- 1Department of Head and Neck Surgery, Fudan University Shanghai Cancer Center, Shanghai, China
- 2Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, China
- 3Department of Pathology, Fudan University Shanghai Cancer Center, Shanghai, China
- 4Department of Technology, Zhejiang Topgen Clinical Laboratory Co, Ltd., Huzhou, China
- 5Endocrine Surgery Department, N.I. Pirogov Clinic of High Medical Technologies, Saint-Petersburg State University, Saint-Petersburg, Russia
- 6Department of Medical Oncology, Fudan University Shanghai Cancer Center, Shanghai, China
Background: Though fine-needle aspiration (FNA) improved the diagnostic methods of thyroid nodules, there are still parts of nodules that cannot be determined according to cytology. In the Bethesda system for reporting thyroid cytopathology, there are two uncertain cytology results. Thanks to the development of next-generation sequencing technology, it is possible to gain the genetic background of pathological tissue efficiently. Therefore, a combination of the cytology and genetic background may enhance the accuracy of diagnosis in thyroid nodules.
Methods: DNA from 73 FNA samples of thyroid nodules belonging to different cytology types was extracted and exome sequencing was performed by the ThyroLead panel. Test for BRAF mutation was also performed by ARMS-qPCR. Information including age, sex, preoperative cytology, BRAF mutation status tested by ARMS-qPCR, and surgical pathology was collected in electronic medical record system.
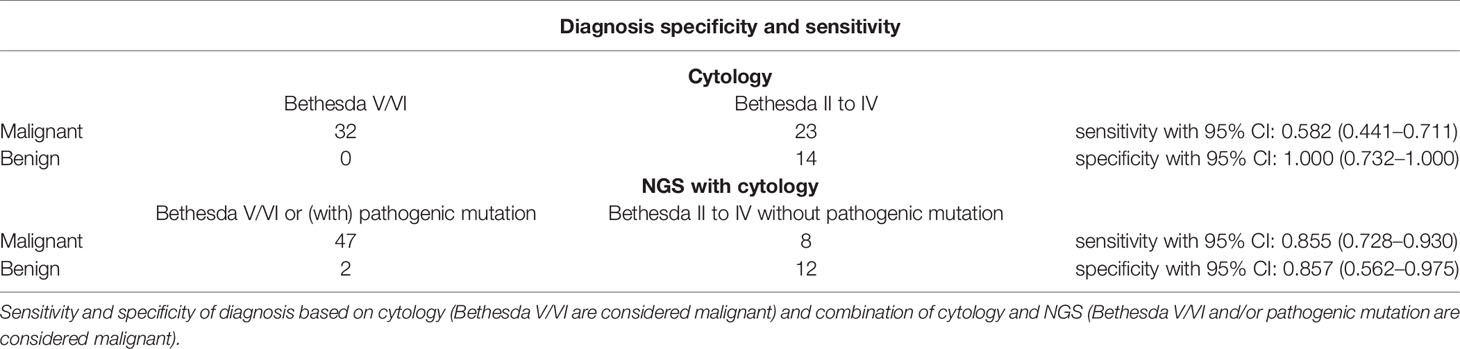
Results: A total of 71 single nucleotide variants, three fusion gene, and two microsatellite instability-high status were detected in 73 FNA samples. BRAF V600E mutation is the most common mutation in these malignant thyroid nodules. After combining the cytology and genetic background detected by next-generation sequencing, the diagnosis sensitivity was increased from 0.582 (95% CI: 0.441–0.711) to 0.855 (95% CI: 0.728–0.930) (P < 0.001) in our group, while the specificity, 1,000 (95% CI: 0.732–1.000) compared to 0.857 (95% CI: 0.562–0.975) (P = 0.25), did not get affected.
Conclusions: Next-generation sequencing in thyroid nodules can enhance the preoperative diagnosis sensitivity by fine-needle aspiration alone. It can also provide genetic background for direction of medication. It is possible for clinicians to combine cytology with genetic alterations for a more precise diagnosis strategy of thyroid nodules.
Introduction
Thyroid cancer came to the 11th place of most common cancer worldwide and 5th in adult women (1), ranking first among endocrine malignancies. Papillary thyroid carcinoma (PTC) and follicular thyroid carcinoma (FTC) known as differentiated thyroid carcinoma (DTC), contribute to more than 95% thyroid malignancies (2). This dramatic increase in the incidence of these cancers is partly due to the improvement of screening methods, such as high-resolution ultrasonic scanning and fine-needle aspiration (FNA) cytopathology, for detecting subclinical thyroid cancers (3). Though thyroid cancer has favorable overall survival, early and accurate diagnosis is crucial for treatment and disease management (4).
Almost all thyroid neoplasms behave as thyroid nodules when first examined, and FNA can provide a robust and rapid evidence to define the characteristics of doubtful nodules after ultrasonic scanning (5). The Bethesda System for Reporting Thyroid Cytopathology (TBSRTC) contains two uncertain cytology between the benign and malignant types. One is Bethesda category III, which contains Atypia of Undetermined Significance (AUS) and Follicular Lesion of Undetermined Significance (FLUS), and another is Bethesda category IV containing Follicular Neoplasm (FN) and Suspicious for a Follicular Neoplasm (SFN) (6). The frequency of incidence of the two uncertain types cytology can reach about 10% respectively (5), leading to a dilemma for both clinicians and patients.
To cope with this predicament, some studies proposed strategies that combine cytopathology with the molecular pattern of aspiration samples to enhance the accuracy of diagnosis (7–9). However, to the best of our knowledge, no research has focused on the difference in genetic change between the nodule of Bethesda III or IV and malignant nodules. Therefore, in this study, we selected 73 samples from patients with thyroid nodules of Bethesda II to VI who underwent FNA in our center to perform target sequencing of thyroid cancer-related genes.
Materials and Methods
Sample Collection and Disposal
We first retrospectively searched FNA samples recorded from 2016 to 2020 and then collected 73 snap-frozen tissue of FNA samples of thyroid nodules at the Department of Pathology, Fudan University Shanghai Cancer Center. DNA was isolated using QIAamp DNA kit (Qiagen, Dusseldorf, Germany).
ARMS-qPCR for BRAF Mutation
BRAF mutation detection assays were routinely performed in our center using BRAF V600E Diagnostic Kit (AmoyDx, China) 145 on an ABI 7500 Real-time PCR system (Life Technologist, 146 USA) according to the instructions of the manufacturer. Genomic DNA, 5–10 ng, was added in a 50 μl system in each assay. The 147 amplification conditions included: 1 cycle at 95°C for 5 min, 15 cycles at 95°C for 25 s, 64°C for 20 s, and 72°C for 20 s, 31 cycles at 93°C for 25 s, 60°C for 35 s, and 72°C for 20 s, and the fluorescent signals were recorded at 60°C. 149 mutation status classification was identified following the manuscripts of kits.
