
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Oncol. , 08 July 2021
Sec. Hematologic Malignancies
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.706798
 Huan-Ping Wang1,2†
Huan-Ping Wang1,2† Jun-Jun He3†
Jun-Jun He3† Qiao-Yun Zhu4†
Qiao-Yun Zhu4† Lin Wang3
Lin Wang3 Jian-Hu Li1
Jian-Hu Li1 Jian-Song Huang1,2
Jian-Song Huang1,2 Wan-Zhuo Xie1
Wan-Zhuo Xie1 Hong-Hu Zhu1,2*
Hong-Hu Zhu1,2* Jie Jin1,2*
Jie Jin1,2*The NUP214-ABL1 fusion gene is a constitutively active tyrosine kinase that can be detected in 6% of T-cell acute lymphoblastic leukemia (T-ALL) patients, and it can also be found in B-cell acute lymphoblastic leukaemia (B-ALL). However the NUP214-ABL1 fusion in acute myeloid leukemia (AML) has not yet been reported. Up to now, the sensitivity of NUP214-ABL1-positive patients to tyrosine kinase inhibitor (TKI) is still controversial. Here we report the first case of an AML patient carrying NUP214-ABL1 fusion gene. The conventional AML chemotherapy regimen for the patient was successful. Identification of additional AML patients with NUP214-ABL1 fusion gene will provide treatment experience and prognostic evaluation.
Identifying genetically targetable abnormalities that may respond to targeted therapy is clinically important in the context of precision medicine. Among all the fusion genes involving ABL1 rearrangement, the NUP214-ABL1 is the second common fusion in haematologic malignancies. About 6% of T-cell acute lymphoblastic leukaemia (T-ALL) patients carry this fusion gene (1). It can also be found in B-cell acute lymphoblastic leukaemia (B-ALL) (2, 3). However, as far as we know, the NUP214-ABL1 fusion gene has never been reported in acute myeloid leukaemia (AML).
NUP214-ABL1, a constitutively active tyrosine kinase, is a potential target of tyrosine kinase inhibitors (TKIs) (4, 5). However, the therapeutic effect of TKI for the NUP214-ABL1-positive patients is controversial due to limited clinical experience (6–11). Some studies have shown that TKI monotherapy or combined chemotherapy is effective for NUP214-ABL1-positive patients (6, 10–13). However, other studies have shown that NUP214-ABL1-positive patients had no response to TKI therapy or developed resistance to TKIs post relapse (3, 7–9).
Given that many NUP214-ABL1-positive patients were diagnosed retrospectively and had already received classical treatment (14), more NUP214-ABL1 cases are needed to guide clinicians caring for this subgroup of patients. Here, we describe the first case of an AML patient with the NUP214-ABL1 fusion gene detected by next-generation sequencing (NGS).
In January 2021, a 42-year-old male patient was admitted to our hospital due to skin bleeding. The patient’s blood count was as follows: white blood cells (WBCs) 82.5×109/L (differential: neutrophils 2.7%, eosinophils 0.5%, basophils 0.1%, lymphocytes 13.7%, monocytes 83%), haemoglobin of 92 g/l, and platelets 10×109/l. Lactate dehydrogenase (LDH) was increased to 2080 U/l (reference <245U/l). D-dimer was quantified at 1730 ug/L (reference range 0-700 ug/L). Ultrasonography revealed lymphadenopathy and splenomegaly. Bone marrow (BM) aspiration revealed 78% blast cells (Figure 1A). The blast cells were positive for cytoplasmic MPO, CD7, CD13, CD33, CD34, CD38 (dim), CD56 (partial), CD117, CD123, and HLA-DR, and negative for cytoplasmic CD3, cytoplasmic CD79a, CD1a, CD3, CD4, CD5, CD8, CD10, CD11b, CD14, CD15, CD16, CD19 and CD64 by flow cytometry (Figure 1B). Based on morphologic and immunophenotypical results, he was diagnosed with AML. Cytogenetic analyses revealed a normal male karyotype 46, XY.
