- 1Division of Hematology and Medical Oncology, West Virginia University Cancer Institute, Schiffler Cancer Center, Wheeling, WV, United States
- 2Division of Hematology, Department of Internal Medicine, The Ohio State University Comprehensive Cancer Center, Columbus, OH, United States
- 3The Ohio State University Comprehensive Cancer Center, Columbus, OH, United States
MicroRNAs (miRs) are short non-coding RNAs, typically 18-25 nucleotides in length, that are critically important, through their direct effects on target mRNAs, in a variety of cellular processes including cell differentiation, proliferation and survival. Dysregulated miR expression has been identified in numerous cancer types including acute myeloid leukemia (AML). From a clinical standpoint, several miRs have been shown to associate with prognosis in AML patients. Furthermore, they also carry the potential to be used as biomarkers and to inform medical decision making. In addition, several preclinical studies have provided strong rationale to develop novel therapeutic strategies to target miRs in AML. This review will focus on potential clinical applications of miRs in adult AML and will discuss unique miR signatures in specific AML subtypes, their role in prognostication and response to therapy, as well as miRs that are promising therapeutic targets and ongoing clinical trials directed towards targeting clinically relevant miRs in AML that could allow for improvements in current treatment strategies.
Introduction
Acute myeloid leukemia (AML) is a clinically and biologically heterogeneous disease characterized by a number of recurring, sequential genetic alterations that result in a block in differentiation and expansion of immature myeloid precursors. In addition to the cytogenetic and molecular changes that are common in AML (1, 2), dysregulated microRNAs (miRs) have also been identified to play a critical role in leukemogenesis (3–5).
MicroRNAs are small non-coding RNAs, typically 18-25 nucleotides in length that affect the post-transcriptional function of specific mRNAs and their resultant protein targets (6, 7). Previous work has revealed that dysregulated expression of even of a single miR targets multiple mRNAs and modulates the function of numerous cellular pathways (8). Clinically, miR expression patterns have the potential to inform medical decision making. For example, miR expression can be used to differentiate between acute leukemias of ambiguous lineage (9, 10), refine current AML prognostic classification systems (5, 11), potentially detect progression of myelodysplastic syndrome (MDS) to AML (12) and to detect measurable residual disease (MRD) (13, 14).
Over the past decade, several miRs have been shown to be aberrantly under or overexpressed in AML. Although miR expression signatures have been shown to be distinct in specific subtypes of AML and to correlate with prognosis (5, 11, 15–18), miR expression profiling is not yet incorporated into routine clinical practice (19). Additionally, several pre-clinical studies have provided proof-of-concept that miRs are actionable therapeutic targets. However, while miR-directed therapies have proven to be successful in other disease types, most notably in hepatocellular carcinoma (20), cutaneous T-cell lymphoma (21) and diffuse large B-cell lymphoma (22), these therapies remain in an early stage of translation in AML. This review will summarize key clinical applications of microRNAs as they pertain to the management of patients with AML (Table 1) and will also discuss the current status of miR-directed therapies and barriers to implementation.
MicroRNAs and Their Key Functions in Acute Myeloid Leukemia
MicroRNAs modulate a large variety of cellular pathways that are critical for leukemogenesis such as cell differentiation, proliferation, epigenetic regulation, and stem cell function and survival (6). MiRs exert their effects at the post-transcriptional level by binding to the complementary 3’ untranslated region (3’ UTR) of target mRNAs and marking them for cleavage or destruction thereby inhibiting translation (6, 7, 44, 45). Under normal physiological conditions, miRs are essential for maintaining hematopoiesis (46) including stem cell function and lineage commitment (47, 48). Thus, it is not surprising that miR dysregulation plays a critical role in the initiation and maintenance of leukemogenesis.
