- 1The First School of Clinical Medicine, Shandong University of Traditional Chinese Medicine, Jinan, China
- 2Department of Thyroid and Breast Surgery, The 960th Hospital of the Chinese People’s Liberation Army, Jinan, China
- 3Department of Pathology, The 960th Hospital of the Chinese People’s Liberation Army, Jinan, China
Thyroid carcinoma (TC) is one of the most prevalent primary endocrine tumors, and its incidence is steadily and gradually increasing worldwide. Accumulating evidence has revealed the critical functions of long noncoding RNAs (lncRNAs) in the tumorigenesis and development of TC. Many TC-associated lncRNAs have been documented to be implicated in TC malignant behaviors, including abnormal cell proliferation, enhanced stem cell properties and aggressiveness, and resistance to therapeutics, through interaction with proteins, DNA, or RNA or encoding small peptides. Therefore, further elucidating the lncRNA dysregulation sheds additional insights into TC tumorigenesis and progression and opens new avenues for the early diagnosis and clinical therapy of TC. In this review, we summarize the abnormal expression of lncRNA in TC and the fundamental characteristics in TC tumorigenesis and development. Additionally, we introduce the potential prognostic and therapeutic significance of lncRNAs in TC.
Introduction
Thyroid carcinoma (TC) is the most prevalent malignancy of the endocrine system and accounts for about 1% of all malignancies. The yearly incidence of TC in 2019 was 15.94 per 100,000 population (1). Currently, TC ranks the fourth most common cancer for women (2, 3). Early TC-associated investigation mainly focused on the biological functions of protein-coding genes (PCGs) for their fundamental functions in the regulation of signaling transduction and various biological activities. Therefore, elucidating the functional roles and molecular mechanisms of noncoding RNAs (ncRNAs) in TC tumorigenesis is of critical significance. It has been well accepted that less than 2% of genome sequence encodes proteins, whereas the remaining 98% genome sequences do not have coding potential and their transcriptional products (ncRNAs) have not been functionally characterized. Moreover, in the past, this was once considered to be “transcriptional noise” (4–6). Based on the length of transcripts, ncRNAs can be subdivided into small ncRNAs (<200 base pairs) or long ncRNAs (>200 base pairs, lncRNAs). The small ncRNAs that play crucial roles in tumorigenesis could be further classified as miRNAs, tsRNAs, and piRNAs. LncRNAs are transcripts like mRNA in length ranging from 200 nt to ~100 kilobases (kb) lacking significant open reading frames and have no coding potential. LncRNAs include subclasses such as pseudogenes and circRNAs (7–9). At present, human ncRNAs include more than 10,000 small ncRNA genes, approximately 14,500 pseudogenes, and almost 16,000 lncRNA genes according to the GENCODE Release 24 annotation (10). LncRNAs are characterized by extremely weak expression, worse conservation in different species, and specifically expressed in different tissues and developmental stages (11). Recently, a growing body of evidence suggests that ncRNAs, particularly lncRNA, have emerged as important regulators of gene expression during signaling transduction and diverse physiological and pathological processes through interacting with RNA, DNA, and protein and forming RNA–RNA, RNA–DNA, and RNA–protein complexes, thereby regulating gene expression via various molecular mechanisms, including modulation of transcription, mRNA stability, and translation (12, 13).
The first lncRNA was identified in fetal liver tissue in 1990 (14). The following year, X chromosome inactivation mediated by one of the most famous lncRNA XIST was found (15). To date, more than 50,000 genes have been found to transcribe lncRNA (16), and this number is still steadily and rapidly increasing. Emerging evidence has been well accepted that lncRNAs are involved in gene regulation at the transcriptional or posttranscriptional level and play critical roles in signaling transduction and multiple physiological and pathological disease initiation and processes. LncRNAs were characterized by differentially expressing different organ types and cancers including TC (17–19). Currently, the functional roles of lncRNAs in TC tumorigenesis and development remain largely unknown, and only a small part of lncRNAs has been extensively studied (3, 20). Interestingly, many lncRNAs were identified in body fluids, which were easily detected and analyzed, making them hold the potential to act as attractive biomarkers in liquid biopsy of TC. For example, lncRNA HOTAIR is overexpressed in the serum and is an independent prognostic marker to predict lymph node metastasis of papillary thyroid carcinoma (PTC) (21). The unique and pivotal characteristics of lncRNA reveal their attractive clinical significance in the TC diagnosis and targeted therapy. LncRNAs bear a tremendous potential to improve our understanding of the steps involved in TC tumorigenesis. In this review, we summarize the aberrant expression of lncRNA in TC and generally introduce the functional roles of lncRNA-mediated pathogenesis in TC tumorigenesis by functioning as tumor-suppressor gene or oncogene. Additionally, we discuss the prognostic, diagnostic, and therapeutic potentials of lncRNA in TC.
Classification of Human TC
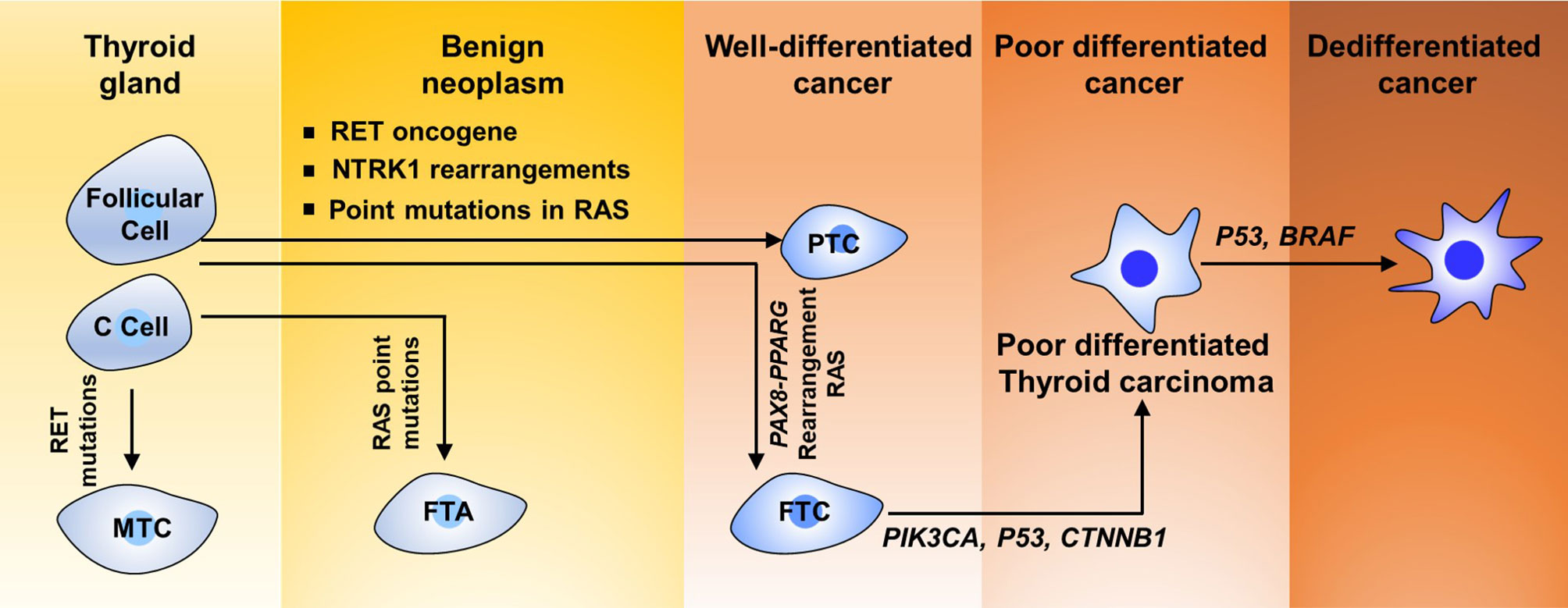
TC includes multiple histological types, such as PTC, follicular thyroid carcinoma (FTC), Hurthle cell carcinoma, poorly differentiated thyroid carcinoma, anaplastic thyroid carcinoma, squamous cell carcinoma, and medullary thyroid carcinoma (Figure 1). TC derived from follicular cells could be further divided into three classes based on the diverse histological and pathological elements: well-differentiated TC, poorly differentiated TC, and highly aggressively dedifferentiated TC or ATCs. Well-differentiated TC is composed of PTC and FTC subtypes. Approximately 80% of TCs are diagnosed as PTCs and 10% as FTCs in clinical trials worldwide (22–25). According to surveillance epidemiology and end results (SEER) data from the USA, 5%–10% of TCs originate from calcitonin-producing parafollicular cells (also known as C cells) within the thyroid gland and are known as medullary TC (MTCs) (26–29).
Papillary Thyroid Carcinoma
Having a papillary architecture and containing carcinoma cells with classic nuclear changes is the unique feature of PTCs (30). PTC incidence is considered to three to six times higher in women than in men among the regions in the world. Exposure to ionizing radiation is considered as an awfully risk element for PTC, as revealed by the higher incidence of PTC among the survivors of the Hiroshima and Nagasaki atomic bombing and among children in Belarus and Ukraine after the Chernobyl accident (31, 32).
Follicular Thyroid Carcinoma
FTCs are routinely well-differentiated, unifocal, and encapsulated, but they trend toward aggressiveness and extend to blood vessels and invade systemically through the bloodstream. The incidence of FTC is significantly superior in endemic goiter regions, indicating a close correlation between FTC tumorigenesis and iodine deficiency (33–35).
Anaplastic Thyroid Carcinoma
ATCs are highly undifferentiated and singularly invasive. ATCs are uncommon carcinoma and approximately account for 2%–5% of all TCs (36). ATC cells lack the ability to seize iodine and form thyroglobulin. A large proportion of ATCs originate from pre-existing PTCs and FTCs, but the others seem to derive from de novo. The median survival time of ATC patients is only 6 months.
Medullary Thyroid Carcinoma
MTCs display well-defined carcinoma, which are characterized by lacking a well-formed capsule. High migration ability is the feature of MTCs. MTCs is frequently found to invade outside the thyroid, migrate to lymph nodes and vasculature. Nevertheless, most MTCs are sporadic, and 25% of the incidence of MTCs is hereditary. This familial disease is inherited in an autosomal dominant manner and composed of three subtypes: MEN2A, MEN2B, and familial MTC.
Genetic Lesions in Thyroid Carcinoma
Like other tumor types, TC tumorigenesis and development require increasing accumulation of diverse genetic and/or epigenetic transformation. Many genetic factors are implicated in TC tumorigenesis. Genetic mutations regulating the biological functions of oncogenic or tumor-suppressive genes have been fully established in thyroid neoplasias. More than one-quarter of MTC cases are hereditary. Approximately 3.5%–6.2% of TCs are idiopathic forms of familial non-MTC (36, 37). In addition, TC develops in patients with cancer syndromes derive from germline mutations (38). Fusion protein generated from rearrangements including NTRK1 has been revealed in 10% of cases of PTCs (39). 1799T>A mutation in BRAF, which results in the switch of V600E at the protein level, was observed in about 40% of PTCs and 20% of ATCs (Figure 1) (40). As shown in Figure 1, more than 70% of PTCs contain crucial mutations in genes encoding core components of the MAPK signaling pathway, namely, the proto-oncogene RET, which is also known as TRK, RAS, and BRAF (41–43). The RET gene encodes a transmembrane receptor protein for neurotrophic growth factor binding and frequent rearrangement between the conserved domain of tyrosine kinase of RET and the 5′ region of their partner genes leading to formulate the RET/PTC oncogene in about 30% of PTCs (44).
