
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 25 May 2021
Sec. Molecular and Cellular Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.690800
This article is part of the Research TopicEpigenetic Reprogramming and Cancer Development Volume IIView all 5 articles
Hepatocellular carcinoma (HCC) is a type of primary liver cancer with a high incidence and mortality rate. HCC develops insidiously, and most newly diagnosed cases are in the middle and advanced stages. The epithelial-mesenchymal transition (EMT) is a vital mechanism underlying metastasis in patients with advanced HCC. EMT is a multistep and complex procedure. The promotion and inhibition of EMT directly affect the migration and invasion of HCC. LncRNAs are involved in the epigenetic modification of genes, regulation of gene transcription, and posttranslational modification of proteins. LncRNAs also play important roles in regulating EMT progression in HCC and are promising biomarkers and therapeutic targets. This review focused on summarizing the mechanism by which lncRNAs regulate EMT in HCC. In particular, lncRNAs were reported to primarily act as RNA sponges, and the regulation of EMT involves major signaling pathways. Finally, we reviewed the mechanisms by which lncRNAs are involved in drug resistance and discussed the clinical prospects and potential challenges of utilizing lncRNAs to treat HCC.
Primary liver cancer was the sixth-highest morbidity and third-highest cancer-related mortality worldwide, with 905,677 (4.7% morbidity) new cases and 830,180 (8.3% mortality) deaths according to the Global Cancer Observatory (GCO) (https://gco.iarc.fr/) (1) in 2020. Hepatocellular carcinoma (HCC) is the most common primary liver cancer, accounting for 75-85% of cases (2). The low survival rate of HCC is due to the asymptomatic onset of HCC in the early stage and the loss of the optimal treatment time when it is newly diagnosed in the middle and late stages (3). Middle- and advanced-stage HCC is prone to metastasis, primarily intrahepatic metastasis, which is also a major reason for the poor effect of local treatment or systemic treatment. Epithelial–mesenchymal transition (EMT) plays a vital role in the development, invasion, and metastasis of growing tumors, including HCC (4). Consequently, elucidating the molecular mechanism underlying the role played by EMT in the progression of HCC may contribute to the diagnosis and therapy of cancer and improve the survival of patients.
EMT is a dynamic transformation process driven by induced signals, transcriptional regulators, and downstream effectors (5). Long noncoding RNA (lncRNA), as a regulatory factor, plays an important role in the process of EMT in tumor tissues, which has been revealed in prior research (6). However, there has been no systematic report on the role of lncRNAs in the process of EMT in HCC. This review discusses and summarizes recent work on the mechanism by which lncRNAs affect EMT in HCC.
EMT refers to the morphological transformation of epithelial cells into fibroblasts or mesenchymal cells, resulting in the loss of cell polarity, cytoskeletal rearrangement, and increased mobility. Multiple steps, reversibility, plasticity, and heterogeneity are the main characteristics of EMT (7, 8). From the embryonic stage to death, a variety of physiological and pathological EMTs occur in the body (5, 9). EMT is also involved in the pathogenesis of tumorigenesis, progression, metastasis, and drug resistance, which is a kind of pathological EMT (10).
The process of EMT involves downregulation of the epithelial cell phenotype and upregulation of the mesenchymal cell phenotype (Figure 1). These altered phenotypes are commonly used as molecular markers for EMT; these markers can be divided into epithelioid cell markers and mesenchymal cell markers. Epithelioid cell markers include E-cadherin, β-catenin, claudin-1, and zona occludens 1 (ZO-1), among which E-cadherin is a Ca2+-dependent transmembrane glycoprotein closely related to intercellular adhesion that is mainly distributed on the membrane surface of epithelial cells and is a typical epithelial cell marker. Markers of mesenchymal-like cells include vimentin, N-cadherin, snail1/2, twist1/2, zinc finger E-box-binding homeobox 1 and 2 (ZEB1/2), and α-smooth muscle actin (α-SMA), among which vimentin is mainly expressed in mesenchymal-origin cells, such as endothelial cells, and is considered an important marker of mesenchymal cells (11, 12). Besides, Müller et al. illustrated a key role of iron as a rate-limiting regulator of epigenetic plasticity during epithelial-mesenchymal transition (13).
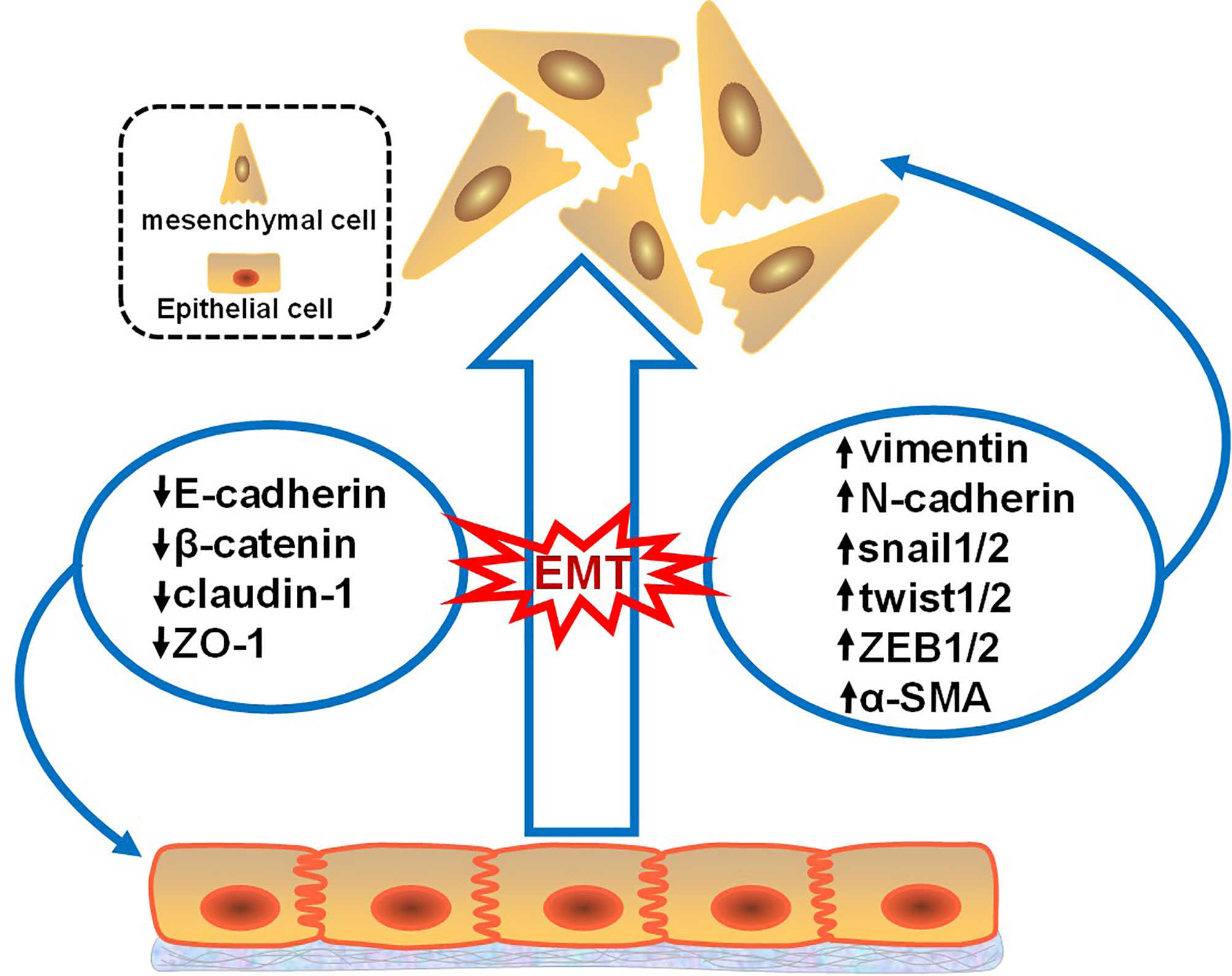
Figure 1 Changes in the EMT markers. This figure illustrates that when tumor epithelial cells develop EMT, epithelial markers decrease and mesenchymal markers increase. Epithelioid cell markers include E-cadherin, β-catenin, claudin-1, and zona occludens 1 (ZO-1). Mesenchymal cell markers include vimentin, N-cadherin, snail1/2, twist1/2, zinc finger E-box-binding homeobox 1 and 2 (ZEB1/2), and α-smooth muscle actin (α-SMA).
