- 1Department of Pathology, Wuhan No.1 Hospital, Wuhan, China
- 2Department of Urology, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
Objective: Tumor metabolism has always been the focus of cancer research. SLC16A1, as a key factor in catalysis of monocarboxylate transport across the plasma membrane, has been found to be associated with the occurrence and metastasis of a variety of cancers, but its prognostic significance and mechanism in different tumors are still unclear.
Methods: Based on the gene expression matrix and clinical information of human cancer tissues acquired from TCGA and GTEX databases, the differential expression of SLC16A1 in different tumors and normal tissues was analyzed. To confirm the association between its expression, the mutation of MMRS gene, and the expression level of DNMTs. Univariate Cox regression was applied to analyze the association between SLC16A1 expression and patient prognosis. The effect of SLC16A1 expression on patient survival was examined by Kaplan Meier analysis. GSEA was used to identify related signaling pathways.
Results: The expression of SLC16A1 was differentially expressed in most tumors, especially in the urinary tract where it is commonly highly expressed, and differential expression of SLC16A1 in different clinical stages. SLC16A1 expression was significantly positively correlated with MMRS gene mutation and DNMTS expression. Moreover, high SLC16A1 expression was associated with poorer overall survival (OS) and progression-free survival (PFS) in urological cancers. In particular, the results of the enrichment analysis showed that SLC16A1 was associated with processes such as cell adhesion and many signaling pathways affecting cell cycle were significantly enriched in the group with high-expressed SLC16A1.
Conclusion: SLC16A1 expression was upregulated in urological cancer. SLC16A1 may promote tumor development by regulating the epigenetic process of urological cancer and demonstrated a great potential as a prognostic biomarker of urological cancer patients.
Introduction
Urological cancer is a highly prevalent neoplastic disease with poor prognosis. Kidney cancer, bladder cancer and prostate cancer, which are the most common urological cancers, account for more than 33% of all male malignant tumors (1, 2) Different urological cancers show different morphological or genetic characteristics, which pose difficulties to the diagnosis and treatment in clinical practice (3, 4). Clinical symptoms of most urological cancers are non-specific, and a late diagnosis will increase the possibility of metastasis and adverse effects on the clinical outcome of patients (5). Although great progress had been made in the molecular study of urological tumors, the understanding of its pathological mechanism is still not satisfied (6). Identifying of certain genes and potential biological markers could improve the evaluation of the efficacy of late treatment (1) and the management of urological cancers.
Study demonstrated an important correlation among urinary system diseases, metabolic syndrome, and metabolic syndrome in endocrine system. Vascular mechanisms affect male urinary system, high insulin hematic disease, in which obesity is often involved. Therefore, discovering metabolism-related genes might provide the corresponding potential targets and improve the treatment of urinary system diseases (7). SLC16 family consists of 14 different monocarboxylate transporters, which play crucial roles in cell metabolism, nutrient transport, and pH regulation. Changes in the expression and function of some members are often reflected in serious diseases, such as cancer and nervous system diseases (8). SLC16A1 is a widely studied member of the SLC16A family. It has been found that SLC16A1 is distributed in almost all tissues in the human body, and is overexpressed in many cancers, moreover, upregulated expression of SLC16A1 is associated with the deterioration of prognosis of many cancers (9, 10).
Pan-cancer analysis is a novel bioinformatics method used to search commonalities across tumor types and organs, and could provide new adaptations for biomarkers across tumors (11). The Cancer Genome Atlas Project (TCGA) and Genotype Tissue Expression (GTEX) is currently the largest available databases for tumor genome analysis (12, 13). Based on TCGA and GTEX databases, this study applied pan-cancer analysis to detect the expression of SLC16A1 in different tumors and explored its prognostic significance. This study provided a potential molecular mechanism of the key role of SLC16A1 in urological cancer.
Materials and Method
Data Source
The TCGA database (https://www.cancer.gov/) contained gene expression profiles of tumors and adjacent normal tissues from different human cancers. In addition to GTEx (https://gtexportal.org/home/) and normal tissue expression, the TIMER 2.0 (http://timer.cistrome.org/) analysis on more than ten thousand samples of the RNA - seq TCGA database data was used and integrated into the TCGA expression. Gene expression matrices were obtained from the CCLE database (https://portals.broadinstitute.org/ccle) for cell lines of different origins, using the Human Protein Atlas (HPA) (https://www.proteinatlas.org/) database as well as the UALCAN (http://ualcan.path.uab.edu/) online website to validate the protein expression of SLC16A1 and its gene expression levels in different clinical features.
Difference Analysis
Statistical differences in SLC16A1 expression levels between groups were calculated using the Wilcoxon test as well as the Kruskal-Wallis H-test. The chi-square test, as well as the t-test, were used to compare previous associations between gene expression and cultural characteristics and were presented visually using the R package ggplot.
Correlation Analysis Between Gene Expression and Epigenetic Regulation
Epigenetic processes are considered to be an important factor affecting gene expression, and DNA replication and DNA methylation are markers in epigenetics (14). DNA mismatch repair (MMRs) plays crucial role in maintaining DNA replication and its structural integrity and stability (15). Changes in DNA methyltransferase (DNMTs) activity could affect gene expression and DNA repair mechanism, and dysfunction of MMRs and DNMTs is the initial conditions for human cancer development (16). Based on the expression profile of TCGA, Pearson test was performed to analyze the correlation between SLC16A1 expression and MMRS gene (MLH1, MSH2, MSH6, PMS2, EpCAM) mutation and DNMTs (DNMT1, DNMT2, DNMT3A, DNMT3B) expression. When R > 0.2 and P< 0.5, SLC16A1 expression was considered to be positively correlated with MMRS and methyltransferase expression.
Survival Analysis
Univariate Cox regression analysis was performed on SLC16A1 expression level in different tumors using the survival package R, and 95% confidence intervals and corresponding hazard ratio (HR) were calculated. Forest plots were drew using the survival package Forestplot. Kaplan-Meier and timeROC was used to examine the relationship between SLC16A1 expression and the survival of urological cancer patients.
Enrichment Analysis
According to the expression of SLC16A1, the samples from TCGA database were divided into high-expression and low-expression groups. Functional enrichment analysis was performed using the R package ClusterProfile based on Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). A list of abs(log2FC) was generated based on two groups of high and low expression and this gene list was mapped GO, KEGG gene set for biological pathway analysis.
