
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 13 July 2021
Sec. Gastrointestinal Cancers
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.704042
This article is part of the Research TopicCholangiocarcinoma: A New Multimodality ApproachView all 8 articles
 Yang Gao1†
Yang Gao1† Rong Zhou2†
Rong Zhou2† Jun-Feng Huang3†
Jun-Feng Huang3† Bo Hu1
Bo Hu1 Jian-Wen Cheng1
Jian-Wen Cheng1 Xiao-Wu Huang1
Xiao-Wu Huang1 Peng-Xiang Wang1
Peng-Xiang Wang1 Hai-Xiang Peng4,5
Hai-Xiang Peng4,5 Wei Guo6
Wei Guo6 Jian Zhou1,7
Jian Zhou1,7 Jia Fan1,7
Jia Fan1,7 Xin-Rong Yang1*
Xin-Rong Yang1*Background: Intrahepatic cholangiocarcinoma (ICC) remains one of the most intractable malignancies. The development of effective drug treatments for ICC is seriously hampered by the lack of reliable tumor models. At present, patient derived xenograft (PDX) models prove to accurately reflect the genetic and biological diversity required to decipher tumor biology and therapeutic vulnerabilities. This study was designed to investigate the establishment and potential application of PDX models for guiding personalized medicine and identifying potential biomarker for lenvatinib resistance.
Methods: We generated PDX models from 89 patients with ICC and compared the morphological and molecular similarities of parental tumors and passaged PDXs. The clinicopathologic features affecting PDX engraftment and the prognostic significance of PDX engraftment were analyzed. Drug treatment responses were analyzed in IMF-138, IMF-114 PDX models and corresponding patients. Finally, lenvatinib treatment response was examined in PDX models and potential drug resistance mechanism was revealed.
Results: Forty-nine PDX models were established (take rate: 55.1%). Successful PDX engraftment was associated with negative HbsAg (P = 0.031), presence of mVI (P = 0.001), poorer tumor differentiation (P = 0.023), multiple tumor number (P = 0.003), presence of lymph node metastasis (P = 0.001), and later TNM stage (P = 0.039). Moreover, patients with tumor engraftment had significantly shorter time to recurrence (TTR) (P < 0.001) and worse overall survival (OS) (P < 0.001). Multivariate analysis indicated that PDX engraftment was an independent risk factor for shortened TTR (HR = 1.84; 95% CI, 1.05–3.23; P = 0.034) and OS (HR = 2.13; 95% CI, 1.11–4.11; P = 0.024). PDXs were histologically and genetically similar to their parental tumors. We also applied IMF-138 and IMF-114 PDX drug testing results to guide clinical treatment for patients with ICC and found similar treatment responses. PDX models also facilitated personalized medicine for patients with ICC based on drug screening results using whole exome sequencing data. Additionally, PDX models reflected the heterogeneous sensitivity to lenvatinib treatment and CDH1 might be vital to lenvatinib-resistance.
Conclusion: PDX models provide a powerful platform for preclinical drug discovery, and potentially facilitate the implementation of personalized medicine and improvement of survival of ICC cancer patient.
Intrahepatic cholangiocarcinoma (ICC) is the second most common primary liver tumor, accounting for approximately 10%-15% of primary hepatic malignancies (1, 2). Moreover, the overall incidence of ICC shows a significant increasing trend worldwide with an annual percentage increase of 2.30% in the past four decades (3, 4). Surgical resection remains the only potentially curative option for patients with ICC, but less than 25% of patients with ICC are eligible for resection (5). Most patients (approximately 70%) are diagnosed at late stages and systematic therapies are recommended (5), while the lack of effective treatment regimens results in dismal outcomes for those patients (6). Despite considerable advances in understanding ICC complexity of this kind of tumor, the development of an effective antitumor drug for ICC patients hasn’t progressed due to a lack of suitable preclinical models that recapitulate the pathologic, biological, and genetic features of ICC.
At present, the use of patient derived xenograft (PDX) models has become an attractive platform for drug development and translational cancer research (7, 8). PDX models are established by directly engrafting fresh tumor tissue during surgery or biopsy into immunodeficient mice, which can recapitulate human tumor biology more accurately than traditional cell line-derived xenografts (9). Cancer organoid is an in vitro 3D culture model from patient tumor and we also some liver cancer organoid model, which potentially enables drug screening and individualized treatment regimen and was regarded as an intermediate modeling platform in precision oncology (10–12). PDX is a kind of in vivo tumor model and has unique advantage in basic research and translational medicine. More importantly, the consistency of treatment responses between PDX models and corresponding patients has been confirmed in several types of solid tumors, including our recent research (13–16). Therefore, PDX model provides an alternative preclinical model for the discovery of novel treatment regimens for patients with malignant tumors. Recently, Wang et al. constructed a cholangiocarcinoma PDX biobank with an engraftment rate of 83.3% (30 of 36) and used these models to reveal the chemosensitivity of proteasome inhibition in cholangiocarcinoma, which further broadened the application in medical research (17). The first report on ICC PDX model was published in 2016 by Giuliana et al., who successfully established and confirmed the histological and genetic similarity between primary tumor and PDX model (18). However, the tumor take rate was extremely low, and only one out of 17 tumors (5.8%) was successfully engrafted (18). Furthermore, the clinical significance of PDX engraftment and its potential applications in guiding personalized medicine still need to be further explored.
Here, we report the establishment of a unique bank of serially transplantable tumor grafts that retain crucial characteristics of the original tumor specimens from ICC patients based on our previous independently developed technique for PDX model engraftment in HCC (15). We found that successful PDX engraftment was significantly associated with presence of microvascular invasion (mVI), poorer tumor differentiation, multiple tumor number, presence of lymph node metastasis, and advanced tumor stage in patients with ICC. In addition, successful tumor engraftment indicated shorter time to recurrence (TTR), and overall survival (OS). This PDX model shows great promise in a preclinical setting for biomarker development, and will aid in understanding the mechanisms of drug resistance, drug screening as well as personalized medicine applications for patients with ICC.
From June 2014 to December 2019, 89 patients with ICC receiving treatment in Zhongshan Hospital of Fudan University were included in this study. ICC was defined based on pathologic diagnosis from formalin-fixed, paraffin-embedded tissue or biopsy tissue. All patients received curative resection (defined as complete excision with negative tumor margins) (19). Tumor staging was based on the Seventh American Joint Committee on Cancer (AJCC) TNM staging system (20). This study was approved by the Ethics Board of the Liver Cancer Institute of Zhongshan Hospital, Fudan University (Shanghai, China) and informed written consent was obtained from each patient. TTR was defined as the interval between the date of surgery and the date of clinical diagnose of tumor recurrence. OS was defined as the interval between the date of surgery and the date of death from any cause or the last follow-up visit. Follow up was terminated on September 1, 2020.
All mouse experiments were approved by the Animal Care and Use Committee of Zhongshan Hospital of Fudan University. Three-week-old male, nonobese diabetic/severe combined immunodeficient (NOD/SCID) or NOG (NOD/Shi-scid, IL-2Rγnull) mice were used for tumor engraftment. Following surgery, tumor samples were collected in serum-free DMEM media (Dulbecco’s Modified Eagle’s Medium, Gibco, United States) supplied with 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco, United States) for engraftment within 1 hour after surgery. Tumor samples (2×4×4 mm3) were subcutaneously implanted into the right flank of each mouse. Tumor growth was measured every 3 days using calipers. To verify the establishment of PDX in each mouse, the tumors were monitored for at least 3 months. When the tumor volumes reached 1000 mm3, the mice were sacrificed and tumors were excised into small fragments and implanted into another cohort of mice, frozen for morphological, genetic expression analysis or whole exome sequencing.
Patient tumor specimens and PDX tissues were formalin fixed, paraffin-embedded, cut into sections (4 μm thick), and stained with hematoxylin and eosin (H&E). The biliary cancer marker cytokeratin (CK) 7 and CK19 (21), endothelial marker CD34 (22) and nuclear proliferation biomarker Ki67 (23) were evaluated by immunohistochemical analysis (IHC) to compare the morphological similarity among tumor tissues and passaged PDXs. Primary antibodies CK7 (1:200), CK19 (1:300), CD34 (1:100), and Ki67 (1:200) were purchased from Abcam (Canbridge, MA, USA). Immunohistochemistry (IHC) was performed using a two-step protocol as previously described (24). Pathological examinations were performed under light microscopy by two pathologists blinded to the clinical information of patients.