Transdifferentiation of epithelial cells is performed by EMT-activating transcription factors (EMT-TFs), mainly snail1/2, twist1/2, and ZEB1/2 (11). As mentioned above, EMT-TFs are also markers of mesenchymal-like cells, but they are a special category. EMT-TFs can bind to the E-box proximal to the E-cadherin gene promoter and inhibit the expression of E-cadherin, thereby inducing the EMT phenomenon in cells (14). EMT-TFs can be induced by certain signaling pathways, such as the Wnt/β-catenin signaling pathway, JAK2/STAT3 signaling pathway, MEK/ERK signaling pathway, and PI3K/AKT/mTOR signaling pathway (15). Increasing evidence shows that lncRNAs play a vital role in the regulation of EMT signaling pathways in tumor tissue. For example, lncRNA NR027113 promotes the process of EMT in gastric cancer through the PI3K/AKT signaling pathway (16).
LncRNAs, which measure over 200 kb in length, are transcribed by polymerase 2 and have a transcription process similar to that of mRNAs. Previously believed to be merely transcriptional noise, lncRNAs have been increasingly recognized to play critical roles in cellular processes (17). LncRNAs are generally considered functional regulatory factors, but recent studies have shown that lncRNAs can encode certain small peptides and thus play biological roles in a subset of tissues, including tumors (18). According to the position of lncRNAs in the genome relative to protein-coding genes, lncRNAs can be divided into five categories (Figure 2A), including 1) sense lncRNAs overlapping with coding mRNAs on the coding DNA strand, 2) antisense lncRNAs overlapping with coding mRNAs on the noncoding DNA strand, 3) bidirectional lncRNAs sharing their transcription initiation sites with noncoding DNA on the contrary strand, 4) intronic lncRNAs transcribed from an intronic region of coding DNA, and 5) intergenic lncRNAs located between coding DNA sites (19). However, as an increasing number of functions of lncRNAs have been discovered, lncRNAs have been classified in another way according to their functions (Figure 2B), including 1) decoy lncRNAs, which sequester proteins to prevent them from binding with target genes; 2) guide lncRNAs, which recruit chromatin remodeling agents to precise gene loci; 3) scaffold lncRNAs, which facilitate protein complex formation; 4) stabilizing lncRNAs, which bind target mRNA transcripts to stabilize them and to promote their translation; and 5) competitive endogenous-lncRNAs (ceRNAs), or RNA sponges, which bind and sequester miRNAs to restrict their effects on mRNA targets (20, 21).
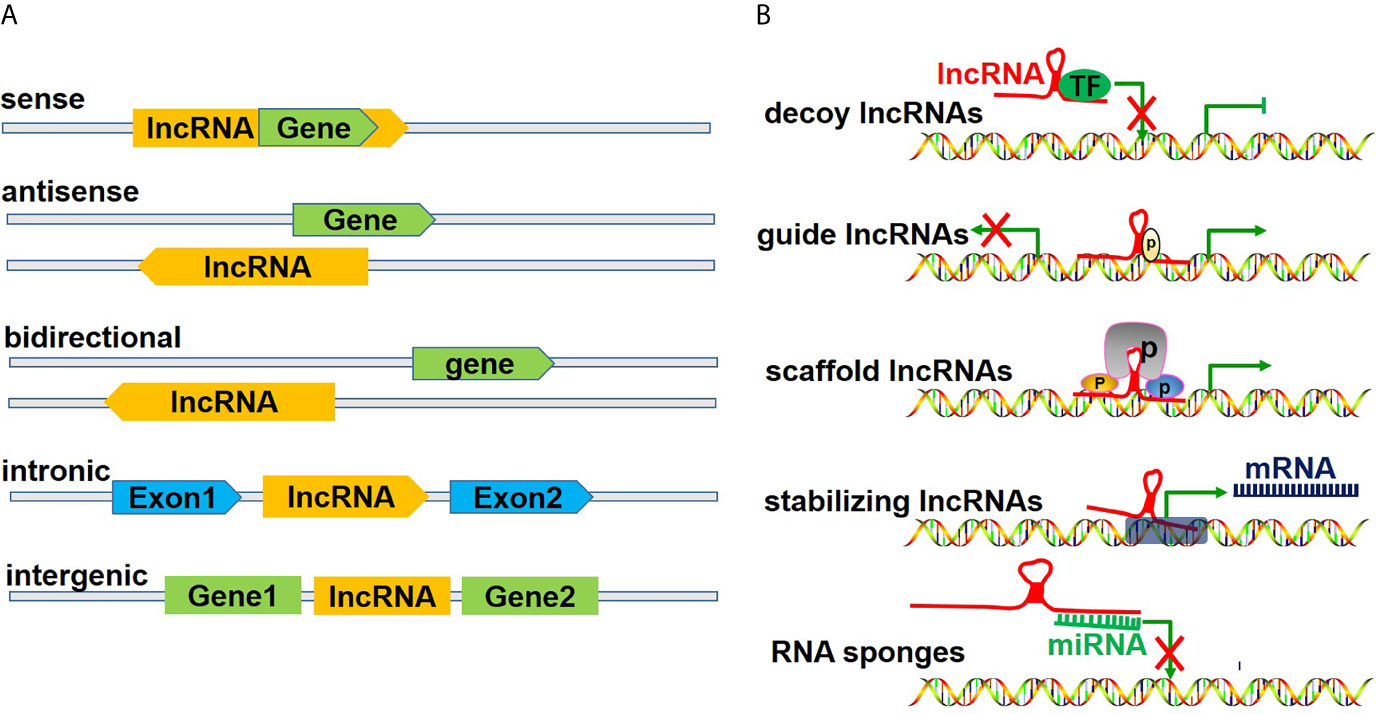
Figure 2 Classification of lncRNAs. (A) illustrates the classification of lncRNAs according to the position of lncRNAs in the genome relative to the coding protein gene, including 1) sense lncRNAs, overlapping with coding mRNAs on the coding DNA strand, 2) ant-sense lncRNAs, overlapping with coding mRNAs on the noncoding DNA strand, 3) bidirectional lncRNAs, sharing their transcription initiation sites with noncoding DNA on the contrary strand, 4) intronic lncRNAs, being transcribed from an intronic region of coding DNA, and 5) intergenic lncRNAs, being located between coding DNA. (B) shows the classification of lncRNAs according to their functions, including 1) decoy lncRNAs, sequestering proteins to restrict combination with target genes, 2) guide lncRNAs, recruiting chromatin remodeling agents to precise gene loci, 3) scaffold lncRNAs, facilitating protein complex formation, 4) stabilizing lncRNAs, binding target mRNA transcripts and stabilizing and promoting their translation, and 5) competitive endogenous lncRNAs (ceRNAs) or RNA sponges, binding and sequestering miRNAs to restrict their impact on mRNA targets.
LncRNAs are abnormally expressed in tumor cells and play an important regulatory role in tumor proliferation in such cancers as hepatocellular carcinoma (22), breast cancer (23), and gastric cancer (24). In addition, a large number of recent reviews have discussed the role and significance of lncRNAs in the EMT of tumors (6, 20, 25). However, to the best of our knowledge, there has not been a review on the role of lncRNAs in EMT in HCC.
Accumulating evidence demonstrates that EMT is a crucial mechanism and link in the development and progression of HCC, especially in the process of migration and invasion (26–28). Because of the stimulation of the tumor microenvironment, the epithelial phenotypes of HCC cells acquire the ability to migrate and invade, facilitating the localization of cancer cells to other tissues and organs or other regions within tissues. This process is often referred to as the epithelial-mesenchymal transition, or EMT. In this process, a series of complex reactions are caused, leading to an increase in EMT-TFs. Upregulated EMT-TFs inhibit the expression of epithelial cell markers and promote the expression of mesenchymal cells. In HCC cells, the silencing or deletion of the common epithelial cell markers E-cadherin and ZO-1 results in the loss of adhesion of epithelial cells and a decrease in cell-cell junction tightness, which can easily leave epithelial cells in a free state. Common markers of mesenchymal cells are N-cadherin and vimentin, among which N-cadherin is transformed from E-cadherin, which can increase the motility of cells and the ability of invasion and metastasis. Additionally, snail1/2, twist1/2, and zeb1/2 are biomarkers of mesenchymal cells and also pivotal transcription factors. In the process of transition of tumor cells, a subset of epithelial cells may not be completely transformed into mesenchymal cells. There are both epithelial and mesenchymal cell markers in the cells, which exhibit an intermediate state, or partial EMT. When transformed epithelial cells migrate to other areas due to their mesenchymal ability, they are stimulated by the local environment, which triggers transformed epithelial cells to re-transform into epithelial cells, thereby facilitating cell proliferation and permanently fixing cancer cells. This process is usually called mesenchymal-epithelial transition (MET) (29–31).
