- 1National Clinical Research Center for Hematologic Diseases, Jiangsu Institute of Hematology, The First Affiliated Hospital of Soochow University, Suzhou, China
- 2Department of Hematology, The Affiliated Suzhou Hospital of Nanjing Medical University (Main part of Suzhou Municipal Hospital), Suzhou, China
- 3Institute of Blood and Marrow Transplantation, Collaborative Innovation Center of Hematology, Soochow University, Suzhou, China
To define the fusion genes in T/myeloid mixed-phenotype acute leukemia (T/M MPAL), we performed transcriptome sequencing of diagnostic bone marrow samples from 20 adult patients. Our analysis identified a second instance of a recurrent MED14-HOXA9 chimeric gene resulting from the in-frame fusion of exon 23 of MED14 and exon 1 of HOXA9, the first in an adult patient. The MED14-HOXA9 fusion gene was detected in both the diagnostic and relapsed blasts with reverse transcription-polymerase chain reaction and Sanger sequencing. The patient received combined conventional chemotherapy but suffered relapse at 11 months and died of disease progression one year after the initial diagnosis. Our data suggest that MED14-HOXA9 is a cryptic recurrent aberration in T/M MPAL, which might indicate an aggressive clinical course and inferior outcome after conventional chemotherapy. Further studies will be carried out to reveal the effects of the MED14-HOXA9 fusion on the differentiation and proliferation of leukemia stem cells, as well as suitable treatment strategies for this emerging entity.
Introduction
T/myeloid mixed-phenotype acute leukemia (T/M MPAL) is a rare malignancy responsible for approximately 1% of all leukemia cases and is characterized by leukemic blasts presenting both T lineage and myeloid markers (1). Only a few patients with T/M MPAL have been identified with recurrent genetic aberrations, such as the t(9;22)(q34.1;q11.2)/BCR-ABL1 and t(v;11q23.3)/KMT2A rearrangements (2). Most T/M MPAL cases carry nonspecific clonal chromosomal abnormalities, lack uniform treatment strategies and carry unfavorable prognoses (3, 4). Using next-generation and transcriptome sequencing, Takahashi and Alexander et al. elucidated the genetic and epigenetic heterogeneity of MPAL in a case series (5, 6), but only a few of the patients had T/M MPAL. For a better understanding of the genomic landscape of T/M MPAL, we performed transcriptome sequencing of diagnostic blasts from 20 adult patients with normal karyotype or karyotype failure and, thereby, detected a cryptic cytogenetic aberration involving chromosomes X and 7 effecting the novel occurrence of a chimeric fusion MED14-HOXA9 in an adult patient.
Materials and Methods
Patient Characteristics
From March 2008 to November 2019, a total of 37 T/M MPAL patients were enrolled. Their median age was 45 years-old (range 17-84 years), comprising 22 males and 15 females. The study was approved by the Ethics Committee of the First Affiliated Hospital of Soochow University in accordance with the Declaration of Helsinki. Written informed consents were obtained from all patients.
R-Banding Karyotype Analysis
Bone marrow (BM) samples were taken at diagnosis. Mononuclear cells were harvested, R-banded according to the routine institutional protocols and 20 metaphases were analyzed for each sample if possible. Chromosomal abnormalities were described according to the International System for Human Cytogenomic Nomenclature (ISCN, 2016) (7).
RNA Sequencing and RT-PCR
Total RNA from BM samples taken at diagnosis was extracted using a RNeasy Mini Kit (QIAgen, Hilden, Germany). RNA sequencing libraries were prepared using 20-100 ng total RNA of BM samples with the TruSeq RNA library preparation kit v2 (Illumina, CA, USA). Paired-end sequencing with a read length of 150bp was performed on Illumina NovaSeq platforms to at least 12G raw data per sample according to the manufacturer’s protocol. Read pairs were aligned to human reference genome (hg38) using the Spliced Transcripts Alignment to a Reference (STAR, version 2.5) (8). The FusionCatcher software was used to find fusion genes. SNVs/indels were analyzed by following the GATK best practices for variant calling on RNA sequencing. The forward and reverse primer sequences used for detection of the MED14-HOXA9 fusion by reverse transcription-polymerase chain reaction (RT-PCR) were 5’-AAGGTCTGTAAATGAGGACG-3’ and 5’-TCGTCTTTTGCTCGGTCTT-3’, respectively.