Results
Expression of SLC16A1 in Urological Cancers and Other Tumors
We found that the expression level of SLC16A1 was different in most urological cancers (Figure 1A). The expression level of SLC16A1 in tumor tissues of BRCA, CHOL, COAD, UCEC and other cancer patients was lower than that in their corresponding paracancerous normal tissues (P<0.001). SLC16A1 expression was upregulated in metastatic SKCM when compared with carcinoma in situ (P<0.01). However, SLC16A1 was significantly overexpressed in tumor tissues of most other cancers (ESCA, GBM, HNSC, etc.) (P<0.05), and this feature was more strongly present in urinary tract tumors (Kich, Kirc, Prad) (P<0.05). However, normal samples in the TCGA database were generally few and even absent in some cancers. We also analyzed the normal samples in GTEX and tumor samples in TCGA and found that the high expression pattern of SLC16A1 was more obvious in urological cancers (ACC,KICH,Kirc,PRAD,TGCT) (Figure 1B). As shown in Figure 1C, SLC16A1 expression was present in cell lines of renal and prostate as well as bladder urogenital origin in the CCLE database. HPA for immunohistochemical staining on SLC16A1 protein in testicular cancer (Figure 1D), and prostate (Figure 1E) were found to have a high signal intensity, and this was also the same in Renal cancer (Figure 1F), uroepithelial cancer (Figure 1G). It is also shown that in comparison with the corresponding normal tissues, SLC16A1 was upregulated in the urinary diseases.
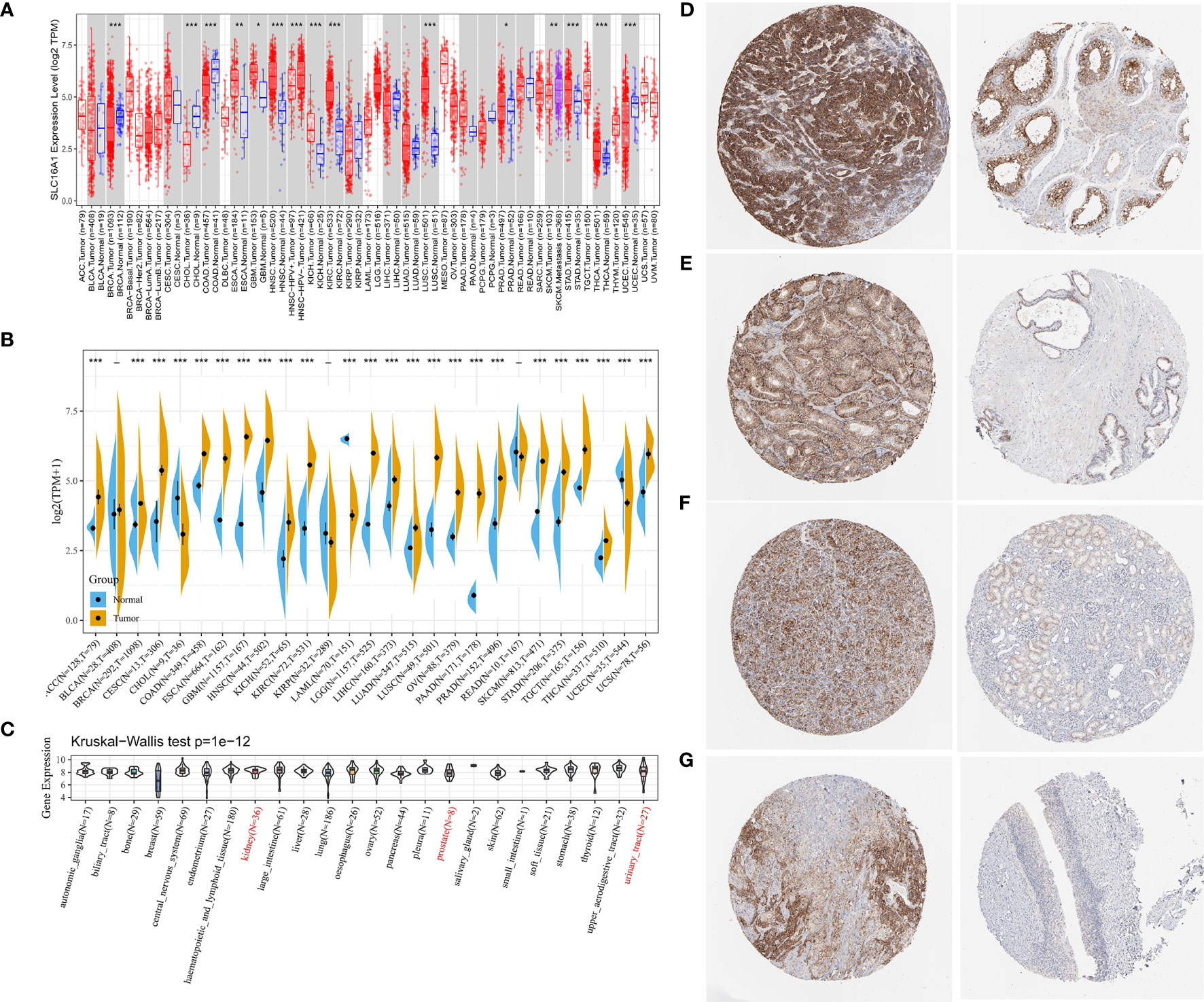
Figure 1 SLC16A1 expression levels in different types of human cancers. (A) Based on the TCGA database, the expression level of SLC16A1 between different cancers was analyzed using Timer 2.0; (B) The expression data of normal tissues in GTEX database were supplemented to analyze the expression levels of SLC16A1 in tumor tissues and normal tissues of different cancers. (C) SLC16A1 expression in each cell line obtained through the CCLE database. Immunohistochemical staining of SLC16A1 protein in (D) testicular cancer; (E) prostate cancer; (F) kidney cancer; (G) bladder uroepithelial cancer tissues and corresponding normal tissues was obtained from the HPA database. TCGA, Cancer Genome Atlas Project; GTEX, genotype tissue expression; HPA, Human Protein Atlas. *P < 0.05, **P < 0.01, ***P < 0.001.