Genomic DNA were extracted from tumor tissues by means of the Allprep DNA FFPE extraction kit (Qiagen, Germantown, MD), according to the manufacturer’s instructions. After purification, DNA was eluted in 30 µL of water, and yield was determined by the Qubit DNA HS Assay (from Thermo Fisher Scientific, Waltham, MA), according to the manufacturer’s recommendation. A predefined yield of 300 ng of DNA was used as acceptance criteria to ensure adequate library preparation. DNA-seq libraries were constructed using the TruSeq DNA exome Kit (Illumina, Inc., San Diego, CA, United States) following the manufacturer’s recommendations. After library quality and quantity assessment, DNA-seq samples were sequenced using an Illumina HiSeq x10 instrument (pair-end 150 cycle reactions). First, sequencing reads were mapped/aligned to reference genome hg19 using the Burrows-Wheeler Aligner (BWA). Second, mapped/aligned reads were sorted and indexed with Samtools 0.1.19. Third, Picard-tools were used to fix mate pair and remove duplicate reads. Fourth, INDEL realignment was conducted with the Genome Analysis Toolkit (GATK 4.1.1). Fifth, base quality was recalibrated by GATK using dbSNP151. Then, GATK Mutect2 was used to detect SNP/Indels mutations with tumor-normal paired mode. During this step, gnomAD v2.1.1 was used to filter germline mutations. False positive mutations were filtered using GATK FilterMutectCalls, and only “PASS”-labeled mutations remained. The functional effect of variants called were annotated by ANNOVAR (25).
Passage 3– 4 PDX models were used to evaluate the efficiency of various treatment regimens for ICC. When the tumor volumes reached approximately 100–150 mm3, mice were randomly divided into groups with 5 mice/group. Groups were treated as follows: (1) control group received weekly intraperitoneal (i.p.) injection of 100 μl phosphate buffered saline; (2) GEMOX group received gemcitabine (50 mg/kg, i.p. once per week) + oxaliplatin (5 mg/kg, i.p. once per week); (3) XELOX group received capecitabine (200 mg/kg, orally, once per day) + oxaliplatin (5 mg/kg, i.p. once per week); (4) FOLFOX group received oxaliplatin (5 mg/kg, i.p. once per week) + 5-FU (25 mg/kg, i.p. once per week); (5) sorafenib group received sorafenib (30 mg/kg orally, once per day); (6) lenvatinib group received lenvatinib (30 mg/kg, orally, once per day); (7) alpelisib group received alpelisib (50 mg/kg, orally, once per day); (8) abemaciclib group received abemaciclib (50 mg/kg, orally, once per day); (9) copanisib received copanisib (6 mg/kg, orally, once per day); and (10) trametinib received trametinib (1 mg/kg, orally, once per day). All drugs were purchased from MedChemExpress Co., Ltd. (Monmouth Junction, NJ, United States). Animals were treated for 4 weeks. All animals were weighed at the time of tumor measurement and were observed daily for physical appearance, behavior, and clinical changes. Tumor volumes were calculated using the formula: V = ½ × length × (width)2, where length = the greatest longitudinal diameter and width = the greatest transverse diameter. Tumor growth inhibition (TGI) was calculated by relative treated group tumor growth (ΔT) divided by relative control group tumor growth (ΔC), which can be expressed by: TGI = ΔT/ΔC (26). We adopted the Division of Cancer Treatment of the National Cancer Institute (NCI) criteria in this study and defined tumor response as 0%–20% TGI, tumor stability as 21%–50% TGI, and tumor progression > 50% TGI (27).
Statistical analysis was performed using SPSS 19.0 software (IBM SPSS Inc., Chicago, IL, USA). χ2 and Fisher’s exact probability tests were used to compare the association between clinicopathological factors and engraftment. Survival analyses were performed using the Kaplan–Meier method and survival curves were compared by log-rank test. Variables of interest were tested using Cox proportional hazards regression analysis and significant variables obtained identified by univariate analysis were subjected to multivariate analysis (28). All statistical analyses were two-sided and a P-value < 0.05 was considered statistically significant.
Patient characteristics are shown in Table 1. The patient cohort included 43 men and 46 women with a median age of 59 years (range, 34–85 years). Finally, 49 of the 89 engrafted tumors were successfully established as PDX models (take rate: 55.1%). We compared clinical characteristics and successful PDX establishment and found that the PDX engraftment rate was associated with negative HbsAg (P = 0.031), presence of mVI (P = 0.001), poorer tumor differentiation (P = 0.023), multiple tumor number (P = 0.003), presence of lymph node metastasis (P=0.001), and later TNM stage (P = 0.039) (Table 1).
The histologic, molecular, and genomic relevance of the engrafted tumors were compared with those of the primary tumors (between F0, F2, F4, and F6). F0 indicated primary tumor and F2, F4 and F6 indicated the second, fourth and sixth passaged xenograft in mice The morphological features of serial passages, as shown by H&E-staining, were well maintained, and tumors of PDX models resembled the original tumors at the cellular and structural levels. The molecular features of the tumors were assessed by immunohistochemical analysis of currently used diagnostic markers. CK7 and CK19 expression in PDX models were mostly consistent with that in the parental primary tumors. In contrast, Ki67 expression was weak in the earlier PDX passages and became stronger in the later PDX passages. CD34 expression also gradually decreased, implying that human stromal elements are replaced by murine stroma as the tumor was engrafted in the new microenvironment (Figure 1A).
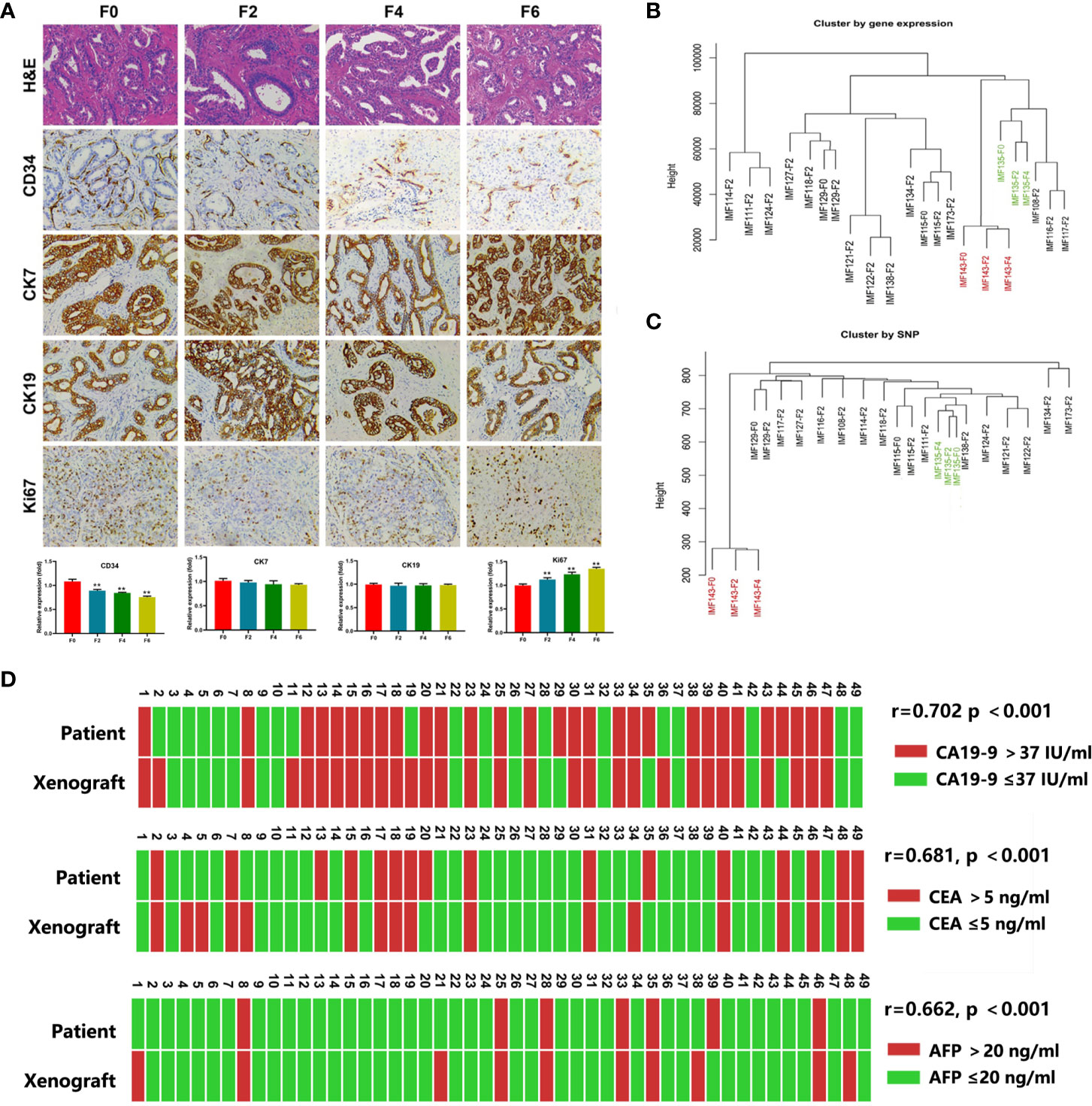
Figure 1 The PDX models recapitulated the characteristics of the parental tumors. (A) Representative H&E sections and immunohistochemical profiles of CD34, CK7, CK19 and Ki67 in serial PDXs and their parental primary tumors (200×). F0, primary tumor; F2, the second passaged xenograft in mice; F4 and F6, the fourth and sixth passaged xenograft; **P < 0.01 (B, C), The dendograms showed unsupervised clustering of samples according to gene expression pattern by Human Genome U133 Plus 2.0 Array (B) and SNP by SNP 6.0 arrays (C); (D) Comparison of serum CA19-9, CEA and AFP in PDX models and corresponding patient.
