- 1Department of Urology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2Department of Breast Surgery, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 3Department of Pathology, Affiliated Hospital of Qingdao University, Qingdao, China
- 4Department of Pathology, Qilu Hospital, Shandong University, Qingdao, China
- 5Clinical College, Qingdao University, Qingdao, China
Background: Bladder cancer is the 10th most common cancer and most common urothelial malignancy worldwide. Prognostic biomarkers for bladder cancer patients are required for individualized treatment. Monocarboxylate transporter 4 (MCT4), encoded by SLC16A3 gene, is a potential biomarker for bladder cancer because of its crucial role in the lactate efflux in the aerobic glycolysis process. We aimed to study the association between MCT4 expression and the overall survival (OS) of bladder cancer patients.
Methods: The published single-cell RNA sequencing data of 49,869 bladder cancer cells and 15,827 normal bladder mucosa cells and The Cancer Genome Atlas (TCGA) bladder cancer cohort data were used to explore the mRNA expression of SLC16A3 in bladder cancer. Eighty-nine consecutive bladder cancer patients who had undergone radical cystectomy were enrolled as a validation cohort. The expression of MCT4 proteins in bladder cancer specimens was detected using immunohistochemistry staining. The Kaplan–Meier survival analysis and Cox regression were performed to analyze the association between MCT4 protein expression and OS in bladder cancer patients.
Results: SLC16A3 mRNA was upregulated in bladder cancer cells. The upregulated genes in SLC16A3-positive epithelial cells were enriched in the glycolysis process pathway and monocarboxylic acid metabolic process pathway. Patients with high SLC16A3 mRNA expression showed significantly poor OS (p = 0.016). High MCT4 protein expression was also found to be an independent predictor for poor OS in bladder cancer patients (HR: 2.462; 95% CI: 1.202~5.042, p = 0.014). A nomogram was built based on the results of the multivariate Cox analysis.
Conclusion: Bladder cancer with high SLC16A3 mRNA expression has a poor OS. High MCT4 protein expression is an independent prognostic factor for bladder cancer patients who had undergone radical cystectomy.
Introduction
Bladder cancer is the 10th most common cancer and most common urothelial malignancy worldwide, with approximately 573,000 new cases and 213,000 deaths per year (1). Radical cystectomy is the first-line treatment for resectable muscle invasive bladder cancer (MIBC) and is also used as radical treatment for high-risk non-MIBC (NMIBC) (2). However, 26.9%–37.5% of patients who had undergone radical cystectomy showed tumor recurrence or metastasis after surgery (3–5). Prognostic biomarkers for bladder cancer patients are needed for more individualized surveillance and intervention after surgery.
In contrast to normal cells, which rely on oxidative phosphorylation to generate the energy needed for cellular processes, cancer cells rely on glycolysis for the energy needed for survival and proliferation (6). This phenomenon is termed aerobic glycolysis or the Warburg effect, which is common among human malignancies (6, 7). Aerobic glycolysis could help tumor cells generate ATPs quickly and acquire substrates for anabolism, which is critical for tumor cell proliferation (8). Additionally, the lactate generated from aerobic glycolysis can be transported into the tumor microenvironment by membrane monocarboxylate transporters, which can re-program the infiltrated immune cells and attenuate their anti-tumor immune response (9). The intensity of tumor aerobic glycolysis may be closely related to a high malignant potential and poor survival (10).
Monocarboxylate transporter 4 (MCT4) is a member of solute carrier family 16 encoded by SLC16A3 gene (11). It is widely expressed, particularly in tissues that rely on glycolysis for energy metabolism, such as tumor cells, immune cells, and astrocytes (12). In tumor cells, the efflux of lactate mediated by MCT4 is essential to maintain cytoplasmic pH (12, 13). The expression of MCT4 is correlated with the clinical outcome of several urologic cancers. Choi et al. (14) found that elevated MCT4 expression is associated with the incidence of castration-resistant prostate cancer and an earlier time to relapse. Fisel et al. (15) reported that the MCT4 mRNA is upregulated in renal cancer and that high MCT4 protein expression is correlated with poor overall survival (OS) in renal cancer. Previous studies have also shown that high MCT4 mRNA expression is associated with poor OS in bladder cancer patients (16). The expression of MCT4 protein was detected in several bladder cancer cell lines, and selective inhibition of MCT4 inhibited the viability of bladder cancer cells and reduced the tumor diameter of an orthotopic xenograft bladder cancer model (16). These studies highlighted the potential of MCT4 as a potential biomarker for bladder cancer.
In the present study, we first evaluated the expression of SLC16A3 mRNA in published single-cell RNA sequencing data of bladder cancer. Furthermore, the influence of SLC16A3 mRNA and MCT4 protein expression on the OS of bladder cancer patients was explored using the clinical data and MCT4 expression data of bladder cancer patients in our affiliations and The Cancer Genome Atlas (TCGA) database.
Materials and Methods
Acquisition of Publicly Available RNA Sequencing Data
The single-cell RNA sequencing data of eight bladder cancer samples and three normal bladder mucosa (NBM) samples were downloaded from the ENA database (https://www.ebi.ac.uk/ena/, accession ID: PRJNA662018) and Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/, accession ID: GSE145140) (17, 18). The transcriptome data and survival data of 402 urothelial bladder cancer patients were obtained from TCGA database (https://tcga-data.nci.nih.gov/tcga/).
Clustering and Cell-Type Annotation
Raw sequencing data were mapped to the human genome, and data cleaning and integration were performed using the Seurat package (19–21). Clustering analysis was performed using the “FindClusters” function of the Seurat package. Clusters were annotated to known cell types according to canonical cell-type-specific markers (22).
Differential Analysis of Single-Cell Sequencing Data
The proportion of each epithelial cell cluster in all cells was calculated using the “prop.table” function of R software. The statistical significance of alterations in epithelial cell proportions was analyzed using one-way ANOVA and displayed with a histogram. The differential expression of SLC16A3 between epithelial cells of NMIBC and MIBC, as well as in NBM epithelial cells, was displayed using violin plots and t-SNE plots. The differentially expressed genes (DEGs) between SLC16A3-positive epithelial cells and SLC16A3-negative epithelial cells were screened using the “FindMarkers” function of the Seurat package with the default parameters. The DEGs were then submitted to Metascape software (https://metascape.org) for further Gene Ontology (GO) enrichment analyses.
Patients and Samples of the Validation Cohort
From October 2010 to March 2015, 89 consecutive bladder cancer patients who had undergone radical cystectomy at Peking Union Medical College Hospital or Qingdao University Affiliated Hospital were enrolled. Formalin-fixed paraffin-embedded tumor samples were retrieved for further immunohistochemistry (IHC) analyses.
The inclusion criteria were as follows: 1) patients with urothelial cell carcinomas identified by pathologic examination and 2) patients who had undergone radical cystectomy for MIBC or high-risk NMIBC.
The exclusion criteria were as follows: 1) patients with other histological types of bladder tumors, such as squamous cell carcinoma or adenocarcinoma; 2) patients with insufficient tumor tissues for IHC analysis; and 3) patients with a history of cancers from other organs or systems.
Formalin-fixed paraffin-embedded tumor samples were retrieved for MCT4 IHC staining. The MCT4 expression of samples was scored according to the intensity of staining and ratio of positive cells (Supplementary Method).
