
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 13 May 2021
Sec. Hematologic Malignancies
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.672287
 Federica Loscocco1*
Federica Loscocco1* Giuseppe Visani1
Giuseppe Visani1 Annamaria Ruzzo2
Annamaria Ruzzo2 Irene Bagaloni2
Irene Bagaloni2 Fabio Fuligni3
Fabio Fuligni3 Sara Galimberti4
Sara Galimberti4 Antonello Di Paolo4
Antonello Di Paolo4 Fabio Stagno5
Fabio Stagno5 Patrizia Pregno6
Patrizia Pregno6 Mario Annunziata7
Mario Annunziata7 Antonella Gozzini8
Antonella Gozzini8 Sara Barulli1
Sara Barulli1 Elisa Gabucci1
Elisa Gabucci1 Mauro Magnani2
Mauro Magnani2 Alessandro Isidori1
Alessandro Isidori1Tyrosine kinase inhibitors (TKIs) have radically changed the outcome of chronic myeloid leukemia (CML) patients in the last 20 years. Moreover, the advent of second generation TKIs, namely nilotinib and dasatinib, have largely increased the number of CML patients achieving deep and sustained molecular responses. However, the possible mechanisms capable of influencing the maintenance of the long-term molecular response are not yet fully known and understood. In this light, polymorphisms in MDR-ABC transporters may influence the efficacy and safety of TKIs. In this study, we examined seven single nucleotide polymorphisms (SNPs) in four ABC transporter genes: ABCC1 rs212090 (5463T>A), ABCC2 rs3740066 (3972C>T), ABCC2 rs4148386 G>A, ABCC2 rs1885301 (1549G>A), ABCG2 rs2231137 (34G>A), ABCG2 rs2231142 G>C, ABCB1 rs1045642 (3435C>T), to determine their effect on the achievement and/or loss of molecular response in 90 CML patients treated with nilotinib. We found that ABCC2 rs3740066 CC and CT as well as the ABCB1 rs1045642 TT genotypes correlated with a higher probability to achieve MR3 in a shorter time (p=0.02, p=0.004, and p=0.01), whereas ABCG2 rs2231137 GG was associated with lower probability of MR3 achievement (p=0.005). Moreover, ABCC2 rs3740066 CC genotype, the ABCB1 rs1045642 CC and TT genotypes were positively correlated with MR4 achievement (p=0.02, p=0.007, and p=0.003). We then generated a predictive model incorporating the information of four genotypes, to evaluate the combined effect of the SNPs. The combination of SNPs present in the model affected the probability and the time to molecular response. This model had a high prognostic significance for both MR3 and MR4 (p=0.005 and p=0.008, respectively). Finally, we found ABCG2 rs2231142 GG genotype to be associated with a decrease risk of MR3 loss. In conclusion, MDR-transporters SNPs may significantly affect the achievement and loss of molecular response in CML patients treated with nilotinib.
Targeted therapy with the selective inhibitors of BCR-ABL tyrosine kinase activity has dramatically changed the treatment of chronic myeloid leukemia (CML) in the last 20 years (1). As a result, patients treated with currently available tyrosine kinase inhibitors (TKIs) can now enjoy a near normal life-expectancy (2).
Imatinib mesylate (IM), a first generation TKI, has proven to be highly active and safe, even in the long run, in CML patients. Accordingly, it has become in recent years the golden standard for front-line CML treatment (3). Despite excellent results in terms of response, leukemic residual cells are still detectable in most patients with CML even after years of IM treatment. Some of these patients, especially those with advanced-phase CML, will eventually develop resistance to IM therapy, and need a second or a third line treatment (3, 4). The mechanisms of drug resistance have been extensively studied and can be classified as primary (also referred to as “refractoriness”), in which TKIs fail to induce an adequate response from the start of therapy, or acquired, developed after the initial achievement of some degree of response to IM, lasting for some time (4, 5). Second-generation BCR-ABL inhibitors (2GTKIs) have been initially developed to overcome acquired resistance to IM. Subsequently, 2GTKIs were tested frontline (2), becoming an alternative to frontline IM in intermediate and high-risk patients. In the path to cure for CML patients, a relevant problem is represented by the high discontinuation rate during treatment with 2GTKIs after IM failure. In the clinical setting, more than 50% of patients will eventually discontinue 2GTKIs treatment during their lifetime, due to lack/loss of response, toxicity or secondary resistance. Although point mutations in BCR-ABL kinase domain are the major mechanism of resistance to TKIs, the bio-availability of TKIs in leukemic stem cells is also an important pharmacokinetic factor that may contribute to its development (6).