Results
After R-banding, 17 patients (17/37, 45.9%) evidenced abnormal karyotypes, 17 tested normal (17/37, 45.9%), and 3 failed analysis. RNA sequencing was performed on the 20 patients with normal karyotypes or who failed karyotype analysis. In 19 patients, RNA sequencing failed to detect any clinically relevant fusion genes. In one patient with a normal karyotype, however, the MED14-HOXA9 fusion transcript was detected using RNA sequencing: accordingly, exon 23 of MED14 was fused in frame with exon 1 of HOXA9 (Figure 1A and Supplementary Table 1), which was further confirmed with RT-PCR and Sanger sequencing (Figures 1B, C). Moreover, the MED14-HOXA9 fusion was also detected in the blasts at relapse with RT-PCR (Figure 1D). The 1153 amino acids encoded by the MED14-HOXA9 fusion transcript cover the entire Med14 and homeobox domains of MED14 and HOXA9, respectively (Figure 1E). Though fusion genes are frequently resulted from balanced chromosome translocations, results of RNA sequencing and RT-PCR revealed no reciprocal HOXA9-MED14 fusion transcripts in this patient (data not shown).
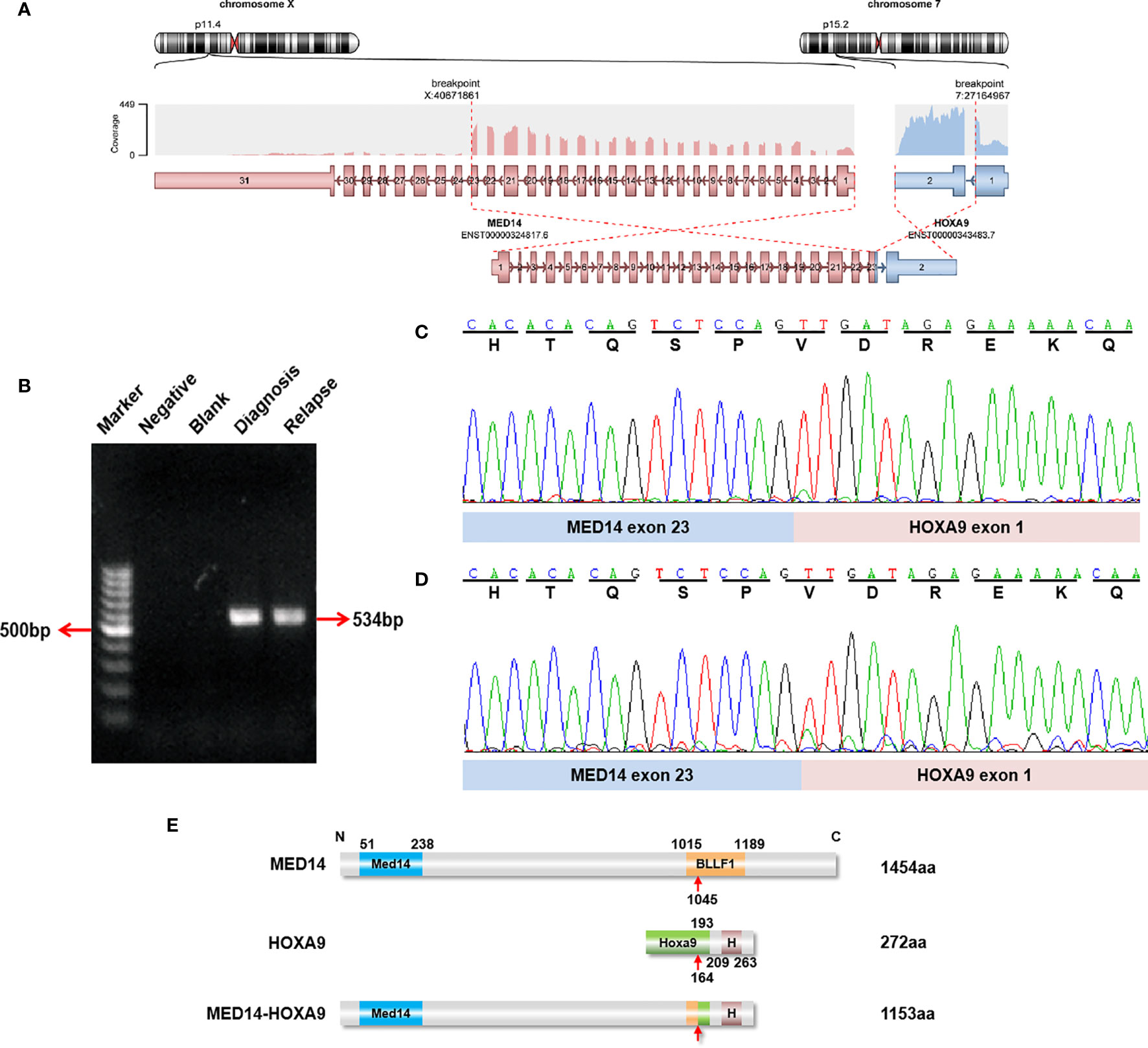
Figure 1 Identification of recurrent MED14-HOXA9 fusion transcripts. (A) RNA sequencing analysis revealed one breakpoint in exon 23 of the MED14 gene and one breakpoint in exon 1 of the HOXA9 gene. (B) A product of 534 bp was detected by RT-PCR in samples taken at diagnosis and relapse. Marker: GeneRuler 100 bp DNA ladder; negative control (with cDNA sample with the JIH-5 cell line); blank control (without cDNA template). (C, D) Sequence alignment of the amplified product revealed breakpoints between exon 23 of MED14 and exon 1 of HOXA9 at diagnosis (C) and relapse (D). (E) Schematic diagram of the MED14 and HOXA9 proteins and the MED14-HOXA9 fusion protein. The breakpoint is indicated by a red arrow. Med14, Mediator complex subunit MED14; BLLF1, Herpes virus major outer envelope glycoprotein (BLLF1); HoxA9, HoxA9 activation region; H, Homeobox domain.