Statistical Analysis
Categorical variables were described with numbers and percentages in all cohorts. Continuous variables fitting a normal distribution were described with the mean ± standard deviation (SD). The association of MCT4 expression with categorical variables was assessed using chi-squared tests. Differences in continuous variables between two groups were analyzed using Student’s t-test. The statistical significance of alterations in epithelial cell proportions was analyzed using one-way ANOVA. The Kaplan–Meier (KM) method and log-rank test were used to analyze the influence of MCT4 expression on OS. Univariate Cox regression model was used to estimate the prognostic significance of each clinical characteristic and MCT4 expression. Variables that showed p < 0.1 in the univariate analysis were included in the multivariate Cox regression analysis. Variables that showed p < 0.05 in the multivariate analysis were independent prognostic factors for survival. The “nomogramEx” package of R software was used to draw a nomogram based on the results of the multivariate Cox regression. All p-values were two-sided, and statistical significance was defined as a p < 0.05. Unless otherwise noted, statistical analyses were performed using SPSS software (version 24.0).
Results
Clustering and Annotation of Single-Cell RNA Sequencing Data
After preprocessing, 65,723 filtered cells were used for bioinformatics analysis, including 49,869 bladder cancer cells and 15,827 cells from the NBM (Figure 1A). Clustering analysis identified 22 cell clusters (Figure 1B), all of which were annotated according to the canonical cell-type markers (Figure 1C). Nine classical cell types were identified, including epithelial cells, endothelial cells, T cells, regulatory T cells (Tregs), fibroblasts, plasma cells, macrophages, granulocytes, and B cells (Figure 1D). Five cell clusters, including clusters 0, 1, 3, 4, and 16, were recognized as epithelial cells by high expression of epithelial cell adhesion molecule (EPCAM). Endothelial cells were marked by FLT1 expression. T cells were labeled according to their expression of CD3D. Tregs were identified by high CD3D and FOXP3 expression. Fibroblasts were labeled by ACTA2 expression. B cells and plasma cells were identified by CD38 and CD79A expression. Granulocytes were identified by GATA2, and macrophages were marked by CD68 expression (Figure 1C).
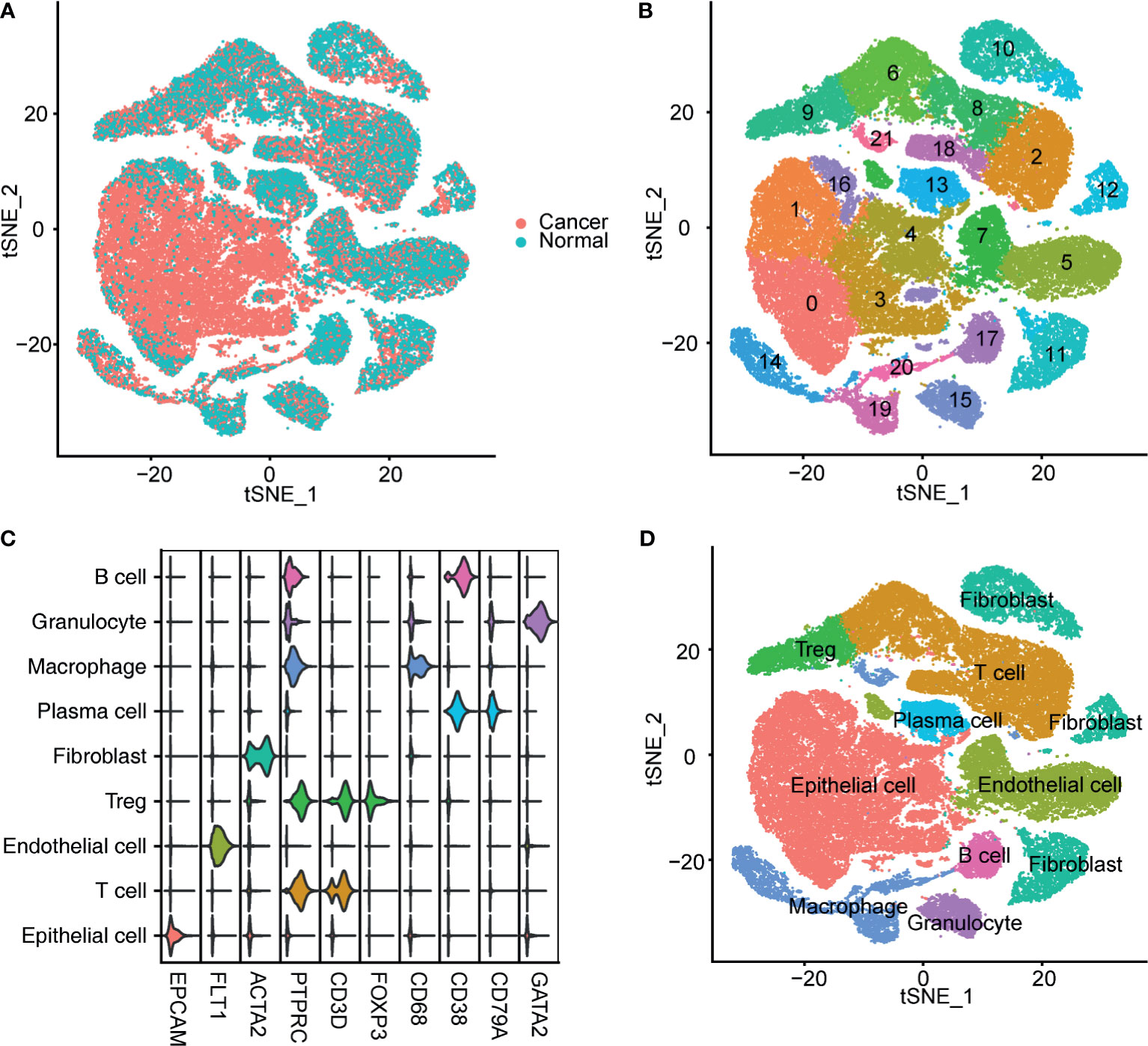
Figure 1 Identification of bladder cancer cells in single-cell sequencing data. (A) T-SNE plot of the integrated bladder cancer single-cell data. Each point represents a cell. Points were colored according to the pathological tissue type. (B) T-SNE plot of the cell clusters identified by the clustering algorithm. Points were colored according to the cluster identity. (C) Violin plot of the expression of classical markers of each cell type. (D) T-SNE plot of the annotated cell clusters. Each point represents a cell. Points were colored according to the cell type.
Alteration of Epithelial Cells Between Bladder Cancer and the Normal Bladder Mucosa
The percentages of epithelial cells in the NBM, NMIBC, and MIBC were 7.87%, 13.53%, and 73.20%, respectively. Clusters 0, 1, and 3 were significantly increased in MIBC, compared with those in the NMIBC and the NBM (Figure 2A). No statistically significant difference was found between the proportions of epithelial cell clusters of the NBM and NMIBC (Figure 2A).