RAS proto-oncogene mutations have been considered to be closely correlated with FTC and have been suggested in about 30% of tumors of the follicular variant of PTC (PTC-FV) (45). Genetic alteration of the RAS gene is also the primary molecular incident indicated in poorly differentiated TC (46). Human RAS genes, which can be further classified into HRAS, KRAS, and NRAS, encode highly evolutionarily conserved G proteins that reside at the intracellular and transduce signaling transmitted from receptor tyrosine kinases and the downstream signaling along the MAPK and PI3K–AKT (Figure 2). Point mutations that activate these signaling pathways typically are codons 12, 13, and 61 of the RAS genes (47–49). In TC, codon 61 mutations in NRAS and HRAS are most the frequent. RAS mutations are commonly found in various TCs, including 10%–20% of PTCs, 40%–50% of FTCs, and 20%–40% of ATCs (50). Conversely, mutations in MAPK signaling effector genes are correlated with early stages of TC tumorigenesis. Interestingly, mutations in AKT1 and BRAF, but not in PIK3CA, have been revealed in poorly differentiated TC (51). Mutations that repress p53 effects are commonly validated in ATCs (Figure 1) (52, 53). Abnormal activation of Wnt/β-catenin (54–56) or STAT3 (55, 57, 58) signaling has been commonly observed in TC initiation and progression and predicts poor survival with TC patients. However, other signaling pathways involved in TC initiation and progression are poorly recognized. Thus, a better understanding of the molecular basis and signaling events associated with TC development not only elucidates the mechanism of the TC process but also offers a new window for diagnostic and prognostic prediction and a profound effect on the targeted therapy for patients with TC.
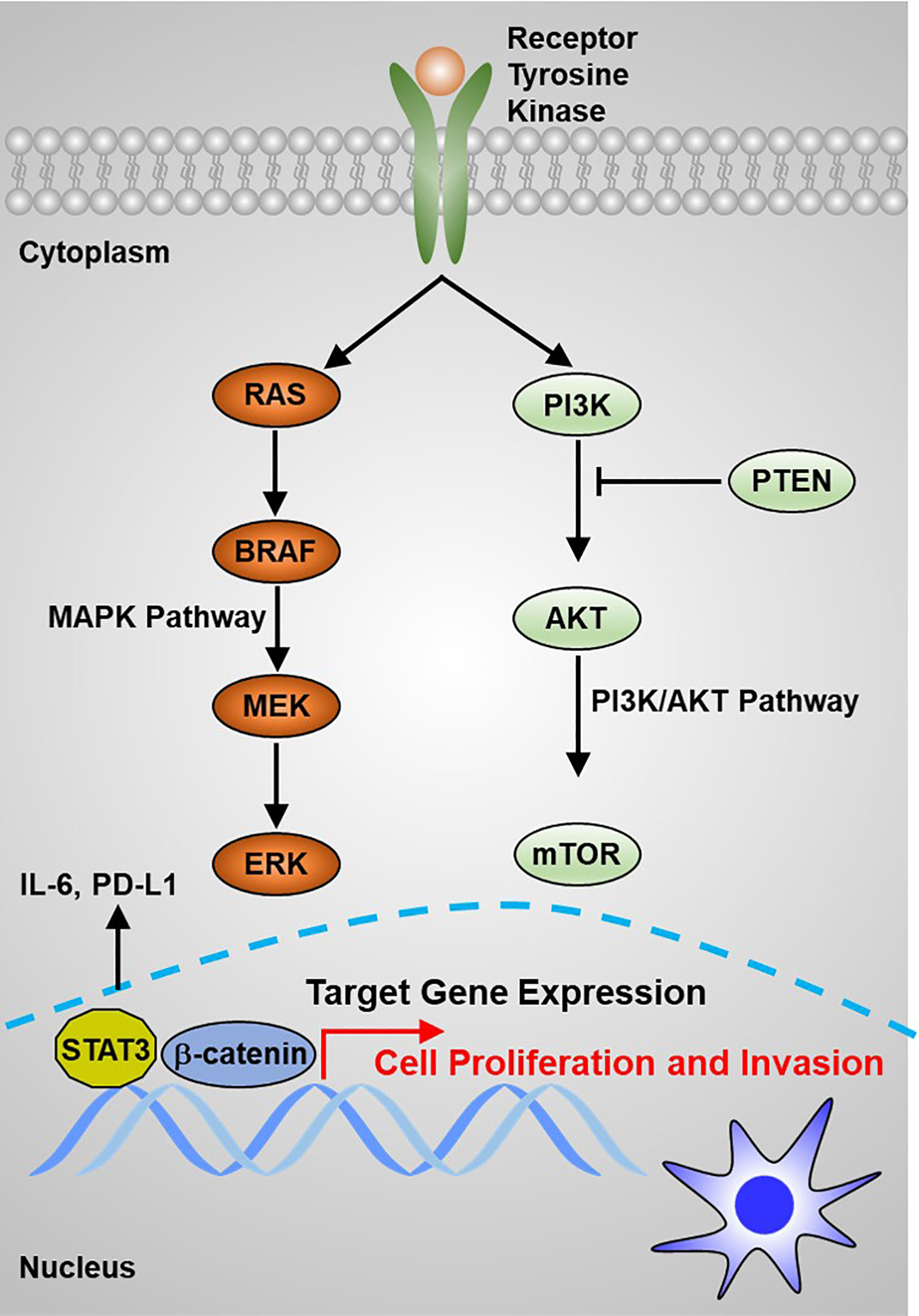
Figure 2 The main signaling pathways that contribute to thyroid carcinoma tumorigenesis are the MAPK and PI3K/AKT pathways.
These genetic alterations open new avenues for rational principles of TC tumorigenesis and have been conventionally considered as biomarkers for early diagnosis and prognosis or therapeutic targets for patients with TC. However, increasing studies on the aberrant expression of lncRNA in thyroid neoplasias might contribute to deep elucidation of the genetic basis of TC and shed additional insights into TC diagnosis, prognosis, and therapy.
Functional Mechanism of lncRNA
LncRNAs are defined as transcripts without coding potential longer than 200 base pairs in length. Like PCGs, lncRNA genomic regions are characterized by the transcriptional start site (TSS) of H3K4 trimethylated abundantly. LncRNA transcripts are composed of several exons that are spliced through canonical mechanisms into a mature transcript and frequently include 5′ caps and 3′ poly(A) tails. However, lncRNAs commonly have fewer exons and are expressed at very weaker levels generally compared with PCGs. Similar to PCGs, lncRNAs are transcribed by RNA polymerase II from independent promoters, but unlike mRNAs, lncRNAs preferentially localize in the nucleus (59, 60), which exert diverse functions, such as regulation of gene expression in cis or in trans, regulation of splicing, and nucleation of subnuclear domain (61, 62). LncRNAs exert their regulatory effects in regulating signaling transduction and cell fate determination through diverse molecular ways, including interaction with DNA, RNA, and proteins, as well as encoding small peptides (6, 63). Firstly, binding to DNA enables lncRNAs to alter chromatin structure and be involved in the modulation of epigenetic modifications, thereby affecting the expression of target genes. Secondly, lncRNAs function as a molecular sponge to bind to mRNAs or miRNAs, resulting in regulating the stability and translation efficiency of mRNAs or the binding of miRNAs with their own targets.
MiRNA Sponging
LncRNA–miRNA interaction displays a common molecular mechanism involved in regulating gene expression and signaling transduction through complementary base pairing (Figure 3A). It has been well accepted that miRNAs interact with mRNAs to mediate mRNA turnover, thereby regulating bound mRNA expression (64–66). LncRNAs can interact with both miRNAs and mRNAs, leading to RNA expression regulation being more complicated. In general, lncRNA–mRNA physical interaction affects the degradation and intracellular localization of the target mRNAs, whereas lncRNA–miRNA binding suppresses the competitive interaction of miRNAs binding to their own targets. To date, there are more than 16,000 lncRNAs in the human genome, and some of them were found to exert their functions through acting as miRNA sponge, therefore also known as competitive endogenous RNA (ceRNA) (67–69). LncRNA–miRNA interaction results in the re-expression of target genes inhibited by miRNA. For example, Lei et al. revealed that TUG1 promotes the development of TC cells via acting as a ceRNA to sponge miR-145 (70). It is worthy to note that multiple miRNA species might simultaneously bind to a single lncRNA, indicating that the interaction of lncRNAs with other RNA molecules could be more effective and multifunctional (71, 72).
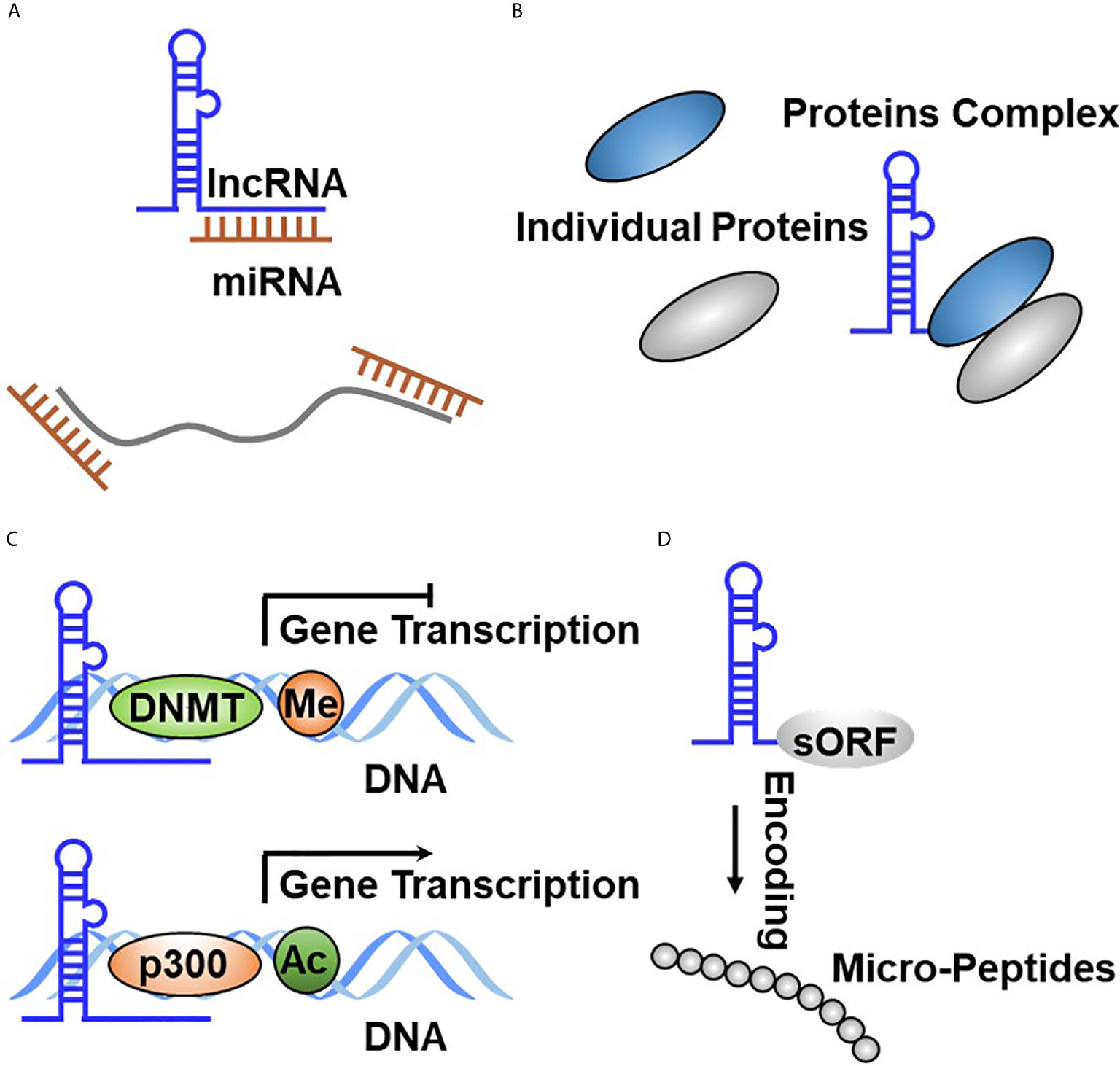
Figure 3 Functional principles of lncRNA in TC. (A) LncRNA directly binds to miRNA through complementary base pairing. (B) LncRNA binds to protein to form the RNA–protein complex. (C) LncRNA regulates gene expression at the transcriptional through binding to DNA through complementary base pairing. (D) LncRNA encodes the small peptide.