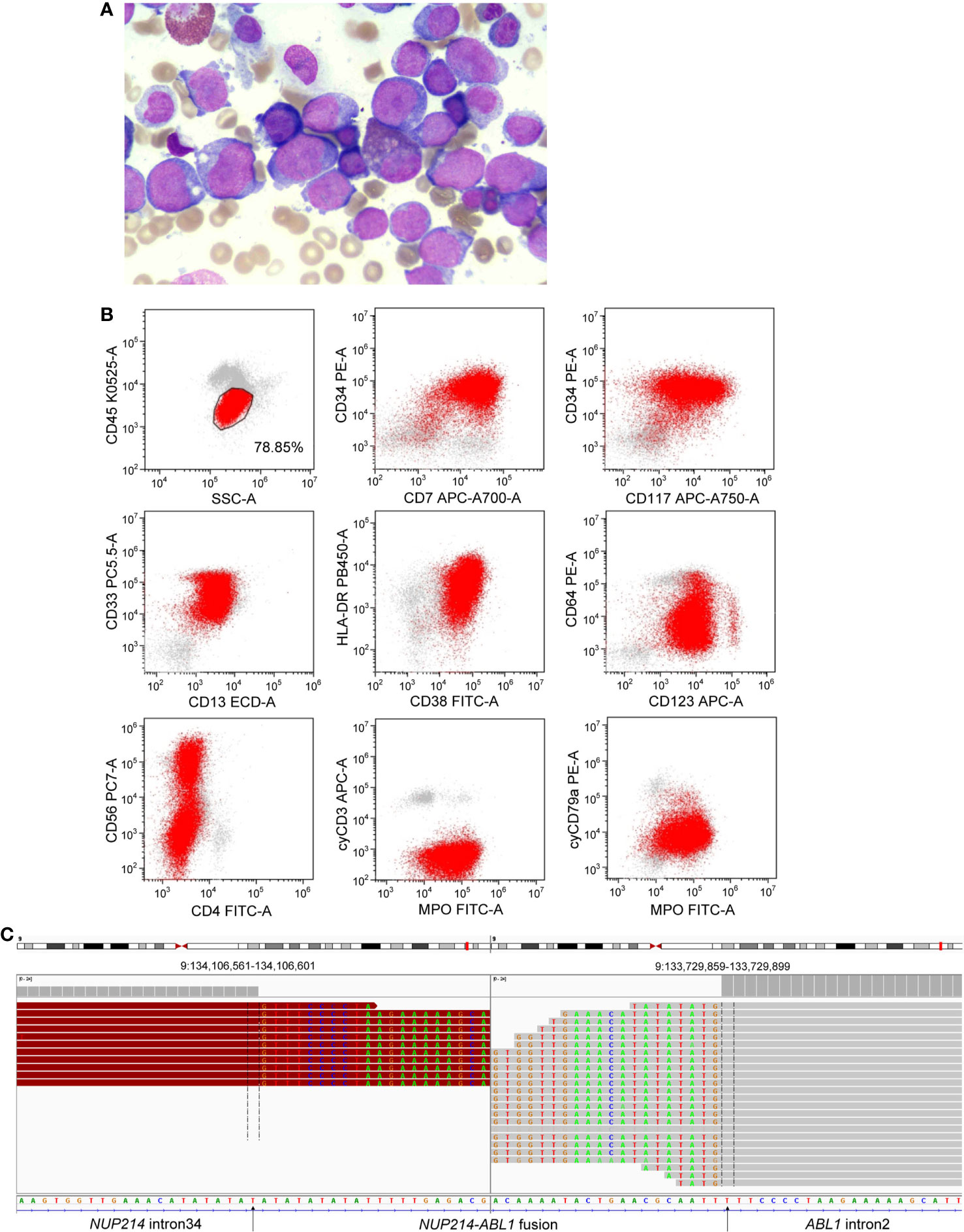
Figure 1 (A) Morphological observation showed blasts in a bone marrow smear. (B) Immunophenotype of AML was determined by flow cytometry. (C) Next-generation sequencing revealed a breakpoint in intron 34 of NUP214 and a breakpoint in intron 2 of ABL1, resulting in the NUP214-ABL1 fusion gene.