With the development of next-generation sequencing technology (NGS), especially transcriptome sequencing technology (RNA-Seq), a large number of lncRNAs have been identified and determined to be involved in the epigenetic modification of genes, posttranscriptional regulation, protein translation, and modification (21, 32, 33). Since the EMT process involves gene expression and modification, as well as transcription regulation, experts and scholars worldwide are devoting increasing attention to the role of lncRNAs as important regulatory factors in this process. LncRNAs ultimately enhance (34, 35) or inhibit (27, 36) the EMT process by directly or indirectly targeting EMT-TFs or EMT markers. As mentioned above, there are five functional categories of lncRNAs, and the most typical category observed in EMT is RNA sponges or competitive endogenous RNA (ceRNA) (Table 1), which can competitively bind with miRNAs and upregulate the expression of target genes to regulate EMT. In addition, lncRNAs may regulate the EMT process through other functional mechanisms, but reports on this topic are relatively scarce, and further research is warranted.
Notably, the same lncRNA may play a role in EMT through multiple mechanisms. Overexpression of lncRNA H19 could promote the process of EMT by three pathways: 1) the H19/miRNA-22/EMT axis (44); 2) the H19/miR-15b/CDC42/PAK1 axis (45); and 3) the H19/miR-193b/MAPK1 (mitogen-activated protein kinase 1) axis (46). LncRNA H19 acted as an RNA sponge in these three pathways. In addition, Li et al. demonstrated (55) that overexpression of long stress-induced noncoding transcripts 5 (LSINCT5) could promote the EMT process through two functional mechanisms: 1) LSINCT5, as an RNA sponge, promoted EMT by regulating the LSINCT5/miR-4516/STAT3/Bclxl axis; and 2) LSINCT5, as a stabilizing lncRNA, promoted EMT via the LSINCT5/HMGA2 (high-mobility group AT-hook 2) axis.
Taken together, these results indicated that lncRNAs exert their functions through lncRNA-gene interactions, lncRNA-miRNA interactions, lncRNA-protein interactions, and mRNA stabilization (56). More importantly, lncRNAs also require the participation of signaling pathways.
The positive feedback loop is a special signaling pathway, accurately described as the signal loop, which refers to the cascading reaction of the signal factors and, finally, the activation of the original signal factors to achieve a self-amplifying effect. The formation of a positive feedback loop is conducive to the proliferation, differentiation, migration, and invasion of tumors independent of the external environment, which is a particularly important mechanism for the occurrence and development of tumors. There are positive feedback loops in a wide variety of tumors, such as bladder cancer (57), breast cancer (58), and lung cancer (59). Positive feedback pathways related to lncRNA regulation of EMT in HCC have also been described.
LncRNAs can enhance the EMT process through a positive feedback loop (Figure 3A). Lin et al. illustrated (47) that the disc large (Drosophila) homolog-associated protein 1 antisense RNA 1 (DLGAP1-AS1) was upregulated in HCC cell lines and promoted the progression of EMT. The regulatory mechanism was divided into two parts. First, DLGAP1-AS1 was an RNA sponge to sequester the HCC-inhibitory miRNAs miR-26a-5p and miR-26b-5p, thereby raising the level of the cytokine interleukin 6 (IL-6). IL-6 stimulated the JAK2/STAT3 signaling pathway and conversely promoted the transcription of DLGAP1-AS1, thereby forming a positive feedback loop. Additionally, DLGAP1-AS1 activated the Wnt/β-catenin pathway by upregulating cyclin-dependent kinase 8 (CDK8) and lipoprotein receptor-related protein 6 (LRP6), downstream elements of miR-26a/b-5p. Lin et al. confirmed (60) that HCCL5, a cytoplasmic lncRNA, accelerated the EMT phenotype by upregulating the expression of transcription factors Snail, Slug, ZEB1, and Twist1 and, in turn, was transcriptionally driven by ZEB1 via a superenhancer. In combination, these processes formed a positive feedback loop. Nevertheless, lncRNAs could suppress the EMT process through a positive feedback loop (Figure 3A). LncRNA neighboring enhancer of FOXA2 (lncRNA-NEF), a novel lncRNA, was expressed at low levels in HCC. Liang et al. indicated (61) that lncRNA-NEF was transcriptionally activated by EMT suppressor Forkhead box protein A2 (FOXA2) and, in turn, activated its neighboring gene FOXA2. Moreover, lncRNA-NEF interacted with β-catenin to increase the binding of glycogen synthase kinase-3β (GSK3β) with β-catenin and thus promoted the inhibitory phosphorylation of β-catenin, thereby leading to the suppression of Wnt/β-catenin signaling to suppress EMT.
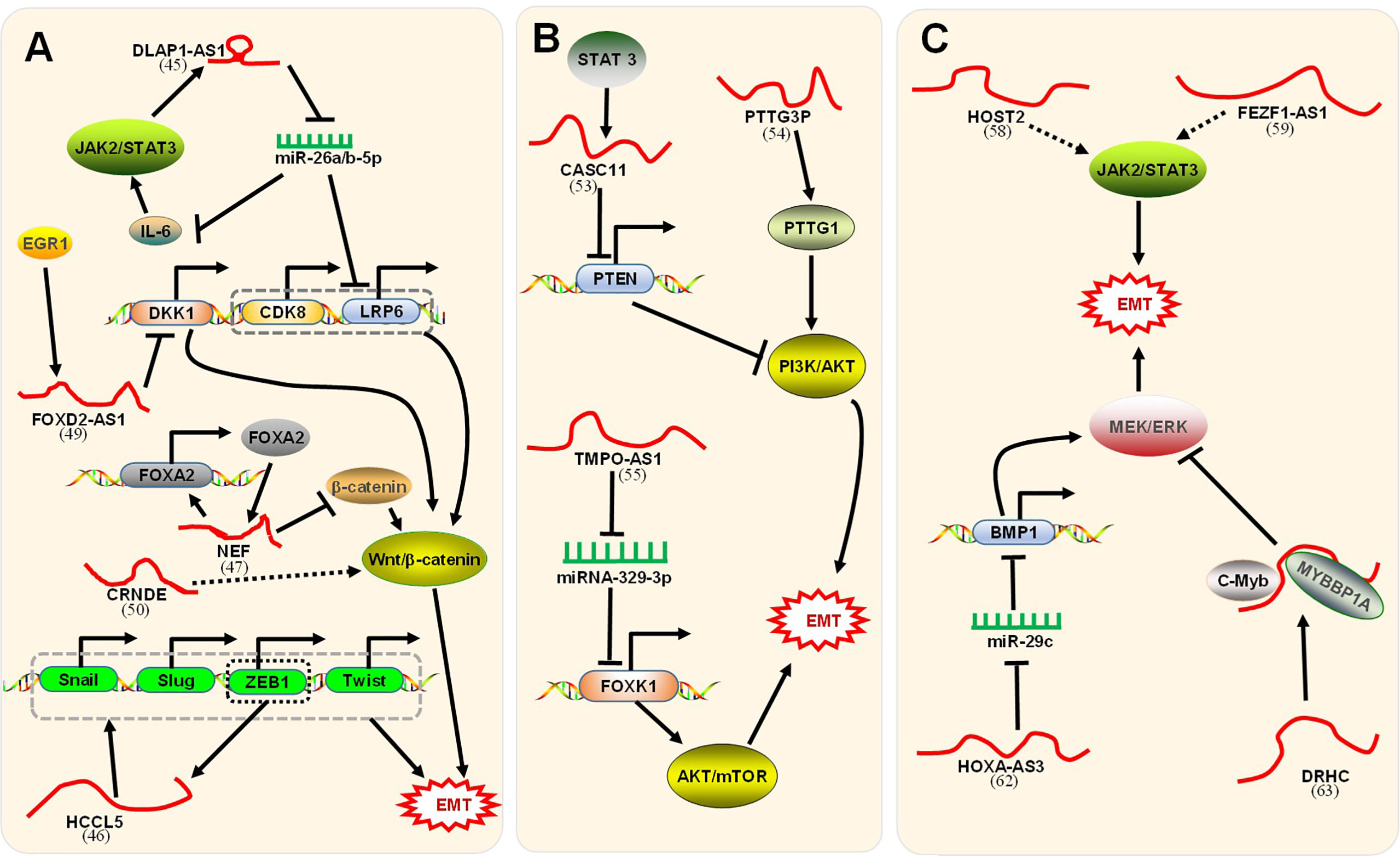
Figure 3 Signaling pathways associated with lncRNAs regulation of EMT. (A) The positive feedback loop and Wnt/β-catenin signaling pathway. First, DLGAP1-AS1 (47) promoted EMT through a positive feedback loop. Mechanistically, 1) DLGAP1-AS1 sponged and sequestered miR-26a-5p and miR-26b-5p to upregulate IL-6. IL-6 stimulated the JAK2/STAT3 signaling pathway, which conversely promoted DLGAP1-AS1. 2) DLGAP1-AS1 activated the Wnt/β-catenin pathway by upregulating the downstream CDK8 and LRP6 of miR-26a/b-5p. Second, HCCL5 (60) accelerated EMT by upregulating the expression of Snail, Slug, ZEB1, and Twist1. In turn, HCCL5 was transcriptionally driven by ZEB1. Third, lncRNA-NEF (61) was transcriptionally activated by FOXA2 and, in turn, activated its neighboring gene FOXA2. LncRNA-NEF interacted with β-catenin to increase the binding of GSK3β with β-catenin, leading to the suppression of Wnt/β-catenin signaling. Fourth, lncRNA FOXD2-AS1 (62) induced by EGR1 promoted the EMT process by binding with EZH2 to epigenetically silence the DKK1 gene and activating the Wnt/β-catenin axis. Fifth, CRNDE-promoted Wnt/β-catenin signaling pathway activity was inhibited by CRNDE (63) knockdown. Unfortunately, the paper did not clarify how CRNDE reduces the level of signaling pathway-related proteins. (B) The PI3K/AKT/mTOR signaling pathway. First, STAT3-induced lncRNA CASC11 (64) promotes EMT by binding with the enhancer of EZH2 to epigenetically silence PTEN and activate the PI3K/AKT axis. Second, lncRNA PTTG3P (65) promotes EMT by upregulating PTTG1 and activating the PI3K/AKT axis. Third, TMPO-AS1 (42) upregulated the oncogene FOXK1 by sponging miR-329-3p to activate the AKT/mTOR signaling pathway. (C) The JAK2/STAT3 signaling pathway and MEK/ERK signaling pathway. First, HOST2 (66) enhanced EMT by upregulating the JAK2-STAT3 signaling pathway. Second, FEZF1-AS1 (67) increased EMT via the JAK2/STAT3 signaling pathway. Third, HOXA-AS3 (68) upregulated BMP1 by sponging miR-29c, thus activating the MEK/ERK signaling pathway to enhance EMT. Fourth, the DRHC/MYBBP1A/C-Myb (69) complex with C-Myb inhibited the MEK/ERK signaling pathway and finally inhibited EMT. The corresponding references were also indicated under the lncRNA names on the figure.