We next evaluated the maintenance of gene expression and single nucleotide polymorphism (SNP) using Human Genome U133 Plus 2.0 Array and SNP 6.0 arrays, respectively and some pairs of parental primary tumors and PDX models (F2–F4 xenograft tumors) were compared. Unsupervised clustering of gene expression and SNP analysis showed that all the PDXs could be tightly clustered with their corresponding primary tumors (Figures 1B, C).
CA19-9 is routinely used in ICC diagnosis and follow-up. Therefore, we also explored the relationship between CA19-9 levels in PDX models and corresponding patients. We found elevated CA19-9 in 60.4% (29/48) of PDXs (median, 86.3 U/ml; range, 0.6 to > 10,000 U/ml), which was consist with corresponding levels in patients (median, 51.7 U/ml; range, 0.6 to > 10,000 U/ml; r = 0.702, p < 0.001). Similar AFP and CEA levels were observed in PDXs and corresponding patients (Figure 1D).
To determine the influence of engraftment ability on clinical outcomes, we assessed TTR and OS in patients with tumors used to create PDX models. Tumor engraftment was significantly associated with shorter TTR (median, 7.53 months versus 15.67 months; P < 0.001; Figure 2A) and OS (median, 13.97 versus 27.50 months; P < 0.001; Figure 2B). Furthermore, univariate analysis revealed that the ability to engraft was a prognostic factor for TTR (HR = 2.36; 95% CI, 1.40–3.98; P = 0.001) and OS (HR = 2.65; 95% CI, 1.48–4.72; P = 0.001) (Table 2). Multivariate analysis was conducted and the ability to generate a stable engraftment was an independent prognostic factor for TTR (HR = 1.84; 95% CI, 1.05–3.23; P = 0.034) and OS (HR = 2.13; 95% CI, 1.11–4.11; P = 0.024).
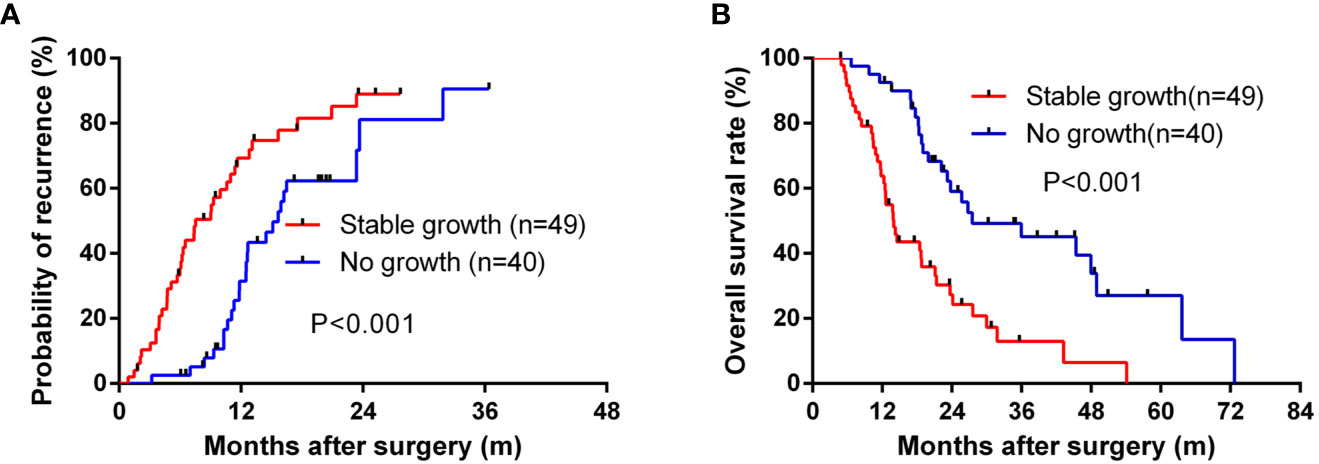
Figure 2 The Kaplan–Meier analysis of TTR and OS for the stable growth of grafts. (A) for TTR and (B) for OS.
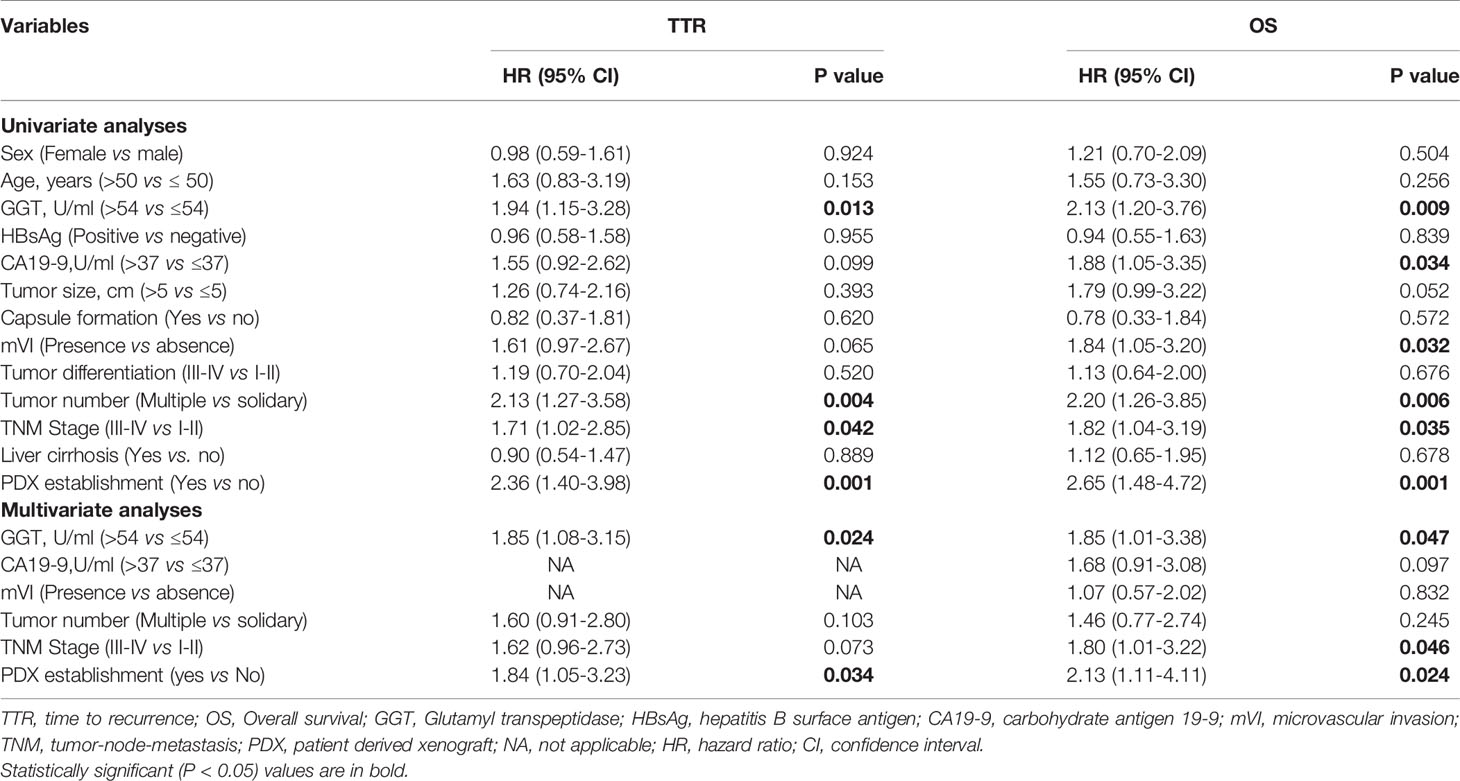
Table 2 Univariate and multivariate analyses showed PDX establishment as an independent predictor for TTR and OS.