NGS (a panel of 88 genes) revealed that CEBPA (NM_004364.3) had double mutations (p.K304_Q305insL, and p.D75Gfs*33) and NRAS (NM_002524.5) had a point mutation (p.G13D). NGS also showed the presence of an NUP214 (NM_005085.4)-ABL1 (NM_007313.2) fusion gene (fusion of NUP214 exon 34 and ABL1 exon 3) (Figure 1C). Reverse transcription-polymerase chain reaction (RT-PCR) (primers: NUP214-E34-F GAGCAGCAGCAACACC, ABL1-E3-R TCACGCACCAAGAAGC) and sequencing of PCR products further confirmed NUP214-ABL1 fusion (Figures 2A, B). The fusion protein of NUP214-ABL1 was shown in Figure 2C.
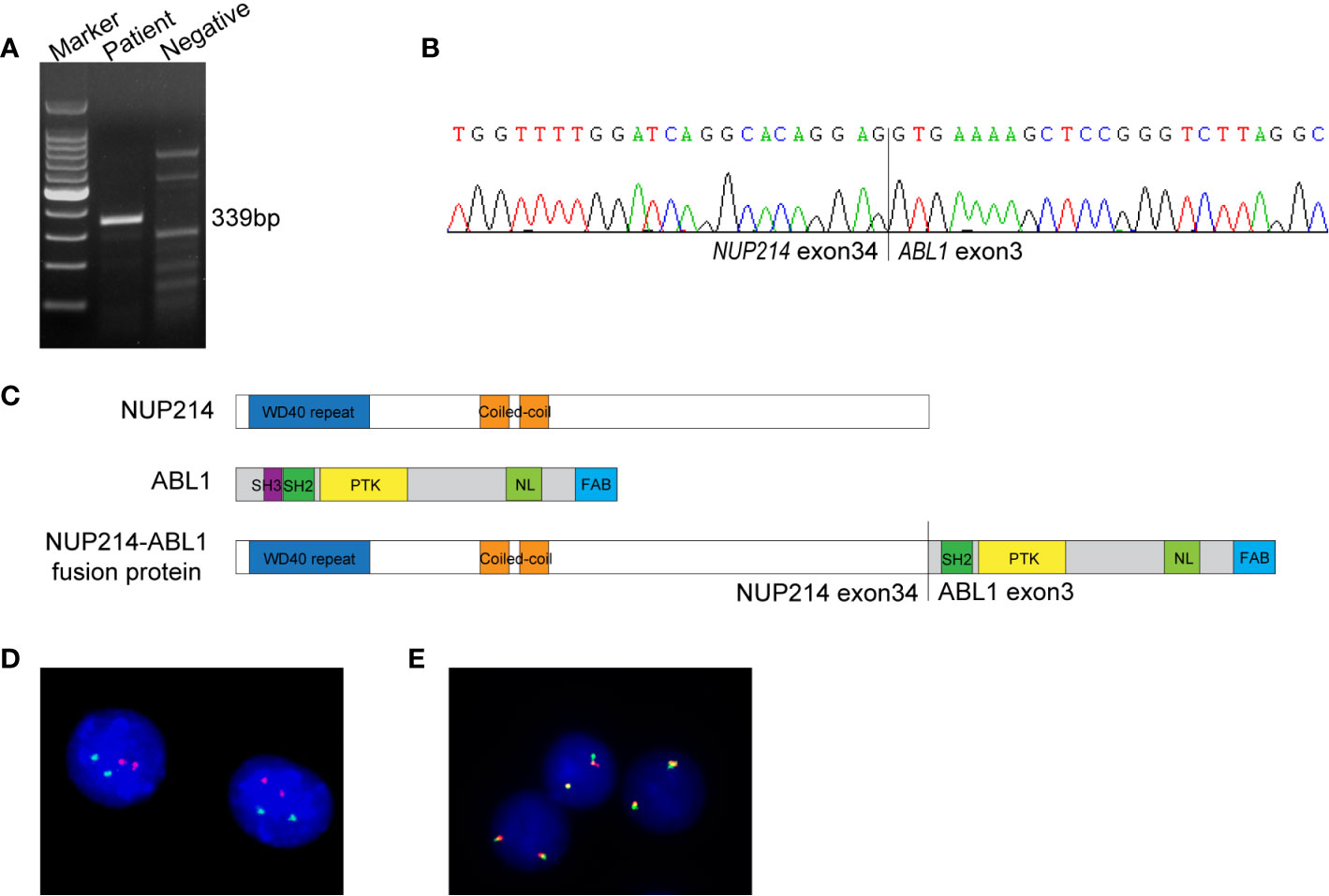
Figure 2 (A) RT-PCR for the NUP214-ABL1 fusion transcript. The expected product size was 339 bp. (B) Sanger sequencing of the PCR product confirmed the NUP214-ABL1 fusion gene of exon 34 of NUP214 to exon 3 of ABL1. (C) The structure of NUP214-ABL1 fusion protein. PTK, protein tyrosine kinase; NL, nuclear localization; FAB, F-actin binding. (D) FISH showed normal signal pattern using BCR/ABL1 dual-color dual-fusion probe. (E) FISH showed two intact ABL1 signals using an ABL1 break-apart probe.