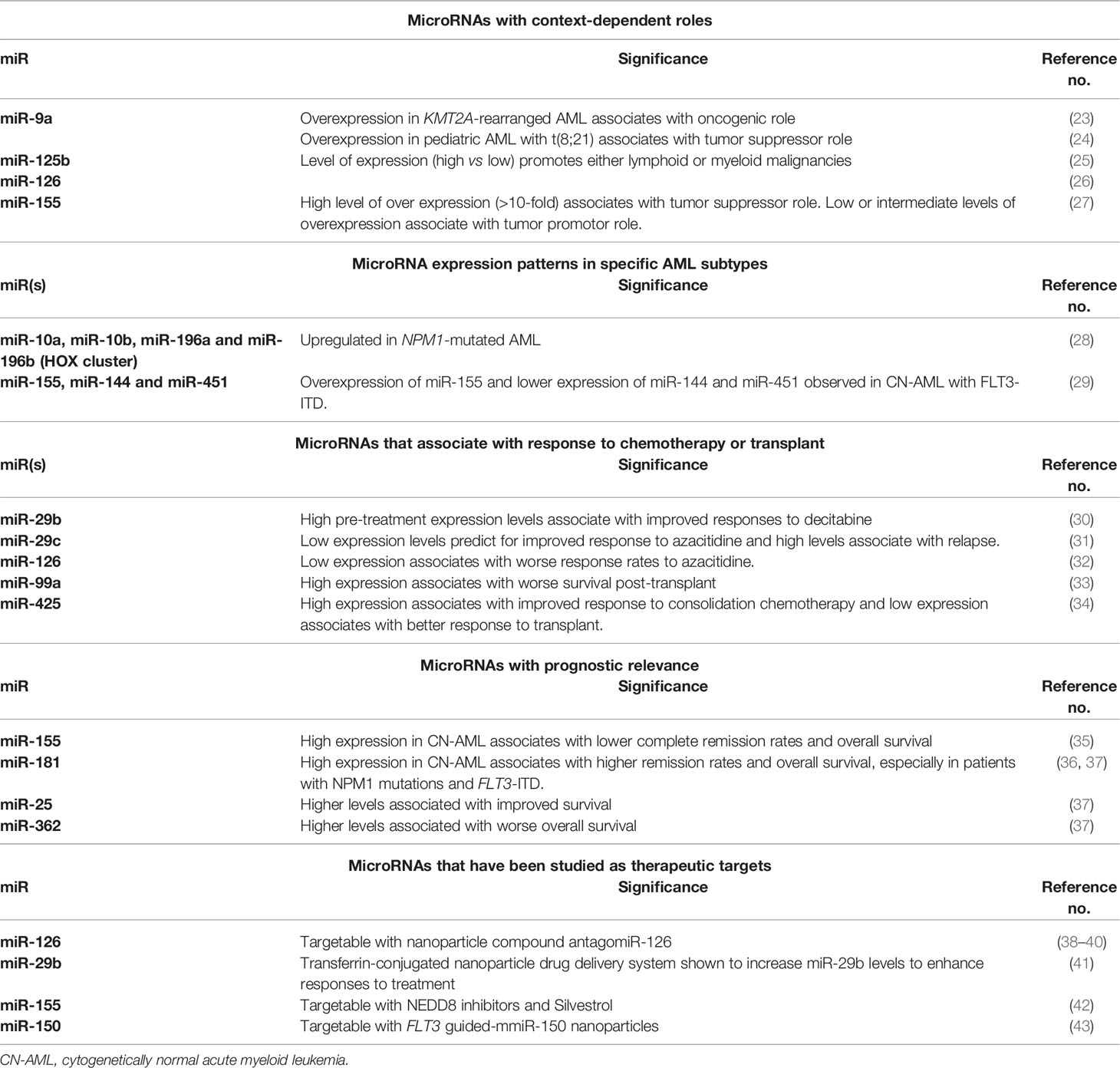
Alterations of miR expression in AML result directly from genomic deletions, translocations, amplifications, and/or from epigenetic alterations through aberrant transcription factors, or oncogenic fusion proteins and global/specific chromatin accessibility changes. Dysregulated miRs can either function as oncogenes (oncomiRs) or as tumor suppressors. Interestingly, it is not uncommon for a specific miR to play opposing roles depending on the cellular context and type of leukemia. In KMT2A-rearranged AML, for instance, miR-9a overexpression has been reported to act as an oncogene (23), but, in pediatric patients with t(8;21), overexpression of miR-9a plays a tumor suppressor role (24).
In addition to this context-dependent duality in function, several studies have demonstrated that the magnitude of expression levels of the same miR (high vs low) can lead to different outcomes when miR expression levels are artificially controlled. For instance, miR-125b promotes either lymphoid or myeloid malignancies depending on its level of expression (25) and dysregulated miR-126 has been shown to promote AML in mouse models due to both over expression and loss of function in concert with t (8, 21) fusion genes (26). Additionally, miR-155 has been shown to function as a tumor suppressor when there is a high level of over expression (>10-fold) and as a tumor promoter when overexpressed to low (< 5 fold) or intermediate levels (5-10 fold) (27). Thus, cellular context is an important consideration when considering the biological functions of miRs in AML.
MicroRNA Expression Patterns Define Specific Subtypes of AML and Associate With Outcome
MicroRNA profiling has been assessed in several AML subtypes and characteristic miR signatures have been observed in specific AML cytogenetic and molecular subgroups (3, 4, 11, 16, 49). One of the first studies to perform comprehensive miR profiling, using a micro-array based assay, on 122 AML patient samples from specific cytogenetic and molecular subgroups identified distinct miR expression profiles associated with cytogenetics and recurrent molecular alterations. Furthermore, this same study demonstrated that overexpression of specific miRs (miR-191 and miR-199a) correlated with prognosis (11). Since then, several groups reported miR expression signatures in several other cytogenetic or molecular subgroups. Older (>60 years) and younger AML patients with NPM1 mutations, displayed upregulation of HOX genes and their associated miRs embedded within the HOX cluster, including miR-10a, miR-10b, miR-196a and miR-196b (28). Cytogenetically normal AML (CN-AML) patients with FLT3-ITD, were also found to have a distinct miR signature, which included overexpression of miR-155 and lower expression of miR-144 and miR-451 (29). Another study profiled miRs in 215 newly diagnosed AML cases using reverse-transcription-polymerase chain reaction (RT-PCR-based) and identified unique miR expression patterns in AML patients with t(8;21), t(15;17), inv(16), NPM1, and CEBPA mutations (3). This, subsequently allowed for identification of specific miRs that were differentially expressed within these subtypes, suggesting that miR expression could potentially be used to classify and characterize AML on a deeper and more comprehensive level than with cytogenetic or molecular data alone (3). In a more recent study, approximately 1000 miRs were sequenced from AML samples and compared to peripheral blood samples collected from control subjects which revealed a higher number of aberrantly expressed miRs in NPM1-mutated and FLT3-mutated AML patients compared to control subjects (50). Several of these miRs had not been previously described in association with these leukemia subtypes (50).
It is worth noting that, although the roles of numerous miRs have been evaluated in AML, only a few have been validated across multiple studies, such as the upregulation of miR-10a, miR-10b and miR-155 in NPM1 mutated and FLT3-ITD AML, respectively.