Association Between SLC16A1 Expression and Clinical Features of Urological Tumors
To further compare the expression of SLC16A1 in urological cancers, patients were grouped according to clinical characteristics such as age, gender, race, TNM stage and clinical stage. As shown in Table 1, SLC16A1 overexpression was more prevalent in female patients in ACC, KICH, and KIRP, and that Caucasians had a higher proportion of SLC16A1 overexpression than other races in BLCA. High SLC16A1 expression was associated with higher T-staging in BLCA and KIRP, and with lymphatic metastases in ACC, KICH, KIRP, TGCT, and distant metastases in ACC and TGCT. Remarkably, SLC16A1 is closely associated with the clinical staging of most urological cancers. Differential expression of SLC16A1 in urological cancers of different clinical stages was also verified by the UALCAN database, as PRAD was missing clinical stage data, but significant differences in SLC16A1 expression were also found in patients with different Gleason scores, as detailed in Figure 2.
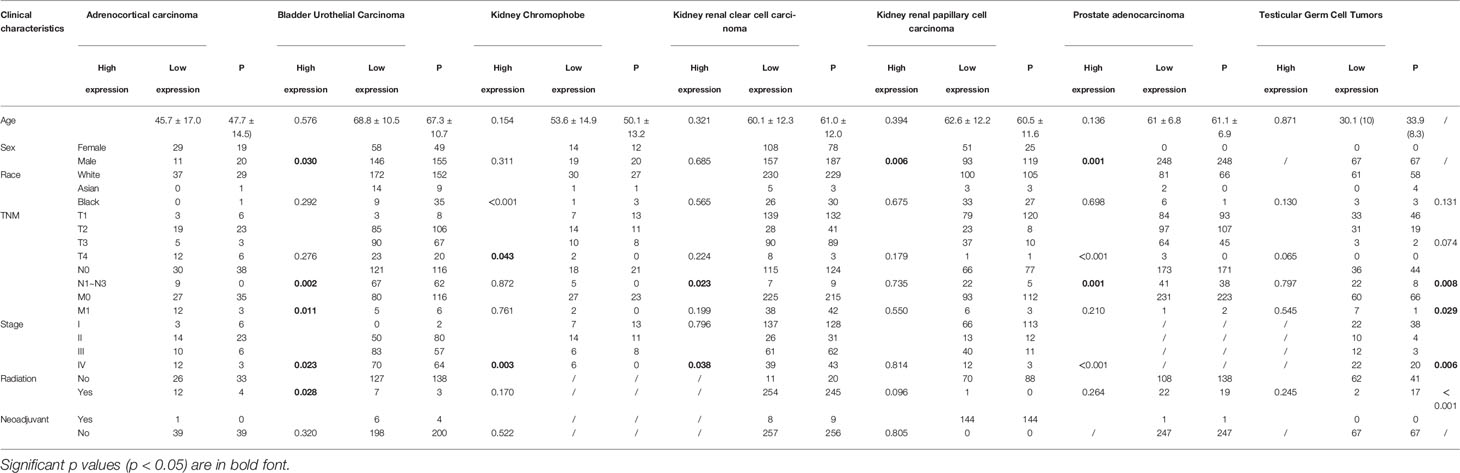
Table 1 Comparison of clinical characteristics of urological cancers patients with high and low expression of SLC16A1.
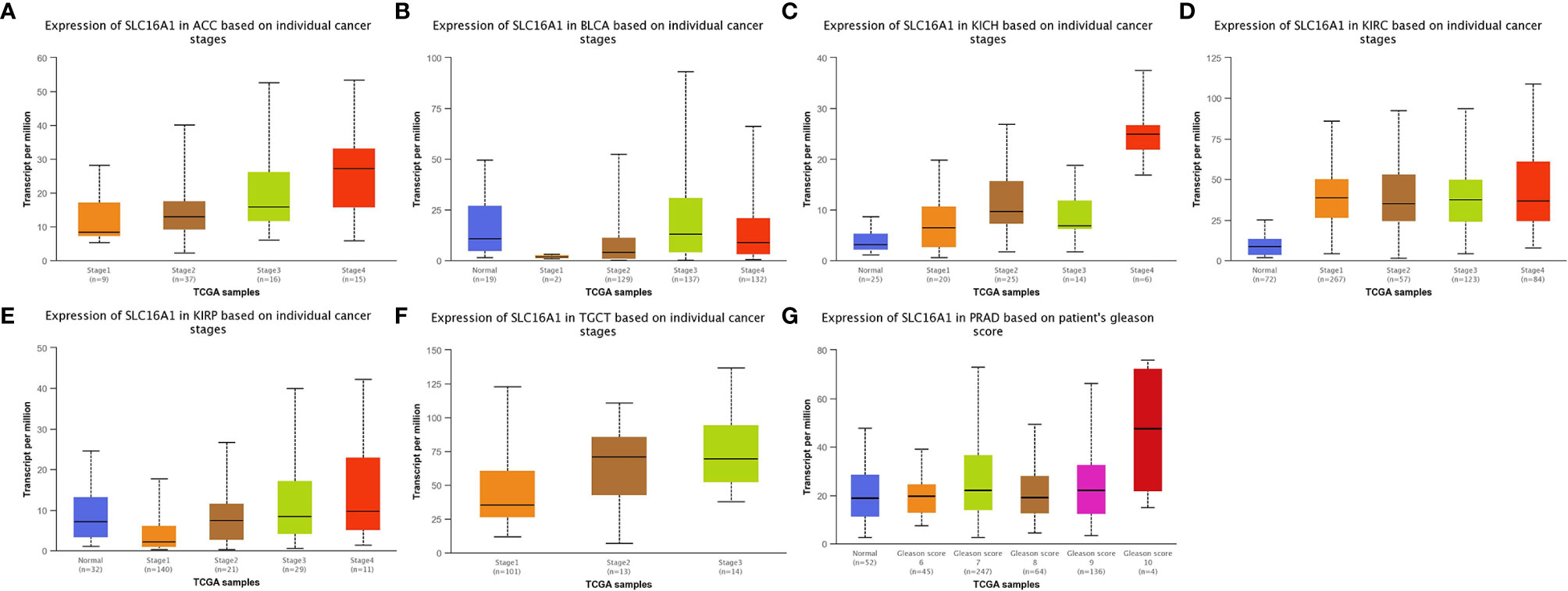
Figure 2 SLC16A1 expression correlates with different periods of urological cancer. SLC16A1 expression in (A) ACC, (B) BLCA, (C) KICH, (D) KIRC, (E) KIRP, (F) TGCT in different clinical stages and (G) PRAD in different Gleason.