To further demonstrate the value of the PDX model in clinical individualized treatment for patients with ICC, two typical ICC patients with established corresponding PDX models were assessed. Patient IMF-138 was initially diagnosed with hepatic lesion and received curative liver resection plus lymphadenectomy. Pathological and immunohistological findings confirmed a moderately differentiated adenocarcinoma, leading to a final diagnose of ICC (pT3N0M0 pStage III)s. After the operation, the patient received adjuvant systemic treatment (GEMOX regimen). However, the patient suffered tumor recurrence during GEMOX treatment (Figure 3A). In parallel, we tested the PDX model with some commonly used regimens (GEMOX, XELOX, FOLFOX, sorafenib, and lenvatinib) and found that the PDX model showed much better response to lenvatinib than other regimens. Consistent with the observed treatment response in patient IMF-138 in clinical practice, the model was not very sensitive to GEMOX regimen (Figure 3B). CA19-9 level was assessed as a surrogate marker of ICC, and was observed to be increased when tumor recurrence occurred (Figure 3C). Based on PDX model drug sensitivity results, lenvatinib was administered to the patient and resulted in tumor shrinkage and stable disease until the last visit was achieved (Figure 3D).
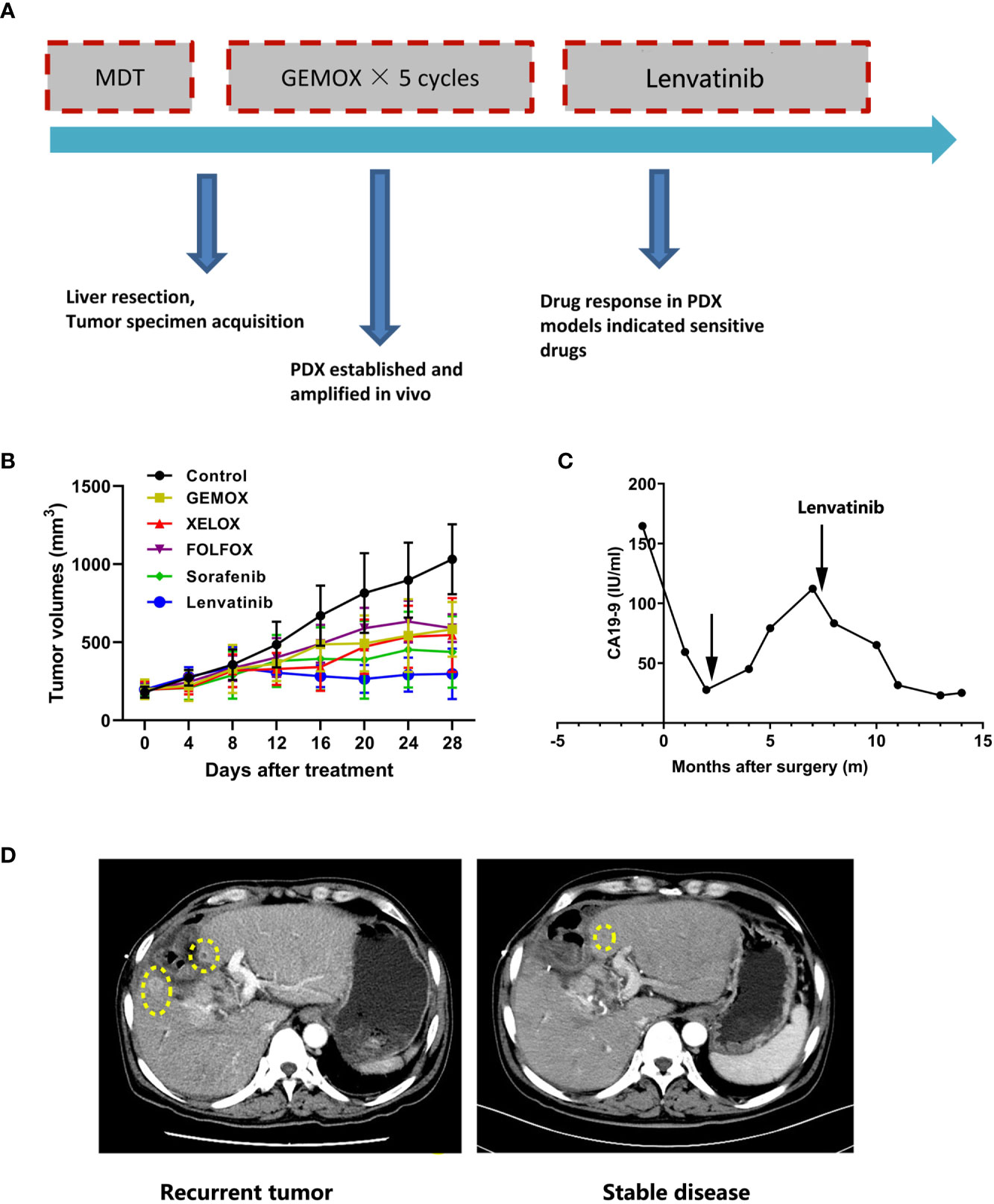
Figure 3 The PDX model guide personalized treatment for ICC patients. (A) Summary of clinical history for IMF-138; (B) PDX models showed diverse treatment response to various treatment regimens; (C) Human CA19-9 levels were observed during the clinical course; (D) The representative CT images for patient who received lenvatinib treatment.
Patient IMF-114 was initially diagnosed with malignant liver tumor and received partial liver resection plus lymphadenectomy. Pathological examination showed a poorly differentiated adenocarcinoma, leading to a final diagnose of ICC (pT3N1M1 pStage IV) (Figure 4A). The tumor tissues were subjected to whole exome sequencing (WES) and four mutations with potential clinical significance were identified: CDKN2A deletion, CDK6 amplification, PIK3CA amplification, and KRAS G12V mutation (Figure 4B). Additionally, PDX models (F2 xenograft tumor) showed a similar mutation landscape to that of the F0 tumor, further validating that the PDX model recapitulated the genomic characteristics of the original tumor. After construction of the matched PDX model, we used it to test treatment responses to the potential drugs based on WES. We found that trametinib produced the best TGI in the PDX model (Figure 4C). During follow-up, 8 months after surgery, patient IMF-114 developed intrahepatic recurrence and lung metastasis during adjuvant GEMOX regimen administration (Figures 4D, E). Based on the drug test results, we decided to try treatment with trametinib in this patient. After trametinib administration, intrahepatic metastasis was suppressed, lung metastasis was stable for 7 months, and CA19-9 decreased to normal levels (Figures 4D, E). At the time of this manuscript preparation the patient is still alive and has survived for 22 months from the date of surgery.
