COX regression analysis with the Lasso method for the 13 H. pylori infection–related lncRNA prognostic signature.
Utilizing Kaplan-Meier analysis, we identified that the high-risk lncRNA signatures were associated with poor survival condition (P value < 0.0001, Figure 4A). And the patient risk survival status plot demonstrated that the risk-scores were proportional to the survival of patients inversely (Figure 4B). Using ROC curve plotting, it was present that the AUC values for the high-risk lncRNA signatures at 1-, 2-, and 3-year survival rates were 0.712, 0.699, and 0.656, respectively (Figure 4C). It was a heatmap for the risk scores (Figure 4D).
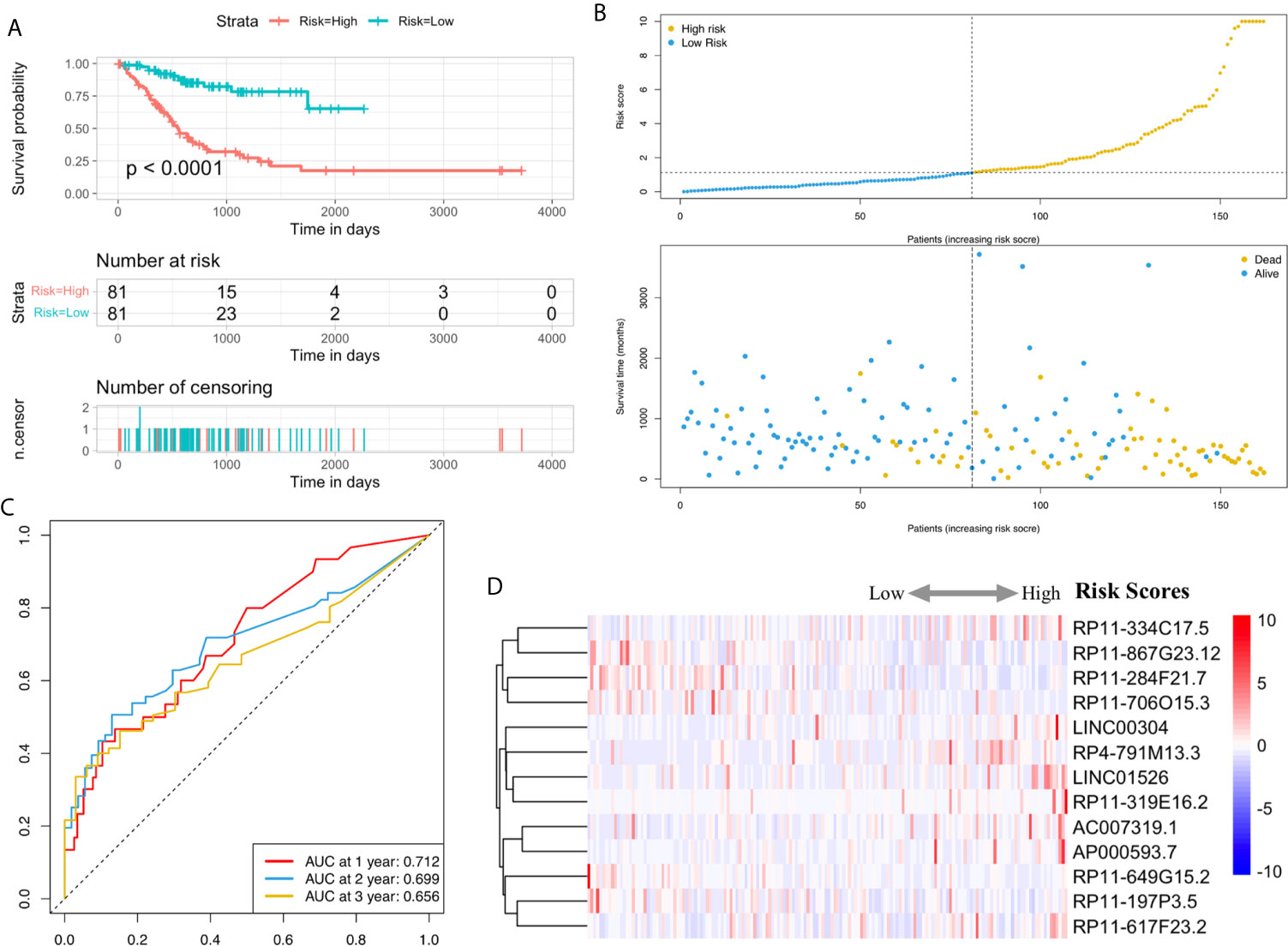
Figure 4 H. pylori infection–related high-risk lncRNA signature. (A) The Kaplan-Meier curves. (B) The risk survival status plot. (C) The ROC curves for the prediction of 1-, 2-, and 3-year survival rate of gastric cancer. (D) Heatmap for the risk scores.