The affected patient was a 37-year-old male referred to our hospital in June 2016 because of mild fever. Physical examination revealed enlarged lymph nodes of the right neck, without hepatomegaly or splenomegaly. Complete blood cell counts showed a white blood cell count of 11.09×109/L, hemoglobin of 10.9 g/dL and a platelet count of 84×109/L (Supplementary Table 2). Differentiation analysis of white blood cells showed 59% blasts. BM aspirates revealed 71% blasts, which were variable in cell and nucleolus size and negative for peroxidase staining (Figure 2A). The blasts were positive for myeloid markers, including CD13 (97.8%), CD15 (68.9%), CD117 (63.2%) and weak expression of MPO (36.1%), as well as the T lineage markers cCD3 (46.1%) and TdT (23.0%) by flow cytometry (Figure 2B and Supplementary Figure 1A). No fusion genes were detected using a multiplex RT-PCR panel covering 43 fusion genes commonly seen in acute leukemia (Supplementary Table 3). Cytogenetic analysis of the BM indicated a normal karyotype. Next generation sequencing (NGS) was not feasible due to sample non-availability. CREBBP-p.Pro583Ser, EED-p.Arg216Ter, NOTCH1-p.Val1578_Leu1579del and PTPN11-p.Ser506Leu were detected by RNA sequencing (Supplementary Table 4). Accordingly, a diagnosis of T/M MPAL, NOS was made according to revised WHO 2016 criteria (2).
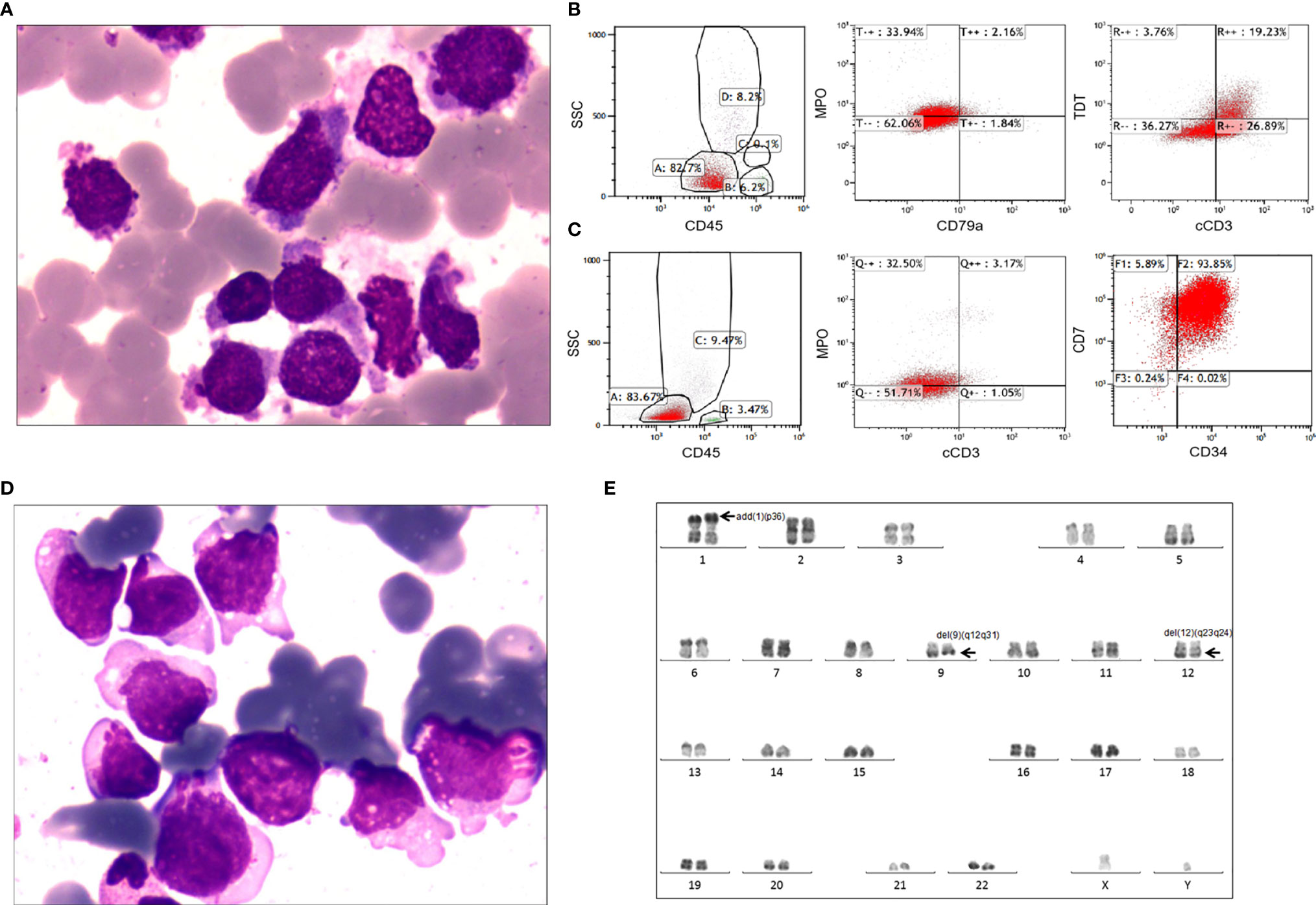
Figure 2 Laboratory characteristics of the adult patient with T/M MPAL with the MED14-HOXA9 fusion. (A) Bone marrow aspirate at diagnosis. Morphological analysis showed hypercellular marrow with increased blasts variable in both cell and nucleolus sizes and negative for POX staining. (Wright’s staining ×1,000). (B) Immunophenotyping of bone marrow cells at diagnosis. Flow cytometric analyses of leukemic cells revealed that the blast cells were positive for TDT, cCD3 and weak positive for myeloperoxidase (MPO), but negative for cCD79a. (C) Immunophenotyping of bone marrow cells at relapse. The blast cells were positive for CD34 and CD7 and weakly positive