MicroRNAs Can Help Predict Response to Chemotherapy
MicroRNA expression has also been used to determine the effects of response to hypomethylating agents (HMAs) in AML patients (32, 51). Blum and colleagues, for instance, were able to demonstrate, in a pivotal phase 2 study, that older AML patients with higher pre-treatment levels of miR-29b, which is known to target DNA methyltransferases, were more likely to achieve a clinical response following induction chemotherapy with 10 days of the HMA, decitabine (30). A subsequent preclinical study using the histone deacetylase (HDAC) inhibitor, AR-42, in combination with decitabine revealed that AR-42 priming was able to increase miR-29b expression levels and enhance the antileukemic activity of decitabine in AML cell lines (52). However, the phase I clinical study that followed was unable to show improved responses using this approach (53).
Azacitidine, another important HMA employed for the treatment of AML in older and/or unfit adults has also been evaluated within the context of miR profiling. Another member of the miR-29 family, miR-29c, has been reported to be predictive of favorable responses to azacitidine at low expression levels, whereas upregulated miR-29c, was associated with higher rates of relapse (31). Several other groups have demonstrated that azacitidine responders have differing miR expression patterns compared to azacitidine non-responders (32, 51). Solly and colleagues compared differences in expression between 754 miRs in azacitidine-resistant and azacitidine-sensitive cell lines and were able to show that low expression levels of miRs that affected the function of the DNA methyltransferase, DNMT1, could potentially account for azacitidine resistance in AML patients. Low expression levels of miR-126, an anti-DNMT1 miR, were found to have the most adverse impact on response rates (32).
In addition, certain miRs have associated with clinical responses following allogeneic hematopoietic stem cell transplant (alloHSCT). For example, overexpression of miR-99a, which correlates with inferior prognosis in AML patients (17) was also studied in 74 AML patients who received alloHSCT. In this setting, high miR-99a expression associated with worse event-free survival (EFS) and overall survival (OS). Furthermore, it was identified as an independent risk factor for inferior EFS and OS in AML patients who received transplant, suggesting that miR-99a expression could be used to predict for unfavorable outcome (33). In another study, miR-425 expression levels and impact on EFS and OS were studied in 162 AML patients who received either consolidative chemotherapy or transplant (34). In this report, AML patients younger than age 60 years with high miR-425 expression levels had improved EFS (P=0.001) and OS (P=0.001) compared to low miR-425 expressers whereas low expressers had improved responses to alloHSCT (P<0.001), thereby supporting the role of miR-425 expression levels in order to select the most effective consolidation therapy in younger patients.
MicroRNAs Carry Prognostic Relevance in AML
Many studies have demonstrated that under or overexpression of specific miRs correlates with prognosis in AML patients, particularly in patients with CN-AML. In a seminal study by Marcucci and colleagues, microRNA expression profiling in younger (below age 60 years) CN-AML patients identified 12 miRs, five of which were in the miR-181 family, from which a weighted miR summary value could be derived that inversely associated with EFS (5). In a separate study of 363 patients with CN-AML, high expression of miR-155 correlated with inferior outcomes compared to patients with low miR-155 expression (35). This study was also the first to demonstrate that miR-155 overexpression was an independent prognostic predictor of lower complete remission (CR) rate and shorter OS (35). Similarly, miR-181a expression was also studied in a cohort of younger CN-AML patients and was found to associate with improved CR rates, OS and disease-free survival (DFS). Notably, patients with FLT3-ITD or wild-type NPM1 with high miR-181a expression experienced higher CR rates and improved DFS and OS (36). Since these studies were published, numerous other miRs have been studied and their prognostic impact has been well described in other reviews (4, 45, 54). In a more recent study, TCGA data was analyzed to identify miRs with the greatest prognostic value in 179 non-M3 AML patients. Out of 705 miRs that were studied, miR-181a-2, miR-25 and miR-362 expression levels correlated the most with prognosis (37).