Furthermore, COX analysis results have shown that the H. pylori infection–related lncRNA signatures (HR, 1.401; 95% CI, 1.243–1.579), AJCC stages (HR, 2.311; 95% CI, 1.293–4.129) as well as TNM stages were independent prognosis factors of OS of H. pylori–infected gastric cancer patients (Table 4). Also, the correlations among H. pylori infection–related lncRNA prognostic signatures and clinical factors were also exhibited using heatmap (Figure 5). Finally, using hybrid nomogram, which merged the clinical characteristics, we verified that the H. pylori infection–related lncRNA prognostic signatures were accurate and relatively stable (Figure 6). Hence, these lncRNA signatures may be used in the prognosis evaluation for H. pylori–infected gastric cancer patients.
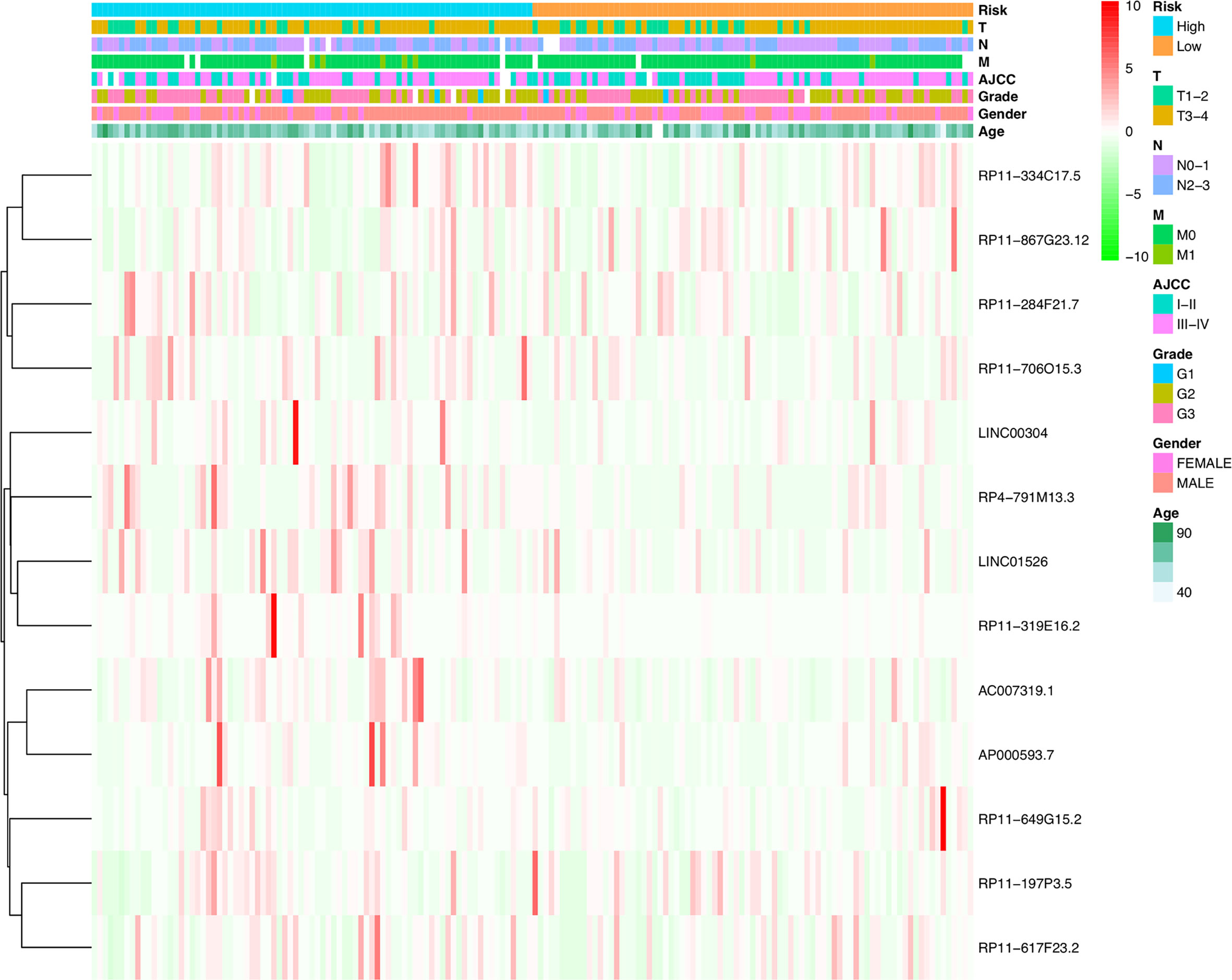
Figure 5 Heatmap for H. pylori infection–related lncRNAs prognostic signature and clinical features.
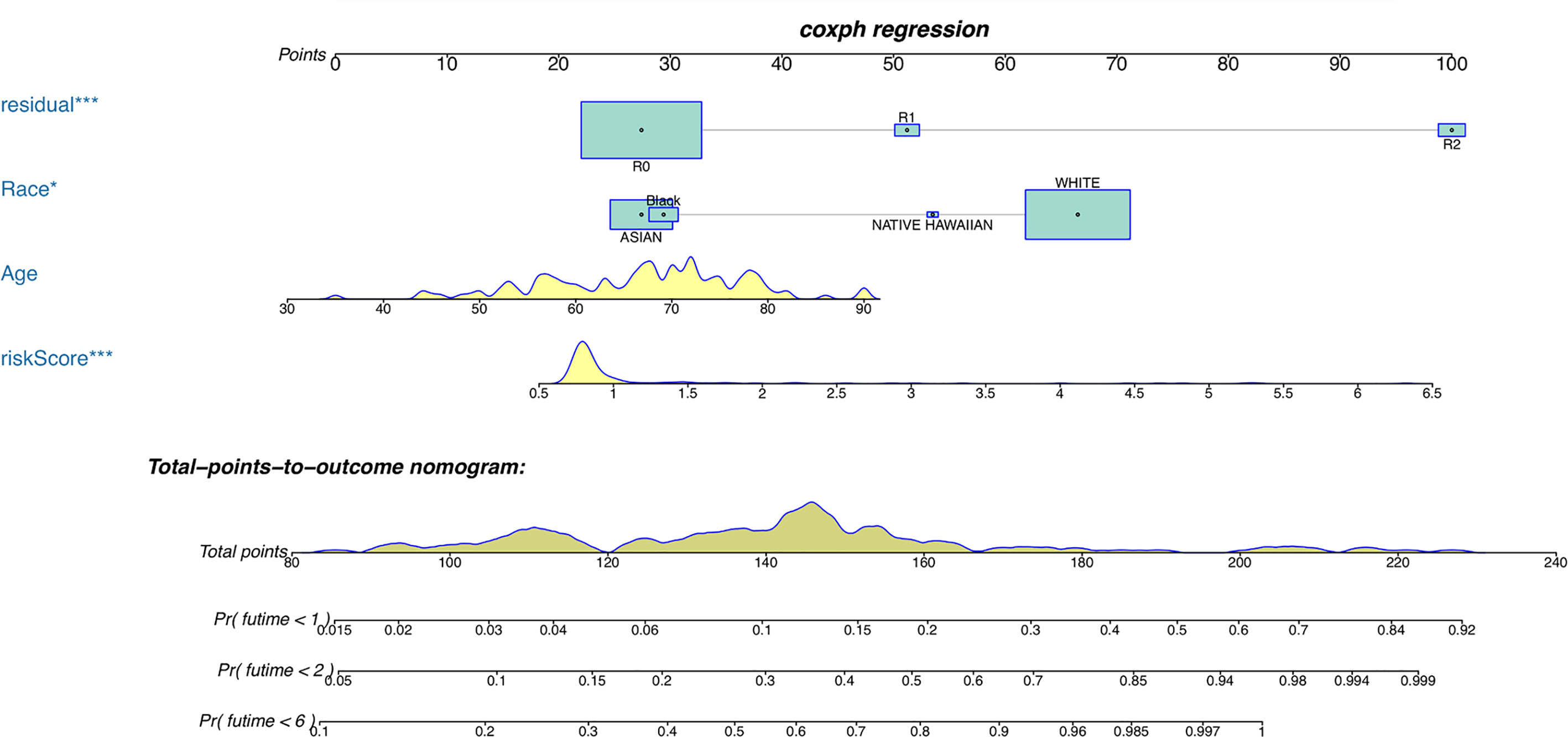
Figure 6 A nomogram for both clinical features and H. pylori infection–related lncRNAs prognostic signatures. *p < 0.05, ***p < 0.001.
The major risk factor associated with gastric adenocarcinoma is infection by H. pylori, and the attributable proportion of H. pylori–infected gastric cancer patients has been estimated to be about 70% (7). Thus, it is particularly critical to explore accurate