To further investigate the NUP214-ABL1 fusion and explore whether there is amplification of the ABL1 gene, interphase fluorescence in situ hybridization (FISH) was performed using the BCR/ABL1 dual-colour dual-fusion probe and the ABL1 break-apart probe (Zytovision, Germany). Both probes demonstrated normal signal patterns. No amplification or the split signal of ABL1 was detected (Figures 2D, E).
With one cycle of conventional induction chemotherapy, which includes idarubicin and cytarabine, the patient achieved complete morphologic remission (CR) with a minimal residual disease (MRD) under 0.01%. After that, he was treated with high-dose cytarabine for consolidation therapy and is currently in his third phase of consolidation therapy. To date, he was in CR status.
In this study, we demonstrated the NUP214-ABL1 fusion gene detected by NGS in a patient with AML. As far as we know, this is the first report of an AML patient with NUP214-ABL1 fusion.
Identifying genetically targetable abnormalities is extremely important due to the proposal of targeted therapy in combination with chemotherapy and improved survival of patients. However, cryptic ABL1 translocation is difficult to detect through conventional cytogenetic and FISH analysis, such as the NUP214-ABL1 fusion in our case. The major cause might be that both NUP214 and ABL1 are located at the edges of 9q34, and FISH may not successfully detect the fusion of these two genes, as reported in a previous study (2). In this case, we revealed the presence of the NUP214-ABL1 fusion through NGS and confirmed it by RT-PCR. Therefore, RT-PCR and some high-resolution sequencing, such as NGS, appear to be a very useful method to identify NUP214-ABL1 fusions (10, 15). In addition, our case did not have extrachromosomal ABL1 amplification, which is similar to the results of previous studies detected by FISH (2, 14). Notably, most NUP214-ABL1 fusions were different exons of NUP214 (from exons 23 to 34) fused to exon 2 of ABL1 (1). However, the case described here involves ABL1 exon 3, which is consistent with the other’s report (12).
Identification of molecular abnormalities by NGS could provide important prognostic and treatment information for AML patients, which has become a part of the clinical workup. Several molecular mutations, including CEBPA, NPM1, FLT3, IDH1/IDH2, c-KIT, ASXL1, RUNX1, and TP53, can refine prognostics groups, especially in patients with a normal karyotype (16). In this case, the patient had a normal karyotype, which is associated with intermediate risk of survival outcomes according to cytogenetic category in AML. He had CEBPA double mutations, NRAS point mutation, and NUP214-ABL1 fusion gene, and he had no other adverse-risk genetic lesions. As we know, double CEBPA mutations are associated with favorable prognosis in patients with AML (17). Combining the results of cytogenetics and molecular mutations, the patient belongs to the group with favorable prognosis. Although the patient had the NUP214-ABL1 fusion, the impact of this fusion on survival has not been determined. However, a promising test result was that the patient had no episomal amplification detected by FISH because previous studies reported that the presence of episomes implies a more radical disease process and a poorer prognosis in T-ALL (7, 8). Based on these clinical data, this patient may have a favorable or intermediate prognosis. Until today, the patient is still in remission with no signs of relapse.