for MPO and cCD3. (D) Bone marrow aspirate at relapse. Morphological analysis showed 72.5% blasts in the bone marrow smear. (Wright’s staining ×1,000). (E) Karyotype analysis at relapse. R-banding analysis using the bone marrow sample revealed a karyotype of 46,XY,add(1)(p36),del(9)(q12q31),del(12)(q23q24)[16]/46,idem,add(8)(q24)[2]/46,XY[2].
After induction chemotherapy, consisting of idarubicin, vinorelbine and prednisone, the patient achieved complete remission, which was maintained during the subsequent 5 cycles of consolidation chemotherapy. He refused allogenic stem cell transplantation due to concerns regarding transplantation-related mortality. Unfortunately, relapse occurred 11 months after diagnosis when he presented with high fever and general soreness. At relapse, the complete blood cell count showed a white blood cell count of 16.04×109/L, a hemoglobin level of 12.5 g/dL, and a platelet count of 61×109/L, with 26% blasts in the peripheral blood. Morphology analysis revealed 72.5% blasts in the BM (Figure 2D). Flow cytometry showed that the blasts were positive for CD7 (99.7%), CD13 (67.0%), CD33 (99.9%) and CD34 (93.9%), had weak expression of both MPO (35.7%) and cCD3 (4.2%) (Figure 2C and Supplementary Figure 1B), confirming the diagnosis of relapse. Cytogenetic analysis of the BM at relapse showed a karyotype of 46,XY,add(1)(p36),del(9)(q12q31),del (12)(q23q24)[16]/46,idem,add (8)(q24)[2]/46,XY[2] (Figure 2E). NGS of the relapse BM samples using a targeted 222-gene panel revealed mutations, including CREBBP-p.Pro583Ser, EED-p.Arg216Ter, NOTCH1-p.Val1578Glu, PTPN11-p.Ser506Leu, ZNF292-p.Asp829fs and ZNF292-p.Met1243fs (Supplementary Tables 4, 5). The patient failed to respond to the initial induction chemotherapy regimen (idarubicin, vinorelbine, prednisone) combined with cyclophosphamide. He was also refractory to another chemotherapy regimen comprising homoharringtonine and cytarabine. The patient died of pulmonary infection at 34 days after relapse. Timeline is shown in Supplementary Figure 2.