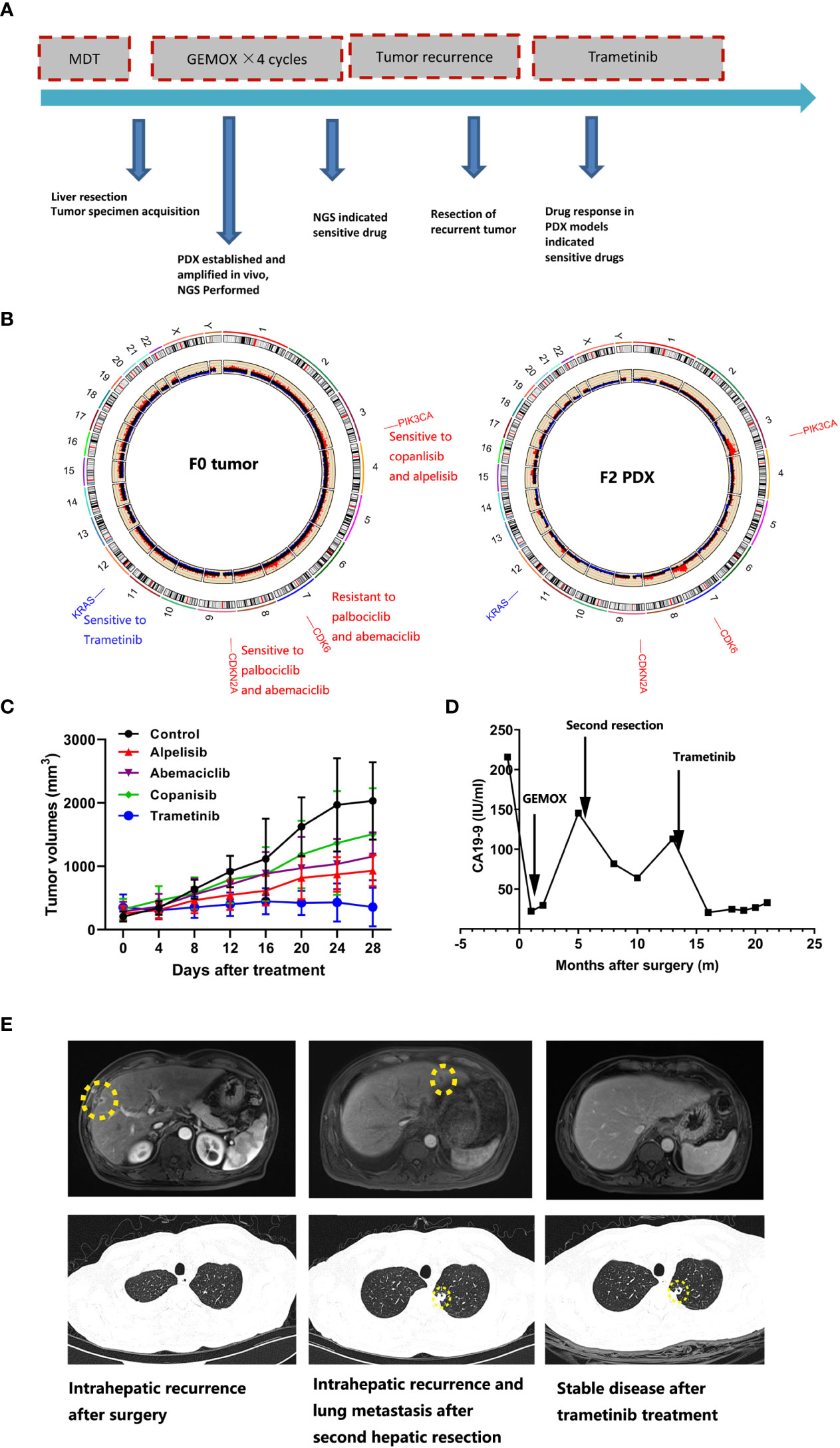
Figure 4 The PDX model combined with the whole exome sequencing data guide drug selection for ICC patients. (A) Summary of clinical history for IMF-11; (B) Whole exome sequencing identified gene mutations with potential clinical value; right figure indicated F0 tumor and left figure indicated F2 xenograft tumor; (C) PDX models showed diverse treatment response to various treatment regimens; (D) Human CA19-9 levels were observed during the clinical course; (E) The representative hepatic MRI and lung CT images for patient who received trametinib treatment.
We explored the mechanism of lenvatinib drug resistance in ICC, by testing 12 established PDX models with lenvatinib (30 mg/kg/day) for 28 days. The PDX models reflected diverse treatment responses to lenvatinib administration. Five PDX models (IMF-138, IMF-122, IMF-111, IMF-129, and IMF-243) were sensitive to lenvatinib treatment, while another four PDX models (IMF-143, IMF-134, IMF-118, and IMF-135) were resistant to the same treatment regimen (Figure 5A). These results indicate that the PDX models reflect the heterogeneity of lenvatinib response observed in patients with ICC.
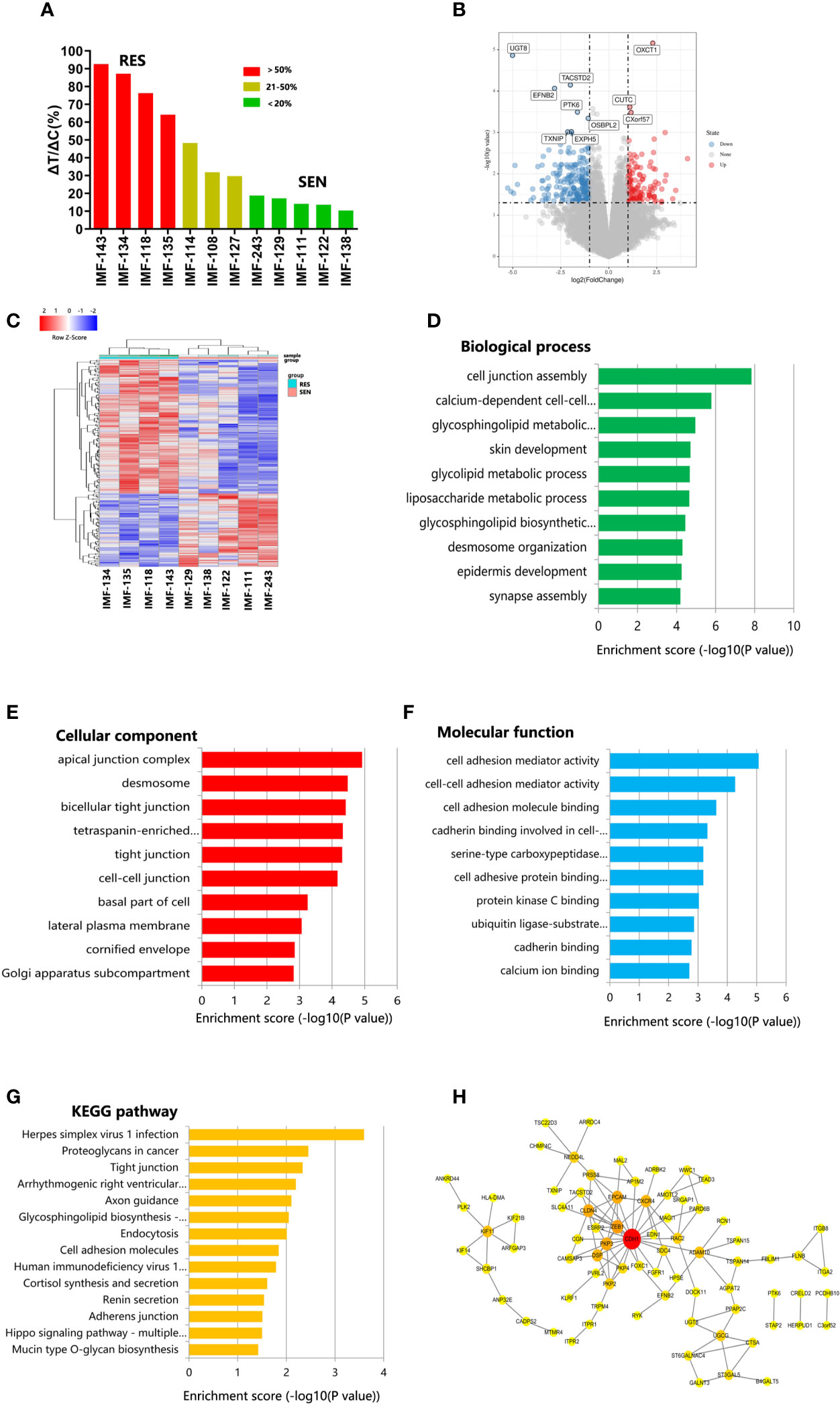
Figure 5 PDX models revealed potential mechanism of lenvatinib resistance. (A) Waterfall plot of lenvatinib response after 4 weeks of treatment in 12 cases. Resistant, stable and sensitive cases are shaded in red, yellow and green, respectively. RES, resistant; SEN, sensitive; (B) Volcano plot represents significantly differentially expressed genes in lenvatinib-sensitive group and lenvatinib-resistant. Selected top up- and downregulated genes are labeled; (C) Heat map of differentially expressed gene between lenvatinib-sensitive and lenvatinib-resistant PDXs. (D–F) GO analysis of differentially expressed gene according to biological process, cellular component and molecular function, respectively; (G) Pathway analysis based on the KEGG database; (H) Protein–protein interaction (PPI) networks for differentially expressed proteins. The gene network was constructed using Cytoscape software based on the STRING database.