Next-Generation Sequencing
For targeted next-generation sequencing analysis, the ThyroLead panel (Topgen, China) covers the coding exons of 16 genes (BRAF, HRAS, KRAS, NRAS, RET, PIK3CA, CTNNB1, TP53, PTEN, IDH1, DICER1, MEN1, MTOR, TSHR, CDC73, CDKN1B), a 1,000-bp region in the Promoter region of TERT, and both coding exons and 12 introns of four frequently rearranged genes(BRAF, NTRK1, RET and PPARG). At least 30 ng genomic DNA was added and sheared using the Covaris E220 instrument (Covaris, Woburn, MA). Sequence libraries were prepared using KAPA HyperPlus Library Preparation Kit by first producing blunt-ended, 5’-phosphorylated fragments. To the 3’ ends of the dsDNA library fragments, dAMP was added (A-tailing). Next, dsDNA adapters with 3’-dTMP were ligated to the A-tailed library fragments. Library fragments with appropriate adapter sequences were amplified via ligation-mediated pre-capture PCR. Library capture was conducted using Igenetech custom probes system and were biotinylated to allow for sequence enrichment by capture using streptavidin-conjugated beads (Thermo Fisher). Pooled libraries containing captured DNA fragments were subsequently sequenced using the NextSeq 500/550 High Output Kit v2.5 (300 cycles) on the Illumina NextSeq 500 system as 2 × 150-bp paired-end reads.
Data Analysis
Illumina software bcl2fastq was used to generate FastQ files; the number of mismatches was set to 1. The generated FastQ files were processed by software Fastp to remove adaptor sequences as well as poor-quality reads (with more than 40% of bases have phred-scale quality of less than 15) (10). Remaining reads were mapped to hg19 reference genome using BWA-MEM algorithm (11), followed by sorting and duplicate-marking using SAMtools v1.9 and Picard v1.76 respectively. Next, commercial software Sentieon TNseq (version 20180808) was adopted to infer short somatic variants with default parameters (12). Software GeneFuse was used to detect fusions (13). All reported variants were manually checked on IGV to ensure their reliability (14). ANNOVAR (2019Oct24) was used to annotate reported SNVs and small InDels (15). As germline sequencing data were not available, we adopted a strict filtering strategy to remove potential germline variants (16). Variants presented in one or more population databases (ESP, 1000Genome, ExAC, gnomAD) with MAF >= 1% were excluded (17–20). Variants listed as “benign” in the ClinVar database were discarded (21).
Sequence variant confirmation was performed by conventional techniques including Sanger sequencing and real-time PCR. The calculation of analytical accuracy, limits of detection, and assay reproducibility was performed using MedCalc Statistical Software version 9.6. The waterfall plot of SNVs was drawn by R package GenVisR (22).
Statistical Analysis
Fisher’s exact test for categorical variables in paired fourfold table was used. A P-value <0.05 was considered statistically significant.
Results
Basic Information of the Patients Included in This Study
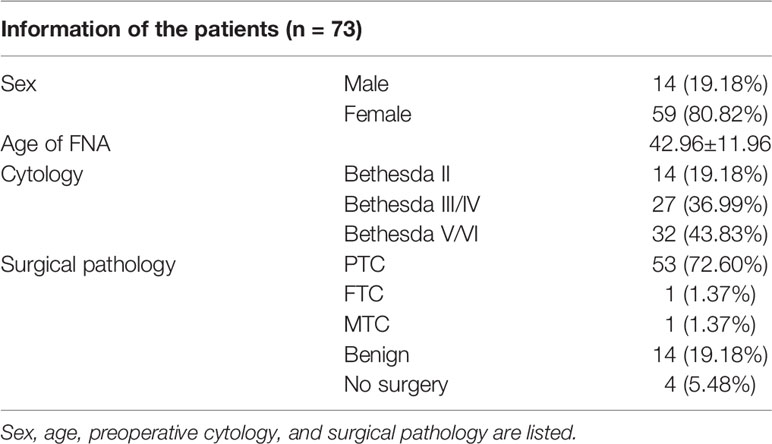
Clinical and demographic data of the patients whose samples were collected for this study were retrieved from the electronic medical record system of our institution. Data on sex, age, Bethesda cytology, and surgical pathology of the study population are provided in Table 1. Among the 73 samples, 14, 27, and 32 were classified as Bethesda II, III/IV, and V/VI respectively. A total of 69 out of 73 patients received surgery in our center and 55 were diagnosed as malignant thyroid tumor, including 53 PTC, 1 FTC, and 1 medullary thyroid tumor (MTC).