Discussion
T/M MPAL has both of myeloid and T lineage features immunophenotypically. Some studies applied NGS on the sorted blasts by flowcytometry to unravel the driver molecular lesions of T/M MPAL in the literature. The study of Xiao et al. showed that PHF6 mutations were present in all blast compartments regardless of lineage differentiation, implicating that PHF6 was an early mutation in T/M MPAL pathogenesis (9). Alexander et al. reported that biallelic WT1 alterations were common in T/M MPAL (5). Because of the low incidence of T/M MPAL, these findings should be confirmed in larger scale studies using flowcytometry, NGS and single cell RNA sequencing. In this study, we performed RNA sequencing on diagnostic BM samples from 20 adult patients in order to explore the genomic landscape of T/M MPAL.
Here, we present the first case of MED14-HOXA9 fusion to be reported in an adult T/M MPAL patient, confirming this fusion to be a rare but recurrent abnormality in T/M MPAL. MED14 and HOXA9 locate in chromosomes X and 7, respectively. Because there were no visible signs of abnormality in chromosomes X and 7 both at diagnosis and at relapse, we suggest that the MED14-HOXA9 fusion resulted from a cryptic chromosomal rearrangement involving Xp11.4 and 7p15. MED14-HOXA9 was detected in both the diagnostic and relapsed BM samples, therefore we believe that it might be one of the driver mutations responsible for leukemogenesis in this case. Because the patient presented with a normal karyotype at diagnosis but had acquired additional abnormalities at relapse, we reason that the structural aberrations involving chromosomes 1, 8, 9 and 12 observed at relapse were secondary to MED14-HOXA9, whether randomly occurring or contributory to disease progression.
HOXA9, located on chromosome 7p15, encodes a homeobox domain-containing and DNA-binding transcription factor, which plays an important role in regulating morphogenesis, differentiation and expansion of hematopoietic stem cells (10, 11). HOXA9 is overexpressed in more than 50% of acute myeloid leukemia (AML) cases and is associated with poor prognoses (12, 13). A variety of upstream genetic alterations, such as MLL translocations, NUP98 fusions, NPM1 mutations and translocations involving HOXA9 itself, have been demonstrated to cause overexpression of HOXA9 (14). HOXA9 reportedly forms translocations with 5 partner genes in myeloid malignancies: ANGPT1, GATA2 and NIPBL in pediatric acute megakaryoblastic leukemia (15, 16); NUP98 mostly in patients with AML and occasionally in patients with chronic myelogenous leukemia (CML) in blast crisis (17–19); and MSI2 in a patient with CML in the accelerated phase (20). Other rearrangement partners of HOXA9 have been reported in lymphoid malignancies, such as TRB in T cell acute lymphoblastic leukemia (T-ALL) patients (21) and MED12 in a pediatric patient with B cell acute lymphoblastic leukemia (B-ALL) (22) (Supplementary Table 6). This report adds a new case, with a novel HOXA9 rearrangement in T/M MPAL.
In previously reported HOXA9-related fusions, all HOXA9 homeodomains were retained. HOXA9-ANGPT1 apart, all other reported fusions comprise the N-terminus of the partner gene and exon 1 of HOXA9 (located in the C-terminus). In this MED14-HOXA9 case, the breakpoint also occurred in exon 1 of HOXA9, and the HOXA9 segment was identical to that reported in other fusions, which indicates that the homeodomain of HOXA9 is essential for leukemogenesis. HOXA9 was shown to be upregulated by upstream fusions such as NUP98-HOXA9, which inhibits the differentiation of hematopoietic stem cells and increases the self-renewal of hematopoietic stem or progenitor cells (23). Collectively, these findings support the view that HOXA9 is also aberrantly activated by the MED14-HOXA9 fusion, which may play critical roles in the leukemogenesis of T/M MPAL.