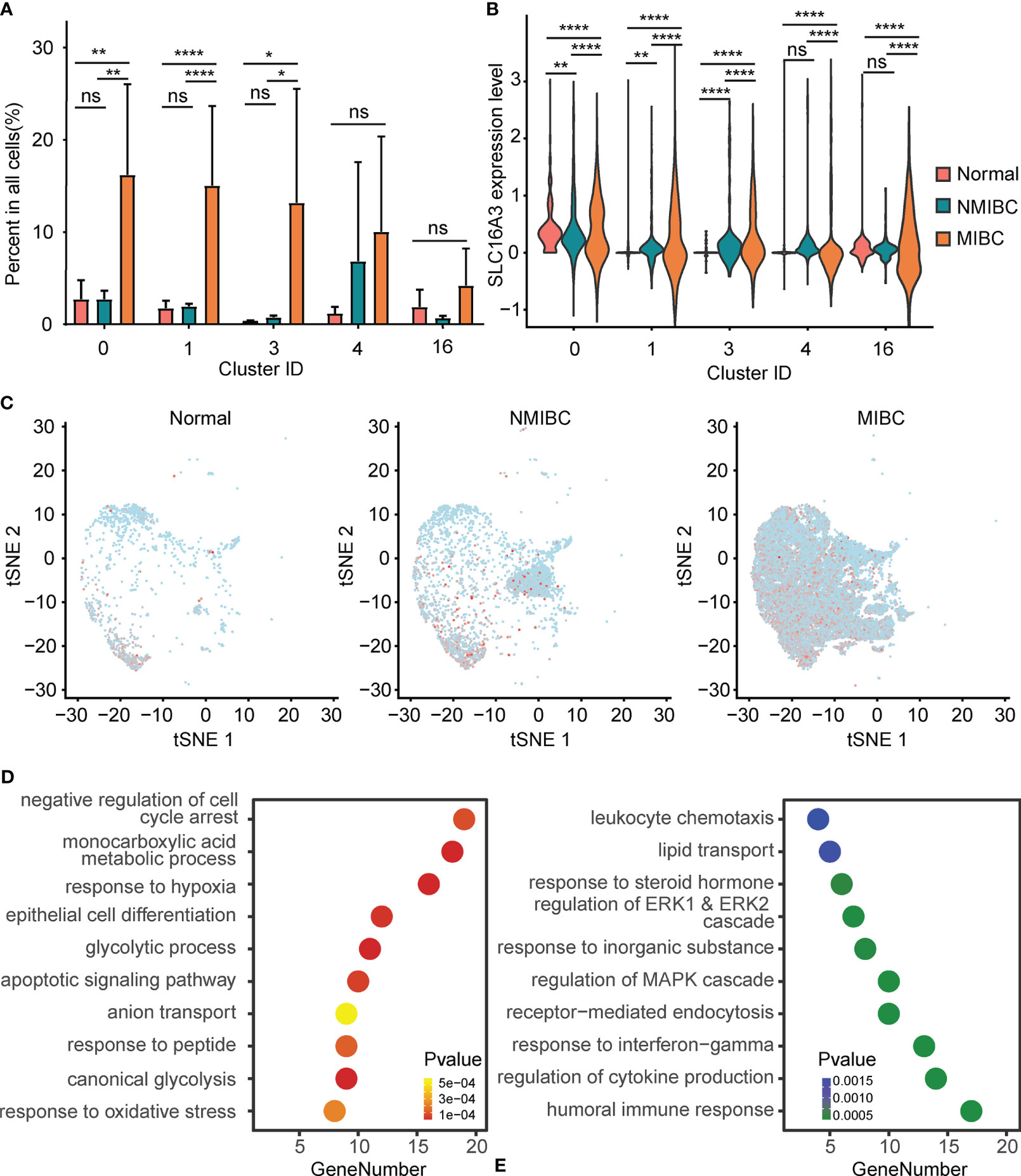
Figure 2 Alteration of PFKFB3 in bladder cancer cells. (A) Histogram of the percentage of epithelial cell adhesion molecule (EPCAM)-positive epithelial cell clusters in non-muscle invasive bladder cancer (NMIBC), MIBC, and the normal bladder mucosa. NS, not significant; *p < 0.05 **p < 0.01; ****p < 0.0001. (B) Violin plot of the expression of SLC16A3 mRNA in epithelial cell clusters of NMIBC, MIBC, and the normal bladder mucosa. NS, not significant; **p < 0.01; ****p < 0.0001. (C) T-SNE plot of the expression of SLC16A3 in epithelial cells in NMIBC, MIBC, and normal bladder mucosa. Red points represent cells with positive PFKFB3 expression, and gray points represent cells with no PFKFB3 expression. (D) Gene set enrichment analysis of the upregulated differentially expressed genes (DEGs) in SLC16A3-positive epithelial cells. The y-axis shows significantly enriched Gene Ontology (GO) terms, and the x-axis shows the number of DEGs enriched in a GO term. (E) Gene set enrichment analysis of the downregulated DEGs in SLC16A3-positive epithelial cells.
Furthermore, compared with the NMIBC and the NBM, MIBC showed significantly elevated SLC16A3 mRNA in all epithelial cell clusters. Compared with the NBM, the NMIBC showed significantly elevated SLC16A3 mRNA in epithelial cell clusters 0, 1, and 3 (Figure 2B). Overall, a gradual increase could be found in the SLC16A3 mRNA expression of all epithelial cells in the NBM, NMIBC, and MIBC (Figure 2C).
Additionally, we screened DEGs between the SLC16A3-positive and SLC16A3-negative epithelial cells and found that 102 genes were upregulated and 66 genes were downregulated in SLC16A3-positive epithelial cells. The upregulated DEGs were mainly enriched in the glycolysis process pathway, response to hypoxia pathway, monocarboxylic acid metabolic process pathway, and epithelial cell differentiation pathway (Figure 2D). The downregulated DEGs were mainly enriched in regulation of humoral immune response pathway and MAPK cascade pathway (Figure 2E).
Prognostic Significance of SLC16A3 mRNA Expression in Bladder Cancer
To explore the influence of SLC16A3 mRNA expression on the survival of bladder cancer patients, we downloaded the survival and transcriptome sequencing data of 402 patients in TCGA database, and the KM survival analysis showed that the patients with high SLC16A3 mRNA expression showed a significantly poorer OS than those with low SLC16A3 mRNA expression (p = 0.012; Figure 3A). Furthermore, subgroup analysis of 234 primary MIBC patients showed that primary MIBC with high SLC16A3 mRNA expression revealed a significantly worse OS (p < 0.001; Figure 3B). Subgroup analysis of 168 subsequent MIBC with an NMIBC history showed that tumors with different SLC16A3 mRNA expression levels did not show a significant difference (p = 0.204, Figure 3C).