Next-Generation Sequencing of FNA Samples
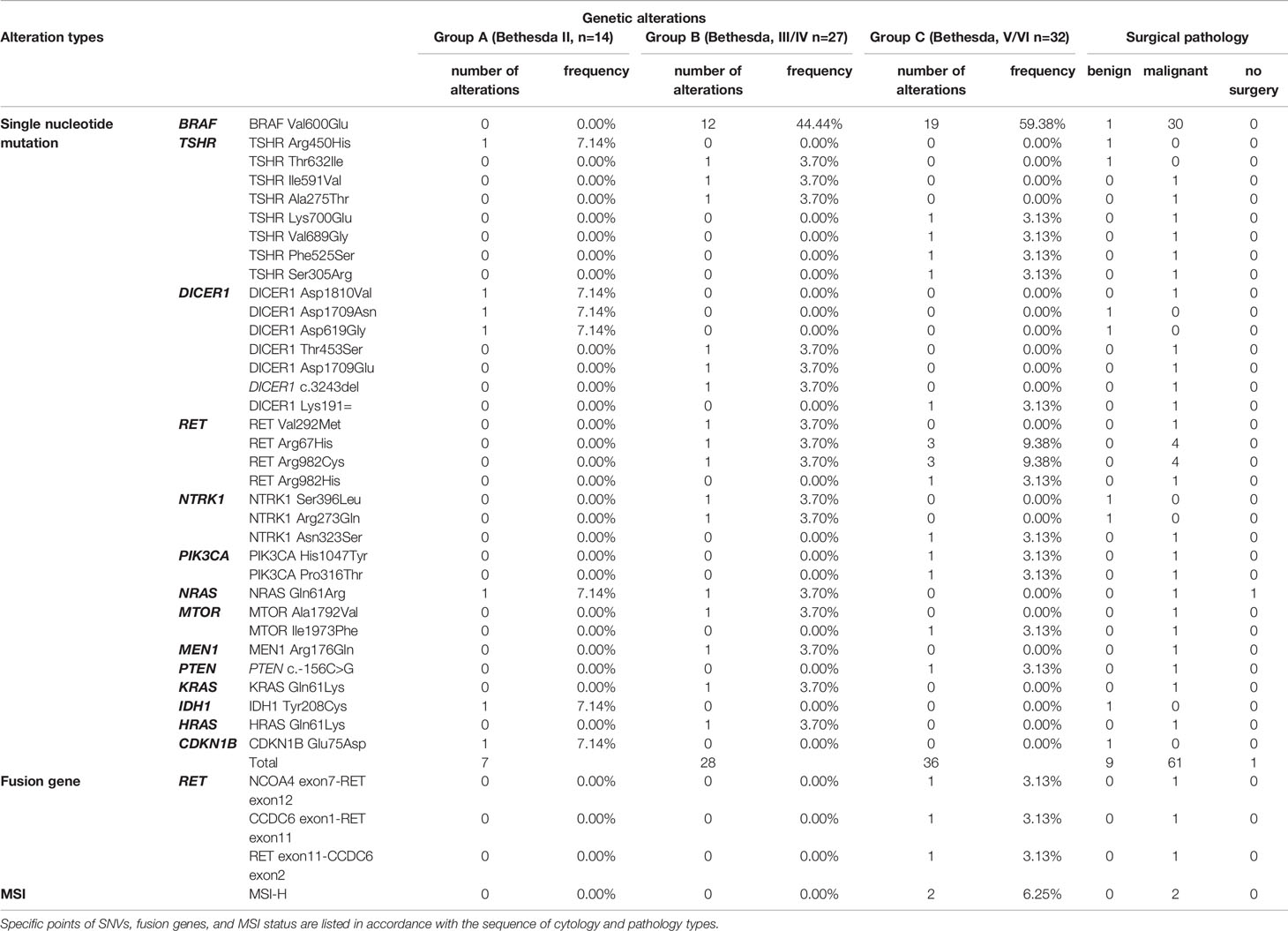
All the FNA samples were tested with the ThyroLead for target sequencing (see details in Methods and Materials), and all quality control (QC) information was listed in Supplementary Table S2. Mutation mapping of single nucleotide variants, fusion genes, and microsatellite instability status (MSI) were shown in Figure 1. A total of 71 single nucleotide mutations were detected unsing ThyroLead (Table 2). BRAF V600E was the most common mutation in thyroid cancer (23), and it all occurred in Bethesda III or higher grade nodules which were categorized by TBSRTC. RET fusion genes, another hallmarker in BRAF-like thyroid cancer (24), were detected in three Bethesda V/VI nodules, and all of these nodules were diagnosed as thyroid cancer after surgery. In addition, four types of RET single nucleotide mutations with uncertain significance were found in Bethesda III/IV and Bethesda V/VI nodules. NRAS, KRAS, and HRAS mutations were found in two, one, and one sample(s) respectively. Three of these four patients underwent surgery, and their samples were determined to be malignant.
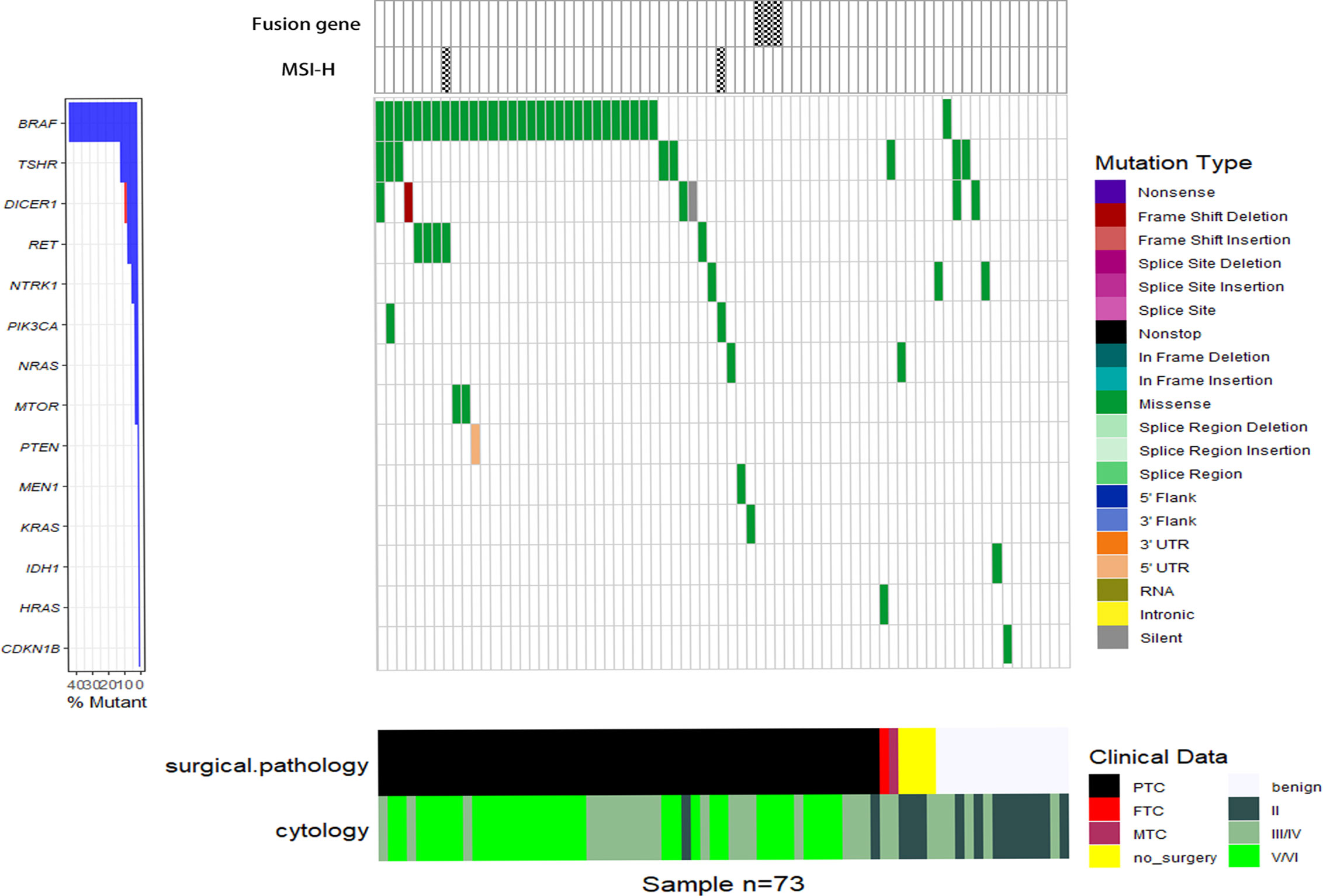
Figure 1 Waterfall plot of SNVs, fusion genes, and MSI-H status detected by NGS. Various types of SNVs, fusion genes and MSI-H status of 73 FNA samples (each vertical column represents a sample) tested by the ThyroLead panel are displayed and the corresponding cytology/pathology of each sample was listed in the bottom. Mutations of different types are marked with respective colors and the spoted grids on the top mean positive fusion genes or MSI-H status.