MED14, mediator (MED) complex 14, plays an important role in coactivating RNA polymerase II (Pol II)-mediated transcription (24). In the MED14-HOXA9 fusion gene, the main transcriptional domain of MED14 was retained, which suggests an important role in MED14-mediated transactivation of HOXA9 in this fusion. MSI2, another fusion partner of HOXA9, is also a putative RNA-binding protein. Thus, the MED14-HOXA9 fusion adds another fusion gene in human leukemia encoding proteins with RNA-binding properties. A further MED-family gene, MED12, forms a fusion gene with HOXA9 in B-ALL (22, 25). Mechanisms underlying the diversity of cellular settings with different HOXA9 translocations might be attributable to the functions of the various translocation partners.
MED14-HOXA9 fusion has been previously reported in a pediatric patient with T/M MPAL (5). Both the previously reported pediatric patient and the patient in this case were males diagnosed with T/M MPAL, had PTPN11 mutations, relapsed with complex karyotypes, failed ALL chemotherapy and had short overall survivals (Supplementary Table 7). These shared characteristics indicate that patients with MED14-HOXA9 fusion may: 1) be accompanied by PTPN11 mutation serving as a critical alteration; and 2) follow an aggressive clinical course and confer an inferior outcome after conventional chemotherapy. Different NOTCH1 mutations were detected in both diagnostic (p.Val1578_Leu1579del) and relapsed (p.Val1578Glu) BM samples, which both locate in the same hotspot in the heterodimerization domain of NOTCH1 (Supplementary Table 4 and Supplementary Figure 3). The different NOTCH1 mutations at different disease timepoints suggest outgrowth of an alternate leukaemic clone carrying the missense mutation potentially due to some selective growth advantage or in response to therapy. Taken together, we speculated that in addition to the MED14-HOXA9 fusion gene, mutations in PTPN11 and NOTCH1 co-operate and could be important for disease pathogenesis and potentially targeted treatments.
In summary, we report the first case of MED14-HOXA9 in an adult T/M MPAL patient accompanied by precise clinical data, and confirm the presence of MED14-HOXA9 fusion gene in both diagnostic and relapsed BM samples. Further studies will be carried out to reveal the mechanisms underlying the effects of the MED14-HOXA9 fusion on the differentiation and proliferation of leukemia stem cells, as well as suitable treatment strategies for this rare disease.
Data Availability Statement
The data generated by RNA-seq (raw FASTQ files) have been deposited to NCBI Sequence Read Archive (SRA) and have been assigned a BioProject accession number PRJNA726744.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the First Affiliated Hospital of Soochow University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author Contributions
QW, LZ and M-qZ analyzed the clinical data and wrote the manuscript. ZZ and J-dX performed molecular studies and analyses. B-zF and NW performed the analysis of bone marrow smear and flow cytometry data. J-lP and C-xW performed karyotype analyses. RZ, S-nC and H-pD conceived and organized the work, and were major contributors in writing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grant from the National Key R&D Program of China (2019YFA0111000), the National Natural Science Foundation of China (81000222, 81200370, 81700140, 81873449, 81970142, 81900130, 81970136, 82000132), the Natural Science Foundation of the Jiangsu Higher Education Institution of China (18KJA320005), the Natural Science Foundation of Jiangsu Province (BK20190180), China Postdoctoral Science Foundation (2018M632372), the Priority Academic Program Development of Jiangsu Higher Education Institution, Translational Research Grant of NCRCH (2020WSB11, 2020WSB13).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
All the samples were from Jiangsu Biobank of Clinical Resources.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.690218/full#supplementary-material
Supplementary Figure 1 | Immunophenotyping of bone marrow cells at diagnosis (A) and at relapse (B), respectively.
Supplementary Figure 2 | Timeline of the treatment regimens and responses. I, idarubicin; V, vinorelbine; P, prednisone; C, cyclophosphamide; H, homoharringtonine; A, cytarabine.
Supplementary Figure 3 | Gene mutations and karyotype results in newly diagnostic and relapsed samples.
Supplementary Table 1 | The result of RNA sequencing for MED14-HOXA9 fusion gene.
Supplementary Table 2 | Clinical characteristics of the patient at diagnosis and at the time of relapse.
Supplementary Table 3 | Detection of 43 fusion genes by a multiplex RT-PCR panel.
Supplementary Table 4 | Gene mutations detected by NGS and RNA sequencing.
Supplementary Table 5 | Detection of 222 genes by a NGS panel.
Supplementary Table 6 | Reported cases of HOXA9 gene rearrangements in AML, ALL, CML, and MPAL.
Supplementary Table 7 | Clinical characteristics of two patients with the MED14-HOXA9 fusion gene.