The Wnt/β-catenin signaling pathway refers to the activation of Wnt ligands combined with Frizzled receptors, which triggers a series of signaling reactions and leads to the aggregation of β-catenin in the cytoplasm. Aggregated β-catenin migrates to the nucleus to form nuclear β-catenin, which acts as a transcription cofactor that activates EMT-TFs but is also negatively regulated by E-cadherin (14, 15). The level of β-catenin in HCC cells is regulated by a set of factors, of which lncRNAs are among the important regulatory factors. As mentioned above, in the positive feedback pathway, lncRNA-NEF inhibited EMT by inactivating the Wnt/β-catenin signaling pathway. However, other lncRNAs could stimulate the signaling pathway and promote EMT (Figure 3A). As an oncogene, lncRNA FOXD2-AS1 is dysregulated in tumor cells and can be used as a prognostic factor (70). Lei et al. demonstrated (62) that lncRNA FOXD2-AS1 induced by the transcription factor EGR1 promotes the process of EMT by binding with EZH2 (enhancer of zeste homolog 2) to epigenetically silence the gene DKK1 (an inhibitor of the Wnt/β-catenin axis) and activate the Wnt/β-catenin axis. Moreover, Zhu et al. confirmed (63) that colorectal neoplasia differentially expressed (CRNDE), a novel lncRNA, was significantly upregulated in HCC tissue and promoted EMT. In vitro cell experiments confirmed that Wnt/β-catenin signaling pathway activity was inhibited by CRNDE knockdown. Unfortunately, this paper did not clarify how CRNDE reduces the levels of signaling pathway-related proteins.
The PI3K/AKT/mTOR signaling pathway is relatively common in most cancers. This process involves the phosphorylation and dephosphorylation of related molecules (71). PTEN is considered to be a key negative regulator of the signaling pathway (72). Currently, studies have identified several lncRNAs that function through the PI3K/AKT/mTOR signaling pathway (Figure 3B). For example, Han et al. revealed (64) that lncRNA CASC11 induced by STAT3 promotes EMT in HCC by binding with the enhancer of zeste homolog 2 (EZH2) to epigenetically silence PTEN and activate the PI3K/AKT axis. Huang et al. illustrated (65) that lncRNA PTTG3P (pituitary tumor-transforming 3, pseudogene) promotes EMT in HCC by upregulating PTTG1 (pituitary tumor-transforming 1) and activating the PI3K/AKT axis. Moreover, Guo et al. confirmed (42) that abnormal expression of lncRNA TMPO antisense RNA 1 (TMPO-AS1) promotes EMT in HCC. The mechanism in this process was that TMPO-AS1 upregulated the oncogene FOXK1 by sponging miR-329-3p, thereby activating the AKT/mTOR signaling pathway.
The Janus kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) axis has been demonstrated to play a crucial role in the progression of HCC and is involved in the process of tumor metastasis (73, 74). Moreover, lncRNAs regulated the JAK2/STAT3 signaling pathways (Figure 3C). Wu et al. proved (66) that human ovarian cancer-specific transcript 2 (HOST2), abnormal regulation of lncRNA in HCC, enhanced EMT by upregulating the JAK2-STAT3 signaling pathway. Cell experiments in vitro confirmed that HOST2 increased the expression of EMT-TFs and vimentin by upregulating the protein levels of JAK2 and STAT3 but decreased the expression of E-cadherin. In addition, Wang et al. confirmed (67) that FEZ family zinc finger 1 antisense RNA 1 (FEZF1-AS1), a novel lncRNA measuring 2653 nucleotides in length, was upregulated in HCC and increased EMT. In in vitro cell experiments, FEZF1-AS1 knockdown significantly downregulated the JAK2/STAT3 signaling pathway, and JAK2 overexpression significantly reversed the response to EMT inhibition caused by AS1 knockdown. Taken together, these results indicated that FEZF1-AS1, as an oncogene, could promote EMT via the JAK2/STAT3 signaling pathway. Furthermore, the JAK2-STAT3 signaling pathway also participated in the above positive feedback loop.
Activation of the mitogen-activated protein kinase/extracellular regulated protein kinase (MEK/ERK) axis has been verified to be fundamental in tumor progression (75) and involved in the process of EMT (76). Moreover, lncRNAs regulated the MEK/ERK signaling pathways (Figure 3C). Tong et al. demonstrated (68) that HOXA-AS3, a novel lncRNA, was abnormally regulated and stimulated EMT in HCC. HOXA-AS3, as a ceRNA, upregulated the expression of bone morphogenetic protein 1 (BMP1) by sponging miR-29c. BMP1, the downstream target gene of miR-29c, is an oncogene whose overexpression can stimulate the MEK/ERK signaling pathway and promote EMT. These findings indicated that HOXA-AS3 upregulated BMP1 by sponging miR-29c, thereby activating the MEK/ERK signaling pathway to enhance EMT. However, lncRNAs could inhibit EMT by inactivating the MEK/ERK signaling pathway. Zhuang et al. demonstrated (69) that CTC-505O3 (lncRNA DRHC), a novel lncRNA, was underexpressed in HCC and has been shown to inhibit EMT. LncRNA DRHC guided the binding of MYBBP1A and C-Myb to form the lncRNA DRHC/MYBBP1A/C-Myb complex by binding to MYBBP1A. MYBBP1A, a transcriptional coregulator, could bind specifically to and inhibit C-Myb transcription. C-Myb is a proto-oncogene that regulates the MEK/ERK signaling pathway. Taken together, these findings indicated that the lncRNA DRHC bound to MYBBP1A and then formed a lncRNA/DRHC/MYBBP1A/c-Myb complex with c-Myb to inhibit the MEK/ERK signaling pathway and finally inhibit EMT.