MicroRNAs as Therapeutic Targets in AML
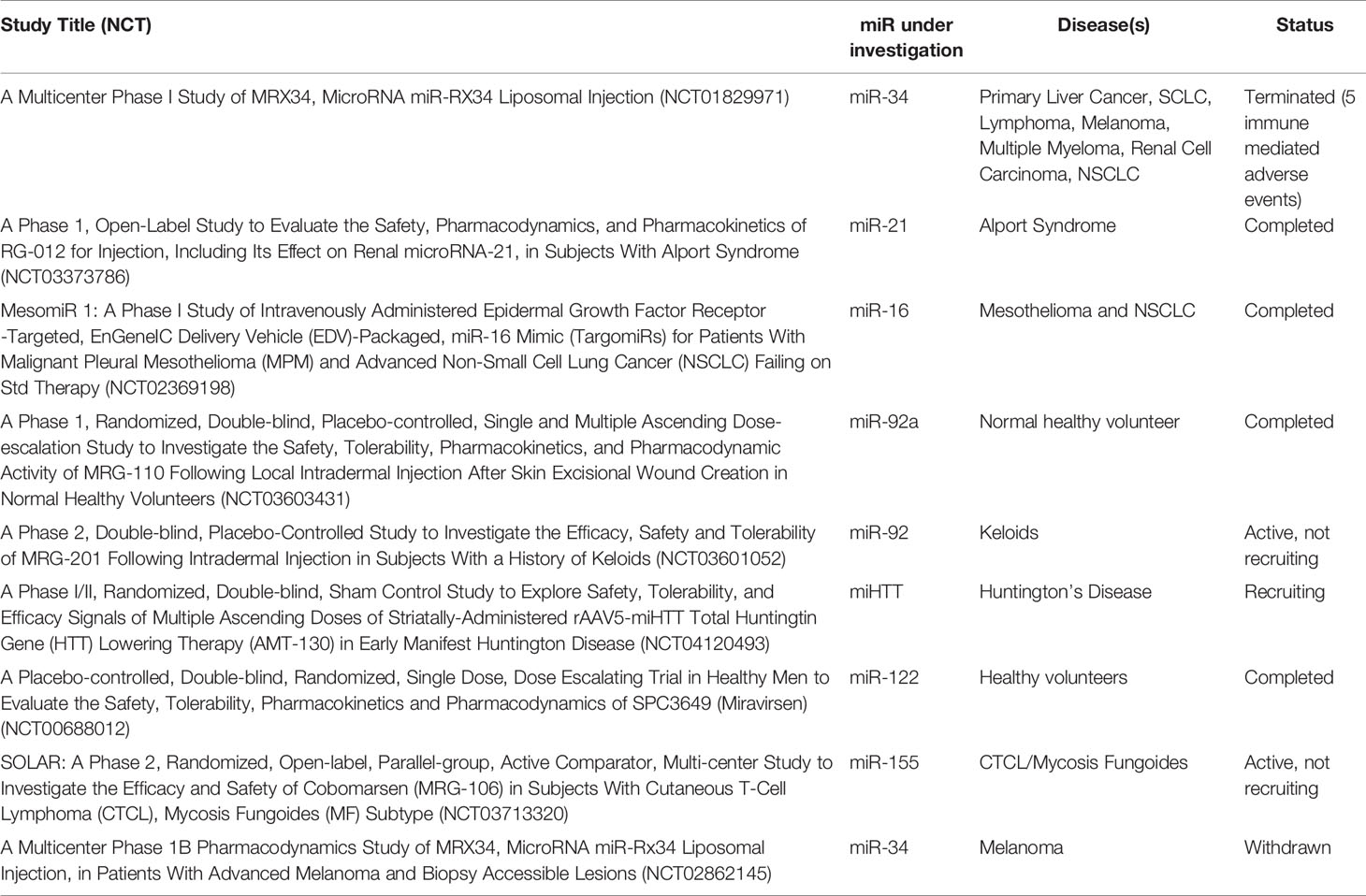
The field of miR therapeutics was originally pioneered in patients with chronic hepatitis C virus (HCV) infection. Miravirsen, an anti-miR-122 locked nucleic acid (LNA) naked oligonucleotide was evaluated in a landmark phase 2 study of 36 patients with chronic HCV and demonstrated a dose-dependent reduction in viral RNA titers and no evidence of viral resistance (20). These findings paved the way for miR-directed therapies in cancer. For example, MRG-106, the LNA miR antagonist to miR-155, was the first of such therapies to be successfully employed in patients with cutaneous T-cell lymphoma (21).
In addition, several miRs serve as potential targets for novel therapeutic approaches in a broad array of diseases. At present, a review of the ClinicalTrials.gov site shows that over 850 studies have been registered that incorporate miRs as biomarkers or as therapeutic targets. Table 2 provides investigational clinical studies that utilize specific therapeutics to target specific miRs.
There are mainly two strategies to silence an oncogenic overexpressed miR (oncomiR): 1) utilization of antisense oligonucleotides (ASOs) and 2) indirect targeting of the oncomiR by using small molecules or other agents that target the transcription or processing of the miR itself. With respect to the first approach, ASOs are oligonucleotides that are complementary to the target sequence and degrade or block the transcript by base-pairing. The initial hurdles in ASO development include the short life, and degradation of ASOs. To overcome this problem chemical modifications such as the addition of 2’-O-methyl groups and LNAs have greatly improved the stability of ASOs as well as their binding affinity and nuclease resistance. Another important issue is addressing optimal delivery of the ASOs to the target cells, more specifically, the AML blasts that are in blood and bone marrow. Over the past several years there has been a push to develop nanoparticle-based drug delivery systems for ASOs, with some success, including the relatively recent Food and Drug Administration approval of the RNA-inhibitor (RNAi) based therapy patisiran in 2018.
The second approach to target an oncomiR is to use small molecules or other agents that affect the transcription or stability of the miR. These indirect strategies are nonspecific since they act on transcription factors that regulate the miR or the epigenetic machinery. For example, one approach that our own group evaluated in a phase 1 study, based on promising pre-clinical data (55) was to use bortezomib and sorafenib priming prior to decitabine therapy in order to increase miR-29b expression in AML blasts to and subsequently improve disease responses (ClinicalTrials.gov Identifier: NCT01861314).
The strategies to restore the expression of a tumor suppressor miR include also an indirect approach using small molecules and other agents and a direct strategy using miR mimics delivered by adenovirus or nanoparticles. In hepatocellular carcinoma, peptide-based nanoparticles have been manufactured to deliver miR-199a-3p, a tumor suppressor, successfully in animal models (56).
Given their pleotropic nature, miR directed therapies offer an attractive treatment approach in AML as well. Several pre-clinical studies have identified candidate miRs that may be amenable to future targeted therapies. Dorrance and colleagues, for instance, were able to confirm that high miR-126 expression levels correlated with adverse outcomes in older CN-AML patients (38). Additionally, miR-126 overexpression was found to associate with a leukemia stem cell (LSC) gene expression profile, suggesting that miR-126 directed therapies contain potential to eradicate LSCs and improve disease responses (38–40). In vivo studies from the same group demonstrated the feasibility of direct targeting of miR-126 using the novel nanoparticle compound antagomiR-126 (38).