Protein Binding
The binding of lncRNAs with proteins plays crucial roles in affecting protein stability or subcellular localization, as well as protein complex formation or the release of proteins from their own bound partners, resulting in effecting their biological function. Apart from miRNAs, many lncRNAs have been reported to bind to proteins, thereby regulating their turnover. These binding proteins primarily cover RNA-binding proteins (RBPs), which are routinely accepted as interacting with mRNA (73, 74). Due to the existence of remarkable similarities between lncRNAs and mRNAs, several RBPs have been reported to orchestrate lncRNA functional roles by physical interaction. The binding between lncRNAs and proteins affects protein intracellular localization or degradation (Figure 3B). For example, Yuan et al. reported that SLC26A4-AS1 simultaneously binds to DDX5 and the E3 ligase TRIM25, thereby accelerating DDX5 degradation through the ubiquitin–proteasome pathway, resulting in the inhibition of TC cell migration and metastasis (75). Meng et al. defined that LINC00673 physically interacts with EZH2 and DNMT1, which promotes TC progression by inhibiting p53 expression (76). Gou et al. reported that lncRNA AB074169 binds to KHSRP, which decreases KHSRP expression and promotes TC cell proliferation and migration (77).
DNA Binding and Transcriptional Regulation
Nuclear lncRNAs are diffusely accepted to bind with DNA, including the noncoding or protein-coding region through complementary base pairing. This RNA–DNA interaction resulted in lncRNAs regulating target gene expression in cis (lncRNA gene sequence and target gene sequence are in the same chromosome) or in trans (lncRNA gene sequence and target gene sequence are in different chromosomes) (Figure 3C). A fully studied DNA-interacting lncRNA is XIST, which is involved in regulating the inactivation of X chromosome during early embryonic development (15). To date, a growing body of evidence has reported that lncRNA is documented to bind to DNA. Regrettably, only a subfraction has been functionally determined. FTNRC6C-AS1 was revealed to specifically bind with the promoter of STK4, which significantly promotes STK4 methylation and results in inhibiting STK4 expression in TC (78). LINC00313 was observed to recruit DNMT1 and DNMT3B to bind to the promoter region of ALX4 and significantly promoted methylate ALX4 promoter, resulting in repressing ALX4 expression (79). Pellecchia et al. characterized that PAR5 binds to the promoter of E-cadherin, which relieved negative regulation of E-cadherin by EZH2 (80). LINC00511 was reported to be overexpressed in TC tissues and promoted radiotherapy by binding with TAF1, which activated JAK2 expression at the transcriptional level (81). Xu et al. identified that CCAT1 specifically binds to the promoter of and promotes ASH1L expression in APC and boosts the growth and migration of APC cells (82).
Encoding Small Peptides
LncRNAs are defined as long transcripts without coding potential. Interestingly, shreds of evidence represented that diverse lncRNAs contain small open reading frames (sORFs) indeed and execute their functions by coding functional peptides (10, 83, 84) (Figure 3D). These novel findings broaden the complexity of lncRNA function. The first lncRNA that was identified to have coding potential is ENOD40, which is derived from plants and was found to encode two small peptides that interact with sucrose synthase (85). Subsequently, Rohrig et al. further revealed that the peptide encoded by ENOD40 binds sucrose synthase and elevates the sucrose cleavage activity of sucrose synthase (86). Currently, no lncRNAs that translate into small peptides were identified in TC tumorigenesis and development. The lack of related research may be attributed to several causes. Firstly, the small peptides encoded by lncRNAs are too insecure to monitor. Secondly, a fair portion of these small peptides has no biological function. Therefore, a better understanding is needed to fully recognize the biological function of small peptides encoded by lncRNA in TC initiation and development.
The Biological Function oF lncRNAs in TC Hallmarks
Different types of cancer share similar hallmarks (87, 88). The crucial roles of lncRNAs in TC initiation and development are evident by the fact that lncRNAs are closely related to TC malignant phenotypes, including sustaining uncontrolled cell growth, having resistance to cellular apoptosis, enhancing invasive ability, and acquiring cancer stem cell-like features. TC-associated lncRNAs facilitate or suppress these malignant behaviors by interacting with their binding macromolecules, indicating rational therapeutic treatment for the therapy of TC.
Maintaining Proliferative Signaling
Tumor cell uncontrolled proliferation is activated by sustaining proliferative signaling (87, 88). Normal cells restrict cell growth to precisely regulate an appropriate architecture of organisms, whereas transforming cells lead to tumorigenesis by constitutively activating RTK signaling or bypassing cell cycle checkpoints. For example, excessive activation of RTKs has been documented to play essential roles in maintaining TC growth (Figure 2). Emerging evidence indicates that lncRNA plays a critical role in supporting TC cell uncontrolled proliferation. For example, Chen et al. reported that lncRNA DGCR5 functions as a novel tumor suppressor in PTC via sponging miR-2861 to inhibit PTC cell proliferation and invasion (89). Chen et al. provided evidence that lncRNA GAS8-AS1 represses PTC cell proliferation by the miR-135b-5p/CCND2 signaling axis (90). Pan et al. further proved that GAS8-AS1 holds potential as a novel applicable diagnostic and therapeutic target (91) (Figure 4A). Ding et al. proved that SNHG12 promotes cellular proliferation and metastasis of PTC cells through activating the Wnt/β-catenin signaling (92). Duan et al. identified that SNHG3 acts as a new tumor suppressor and attenuates PTC cellular proliferation and metastasis through the AKT/mTOR/ERK signaling pathway (93). Interestingly, another study defined that SNHG3 exerts oncogenic effects in PTC through the miR-214-3p/PSMD10 signaling axis (94), suggesting that the biological function of SNHG3 is controversial. SNHG15 was proved to serve as a ceRNA to sponging miR-200a-3p to activate YAP1–Hippo signaling (95). He et al. identified that NORAD has an oncogenic role to promote PTC tumorigenesis by facilitating cellular proliferation and invasion by sponging and inhibiting miR-202-5p expression (96). Le et al. characterized that WT1-AS promotes PTC cell growth by sponging miR-203 (97). Li et al. identified that SOX2 directly binds to the LINC01510 promoter and promotes its expression at the transcriptional level. LINC01510 executes its function as a ceRNA by specifically binding miR-335, leading to inhibiting miR-335 expressions and accelerating PTC cell growth and migration (98). Liang et al. proved that HCP5 was highly expressed in FTC and promoted the proliferation, invasive ability, and angiogenic capability of FTC cells by acting as a ceRNA sponge for miR-22-3p, miR-186-5p, and miR-216a-5p, which promotes ST6GAL2 (99). Liu et al. identified that lncRNA BANCR promotes TC cell proliferation through activating thyroidal thyrotropin (TSH) receptors and downstream oncogenic signaling (100). Liu et al. recognized that XIST promotes TC cell growth and tumor growth in vivo through the miR-34a–MET–PI3K–AKT signaling axis (101). Liu et al. verified that lncRNA UCA1 exerts oncogenic activity by regulating the miR-204/IGFBP5 axis in PTC (102). Meng et al. elucidated that LINC00673 cells dramatically increase the proliferative rate of PTC cells (76). Xia et al. reported that LINC00673 potentiates PTC cell migration and invasion through KLF2 expression (103). Pang et al. defined that targeting of DUXAP8 induces cell apoptosis and attenuates cell proliferation (104). Li et al. reported that TPTE2P1 is required for PTC cell proliferation by sponging and inhibiting miR-520c-3p expression (105). Zhang et al. characterized that BISPR promotes PTC cell proliferation and invasion by depressing miR-21-5p expression (106). Another group further confirmed that NEAT1 increases the resistance effects of ATC cells to cisplatin treatment through sponging miR95p and regulating SPAG9 expression (107). UCA1 attenuates the lactiferous effects exhibited by cytotoxic CD8+ T cells on ATC cells via the miR-148a/PD-L1 signaling pathway (108).
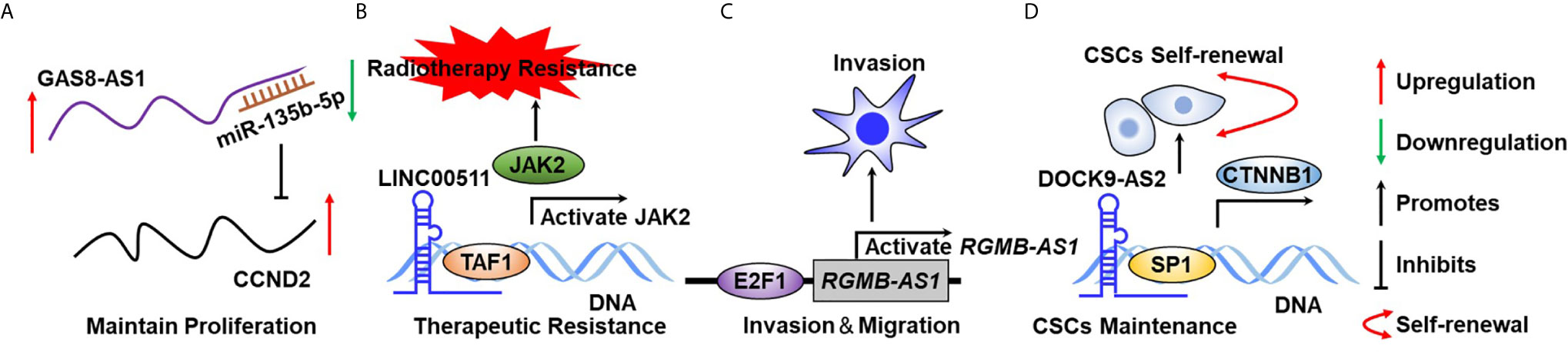
Figure 4 The biological function of lncRNA in TC hallmarks. (A) GAS8-AS1 sustains TC cell growth through binding to miR-135b-5p in a complementary base pairing manner. (B) LncRNA00511 activates JAK2 expression at the transcriptional level through binding to DNA. (C) E2F1 activates RGMB-AS1 expression at the transcriptional level to maintain TC cell migration. (D) DOCK9-AS2 binds to SP1 by serving as a scaffold protein to sustain CSC self-renewal. .
Therapeutic Resistance
To date, the therapeutic strategies routinely applied for TC clinical therapy are surgery, radiotherapy, and chemotherapy (109). However, TC cells can gradually develop resistance to therapeutics, particularly chemotherapy, during clinical treatment which can diffusely be attributed to but not limited to the following: 1) considerably enhanced certain transport protein expression that reduces the intracellular concentration of therapeutic drugs, resulting in decreasing chemotherapy efficacy; 2) genetic or epigenetic alterations that enable TC cells to become resistant to cellular death; and 3) augmentation in DNA repair ability (110, 111). Consequently, resistance to chemotherapy is still a major challenge for the clinical therapy of patients with TC because it frequently causes therapeutic failure. Overall, the underlying principles of TC patients resistant to chemotherapeutics remain to be functionally elucidated.