Ma et al. reported (77) that a novel chromatin-enriched lncRNA, known as metabolism-induced tumor activator 1 (MITA1), induced energy stress, which was upregulated in HCC and conducive to metastasis in the absence of energy. MITA1 accelerated the EMT process largely by increasing Slug (snail2) transcription. In addition, the remaining lncRNAs regulated EMT through various pathways (Table 2). LINC00473, IHS, and ANCR were overexpressed in HCC and promoted EMT. However, ELF209 and ID2-AS1 were underexpressed in HCC and suppressed EMT.
To date, little evidence regarding the clinical application of lncRNAs has been presented. However, studies on the role of lncRNAs in drug resistance and the mechanism of drug effects have been conducted. As an example, lncRNAs were involved in the drug resistance of sorafenib. Zhang et al. proved (83) that the overexpression of small nucleolar RNA host gene 3 (SNHG3) induced EMT and sorafenib resistance by regulating miR‐128/CD151/AKT/PI3K feedback loop signaling. Fan et al. revealed (84) that the overexpression of metastasis associated with lung adenocarcinoma transcript-1 (MALAT1) upregulated Aurora-A expression by sponging miR-140-5p and consequently increasing sorafenib resistance in HCC. Chen et al. illustrated (85) that the overexpression of lncRNA‐POIR accelerated EMT progression and synchronously inhibited sorafenib sensitivity by sponging miR‐182‐5p. Additionally, a large number of agents could inhibit EMT by promoting lncRNA-related pathways. Fan et al. suggested (86) that arsenic trioxide inhibited EMT by promoting lncRNA MEG3 (maternal expression gene 3) upregulation in in vitro experiments. Wang et al. showed (87) that melatonin upregulated the transcription of lncRNA-CPS1 intronic transcript 1 (CPS1- IT1) by enhancing the expression of FOXA2, which weakened HIF-1α activity and accordingly inhibited EMT in HCC.
The crucial factors for the poor prognosis of HCC are drug resistance, metastasis, and limited therapeutic targeting options. As mentioned above, lncRNAs were observed to promote EMT and simultaneously enhance sorafenib resistance or reduce sorafenib sensitivity. Consequently, lncRNAs have emerged as promising predictive factors of sorafenib response in HCC. Moreover, lncRNAs can be designed as sorafenib sensitizers to weaken sorafenib resistance and enhance synergistic anticancer effects. EMT is a fundamental biological program of malignant tumor metastasis. In particular, lncRNAs play a key regulatory role in the EMT process of HCC. The expression of lncRNAs that promote EMT is positively correlated with EMT. Hence, lncRNAs may represent an excellent therapeutic target for the treatment of advanced HCC. Additionally, lncRNAs may also serve as new potential biomarkers to predict the prognosis of HCC.
Although the current work is increasingly inclined toward the feasible clinical application of lncRNAs, a number of questions and challenges still exist. First, most of the current studies remain at the basic research stage, primarily at the cellular level, and thus overlook the interaction between cells and the tumor microenvironment. Further clinical trials should be performed to validate the clinical significance of this interaction. Second, with the progress made in sequencing technology, a large number of functional lncRNAs have been identified, and they are gradually increasing. However, the transcriptional amount of each lncRNA may not be uniform in each cancer patient, which may reduce the efficacy of targeted therapy or the sensitivity of lncRNA markers. Third, the upstream regulatory mechanism governing lncRNAs should be further explored. With the progress of science and technology, the utilization of lncRNAs may ultimately enable doctors to make precise clinical decisions from diagnosis to treatment to prognosis evaluation.
In conclusion, the findings described in this review indicate that the regulation of EMT by lncRNAs in HCC is a multistep and complex process that is influenced and regulated by the tumor itself, as well as the tumor environment. With the development of science and technology and the improvement of medical techniques, I believe that the difficulties and challenges associated with this research will be resolved, and lncRNAs will eventually be utilized in the clinic to serve patients.
XH, XG, and YY designed the concept of this manuscript. XG, YY, JL, and ZL collected the related paper. XG wrote the manuscript. XH reviewed this manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
HCC, hepatocellular carcinoma; EMT, epithelial–mesenchymal transition; PI3K, phosphoinositide-3-kinase; mTOR, mammalian target of rapamycin; AKT, serine-threonine kinase; JAK2, Janus kinase 2; STAT3, signal transducer and activator of transcription 3; ERK, extracellular-signal-regulated kinase; MEK, mitogen-activated protein kinase; CFL1, cofilin1; E-cad, E‐cadherin; N‐cad, N‐cadherin; Vim, vimentin; ZO-1, zona occludens 1; ZEB1/2, zinc finger E-box-binding homeobox 1 and 2; α-SMA, α-smooth muscle actin; NGS, next-generation sequencing technology; RNA-Seq, transcriptome sequencing technology; DLGAP1-AS1, discs, large (Drosophila) homolog-associated protein 1 antisense RNA 1; CDK8, cyclin-dependent Kinase 8; LRP6, lipoprotein receptor-related protein 6; lncRNA-NEF, lncRNA neighboring enhancer of FOXA2; FOXA2, forkhead box protein A2; GSK-3β, glycogen synthase kinase-3β; EZH2, enhancer of zeste homolog 2; DKK1, dickkopf-1; CRNDE, colorectal neoplasia differentially expressed; PTEN, phosphatase and tensin homolog; PTTG3P, pituitary tumor-transforming 3, pseudogene; PTTG1, pituitary tumor-transforming 1; TMPO-AS1, TMPO antisense RNA 1; FOXK1, forkhead box K1; HOST2, human ovarian cancer-specific transcript 2; FEZF1-AS1, FEZ family zinc finger 1 antisense RNA 1; BMP1, bone morphogenetic protein 1; MYBBP1A, MYB Binding Protein 1a; MITA1, metabolism-induced tumor activator 1; SNHG 3, small nucleolar RNA host gene 3; MALAT1, metastasis associated with lung adenocarcinoma transcript-1; MEG3, maternal expression gene 3; CPS1- IT1, lncRNA-CPS1 intronic transcript 1; HIF-1α, hypoxia-inducible factor 1α; MAPK1, mitogen-activated protein kinase 1; LSINCT5, long stress Induced non-coding transcripts 5; HMGA2, high-mobility group AT-hook 2.
1. Ferlay J, Ervik M, Lam F, Colombet M, Mery L, et al. Global Cancer Observatory: Cancer Today (2020). Lyon, France: International Agency for Research on Cancer. Available at: https://gco.iarc.fr/today (Accessed 08 May 2021).