biomarkers for predicting prognosis of H. pylori–infected gastric cancer patients. In this study, we first identified a novel 13-lncRNA risk signature that was highly associated with the OS of H. pylori–infected gastric cancer patients based on the TCGA.
Overall, we identified 115 H. pylori–related differentially expressed genes in GC patients. KEGG analyses further revealed that the differentially expressed genes mainly participated in GO and identified that these genes participated in the cellular response to molecule of bacterial origin, regulation of leukocyte migration, and leukocyte cell-cell adhesion. KEGG analyses revealed that the genes mainly participated in epithelial cell signaling in H. pylori infection, viral protein interaction with cytokine and cytokine receptor, NF-kappa B signaling, and MAPK signaling pathway. So, these items imply that the H. pylori infection can induce and activate the cancer-promoting signaling pathway and affect the occurrence and outcome of gastric cancer. In addition, we also identified 13 differentially expressed lncRNAs associated with prognosis for gastric cancer patients together with H. pylori infection. Among them, Zhang et al. indicated that LINC00304 plays a tumor-promotive functional role and promotes PCa cell proliferation and cell cycle by upregulating CCNA1 expression (8). Cheng et al. showed that LINC01526 was among the noninvasive prognostic signatures, which was a preoperative prediction of disease-free survival in GC patients (9). However, the molecular mechanism and effect of other lncRNAs in our 13-lncRNA signature have not been explored and are still uncertain. Increasing studies showed that lncRNAs play important roles in tumorigenesis and metastasis by regulating the expression of protein-coding genes through transcriptional, posttranscriptional, and posttranslational regulation (10–13). Also, several reports indicated that LncRNAs may show oncogenic and tumor-suppressive activities in the initiation and progression of GC (14). Yang et al. reported that LINC00152 and H19 were significantly increased in the serum and cancer tissues of patients with GC and may be possible biomarkers for diagnosis and prognosis of GC (15). Another study showed that non-overlapping signatures of a few lncRNAs were abnormally expressed in gastric epithelial cells infected by H. pylori, and these lncRNAs may have a function in the immune system response against H. pylori, which in turn may lead to the occurrence of gastric cancer with H. pylori (16). Several studies indicated the influence of H. pylori infection on the dysregulation of lncRNA expression profiles, including Lnc-SGK1, THAP9-AS1, NR-026827, and AF147447 (17–20).