To reveal the potential mechanism of lenvatinib resistance in ICC, we compared SNP and gene expression in five lenvatinib-sensitive PDXs and four lenvatinib-resistant PDXs. Using a P-value < 0.05 and fold change > 2 to compare lenvatinib-sensitive PDXs lenvatinib-resistant PDXs, we identified 64 and 117 genes that were up- and down-regulated (Figures 5B, C). Differentially expressed genes were subjected to gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) analyses. GO analysis provides information about biological processes, cellular components, and molecular function (Figures 5D–F). With respect to molecular functions, the most highly enriched GO terms in the differentially expressed transcripts included cell adhesion mediator activity, cell–cell adhesion mediator activity, cell adhesion molecule binding, cadherin binding involved in cell–cell adhesion, and serine-type carboxypeptidase activity. Biological processes were mainly enriched in cell junction assembly, calcium-dependent cell–cell adhesion via plasma membrane cell adhesion molecules, glycosphingolipid metabolic process, skin development, and glycolipid metabolic process. GO cellular components show that the differentially expressed genes were significantly located in the apical junction complex, desmosome, bicellular tight junction, tetraspanin-enriched microdomain, and tight junctions. KEGG pathway analysis of differentially expressed genes showed that they were enriched in proteoglycans in cancer, tight junctions, cell adhesion molecules, and the Hippo signaling pathway, which may harbor significance in mediating lenvatinib resistance in ICC (Figure 5G).
To better understand the role of the differentially expressed genes in lenvatinib resistance, protein–protein interaction (PPI) analysis was also performed. All DEGs were submitted to STRING 11.0 (29), and we obtained 77 PPI nodes and 117 edges. Finally, PPI analysis revealed that CDH1 potentially plays a vital role in lenvatinib resistance in ICC (Figures 5B, H).
The PDX model has become a prevalent platform for pre-clinical drug screening due to the ability to recapitulate tumor biology. Moreover, this tool has several advantages when compared to the use of conventional cells lines (30). Therefore, in the era of precision medicine, PDX models have become a valuable tool. In this study, we successfully established PDX models derived from 49 patients with ICC, with an engraftment rate of 55.1%. PDX establishment was associated with negative HBsAg, the presence of mVI, poorer tumor differentiation, multiple tumor number, presence of lymph node metastasis and later TNM stage. Patients with PDX establishment had a higher propensity to relapse and a lower survival rate after surgery. Furthermore, the established PDX models can be used to predict treatment response and to identify potential mechanisms of drug resistance. Therefore, the PDX platform will contribute to advancing precision medicine for the management of ICC.
We characterized the preserved histological and genetic features of PDXs. The results demonstrated that histological morphology and ICC markers (CK7 and CK19) were well maintained during passages. The consistency of these markers was important for evaluating the utility of selected therapeutic drugs, which represent the pathological similarity between PDXs and primary tumors (31). Noticeably, Ki 67 expression was relatively low in the PDX F2 passage, and Ki67 expression increased in following F4 and F6 passages. This may account for the phenomenon that the latency period becomes shorter during the following PDX passages (32, 33). Additionally, expression of human-derived CD34 gradually decreased, implying that human stromal elements can change as the engrafted tumors settle into their new tumor microenvironment (34). The speed and extent of the drift within stromal components remains elusive, and the impact of drug tests remain controversial. Therefore, PDX models within 10 passages are considered appropriate for drug testing in light of the preservation of parental tumor histopathological and genetic information (35, 36). Our examination of gene expression and SNP profiles revealed that the PDX models could maintain the heterogeneity of the primary tumor and the homogeneity of the patients during passage, which is consistent with results previously reported (30, 36, 37).
In this study, we found that presence of mVI, poorer tumor differentiation, multiple tumor number, presence of lymph node metastasis, and later TNM stage were associated with higher PDX establishment rate. These factors are also indicative of aggressive biological characteristics and poor outcome (5). Moreover, successful engraftment indicated reduced TTR and OS. Therefore, PDX formation from resected tumor specimens without excessive in vitro manipulation reflects the authentic tumor biology of the most aggressive tumors (34). To the best of our knowledge, this is the first study to evaluate the relationship between tumor engraftment in PDX models and clinical outcomes in ICC patients (18). PDX establishment may help identify the most malignant tumors and treatment strategies based on the evaluation of drug sensitivity in PDX models could be used to improve patient clinical outcomes.
The prerequisite step for downstream application and analysis is to obtain stable and high take rates for facilitating the broad application of PDX models. Several technical aspects of PDX establishment should be considered. In this study, tissue mince was incubated in growth factors (basic fibroblast growth factors and human epidermal growth factor) and then incubated with Matrigel™. A transplant needle was used to implant these tissues into the subcutaneous area of right flanks. We obtained a relative high take rate of 55.1%. The addition of growth factors and collagen may promote the survival of tumor cells (38, 39). Additionally, the use of transplant needles simplified implantation procedures, and reduced the failure rate caused by potential infection (40). Furthermore, we also managed to establish PDX models derived from metastatic lesions. The most commonly used tissues derived from surgical specimens and other more convenient approaches, including fine-needle aspiration and endoscopic biopsy, should be investigated to establish PDXs to expand their practical application. Recent literature reports show that PDXs have been established using gastroscopic biopsies in gastric cancer, and fine needle aspirate in primary pancreatic ductal adenocarcinoma and metastatic sites, which largely promote application of PDX engraftment in various resectable and unresectable malignancies (32, 41).
Lenvatinib has been approved as first-line treatment for advanced hepatocellular carcinoma in the US, China, Japan, and many other countries (42). As a counterpart of hepatocellular carcinoma, treatment efficacy of lenvatinib plus PD-1 antibody has been observed in a subpopulation of patients with ICC (43). In this study, gene expression and SNP data based on the lenvatinib-sensitive and resistant PDX models indicate that CDH1 is likely to regulate lenvatinib resistance. However, the specific mechanism through which this occurs requires further study. Because PDX models authentically reflect the histological and genomic characteristics of original tumors, they may represent a powerful modality to replicate patients’ treatment response and clinical efficiency. One large-scale study that included tumor samples from 1163 patients with a variety of advanced solid tumors revealed a sensitivity of 96% and specificity of 70% in drug screening (44). These results indicate that PDX models are capable of guiding oncologists to the most efficient treatments (44). For example, patients IMF-138 and IMF-114 achieved satisfactory treatment efficiency following the drug screening results of PDX models. As a functional diagnosis platform, PDX can produce convincing results when next generation sequencing results are unclear or controversial with respect to drug administration, emphasizing the role of functional diagnostic models (45).
PDX models provide an opportunity for preclinical drug-screening and personalized cancer treatment. Several obstacles should be overcome before PDX models are a routine part of clinical practices. One major challenge is time, with the average time between engraftment and drug exposure across various implanted tumors being 16 weeks. During this period, some patients may lose the opportunity to use the resultant treatment regimens, which undermines the practical value of this technique in clinical decision-making. Therefore, PDX- or primary tumor-based rapid drug screening assays should be developed (44, 46). Another challenge is that engraftment failure is still high for ICC. A take rate of at least 60%–70% is required for personalized medicine strategies, and this aspect of PDX development requires further improvement (8).