Recently, lncRNAs have emerged as new regulators in chemotherapeutic resistance in TC clinical treatment and have achieved great attention in the field of TC research. Many studies on lncRNA and resistance to therapeutic drugs have been documented over the last decade. For example, Liu et al. identified that MEG3 is weakly expressed in TC tumor tissues, and validated that MEG3 acts as a ceRNA sponge for miR-182, resulting in enhancement of radiotherapy effects (112). Song et al. characterized that GAS5 represses TC cell growth and resistance to doxorubicin treatment in ATC through inhibiting miR-96 expression (113). Another group further confirmed the tumor-suppressor roles of GAS5 in PTC progression (114). Xiang et al. suggested that SLC6A9 was considerably weakly expressed in 131I-resistant TC cell lines and overexpression of SLC6A9-5:2 dramatically sensitizes TC cells to 131I (115). Chen et al. identified that LINC00511 was markedly overexpressed in TC and silencing LINC00511 promoted the radiosensitivity of TC cells by binding to TAF1 and further promoted JAK2 expression (81) (Figure 4B). Xia et al. proved that CCND2-AS1 was significantly highly expressed in PTC and targeting CCND2-AS1 suppressed cell proliferation, migration, and invasion (116). Esposito et al. proved that silencing COMET significantly potentiated the sensitivity of TC cells to vemurafenib, which is an ordinary inhibitor of mutated B-raf (117).
Activating Migration and Invasion
Tumor metastasis is the real cause of the vast majority of cancer-related deaths (118–120). In brief, the nature of metastasis of TC shares remarkable similarities with other cancer types, whereas epithelial–mesenchymal transition program and increased angiogenesis capability have been considered as critical incidents driving TC metastasis. Many lncRNAs are well established to implicate in the regulation of TC cell invasion and migration, leading to affect TC metastasis. For example, Chen et al. reported that lncRNA TTTY10 enhanced PTC cell migration ability and was defined as novel potential prognostic biomarkers for predicting recurrence of PTC patients (121). Chen et al. revealed that lncRNA ZFAS1 inhibits TC cell proliferation and invasion via sponging miR3023p and decreasing cyclin D1 (122). Cui et al. proved that lncRNA-ATB, transcriptionally activated by transforming growth factor beta1 (TGF-β1), was involved in facilitating PTC cell migration and invasion (123). Shen et al. characterized that PROX1-AS1 was overexpressed in PTC and played an important role in PTC proliferation, invasion, and migration (124). NEAT1 was found to be overexpressed in both ATC tissues and cells and was induced in the hypoxia niche. NEAT1 depletion inhibits ATC cell invasion and glycolysis by exerting as a ceRNA sponge of miR-206 and miR-599 under hypoxia (125). HOTAIR was found to promote PTC cell proliferation and invasion via implicating the miR-488-5p/NUP205 signaling axis (21, 126). RGMB-AS1 is transcriptionally promoted by E2F1 and activated PTC cell proliferation and invasion (127) (Figure 4C). CCAT1 has been reported to promote PTC cell proliferation and migration through inhibiting miR-143 expression (128).
Sustaining Stem Cell-Like Characteristics
Cancer stem cells (CSCs) are a small subpopulation of cells that reside in tumors, which are well investigated being accounted for tumor initiation, therapeutic resistance, and tumor relapse (129–131). Emerging evidence suggested that Wnt/β-catenin, JAK/STAT, and Hippo/YAP have emerged as crucial players in affecting the stemness of TC CSCs (129, 132). A growing body of evidence validated that lncRNAs participate in CSC formation and maintenance in human cancers, including TC (133, 134). In the past several decades, the biological function of protein-coding genes in TC tumorigenesis and CSC self-renewal maintenance was well established, whereas the roles of lncRNAs have not been fully elucidated. Several pieces of evidence have demonstrated that lncRNA plays a crucial role in sustaining TC CSC self-renewal. For example, Dai et al. proved that lncRNA DOCK9-AS2 interacted with SP1 and activated CTNNB1 expression at the transcriptional level promoting PTC CSC maintenance (135) (Figure 4D). LncRNA-H19 accelerates cancer stem-like properties in PTC by activating estrogen receptor β expression (136). PTCSC3 was found to play critical roles in inhibiting PTC stem cell self-renewal (137). In addition, another group further validated that PTCSC3 suppresses PTC cell proliferation and migration by sponging miR-574-5p and represses β-catenin signaling (138).
Clinical Relevance of lncRNA in TC
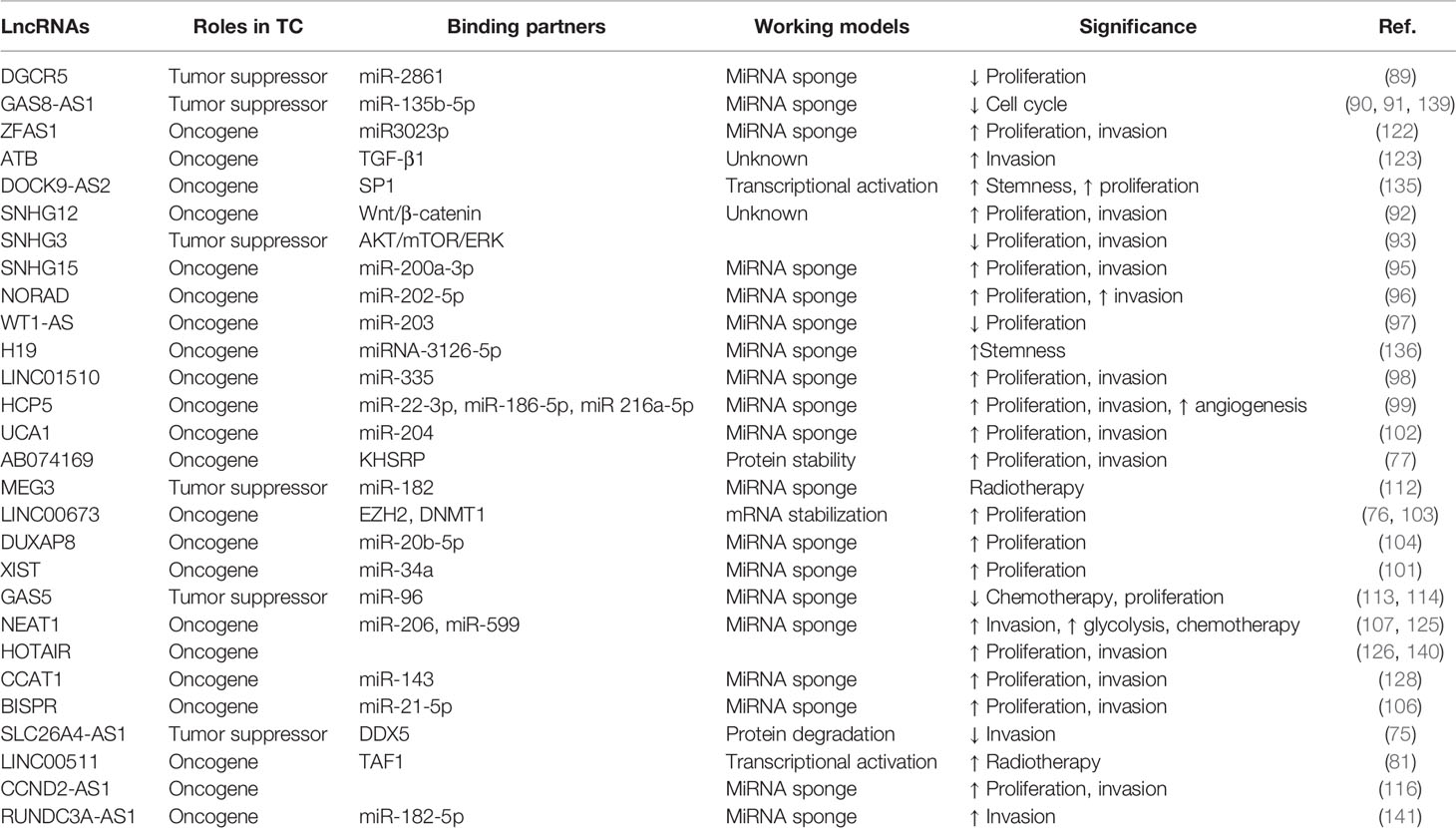
Several lncRNAs were identified to be dysregulated in TC and lncRNAs have been proved to hold the potential to function as biomarkers in TC diagnosis and prognosis. Table 1 summarizes that lncRNAs are dysregulated in TC and have oncogenic and tumor-suppressive roles in TC tumorigenesis and development, given the various regulatory molecular mechanisms and the diverse downstream signaling cascades affected. It is not surprising that several lncRNAs, including many of those emphasized above, have been observed through in vivo experiments to be essential players in TC tumorigenesis and progression and display attractive targeted therapeutics (Table 1). Given the poor conservation in different species, most of the lncRNA-related studies relied on regulating lncRNA expression in xenograft tumors derived from human cancer cell lines in mice. Similar to miRNAs, lncRNAs have been documented to execute their function either as tumor suppressors or oncogenes or display cellular context-dependent functions (Table 1). WT1-AS was inversely associated with the overall survival of patients with TC (97). Overexpression of lncRNA HOTAIR is correlated with worse clinical outcomes in TC (142). Pan et al. defined lncRNA GAS8-AS1 and LPAR4 holds great potential as diagnostic and therapeutic targets (91). Notably, several clinical and analytical factors could influence the final results of the sequencing results (143).
LncRNA as Cancer Biomarkers
Currently, TC diagnosis mainly depends on ultrasonography imaging. In addition, very limited therapeutic alternatives have been applied to TC-related deaths for lacking prognostic biomarkers or therapeutic targets. Thus, therapeutic targets and novel biomarkers are urgently required for developing the clinical therapies of TC. LncRNA can be easily detected from the serum and urine, and many lncRNAs have been reported to secrete from tumor cells into the circulatory system (144). Recent reports have documented that many lncRNAs are stable enough to be detected in the serum or urine of patients with cancer (53). Therefore, examination of the existence of circulating lncRNAs in serum or urine can be employed as diagnostic biomarkers for thyroid cancer diagnosis and prognosis in a noninvasive manner. LncRNA is a useful diagnostic biomarker because dysregulated lncRNA expression is observed across diverse cancers including TC and can be easily detected. Ge et al. identified 795 differentially expressed lncRNAs using the Agilent lncRNA microarray chips approach (145). Teng et al. characterized that eight genes and two novel lncRNAs were identified to correlate with the aggressive and disease-free survival of patients with PTC (146). Song et al. identified 1,878 abnormally expressed lncRNA, of which 1,449 were downregulated and 429 were upregulated, by performing microarray analysis in 86 PTC samples and adjacent noncancerous thyroid tissues (147). Additionally, Yang et al. defined that 751 mRNAs and 675 lncRNAs were dysregulated in the three PTC tissues compared with normal noncancerous thyroid tissues by performing lncRNA microarray analysis (148).
Diagnostic and Therapeutic Potentials of lncRNA in TC
Many inhibitors have been reported to induce cancer regression in a different type of cancer, yet only a small portion of them have been applied in clinical trials (149–151). Recently, the characterization and application of antisense oligonucleotide (ASO) opens a new avenue for the development of cancer therapeutics, and many of them have been implemented in different clinical trials (152–154). For instance, a phase I study of ASO specifically targeting vascular endothelial growth factor dramatically inhibits TC growth (155). However, no evidence has shown that ASO directly targets lncRNAs to display inhibitory roles in repressing TC, and no ASO targets to lncRNAs have been entered into clinical trials in TC, suggesting that there is an urgent need to define novel ASO directly targeting crucial lncRNAs required for sustaining TC malignant behaviors and to translate them into clinical therapeutic for patients with TC. Prognostically, as mentioned before, many lncRNAs have been reported to be differentially expressed in the progression of TC. For instance, Wen et al. defined that a higher level of ABHD11-AS1 correlated with worse clinical outcomes for PTC patients (156). Pan et al. verified that GAS8-AS1 and LPAR4 hold potential as diagnostics biomarkers or therapeutic targets through characterizing an exome mutational spectrum of PTC patients in a Chinese population (91). Zhou et al. showed that the genetic mutation of GAS8-AS1 could be employed as a diagnosis biomarker for PTC screening (139). Thus, lncRNAs hold great potential to function as novel biomarkers for the early diagnosis and prognosis of patients with TC. Additionally, it is of importance to characterize more dysregulated lncRNAs in TC and translate them into useful biomarkers for diagnostic and prognostic potential of TC patients.