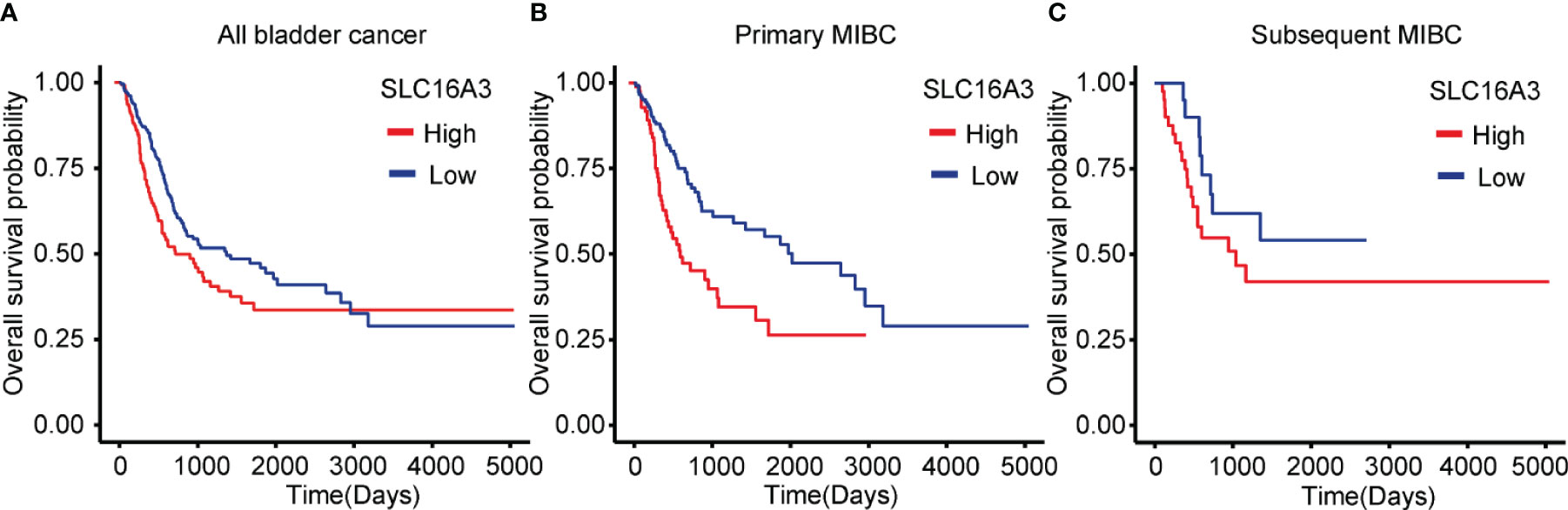
Figure 3 Influence of SLC16A3 expression on the survival of bladder cancer patients. (A) Bladder cancer patients with high SLC16A3 mRNA expression had worse overall survival rates than those with low SLC16A3 mRNA expression (p = 0.016). (B) Primary muscle invasive bladder cancer (MIBC) patients with high SLC16A3 mRNA expression showed worse overall survival rates than those with low SLC16A3 mRNA expression (p < 0.001). (C) Subsequent MIBC patients with high SLC16A3 mRNA expression showed no significant difference from patients with low SLC16A3 mRNA expression (p = 0.204).
Clinical Characteristics and MCT4 Expression of the Validation Cohort
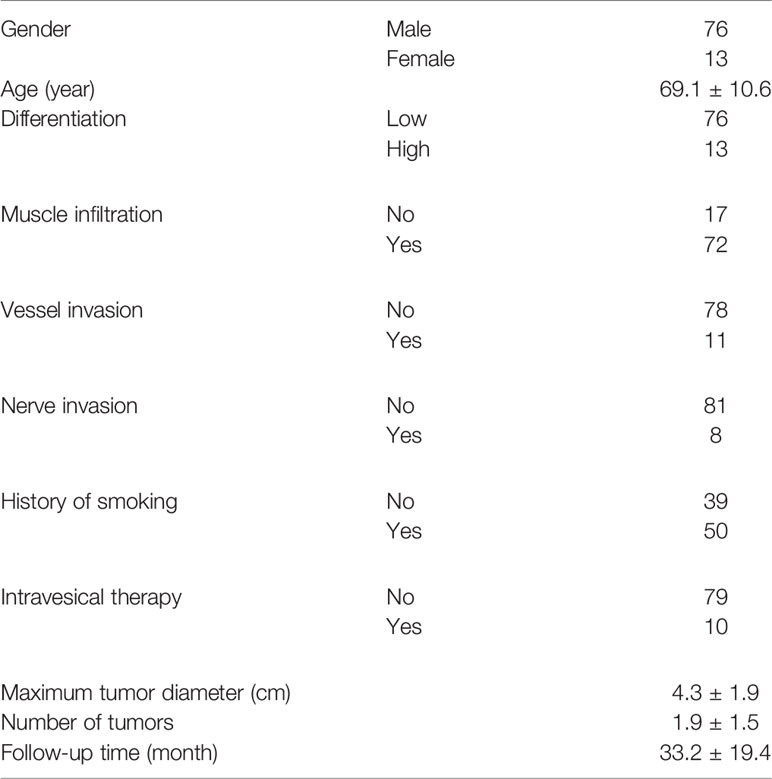
We further enrolled a validation cohort including 89 consecutive bladder cancer patients. The average age of these patients was 69.1 years, and 50 of them had a smoking history. Seventy-two of them had MIBC, and the remaining 17 patients had high-risk NMIBC. Ten patients had previously received intravesical pirarubicin or Bacillus Calmette–Guérin vaccine therapy. Fifty-nine of them had one tumor focus. The other 30 patients had multiple tumors. The average number of tumors was 1.9, and the average maximum tumor diameter was 4.3 cm. Seventy-six tumors showed low pathological differentiation, while 13 tumors showed high differentiation. Pathological vessel or nerve invasion was found in tumors from 11 and eight patients, respectively. The median follow-up time was 33.2 months (Table 1). No patient died from preexisting chronic disorders during the follow-up.
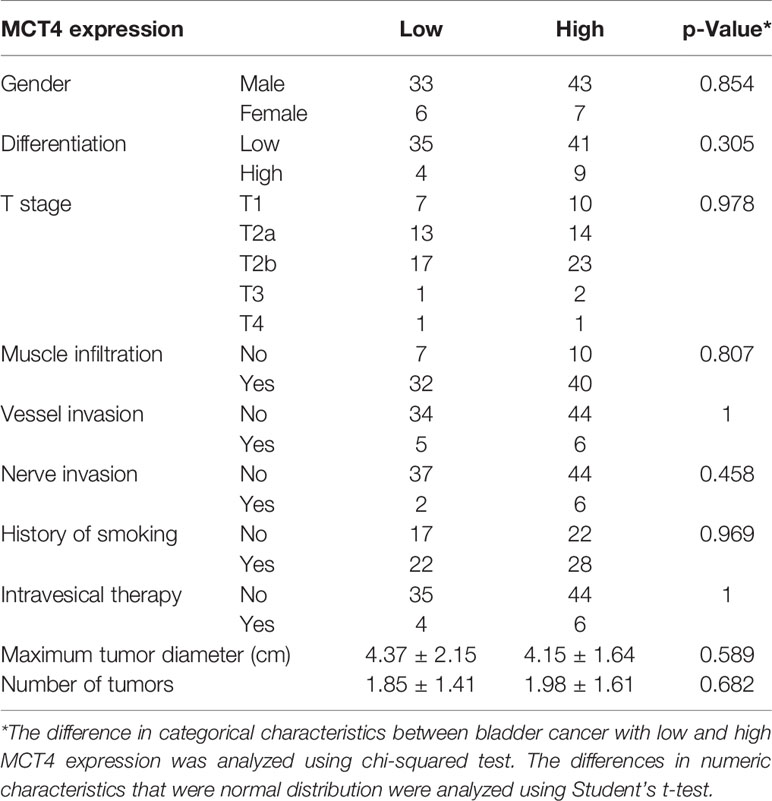
IHC staining was used to examine the expression of MCT4 protein in bladder cancer samples. Positive staining was mainly found in the membrane of the cancer cells, while the nuclei were not stained (Figures 4A–D). Fifty tumor tissues showed high MCT4 expression, and the remaining 39 tumors showed low MCT4 expression. Patients with different gender, pathological characteristics, smoking history, intravesical therapy, tumor diameter, or tumor numbers showed no significant difference in MCT4 expression (Table 2).