Eight different types of TSHR mutations were identified. The TSHR Thr632Ile, which was previously detected in thyroid cancer with uncertain significance (25–27), occurred in one Bethesda III/IV nodule. However, it was found to be associated with a benign lesion. The other seven undefined TSHR mutations were sporadically present in various types of cytology.
Mutation of the tumor suppressor gene, DICER1, is related to DICER1 syndrome, which often manifests in an autosomal dominant manner (28, 29). Different from most tumor suppressor genes, loss of one DICER1 allele is sufficient to induce tumorgenesis (30), while homozygous deletion is rare in DICER1-related diseases (31). Furthermore, direct correlation between DICER1 mutations and thyroid cancer has been reported previously (32, 33). Three pathogenic substitutions (two in D1709, one in D1810) and one frameshift deletion of DICER1 were found among the three samples, and two of them were verified as PTC by surgical pathology. However, we were unable to ascertain whether the alterations were germline mutations due to a lack of blood samples.
MSI results from defects in the DNA mismatch repair system (MMR) (34), which is related to a wide range of types, including thyroid cancer (35–37). Through NGS, we detected two samples with MSI-high (MSI-H) status, and they were determined to be papillary thyroid microcarcinoma through surgical pathology.
In addition, pathogenic mutations, including PIK3CA H1047 (38, 39), and mutations with uncertain significance including NTRK1, MTOR, PTEN, IDH1, MEN1, PIK3CA, and CDKN1B were detected. The co-existence of BRAF V600E and DICER1 D1709E along with DICER1 frameshift deletion, BRAF V600E with PIK3CA H1047Y, and BRAF V600E with MSI-H was found in three samples, respectively.
ARMS-qPCR Was More Sensitive Than NGS in BRAF V600E Detection
We have reported that ARMS-qPCR has a higher sensitivity than Sanger sequencing in detecting the BRAF V600E mutation (40), but the efficiency of NGS in BRAF V600E detection was unknown. Therefore, we compared the BRAF V600E mutation frequencies, which were determined by ThyroLead and ARMS-qPCR in our samples. We found that BRAF V600E mutation was detected in 39 of 73 (53.42%) samples and 31 of 73 (42.47%) samples by ARMS-qPCR and NGS, respectively (Supplementary Table S1), and all the positive samples tested by NGS were also positive by ARMS-qPCR. Therefore, the sentivity of ARMS-qPCR in detecting BRAF V600E mutation was higher than that of NGS.
Combination of Cytology With NGS Improved the Rate of Detection of Malignant Nodules
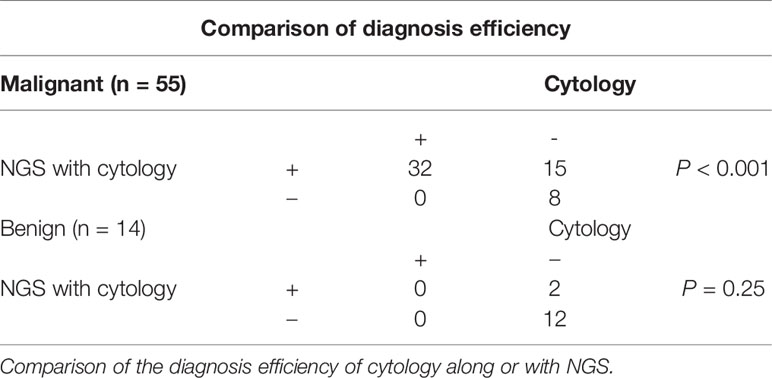
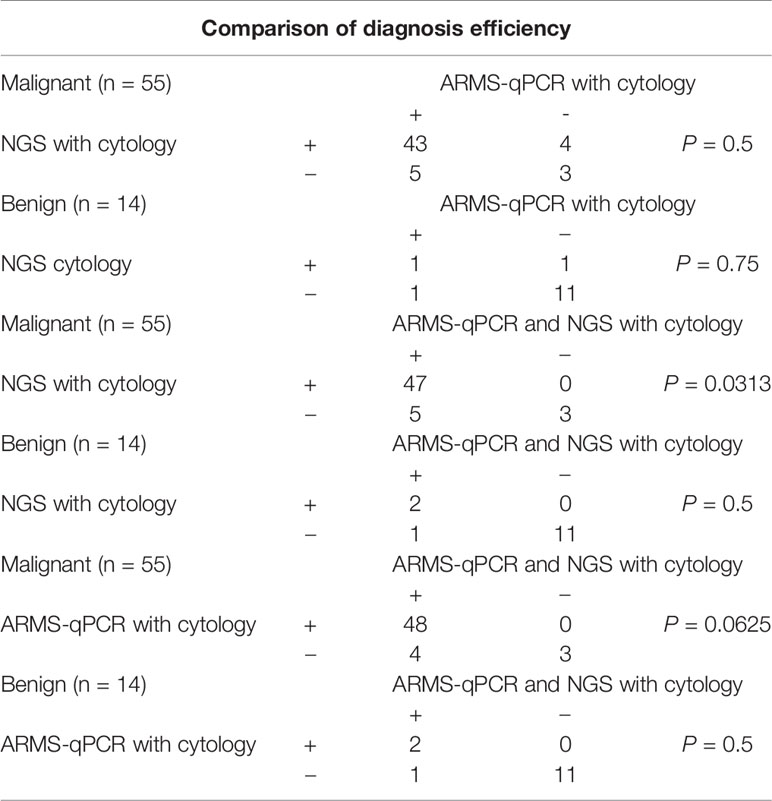
To determine the diagnostic efficiency of NGS for malignant nodules, we compared the diagnostic sensitivity and specificity of cytology, along with a combination of cytology and NGS. Among the 73 patients involved in this study, 69 patients received thyroidectomy and got final surgical pathology. After combining cytology with NGS data, diagnostic sensitivity increased from 0.582 (0.441–0.711) to 0.855 (0.725–0.930) (P < 0.001), while specificity did not show statistically significant change (P = 0.25, Tables 3, 4). Thus, we concluded that the combination of cytology and NGS can markedly improve the detection rate of malignant thyroid nodules compared to that by cytology alone.