Similarly, Huang and colleagues, were able to demonstrate that a novel transferrin-conjugated nanoparticle drug delivery system could be effectively utilized to increase miR-29b levels and that priming AML cells with this agent enhanced responses to decitabine (41).
Preclinical studies evaluating the activity of NEDD8 activating enzyme (NAE) inhibition on miR-155 expression in AML cell lines demonstrated that miR-155 could be downregulated through disruption of binding of NF-KB to the miR-155 promoter, suggesting that NEDD8 inhibitors, such as pevonedistat, may be novel treatment options for AML associated with high miR-155 expression (42). Another preclinical study evaluated the activity of a natural compound, silvestrol, in FLT3-ITD and FLT3-wild type AML and demonstrated potent antileukemic activity and marked downregulation of miR-155, which is typically concurrently regulated in patients with AML and FLT3-ITD (43).
Another miR that is potentially targetable is miR-150, a tumor suppressor and negative regulator of FLT3. MiR-150 has also been described as a promoter of myeloid differentiation, therefore, low or absent expression of miR-150 leads to maturation arrest in AML cells (57). FLT3 guided-mmiR-150 nanoparticles, in preclinical studies, were able to penetrate the bone marrow and suppress the growth of FLT3-mutated AML cells (58).
Taken together, these pre-clinical studies provide important proof-of-concept that miRs can be targeted and their function can be altered with miR-directed therapies. However, translation of these findings to AML patients has been met with significant challenges.
Discussion
Essentially, over the past 15 years, the discovery of miRs and their numerous functions under both normal and pathogenic conditions have provided important biologic insights pertaining to their roles in the development of both malignant and non-malignant disease states.
In AML, specifically, several elegant studies have provided strong rationale to utilize miR expression profiling, in addition to current genetic and molecular testing, in order to better characterize an individual’s specific leukemia. Other groups have also aptly demonstrated that specific miRs can help predict responses to commonly used AML-directed chemotherapy regimens or allogeneic stem cell transplant (30–34, 51). Furthermore, Shivarov and colleagues have suggested that certain overexpressed miRs (miR-19a, miR-181a, miR-17, miR-181b, miR-221, miR-326, and miR-222) can potentially be used for PCR-based MRD detection following intensive induction chemotherapy, though larger studies are needed to validate these findings (14). Haferlach and colleagues led an in international effort, through the Microarray Innovations in Leukemia study, and were able to demonstrate the feasibility of performing whole genome expression profiling in over 3,000 patients with acute leukemia and MDS and also reported a median sensitivity exceeding 99% in classifying at least 14 subtypes of leukemia (26).
Despite these results as well as the unequivocally impactful role of miR expression profiling in the management of AML patients, this platform is not yet routinely incorporated into clinical practice, nor has it been included in commonly used AML risk stratification systems (2, 59). Many of the barriers precluding widespread use and integration are largely related to lack of standardized approaches regarding the optimal sample type as well as the variability in platforms used for miR profiling (array based, RT-PCR or NGS) and the sensitivity of each of these techniques (13, 60). Novel strategies such as direct measurement of the miR molecules like nanoString may circumvent this problem. As miRs continue to be explored and validated in AML, it is likely that, over time, the most optimal way to integrate them into clinical management will become better defined and that miR expression profiling will, at some point, become a more standard aspect of disease classification.
Unfortunately, although several miRs are promising pharmacologic targets in AML, miR-directed treatments have not yet been studied in clinical trials yet for AML patients. There are still several barriers to drug development in this area including concerns for off target effects, toxicity and target delivery to blasts. Hopefully, with proper design of ASOs or mimics and with better delivery vehicles there will soon be phase 1 trials in AML targeting miRs.
Author Contributions
BB and RG wrote, revised, and edited this manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment Cytogenetic Abnormalities are Predictive of Induction Success, Cumulative Incidence of Relapse, and Overall Survival in Adult Patients With De Novo Acute Myeloid Leukemia: Results From Cancer and Leukemia Group B (CALGB 8461). Blood (2002) 100(13):4325–36. doi: 10.1182/blood-2002-03-0772
2. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood (2016) 127(20):2391–405. doi: 10.1182/blood-2016-03-643544
3. Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Löwenberg B. MicroRNA Expression Profiling in Relation to the Genetic Heterogeneity of Acute Myeloid Leukemia. Blood (2008) 111(10):5078–85. doi: 10.1182/blood-2008-01-133355
4. Marcucci G, Mrózek K, Radmacher MD, Garzon R, Bloomfield CD. The Prognostic and Functional Role of microRNAs in Acute Myeloid Leukemia. Blood (2011) 117(4):1121–9. doi: 10.1182/blood-2010-09-191312
5. Marcucci G, Radmacher MD, Maharry K, Mrózek K, Ruppert AS, Paschka P, et al. MicroRNA Expression in Cytogenetically Normal Acute Myeloid Leukemia. N Engl J Med (2008) 358(18):1919–28. doi: 10.1056/NEJMoa074256
6. Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell (2004) 116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5
7. Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell (2009) 136(2):215–33. doi: 10.1016/j.cell.2009.01.002
8. Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-Target Recognition. PloS Biol (2005) 3(3):e85. doi: 10.1371/journal.pbio.0030085
9. Merkerova M, Belickova M, Bruchova H. Differential Expression of microRNAs in Hematopoietic Cell Lineages. Eur J Haematol (2008) 81(4):304–10. doi: 10.1111/j.1600-0609.2008.01111.x
10. de Leeuw D, van den Ancker W, Denkers F, de Menezes RX, Westers TM, Ossenkoppele GJ, et al. MicroRNA Profiling Can Classify Acute Leukemias of Ambiguous Lineage as Either Acute Myeloid Leukemia or Acute Lymphoid Leukemia. Clin Cancer Res (2013) 19(8):2187–96. doi: 10.1158/1078-0432
11. Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, et al. MicroRNA Signatures Associated With Cytogenetics and Prognosis in Acute Myeloid Leukemia. Blood (2008) 111(6):3183–9. doi: 10.1182/blood-2007-07-098749
12. Guo Y, Strickland SA, Mohan S, Li S, Bosompem A, Vickers KC, et al. MicroRNAs and tRNA-Derived Fragments Predict the Transformation of Myelodysplastic Syndromes to Acute Myeloid Leukemia. Leuk Lymphoma (2017) 58(9):1–15. doi: 10.1080/10428194.2016.1272680
13. Grasedieck S, Sorrentino A, Langer C, Buske C, Döhner H, Mertens D, et al. Circulating microRNAs in Hematological Diseases: Principles, Challenges, and Perspectives. Blood (2013) 121(25):4977–84. doi: 10.1182/blood-2013-01-480079
14. Shivarov V, Stoimenov A, Spssov B, Angelova S, Niagolov M, Ivanova M. Patient-Specific microRNA Expression Profiles as a Marker for Minimal Residual Disease in Acute Myeloid Leukemia. Hematology (2014) 19(1):18–21. doi: 10.1179/1607845413y.0000000089
15. Dixon-McIver A, East P, Mein CA, Cazier JB, Molloy G, Chaplin T, et al. Distinctive Patterns of microRNA Expression Associated With Karyotype in Acute Myeloid Leukaemia. PloS One (2008) 3(5):e2141. doi: 10.1371/journal.pone.0002141
16. Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, et al. Distinctive microRNA Signature of Acute Myeloid Leukemia Bearing Cytoplasmic Mutated Nucleophosmin. Proc Natl Acad Sci USA (2008) 105(10):3945–50. doi: 10.1073/pnas.0800135105
17. Si X, Zhang X, Hao X, Li Y, Chen Z, Ding Y, et al. Upregulation of miR-99a Is Associated With Poor Prognosis of Acute Myeloid Leukemia and Promotes Myeloid Leukemia Cell Expansion. Oncotarget (2016) 7(47):78095–109. doi: 10.18632/oncotarget.12947
18. Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, et al. Distinct microRNA Expression Profiles in Acute Myeloid Leukemia With Common Translocations. Proc Natl Acad Sci USA (2008) 105(40):15535–40. doi: 10.1073/pnas.0808266105
19. Foran JM. New Prognostic Markers in Acute Myeloid Leukemia: Perspective From the Clinic. Hematol Am Soc Hematol Educ Program (2010) 2010:47–55. doi: 10.1182/asheducation-2010.1.47
20. Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV Infection by Targeting microRNA. N Engl J Med (2013) 368(18):1685–94. doi: 10.1056/NEJMoa1209026
21. Querfeld C, Pacheco T, Foss FM, Halwani AS, Porcu P, Seto AG, et al. Preliminary Results of a Phase 1 Trial Evaluating MRG-106, A Synthetic microRNA Antagonist (LNA Antimir) of microRNA-155, in Patients With CTCL. Blood (2016) 128(22):1829–. doi: 10.1182/blood.V128.22.1829.1829
22. Anastasiadou E, Seto AG, Beatty X, Hermreck M, Gilles ME, Stroopinsky D, et al. Cobomarsen, An Oligonucleotide Inhibitor of miR-155, Slows DLBCL Tumor Cell Growth In Vitro and In Vivo. Clin Cancer Res (2021) 27(4):1139–49. doi: 10.1158/1078-0432.Ccr-20-3139
23. Chen P, Price C, Li Z, Li Y, Cao D, Wiley A, et al. miR-9 is an Essential Oncogenic microRNA Specifically Overexpressed in Mixed Lineage Leukemia-Rearranged Leukemia. Proc Natl Acad Sci USA (2013) 110(28):11511–6. doi: 10.1073/pnas.1310144110