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
3. Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular Carcinoma. Nat Rev Dis Primers (2016) 2:16018. doi: 10.1038/nrdp.2016.18
4. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. Cell (2009) 139(5):871–90. doi: 10.1016/j.cell.2009.11.007
5. Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, et al. Guidelines and Definitions for Research on Epithelial-Mesenchymal Transition. Nat Rev Mol Cell Biol (2020) 21(6):341–52. doi: 10.1038/s41580-020-0237-9
6. Gugnoni M, Ciarrocchi A. Long Noncoding RNA and Epithelial Mesenchymal Transition in Cancer. Int J Mol Sci (2019) 20(8):1924. doi: 10.3390/ijms20081924
7. Jolly MK, Celia-Terrassa T. Dynamics of Phenotypic Heterogeneity Associated With EMT and Stemness During Cancer Progression. J Clin Med (2019) 8(10):1542. doi: 10.3390/jcm8101542
8. Navas T, Kinders RJ, Lawrence SM, Ferry-Galow KV, Borgel S, Hollingshead MG, et al. Clinical Evolution of Epithelial-Mesenchymal Transition in Human Carcinomas. Cancer Res (2020) 80(2):304–18. doi: 10.1158/0008-5472
9. Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell (2016) 166(1):21–45. doi: 10.1016/j.cell.2016.06.028
10. Iwatsuki M, Mimori K, Yokobori T, Ishi H, Beppu T, Nakamori S, et al. Epithelial-Mesenchymal Transition in Cancer Development and its Clinical Significance. Cancer Sci (2010) 101(2):293–9. doi: 10.1111/j.1349-7006.2009.01419.x
11. Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in Cancer. Nat Rev Cancer (2018) 18(2):128–34. doi: 10.1038/nrc.2017.118
12. Lamouille S, Xu J, Derynck R. Molecular Mechanisms of Epithelial-Mesenchymal Transition. Nat Rev Mol Cell Biol (2014) 15(3):178–96. doi: 10.1038/nrm3758
13. Muller S, Sindikubwabo F, Caneque T, Lafon A, Versini A, Lombard B, et al. CD44 Regulates Epigenetic Plasticity by Mediating Iron Endocytosis. Nat Chem (2020) 12(10):929–38. doi: 10.1038/s41557-020-0513-5
14. Gonzalez DM, Medici D. Signaling Mechanisms of the Epithelial-Mesenchymal Transition. Sci Signal (2014) 7(344):re8. doi: 10.1126/scisignal.2005189
15. Dongre A, Weinberg RA. New Insights Into the Mechanisms of Epithelial-Mesenchymal Transition and Implications for Cancer. Nat Rev Mol Cell Biol (2019) 20(2):69–84. doi: 10.1038/s41580-018-0080-4
16. Chen QF, Hu CF, Sun K, Lang YP. LncRNA NR027113 Promotes Malignant Progression of Gastric Carcinoma Via EMT Signaling Pathway. Eur Rev Med Pharmacol Sci (2019) 23(11):4746–55. doi: 10.26355/eurrev_201906_18056
17. Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell (2009) 136(4):629–41. doi: 10.1016/j.cell.2009.02.006
18. Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, et al. Emerging Role of Tumor-Related Functional Peptides Encoded by lncRNA and CircRNA. Mol Cancer (2020) 19(1):22. doi: 10.1186/s12943-020-1147-3
19. Heery R, Finn SP, Cuffe S, Gray SG. Long Non-Coding RNAs: Key Regulators of Epithelial-Mesenchymal Transition, Tumour Drug Resistance and Cancer Stem Cells. Cancers (2017) 9(4):38. doi: 10.3390/cancers9040038
20. McCabe EM, Rasmussen TP. lncRNA Involvement in Cancer Stem Cell Function and Epithelial-Mesenchymal Transitions. Semin Cancer Biol (2020) S1044-579X(20):80272–8. doi: 10.1016/j.semcancer.2020.12.012
21. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell (2018) 172(3):393–407. doi: 10.1016/j.cell.2018.01.011
22. Liu LX, Liu B, Yu J, Zhang DY, Shi JH, Liang P. SP1-Induced Upregulation of lncRNA CTBP1-AS2 Accelerates the Hepatocellular Carcinoma Tumorigenesis Through Targeting CEP55 Via Sponging Mir-195-5p. Biochem Biophys Res Commun (2020) 533(4):779–85. doi: 10.1016/j.bbrc.2020.09.080
23. Yang F, Shen Y, Zhang W, Jin J, Huang D, Fang H, et al. An Androgen Receptor Negatively Induced Long Non-Coding RNA ARNILA Binding to miR-204 Promotes the Invasion and Metastasis of Triple-Negative Breast Cancer. Cell Death Differ (2018) 25(12):2209–20. doi: 10.1038/s41418-018-0123-6
24. Zhang K, Lu C, Huang X, Cui J, Li J, Gao Y, et al. Long Noncoding RNA AOC4P Regulates Tumor Cell Proliferation and Invasion by Epithelial-Mesenchymal Transition in Gastric Cancer. Therap Adv Gastroenterol (2019) 12:1756284819827697. doi: 10.1177/1756284819827697
25. Mondal P, Meeran SM. Long Non-Coding RNAs in Breast Cancer Metastasis. Noncoding RNA Res (2020) 5(4):208–18. doi: 10.1016/j.ncrna.2020.11.004
26. Hu YL, Feng Y, Ma P, Wang F, Huang H, Guo YB, et al. HAX-1 Promotes the Migration and Invasion of Hepatocellular Carcinoma Cells Through the Induction of Epithelial-Mesenchymal Transition Via the NF-kappaB Pathway. Exp Cell Res (2019) 381(1):66–76. doi: 10.1016/j.yexcr.2019.04.030
27. Song S, Qiu X. LncRNA miR503HG Inhibits Epithelial-Mesenchymal Transition and Angiogenesis in Hepatocellular Carcinoma by Enhancing PDCD4 Via Regulation of miR-15b. Digest Liver Dis Off J Ital Soc Gastroenterol Ital Assoc Study Liver (2021) 53(1):107–16. doi: 10.1016/j.dld.2020.09.008
28. Xiao S, Hu J, Hu N, Sheng L, Rao H, Zheng G. Identification of a Novel Epithelial-to-Mesenchymal-Related Gene Signature in Predicting Survival of Patients With Hepatocellular Carcinoma. Combinatorial Chem High Throughput Screening (2021). doi: 10.2174/1386207324666210303093629
29. Chaw SY, Abdul Majeed A, Dalley AJ, Chan A, Stein S, Farah CS. Epithelial to Mesenchymal Transition (EMT) Biomarkers–E-Cadherin, Beta-Catenin, APC and Vimentin–in Oral Squamous Cell Carcinogenesis and Transformation. Oral Oncol (2012) 48(10):997–1006. doi: 10.1016/j.oraloncology.2012.05.011
30. Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of Epithelial to Mesenchymal Transition in Hepatocellular Carcinoma. J Hepatol (2016) 65(4):798–808. doi: 10.1016/j.jhep.2016.05.007
31. Pei D, Shu X, Gassama-Diagne A, Thiery JP. Mesenchymal-Epithelial Transition in Development and Reprogramming. Nat Cell Biol (2019) 21(1):44–53. doi: 10.1038/s41556-018-0195-z
32. Statello L, Guo CJ, Chen LL, Huarte M. Gene Regulation by Long non-Coding RNAs and its Biological Functions. Nat Rev Mol Cell Biol (2021) 22(2):96–118. doi: 10.1038/s41580-020-00315-9
33. Bridges MC, Daulagala AC, Kourtidis A. Lnccation: lncRNA Localization and Function. J Cell Biol (2021) 220(2):e202009045. doi: 10.1083/jcb.202009045
34. Yang L, Deng WL, Zhao BG, Xu Y, Wang XW, Fang Y, et al. FOXO3-Induced lncRNA LOC554202 Contributes to Hepatocellular Carcinoma Progression Via the miR-485-5p/BSG Axis. Cancer Gene Ther (2021). doi: 10.1038/s41417-021-00312-w
35. Ma W, Chen X, Wu X, Li J, Mei C, Jing W, et al. Long Noncoding RNA Spry4-IT1 Promotes Proliferation and Metastasis of Hepatocellular Carcinoma Via Mediating TNF Signaling Pathway. J Cell Physiol (2020) 235(11):7849–62. doi: 10.1002/jcp.29438
36. Xu D, Liu X, Wu J, Wang Y, Zhou K, Chen W, et al. Lncrna WWOX-AS1 Sponges miR-20b-5p in Hepatocellular Carcinoma and Represses its Progression by Upregulating WWOX. Cancer Biol Ther (2020) 21(10):927–36. doi: 10.1080/15384047.2020.1806689