Diagnostic Efficiency of Combined Data of ARMS-qPCR and Genetic Alterations
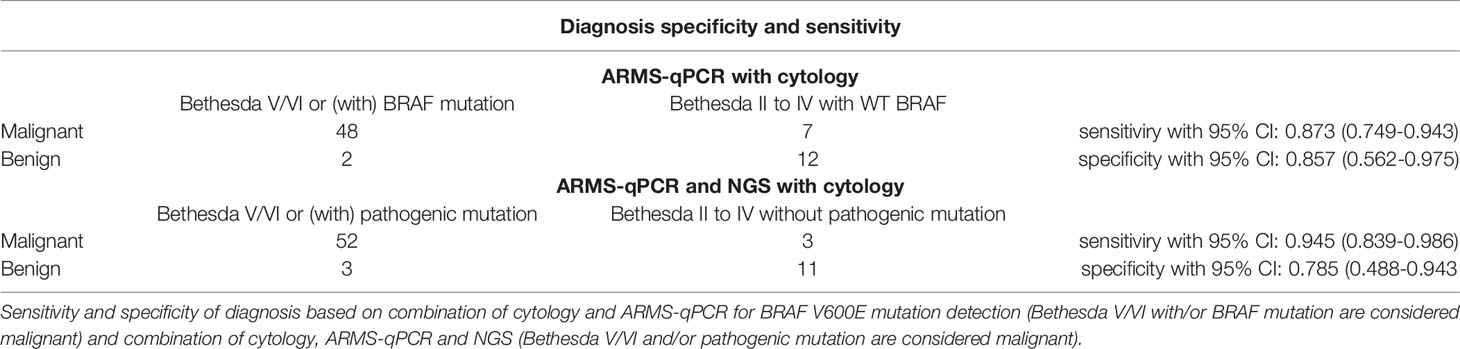
Although BRAF V600E has a favorable diagnostic value in thyroid cancer (23), it only provides evidence for PTC or PTC-derived poorly differentiated thyroid carcinoma (41). To explore whether NGS can increase the accuracy of diagnosis compared with methods based on the detection of BRAF mutations, we analyzed the diagnostic specificity and sensitivity of thyroid cancer by FNA cytology combined with ThyroLead or with ARMS-qPCR for BRAF mutation merely. As to combination of cytology with ARMS-qPCR, the diagnotic sensitivity and specificity were 0.873 (95% CI: 0.749–0.943) and 0.857 (95% CI: 0.562–0.975), respectively (Table 5) and showed no statistical significance compared to that obtained by combination of cytology and NGS (Table 6). Four thyroid carcinomas with wildtype BRAF were classified as malignant nodules by NGS, and five BRAF V600E mutations were detected by ARMS-qPCR and missed by NGS.
We then combined the ARMS-qPCR and specific pathogenic mutations obtained by ThyroLead and recalculated the diagnostic sensitivity and specificity. We found that 52 of the 55 cancerous nodules showed malignant cytology characteristics or/with pathogenic mutation, with a sensitivity of 0.9454 (95% CI: 0.839–0.986) and the specificity of 0.785 (95% CI: 0.488–0.943) (Table 5).
The sensitivity of the combination group was higher than that of the NGS group (P = 0.0313) but showed no statistical significance compared with the ARMS-qPCR group (P = 0.0625). The specificity between the three groups did not differ significantly (P = 0.5). Comparison of the diagnotic efficiencies is shown in Table 6.
Discussion
Standard and advanced preoperative thyroid biopsy methods, which enable patients to receive optimal treatment and avoid unnecessary surgery, have been employed for early and precise diagnosis of thyroid cancer (42). Nonetheless, a considerable proportion of nodules remain ambiguous after FNA, necessitating the need for more accurate diagnostic approaches. BRAF V600E is the earliest and most widely used molecular marker for PTC diagnosis (43). However, there are no such established molecular markers for the remaining pathological types of thyroid cancer, especially FTC (44, 45), often complicating the preoperative diagnosis. Moreover, the prognosis of these non-PTC thyroid cancers is commonly worse because of severe local invasion or distant metastasis (46). This means that it is challenging to diagnose highly malignant cancers preoperatively. NGS technology has revealed genetic alterations underlying thyroid cancer with high efficiency (47). The findings of this study also revealed increase in sensitivity of diagnosis by using NGS to detect cancer-related mutation in thyroid FNA samples including non-PTC cancers.
The Cancer Genome Atlas (TCGA) Research Network described the genomic landscape of PTC in 2014 (24) and revealed the most comprehensive genetic alteration behind PTC. In this large sample atlas, 3.5% of the tumor samples lacked obvious cancer drivers (point mutations, fusion genes, and somatic copy number alterations). In our group, nine of 53 (16.98%) PTC patients showed no cancer drivers (point mutations, fusion genes, and MSI-H), indicating that a more comprehensive target panel could be taken into consideration to enhance the sensitivity of diagnosis.
According to previous reports, BRAF V600E is rarely found in benign thyroid lesion (48). Incidentally, two nodules with BRAF mutations and AUS cytology were surgically proven to be benign in this study. The pathological features represented Hashimoto thyroiditis with dysplasia in individual follicular epithelium for one and adenomatous nodules for another. In addition, the pathogenic TSHR Thr632Ile (25, 26) mutation was found in one sample of AUS cytology, and the final pathology indicated a nodular goiter with fibrosis and calcification. It is unclear whether this group of nodules with pathogenic mutations but benign pathological features is precancerous or just benign lesions. Thus, further studies on the transformation of normal epithelium to a malignant one driven by pathogenic mutations are warranted.
DICER1 functions as an enzyme, which participates in the production of miRNAs (49). Most frequently, germline loss-of-function mutations in DICER1 combined with a somatic mutation in hotspot (D1705, D1709, G1809, D1810, and E1813) of the RNase IIIb region contribute to the DICER1 syndrome, manifesting as susceptibility to various tumors (50). Plenary studies have shown a correlation between DICER1 syndrome and thyroid cancer (32, 33), and studies revealed that the loss expression of DICER1 was correlated with the malignant status of thyroid cancer cells in a miRNA-dependent manner both in vitro and in vivo, and predicted a worse outcome in patients with thyroid cancers (51, 52); however, whether a mere somatic mutation can lead to thyroid cancer is uncertain.
We hypothesize that the development of NGS technology in diagnosis will lead to the discovery of more nodules, which can advance our understanding of thyroid tumorigenesis.