The Expression of SLC16A1 in Urological Cancers Promoted MMRS Mutations and DNMTs Activity
SLC16A1 expression was significantly positively correlated with gene mutations of MMRs in majority of tumors (Figure 3A). In particular, the three MMR genes (MLH1, MSH2, and MSH6) were significantly associated with almost all urinary tumors (ACC, BLCA, KICH, Kirc, Kirp, PRAD, TGCT). As shown in Figure 3B, SLC16A1 expression was positively correlated with DNMTs expression in all human tumors. KICH, Kirc, and Prad were significantly correlated with the expression of four DTMTs (Figure 3B). The results showed that SLC16A1 could promote the occurrence and development of tumor by inducing MMRS mutation and up-regulating the activity of DNMTs.
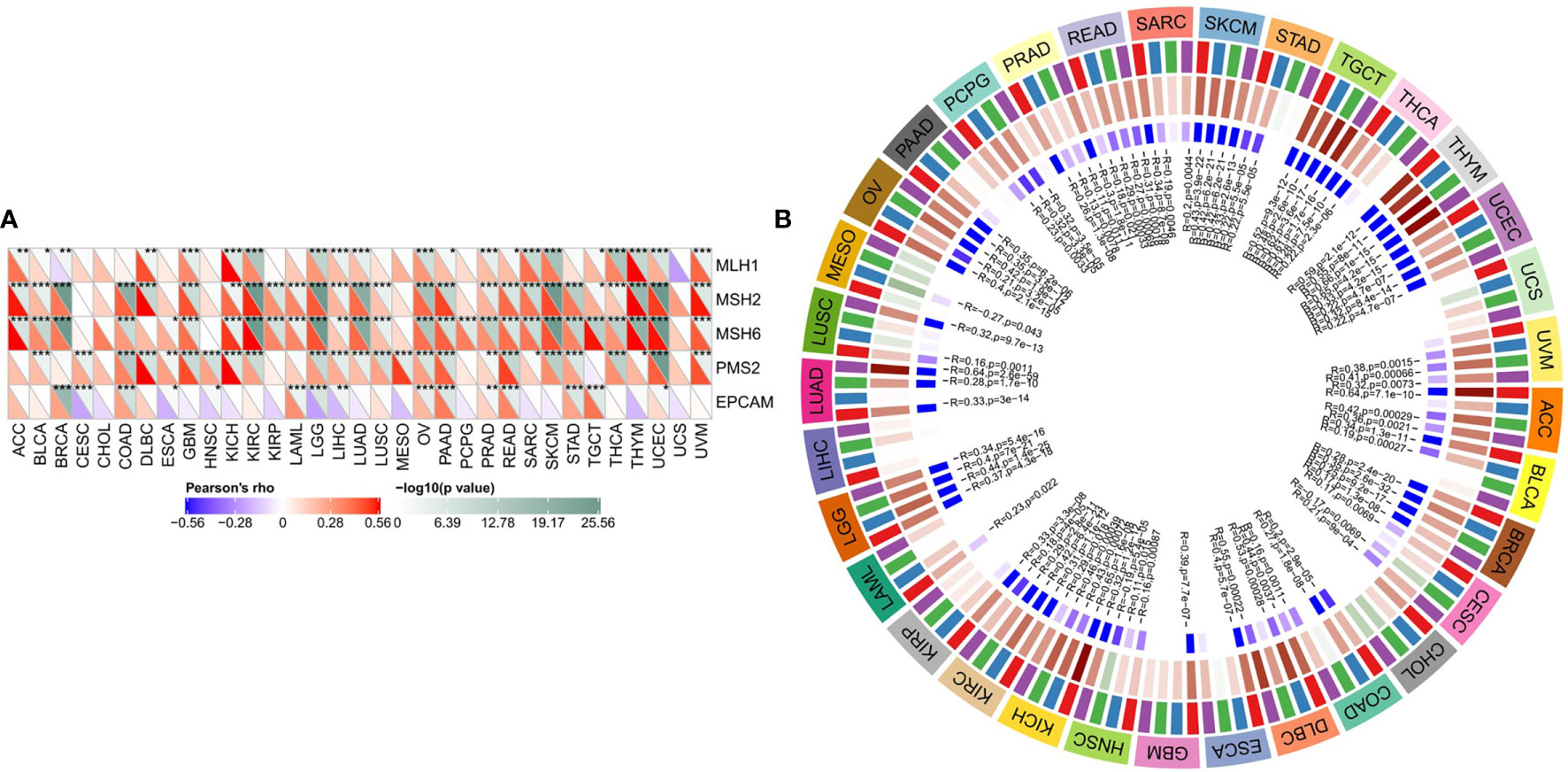
Figure 3 SLC16A1 expression in urological cancers promotes MMRS mutations and DNMTs activity. SLC16A1 expression and (A) ML H1, MSH2, MSH6, PMS2, EpCAM mutations; (B) Correlation between DNMT1, DNMT2, DNMT3A and DNMT3b expression. Note: MMRS: DNA mismatch repair; DNMTs: DNA methyltransferase. *P < 0.05, **P < 0.01, ***P < 0.001.
High Expression of SLC16A1 Suggested a Poor Prognosis of Urological Cancers
The association between SLC16A1 expression and overall survival (OS) in urological cancers was explored as shown in Figure 4A. SLC16A1 expression is a prognostic risk factor for ACC, KICH, and KIRP. To further analyze the impact of disease progression on prognostic outcomes, we also analyzed the effect of SLC16A1 expression on progression-free survival (PFS) of patients. SLC16A1 expression was significantly associated with ACC and PFS in the three renal cancers (Figure 4B). The prognostic value was also tested using Kaplan Meier as well as timeROC analysis. The results showed that overall survival was lower in urological patients with high SLC16A1 expression than in those with its low expression, except for two cancers, PRAD and TGCT (Figures 4C–I). Notably, SLC16A1 expression had a higher degree of impact on PFS in these urological patients (Figures 4J–P), and high SLC16A1 expression may suggest a poor prognosis for urological cancer patients.