24. Emmrich S, Katsman-Kuipers JE, Henke K, Khatib ME, Jammal R, Engeland F, et al. miR-9 Is a Tumor Suppressor in Pediatric AML With T(8;21). Leukemia (2014) 28(5):1022–32. doi: 10.1038/leu.2013.357
25. So AY, Sookram R, Chaudhuri AA, Minisandram A, Cheng D, Xie C, et al. Dual Mechanisms by Which miR-125b Represses IRF4 to Induce Myeloid and B-Cell Leukemias. Blood (2014) 124(9):1502–12. doi: 10.1182/blood-2014-02-553842
26. Li Z, Chen P, Su R, Li Y, Hu C, Wang Y, et al. Overexpression and Knockout of miR-126 Both Promote Leukemogenesis. Blood (2015) 126(17):2005–15. doi: 10.1182/blood-2015-04-639062
27. Narayan N, Morenos L, Phipson B, Willis SN, Brumatti G, Eggers S, et al. Functionally Distinct Roles for Different miR-155 Expression Levels Through Contrasting Effects on Gene Expression, in Acute Myeloid Leukaemia. Leukemia (2017) 31(4):808–20. doi: 10.1038/leu.2016.279
28. Becker H, Marcucci G, Maharry K, Radmacher MD, Mrózek K, Margeson D, et al. Favorable Prognostic Impact of NPM1 Mutations in Older Patients With Cytogenetically Normal De Novo Acute Myeloid Leukemia and Associated Gene- and microRNA-Expression Signatures: A Cancer and Leukemia Group B Study. J Clin Oncol (2010) 28(4):596–604. doi: 10.1200/jco.2009.25.1496
29. Whitman SP, Maharry K, Radmacher MD, Becker H, Mrózek K, Margeson D, et al. FLT3 Internal Tandem Duplication Associates With Adverse Outcome and Gene- and microRNA-Expression Signatures in Patients 60 Years of Age or Older With Primary Cytogenetically Normal Acute Myeloid Leukemia: A Cancer and Leukemia Group B Study. Blood (2010) 116(18):3622–6. doi: 10.1182/blood-2010-05-283648
30. Blum W, Garzon R, Klisovic RB, Schwind S, Walker A, Geyer S, et al. Clinical Response and miR-29b Predictive Significance in Older AML Patients Treated With a 10-Day Schedule of Decitabine. Proc Natl Acad Sci USA (2010) 107(16):7473–8. doi: 10.1073/pnas.1002650107
31. Butrym A, Rybka J, Baczyńska D, Poręba R, Kuliczkowski K, Mazur G. Clinical Response to Azacitidine Therapy Depends on microRNA-29c (miR-29c) Expression in Older Acute Myeloid Leukemia (AML) Patients. Oncotarget (2016) 7(21):30250–7. doi: 10.18632/oncotarget.7172
32. Solly F, Koering C, Mohamed AM, Maucort-Boulch D, Robert G, Auberger P, et al. An miRNA-DNMT1 Axis Is Involved in Azacitidine Resistance and Predicts Survival in Higher-Risk Myelodysplastic Syndrome and Low Blast Count Acute Myeloid Leukemia. Clin Cancer Res (2017) 23(12):3025–34. doi: 10.1158/1078-0432.Ccr-16-2304
33. Cheng Z, Zhou L, Hu K, Dai Y, Pang Y, Zhao H, et al. Prognostic Significance of microRNA-99a in Acute Myeloid Leukemia Patients Undergoing Allogeneic Hematopoietic Stem Cell Transplantation. Bone Marrow Transplant (2018) 53(9):1089–95. doi: 10.1038/s41409-018-0146-0
34. Zhang J, Shi J, Zhang G, Zhang X, Yang X, Yang S, et al. MicroRNA-425 Upregulation Indicates Better Prognosis in Younger Acute Myeloid Leukemia Patients Undergoing Chemotherapy. Oncol Lett (2019) 17(6):5793–802. doi: 10.3892/ol.2019.10217
35. Marcucci G, Maharry KS, Metzeler KH, Volinia S, Wu YZ, Mrózek K, et al. Clinical Role of microRNAs in Cytogenetically Normal Acute Myeloid Leukemia: miR-155 Upregulation Independently Identifies High-Risk Patients. J Clin Oncol (2013) 31(17):2086–93. doi: 10.1200/jco.2012.45.6228
36. Schwind S, Maharry K, Radmacher MD, Mrózek K, Holland KB, Margeson D, et al. Prognostic Significance of Expression of a Single microRNA, miR-181a, in Cytogenetically Normal Acute Myeloid Leukemia: A Cancer and Leukemia Group B Study. J Clin Oncol (2010) 28(36):5257–64. doi: 10.1200/jco.2010.29.2953
37. Xue Y, Ge Y, Kang M, Wu C, Wang Y, Rong L, et al. Selection of Three miRNA Signatures With Prognostic Value in Non-M3 Acute Myeloid Leukemia. BMC Cancer (2019) 19(1):109. doi: 10.1186/s12885-019-5315-z
38. Dorrance AM, Neviani P, Ferenchak GJ, Huang X, Nicolet D, Maharry KS, et al. Targeting Leukemia Stem Cells In Vivo With antagomiR-126 Nanoparticles in Acute Myeloid Leukemia. Leukemia (2015) 29(11):2143–53. doi: 10.1038/leu.2015.139
39. Lechman ER, Gentner B, Ng SW, Schoof EM, van Galen P, Kennedy JA, et al. miR-126 Regulates Distinct Self-Renewal Outcomes in Normal and Malignant Hematopoietic Stem Cells. Cancer Cell (2016) 29(2):214–28. doi: 10.1016/j.ccell.2015.12.011
40. de Leeuw D, Denkers F, Olthof MC, Rutten AP, Pouwels W, Schuurhuis GJ, et al. Attenuation of microRNA-126 Expression That Drives CD34+38- Stem/Progenitor Cells in Acute Myeloid Leukemia Leads to Tumor Eradication. Cancer Res (2014) 74(7):2094–105. doi: 10.1158/0008-5472
41. Huang X, Schwind S, Yu B, Santhanam R, Wang H, Hoellerbauer P, et al. Targeted Delivery of microRNA-29b by Transferrin-Conjugated Anionic Lipopolyplex Nanoparticles: A Novel Therapeutic Strategy in Acute Myeloid Leukemia. Clin Cancer Res (2013) 19(9):2355–67. doi: 10.1158/1078-0432.Ccr-12-3191