References
1. Weinberg OK, Arber DA. Mixed-Phenotype Acute Leukemia: Historical Overview and a New Definition. Leukemia (2010) 24:1844–51. doi: 10.1038/leu.2010.202
2. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood (2016) 127:2391–405. doi: 10.1182/blood-2016-03-643544
3. Matutes E, Pickl WF, Van’t Veer M, Morilla R, Swansbury J, Strobl H, et al. Mixed-Phenotype Acute Leukemia: Clinical and Laboratory Features and Outcome in 100 Patients Defined According to the WHO 2008 Classification. Blood (2011) 117:3163–71. doi: 10.1182/blood-2010-10-314682
4. Maruffi M, Sposto R, Oberley MJ, Kysh L, Orgel E. Therapy for Children and Adults With Mixed Phenotype Acute Leukemia: A Systematic Review and Meta-Analysis. Leukemia (2018) 32:1515–28. doi: 10.1038/s41375-018-0058-4
5. Alexander TB, Gu Z, Iacobucci I, Dickerson K, Choi JK, Xu B, et al. The Genetic Basis and Cell of Origin of Mixed Phenotype Acute Leukaemia. Nature (2018) 562:373–9. doi: 10.1038/s41586-018-0436-0
6. Takahashi K, Wang F, Morita K, Yan Y, Hu P, Zhao P, et al. Integrative Genomic Analysis of Adult Mixed Phenotype Acute Leukemia Delineates Lineage Associated Molecular Subtypes. Nat Commun (2018) 9:2670. doi: 10.1038/s41467-018-04924-z
7. McGowan-Jordan J, Simons A, Schmid R eds. An International System for Human Cytogenomic Nomenclature. S. Karger, Basel (2016). [Reprint of Cytogenet and Genome Res 149(1–2)]
8. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics (2013) 29:15–21. doi: 10.1093/bioinformatics/bts635
9. Xiao W, Bharadwaj M, Levine M, Farnhoud N, Pastore F, Getta BM, et al. PHF6 and DNMT3A Mutations are Enriched in Distinct Subgroups of Mixed Phenotype Acute Leukemia With T-Lineage Differentiation. Blood Adv (2018) 2:3526–39. doi: 10.1182/bloodadvances.2018023531
10. Abramovich C, Pineault N, Ohta H, Humphries RK. Hox Genes: From Leukemia to Hematopoietic Stem Cell Expansion. Ann N Y Acad Sci (2005) 1044:109–16. doi: 10.1196/annals.1349.014
11. Ferrell CM, Dorsam ST, Ohta H, Humphries RK, Derynck MK, Haqq C, et al. Activation of Stem-Cell Specific Genes by HOXA9 and HOXA10 Homeodomain Proteins in CD34+ Human Cord Blood Cells. Stem Cells (2005) 23:644–55. doi: 10.1634/stemcells.2004-0198
12. Collins CT, Hess JL. Role of HOXA9 in Leukemia: Dysregulation, Cofactors and Essential Targets. Oncogene (2016) 35:1090–8. doi: 10.1038/onc.2015.174
13. Lambert M, Alioui M, Jambon S, Depauw S, Van Seuningen I, David-Cordonnier MH. Direct and Indirect Targeting of HOXA9 Transcription Factor in Acute Myeloid Leukemia. Cancers (2019) 11:837. doi: 10.3390/cancers11060837
14. De Braekeleer E, Douet-Guilbert N, Basinko A, Le Bris MJ, Morel F, De Braekeleer M. Hox Gene Dysregulation in Acute Myeloid Leukemia. Future Oncol (2014) 10:475–95. doi: 10.2217/fon.13.195
15. Gruber TA, Larson Gedman A, Zhang J, Koss CS, Marada S, Ta HQ, et al. An Inv(16)(p13.3q24.3)-Encoded CBFA2T3-GLIS2 Fusion Protein Defines an Aggressive Subtype of Pediatric Acute Megakaryoblastic Leukemia. Cancer Cell (2012) 22:683–97. doi: 10.1016/j.ccr.2012.10.007
16. De Rooij JD, Branstetter C, Ma J, Li Y, Walsh MP, Cheng J, et al. Pediatric non-Down Syndrome Acute Megakaryoblastic Leukemia Is Characterized by Distinct Genomic Subsets With Varying Outcomes. Nat Genet (2017) 49:451–6. doi: 10.1038/ng.3772