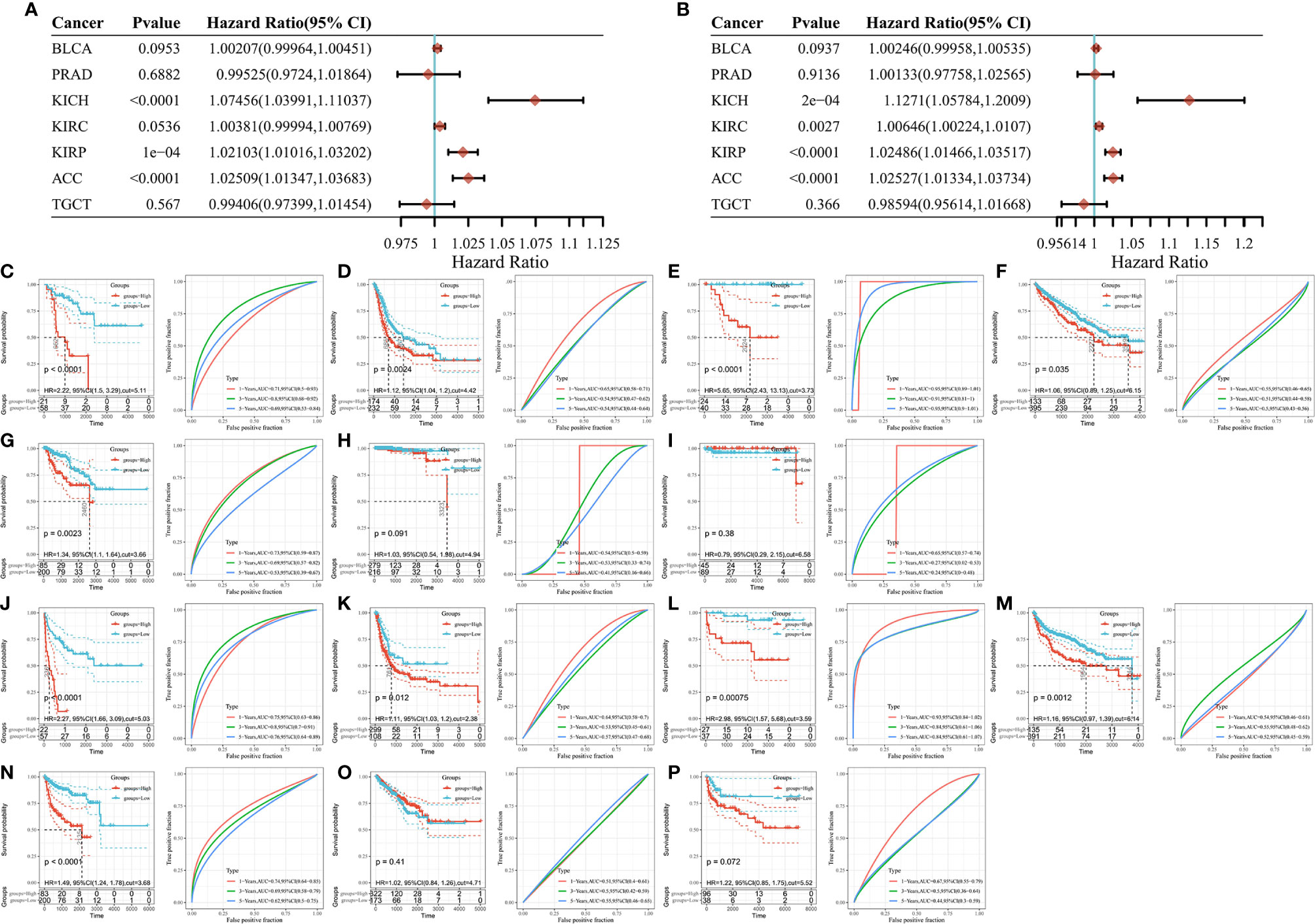
Figure 4 High SLC16A1 expression suggests a poor prognosis for urological cancer. Univariate Cox analysis of patients with different tumors (A) OS and (B) PFS; Kaplan Meier and timeROC analysis to detect the effect of SLC16A1 expression on OS in (C) ACC, (D) BLCA), (E) KICH, (F) KIRC, (G) KIRP, (H) PRAD, (I)TGCT patients and PFS in (J) ACC, (K) BLCA, (L) KICH, (M) KIRC, (N) KIRP, (O) PRAD, (P) TGCT patients. OS, total survival; PFS, progression-free survival; HR, Hazard ratio.
SLC16A1 Expression Was Associated With Carcinogenic Signaling Pathways
GOChord diagram visualizing the interconnection between GO terms and genes. As shown in Figure 5A, SLC16A1 was associated with cell activation, biological processes such as cell adhesion (BP), and was significantly enriched in intercellular-related cellular components such as focal adhesion (CC) (Figure 5B) and binding-related molecular functions such as glycosaminoglycan binding (MF) (Figure 5C). The results of KEGG analysis showed that slc16a1 expression was correlated with signals including cytokine cytokine receptor interaction, proteoglycans in cancer, and cell adhesion molecules. Using GSEA to look for signals that are activated in cancer, the most significantly enriched GO and KEGG signaling pathways have been listed in the top right corner of Figures 5E, F. High SLC16A1 expression was associated with cell cycle, PI3K-Akt and other signaling pathways, which may suggest that high SLC16A1 expression may be involved in tumor progression through regulation of the cell cycle.