Conclusions and Perspectives
Numerous studies have demonstrated that lncRNAs can function as regulatory molecules involved in a variety of biological processes. This review highlights the differentially expressed lncRNAs in TC initiation and progression, showing the great potential of lncRNAs as a regulatory molecule, as well as its diagnostic, prognostic, and therapeutic potential in TC studies. LncRNAs have been reported to be involved in various complex functions and molecular mechanisms; however, only a small fraction of lncRNAs and their functions in TC initiation and development have been investigated. Significant improvement has been achieved in uncovering lncRNAs and in understanding their functional roles and molecular mechanism in TC tumorigenesis and development. However, several fundamental issues remain to be addressed so as to obtain a deeper understanding of the biological function and clinical significance of lncRNAs in TC tumorigenesis and development. The functions of the few known lncRNAs still need to be characterized in detail and additional molecular mechanisms of lncRNAs need to be defined. Since many lncRNAs exert diverse functions in affecting multiple cellular processes in many tissues, the elaborate functional principles of lncRNA biological functions require to be understood on a case-by-case basis. In addition, it is indispensable to define whether the sensitivity and reliability of lncRNAs are enough for their substantial clinical application as biomarkers. Whether lncRNAs have advantages compared with other biomarkers used for TC diagnosis needs to be further elucidated. We are optimistic that the application of the new sequencing strategies will decisively determine the functional mechanism of lncRNAs implicated in TC tumorigenesis and development, which will eventually expedite the clinical application of lncRNAs for their use in early diagnosis, clinical therapy, and prognosis evaluation. The vast majority of PTC patients can be managed effectively; however, mortality associated with advanced and iodine-refractory TC remains high. Meanwhile, ATC displays an extraordinary worse prognosis. Whether lncRNAs hold great potential in better understanding the pathogenesis or serve as biomarkers for these advanced TC early diagnosis and effective treatment still needs to be ulteriorly investigated. Epigenetic regulation of lncRNAs bears an exceedingly superb potential in mediating TC initiation and development and should focus on investigating not only through achieving a better understanding of TC but also acquiring therapeutic successes.
Overall, a growing body of evidence demonstrates that specifically targeting lncRNAs might be a novel strategy for the clinical therapy of patients with TC. However, whether lncRNAs could be applied in deeper understanding of the pathogenesis or if it can be potentially used for the biomarkers for TC patients’ early diagnosis and advanced research is needed to further characterize. Additionally, it is of significance to decipher the biological functions of supernumerary lncRNAs correlated with TC tumorigenesis and translate them into clinical biomarkers applied for TC early diagnosis and therapeutic target.
Author Contributions
QH wrote the manuscript. PL searched the literature and made the figures and tables. JZ was the major contributor in preparing and writing the manuscript. CL, DW, XC, SW, YL, and JW provided help in editing and writing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the Jinan Technology and Innovation Development Scheme (No. 202019024) and National Special Project Funding Project of the Key R&D Program of the Ministry of Science and Technology (No. 2019YFC0119205).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all the scientists for their excellent contributions reviewed here and apologize to colleagues whose scientific work was not reviewed due to space constraints.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2019. CA Cancer J Clin (2019) 69:7–34. doi: 10.3322/caac.21551
2. Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, et al. Cancer Treatment and Survivorship Statistics, 2019. CA Cancer J Clin (2019) 69:363–85. doi: 10.3322/caac.21565
3. Xia F, Zhang Z, Li X. Emerging Roles of Circular RNAs in Thyroid Cancer. Front Cell Dev Biol (2021) 9:636838. doi: 10.3389/fcell.2021.636838
4. Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, et al. ENCODE Data in the UCSC Genome Browser: Year 5 Update. Nucleic Acids Res (2013) 41:D56–63. doi: 10.1093/nar/gks1172
5. Meller VH, Joshi SS, Deshpande N. Modulation of Chromatin by Noncoding RNA. Annu Rev Genet (2015) 49:673–95. doi: 10.1146/annurev-genet-112414-055205
6. Rinn JL, Chang HY. Long Noncoding RNAs: Molecular Modalities to Organismal Functions. Annu Rev Biochem (2020) 89:283–308. doi: 10.1146/annurev-biochem-062917-012708
7. Slack FJ, Chinnaiyan AM. The Role of Non-Coding RNAs in Oncology. Cell (2019) 179:1033–55. doi: 10.1016/j.cell.2019.10.017
8. Zhan H, Tu S, Zhang F, Shao A, Lin J. MicroRNAs and Long Non-Coding RNAs in C-Met-Regulated Cancers. Front Cell Dev Biol (2020) 8:145. doi: 10.3389/fcell.2020.00145
9. Chen R, Lei S, Jiang T, She Y, Shi H. Regulation of Skeletal Muscle Atrophy in Cachexia by MicroRNAs and Long Non-Coding RNAs. Front Cell Dev Biol (2020) 8:577010. doi: 10.3389/fcell.2020.577010
10. Wang Y, Hou J, He D, Sun M, Zhang P, Yu Y, et al. The Emerging Function and Mechanism of ceRNAs in Cancer. Trends Genet (2016) 32:211–24. doi: 10.1016/j.tig.2016.02.001
11. Yuan L, Xu ZY, Ruan SM, Mo S, Qin JJ, Cheng XD. Long Non-Coding RNAs Towards Precision Medicine in Gastric Cancer: Early Diagnosis, Treatment, and Drug Resistance. Mol Cancer (2020) 19:96. doi: 10.1186/s12943-020-01219-0
12. Gil N, Ulitsky I. Regulation of Gene Expression by Cis-Acting Long Non-Coding RNAs. Nat Rev Genet (2020) 21:102–17. doi: 10.1038/s41576-019-0184-5
13. Yao RW, Wang Y, Chen LL. Cellular Functions of Long Noncoding RNAs. Nat Cell Biol (2019) 21:542–51. doi: 10.1038/s41556-019-0311-8
14. Brannan CI, Dees EC, Ingram RS, Tilghman SM. The Product of the H19 Gene may Function as an RNA. Mol Cell Biol (1990) 10:28–36. doi: 10.1128/MCB.10.1.28
15. Brown CJ, Lafreniere RG, Powers VE, Sebastio G, Ballabio A, Pettigrew AL, et al. Localization of the X Inactivation Centre on the Human X Chromosome in Xq13. Nature (1991) 349:82–4. doi: 10.1038/349082a0
16. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The Landscape of Long Noncoding RNAs in the Human Transcriptome. Nat Genet (2015) 47:199–208. doi: 10.1038/ng.3192
17. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell (2016) 29:452–63. doi: 10.1016/j.ccell.2016.03.010
18. Flynn RA, Chang HY. Long Noncoding RNAs in Cell-Fate Programming and Reprogramming. Cell Stem Cell (2014) 14:752–61. doi: 10.1016/j.stem.2014.05.014
19. Batista PJ, Chang HY. Long Noncoding RNAs: Cellular Address Codes in Development and Disease. Cell (2013) 152:1298–307. doi: 10.1016/j.cell.2013.02.012
20. Shen F, Gan X, Zhong R, Feng J, Chen Z, Guo M, et al. Identifying Thyroid Carcinoma-Related Genes by Integrating GWAS and eQTL Data. Front Cell Dev Biol (2021) 9:645275. doi: 10.3389/fcell.2021.645275
21. Wu L, Shi Y, Liu B, Zhao M. Expression of lncRNA-HOTAIR in the Serum of Patients With Lymph Node Metastasis of Papillary Thyroid Carcinoma and its Impact. Oncol Lett (2020) 20:907–13. doi: 10.3892/ol.2020.11620
22. Titov SE, Kozorezova ES, Demenkov PS, Veryaskina YA, Kuznetsova IV, Vorobyev SL, et al. Preoperative Typing of Thyroid and Parathyroid Tumors With a Combined Molecular Classifier. Cancers (Basel) (2021) 13:237. doi: 10.3390/cancers13020237
24. Nikiforov YE, Nikiforova MN. Molecular Genetics and Diagnosis of Thyroid Cancer. Nat Rev Endocrinol (2011) 7:569–80. doi: 10.1038/nrendo.2011.142
25. Nikiforova MN, Nikiforov YE. Molecular Genetics of Thyroid Cancer: Implications for Diagnosis, Treatment and Prognosis. Expert Rev Mol Diagn (2008) 8:83–95. doi: 10.1586/14737159.8.1.83
26. Carcangiu ML, Zampi G, Rosai J. Poorly Differentiated (“Insular”) Thyroid Carcinoma. A Reinterpretation of Langhans’ “Wuchernde Struma”. Am J Surg Pathol (1984) 8:655–68. doi: 10.1097/00000478-198409000-00005
27. Capurso G, Festa S, Valente R, Piciucchi M, Panzuto F, Jensen RT, et al. Molecular Pathology and Genetics of Pancreatic Endocrine Tumours. J Mol Endocrinol (2012) 49:R37–50. doi: 10.1530/JME-12-0069
28. Kondo T, Ezzat S, Asa SL. Pathogenetic Mechanisms in Thyroid Follicular-Cell Neoplasia. Nat Rev Cancer (2006) 6:292–306. doi: 10.1038/nrc1836
29. LiVolsi VA, Asa SL. The Demise of Follicular Carcinoma of the Thyroid Gland. Thyroid (1994) 4:233–6. doi: 10.1089/thy.1994.4.233
30. Lloyd RV, Erickson LA, Casey MB, Lam KY, Lohse CM, Asa SL, et al. Observer Variation in the Diagnosis of Follicular Variant of Papillary Thyroid Carcinoma. Am J Surg Pathol (2004) 28:1336–40. doi: 10.1097/01.pas.0000135519.34847.f6
31. Williams D. Cancer After Nuclear Fallout: Lessons From the Chernobyl Accident. Nat Rev Cancer (2002) 2:543–9. doi: 10.1038/nrc845
32. Lubitz CC, Economopoulos KP, Pawlak AC, Lynch K, Dias-Santagata D, Faquin WC, et al. Hobnail Variant of Papillary Thyroid Carcinoma: An Institutional Case Series and Molecular Profile. Thyroid (2014) 24:958–65. doi: 10.1089/thy.2013.0573
33. Molinaro E, Romei C, Biagini A, Sabini E, Agate L, Mazzeo S, et al. Anaplastic Thyroid Carcinoma: From Clinicopathology to Genetics and Advanced Therapies. Nat Rev Endocrinol (2017) 13:644–60. doi: 10.1038/nrendo.2017.76
34. Yang J, Barletta JA. Anaplastic Thyroid Carcinoma. Semin Diagn Pathol (2020) 37:248–56. doi: 10.1053/j.semdp.2020.06.005
35. De Luca R, Davis PJ, Lin HY, Gionfra F, Percario ZA, Affabris E, et al. Thyroid Hormones Interaction With Immune Response, Inflammation and Non-Thyroidal Illness Syndrome. Front Cell Dev Biol (2020) 8:614030. doi: 10.3389/fcell.2020.614030
36. Pallante P, Battista S, Pierantoni GM, Fusco A. Deregulation of microRNA Expression in Thyroid Neoplasias. Nat Rev Endocrinol (2014) 10:88–101. doi: 10.1038/nrendo.2013.223
37. Lupoli G, Vitale G, Caraglia M, Fittipaldi MR, Abbruzzese A, Tagliaferri P, et al. Familial Papillary Thyroid Microcarcinoma: A New Clinical Entity. Lancet (1999) 353:637–9. doi: 10.1016/S0140-6736(98)08004-0
38. Lindor NM, Greene MH. The Concise Handbook of Family Cancer Syndromes. Mayo Familial Cancer Program. J Natl Cancer Inst (1998) 90:1039–71. doi: 10.1093/jnci/90.14.1039
39. Pierotti MA, Bongarzone I, Borrello MG, Mariani C, Miranda C, Sozzi G, et al. Rearrangements of TRK Proto-Oncogene in Papillary Thyroid Carcinomas. J Endocrinol Invest (1995) 18:130–3. doi: 10.1007/BF03349721
40. Xing M. BRAF Mutation in Thyroid Cancer. Endocr Relat Cancer (2005) 12:245–62. doi: 10.1677/erc.1.0978
41. Frattini M, Ferrario C, Bressan P, Balestra D, De Cecco L, Mondellini P, et al. Alternative Mutations of BRAF, RET and NTRK1 Are Associated With Similar But Distinct Gene Expression Patterns in Papillary Thyroid Cancer. Oncogene (2004) 23:7436–40. doi: 10.1038/sj.onc.1207980
42. Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High Prevalence of BRAF Mutations in Thyroid Cancer: Genetic Evidence for Constitutive Activation of the RET/PTC-RAS-BRAF Signaling Pathway in Papillary Thyroid Carcinoma. Cancer Res (2003) 63:1454–7.