The sensitivity of patients with NUP214-ABL1 fusion to TKIs is controversial, as these subgroups of patients are rare (7–9, 14). Some reports have shown that TKIs might be effective for patients with NUP214-ABL1 fusion (6, 10, 12, 13); however, they may be more suitable for use either in combination with other drugs (11–13) or as maintenance therapy after allo-HSCT (10). Some studies have demonstrated that NUP214-ABL1-positive patients show an initial favorable response to TKIs post relapse but can develop resistance to TKIs (8, 9). In this report, the patient received conventional AML-type chemotherapy, and he achieved a CR at the end of induction. Additionally, similar to our case, some patients with NUP214-ABL1 fusion are treated by conventional chemotherapy without using TKIs and achieve a CR at the end of induction (9, 15). However, if this patient experiences relapse, we may consider adding TKIs, as previous studies suggested that NUP214-ABL1-positive patients could benefit from TKIs post relapse (12, 13). However, it remains unknown to what extent NUP214-ABL1 is similar in ALL and AML and whether TKIs are also applicable to AML patients with the NUP214-ABL1 fusion, as there is currently no report on this aspect. We look forward to more clinical experience to determine the sensitivity of NUP214-ABL1-positive AML to TKIs.
To our knowledge, we describe the first case of NUP214-ABL1 fusion gene in a patient with AML. This study emphasizes the need to detect NUP214-ABL1 fusion gene in AML. The good result of this patient with conventional AML treatment regimen made it impossible to determine the sensitivity of NUP214-ABL1 to TKIs. More case reports are needed to better study the sensitivity of NUP214-ABL1 fusion protein to TKIs in AML.
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2017) in National Genomics Data Center (Nucleic Acids Res 2021), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences. The accession number is: HRA000867. Please access it from the following link: https://bigd.big.ac.cn/gsa-human/browse/HRA000867.
The studies involving human participants were reviewed and approved by the Research Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University. The patients/participants provided their written informed consent to participate in this study.
H-PW wrote the manuscript. H-PW, J-JH, Q-YZ, LW, J-HL, and J-SH performed the research and analyzed the data. W-ZX provided samples and clinical data. JJ and H-HZ critically revised the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, et al. Fusion of NUP214 to ABL1 on Amplified Episomes in T-Cell Acute Lymphoblastic Leukemia. Nat Genet (2004) 36(10):1084–9. doi: 10.1038/ng1425
2. Tsujimoto SI, Nakano Y, Osumi T, Okada K, Ouchi-Uchiyama M, Kataoka K, et al. A Cryptic NUP214-ABL1 Fusion in B-Cell Precursor Acute Lymphoblastic Leukemia. J Pediatr Hematol Oncol (2018) 40(6):e397–9. doi: 10.1097/MPH.0000000000001007
3. Eyre T, Schwab CJ, Kinstrie R, et al, McGuire AK, Strefford J, Peniket A. Episomal Amplification of NUP214-ABL1 Fusion Gene in B-Cell Acute Lymphoblastic Leukemia. Blood (2012) 120(22):4441–3. doi: 10.1182/blood-2012-09-456517
4. De Keersmaecker K, Rocnik JL, Bernad R, Lee BH, Leeman D, Gielen E, et al. Kinase Activation and Transformation by NUP214-ABL1 Is Dependent on the Context of the Nuclear Pore. Mol Cell (2008) 31(1):134–42. doi: 10.1016/j.molcel.2008.05.005
5. De Keersmaecker K, Versele M, Cools J, Superti-Furga G, Hantschel O. Intrinsic Differences Between the Catalytic Properties of the Oncogenic NUP214-ABL1 and BCR-ABL1 Fusion Protein Kinases. Leukemia (2008) 22(12):2208–16. doi: 10.1038/leu.2008.242