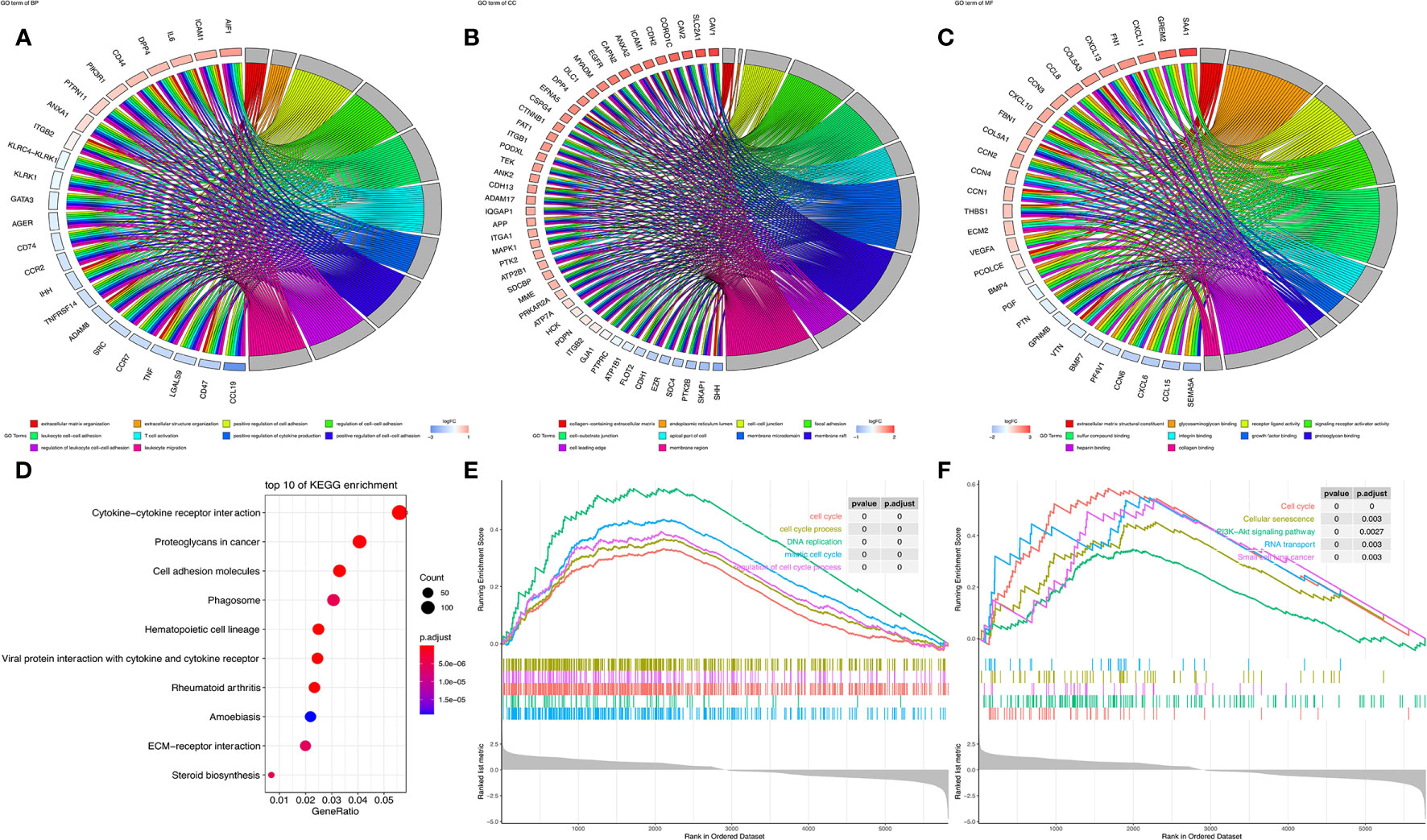
Figure 5 SLC16A1 expression is associated with carcinogenic signaling pathways. In GOChord plots showing the top 10 GO terms enriched in (A) BP, (B) CC, (C) MF and genes positively associated with SLC16A1; (D) bubble plots to show the top 10 pathways in KEGG analysis; GSEA analysis showing the set of (E) GO and (F) KEGG genes associated with SLC16A1 expression. GSEA, single gene set enrichment analysis; GO, Gene Ontology; BP, Bioengineering; CC, Cellular Component; MF, Molecular Function; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
Tumor cells show a much higher growth activity than normal cells. In recent years, it has been suggested that tumor cells metastasize to escape metabolic stress, which is a process that often involves metabolic reprogramming. Rewriting of energy metabolism pattern will confer tumor cells the ability to escape normal apoptotic process and to grow, proliferate and migrate (17). Since Warburg found that cancer cells tend to produce energy through glycolysis, many studies have been carried out on the energy metabolism of tumors. Studies have found that cancer cells still obtain energy through glycolysis despite the presence of oxygen (18). Lactic acid as the main product of anaerobic metabolism is considered to be one of the most important energies. The continuous production and accumulation of lactic acid can lead to metabolic gene expression changes, result in abnormal cell signal, weaken the function of mitochondria, upregulate SLC16 protein expression, thereby leading to cell migration and immune escape, and the promotion of cancerous cell process (19). SLC16A1 plays an important role in cancer metabolism. Lactic acid itself can rapidly upregulate SLC16A1 expression, which maintains glycolysis efficiency by regulating pH value in cells and interstitium. SLC16A1 plays a major role in lactate inflow, and cancer cells transport lactic acid inward through overexpression of SLC16A1, increasing carcinogenicity and invasion. Thus, targeting SLC16A1 may be a promising therapeutic strategy for some cancers (20, 21).
Based on TCGA and GTEX databases, we compared SLC16A1 expression in 27 different human cancers using TIMER and differential analysis. The results showed that SLC16A1 was differentially expressed in most solid cancers, particularly concentrated in urological cancers. This is also supported by the results of the CCLE database: SLC16A1 is significantly expressed in cell lines of urological origin. Further analysis of SLC16A1 expression in urological cancers showed that SLC16A1 protein expression was significantly stronger in pathological tissues of the kidney, prostate, and bladder than in the corresponding normal tissues, and that there was an association between high and low SLC16A1 expression and the different clinical features of urological cancers. SLC16A1 was differentially expressed between the different clinical stages of seven different urological cancers, and it is therefore conjectured that SLC16A1 may be involved in the disease progression of urological cancers.
Aberrations in epigenetic mechanisms have now been recognized as an important cause of human cancer progression (22). The continuous accumulation of MMRS gene mutations throughout the genome will allow the malignant transformation of cell signals, and promote the carriers of MMRS mutation to develop cancer (23). DNA methylation is critical for maintaining cellular phenotypes during DNA replication and is involved in defining when and where genes are expressed in normal and disease environments (24, 25). Currently, DNMTs inhibitors are the most widely studied compounds that inhibit epigenetic processes and have also been used in the treatment of cancer (26). Our results showed that SLC16A1 expression was significantly positively correlated with MMRs mutation and expression of DNMTs in urological tumors, indicating that SLC16A1 was related to the occurrence and development of urological tumors.
Besides, high expression in urological cancers appears to be with an increased prognostic risk. Our study found that SLC16A1 could serve as a biomarker for the prediction of OS and PFS of urological cancers and identifying cancer subtypes with higher prognostic risk, especially in several cancers of ACC, KIRC and KICH. SLC16A1 plays a key role in promoting cancer progression and metastasis. It has been found that SLC16A1 and SLC16A1-AS1 may together form a “head-to-head” complex unit with E2F1 promoter in muscle-infiltrating bladder cancer cells. E2F1 activates SLC16A1 and SLC16A1-AS1 to cooperatively regulate the corresponding targets related to cell migration and promote metabolic reprogramming and cell migration (27). High-expressed SLC16A1 has also been found to be associated with lymph node metastasis and distant metastasis of bladder cancer, thereby showing a negative impact on the overall survival of patients. Inhibiting SLC16A1 can significantly suppress the proliferation, migration and invasion of bladder cancer cells, and SLC16A1 promotes the progression of bladder cancer by affecting epithelial mesenchymal transformation and glycolysis (28). Fuhrman grade is a histological grade of renal cancer based on tumor nuclear morphology, and has been widely applied as the most effective prognostic parameter for predicting DSS (29). Ambrosetti et al. (30) conducted a semi-quantitative and qualitative analysis on 45 Kirc cases, and found that SLC16A1 is positively correlated with higher Fuhrman grade. All the above studies suggested that SLC16A1 is a risk factor for evaluating the prognosis of urological cancers.