In summary, we have developed a panel of PDXs from patients with ICC that authentically recapitulate the parental tumors and demonstrate that PDX establishment can function as an independent indicator of clinical outcomes. Additionally, in vitro patient derived cell line of patient derived organoid drug screening followed by in vivo PDX validation will provide a practical framework for cancer drug discovery. Although some factors still hinder widespread clinical application, including the long timeframe for engraftment and lack of immune components, extensive research will eventually facilitate the implementation of PDXs in personalized medicine and to improve survival rates in patients with cancer.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI and accession number PRJNA729191 (https://www.ncbi.nlm.nih.gov/sra/PRJNA729191).
The studies involving human participants were reviewed and approved by Ethics Board at the Liver Cancer Institute of Zhongshan hospital, Fudan University. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Animal Care and Use Committee of Zhongshan Hospital of Fudan University.
YG, RZ, J-FH, and H-XP performed the experiments and analyzed the data. BH, P-XW, and RZ contributed reagents and materials. YG, X-RY, J-WC, and X-WH conceived and designed the experiments. WG, JZ, and JF discussed the manuscript. JF and X-RY critically reviewed the manuscript. YG contributed to drafting and revising the manuscript. All authors contributed to the article and approved the submitted version.
This study was jointly supported by the National Key R&D Program of China (2019YFC1315800, 2019YFC1315802), the State Key Program of National Natural Science of China (81830102), National Natural Science Foundation of China (81772578, 81772551, 81872355 and 82072715), the Shanghai Municipal Health Commission Collaborative Innovation Cluster Project (2019CXJQ02), Shanghai “Rising Stars of Medical Talent” Youth Development Program (Outstanding Youth Medical Talents), Zhong-shan Hospital Science Foundation (2020ZSQN87), the Projects from the Shanghai Science and Technology Commission (19441905000), Shanghai Municipal Key Clinical Specialty.
Author H-XP was employed by companies Shanghai Dunwill Medical Technology Co., Ltd and Shanghai Epione Medlab Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.704042/full#supplementary-material
1. Dodson RM, Weiss MJ, Cosgrove D, Herman JM, Kamel I, Anders R, et al. Intrahepatic Cholangiocarcinoma: Management Options and Emerging Therapies. J Am Coll Surg (2013) 217:736–50.e734. doi: 10.1016/j.jamcollsurg.2013.05.021
2. Ross JS, Wang K, Gay L, Al-Rohil R, Rand JV, Jones DM, et al. New Routes to Targeted Therapy of Intrahepatic Cholangiocarcinomas Revealed by Next-Generation Sequencing. Oncologist (2014) 19:235–42. doi: 10.1634/theoncologist.2013-0352
3. Saha SK, Zhu AX, Fuchs CS, Brooks GA. Forty-Year Trends in Cholangiocarcinoma Incidence in the U.S.: Intrahepatic Disease on the Rise. Oncologist (2016) 21:594–9. doi: 10.1634/theoncologist.2015-0446
4. Aljiffry M, Abdulelah A, Walsh M, Peltekian K, Alwayn I, Molinari M. Evidence-Based Approach to Cholangiocarcinoma: A Systematic Review of the Current Literature. J Am Coll Surg (2009) 208:134–47. doi: 10.1016/j.jamcollsurg.2008.09.007
5. Banales JM, Marin JJG, Lamarca A, Rodrigues PM, Khan SA, Roberts LR, et al. Cholangiocarcinoma 2020: The Next Horizon in Mechanisms and Management. Nat Rev Gastroenterol Hepatol (2020) 17:557–88. doi: 10.1038/s41575-020-0310-z
6. Sirica AE, Gores GJ, Groopman JD, Selaru FM, Strazzabosco M, Wei Wang X, et al. Intrahepatic Cholangiocarcinoma: Continuing Challenges and Translational Advances. Hepatology (2019) 69:1803–15. doi: 10.1002/hep.30289
7. Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-Derived Tumour Xenografts as Models for Oncology Drug Development. Nat Rev Clin Oncol (2012) 9:338–50. doi: 10.1038/nrclinonc.2012.61
8. Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, et al. Patient-Derived Xenograft Models: An Emerging Platform for Translational Cancer Research. Cancer Discov (2014) 4:998–1013. doi: 10.1158/2159-8290.CD-14-0001
9. Gu Q, Zhang B, Sun H, Xu Q, Tan Y, Wang G, et al. Genomic Characterization of a Large Panel of Patient-Derived Hepatocellular Carcinoma Xenograft Tumor Models for Preclinical Development. Oncotarget (2015) 6:20160–76. doi: 10.18632/oncotarget.3969
10. Hill SJ, Decker B, Roberts EA, Horowitz NS, Muto MG, Worley MJ Jr., et al. Prediction of DNA Repair Inhibitor Response in Short-Term Patient-Derived Ovarian Cancer Organoids. Cancer Discov (2018) 8:1404–21. doi: 10.1158/2159-8290.CD-18-0474
11. Roy V, Magne B, Vaillancourt-Audet M, Blais M, Chabaud S, Grammond E, et al. Human Organ-Specific 3D Cancer Models Produced by the Stromal Self-Assembly Method of Tissue Engineering for the Study of Solid Tumors. BioMed Res Int (2020) 2020:6051210. doi: 10.1155/2020/6051210
12. Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarro LM, Bradshaw CR, et al. Human Primary Liver Cancer-Derived Organoid Cultures for Disease Modeling and Drug Screening. Nat Med (2017) 23:1424–35. doi: 10.1038/nm.4438
13. Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-Throughput Screening Using Patient-Derived Tumor Xenografts to Predict Clinical Trial Drug Response. Nat Med (2015) 21:1318–25. doi: 10.1038/nm.3954
14. Byrne AT, Alferez DG, Amant F, Annibali D, Arribas J, Biankin AV, et al. Interrogating Open Issues in Cancer Precision Medicine With Patient-Derived Xenografts. Nat Rev Cancer (2017) 17:254–68. doi: 10.1038/nrc.2016.140
15. Hu B, Li H, Guo W, Sun YF, Zhang X, Tang WG, et al. Establishment of a Hepatocellular Carcinoma Patient-Derived Xenograft Platform and Its Application in Biomarker Identification. Int J Cancer (2020) 146:1606–17. doi: 10.1002/ijc.32564
16. He S, Hu B, Li C, Lin P, Tang WG, Sun YF, et al. PDXliver: A Database of Liver Cancer Patient Derived Xenograft Mouse Models. BMC Cancer (2018) 18:550. doi: 10.1186/s12885-018-4459-6
17. Jiang TY, Pan YF, Wan ZH, Lin YK, Zhu B, Yuan ZG, et al. PTEN Status Determines Chemosensitivity to Proteasome Inhibition in Cholangiocarcinoma. Sci Transl Med (2020) 12:1–18. doi: 10.1126/scitranslmed.aay0152
18. Cavalloni G, Peraldo-Neia C, Sassi F, Chiorino G, Sarotto I, Aglietta M, et al. Establishment of a Patient-Derived Intrahepatic Cholangiocarcinoma Xenograft Model With KRAS Mutation. BMC Cancer (2016) 16:90. doi: 10.1186/s12885-016-2136-1
19. Muller SA, Mehrabi A, Rahbari NN, Warschkow R, Elbers H, Leowardi C, et al. Allogeneic Blood Transfusion Does Not Affect Outcome After Curative Resection for Advanced Cholangiocarcinoma. Ann Surg Oncol (2014) 21:155–64. doi: 10.1245/s10434-013-3226-9
20. Byrd DR, Carducci MA, Compton CC, Fritz AG, Greene FL. AJCC Cancer Staging Manual. New York: Springer (2010). p. 201.