17. Ahuja HG, Popplewell L, Tcheurekdjian L, Slovak ML. NUP98 Gene Rearrangements and the Clonal Evolution of Chronic Myelogenous Leukemia. Genes Chromosomes Cancer (2001) 30:410–5. doi: 10.1002/1098-2264(2001)9999:9999<::aid-gcc1108>3.0.co;2-9
18. Yamamoto K, Nakamura Y, Saito K, Furusawa S. Expression of the NUP98/HOXA9 Fusion Transcript in the Blast Crisis of Philadelphia Chromosome-Positive Chronic Myelogenous Leukaemia With T (7,11)(P15;P15). Br J Haematol (2000) 109:423–6. doi: 10.1046/j.1365-2141.2000.02003.x
19. Borrow J, Shearman AM, Stanton VP Jr., Becher R, Collins T, Williams AJ, et al. The T (7,11(P15;P15) Translocation in Acute Myeloid Leukaemia Fuses the Genes for Nucleoporin NUP98 and Class I Homeoprotein HOXA9. Nat Genet (1996) 12:159–67. doi: 10.1038/ng0296-159
20. Barbouti A, Höglund M, Johansson B, Lassen C, Nilsson PG, Hagemeijer A, et al. A Novel Gene, MSI2, Encoding a Putative RNA-Binding Protein Is Recurrently Rearranged at Disease Progression of Chronic Myeloid Leukemia and Forms a Fusion Gene With HOXA9 as a Result of the Cryptic T (7,17)(P15;Q23). Cancer Res (2003) 63:1202–6.
21. Le Noir S, Ben Abdelali R, Lelorch M, Bergeron J, Sungalee S, Payet-Bornet D, et al. Extensive Molecular Mapping of Tcrα/δ- and Tcrβ-Involved Chromosomal Translocations Reveals Distinct Mechanisms of Oncogene Activation in T-ALL. Blood (2012) 120:3298–309. doi: 10.1182/blood-2012-04-425488
22. Lilljebjörn H, Henningsson R, Hyrenius-Wittsten A, Olsson L, Orsmark-Pietras C, von Palffy S, et al. Identification of ETV6-RUNX1-Like and DUX4-Rearranged Subtypes in Paediatric B-Cell Precursor Acute Lymphoblastic Leukaemia. Nat Commun (2016) 7:11790. doi: 10.1038/ncomms11790
23. Gough SM, Slape CI, Aplan PD. NUP98 Gene Fusions and Hematopoietic Malignancies: Common Themes and New Biologic Insights. Blood (2011) 118:6247–57. doi: 10.1182/blood-2011-07-328880
24. Cevher MA, Shi Y, Li D, Chait BT, Malik S, Roeder RG. Reconstitution of Active Human Core Mediator Complex Reveals a Critical Role of the MED14 Subunit. Nat Struct Mol Biol (2014) 21:1028–34. doi: 10.1038/nsmb.2914
Keywords: mixed-phenotype acute leukemia, MED14-HOXA9, fusion gene, PTPN11, NOTCH1
Citation: Wang Q, Zhang L, Zhu M-q, Zeng Z, Fang B-z, Xie J-d, Pan J-l, Wu C-x, Wu N, Zhang R, Chen S-n and Dai H-p (2021) A Recurrent Cryptic MED14-HOXA9 Rearrangement in an Adult Patient With Mixed-Phenotype Acute Leukemia, T/myeloid, NOS. Front. Oncol. 11:690218. doi: 10.3389/fonc.2021.690218
Received: 02 April 2021; Accepted: 07 July 2021;
Published: 22 July 2021.
Edited by:
Gurvinder Kaur, All India Institute of Medical Sciences, IndiaReviewed by:
Michael Diamantidis, University Hospital of Larissa, GreeceLaura N. Eadie, South Australian Research and Development Institute, Australia
Copyright © 2021 Wang, Zhang, Zhu, Zeng, Fang, Xie, Pan, Wu, Wu, Zhang, Chen and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ri Zhang, emhhbmdyaV9zekAxNjMuY29t; Su-ning Chen, Y2hlbnN1bmluZ0BzaW5hLmNvbQ==; Hai-ping Dai, ZGFpaGFpcGluZzhAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Qian Wang
Qian Wang Ling Zhang
Ling Zhang Ming-qing Zhu
Ming-qing Zhu Zhao Zeng
Zhao Zeng Bao-zhi Fang
Bao-zhi Fang Jun-dan Xie
Jun-dan Xie Jin-lan Pan
Jin-lan Pan Chun-xiao Wu
Chun-xiao Wu Ni Wu
Ni Wu Ri Zhang
Ri Zhang Su-ning Chen
Su-ning Chen Hai-ping Dai
Hai-ping Dai