Here, the differentially expressed H. pylori–associated lncRNAs in our study were stratified into two groups of high- and low-risk scores to explore their potential roles in H. pylori–infected gastric cancer patients. Our study indicated that patients with H. pylori infection in the high-risk group had a shorter survival than those in the low-risk group, and the signature also displayed a high prediction sensitivity and specificity. In this study, we constructed a nomogram by combining the 13-lncRNA signature and clinicopathological factors in 162 patients from the TCGA, and the nomogram is an effective predictive tool for patients with H. pylori infection.
A large number of studies have explored the prognostic factors of gastric cancer from the level of genes and proteins. The expression of HOXC9 mRNA and HOXC9 protein in gastric cancer tissue was significalntly higher than that in non-gastric cancer tissue (21). CD40, CAP2 proteins, and mRNAs expressions are closely related to prognosis, distant metastasis, and stage of patients with gastric cancer (22, 23). The overall survival rate of patients with up-regulated SLC22A16 expression was worse than that of patients without up-regulated SLC22A16 expression (24). CXCR7 may be a prognostic marker for gastric cancer with peritoneal metastasis; however, the number of cases in this study were small, and the prognostic significance of CXCR7 was not confirmed (25). Wang et al. indicated that the high expression of SCD1 might predict poor prognosis in gastric cancer patients, but the AUC value at 1-, 3-, and 5-year survival rates were 0.557, 0.569, and 0.595, respectively (26), were significantly lower than the results of our study. In addition, the AUC’s value of another predictive biomaker (linc-ROR) was 0.6495 (27), which was also lower than ours. So, we proposed that our findings improved the prediction accuracy for the prognosis of gastric cancer patients.
H. pylori is considered as a precancerous lesion of gastric cancer and is closely related to the occurrence and development of gastric cancer (28). The eradication of H. pylori can reduce the incidence of gastric cancer (29, 30), but the prevalence of H. pylori is high, especially in economically underdeveloped areas (31). It has been reported that the prognostic factors of patients with gastric cancer infected with H. pylori, for example, miR-490-3p is associated with poor clinical prognosis. Low expression of miR-490-3p is associated with poor prognosis in H. pylori–infected gastric cancer patients. However, the molecular functions of miR-490-3p are very diverse, and miR-490-3p can also promote the development of some cancers. The function of miR-490-3p is closely related to the tumor microenvironment (32). The expression levels of anti-H. pylori antibody, CA724, CA19-9, and CEA in H. pylori–infected gastric cancer patients were positively correlated with tumor stage, tumor size, and lymph node (33).However, Tas and Chae pointed out that the levels of CA724, CA19-9, CEA, and other markers had no significance in evaluating the prognosis of gastric cancer patients (34, 35). In addition, this study was a retrospective study with a small sample size and the same hospital. Therefore, the accuracy of anti-H. pylori antibody, CA724, CA19-9, and CEA in predicting H. pylori–infected gastric cancer patients is still to be discussed. DNA methylation can induce the occurrence of gastric cancer in H. pylori–infected gastric cancer patients (36). However, this study did not establish a prediction model and could not accurately predict the patients.
Our research has some obvious strengths. First, the effect of lncRNA on the prognosis of H. pylori–infected gastric cancer patients has never been reported, and our research is the first to present this. Second, in this study, a model affecting the prognosis of H. pylori–infected gastric cancer patients was established based on the level of lncRNA, and a nomogram was constructed, which is conducive to more accurate prediction of the prognosis of H. pylori–infected gastric cancer patients and improvement of the survival rate of patients. Also, the signature and nomogram is convenient to apply in clinical practice for doctors. In addition, there are several limitations. First, the number of patients included in the study is relatively insufficient. Second, the 13-lncRNA signature and predicted nomogram were not validated in both internal and external cohorts and not compared with other models. Third, there is still a lack of experimental researches about the molecular function of these lncRNAs, and we will collect specimens from H. pylori (+) and H. pylori (−) GC patients and carry out experimental research.
In conclusion, we successfully constructed an H. pylori–related lncRNA risk signature and nomogram associated with H. pylori–infected gastric cancer patients prognosis in TCGA. The results indicated that the signature and nomogram can predict the prognosis of these patients. Furthermore, we should carry out related molecular research to clarify the mechanism of related lncRNA in the future.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
ZX, NZ, and HX came up with the design and critical revision of the manuscript. The data analysis were conducted by YW, YZ, YS, and WZ. The original writing of the draft and its editing were conducted by LZ, ML, LS, and NX. All authors contributed to the article and approved the submitted version.