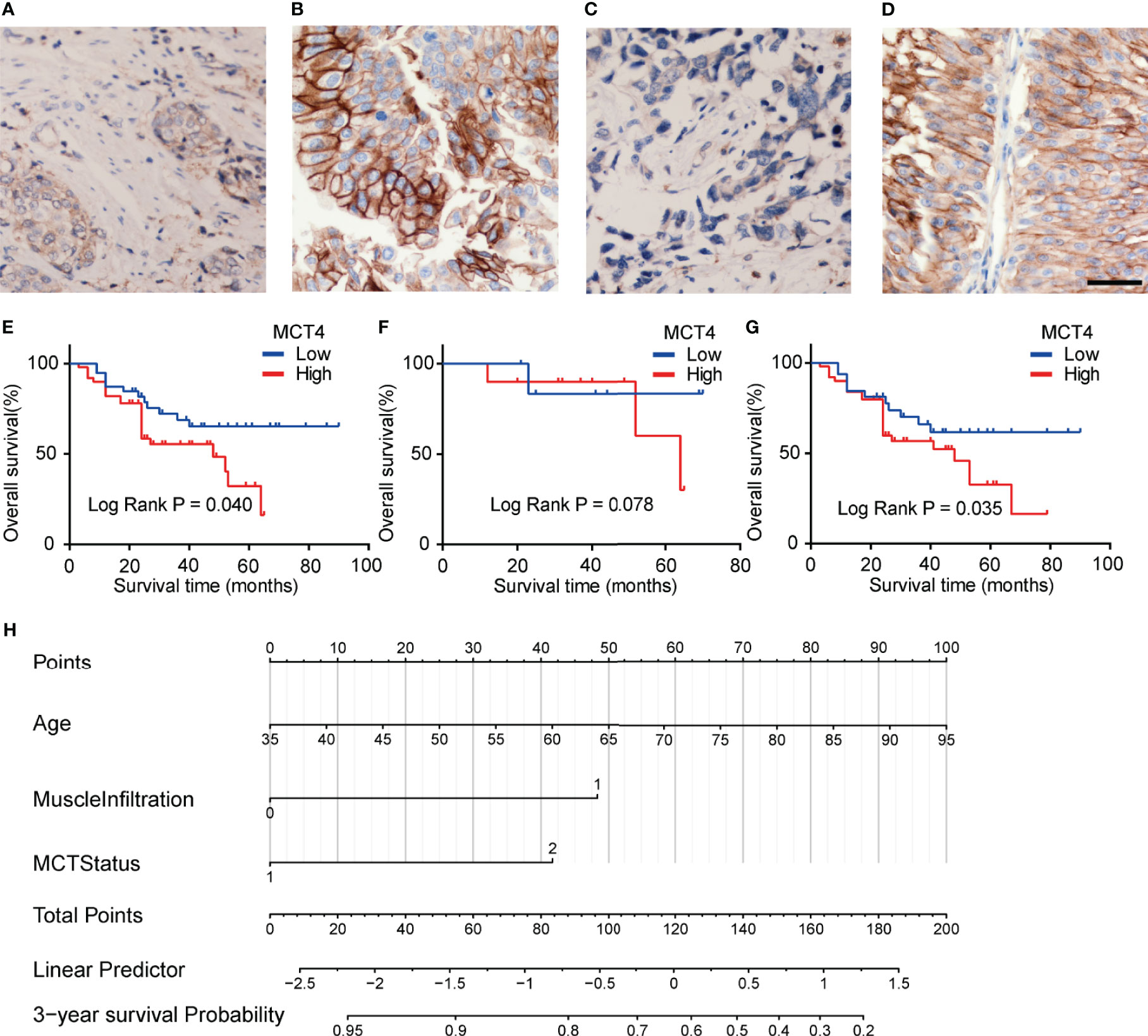
Figure 4 Expression of MCT4 protein in bladder cancer. Low MCT4 (A) and high MCT4 (B) expression in non-muscle invasive bladder cancer (NMIBC). Low MCT4 (C) and high MCT4 (D) expression in MIBC. Magnification: ×400. Scale bar: 10 μm. (E) High MCT4 protein expressions were associated with poor overall survival (OS) in all bladder cancer patients who received radical cystectomy (log-rank test, p = 0.040). (F) No significant difference was found between the OS of NMIBC patients with high MCT4 protein expression and NMIBC patients with low MCT4 expression (log-rank test, p = 0.078). (G) MIBC patients with high MCT4 protein expressions have worse OS rates than those with low MCT4 expression (log-rank test, p = 0.035). (H) Nomogram to predict the postcystectomy survival of bladder cancer patients. Patients were scored according to age (0–100 scores), existence of MIBC (48 for MIBC and 0 for NMIBC), and MCT4 expression (0 for low MCT4 expression and 42 for high MCT4 expression). The scores were then summed to obtain the total points; and the number in the last line, which is vertically beneath the accumulated total points, is the predicted 3-year survival for this patient.
Prognostic Significance of MCT4 Expression
The KM analysis showed that high MCT4 protein expression was associated with poor OS after cystectomy in bladder cancer patients (p = 0.04; Figure 4E). In the subgroup analysis of NMIBC patients, no significant difference was found in OS between tumors with high and low MCT4 expression (p = 0.078; Figure 4F), while in the MIBC patients, tumors with high MCT4 expression showed significantly worse OS than those with low MCT4 expression (p = 0.035; Figure 4G).
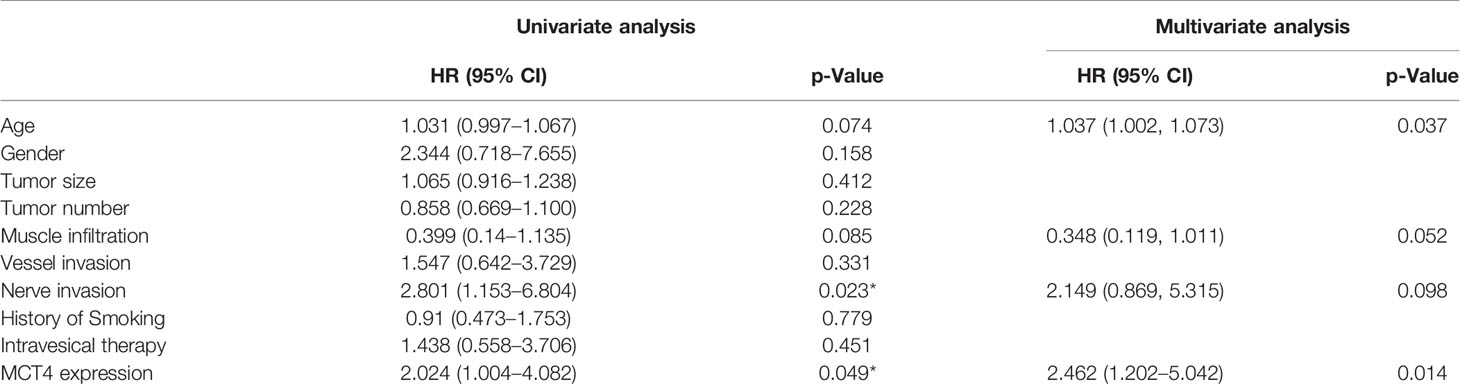
Furthermore, in the univariate Cox regression analysis, high MCT4 expression (HR: 2.024; 95% CI: 1.004–4.082; p = 0.049) and nerve infiltration (HR: 2.801; 95% CI: 1.153–6.804; p = 0.023) showed a significant influence on the OS of bladder cancer patients (Table 3). Characteristics with p-values <0.1, including age, MCT4 expression, nerve invasion, and muscle infiltration, were further analyzed in the multivariate Cox regression model. High MCT4 expression (HR: 2.462; 95% CI: 1.202–5.042; p = 0.014) and age (HR: 1.037; 95% CI: 1.002–1.073; p = 0.037) were independent predictors for the postcystectomy OS of bladder cancer patients (Table 3). To facilitate the use of this model, we built a nomogram for postcystectomy survival prediction using the results of Cox regression analysis (Figure 4H).