Although ARMS-qPCR is more sensitive in detecting BRAF V600E than Sanger (40) and NGS, the result is restricted to a single mutation. Evidence has shown that the co-existence of mutations, including BRAF, TERET, and P53 often predicts poor prognosis in thyroid cancer (53). It is crucial to identify this group of patients to provide early and personalized treatment, which may improve their outcomes. NGS has the capacity to detect multiple mutations simultaneously, aiding clinicians to perform early interventions in patients with worse prognosis due to the co-existence of mutations. The combination of ARMS-qPCR and NGS showed an increase in diagnostic sensitivity, although it did not reach statistical significance when compared to ARMS-qPCR alone, which may be a result of the limited sample size.
The NGS data of FNA provides not only clues for diagnosis, but also the direction for medication. The BRAF inhibitor dabrafenib and MEK inhibitor trametinib have achieved great success in BRAF V600E-mutated anaplastic thyroid carcinoma (ATC) (54). Targeted therapy for other mutations also showed a breakthrough in advanced thyroid cancer (55, 56). Recent studies have shown that several solid tumors with MSI-H status showed good response to PD-1 inhibitors (57–59) and the efficacy of the immune checkpoint inhibitor (ICI) in ATC has been proven (60). Thus, MSI status could be a potential predictive factor for immunotherapy in advanced thyroid cancer.
Our study had some limitations. First, due to the small number of samples in our study, we were unable to associate some mutations with benign or malignant thyroid nodules. Moreover, some potentially instructive mutations were not detected in NGS sequencing. Second, we were unable to evaluate the germline mutation of DICER1. Therefore, further studies with a more complete panel are warranted.
Conclusions
In this study, we performed exome sequencing of FNA samples of thyroid nodules belonging to different cytology types and mapped mutations in the corresponding patients. By analyzing the sequencing data between preoperative cytology types and final pathology after surgery, we concluded that the combination of cytology with genetic alterations contributes to a more precise diagnosis strategy for thyroid nodules.
Data Availability Statement
The data are now openly accessible in China National Center for Bioinformation at https://bigd.big.ac.cn/gsa-human/browse/HRA000846.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Fudan University Shanghai Cancer Center. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conceptualization, NQ and Y-LW. Methodology, D-MJ and L-YL. Writing-original draft preparation, L-CT and W-LL. Investigation, W-LL, L-CT, and LZ. Visualization, L-CT and P-ZH. Writing—review and editing, Y-LW, NQ, D-MJ, YW, and Q-HJ, and AS. Data curation, L-CT, X-LZ, and L-YL. Resources, X-LZ, P-CY, and XS. Supervision, D-MJ. Project administration, NQ and Y-LW. Funding acquisition, NQ and Y-LW. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (82002831 to W-LL, 81772851 and 81972501 to Y-LW, 81702649 to NQ).
Conflict of Interest
Author L-YL was employed by Zhejiang Topgen Clinical Laboratory Co, Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.677892/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Pereira M, Williams VL, Hallanger Johnson J, Valderrabano P. Thyroid Cancer Incidence Trends in the United States: Association With Changes in Professional Guideline Recommendation. Thyroid (2020) 30(8):1132–40. doi: 10.1089/thy.2019.0415
3. Furuya-Kanamori L, Bell KJL, Clark J, Glasziou P, Doi SAR. Prevalence of Differentiated Thyroid Cancer in Autopsy Studies Over Six Decades: A Meta-Analysis. JCO (2016) 34(30):3672–9. doi: 10.1200/JCO.2016.67.7419
4. Lan L, Luo Y, Zhou M, Huo L, Chen H, Zuo Q, et al. Comparison of Diagnostic Accuracy of Thyroid Cancer With Ultrasound-Guided Fine-Needle Aspiration and Core-Needle Biopsy: A Systematic Review and Meta-Analysis. Front Endocrinol (2020) 11:44. doi: 10.3389/fendo.2020.00044
5. Bongiovanni M, Spitale A, Faquin WC, Mazzucchelli L, Baloch ZW. The Bethesda System for Reporting Thyroid Cytopathology: A Meta-Analysis. Acta Cytol (2012) 56(4):333–9. doi: 10.1159/000339959
6. Cibas ES, Ali SZ. The 2017 Bethesda System for Reporting Thyroid Cytopathology. Thyroid (2017) 27(11):1341–6. doi: 10.1089/thy.2017.0500
7. Keutgen XM, Filicori F, Crowley MJ, Wang Y, Scognamiglio T, Hoda R, et al. A Panel of Four miRNAs Accurately Differentiates Malignant From Benign Indeterminate Thyroid Lesions on Fine Needle Aspiration. Clin Cancer Res (2012) 18(7):2032–8. doi: 10.1158/1078-0432.CCR-11-2487
8. Nikiforov YE, Yip L, Nikiforova MN. New Strategies in Diagnosing Cancer in Thyroid Nodules: Impact of Molecular Marker. Clin Cancer Res (2013) 19(9):2283–8. doi: 10.1158/1078-0432.CCR-12-1253
9. Steward DL, Carty SE, Sippel RS, Yang SP, Sosa JA, Sipos JA, et al. Performance of a Multigene Genomic Classifier in Thyroid Nodules With Indeterminate Cytology: A Prospective Blinded Multicenter Study. JAMA Oncol (2019) 5(2):204. doi: 10.1001/jamaoncol.2018.4616
10. Chen S, Zhou Y, Chen Y, Gu J. Fastp: An Ultra-Fast All-In-One FASTQ Preprocessor. Bioinformatics (2018) 34(17):i884–90. doi: 10.1093/bioinformatics/bty560
11. Li H, Durbin R. Fast and Accurate Short Read Alignment With Burrows-Wheeler Transform. Bioinformatics (2009) 25(14):1754–60. doi: 10.1093/bioinformatics/btp324