This work was supported by the Department of Finance of Jilin Province (JLSWSRCZX2020-083), Department of Science and technology of Jilin Province (3D517DF53428, 2020Q051), and the Bethune Project of Jilin University (2020B09).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to acknowledge the reviewers for their helpful comments on this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.709796/full#supplementary-material
Supplementary Figure 1 | KEGG graph. (A) KEGG graph for “hsa04064: NF-kappa B signaling pathway” (B) KEGG graph for “hsa04010: MAPK signaling pathway. The colored molecules were enriched differentially expressed H. pylori infection–related genes. The color represents the log2 (Fold Change) values for each molecule. Red represents the up regulation. Green represents the down regulation.
Supplementary Figure 2 | Immunohistochemical images from HPA. (A, B) The immunohistochemical images of GBP5 in normal stomach tissues and gastric cancer tissues. (C, D) The immunohistochemical images of ATP6V1G2 in normal stomach tissues and gastric cancer tissues. (E, F) The immunohistochemical images of CXCL8 in normal stomach tissues and gastric cancer tissues. (G, H) The immunohistochemical images of DEPDC1 in normal stomach tissues and gastric cancer tissues. (I, J) The immunohistochemical images of MET in normal stomach tissues and gastric cancer tissues.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492
2. Amieva M, Peek RM Jr. Pathobiology of Helicobacter Pylori-Induced Gastric Cancer. Gastroenterology (2016) 150:64–78. doi: 10.1053/j.gastro.2015.09.004
3. Toh JWT, Wilson RB. Pathways of Gastric Carcinogenesis, Helicobacter Pylori Virulence and Interactions With Antioxidant Systems, Vitamin C and Phytochemicals. Int J Mol Sci (2020) 21:6451. doi: 10.3390/ijms21176451
4. Ulitsky I, Bartel DP. lincRNAs: Genomics, Evolution, and Mechanisms. Cell (2013) 154:26–46. doi: 10.1016/j.cell.2013.06.020
5. Dastmalchi N, Khojasteh SMB, Nargesi MM, Safaralizadeh R. The Correlation Between lncRNAs and Helicobacter Pylori in Gastric Cancer. Pathog Dis (2019) 77:ftaa004. doi: 10.1093/femspd/ftaa004
6. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and Protein-RNA Interaction Networks From Large-Scale CLIP-Seq Data. Nucleic Acids Res (2014) 42:D92–7. doi: 10.1093/nar/gkt1248
7. De Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, et al. Global Burden of Cancers Attributable to Infections in 2008: A Review and Synthetic Analysis. Lancet Oncol (2012) 13:607–15. doi: 10.1016/s1470-2045(12)70137-7
8. Zhang P, Lu Y, Kong Z, Zhang Y, Fu F, Su X, et al. Androgen-Responsive Lncrna LINC00304 Promotes Cell Cycle and Proliferation Via Regulating CCNA1. Prostate (2019) 79:994–100689JN. doi: 10.1002/pros.23811
9. Cheng C, Wang Q, Zhu M, Liu K, Zhang Z. Integrated Analysis Reveals Potential Long non-Coding RNA Biomarkers and Their Potential Biological Functions for Disease Free Survival in Gastric Cancer Patients. Cancer Cell Int (2019) 19:123. doi: 10.1186/s12935-019-0846-6
10. Geisler S, Coller J. RNA in Unexpected Places: Long non-Coding RNA Functions in Diverse Cellular Contexts. Nat Rev Mol Cell Biol (2013) 14:699–712. doi: 10.1038/nrm3679