43. Soares P, Trovisco V, Rocha AS, Lima J, Castro P, Preto A, et al. BRAF Mutations and RET/PTC Rearrangements Are Alternative Events in the Etiopathogenesis of PTC. Oncogene (2003) 22:4578–80. doi: 10.1038/sj.onc.1206706
44. Santoro M, Carlomagno F, Melillo RM, Fusco A. Dysfunction of the RET Receptor in Human Cancer. Cell Mol Life Sci (2004) 61:2954–64. doi: 10.1007/s00018-004-4276-8
45. Motoi N, Sakamoto A, Yamochi T, Horiuchi H, Motoi T, Machinami R. Role of Ras Mutation in the Progression of Thyroid Carcinoma of Follicular Epithelial Origin. Pathol Res Pract (2000) 196:1–7. doi: 10.1016/S0344-0338(00)80015-1
46. Volante M, Rapa I, Gandhi M, Bussolati G, Giachino D, Papotti M, et al. RAS Mutations Are the Predominant Molecular Alteration in Poorly Differentiated Thyroid Carcinomas and Bear Prognostic Impact. J Clin Endocrinol Metab (2009) 94:4735–41. doi: 10.1210/jc.2009-1233
47. Hancock JF. Ras Proteins: Different Signals From Different Locations. Nat Rev Mol Cell Biol (2003) 4:373–84. doi: 10.1038/nrm1105
48. Li S, Balmain A, Counter CM. A Model for RAS Mutation Patterns in Cancers: Finding the Sweet Spot. Nat Rev Cancer (2018) 18:767–77. doi: 10.1038/s41568-018-0076-6
49. Downward J. Targeting RAS Signalling Pathways in Cancer Therapy. Nat Rev Cancer (2003) 3:11–22. doi: 10.1038/nrc969
50. Garcia-Rostan G, Costa AM, Pereira-Castro I, Salvatore G, Hernandez R, Hermsem MJ, et al. Mutation of the PIK3CA Gene in Anaplastic Thyroid Cancer. Cancer Res (2005) 65:10199–207. doi: 10.1158/0008-5472.CAN-04-4259
51. Ricarte-Filho JC, Ryder M, Chitale DA, Rivera M, Heguy A, Ladanyi M, et al. Mutational Profile of Advanced Primary and Metastatic Radioactive Iodine-Refractory Thyroid Cancers Reveals Distinct Pathogenetic Roles for BRAF, PIK3CA, and AKT1. Cancer Res (2009) 69:4885–93. doi: 10.1158/0008-5472.CAN-09-0727
52. Donghi R, Longoni A, Pilotti S, Michieli P, Della Porta G, Pierotti MA. Gene P53 Mutations are Restricted to Poorly Differentiated and Undifferentiated Carcinomas of the Thyroid Gland. J Clin Invest (1993) 91:1753–60. doi: 10.1172/JCI116385
53. Fagin JA, Matsuo K, Karmakar A, Chen DL, Tang SH, Koeffler HP. High Prevalence of Mutations of the P53 Gene in Poorly Differentiated Human Thyroid Carcinomas. J Clin Invest (1993) 91:179–84. doi: 10.1172/JCI116168
54. Geng Q, Deng H, Fu J, Cui F. SOX18 Exerts Tumor-Suppressive Functions in Papillary Thyroid Carcinoma Through Inhibition of Wnt/β-Catenin Signaling. Exp Cell Res (2020) 396:112249. doi: 10.1016/j.yexcr.2020.112249
55. Dai W, Jin X, Han L, Huang H, Ji Z, Xu X, et al. Exosomal lncRNA DOCK9-AS2 Derived From Cancer Stem Cell-Like Cells Activated Wnt/β-Catenin Pathway to Aggravate Stemness, Proliferation, Migration, and Invasion in Papillary Thyroid Carcinoma. Cell Death Dis (2020) 11:743. doi: 10.1038/s41419-020-02827-w
56. Bi W, Huang J, Nie C, Liu B, He G, Han J, et al. CircRNA circRNA_102171 Promotes Papillary Thyroid Cancer Progression Through Modulating CTNNBIP1-Dependent Activation of β-Catenin Pathway. J Exp Clin Cancer Res (2018) 37:275. doi: 10.1186/s13046-018-0936-7
57. Kong D, Li A, Liu Y, Cui Q, Wang K, Zhang D, et al. SIX1 Activates STAT3 Signaling to Promote the Proliferation of Thyroid Carcinoma Via EYA1. Front Oncol (2019) 9:1450. doi: 10.3389/fonc.2019.01450
58. Notarangelo T, Sisinni L, Trino S, Calice G, Simeon V, Landriscina M. IL6/STAT3 Axis Mediates Resistance to BRAF Inhibitors in Thyroid Carcinoma Cells. Cancer Lett (2018) 433:147–55. doi: 10.1016/j.canlet.2018.06.038
59. Goodall GJ, Wickramasinghe VO. RNA in Cancer. Nat Rev Cancer (2021) 21:22–36. doi: 10.1038/s41568-020-00306-0
60. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative Annotation of Human Large Intergenic Noncoding RNAs Reveals Global Properties and Specific Subclasses. Genes Dev (2011) 25:1915–27. doi: 10.1101/gad.17446611
61. Engreitz JM, Ollikainen N, Guttman M. Long Non-Coding RNAs: Spatial Amplifiers That Control Nuclear Structure and Gene Expression. Nat Rev Mol Cell Biol (2016) 17:756–70. doi: 10.1038/nrm.2016.126
62. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE V7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res (2012) 22:1775–89. doi: 10.1101/gr.132159.111
63. Huang Z, Zhou JK, Peng Y, He W, Huang C. The Role of Long Noncoding RNAs in Hepatocellular Carcinoma. Mol Cancer (2020) 19:77. doi: 10.1186/s12943-020-01188-4
64. Peng Y, Croce CM. The Role of MicroRNAs in Human Cancer. Signal Transduct Target Ther (2016) 1:15004. doi: 10.1038/sigtrans.2015.4
65. Zhou H, Ni WJ, Meng XM, Tang LQ. MicroRNAs as Regulators of Immune and Inflammatory Responses: Potential Therapeutic Targets in Diabetic Nephropathy. Front Cell Dev Biol (2020) 8:618536. doi: 10.3389/fcell.2020.618536
66. Dexheimer PJ, Cochella L. MicroRNAs: From Mechanism to Organism. Front Cell Dev Biol (2020) 8:409. doi: 10.3389/fcell.2020.00409
67. Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, et al. Endogenous miRNA Sponge lincRNA-RoR Regulates Oct4, Nanog, and Sox2 in Human Embryonic Stem Cell Self-Renewal. Dev Cell (2013) 25:69–80. doi: 10.1016/j.devcel.2013.03.002
68. Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-P21 Suppresses Target mRNA Translation. Mol Cell (2012) 47:648–55. doi: 10.1016/j.molcel.2012.06.027
69. Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB Up-Regulates Long Non-Coding RNA, HULC Expression Through Interaction With microRNA-372 in Liver Cancer. Nucleic Acids Res (2010) 38:5366–83. doi: 10.1093/nar/gkq285
70. Lei H, Gao Y, Xu X. LncRNA TUG1 Influences Papillary Thyroid Cancer Cell Proliferation, Migration and EMT Formation Through Targeting miR-145. Acta Biochim Biophys Sin (Shanghai) (2017) 49:588–97. doi: 10.1093/abbs/gmx047
71. Chen S, Shen X. Long Noncoding RNAs: Functions and Mechanisms in Colon Cancer. Mol Cancer (2020) 19:167. doi: 10.1186/s12943-020-01287-2
72. Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res (2017) 77:3965–81. doi: 10.1158/0008-5472.CAN-16-2634
73. Gstaiger M, Aebersold R. Applying Mass Spectrometry-Based Proteomics to Genetics, Genomics and Network Biology. Nat Rev Genet (2009) 10:617–27. doi: 10.1038/nrg2633
74. Castello A, Horos R, Strein C, Fischer B, Eichelbaum K, Steinmetz LM, et al. System-Wide Identification of RNA-Binding Proteins by Interactome Capture. Nat Protoc (2013) 8:491–500. doi: 10.1038/nprot.2013.020
75. Yuan J, Song Y, Pan W, Li Y, Xu Y, Xie M, et al. LncRNA SLC26A4-AS1 Suppresses the MRN Complex-Mediated DNA Repair Signaling and Thyroid Cancer Metastasis by Destabilizing DDX5. Oncogene (2020) 39:6664–76. doi: 10.1038/s41388-020-01460-3
76. Meng XF, Zhao LY, Chu XF. LncRNA LINC00673 Inhibits P53 Expression by Interacting With EZH2 and DNMT1 in Papillary Thyroid Carcinoma. Eur Rev Med Pharmacol Sci (2019) 23:2075–83. doi: 10.26355/eurrev_201903_17249
77. Gou Q, Gao L, Nie X, Pu W, Zhu J, Wang Y, et al. Long Noncoding RNA AB074169 Inhibits Cell Proliferation Via Modulation of KHSRP-Mediated CDKN1a Expression in Papillary Thyroid Carcinoma. Cancer Res (2018) 78:4163–74. doi: 10.1158/0008-5472.CAN-17-3766
78. Yang LX, Wu J, Guo ML, Zhang Y, Ma SG. Suppression of Long Non-Coding RNA TNRC6C-AS1 Protects Against Thyroid Carcinoma Through DNA Demethylation of STK4 Via the Hippo Signalling Pathway. Cell Prolif (2019) 52:e12564. doi: 10.1111/cpr.12564
79. Zhao X, Hu X. Downregulated Long Noncoding RNA LINC00313 Inhibits the Epithelial-Mesenchymal Transition, Invasion, and Migration of Thyroid Cancer Cells Through Inhibiting the Methylation of ALX4. J Cell Physiol (2019) 234:20992–1004. doi: 10.1002/jcp.28703
80. Pellecchia S, Sepe R, Decaussin-Petrucci M, Ivan C, Shimizu M, Coppola C, et al. The Long Non-Coding RNA Prader Willi/Angelman Region RNA5 (PAR5) Is Downregulated in Anaplastic Thyroid Carcinomas Where It Acts as a Tumor Suppressor by Reducing EZH2 Activity. Cancers (Basel) (2020) 12:235. doi: 10.3390/cancers12010235
81. Chen Y, Bao C, Zhang X, Lin X, Fu Y. Knockdown of LINC00511 Promotes Radiosensitivity of Thyroid Carcinoma Cells Via Suppressing JAK2/STAT3 Signaling Pathway. Cancer Biol Ther (2019) 20:1249–57. doi: 10.1080/15384047.2019.1617569
82. Xu B, Qin T, Yu J, Giordano TJ, Sartor MA, Koenig RJ. Novel Role of ASH1L Histone Methyltransferase in Anaplastic Thyroid Carcinoma. J Biol Chem (2020) 295:8834–45. doi: 10.1074/jbc.RA120.013530