12. Freed D, Aldana R, Weber JA, Edwards JS. The Sentieon Genomics Tools - A Fast and Accurate Solution to Variant Calling From Next-Generation Sequence Data [Internet]. Bioinformatics (2017). doi: 10.1101/115717
13. Chen S, Liu M, Huang T, Liao W, Xu M, Gu J. GeneFuse: Detection and Visualization of Target Gene Fusions From DNA Sequencing Data. Int J Biol Sci (2018) 14(8):843–8. doi: 10.7150/ijbs.24626
15. Wang K, Li M, Hakonarson H. ANNOVAR: Functional Annotation of Genetic Variants From High-Throughput Sequencing Data. Nucleic Acids Res (2010) 38(16):e164–4. doi: 10.1093/nar/gkq603
16. Sukhai MA, Misyura M, Thomas M, Garg S, Zhang T, Stickle N, et al. Somatic Tumor Variant Filtration Strategies to Optimize Tumor-Only Molecular Profiling Using Targeted Next-Generation Sequencing Panel. J Mol Diagnostics (2019) 21(2):261–73. doi: 10.1016/j.jmoldx.2018.09.008
17. Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, et al. Analysis of 6,515 Exomes Reveals the Recent Origin of Most Human Protein-Coding Variants. Nature (2013) 493(7431):216–20. doi: 10.1038/nature11690
18. Genome Aggregation Database Consortium, Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, et al. The Mutational Constraint Spectrum Quantified From Variation in 141,456 Humans. Nature (2020) 581(7809):434–43. doi: 10.1038/s41586-020-2308-7
19. Karczewski KJ, Weisburd B, Thomas B, Solomonson M, Ruderfer DM, Kavanagh D, et al. The ExAC Browser: Displaying Reference Data Information From Over 60 000 Exomes. Nucleic Acids Res (2017) 45(D1):D840–5. doi: 10.1093/nar/gkw971
20. The 1000 Genomes Project Consortium. A Global Reference for Human Genetic Variation. Nature (2015) 526(7571):68–74. doi: 10.1038/nature15393
21. Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: Public Archive of Relationships Among Sequence Variation and Human Phenotype. Nucl Acids Res (2014) 42(D1):D980–5. doi: 10.1093/nar/gkt1113
22. Skidmore ZL, Wagner AH, Lesurf R, Campbell KM, Kunisaki J, Griffith OL, et al. GenVisR: Genomic Visualizations in R. Bioinformatics (2016) 32(19):3012–4. doi: 10.1093/bioinformatics/btw325
23. Nikiforov YE, Nikiforova MN. Molecular Genetics and Diagnosis of Thyroid Cancer. Nat Rev Endocrinol (2011) 7(10):569–80. doi: 10.1038/nrendo.2011.142
24. Agrawal N, Akbani R, Aksoy BA, Ally A, Arachchi H, Asa SL, et al. Integrated Genomic Characterization of Papillary Thyroid Carcinoma. Cell (2014) 159(3):676–90. doi: 0.1016/j.cell.2014.09.050
25. Führer D, Holzapfel H-P, Wonerow P, Scherbaum WA, Paschke R. Somatic Mutations in the Thyrotropin Receptor Gene and Not in the G s α Protein Gene in 31 Toxic Thyroid Nodules 1. J Clin Endocrinol Metab (1997) 82(11):3885–91. doi: 10.1210/jcem.82.11.4382
26. Stephenson A, Lau L, Eszlinger M, Paschke R. The Thyrotropin Receptor Mutation Database Update. Thyroid (2020) 30(6):931–5. doi: 10.1089/thy.2019.0807
27. Dorothea S, Nazy S, Rossella E, Jorge LG, Geraldo M-N, James AF. Structural Studies of the Thyrotropin Receptor and Gs Alpha in Human Thyroid Cancers: Low Prevalence of Mutations Predicts Infrequent Involvement in Malignant Transformation. J Clin Endocrinol Metab (1996) 81(11):3898–901. doi: 10.1210/jcem.81.11.8923835
28. Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, et al. DICER1 Mutations in Familial Pleuropulmonary Blastoma. Science (2009) 325(5943):965–5. doi: 10.1126/science.1174334
29. de Kock L, Priest JR, Foulkes WD, Alexandrescu S. An Update on the Central Nervous System Manifestations of DICER1 Syndrome. Acta Neuropathol (2020) 139(4):689–701. doi: 10.1007/s00401-019-01997-y
30. Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, et al. Dicer1 Functions as a Haploinsufficient Tumor Suppressor. Genes Dev (2009) 23(23):2700–4. doi: 10.1101/gad.1848209
31. Foulkes WD, Priest JR, Duchaine TF. DICER1: Mutations, microRNAs and Mechanisms. Nat Rev Cancer (2014) 14(10):662–72. doi: 10.1038/nrc3802
32. Rutter MM, Jha P, Schultz KAP, Sheil A, Harris AK, Bauer AJ, et al. DICER1 Mutations and Differentiated Thyroid Carcinoma: Evidence of a Direct Association. J Clin Endocrinol Metab (2016) 101(1):1–5. doi: 10.1210/jc.2015-2169
33. Khan NE, Bauer AJ, Schultz KAP, Doros L, Decastro RM, Ling A, et al. Quantification of Thyroid Cancer and Multinodular Goiter Risk in the DICER1 Syndrome: A Family-Based Cohort Study. J Clin Endocrinol Metab (2017) 102(5):1614–22. doi: 10.1210/jc.2016-2954
34. Eshleman JR, Markowitz SD. Mismatch Repair Defects in Human Carcinogenesis. Hum Mol Genet (1996) 5(Supplement_1):1489–94. doi: 10.1093/hmg/5.Supplement_1.1489
35. Boland CR, Goel A. Microsatellite Instability in Colorectal Cancer. Gastroenterology (2010) 138(6):2073–87. e3. doi: 10.1053/j.gastro.2009.12.064
36. Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and Characterization of Microsatellite Instability Across 18 Cancer Types. Nat Med (2016) 22(11):1342–50. doi: 10.1038/nm.4191
37. Genutis LK, Tomsic J, Bundschuh RA, Brock PL, Williams MD, Roychowdhury S, et al. Microsatellite Instability Occurs in a Subset of Follicular Thyroid Cancer. Thyroid (2019) 29(4):523–9. doi: 10.1089/thy.2018.0655
38. Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying Recurrent Mutations in Cancer Reveals Widespread Lineage Diversity and Mutational Specificity. Nat Biotechnol (2016) 34(2):155–63. doi: 10.1038/nbt.3391
39. Kunstman JW, Juhlin CC, Goh G, Brown TC, Stenman A, Healy JM, et al. Characterization of the Mutational Landscape of Anaplastic Thyroid Cancer via Whole-Exome Sequencing. Hum Mol Genet (2015) 24(8):2318–29. doi: 10.1093/hmg/ddu749