6. Deenik W, Beverloo HB, van der Poel-van de Luytgaarde SC, Wattel MM, van Esser JW, Valk PJ, et al. Rapid Complete Cytogenetic Remission After Upfront Dasatinib Monotherapy in a Patient With a NUP214-ABL1-Positive T-Cell Acute Lymphoblastic Leukemia. Leukemia (2009) 23(3):627–9. doi: 10.1038/leu.2008.318
7. Stergianou K, Fox C, Russell NH. Fusion of NUP214 to ABL1 on Amplified Episomes in T-ALL–Implications for Treatment. Leukemia (2005) 19(9):1680–1. doi: 10.1038/sj.leu.2403877
8. Koschmieder S, Burmeister T, Bruggemann M, Berkemeier A, Volpert S, Wieacker P, et al. Molecular Monitoring in NUP214-ABL-Positive T-Acute Lymphoblastic Leukemia Reveals Clonal Diversity and Helps to Guide Targeted Therapy. Leukemia (2014) 28(2):419–22. doi: 10.1038/leu.2013.272
9. Tsurusaki Y, Nagai JI, Fujita S, Sugiyama M, Nakamura W, Hayashi A, et al. Whole-Exome Sequencing Reveals the Subclonal Expression of NUP214-ABL1 Fusion Gene in T-Cell Acute Lymphoblastic Leukemia. Pediatr Blood Cancer (2020) 67(1):e28019. doi: 10.1002/pbc.28019
10. Aldoss I, Pullarkat V. Response to Single Agent Dasatinib Post Allogeneic Transplant in B-Cell Acute Lymphoblastic Leukemia With NUP214-Abl1. Leuk Lymphoma (2019) 60(11):2832–4. doi: 10.1080/10428194.2019.1605510
11. Chen Y, Zhang L, Huang J, Hong X, Zhao J, Wang Z, et al. Dasatinib and Chemotherapy in a Patient With Early T-Cell Precursor Acute Lymphoblastic Leukemia and NUP214-ABL1 Fusion: A Case Report. Exp Ther Med (2017) 14(5):3979–84. doi: 10.3892/etm.2017.5046
12. Duployez N, Grzych G, Ducourneau B, Alarcon Fuentes M, Grardel N, Boyer T, et al. NUP214-ABL1 Fusion Defines a Rare Subtype of B-Cell Precursor Acute Lymphoblastic Leukemia That Could Benefit From Tyrosine Kinase Inhibitors. Haematologica (2016) 101(4):e133–4. doi: 10.3324/haematol.2015.136499
13. Clarke S, O’Reilly J, Romeo G, Cooney J. NUP214-ABL1 Positive T-Cell Acute Lymphoblastic Leukemia Patient Shows an Initial Favorable Response to Imatinib Therapy Post Relapse. Leuk Res (2011) 35(7):e131–3. doi: 10.1016/j.leukres.2011.03.025
14. Graux C, Stevens-Kroef M, Lafage M, Dastugue N, Harrison CJ, Mugneret F, et al. Heterogeneous Patterns of Amplification of the NUP214-ABL1 Fusion Gene in T-Cell Acute Lymphoblastic Leukemia. Leukemia (2009) 23(1):125–33. doi: 10.1038/leu.2008.278
15. Peterson JF, Pitel BA, Smoley SA, Smadbeck JB, Johnson SH, Vasmatzis G, et al. Detection of a Cryptic NUP214/ABL1 Gene Fusion by Mate-Pair Sequencing (MPseq) in a Newly Diagnosed Case of Pediatric T-Lymphoblastic Leukemia. Cold Spring Harb Mol Case Stud (2019) 5(2). doi: 10.1101/mcs.a003533
16. NCCN. Clinical Practice Guidelines in Oncology-Acute Myeloid Leukemia (Version 3.2019) [DB/O]. Available at: http://www.nccn.org.
Keywords: ABL1, NUP214-ABL1, acute myeloid leukemia, tyrosine kinase, next-generation sequencing
Citation: Wang H-P, He J-J, Zhu Q-Y, Wang L, Li J-H, Huang J-S, Xie W-Z, Zhu H-H and Jin J (2021) Case Report: The First Report of NUP214-ABL1 Fusion Gene in Acute Myeloid Leukemia Patient Detected by Next-Generation Sequencing. Front. Oncol. 11:706798. doi: 10.3389/fonc.2021.706798
Received: 08 May 2021; Accepted: 28 June 2021;
Published: 08 July 2021.
Edited by:
Robert Ohgami, University of California, San Francisco, United StatesReviewed by:
Christian Kunder, Stanford Healthcare, United StatesCopyright © 2021 Wang, He, Zhu, Wang, Li, Huang, Xie, Zhu and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Jin, amllajA1MDNAemp1LmVkdS5jbg==; Hong-Hu Zhu, emh1aGhkb2NAMTYzLmNvbQ==
†These authors contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.