21. Lee MJ, Lee HS, Kim WH, Choi Y, Yang M. Expression of Mucins and Cytokeratins in Primary Carcinomas of the Digestive System. Mod Pathol (2003) 16:403–10. doi: 10.1097/01.MP.0000067683.84284.66
22. Pusztaszeri MP, Seelentag W, Bosman FT. Immunohistochemical Expression of Endothelial Markers CD31, CD34, Von Willebrand Factor, and Fli-1 in Normal Human Tissues. J Histochem Cytochem (2006) 54:385–95. doi: 10.1369/jhc.4A6514.2005
23. Leung SCY, Nielsen TO, Zabaglo L, Arun I, Badve SS, Bane AL, et al. Analytical Validation of a Standardized Scoring Protocol for Ki67: Phase 3 of an International Multicenter Collaboration. NPJ Breast Cancer (2016) 2:16014. doi: 10.1038/npjbcancer.2016.14
24. Yang XR, Xu Y, Shi GM, Fan J, Zhou J, Ji Y, et al. Cytokeratin 10 and Cytokeratin 19: Predictive Markers for Poor Prognosis in Hepatocellular Carcinoma Patients After Curative Resection. Clin Cancer Res (2008) 14:3850–9. doi: 10.1158/1078-0432.CCR-07-4338
25. Wang K, Li M, Hakonarson H. ANNOVAR: Functional Annotation of Genetic Variants From High-Throughput Sequencing Data. Nucleic Acids Res (2010) 38:e164. doi: 10.1093/nar/gkq603
26. Mohseni MJ, Amanpour S, Muhammadnejad S, Sabetkish S, Muhammadnejad A, Heidari R, et al. Establishment of a Patient-Derived Wilms’ Tumor Xenograft Model: A Promising Tool for Individualized Cancer Therapy. J Pediatr Urol (2014) 10:123–9. doi: 10.1016/j.jpurol.2013.07.009
27. Teicher BA, Paul AA. Anticancer Drug Development Guide: Preclinical Screening, Clinical Trials, and Approval. New York: Humana Press, Totowa (2004) p. 125–52. doi: 10.1007/978-1-59259-739-0
28. Smyth EC, Fassan M, Cunningham D, Allum WH, Okines AF, Lampis A, et al. Effect of Pathologic Tumor Response and Nodal Status on Survival in the Medical Research Council Adjuvant Gastric Infusional Chemotherapy Trial. J Clin Oncol (2016) 34:2721–7. doi: 10.1200/JCO.2015.65.7692
29. West NP, Watts AM, Smith PK, Zhang P, Besseling-van der Vaart I, Cripps AW, et al. Digital Immune Gene Expression Profiling Discriminates Allergic Rhinitis Responders From Non-Responders to Probiotic Supplementation. Genes (Basel) (2019) 10:1–7. doi: 10.3390/genes10110889
30. Cassidy JW, Caldas C, Bruna A. Maintaining Tumor Heterogeneity in Patient-Derived Tumor Xenografts. Cancer Res (2015) 75:2963–8. doi: 10.1158/0008-5472.CAN-15-0727
31. Zhang T, Zhang L, Fan S, Zhang M, Fu H, Liu Y, et al. Patient-Derived Gastric Carcinoma Xenograft Mouse Models Faithfully Represent Human Tumor Molecular Diversity. PloS One (2015) 10:e0134493. doi: 10.1371/journal.pone.0134493
32. Zhu Y, Tian T, Li Z, Tang Z, Wang L, Wu J, et al. Establishment and Characterization of Patient-Derived Tumor Xenograft Using Gastroscopic Biopsies in Gastric Cancer. Sci Rep (2015) 5:8542. doi: 10.1038/srep08542
33. Pearson AT, Finkel KA, Warner KA, Nor F, Tice D, Martins MD, et al. Patient-Derived Xenograft (PDX) Tumors Increase Growth Rate With Time. Oncotarget (2016) 7:7993–8005. doi: 10.18632/oncotarget.6919
34. DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, et al. Tumor Grafts Derived From Women With Breast Cancer Authentically Reflect Tumor Pathology, Growth, Metastasis and Disease Outcomes. Nat Med (2011) 17:1514–20. doi: 10.1038/nm.2454
35. Izumchenko E, Meir J, Bedi A, Wysocki PT, Hoque MO, Sidransky D. Patient-Derived Xenografts as Tools in Pharmaceutical Development. Clin Pharmacol Ther (2016) 99:612–21. doi: 10.1002/cpt.354
36. Pompili L, Porru M, Caruso C, Biroccio A, Leonetti C. Patient-Derived Xenografts: A Relevant Preclinical Model for Drug Development. J Exp Clin Cancer Res (2016) 35:189. doi: 10.1186/s13046-016-0462-4
37. Dobbin ZC, Katre AA, Steg AD, Erickson BK, Shah MM, Alvarez RD, et al. Using Heterogeneity of the Patient-Derived Xenograft Model to Identify the Chemoresistant Population in Ovarian Cancer. Oncotarget (2014) 5:8750–64. doi: 10.18632/oncotarget.2373
38. Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor Stem Cells Derived From Glioblastomas Cultured in bFGF and EGF More Closely Mirror the Phenotype and Genotype of Primary Tumors Than do Serum-Cultured Cell Lines. Cancer Cell (2006) 9:391–403. doi: 10.1016/j.ccr.2006.03.030
39. Aparicio S, Hidalgo M, Kung AL. Examining the Utility of Patient-Derived Xenograft Mouse Models. Nat Rev Cancer (2015) 15:311–6. doi: 10.1038/nrc3944
40. Chijiwa T, Kawai K, Noguchi A, Sato H, Hayashi A, Cho H, et al. Establishment of Patient-Derived Cancer Xenografts in Immunodeficient NOG Mice. Int J Oncol (2015) 47:61–70. doi: 10.3892/ijo.2015.2997
41. Allaway RJ, Fischer DA, de Abreu FB, Gardner TB, Gordon SR, Barth RJ, et al. Genomic Characterization of Patient-Derived Xenograft Models Established From Fine Needle Aspirate Biopsies of a Primary Pancreatic Ductal Adenocarcinoma and From Patient-Matched Metastatic Sites. Oncotarget (2016) 7:17087–102. doi: 10.18632/oncotarget.7718
42. Zhao Y, Zhang YN, Wang KT, Chen L. Lenvatinib for Hepatocellular Carcinoma: From Preclinical Mechanisms to Anti-Cancer Therapy. Biochim Biophys Acta Rev Cancer (2020) 1874:188391. doi: 10.1016/j.bbcan.2020.188391
43. Chen WX, Li GX, Hu ZN, Zhu P, Zhang BX, Ding ZY. Significant Response to Anti-PD-1 Based Immunotherapy Plus Lenvatinib for Recurrent Intrahepatic Cholangiocarcinoma With Bone Metastasis: A Case Report and Literature Review. Med (Baltimore) (2019) 98:e17832. doi: 10.1097/MD.0000000000017832
44. Izumchenko E, Paz K, Ciznadija D, Sloma I, Katz A, Vasquez-Dunddel D, et al. Patient-Derived Xenografts Effectively Capture Responses to Oncology Therapy in a Heterogeneous Cohort of Patients With Solid Tumors. Ann Oncol (2017) 28:2595–605. doi: 10.1093/annonc/mdx416
45. Letai A. Functional Precision Cancer Medicine-Moving Beyond Pure Genomics. Nat Med (2017) 23:1028–35. doi: 10.1038/nm.4389
Keywords: patient derived xenograft, intrahepatic cholangiocarcinoma, lenvatinib, drug resistance, personalized medicine
Citation: Gao Y, Zhou R, Huang J-F, Hu B, Cheng J-W, Huang X-W, Wang P-X, Peng H-X, Guo W, Zhou J, Fan J and Yang X-R (2021) Patient-Derived Xenograft Models for Intrahepatic Cholangiocarcinoma and Their Application in Guiding Personalized Medicine. Front. Oncol. 11:704042. doi: 10.3389/fonc.2021.704042
Received: 01 May 2021; Accepted: 25 June 2021;
Published: 13 July 2021.
Edited by:
Francesco Giuseppe Carbone, Azienda Provinciale per i Servizi Sanitari (APSS), ItalyReviewed by:
Monique M.A. Verstegen, Erasmus Medical Center, NetherlandsCopyright © 2021 Gao, Zhou, Huang, Hu, Cheng, Huang, Wang, Peng, Guo, Zhou, Fan and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-Rong Yang, eWFuZy54aW5yb25nQHpzLWhvc3BpdGFsLnNoLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.