40. Yu P-C, Tan L-C, Zhu X-L, Shi X, Chernikov R, Semenov A, et al. ARMS-qPCR Improves Detection Sensitivity of Earlier Diagnosis of Papillary Thyroid Cancers With Worse Prognosis Determined by BRAF V600E and TERT Promoter Co-Existing Mutations. Endocrine Practice (2021) 27(7):698–705. doi: 10.1016/j.eprac.2021.01.015
41. Nikiforova MN, Kimura ET, Gandhi M, Biddinger PW, Knauf JA, Basolo F, et al. BRAF Mutations in Thyroid Tumors Are Restricted to Papillary Carcinomas and Anaplastic or Poorly Differentiated Carcinomas Arising From Papillary Carcinoma. J Clin Endocrinol Metab (2003) 88(11):5399–404. doi: 10.1210/jc.2003-030838
42. Feldkamp J, Führer D, Luster M, Musholt TJ, Spitzweg C, Schott M. Fine Needle Aspiration in the Investigation of Thyroid Nodules. Deutsches Ärzteblatt Int (2016) 113(20):353–359. doi: 10.3238/arztebl.2016.0353
43. Zhao C-K, Zheng J-Y, Sun L-P, Xu R-Y, Wei Q, Xu H-X. BRAFV600E Mutation Analysis in Fine-Needle Aspiration Cytology Specimens for Diagnosis of Thyroid Nodules: The Influence of False-Positive and False-Negative Results. Cancer Med (2019) 8(12):5577–89. doi: 10.1002/cam4.2478
44. Castellana M, Piccardo A, Virili C, Scappaticcio L, Grani G, Durante C, et al. Can Ultrasound Systems for Risk Stratification of Thyroid Nodules Identify Follicular Carcinoma? Cancer Cytopathol (2020) 128(4):250–9. doi: 10.1002/cncy.22235
45. Trimboli P, Giovanella L, Valabrega S, Andrioli M, Baldelli R, Cremonini N, et al. Ultrasound Features of Medullary Thyroid Carcinoma Correlate With Cancer Aggressiveness: A Retrospective Multicenter Study. Clin Cancer Res (2014) 33(1):87. doi: 10.1186/s13046-014-0087-4
46. Carling T, Udelsman R. Thyroid Cancer. Annu Rev Med (2014) 65(1):125–37. doi: 10.1146/annurev-med-061512-105739
47. Cha YJ, Koo JS. Next-Generation Sequencing in Thyroid Cancer. J Transl Med (2016) 14(1):322. doi: 10.1186/s12967-016-1074-7
48. Kim SK, Hwang TS, Yoo YB, Han HS, Kim D-L, Song K-H, et al. Surgical Results of Thyroid Nodules According to a Management Guideline Based on the BRAFV600E Mutation Status. J Clin Endocrinol Metab (2011) 96(3):658–64. doi: 10.1210/jc.2010-1082
49. Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a Bidentate Ribonuclease in the Initiation Step of RNA Interference. Nature (2001) 409(6818):363–6. doi: 10.1038/35053110
50. Kock L, Wu MK, Foulkes WD. Ten Years of DICER1 Mutations: Provenance, Distribution, and Associated Phenotypes. Hum Mutat (2019) 40(11):1939–53. doi: 10.1002/humu.23877
51. Yuan X, Mu N, Wang N, Strååt K, Sofiadis A, Guo Y, et al. GABPA Inhibits Invasion/Metastasis in Papillary Thyroid Carcinoma by Regulating DICER1 Expression. Oncogene (2019) 38(7):965–79. doi: 10.1038/s41388-018-0483-x
52. Ramírez-Moya J, Wert-Lamas L, Riesco-Eizaguirre G, Santisteban P. Impaired microRNA Processing by DICER1 Downregulation Endows Thyroid Cancer With Increased Aggressiveness. Oncogene (2019) 38(27):5486–99. doi: 10.1038/s41388-019-0804-8
53. Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, et al. Genomic and Transcriptomic Hallmarks of Poorly Differentiated and Anaplastic Thyroid Cancers. J Clin Invest (2016) 126(3):1052–66. doi: 10.1172/JCI85271
54. Subbiah V, Kreitman RJ, Wainberg ZA, Cho JY, Schellens JHM, Soria JC, et al. Dabrafenib and Trametinib Treatment in Patients With Locally Advanced or Metastatic BRAF V600–Mutant Anaplastic Thyroid Cancer. JCO (2018) 36(1):7–13. doi: 10.1200/JCO.2017.73.6785
55. Ho AL, Grewal RK, Leboeuf R, Sherman EJ, Pfister DG, Deandreis D, et al. Selumetinib-Enhanced Radioiodine Uptake in Advanced Thyroid Cancer. N Engl J Med (2013) 368(7):623–32. doi: 10.1056/NEJMoa1209288
56. Wirth LJ, Sherman E, Robinson B, Solomon B, Kang H, Lorch J, et al. Efficacy of Selpercatinib in RET -Altered Thyroid Cancers. N Engl J Med (2020) 383(9):825–35. doi: 10.1056/NEJMe2024831
57. André T, Shiu K-K, Kim TW, Jensen BV, Jensen LH, Punt C, et al. Pembrolizumab in Microsatellite-Instability–High Advanced Colorectal Cancer. N Engl J Med (2020) 383(23):2207–18. doi: 10.1056/NEJMoa2017699
58. Ke W, Cai De L, Jian Nan S, Chun Nian W, Guo G, Yang Ke H, et al. Primary Hepatic Squamous Cell Carcinoma With MSI-H Shows Good Response to PD-1 Inhibitor as Adjuvant Therapy. Hepatology (2021). doi: 10.1002/hep.31737
59. Raj N, Zheng Y, Kelly V, Katz SS, Chou J, Do RKG, et al. PD-1 Blockade in Advanced Adrenocortical Carcinoma. JCO (2020) 38(1):71–80. doi: 10.1200/JCO.19.01586
Keywords: thyroid nodules, next-generation sequencing, fine-needle aspiration, diagnosis, BRAF mutation
Citation: Tan L-C, Liu W-L, Zhu X-L, Yu P-C, Shi X, Han P-Z, Zhang L, Lin L-Y, Semenov A, Wang Y, Ji Q-H, Ji D-M, Wang Y-L and Qu N (2021) Next-Generation Sequencing Enhances the Diagnosis Efficiency in Thyroid Nodules. Front. Oncol. 11:677892. doi: 10.3389/fonc.2021.677892
Received: 08 March 2021; Accepted: 21 June 2021;
Published: 12 July 2021.
Edited by:
Sergio Giannattasio, Bioenergetica e Biotecnologie Molecolari (IBIOM), ItalyReviewed by:
Matteo Chiara, University of Milan, ItalyCarl Christofer Juhlin, Karolinska Institutet (KI), Sweden
Copyright © 2021 Tan, Liu, Zhu, Yu, Shi, Han, Zhang, Lin, Semenov, Wang, Ji, Ji, Wang and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong-Mei Ji, amlkb25nbWVpMjAwMEBob3RtYWlsLmNvbQ==; Yu-Long Wang, eXVsb25nd2FuZ0BmdWRhbi5lZHUuY24=; Ning Qu, am9uYXRoYW5fcW5AMTYzLmNvbQ==
†These authors have contributed equally to this work
 Li-Cheng Tan
Li-Cheng Tan Wan-Lin Liu
Wan-Lin Liu Xiao-Li Zhu2,3†
Xiao-Li Zhu2,3† Arseny Semenov
Arseny Semenov