37. Guo D, Li Y, Chen Y, Zhang D, Wang X, Lu G, et al. DANCR Promotes HCC Progression and Regulates EMT by Sponging miR-27a-3p Via ROCK1/LIMK1/COFILIN1 Pathway. Cell Prolif (2019) 52(4):e12628. doi: 10.1111/cpr.12628
38. Liu Y, Yan W, Zhou D, Jin G, Cheng X. Long Non−Coding RNA HOXA11−as Accelerates Cell Proliferation and Epithelial−Mesenchymal Transition in Hepatocellular Carcinoma by Modulating the miR−506−3p/Slug Axis. Int J Mol Med (2020) 46(5):1805–15. doi: 10.3892/ijmm.2020.4715
39. Sun C, Huang S, Hou Y, Li Z, Xia D, Zhang L, et al. Long Noncoding RNA AC092171.4 Promotes Hepatocellular Carcinoma Progression by Sponging microRNA-1271 and Upregulating GRB2. Aging (2020) 12(14):14141–56. doi: 10.18632/aging.103419
40. Liu Z, Wang Y, Wang L, Yao B, Sun L, Liu R, et al. Long Non-Coding RNA Agap2-AS1, Functioning as a Competitive Endogenous RNA, Upregulates ANXA11 Expression by Sponging miR-16-5p and Promotes Proliferation and Metastasis in Hepatocellular Carcinoma. J Exp Clin Cancer Res CR (2019) 38(1):194. doi: 10.1186/s13046-019-1188-x
41. Li Y, Guo D, Zhao Y, Ren M, Lu G, Wang Y, et al. Long Non-Coding RNA SNHG5 Promotes Human Hepatocellular Carcinoma Progression by Regulating Mir-26a-5p/GSK3β Signal Pathway. Cell Death Dis (2018) 9(9):888. doi: 10.1038/s41419-018-0882-5
42. Guo X, Wang Y. LncRNA TMPO-AS1 Promotes Hepatocellular Carcinoma Cell Proliferation, Migration and Invasion Through Sponging miR-329-3p to Stimulate FOXK1-Mediated Akt/mTOR Signaling Pathway. Cancer Med (2020) 9(14):5235–46. doi: 10.1002/cam4.3046
43. Ma T, Zhou X, Wei H, Yan S, Hui Y, Liu Y, et al. Long Non-coding RNA SNHG17 Upregulates RFX1 by Sponging miR-3180-3p and Promotes Cellular Function in Hepatocellular Carcinoma. Front Genet (2020) 11:607636. doi: 10.3389/fgene.2020.607636
44. Li L, Han T, Liu K, Lei CG, Wang ZC, Shi GJ. LncRNA H19 Promotes the Development of Hepatitis B Related Hepatocellular Carcinoma Through Regulating microRNA-22 Via EMT Pathway. Eur Rev Med Pharmacol Sci (2019) 23(12):5392–401. doi: 10.26355/eurrev_201906_18208
45. Zhou Y, Fan RG, Qin CL, Jia J, Wu XD, Zha WZ. LncRNA-H19 Activates CDC42/PAK1 Pathway to Promote Cell Proliferation, Migration and Invasion by Targeting miR-15b in Hepatocellular Carcinoma. Genomics (2019) 111(6):1862–72. doi: 10.1016/j.ygeno.2018.12.009
46. Ye Y, Guo J, Xiao P, Ning J, Zhang R, Liu P, et al. Macrophages-Induced Long Noncoding RNA H19 Up-Regulation Triggers and Activates the miR-193b/MAPK1 Axis and Promotes Cell Aggressiveness in Hepatocellular Carcinoma. Cancer Lett (2020) 469:310–22. doi: 10.1016/j.canlet.2019.11.001
47. Lin Y, Jian Z, Jin H, Wei X, Zou X, Guan R, et al. Long non-Coding RNA Dlgap1-AS1 Facilitates Tumorigenesis and Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma Via the Feedback Loop of miR-26a/b-5p/IL-6/JAK2/STAT3 and Wnt/β-Catenin Pathway. Cell Death Dis (2020) 11(1):34. doi: 10.1038/s41419-019-2188-7
48. Zhou F, Lei Y, Xu X, Zhou H, Liu H, Jiang J, et al. LINC00355:8 Promotes Cell Proliferation and Migration With Invasion Via the MiR-6777-3p/Wnt10b Axis in Hepatocellular Carcinoma. J Cancer (2020) 11(19):5641–55. doi: 10.7150/jca.43831
49. Han L, Jia L, Zan Y. Long Intergenic Noncoding RNA Smad7 (Linc-smad7) Promotes the Epithelial-Mesenchymal Transition of HCC by Targeting the miR-125b/SIRT6 Axis. Cancer Med (2020) 9(23):9123–37. doi: 10.1002/cam4.3515
50. Lin Z, Liu J. lncRNA DQ786243 Promotes Hepatocellular Carcinoma Cell Invasion and Proliferation by Regulating the miR15p5p/Wnt3A Axis. Mol Med Rep (2021) 23(5):318. doi: 10.3892/mmr.2021.11957
51. Liu C, Wang H, Tang L, Huang H, Xu M, Lin Y, et al. lncRNA BACE1-AS Enhances the Invasive and Metastatic Capacity of Hepatocellular Carcinoma Cells Through Mediating miR-377-3p/CELF1 Axis. Life Sci (2021) 275:119288. doi: 10.1016/j.lfs.2021.119288
52. Song W, Zhang J, Zhang J, Sun M, Xia Q. Overexpression of lncRNA PIK3CD-AS1 Promotes Expression of LATS1 by Competitive Binding With microRNA-566 to Inhibit the Growth, Invasion and Metastasis of Hepatocellular Carcinoma Cells. Cancer Cell Int (2019) 19:150. doi: 10.1186/s12935-019-0857-3
53. Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang C, et al. Long Non-Coding RNA CASC2 Suppresses Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma Cells Through CASC2/miR-367/FBXW7 Axis. Mol Cancer (2017) 16(1):123. doi: 10.1186/s12943-017-0702-z
54. Wang Y, Liu Z, Yao B, Dou C, Xu M, Xue Y, et al. Long Non-Coding RNA TUSC7 Acts a Molecular Sponge for miR-10a and Suppresses EMT in Hepatocellular Carcinoma. Tumour Biol J Int Soc Oncodevelopmental Biol Med (2016) 37(8):11429–41. doi: 10.1007/s13277-016-4892-6
55. Li O, Li Z, Tang Q, Li Y, Yuan S, Shen Y, et al. Long Stress Induced Non-Coding Transcripts 5 (Lsinct5) Promotes Hepatocellular Carcinoma Progression Through Interaction With High-Mobility Group AT-hook 2 and mir-4516. Med Sci Monit Int Med J Exp Clin Res (2018) 24:8510–23. doi: 10.12659/MSM.911179
56. Mehra M, Chauhan R. Long Noncoding RNAs as a Key Player in Hepatocellular Carcinoma. Biomark Cancer (2017) 9:1179299X17737301. doi: 10.1177/1179299X17737301
57. Wang F, Fan M, Zhou X, Yu Y, Cai Y, Wu H, et al. A Positive Feedback Loop Between TAZ and miR-942-3p Modulates Proliferation, Angiogenesis, Epithelial-Mesenchymal Transition Process, Glycometabolism and ROS Homeostasis in Human Bladder Cancer. J Exp Clin Cancer Res CR (2021) 40(1):44. doi: 10.1186/s13046-021-01846-5
58. Al-Harbi B, Aboussekhra A. Cucurbitacin I (JSI-124)-dependent Inhibition of STAT3 Permanently Suppresses the Pro-Carcinogenic Effects of Active Breast Cancer-Associated Fibroblasts. Mol Carcinog (2021) 60(4):242–51. doi: 10.1002/mc.23287
59. Tong X, Wang S, Lei Z, Li C, Zhang C, Su Z, et al. MYOCD and SMAD3/SMAD4 Form a Positive Feedback Loop and Drive TGF-β-Induced Epithelial-Mesenchymal Transition in non-Small Cell Lung Cancer. Oncogene (2020) 39(14):2890–904. doi: 10.1038/s41388-020-1189-4
60. Peng L, Jiang B, Yuan X, Qiu Y, Peng J, Huang Y, et al. Super-Enhancer-Associated Long Noncoding RNA Hccl5 Is Activated by ZEB1 and Promotes the Malignancy of Hepatocellular Carcinoma. Cancer Res (2019) 79(3):572–84. doi: 10.1158/0008-5472.CAN-18-0367
61. Liang WC, Ren JL, Wong CW, Chan SO, Waye MM, Fu WM, et al. LncRNA-NEF Antagonized Epithelial to Mesenchymal Transition and Cancer Metastasis Via Cis-Regulating FOXA2 and Inactivating Wnt/β-Catenin Signaling. Oncogene (2018) 37(11):1445–56. doi: 10.1038/s41388-017-0041-y
62. Lei T, Zhu X, Zhu K, Jia F, Li S. EGR1-Induced Upregulation of LncRNA FOXD2-AS1 Promotes the Progression of Hepatocellular Carcinoma Via Epigenetically Silencing DKK1 and Activating Wnt/β-Catenin Signaling Pathway. Cancer Biol Ther (2019) 20(7):1007–16. doi: 10.1080/15384047.2019.1595276