Discussion
Bladder cancer is the 10th most common cancer and most common urothelial malignancy worldwide, with approximately 573,000 new cases and 213,000 deaths per year (1). However, 26.9%–37.5% of patients who receive radical treatment show tumor recurrence or metastasis (3–5). Recent advances in prognostic biomarkers provided new methods for more individualized surveillance and treatment of bladder cancer (23, 24). Comprehensive use of cystoscopy, liquid biopsy, and pathology examination could profoundly facilitate the diagnosis and surveillance of bladder cancer and could improve the survival of bladder cancer patients (23, 25).
Despite substantial advances in the field of biomarker detection, new cost-effective markers are still needed to improve diagnostic accuracy and reduce further testing (25). The reprogramming of energy metabolism is a canonical hallmark of cancer cells. Cancer cells rely on glycolysis to generate ATPs even in the presence of oxygen. This phenomenon is termed aerobic glycolysis or the Warburg effect (26). In tumor cells, aerobic glycolysis can rapidly generate ATP and lactate. The lactate produced by aerobic glycolysis plays an important role in the immune evasion and tumor angiogenesis (9, 27). Enhanced lactate shuttling between the cytoplasm and microenvironment is needed to induce immune evasion and tumor angiogenesis and to maintain the stable intracellular pH of tumor cells (28). MCT4 is the major type of transmembrane lactate transporter because of its high preference for lactate. MCT4 inhibition results in intracellular accumulation of lactate, reduction in cell growth and tumor angiogenesis, and induction of reactive oxygen species generation and apoptosis (9, 16). High MCT4 expression is also a signal for poor response to platinum-based chemotherapy (29, 30). In this study, we evaluated the possible use of MCT4 as a potential prognostic biomarker for bladder cancer patients.
The bladder cancer microenvironment is rich in various mesenchymal cells (31). The complex microenvironment is an obstacle for the study of glycolytic tumor biomarkers. In polymerase chain reaction (PCR), bulk RNA sequencing, and Western blotting assays, the upregulation of MCT4 in cancer cells may be covered by other cells with high MCT4 expression, such as immune cells and vascular endothelial cells. Single-cell sequencing is an emerging technique in cancer research that allows examination of the alteration of cancer cells alone. In this study, using single-cell RNA sequencing data, we identified five clusters of epithelial cells, three of which were enriched in MIBCs. Although the proportions of these five clusters were similar between the NMIBC and the NBM, SLC16A3 mRNA was upregulated in three clusters of epithelial cells in the NMIBC compared with the NBM. Additionally, the expression of SLC16A3 was markedly upregulated in all five epithelial cell clusters in MIBC compared with both the NMIBC and the NBM. These results suggest that the upregulation of MCT4 may occur earlier than the massive proliferation of malignant epithelial cells in the development of bladder cancer. More importantly, DEG screening and GO enrichment analysis showed that glycolytic processes and monocarboxylic acid metabolic processes were upregulated in SLC16A3-positive cells, which proves that the expression of SLC16A3 mRNA is closely related to aerobic glycolysis pathways. Therefore, SLC16A3 expression could well reflect the activation status of tumor aerobic glycolysis.
In this study, survival analysis of the transcriptome and the clinical data of bladder cancer patients in TCGA database showed that high SLC16A3 mRNA expression was associated with poor OS in patients with primary MIBC. SLC16A3 mRNA expression showed only a statistically insignificant trend of different OS in subsequent MIBC with NMIBC history. The time needed to progress to MIBC from the NMIBC is highly variable because of the heterogeneity in NMIBC malignancy, likely a reason for this insignificant result.
Furthermore, the KM analysis of the clinical and IHC data of the validation cohort showed that high MCT4 protein expression was significantly associated with poor OS in bladder cancer patients. Subgroup analysis showed that MIBC patients with high MCT4 expression had significantly worse OS than those with low MCT4 expression. In the NMIBC patients, the KM analysis showed insignificant results (p = 0.072), which may be caused by the small NMIBC patient number. However, a trend of poor OS in patients with high MCT4 expression was still found in the KM plot. Choi et al. (32) also reported that MCT4 protein expression negatively correlates with recurrence-free survival in bladder cancer patients in a cohort including all stages of bladder cancer. However, in their study, the therapy for bladder cancer was not specified. Since treatment and tumor malignancy potential can vary considerably based on the TNM stage of bladder cancer, the lack of treatment information may lead to intervention-derived bias of the results. Compared with the cohort reported by Choi et al., we enrolled only patients who had undergone radical cystectomy. Our cohort is more homogeneous in clinical treatment; thus, our results can better support using MCT4 as a prognostic biomarker in MIBC.
Finally, the multivariate Cox regression revealed that high MCT4 expression and age are independent prognostic factors for poor OS in bladder cancer patients undergoing cystectomy. Overall, we found that MCT4 IHC staining can help identify bladder cancer patients with high postcystectomy mortality. We built a nomogram according to the results of Cox analysis. Each patient was scored according to age, the existence of MIBC, and the MCT4 expression status. The scores were then summed to obtain the total points, which were used to predict the 3-year OS for each patient (Figure 4H). Based on the nomogram, the influence of MCT4 expression on the survival of bladder cancer patients can be considered along with other regular prognostic factors, such as age and the depth of invasion. Patients with a poor OS prediction should receive more frequent surveillance after cystectomy, and adjuvant chemotherapy and radiotherapy could be considered based on the prediction of the nomogram and clinical status of the bladder cancer patients. Additionally, targeting MCT4 inhibits the viability of bladder cancer cells and reduces the tumor diameter in a xenograft bladder cancer tumor model (9, 16). Thus, bladder cancer patients with high MCT4 expression may also benefit from MCT4-targeted therapy. Additionally, the use of MCT4 targeted therapy may be combined with functional MRI or PET-CT imaging to screen in vivo drug dynamics (33–37). Furthermore, the combination of MCT4 IHC and previously identified noninvasive peripheral blood biomarkers, such as survivin, circulating tumor cells, and systemic combining inflammatory score, may also help to further improve the clinical management of bladder cancer (38–41). However, further preclinical and clinical studies are needed to further validate the effect of MCT4-targeted therapy in bladder cancer.
Our studies have some limitations. First, all samples were collected during radical cystectomy; thus, we lacked data on MCT4 protein expression in early-stage bladder cancer and metastatic bladder cancer, which are not eligible for radical cystectomy. This finding may limit the use of nomograms in the NMIBC and metastatic bladder cancer patients. Second, chronic disorders such as diabetes mellitus and obesity may also significantly influence the survival of bladder cancer patients (42–44). In this study, the influence of preexisting chronic disorders on the survival of bladder cancer patients could not be ruled out. Further studies with more detailed background information and larger sample sizes are needed to better use MCT4 in the prognosis of bladder cancer.