83. Chew GL, Pauli A, Rinn JL, Regev A, Schier AF, Valen E. Ribosome Profiling Reveals Resemblance Between Long Non-Coding RNAs and 5’ Leaders of Coding RNAs. Development (2013) 140:2828–34. doi: 10.1242/dev.098343
84. Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, et al. A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell (2015) 160:595–606. doi: 10.1016/j.cell.2015.01.009
85. Rohrig H, Schmidt J, Miklashevichs E, Schell J, John M. Soybean ENOD40 Encodes Two Peptides That Bind to Sucrose Synthase. Proc Natl Acad Sci USA (2002) 99:1915–20. doi: 10.1073/pnas.022664799
86. Rohrig H, John M, Schmidt J. Modification of Soybean Sucrose Synthase by S-Thiolation With ENOD40 Peptide A. Biochem Biophys Res Commun (2004) 325:864–70. doi: 10.1016/j.bbrc.2004.10.100
87. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
88. Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell (2000) 100:57–70. doi: 10.1016/S0092-8674(00)81683-9
89. Chen F, Yin S, Zhu J, Liu P, Yang C, Feng Z, et al. lncRNA DGCR5 Acts as a Tumor Suppressor in Papillary Thyroid Carcinoma Via Sequestering miR-2861. Exp Ther Med (2019) 17:895–900. doi: 10.3892/etm.2018.7012
90. Chen N, Yin D, Lun B, Guo X. LncRNA GAS8-AS1 Suppresses Papillary Thyroid Carcinoma Cell Growth Through the miR-135b-5p/CCND2 Axis. Biosci Rep (2019) 39:BSR20181440. doi: 10.1042/BSR20181440
91. Pan W, Zhou L, Ge M, Zhang B, Yang X, Xiong X, et al. Whole Exome Sequencing Identifies lncRNA GAS8-AS1 and LPAR4 as Novel Papillary Thyroid Carcinoma Driver Alternations. Hum Mol Genet (2016) 25:1875–84. doi: 10.1093/hmg/ddw056
92. Ding S, Qu W, Jiao Y, Zhang J, Zhang C, Dang S. LncRNA SNHG12 Promotes the Proliferation and Metastasis of Papillary Thyroid Carcinoma Cells Through Regulating Wnt/Beta-Catenin Signaling Pathway. Cancer Biomark (2018) 22:217–26. doi: 10.3233/CBM-170777
93. Duan Y, Wang Z, Xu L, Sun L, Song H, Yin H, et al. lncRNA SNHG3 Acts as a Novel Tumor Suppressor and Regulates Tumor Proliferation and Metastasis Via AKT/mTOR/ERK Pathway in Papillary Thyroid Carcinoma. J Cancer (2020) 11:3492–501. doi: 10.7150/jca.42070
94. Sui G, Zhang B, Fei D, Wang H, Guo F, Luo Q. The lncRNA SNHG3 Accelerates Papillary Thyroid Carcinoma Progression Via the miR-214-3p/PSMD10 Axis. J Cell Physiol (2020) 235:6615–24. doi: 10.1002/jcp.29557
95. Wu DM, Wang S, Wen X, Han XR, Wang YJ, Shen M, et al. LncRNA SNHG15 Acts as a ceRNA to Regulate YAP1-Hippo Signaling Pathway by Sponging miR-200a-3p in Papillary Thyroid Carcinoma. Cell Death Dis (2018) 9:947. doi: 10.1038/s41419-018-0975-1
96. He H, Yang H, Liu D, Pei R. LncRNA NORAD Promotes Thyroid Carcinoma Progression Through Targeting miR-202-5p. Am J Transl Res (2019) 11:290–9.
97. Le F, Luo P, Ouyang Q, Zhong X. LncRNA WT1-AS Downregulates Survivin by Upregulating miR-203 in Papillary Thyroid Carcinoma. Cancer Manag Res (2020) 12:443–9. doi: 10.2147/CMAR.S232294
98. Li Q, Wang XJ, Jin JH. SOX2-Induced Upregulation of lncRNA LINC01510 Promotes Papillary Thyroid Carcinoma Progression by Modulating miR-335/SHH and Activating Hedgehog Pathway. Biochem Biophys Res Commun (2019) 520:277–83. doi: 10.1016/j.bbrc.2019.09.138
99. Liang L, Xu J, Wang M, Xu G, Zhang N, Wang G, et al. LncRNA HCP5 Promotes Follicular Thyroid Carcinoma Progression Via miRNAs Sponge. Cell Death Dis (2018) 9:372. doi: 10.1038/s41419-018-0382-7
100. Liu C, Lin Y, Xu J, Chu H, Hao S, Liu X, et al. Luteolin Suppresses Tumor Progression Through lncRNA BANCR and Its Downstream TSHR/CCND1 Signaling in Thyroid Carcinoma. Int J Clin Exp Pathol (2017) 10:9591–8.
101. Liu H, Deng H, Zhao Y, Li C, Liang Y. LncRNA XIST/miR-34a Axis Modulates the Cell Proliferation and Tumor Growth of Thyroid Cancer Through MET-PI3K-AKT Signaling. J Exp Clin Cancer Res (2018) 37:279. doi: 10.1186/s13046-018-0950-9
102. Liu H, Li R, Guan L, Jiang T. Knockdown of lncRNA UCA1 Inhibits Proliferation and Invasion of Papillary Thyroid Carcinoma Through Regulating miR-204/IGFBP5 Axis. Onco Targets Ther (2018) 11:7197–204. doi: 10.2147/OTT.S175467
103. Xia E, Bhandari A, Shen Y, Zhou X, Wang O. lncRNA LINC00673 Induces Proliferation, Metastasis and Epithelial-Mesenchymal Transition in Thyroid Carcinoma Via Kruppel-Like Factor 2. Int J Oncol (2018) 53:1927–38. doi: 10.3892/ijo.2018.4524
104. Pang R, Yang S. lncRNA DUXAP8 Inhibits Papillary Thyroid Carcinoma Cell Apoptosis Via Sponging the Mir20b5p/SOS1 Axis. Oncol Rep (2021) 45:64. doi: 10.3892/or.2021.8015
105. Peng L, Gao S, Bai F, An Y, Wan F. LncRNA TPTE2P1 Promotes the Proliferation of Thyroid Carcinoma by Inhibiting miR-520c-3p. Panminerva Med (2020) 62:69–71. doi: 10.23736/S0031-0808.19.03645-0
106. Zhang H, Cai Y, Zheng L, Zhang Z, Lin X, Jiang N. LncRNA BISPR Promotes the Progression of Thyroid Papillary Carcinoma by Regulating miR-21-5p. Int J Immunopathol Pharmacol (2018) 32:2058738418772652. doi: 10.1177/2058738418772652
107. Yan P, Su Z, Zhang Z, Gao T. LncRNA NEAT1 Enhances the Resistance of Anaplastic Thyroid Carcinoma Cells to Cisplatin by Sponging Mir95p and Regulating SPAG9 Expression. Int J Oncol (2019) 55:988–1002. doi: 10.3892/ijo.2019.4868
108. Wang X, Zhang Y, Zheng J, Yao C, Lu X. LncRNA UCA1 Attenuated the Killing Effect of Cytotoxic CD8 + T Cells on Anaplastic Thyroid Carcinoma Via miR-148a/PD-L1 Pathway. Cancer Immunol Immunother (2021) 70:2235–45. doi: 10.1007/s00262-020-02753-y
109. Ethun CG, Bilen MA, Jani AB, Maithel SK, Ogan K, Master VA. Frailty and Cancer: Implications for Oncology Surgery, Medical Oncology, and Radiation Oncology. CA Cancer J Clin (2017) 67:362–77. doi: 10.3322/caac.21406
110. Brown R, Curry E, Magnani L, Wilhelm-Benartzi CS, Borley J. Poised Epigenetic States and Acquired Drug Resistance in Cancer. Nat Rev Cancer (2014) 14:747–53. doi: 10.1038/nrc3819
111. Chen Z, Shi T, Zhang L, Zhu P, Deng M, Huang C, et al. Mammalian Drug Efflux Transporters of the ATP Binding Cassette (ABC) Family in Multidrug Resistance: A Review of the Past Decade. Cancer Lett (2016) 370:153–64. doi: 10.1016/j.canlet.2015.10.010
112. Liu Y, Yue P, Zhou T, Zhang F, Wang H, Chen X. LncRNA MEG3 Enhances (131)I Sensitivity in Thyroid Carcinoma Via Sponging miR-182. BioMed Pharmacother (2018) 105:1232–9. doi: 10.1016/j.biopha.2018.06.087
113. Song CQ, Wang M, Zhang SM, Ma XY. LncRNA GAS5 Inhibits Cell Proliferation and Resistance to Doxorubicin in Anaplastic Thyroid Carcinoma by Regulating miR-96. J Biol Regul Homeost Agents (2020) 34:1787–92. doi: 10.23812/20-420-L
114. Zhang XF, Ye Y, Zhao SJ. LncRNA Gas5 Acts as a ceRNA to Regulate PTEN Expression by Sponging miR-222-3p in Papillary Thyroid Carcinoma. Oncotarget (2018) 9:3519–30. doi: 10.18632/oncotarget.23336
115. Xiang C, Zhang ML, Zhao QZ, Xie QP, Yan HC, Yu X, et al. LncRNA-SLC6A9-5:2: A Potent Sensitizer in 131I-Resistant Papillary Thyroid Carcinoma With PARP-1 Induction. Oncotarget (2017) 8:22954–67. doi: 10.18632/oncotarget.14578
116. Xia E, Bhandari A, Shen Y, Zhou X, Sindan N, Xiang J, et al. LncRNA CCND2-AS1 Promotes Proliferation, Migration, and Invasion in Papillary Thyroid Carcinoma. Biochem Biophys Res Commun (2018) 496:628–32. doi: 10.1016/j.bbrc.2018.01.102
117. Esposito R, Esposito D, Pallante P, Fusco A, Ciccodicola A, Costa V. Oncogenic Properties of the Antisense lncRNA COMET in BRAF- and RET-Driven Papillary Thyroid Carcinomas. Cancer Res (2019) 79:2124–35. doi: 10.1158/0008-5472.CAN-18-2520
118. Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S, et al. YTHDF3 Induces the Translation of M(6)A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell (2020) 38:857–71.e7. doi: 10.1016/j.ccell.2020.10.004
119. Senbanjo LT, Chellaiah MA. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front Cell Dev Biol (2017) 5:18. doi: 10.3389/fcell.2017.00018
120. Tiedemann K, Hussein O, Komarova SV. Role of Altered Metabolic Microenvironment in Osteolytic Metastasis. Front Cell Dev Biol (2020) 8:435. doi: 10.3389/fcell.2020.00435
121. Chen F, Li Z, Deng C, Yan H. Integrated Analysis Identifying New lncRNA Markers Revealed in ceRNA Network for Tumor Recurrence in Papillary Thyroid Carcinoma and Build of Nomogram. J Cell Biochem (2019) 120:19673–83. doi: 10.1002/jcb.29273
122. Chen W, Zhai L, Liu H, Li Y, Zhang Q, Xu D, et al. Downregulation of lncRNA ZFAS1 Inhibits the Hallmarks of Thyroid Carcinoma Via the Regulation of Mir3023p on Cyclin D1. Mol Med Rep (2021) 23:1. doi: 10.3892/mmr.2020.11640