11. Sun M, Xia R, Jin F, Xu T, Liu Z, De W, et al. Downregulated Long Noncoding RNA MEG3 is Associated With Poor Prognosis and Promotes Cell Proliferation in Gastric Cancer. Tumour Biol (2014) 35:1065–73. doi: 10.1007/s13277-013-1142-z
12. Kim T, Cui R, Jeon YJ, Fadda P, Alder H, Croce CM. MYC-Repressed Long Noncoding RNAs Antagonize MYC-Induced Cell Proliferation and Cell Cycle Progression. Oncotarget (2015) 6:18780–9. doi: 10.18632/oncotarget.3909
13. Song W, Liu YY, Peng JJ, Liang HH, Chen HY, Chen JH, et al. Identification of Differentially Expressed Signatures of Long Non-Coding RNAs Associated With Different Metastatic Potentials in Gastric Cancer. J Gastroenterol (2016) 51:119–29. doi: 10.1007/s00535-015-1091-y
14. Wang J, Song YX, Wang ZN. Non-Coding RNAs in Gastric Cancer. Gene (2015) 560:1–8. doi: 10.1016/j.gene.2015.02.004
15. Yang T, Zeng H, Chen W, Zheng R, Zhang Y, Li Z, et al. Helicobacter Pylori Infection, H19 and LINC00152 Expression in Serum and Risk of Gastric Cancer in a Chinese Population. Cancer Epidemiol (2016) 44:147–53. doi: 10.1016/j.canep.2016.08.015
16. Yang L, Long Y, Li C, Cao L, Gan H, Huang K, et al. Genome-Wide Analysis of Long Noncoding RNA Profile in Human Gastric Epithelial Cell Response to Helicobacter Pylori. Jpn J Infect Dis (2015) 68:63–6. doi: 10.7883/yoken.JJID.2014.149
17. Yao Y, Jiang Q, Jiang L, Wu J, Zhang Q, Wang J, et al. Lnc-SGK1 Induced by Helicobacter Pylori Infection and Highsalt Diet Promote Th2 and Th17 Differentiation in Human Gastric Cancer by SGK1/Jun B Signaling. Oncotarget (2016) 7:20549–60. doi: 10.18632/oncotarget.7823
18. Zhou X, Chen H, Zhu L, Hao B, Zhang W, Hua J, et al. Helicobacter Pylori Infection Related Long Noncoding RNA (Lncrna) AF147447 Inhibits Gastric Cancer Proliferation and Invasion by Targeting MUC2 and Up-Regulating Mir-34c. Oncotarget (2016) 7:82770–82. doi: 10.18632/oncotarget.13165
19. Zhong F, Zhu M, Gao K, Xu P, Yang H, Hu D, et al. Low Expression of the Long Non-Coding RNA NR_026827 in Gastric Cancer. Am J Transl Res (2018) 10:2706–11.
20. Jia W, Zhang J, Ma F, Hao S, Li X, Guo R, et al. Long Noncoding RNA Thap9-AS1 Is Induced by Helicobacter Pylori and Promotes Cell Growth and Migration of Gastric Cancer. Onco Targets Ther (2019) 12:6653–63. doi: 10.2147/ott.s201832
21. Zhao XF, Yang YS, Park YK. HOXC9 Overexpression is Associated With Gastric Cancer Progression and a Prognostic Marker for Poor Survival in Gastric Cancer Patients. Int J Clin Oncol (2020) 25:2044–54. doi: 10.1007/s10147-020-01772-0
22. Guo J, Xiao JJ, Zhang X, Fan KX. CD40 Expression and its Prognostic Significance in Human Gastric Carcinoma. Med Oncol (2015) 32:63. doi: 10.1007/s12032-014-0463-0
23. Li L, Fu LQ, Wang HJ, Wang YY. CAP2 Is a Valuable Biomarker for Diagnosis and Prognostic in Patients With Gastric Cancer. Pathol Oncol Res (2020) 26:273–9. doi: 10.1007/s12253-018-0450-4
24. Zhao W, Wang Y, Yue X. SLC22A16 Upregulation Is an Independent Unfavorable Prognostic Indicator in Gastric Cancer. Future Oncol (2018) 14:2139–48. doi: 10.2217/fon-2018-0207