In conclusion, our study revealed that SLC16A3 mRNA is upregulated in bladder cancer cells. Bladder cancer with high SLC16A3 mRNA expression has a poor OS. High MCT4 protein expression is an independent prognostic factor for bladder cancer patients who had undergone radical cystectomy, and a nomogram based on the Cox regression analysis of our data was built to facilitate the use of MCT4 expression in the prognosis of bladder cancer.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the ethics committee of Peking Union College Hospital and the ethics committee of Affiliated Hospital of Qingdao University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YZ, BZ, and Y-SZ analyzed the data and drafted the manuscript. YX, and W-HY performed the IHC staining and scoring. YZ, BZ, Z-HW, W-DW, G-YZ, and Y-SZ collected the tumor samples and performed the follow-up of patients. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the grants from the National Natural Science Foundation of China (grant no. 81602231 and 81670611) and Major Research Program of Shandong Province (grant no. 2017GSF221016).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.704857/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Flaig TW, Spiess PE, Agarwal N, Bangs R, Boorjian SA, Buyyounouski MK, et al. Bladder Cancer, Version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Cancer Netw: JNCCN (2020) 18(3):329–54. doi: 10.6004/jnccn.2020.0011
3. Yuk HD, Jeong CW, Kwak C, Kim HH, Moon KC, Ku JH. Clinical Outcomes of Muscle Invasive Bladder Cancer According to the BASQ Classification. BMC Cancer (2019) 19(1):897. doi: 10.1186/s12885-019-6042-1
4. Yoo SH, Kim H, Kwak C, Kim HH, Jung JH, Ku JH. Late Recurrence of Bladder Cancer Following Radical Cystectomy: Characteristics and Outcomes. Urol Int (2019) 103(3):291–6. doi: 10.1159/000502656
5. Moschini M, Sharma V, Dell’oglio P, Cucchiara V, Gandaglia G, Cantiello F, et al. Comparing Long-Term Outcomes of Primary and Progressive Carcinoma Invading Bladder Muscle After Radical Cystectomy. BJU Int (2016) 117(4):604–10. doi: 10.1111/bju.13146
6. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science (2009) 324(5930):1029–33. doi: 10.1126/science.1160809
7. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
8. Ganapathy-Kanniappan S, Geschwind JF. Tumor Glycolysis as a Target for Cancer Therapy: Progress and Prospects. Mol Cancer (2013) 12:152. doi: 10.1186/1476-4598-12-152
9. Zhao Y, Wang D, Xu T, Liu P, Cao Y, Wang Y, et al. Bladder Cancer Cells Re-Educate TAMs Through Lactate Shuttling in the Microfluidic Cancer Microenvironment. Oncotarget (2015) 6(36):39196–210. doi: 10.18632/oncotarget.5538
10. Xu XD, Shao SX, Jiang HP, Cao YW, Wang YH, Yang XC, et al. Warburg Effect or Reverse Warburg Effect? A Review of Cancer Metabolism. Oncol Res Treat (2015) 38(3):117–22. doi: 10.1159/000375435
11. Halestrap AP, Meredith D. The SLC16 Gene Family-From Monocarboxylate Transporters (MCTs) to Aromatic Amino Acid Transporters and Beyond. Pflugers Archiv: Eur J Physiol (2004) 447(5):619–28. doi: 10.1007/s00424-003-1067-2
12. Halestrap AP. Monocarboxylic Acid Transport. Compr Physiol (2013) 3(4):1611–43. doi: 10.1002/cphy.c130008
13. Chiche J, Le Fur Y, Vilmen C, Frassineti F, Daniel L, Halestrap AP, et al. In Vivo pH in Metabolic-Defective Ras-Transformed Fibroblast Tumors: Key Role of the Monocarboxylate Transporter, MCT4, for Inducing an Alkaline Intracellular pH. Int J Cancer J Int Cancer (2012) 130(7):1511–20. doi: 10.1002/ijc.26125
14. Choi SY, Xue H, Wu R, Fazli L, Lin D, Collins CC, et al. The MCT4 Gene: A Novel, Potential Target for Therapy of Advanced Prostate Cancer. Clin Cancer Res: Off J Am Assoc Cancer Res (2016) 22(11):2721–33. doi: 10.1158/1078-0432.CCR-15-1624
15. Fisel P, Kruck S, Winter S, Bedke J, Hennenlotter J, Nies AT, et al. DNA Methylation of the SLC16A3 Promoter Regulates Expression of the Human Lactate Transporter MCT4 in Renal Cancer With Consequences for Clinical Outcome. Clin Cancer Res: Off J Am Assoc Cancer Res (2013) 19(18):5170–81. doi: 10.1158/1078-0432.CCR-13-1180
16. Todenhofer T, Seiler R, Stewart C, Moskalev I, Gao J, Ladhar S, et al. Selective Inhibition of the Lactate Transporter MCT4 Reduces Growth of Invasive Bladder Cancer. Mol Cancer Ther (2018) 17(12):2746–55. doi: 10.1158/1535-7163.MCT-18-0107
17. Chen Z, Zhou L, Liu L, Hou Y, Xiong M, Yang Y, et al. Single-Cell RNA Sequencing Highlights the Role of Inflammatory Cancer-Associated Fibroblasts in Bladder Urothelial Carcinoma. Nat Commun (2020) 11(1):5077. doi: 10.1038/s41467-020-18916-5
18. Lee HW, Chung W, Lee HO, Jeong DE, Jo A, Lim JE, et al. Single-Cell RNA Sequencing Reveals the Tumor Microenvironment and Facilitates Strategic Choices to Circumvent Treatment Failure in a Chemorefractory Bladder Cancer Patient. Genome Med (2020) 12(1):47. doi: 10.1186/s13073-020-00741-6
19. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating Single-Cell Transcriptomic Data Across Different Conditions, Technologies, and Species. Nat Biotechnol (2018) 36(5):411–20. doi: 10.1038/nbt.4096
20. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell (2019) 177(7):1888–902.e21. doi: 10.1016/j.cell.2019.05.031
21. Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, et al. Dimensionality Reduction for Visualizing Single-Cell Data Using UMAP. Nat Biotechnol (2019) 37:38–44. doi: 10.1038/nbt.4314
22. Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating High-Throughput Genomic Analysis With Bioconductor. Nat Methods (2015) 12(2):115–21. doi: 10.1038/nmeth.3252
23. Ferro M, La Civita E, Liotti A, Cennamo M, Tortora F, Buonerba C, et al. Liquid Biopsy Biomarkers in Urine: A Route Towards Molecular Diagnosis and Personalized Medicine of Bladder Cancer. J Pers Med (2021) 11(3):237. doi: 10.3390/jpm11030237
24. Lodewijk I, Duenas M, Rubio C, Munera-Maravilla E, Segovia C, Bernardini A, et al. Liquid Biopsy Biomarkers in Bladder Cancer: A Current Need for Patient Diagnosis and Monitoring. Int J Mol Sci (2018) 19(9):2514. doi: 10.3390/ijms19092514