123. Cui M, Chang Y, Du W, Liu S, Qi J, Luo R, et al. Upregulation of lncRNA-ATB by Transforming Growth Factor Beta1 (TGF-Beta1) Promotes Migration and Invasion of Papillary Thyroid Carcinoma Cells. Med Sci Monit (2018) 24:5152–8. doi: 10.12659/MSM.909420
124. Shen Y, Xia E, Bhandari A, Wang X, Guo G. LncRNA PROX1-AS1 Promotes Proliferation, Invasion, and Migration in Papillary Thyroid Carcinoma. Biosci Rep (2018) 38:BSR20180862. doi: 10.1042/BSR20180862
125. Tan X, Wang P, Lou J, Zhao J. Knockdown of lncRNA NEAT1 Suppresses Hypoxia-Induced Migration, Invasion and Glycolysis in Anaplastic Thyroid Carcinoma Cells Through Regulation of miR-206 and miR-599. Cancer Cell Int (2020) 20:132. doi: 10.1186/s12935-020-01222-x
126. Xia F, Xia W, Yu X. LncRNA HOTAIR Influences the Growth, Migration, and Invasion of Papillary Thyroid Carcinoma Via Affection on the miR-488-5p/NUP205 Axis. Technol Cancer Res Treat (2020) 19:1533033820962125. doi: 10.1177/1533033820962125
127. Zhang Z, Li SY, Zhang LB. LncRNA RGMB-AS1 Is Activated by E2F1 and Promotes Cell Proliferation and Invasion in Papillary Thyroid Carcinoma. Eur Rev Med Pharmacol Sci (2018) 22:1979–86. doi: 10.26355/eurrev_201804_14725
128. Yang T, Zhai H, Yan R, Zhou Z, Gao L, Wang L. lncRNA CCAT1 Promotes Cell Proliferation, Migration, and Invasion by Down-Regulation of miR-143 in FTC-133 Thyroid Carcinoma Cell Line. Braz J Med Biol Res (2018) 51:e7046. doi: 10.1590/1414-431x20187046
129. Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, et al. Targeting Cancer Stem Cell Pathways for Cancer Therapy. Signal Transduct Target Ther (2020) 5:8. doi: 10.1038/s41392-020-0110-5
130. Klonisch T, Wiechec E, Hombach-Klonisch S, Ande SR, Wesselborg S, Schulze-Osthoff K, et al. Cancer Stem Cell Markers in Common Cancers - Therapeutic Implications. Trends Mol Med (2008) 14:450–60. doi: 10.1016/j.molmed.2008.08.003
131. Pattabiraman DR, Weinberg RA. Tackling the Cancer Stem Cells - What Challenges do They Pose? Nat Rev Drug Discov (2014) 13:497–512. doi: 10.1038/nrd4253
132. Lytle NK, Barber AG, Reya T. Stem Cell Fate in Cancer Growth, Progression and Therapy Resistance. Nat Rev Cancer (2018) 18:669–80. doi: 10.1038/s41568-018-0056-x
133. Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, et al. The Long Noncoding RNA lncTCF7 Promotes Self-Renewal of Human Liver Cancer Stem Cells Through Activation of Wnt Signaling. Cell Stem Cell (2015) 16:413–25. doi: 10.1016/j.stem.2015.03.003
134. Wu J, Zhu P, Lu T, Du Y, Wang Y, He L, et al. The Long Non-Coding RNA LncHDAC2 Drives the Self-Renewal of Liver Cancer Stem Cells Via Activation of Hedgehog Signaling. J Hepatol (2019) 70:918–29. doi: 10.1016/j.jhep.2018.12.015
135. Dai W, Jin X, Han L, Huang H, Ji Z, Xu X, et al. Exosomal lncRNA DOCK9-AS2 Derived From Cancer Stem Cell-Like Cells Activated Wnt/beta-Catenin Pathway to Aggravate Stemness, Proliferation, Migration, and Invasion in Papillary Thyroid Carcinoma. Cell Death Dis (2020) 11:743. doi: 10.1038/s41419-020-02827-w
136. Li M, Chai HF, Peng F, Meng YT, Zhang LZ, Zhang L, et al. Estrogen Receptor Beta Upregulated by lncRNA-H19 to Promote Cancer Stem-Like Properties in Papillary Thyroid Carcinoma. Cell Death Dis (2018) 9:1120. doi: 10.1038/s41419-018-1077-9
137. Wang X, Lu X, Geng Z, Yang G, Shi Y. LncRNA PTCSC3/miR-574-5p Governs Cell Proliferation and Migration of Papillary Thyroid Carcinoma Via Wnt/beta-Catenin Signaling. J Cell Biochem (2017) 118:4745–52. doi: 10.1002/jcb.26142
138. Jendrzejewski J, He H, Radomska HS, Li W, Tomsic J, Liyanarachchi S, et al. The Polymorphism Rs944289 Predisposes to Papillary Thyroid Carcinoma Through a Large Intergenic Noncoding RNA Gene of Tumor Suppressor Type. Proc Natl Acad Sci USA (2012) 109:8646–51. doi: 10.1073/pnas.1205654109
139. Zhou DL, Liu Q, Xu BH, Li Y, Su X, Ye ZL, et al. lncRNA GAS8-AS1 Genetic Alterations in Papillary Thyroid Carcinoma and Their Clinical Significance. Cancer Biomark (2020) 29:255–64. doi: 10.3233/CBM-191071
140. Zhu H, Lv Z, An C, Shi M, Pan W, Zhou L, et al. Onco-lncRNA HOTAIR and its Functional Genetic Variants in Papillary Thyroid Carcinoma. Sci Rep (2016) 6:31969. doi: 10.1038/srep31969
141. Ma D, Zhu Y, Zhang X, Zhang J, Chen W, Chen X, et al. Long Non-Coding RNA RUNDC3A-AS1 Promotes Lung Metastasis of Thyroid Cancer Via Targeting the miR-182-5p/ADAM9. Front Cell Dev Biol (2021) 9:650004. doi: 10.3389/fcell.2021.650004
142. Li HM, Yang H, Wen DY, Luo YH, Liang CY, Pan DH, et al. Overexpression of LncRNA HOTAIR Is Associated With Poor Prognosis in Thyroid Carcinoma: A Study Based on TCGA and GEO Data. Horm Metab Res (2017) 49:388–99. doi: 10.1055/s-0043-103346
143. Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: A Rising Star in Gastric Cancer. Cell Mol Life Sci (2020) 77:1661–80. doi: 10.1007/s00018-019-03345-5
144. Liu K, Gao L, Ma X, Huang JJ, Chen J, Zeng L, et al. Long Non-Coding RNAs Regulate Drug Resistance in Cancer. Mol Cancer (2020) 19:54. doi: 10.1186/s12943-020-01162-0
145. Ge MH, Jiang LH, Wen QL, Tan Z, Chen C, Zheng CM, et al. Preliminary Screening and Analysis of Metastasis-Related lncRNA and Co-Expressed Papillary Thyroid Carcinoma mRNA. Oncol Lett (2018) 16:3715–25. doi: 10.3892/ol.2018.9080
146. Teng H, Mao F, Liang J, Xue M, Wei W, Li X, et al. Transcriptomic Signature Associated With Carcinogenesis and Aggressiveness of Papillary Thyroid Carcinoma. Theranostics (2018) 8:4345–58. doi: 10.7150/thno.26862
147. Song B, Li R, Zuo Z, Tan J, Liu L, Ding D, et al. LncRNA ENST00000539653 Acts as an Oncogenic Factor Via MAPK Signalling in Papillary Thyroid Cancer. BMC Cancer (2019) 19:297. doi: 10.1186/s12885-019-5533-4
148. Yang M, Tian J, Guo X, Yang Y, Guan R, Qiu M, et al. Long Noncoding RNA Are Aberrantly Expressed in Human Papillary Thyroid Carcinoma. Oncol Lett (2016) 12:544–52. doi: 10.3892/ol.2016.4653
149. Liu K, Pu J, Nie Z, Shi Y, Jiang L, Wu Q, et al. Ivacaftor Inhibits Glioblastoma Stem Cell Maintenance and Tumor Progression. Front Cell Dev Biol (2021) 9:678209. doi: 10.3389/fcell.2021.678209
150. Shi Y, Guryanova OA, Zhou W, Liu C, Huang Z, Fang X, et al. Ibrutinib Inactivates BMX-STAT3 in Glioma Stem Cells to Impair Malignant Growth and Radioresistance. Sci Transl Med (2018) 10:eaah6816. doi: 10.1126/scitranslmed.aah6816
151. Li Y, Rogoff HA, Keates S, Gao Y, Murikipudi S, Mikule K, et al. Suppression of Cancer Relapse and Metastasis by Inhibiting Cancer Stemness. Proc Natl Acad Sci USA (2015) 112:1839–44. doi: 10.1073/pnas.1424171112
152. Reilley MJ, McCoon P, Cook C, Lyne P, Kurzrock R, Kim Y, et al. STAT3 Antisense Oligonucleotide AZD9150 in a Subset of Patients With Heavily Pretreated Lymphoma: Results of a Phase 1b Trial. J Immunother Cancer (2018) 6:119. doi: 10.1186/s40425-018-0436-5
153. Buller HR, Bethune C, Bhanot S, Gailani D, Monia BP, Raskob GE, et al. Factor XI Antisense Oligonucleotide for Prevention of Venous Thrombosis. N Engl J Med (2015) 372:232–40. doi: 10.1056/NEJMoa1405760
154. Miller T, Cudkowicz M, Shaw PJ, Andersen PM, Atassi N, Bucelli RC, et al. Phase 1-2 Trial of Antisense Oligonucleotide Tofersen for SOD1 ALS. N Engl J Med (2020) 383:109–19. doi: 10.1056/NEJMoa2003715
155. Levine AM, Tulpule A, Quinn DI, Gorospe G 3rd, Smith DL, Hornor L, et al. Phase I Study of Antisense Oligonucleotide Against Vascular Endothelial Growth Factor: Decrease in Plasma Vascular Endothelial Growth Factor With Potential Clinical Efficacy. J Clin Oncol (2006) 24:1712–9. doi: 10.1200/JCO.2005.03.4801
Keywords: long noncoding RNA, thyroid carcinoma, biomarker, therapeutic target, clinical significance
Citation: Zhu J, Liu C, Wang D, Cao X, Wang S, Liu Y, Wang J, Li P and He Q (2021) The Emerging Landscapes of Long Noncoding RNA in Thyroid Carcinoma: Biological Functions and Clinical Significance. Front. Oncol. 11:706011. doi: 10.3389/fonc.2021.706011
Received: 06 May 2021; Accepted: 22 July 2021;
Published: 10 August 2021.
Edited by:
Liping Jiang, Kunming Institute of Zoology, ChinaReviewed by:
Wang Xiang Peng, The First Affiliated Hospital of Kunming Medical University, ChinaZhao Limin, Nanchang University, China
Copyright © 2021 Zhu, Liu, Wang, Cao, Wang, Liu, Wang, Li and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peifeng Li, bGlwZWlmZW5nMDBAaG90bWFpbC5jb20=; Qingqing He, aGVxaW5ncWluZ0B5ZWFoLm5ldA==
 Jian Zhu1,2
Jian Zhu1,2 Changrui Liu
Changrui Liu Peifeng Li
Peifeng Li