On the other hand, to further study the role of SLC16A1 in tumors, We found by enrichment analysis that SLC16A1 expression is associated with cell activation, cell adhesion and other functions, and may be involved in signaling pathways such as the cell cycle. During tumor progression, metabolic reprogramming is often accompanied by cytoskeletal remodeling and the activation of transduction of mechanical signaling in cancer cells. Cancer cells induce metabolism-related signaling by continuously altering intercellular adhesions and finally by glycolysis to meet the increased motility as well as aggressiveness of cancer cells (31, 32). We speculate that the high expression of SLC16A1 in urological cancers may also suggest a metabolic reprogramming mediated by altered intercellular adhesion.
In Pereira’s study, to maintain a high glycolytic phenotype, prostate cancer is effectively transported to the breast via SLC16A1 and SLC16A4.The expression level of SLC16A1 in metastatic prostate cancer cells is significantly higher than that in more restricted prostate cancer cell lines. Silencing SLC16A1 can significantly inhibit the growth and motor characteristics of cancer cells (33), high expression of SLC16A1 may stimulate prostate cancer cell activation. Feng et al. (34) also speculated that SLC16A1-AS1 might be involved in the occurrence and metastasis of cancer through regulation of cell cycle through three different algorithms of WGCNA, GSEA and GSVA, and verified at the cellular level that SLC16A1-AS1 silencing could inhibit the expression of cyclin D1, promote cell stagnation in G0/G1 phase, and suppress the proliferation of oral squamous cell carcinoma cells. In Addition, we also found that SLC16A1 is associated with signaling pathways such as P13K-Akt, which is also a key factor in the cell cycle, and targeting PI3K-Akt signaling may induce cell cycle arrest (35, 36). This also suggests that SLC16A1 is involved in tumor development from another perspective.
To sum up, this research showed that SLC16A1 expression was present in urological tumors and found that the high expression of SLC16A1 was related to poor prognosis of patients with urological cancer and abnormal epigenetic processes, providing clinically useful biological markers.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author Contributions
HS, Z-SS, Z-SW, Y-LG, C-GX, and LZ: These authors have contributed equally to this work. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Wuhan Health Research Foundation (No.WX21M04), National Natural science Foundation of China (Grant No. 82002722), Hubei Province Health and Family Planning Scientific Research Project (No.WJ2018H209), Natural Science Foundation of Hubei Province (No.2020CFB175), and Research Fund of Wuhan Health and Family Planning Commission (No.WX20Q24).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Falzarano SM, Feely MM. How can Biomarkers Assist the Prognosis of Urologic Malignancies? Expert Rev Mol Diagn (2020) 20(2):131–3. doi: 10.1080/14737159.2019.1665506
2. Manini C, López JI. Insights Into Urological Cancer. Cancers (Basel) (2021) 13(2):204. doi: 10.3390/cancers13020204
3. Kheirollahi A, Hasanvand A, Abbaszadeh S, Moghadasi M. Pathophysiology and Urinary System Cancer: An Overview of the Most Important Herbal Plants and Natural Antioxidants on Kidney and Bladder Disorders. Res J Pharm Technol (2019) 12(2):972–80. doi: 10.5958/0974-360X.2019.00161.6
4. Breshears MA, Confer AW. The Urinary System. In: Pathologic basis of veterinary disease. Mosby (2017). p. 617.
5. Meuten DJ, Meuten TL. Tumors of the Urinary System. In: Tumors in domestic animals. John Wiley & Sons (2016). p. 632–88.
6. Antognelli C, Talesa VN. Glyoxalases in Urological Malignancies. Int J Mol Sci (2018) 19(2):415. doi: 10.3390/ijms19020415
7. Oztekin CV, Kaya-Sezginer E, Yilmaz-Oral D, Gur S. Male Urogenital Disorders and Metabolic Syndrome: Possible Links, Characteristics and Potential Treatment Strategies. Curr Pharm Des (2018) 24(9):1019–33. doi: 10.2174/1381612824666171213102836
8. Bosshart PD, Charles R-P, Garibsingh R-AA, Schlessinger A, Fotiadis D. SLC16 Family: From Atomic Structure to Human Disease. Trends Biochem Sci (2021) 46(1):28–40. doi: 10.1016/j.tibs.2020.07.005
9. Latif A, Chadwick AL, Kitson SJ, Gregson HJ, Sivalingam VN, Bolton J, et al. Monocarboxylate Transporter 1 (MCT1) Is an Independent Prognostic Biomarker in Endometrial Cancer. BMC Clin Pathol (2017) 17:27. doi: 10.1186/s12907-017-0067-7
10. Felmlee MA, Jones RS, Rodriguez-Cruz V, Follman KE, Morris ME. Monocarboxylate Transporters (SLC16): Function, Regulation, and Role in Health and Disease. Pharmacol Rev (2020) 72(2):466–85. doi: 10.1124/pr.119.018762
11. Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, et al. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat Genet (2013) 45(10):1113–20. doi: 10.1038/ng.2764
12. Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): An Immeasurable Source of Knowledge. Contemp Oncol (2015) 19(1A):A68. doi: 10.5114/wo.2014.47136
13. GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) Pilot Analysis: Multitissue Gene Regulation in Humans. Science (2015) 348(6235):648–60. doi: 10.1126/science.1262110