63. Zhu L, Yang N, Du G, Li C, Liu G, Liu S, et al. LncRNA CRNDE Promotes the Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma Cells Via Enhancing the Wnt/β-Catenin Signaling Pathway. J Cell Biochem (2018) 120(2):1156–64. doi: 10.1002/jcb.26762
64. Han Y, Chen M, Wang A, Fan X. STAT3-Induced Upregulation of lncRNA CASC11 Promotes the Cell Migration, Invasion and Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma by Epigenetically Silencing PTEN and Activating PI3K/AKT Signaling Pathway. Biochem Biophys Res Commun (2019) 508(2):472–9. doi: 10.1016/j.bbrc.2018.11.092
65. Huang JL, Cao SW, Ou QS, Yang B, Zheng SH, Tang J, et al. The Long Non-Coding RNA PTTG3P Promotes Cell Growth and Metastasis Via Up-Regulating PTTG1 and Activating PI3K/AKT Signaling in Hepatocellular Carcinoma. Mol Cancer (2018) 17(1):93. doi: 10.1186/s12943-018-0841-x
66. Wu Y, Yuan T, Wang WW, Ge PL, Gao ZQ, Zhang G, et al. Long Noncoding RNA HOST2 Promotes Epithelial-Mesenchymal Transition, Proliferation, Invasion and Migration of Hepatocellular Carcinoma Cells by Activating the JAK2-STAT3 Signaling Pathway. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol (2018) 51(1):301–14. doi: 10.1159/000495231
67. Wang YD, Sun XJ, Yin JJ, Yin M, Wang W, Nie ZQ, et al. Long non-Coding RNA Fezf1-AS1 Promotes Cell Invasion and Epithelial-Mesenchymal Transition Through JAK2/STAT3 Signaling Pathway in Human Hepatocellular Carcinoma. Biomed Pharmacother = Biomed Pharmacother (2018) 106:134–41. doi: 10.1016/j.biopha.2018.05.116
68. Tong Y, Wang M, Dai Y, Bao D, Zhang J, Pan H. LncRNA HOXA-AS3 Sponges miR-29c to Facilitate Cell Proliferation, Metastasis, and EMT Process and Activate the MEK/ERK Signaling Pathway in Hepatocellular Carcinoma. Hum Gene Ther Clin Dev (2019) 30(3):129–41. doi: 10.1089/humc.2018.266
69. Zhuang R, Zhang X, Lu D, Wang J, Zhuo J, Wei X, et al. LncRNA DRHC Inhibits Proliferation and Invasion in Hepatocellular Carcinoma Via C-Myb-Regulated MEK/ERK Signaling. Mol Carcinog (2019) 58(3):366–75. doi: 10.1002/mc.22934
70. Zhu Y, Qiao L, Zhou Y, Ma N, Wang C, Zhou J. Long Non-Coding RNA Foxd2-AS1 Contributes to Colorectal Cancer Proliferation Through its Interaction With microRNA-185-5p. Cancer Sci (2018) 109(7):2235–42. doi: 10.1111/cas.13632
71. Ediriweera MK, Tennekoon KH, Samarakoon SR. Role of the PI3K/AKT/mTOR Signaling Pathway in Ovarian Cancer: Biological and Therapeutic Significance. Semin Cancer Biol (2019) 59:147–60. doi: 10.1016/j.semcancer.2019.05.012
72. Hu M, Zhu S, Xiong S, Xue X, Zhou X. MicroRNAs and the PTEN/PI3K/Akt Pathway in Gastric Cancer (Review). Oncol Rep (2019) 41(3):1439–54. doi: 10.3892/or.2019.6962
73. Teng Y, Ghoshal P, Ngoka L, Mei Y, Cowell JK. Critical Role of the WASF3 Gene in JAK2/STAT3 Regulation of Cancer Cell Motility. Carcinogenesis (2013) 34(9):1994–9. doi: 10.1093/carcin/bgt167
74. Yang S, Luo C, Gu Q, Xu Q, Wang G, Sun H, et al. Activating JAK1 Mutation may Predict the Sensitivity of JAK-STAT Inhibition in Hepatocellular Carcinoma. Oncotarget (2016) 7(5):5461–9. doi: 10.18632/oncotarget.6684
75. Wang H, Li K, Ma L, Wu S, Hu J, Yan H, et al. Berberine Inhibits Enterovirus 71 Replication by Downregulating the MEK/ERK Signaling Pathway and Autophagy. Virol J (2017) 14(1):2. doi: 10.1186/s12985-016-0674-4
76. Tashiro E, Henmi S, Odake H, Ino S, Imoto M. Involvement of the MEK/ERK Pathway in EGF-Induced E-Cadherin Down-Regulation. Biochem Biophys Res Commun (2016) 477(4):801–6. doi: 10.1016/j.bbrc.2016.06.138
77. Ma M, Xu H, Liu G, Wu J, Li C, Wang X, et al. Metabolism-Induced Tumor Activator 1 (MITA1), an Energy Stress-Inducible Long Noncoding RNA, Promotes Hepatocellular Carcinoma Metastasis. Hepatol (Baltimore Md) (2019) 70(1):215–30. doi: 10.1002/hep.30602
78. Chen H, Yang F, Li X, Gong ZJ, Wang LW. Long Noncoding RNA LNC473 Inhibits the Ubiquitination of Survivin Via Association With USP9X and Enhances Cell Proliferation and Invasion in Hepatocellular Carcinoma Cells. Biochem Biophys Res Commun (2018) 499(3):702–10. doi: 10.1016/j.bbrc.2018.03.215
79. Chen Z, Yu W, Zhou Q, Zhang J, Jiang H, Hao D, et al. A Novel Lncrna IHS Promotes Tumor Proliferation and Metastasis in HCC by Regulating the ERK- and AKT/GSK-3β-Signaling Pathways. Mol Ther Nucleic Acids (2019) 16:707–20. doi: 10.1016/j.omtn.2019.04.021
80. Wen Z, Lian L, Ding H, Hu Y, Xiao Z, Xiong K, et al. lncRNA ANCR Promotes Hepatocellular Carcinoma Metastasis Through Upregulating HNRNPA1 Expression. RNA Biol (2020) 17(3):381–94. doi: 10.1080/15476286.2019.1708547
81. Yang Y, Chen Q, Piao HY, Wang B, Zhu GQ, Chen EB, et al. HNRNPAB-Regulated lncRNA-ELF209 Inhibits the Malignancy of Hepatocellular Carcinoma. Int J Cancer (2020) 146(1):169–80. doi: 10.1002/ijc.32409
82. Zhou Y, Huan L, Wu Y, Bao C, Chen B, Wang L, et al. lncRNA ID2-AS1 Suppresses Tumor Metastasis by Activating the HDAC8/ID2 Pathway in Hepatocellular Carcinoma. Cancer Lett (2020) 469:399–409. doi: 10.1016/j.canlet.2019.11.007
83. Zhang PF, Wang F, Wu J, Wu Y, Huang W, Liu D, et al. LncRNA SNHG3 Induces EMT and Sorafenib Resistance by Modulating the miR-128/CD151 Pathway in Hepatocellular Carcinoma. J Cell Physiol (2019) 234(3):2788–94. doi: 10.1002/jcp.27095
84. Fan L, Huang X, Chen J, Zhang K, Gu YH, Sun J, et al. Long Noncoding RNA MALAT1 Contributes to Sorafenib Resistance by Targeting Mir-140-5p/Aurora-a Signaling in Hepatocellular Carcinoma. Mol Cancer Ther (2020) 19(5):1197–209. doi: 10.1158/1535-7163.MCT-19-0203
85. Chen BW, Zhou Y, Wei T, Wen L, Zhang YB, Shen SC, et al. lncRNA-POIR Promotes Epithelial-Mesenchymal Transition and Suppresses Sorafenib Sensitivity Simultaneously in Hepatocellular Carcinoma by Sponging Mir-182-5p. J Cell Biochem (2021) 122(1):130–42. doi: 10.1002/jcb.29844
86. Fan Z, He J, Fu T, Zhang W, Yang G, Qu X, et al. Arsenic Trioxide Inhibits EMT in Hepatocellular Carcinoma by Promoting lncRNA MEG3 Via PKM2. Biochem Biophys Res Commun (2019) 513(4):834–40. doi: 10.1016/j.bbrc.2019.04.081
Keywords: long noncoding RNA, hepatocellular carcinoma, epithelial–mesenchymal transition, signaling pathways, biomarker, therapeutic target
Citation: Ge X, Yao Y, Li J, Li Z and Han X (2021) Role of LncRNAs in the Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma. Front. Oncol. 11:690800. doi: 10.3389/fonc.2021.690800
Received: 04 April 2021; Accepted: 10 May 2021;
Published: 25 May 2021.
Edited by:
Carolina Vicente-Dueñas, University of Salamanca, SpainReviewed by:
Ayse Banu Demir, İzmir University of Economics, TurkeyCopyright © 2021 Ge, Yao, Li, Li and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinwei Han, MTM1OTI1ODM5MTFAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.