42. Khalife J, Radomska HS, Santhanam R, Huang X, Neviani P, Saultz J, et al. Pharmacological Targeting of miR-155 via the NEDD8-Activating Enzyme Inhibitor MLN4924 (Pevonedistat) in FLT3-ITD Acute Myeloid Leukemia. Leukemia (2015) 29(10):1981–92. doi: 10.1038/leu.2015.106
43. Alachkar H, Santhanam R, Harb JG, Lucas DM, Oaks JJ, Hickey CJ, et al. Silvestrol Exhibits Significant In Vivo and In Vitro Antileukemic Activities and Inhibits FLT3 and miR-155 Expressions in Acute Myeloid Leukemia. J Hematol Oncol (2013) 6:21. doi: 10.1186/1756-8722-6-21
44. Ambros V. The Functions of Animal microRNAs. Nature (2004) 431(7006):350–5. doi: 10.1038/nature02871
45. Wallace JA, O'Connell RM. MicroRNAs and Acute Myeloid Leukemia: Therapeutic Implications and Emerging Concepts. Blood (2017) 130(11):1290–301. doi: 10.1182/blood-2016-10-697698
46. Garzon R, Croce CM. MicroRNAs in Normal and Malignant Hematopoiesis. Curr Opin Hematol (2008) 15(4):352–8. doi: 10.1097/MOH.0b013e328303e15d
47. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs Modulate Hematopoietic Lineage Differentiation. Science (2004) 303(5654):83–6. doi: 10.1126/science.1091903
48. Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, A microRNA Expressed in Mature B and T Cells, Blocks Early B Cell Development When Expressed Prematurely. Proc Natl Acad Sci USA (2007) 104(17):7080–5. doi: 10.1073/pnas.0702409104
49. Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Béné MC, et al. Clinical Utility of Microarray-Based Gene Expression Profiling in the Diagnosis and Subclassification of Leukemia: Report From the International Microarray Innovations in Leukemia Study Group. J Clin Oncol (2010) 28(15):2529–37. doi: 10.1200/jco.2009.23.4732
50. Pandita A, Ramadas P, Poudel A, Saad N, Anand A, Basnet A, et al. Differential Expression of miRNAs in Acute Myeloid Leukemia Quantified by Nextgen Sequencing of Whole Blood Samples. PloS One (2019) 14(3):e0213078. doi: 10.1371/journal.pone.0213078
51. Krejcik Z, Belickova M, Hrustincova A, Votavova H, Jonasova A, Cermak J, et al. MicroRNA Profiles as Predictive Markers of Response to Azacitidine Therapy in Myelodysplastic Syndromes and Acute Myeloid Leukemia. Cancer Biomark (2018) 22(1):101–10. doi: 10.3233/cbm-171029
52. Mims A, Walker AR, Huang X, Sun J, Wang H, Santhanam R, et al. Increased Anti-Leukemic Activity of Decitabine via AR-42-Induced Upregulation of miR-29b: A Novel Epigenetic-Targeting Approach in Acute Myeloid Leukemia. Leukemia (2013) 4):871–8. doi: 10.1038/leu.2012.342
53. Liva SG, Coss CC, Wang J, Blum W, Klisovic R, Bhatnagar B, et al. Phase I Study of AR-42 and Decitabine in Acute Myeloid Leukemia. Leuk Lymphoma (2020) 61(6):1484–92. doi: 10.1080/10428194.2020.1719095
54. Liao Q, Wang B, Li X, Jiang G. miRNAs in Acute Myeloid Leukemia. Oncotarget (2017) 8(2):3666–82. doi: 10.18632/oncotarget.12343
55. Blum W, Schwind S, Tarighat SS, Geyer S, Eisfeld AK, Whitman S, et al. Clinical and Pharmacodynamic Activity of Bortezomib and Decitabine in Acute Myeloid Leukemia. Blood (2012) 119(25):6025–31. doi: 10.1182/blood-2012-03-413898
56. Varshney A, Panda JJ, Singh AK, Yadav N, Bihari C, Biswas S, et al. Targeted Delivery of microRNA-199a-3p Using Self-Assembled Dipeptide Nanoparticles Efficiently Reduces Hepatocellular Carcinoma in Mice. Hepatology (2018) 67(4):1392–407. doi: 10.1002/hep.29643
57. Morris VA, Zhang A, Yang T, Stirewalt DL, Ramamurthy R, Meshinchi S, et al. MicroRNA-150 Expression Induces Myeloid Differentiation of Human Acute Leukemia Cells and Normal Hematopoietic Progenitors. PloS One (2013) 8(9):e75815. doi: 10.1371/journal.pone.0075815
58. Jiang X, Bugno J, Hu C, Yang Y, Herold T, Qi J, et al. Eradication of Acute Myeloid Leukemia With FLT3 Ligand-Targeted miR-150 Nanoparticles. Cancer Res (2016) 76(15):4470–80. doi: 10.1158/0008-5472.Can-15-2949
59. Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and Management of AML in Adults: 2017 ELN Recommendations From an International Expert Panel. Blood (2017) 129(4):424–47. doi: 10.1182/blood-2016-08-733196
Keywords: acute myeloid leukemia, AML, miR, microRNA, review
Citation: Bhatnagar B and Garzon R (2021) Clinical Applications of MicroRNAs in Acute Myeloid Leukemia: A Mini-Review. Front. Oncol. 11:679022. doi: 10.3389/fonc.2021.679022
Received: 10 March 2021; Accepted: 13 July 2021;
Published: 11 August 2021.
Edited by:
Yong-mi Kim, Children’s Hospital of Los Angeles, United StatesReviewed by:
Deepshi Thakral, All India Institute of Medical Sciences, IndiaLinda Smit, VU University Medical Center, Netherlands
Copyright © 2021 Bhatnagar and Garzon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bhavana Bhatnagar, QmhhdmFuYS5CaGF0bmFnYXIxQGhzYy53dnUuZWR1
 Bhavana Bhatnagar
Bhavana Bhatnagar Ramiro Garzon2,3
Ramiro Garzon2,3