25. Nambara S, Iguchi T, Oki E, Tan P, Maehara Y, Mimori K. Overexpression of CXCR7 Is a Novel Prognostic Indicator in Gastric Cancer. Dig Surg (2017) 34:312–8. doi: 10.1159/000452977
26. Wang C, Shi M, Ji J, Cai Q, Zhao Q, Jiang J, et al. tearoyl-CoA Desaturase 1 (SCD1) Facilitates the Growth and Anti-Ferroptosis of Gastric Cancer Cells and Predicts Poor Prognosis of Gastric Cancer. Aging (Albany NY) (2020) 12:15374–91. doi: 10.18632/aging.103598
27. Yu X, Ding H, Shi Y, Yang L, Zhou J, Yan Z. Downregulated Expression of linc-ROR in Gastric Cancer and Its Potential Diagnostic and Prognosis Value. Int J Clin Oncol (2020) 2020:7347298. doi: 10.1155/2020/7347298
28. Bakhti SZ, Latifi-Navid S. Helicobacter Pylori-Related Risk Predictors of Gastric Cancer: The Latest Models, Challenges, and Future Prospects. Cancer Med (2020) 9:4808–22. doi: 10.1002/cam4.3068
29. Lee YC, Chiang TH, Chou CK, Tu YK, Liao WC, Wu MS, et al. Association Between Helicobacter Pylori Eradication and Gastric Cancer Incidence: A Systematic Review and Meta-Analysis. Gastroenterology (2016) 150:1113–24.e1115. doi: 10.1053/j.gastro.2016.01.028
30. Sugano K. Effect of Helicobacter Pylori Eradication on the Incidence of Gastric Cancer: A Systematic Review and Meta-Analysis. Gastric Cancer (2019) 22:435–45. doi: 10.1007/s10120-018-0876-0
31. Mentis A, Lehours P, Mégraud F. Epidemiology and Diagnosis of Helicobacter Pylori Infection. Helicobacter (2015) 20(Suppl 1):1–7. doi: 10.1111/hel.12250
32. Qu M, Li L, Zheng WC. Reduced miR-490-3p Expression Is Associated With Poor Prognosis of Helicobacter Pylori Induced Gastric Cancer. Eur Rev Med Pharmacol Sci (2017) 21:3384–8.
33. Gong X, Zhang H. Diagnostic and Prognostic Values of Anti-Helicobacter Pylori Antibody Combined With Serum CA724, Ca19-9, and CEA for Young Patients With Early Gastric Cancer. J Clin Lab Anal (2020) 34:e23268. doi: 10.1002/jcla.23268
34. Chae HD, Kim IH. Prognostic Significance of CEA Expression by RT-PCR in Peritoneal Wash From Patients With Gastric Cancer: Result of a 5-Year Follow-Up After Curative Resection. Scand J Gastroenterol (2016) 51:956–60. doi: 10.3109/00365521.2016.1172339
35. Tas F, Karabulut S, Erturk K, Duranyildiz D. Clinical Significance of Serum Caveolin-1 Levels in Gastric Cancer Patients. Exp Oncol (2018) 40:323–7. doi: 10.31768/2312-8852.2018.40(4):323-327
Keywords: Helicobacter pylori, lncRNA, prognosis prediction, data mining, regulatory pattern
Citation: Xin Z, Zhang L, Liu M, Wang Y, Zhang Y, Zhao W, Sun Y, Shi L, Xu N, Zhang N and Xu H (2021) Helicobacter pylori Infection–Related Long Non-Coding RNA Signatures Predict the Prognostic Status for Gastric Cancer Patients. Front. Oncol. 11:709796. doi: 10.3389/fonc.2021.709796
Received: 14 May 2021; Accepted: 06 July 2021;
Published: 27 July 2021.
Edited by:
Peng Qu, National Institutes of Health (NIH), United StatesReviewed by:
Chunqing Guo, Virginia Commonwealth University, United StatesCopyright © 2021 Xin, Zhang, Liu, Wang, Zhang, Zhao, Sun, Shi, Xu, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Zhang, emhhbmduYW5Aamx1LmVkdS5jbg==; Hong Xu, eF9ob25nQGpsdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.