25. Maas M, Bedke J, Stenzl A, Todenhofer T. Can Urinary Biomarkers Replace Cystoscopy? World J Urol (2019) 37(9):1741–9. doi: 10.1007/s00345-018-2505-2
26. Burk D, Schade AL. On Respiratory Impairment in Cancer Cells. Science (1956) 124(3215):270–2. doi: 10.1126/science.124.3215.267
27. Hu KY, Wang de G, Liu PF, Cao YW, Wang YH, Yang XC, et al. Targeting of MCT1 and PFKFB3 Influences Cell Proliferation and Apoptosis in Bladder Cancer by Altering the Tumor Microenvironment. Oncol Rep (2016) 36(2):945–51. doi: 10.3892/or.2016.4884
28. Jones RG, Thompson CB. Tumor Suppressors and Cell Metabolism: A Recipe for Cancer Growth. Genes Dev (2009) 23(5):537–48. doi: 10.1101/gad.1756509
29. Afonso J, Santos LL, Miranda-Goncalves V, Morais A, Amaro T, Longatto-Filho A, et al. CD147 and MCT1-Potential Partners in Bladder Cancer Aggressiveness and Cisplatin Resistance. Mol Carcinog (2015) 54(11):1451–66. doi: 10.1002/mc.22222
30. Afonso J, Santos LL, Morais A, Amaro T, Longatto-Filho A, Baltazar F. Metabolic Coupling in Urothelial Bladder Cancer Compartments and its Correlation to Tumor Aggressiveness. Cell Cycle (2016) 15(3):368–80. doi: 10.1080/15384101.2015.1121329
31. Liu PF, Cao YW, Jiang HP, Wang YH, Yang XC, Wang XS, et al. Heterogeneity Research in Muscle-Invasive Bladder Cancer Based on Differential Protein Expression Analysis. Med Oncol (2014) 31(9):21. doi: 10.1007/s12032-014-0021-9
32. Choi JW, Kim Y, Lee JH, Kim YS. Prognostic Significance of Lactate/Proton Symporters MCT1, MCT4, and Their Chaperone CD147 Expressions in Urothelial Carcinoma of the Bladder. Urology (2014) 84(1):245 e9–15. doi: 10.1016/j.urology.2014.03.031
33. Del Giudice F, Leonardo C, Simone G, Pecoraro M, De Berardinis E, Cipollari S, et al. Preoperative Detection of Vesical Imaging-Reporting and Data System (VI-RADS) Score 5 Reliably Identifies Extravesical Extension of Urothelial Carcinoma of the Urinary Bladder and Predicts Significant Delayed Time to Cystectomy: Time to Reconsider the Need for Primary Deep Transurethral Resection of Bladder Tumour in Cases of Locally Advanced Disease? BJU Int (2020) 126(5):610–9. doi: 10.1111/bju.15188
34. Del Giudice F, Pecoraro M, Vargas HA, Cipollari S, De Berardinis E, Bicchetti M, et al. Systematic Review and Meta-Analysis of Vesical Imaging-Reporting and Data System (VI-RADS) Inter-Observer Reliability: An Added Value for Muscle Invasive Bladder Cancer Detection. Cancers (2020) 12(10):2994. doi: 10.1002/jmri.27361
35. Del Giudice F, Barchetti G, De Berardinis E, Pecoraro M, Salvo V, Simone G, et al. Prospective Assessment of Vesical Imaging Reporting and Data System (VI-RADS) and Its Clinical Impact on the Management of High-Risk Non-Muscle-Invasive Bladder Cancer Patients Candidate for Repeated Transurethral Resection. Eur Urol (2020) 77(1):101–9. doi: 10.1016/j.eururo.2019.09.029
36. Woo S, Panebianco V, Narumi Y, Del Giudice F, Muglia VF, Takeuchi M, et al. Diagnostic Performance of Vesical Imaging Reporting and Data System for the Prediction of Muscle-Invasive Bladder Cancer: A Systematic Review and Meta-Analysis. Eur Urol Oncol (2020) 3(3):306–15. doi: 10.1016/j.euo.2020.02.007
37. Panebianco V, Pecoraro M, Del Giudice F, Takeuchi M, Muglia VF, Messina E, et al. VI-RADS for Bladder Cancer: Current Applications and Future Developments. J Magn Reson Imaging: JMRI (2020). doi: 10.1002/jmri.27361
38. Busetto GM, Ferro M, Del Giudice F, Antonini G, Chung BI, Sperduti I, et al. The Prognostic Role of Circulating Tumor Cells (CTC) in High-Risk Non-Muscle-Invasive Bladder Cancer. Clin Genitourinary Cancer (2017) 15(4):e661–6. doi: 10.1016/j.urolonc.2019.12.002
39. Nicolazzo C, Busetto GM, Del Giudice F, Sperduti I, Giannarelli D, Gradilone A, et al. The Long-Term Prognostic Value of Survivin Expressing Circulating Tumor Cells in Patients With High-Risk non-Muscle Invasive Bladder Cancer (NMIBC). J Cancer Res Clin Oncol (2017) 143(10):1971–6. doi: 10.1007/s00432-017-2449-8
40. Nicolazzo C, Busetto GM, Gradilone A, Sperduti I, Del Giudice F, Loreni F, et al. Circulating Tumor Cells Identify Patients With Super-High-Risk Non-Muscle-Invasive Bladder Cancer: Updated Outcome Analysis of a Prospective Single-Center Trial. Oncol (2019) 24(5):612–6. doi: 10.1634/theoncologist.2018-0784
41. Ferro M, Di Mauro M, Cimino S, Morgia G, Lucarelli G, Abu Farhan AR, et al. Systemic Combining Inflammatory Score (SCIS): A New Score for Prediction of Oncologic Outcomes in Patients With High-Risk non-Muscle-Invasive Urothelial Bladder Cancer. Trans Androl Urol (2021) 10(2):626–35. doi: 10.21037/tau-20-1272
42. Ferro M, Vartolomei MD, Russo GI, Cantiello F, Farhan ARA, Terracciano D, et al. An Increased Body Mass Index is Associated With a Worse Prognosis in Patients Administered BCG Immunotherapy for T1 Bladder Cancer. World J Urol (2019) 37(3):507–14. doi: 10.1007/s00345-018-2397-1
43. Ferro M, Katalin MO, Buonerba C, Marian R, Cantiello F, Musi G, et al. Type 2 Diabetes Mellitus Predicts Worse Outcomes in Patients With High-Grade T1 Bladder Cancer Receiving Bacillus Calmette-Guerin After Transurethral Resection of the Bladder Tumor. Urol Oncol (2020) 38(5):459–64. doi: 10.1016/j.urolonc.2020.02.016
Keywords: bladder cancer, monocarboxylate transporter 4, solute carrier family 16 member 3, single-cell RNA sequencing, immunohistochemistry
Citation: Zhao Y, Zhao B, Yan W-H, Xia Y, Wang Z-H, Zheng G-Y, Wang W-D and Zhang Y-S (2021) Integrative Analysis Identified MCT4 as an Independent Prognostic Factor for Bladder Cancer. Front. Oncol. 11:704857. doi: 10.3389/fonc.2021.704857
Received: 04 May 2021; Accepted: 02 August 2021;
Published: 26 August 2021.
Edited by:
Matteo Ferro, European Institute of Oncology (IEO), ItalyReviewed by:
Octavian Sabin Tataru, George Emil Palade University of Medicine, Pharmacy, Science and Technology of Târgu Mureş, RomaniaFelice Crocetto, Federico II University Hospital, Italy
Francesco Del Giudice, Sapienza University of Rome, Italy
Copyright © 2021 Zhao, Zhao, Yan, Xia, Wang, Zheng, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Shi Zhang, emhhbmd5dXNoaVBVTUNIQDE2My5jb20=
 Yang Zhao
Yang Zhao Bin Zhao
Bin Zhao Wei-Hua Yan3
Wei-Hua Yan3 Yu-Shi Zhang
Yu-Shi Zhang