14. Gibney E, Nolan C. Epigenetics and Gene Expression. Heredity (2010) 105(1):4–13. doi: 10.1038/hdy.2010.54
15. Wang T, Stadler ZK, Zhang L, Weiser MR, Basturk O, Hechtman JF, et al. Immunohistochemical Null-Phenotype for Mismatch Repair Proteins in Colonic Carcinoma Associated With Concurrent MLH1 Hypermethylation and MSH2 Somatic Mutations. Familial Cancer (2018) 17(2):225–8. doi: 10.1007/s10689-017-0031-9
16. Jasek K, Kubatka P, Samec M, Liskova A, Smejkal K, Vybohova D, et al. DNA Methylation Status in Cancer Disease: Modulations by Plant-Derived Natural Compounds and Dietary Interventions. Biomolecules (2019) 9(7):289. doi: 10.3390/biom9070289
17. Lu J. The Warburg Metabolism Fuels Tumor Metastasis. Cancer Metastasis Rev (2019) 38(1):157–64. doi: 10.1007/s10555-019-09794-5
18. Xu XD, Shao SX, Jiang HP, Cao YW, Wang YH, Yang XC, et al. Warburg Effect or Reverse Warburg Effect? A Review of Cancer Metabolism. Oncol Res Treat (2015) 38(3):117–22. doi: 10.1159/000375435
19. San-Millán I, Brooks GA. Reexamining Cancer Metabolism: Lactate Production for Carcinogenesis Could be the Purpose and Explanation of the Warburg Effect. Carcinogenesis (2017) 38(2):119–33. doi: 10.1093/carcin/bgw127
20. Dell’Anno I, Barone E, Mutti L, Rassl DM, Marciniak SJ, Silvestri R, et al. Tissue Expression of Lactate Transporters (MCT1 and MCT4) and Prognosis of Malignant Pleural Mesothelioma (Brief Report). J Trans Med (2020) 18(1):1–7. doi: 10.1186/s12967-020-02487-6
21. Hashimoto T, Hussien R, Oommen S, Gohil K, Brooks GA. Lactate Sensitive Transcription Factor Network in L6 Cells: Activation of MCT1 and Mitochondrial Biogenesis. FASEB J (2007) 21(10):2602–12. doi: 10.1096/fj.07-8174com
22. Taby R, Issa JPJ. Cancer Epigenetics. CA Cancer J Clin (2010) 60(6):376–92. doi: 10.3322/caac.20085
23. Peltomäki P. DNA Mismatch Repair and Cancer. Mutat Res Rev Mutat Res (2001) 488(1):77–85. doi: 10.1016/S1383-5742(00)00058-2
24. Bird A. DNA Methylation Patterns and Epigenetic Memory. Genes Dev (2002) 16(1):6–21. doi: 10.1101/gad.947102
25. Pechalrieu D, Etievant C, Arimondo PB. DNA Methyltransferase Inhibitors in Cancer: From Pharmacology to Translational Studies. Biochem Pharmacol (2017) 129:1–13. doi: 10.1016/j.bcp.2016.12.004
26. Brueckner B, Kuck D, Lyko F. DNA Methyltransferase Inhibitors for Cancer Therapy. Cancer J (2007) 13(1):17–22. doi: 10.1097/PPO.0b013e31803c7245
27. Jansen BA, Logotheti S, Heckl D, Klusmann J-H, Kozcan D, Taher L, et al. A Conserved E2F1-Activated Gene Regulatory Network Encompassing Monocarboxylic Acid Transporter-1, Its Co-Operating Antisense lncRNA SLC16A1-AS1 and Their Common Downstream Targets Mediates Bladder Cancer Invasiveness. AACR (2017) 77(13). doi: 10.1158/1538-7445.AM2017-1895
28. Zhang G, Zhang Y, Dong D, Wang F, Ma X, Guan F, et al. MCT1 Regulates Aggressive and Metabolic Phenotypes in Bladder Cancer. J Cancer (2018) 9(14):2492. doi: 10.7150/jca.25257
29. Fuhrman SA, Lasky LC, Limas C. Prognostic Significance of Morphologic Parameters in Renal Cell Carcinoma. Am J Surg Pathol (1982) 6(7):655–63. doi: 10.1097/00000478-198210000-00007
30. Ambrosetti D, Dufies M, Dadone B, Durand M, Borchiellini D, Amiel J, et al. The Two Glycolytic Markers GLUT1 and MCT1 Correlate With Tumor Grade and Survival in Clear-Cell Renal Cell Carcinoma. PloS One (2018) 13(2):e0193477. doi: 10.1371/journal.pone.0193477
31. Sousa B, Pereira J, Paredes J. The Crosstalk Between Cell Adhesion and Cancer Metabolism. Int J Mol Sci (2019) 20(8):1933. doi: 10.3390/ijms20081933
32. Elia I, Doglioni G, Fendt S-M. Metabolic Hallmarks of Metastasis Formation. Trends Cell Biol (2018) 28(8):673–84. doi: 10.1016/j.tcb.2018.04.002
33. Pereira-Nunes A, Simões-Sousa S, Pinheiro C, Miranda-Gonçalves V, Granja S, Baltazar F. Targeting Lactate Production and Efflux in Prostate Cancer. Biochim Biophys Acta (BBA)-Mol Basis Dis (2020) 1866(11):165894. doi: 10.1016/j.bbadis.2020.165894
34. Feng H, Zhang X, Lai W, Wang J. Long Non-Coding RNA SLC16A1-AS1: Its Multiple Tumorigenesis Features and Regulatory Role in Cell Cycle in Oral Squamous Cell Carcinoma. Cell Cycle (2020) 19(13):1641–53. doi: 10.1080/15384101.2020.1762048
35. Liang J, Slingerland JM. Multiple Roles of the PI3K/PKB (Akt) Pathway in Cell Cycle Progression. Cell Cycle (2003) 2(4):336–42. doi: 10.4161/cc.2.4.433
Keywords: SLC16A1, cancer in urology, generic cancer, biological marker, prognosis
Citation: Zhang L, Song Z-S, Wang Z-S, Guo Y-L, Xu C-G and Shen H (2021) High Expression of SLC16A1 as a Biomarker to Predict Poor Prognosis of Urological Cancers. Front. Oncol. 11:706883. doi: 10.3389/fonc.2021.706883
Received: 08 May 2021; Accepted: 12 July 2021;
Published: 22 September 2021.
Edited by:
Ye Wang, Qingdao University Medical College, ChinaReviewed by:
Wei Zhang, Beijing Hospital, ChinaJianwei Zhang, Zhengzhou University, China
Huilin Xu, The Fifth Hospital of Wuhan, China
Copyright © 2021 Zhang, Song, Wang, Guo, Xu and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Shen, MjAwOTIwMzAyMDExNUB3aHUuZWR1LmNvbQ==
†These authors have contributed equally to this work
 Ling Zhang
Ling Zhang Zheng-Shuai Song
Zheng-Shuai Song Zhi-Shun Wang2†
Zhi-Shun Wang2†