The pharmacokinetic of TKIs is dependent on gastrointestinal absorption, metabolism and cellular influx and efflux (5). ATP-binding cassette (MDR-ABC) transporters are membrane glycoproteins that play a key role in the efflux of toxic agents, including TKIs, and are involved in the regulation of intracellular drug accumulation. Accordingly, changes in the level of expression and functionality of MDR-ABC transporters influence the efficacy and safety of the drug transported. ATP-binding cassette sub-family B member 1 (also known as P-glycoprotein MDR1) encoded by ABCB1 gene, and ATP-binding cassette transporter G2 (also known as breast cancer resistance protein, BCRP) encoded by ABCG2 gene, are two adenosine triphosphate-binding cassette members, with a prominent role in multidrug resistance (7). They are involved in the transport of bile, lipoprotein, drug substrates and other peptides. They are normally localized in the small intestine, in bile canalicular membranes of hepatocytes in the liver, in renal proximal tubes and in the blood-brain barrier. They are also abundantly expressed in hematopoietic progenitor cells, and overexpressed in cancer stem cells (8). As an efflux pump localized on the cellular plasma membrane, ABCB1, and ABCG2 proteins excrete a variety of endogenous and exogenous compounds including chemotherapeutic agents, such as mitoxantrone and methotrexate (9). Moreover, several recent findings support ABCB1 and ABCG2-mediated efflux of asciminib, a new drug in development in CML, as the mechanism of resistance in these cell lines (10, 11). In addition, multidrug resistance proteins 1 (MPR1) and 2 (MPR2), which are encoded by ABCC1 and ABCC2 genes, respectively are mostly involved in the transport of xenobiotics, and also play a role in excretion of TKIs (9). The overexpression of MDR-ABC transporters genes such as ABCB1, ABCG2, and ABCC2 has been reported to lead to interindividual difference in bioavailability of drugs, and it represents a potential mechanism of primary resistance to anti-cancer agents (5).
Genetic constitution of any individual plays a significant role in interpatient variability to therapeutic response. The genetic variations like single nucleotide polymorphisms (SNPs) can affect gene expression or function in normal and cancer cells causing inherent interindividual differences in the metabolism and in the availability of drugs (12). ABCB1 gene, located at chromosome 7q21 is highly polymorphic; three polymorphisms have been extensively studied, two synonymous SNPs rs1128503 (1236C>T) in the exon 12 and rs1045642 (3435C>T) in exon 26; and one missense SNP rs2032582, (Ser893Ala/Thr 2677G>T/A) in exon 21. These three polymorphisms define a haplotype related to an increased activity in MDR1 (7). Moreover, several ABCG2 DNA variants have been shown to reduce its protein expression through an effect of the drug that interacts with BCRP transporter (9, 12). A number of ABCC2 SNPs have been studied for their potential functional influence on the expression level and transport activity. Few genetic variants −24C>T, 1249G>A and 3972C>T of ABCC2 gene have been reported to lead to differences in pharmacokinetics of various drugs7. In the present study, we hypothesized that the most relevant SNPs in genes encoded for MDR1, BCRP, and MPR2 could predict the outcome of CML patients treated with nilotinib. The aim of this study was to evaluate whether the SNPs in ABCB1, ABCC1, ABCC2, and ABCG2 genes have an impact on the efficacy of nilotinib treatment.
90 Caucasian adult CML patients treated at six Italian sites were enrolled in an observational, multi-institutional, no-profit study (EudraCT: 2011-00637-37) from April 2011 to December 2017. All patients signed informed consent. The study was performed in accordance with the International Conference on Harmonization Good Clinical Practice Guidelines, the Declaration of Helsinki (1996) and it was approved by the Ethics Committees of all sites involved in the study.
To be eligible for inclusion, patients had to be at least 18 years of age, affected by CML in chronic phase requiring treatment with nilotinib either in first or second line. According to International scale13 major molecular response (MMR) was defined as BCR-ABL qPCR transcript level ≤0.1%, whereas MR4 indicated levels of disease that were ≤0.01% BCR-ABL transcript. Minimum follow-up to be included in the study was 12 months after the first dose of nilotinib.
Gene analyses were performed on DNA isolated from peripheral blood samples of patients screened in eight Italian Hematology Division. Blood samples were collected before nilotinib (baseline).
This study examined seven SNPs in four ABC transporter genes: ABCC1 rs212090 (5463T>A), ABCC2 rs3740066 (3972C>T), ABCC2 rs4148386 G>A, ABCC2 rs1885301 (1549G>A), ABCG2 rs2231137 (34G>A), ABCG2 rs2231142 G>C, ABCB1 rs1045642 (3435C>T). The genotyping was performed blind to the patients’ treatment and clinical outcomes. Genomic DNA was isolated from 1 ml of peripheral blood by means of commercially available kit (QIAamp DNA Blood Midi kit, Hilden, Germany) according to the manufacturer’s instructions. Polymorphisms for ABCC1, ABCC2, and ABCG2 genes were detected by PCR high-resolution melting assay (HRM), restriction fragment length polymorphism (RFLP) of PCR products and pyrosequencing assays. The PCR for RFLP and pyrosequencing analyses were performed in a volume of 25 μl using 2 × PCR Master Mix kit (Diatheva, Fano, Italy), 25 ng of genomic DNA and 200 nM of each primer. The HRM assays were performed by using commercial kits according to the manufacturer’s instructions (Diatheva, Fano, Italy). Genotypes for ABCB1 rs1045642 (c.3435C>T) polymorphism were obtained through allele-specific probes using an Applied Biosystems kit on an ABI
Prism 7900HT Sequence Detection System instrument (ThermoFisher). Primer sequences, preparative PCR conditions and additional details are listed in Table 1.
The gene expression levels of BCR-ABL1 were determined using quantitative real-time polymerase chain reaction (qRT-PCR) from total leukocyte RNA of peripheral blood samples. Ratios derived from BCR-ABL/ABL1 were converted to the International Scale (13).
All statistical analyses presented and discussed here were performed using Statistical Package for the Social Sciences SPSS version 20 (SPSS, Chicago, USA). The frequency of each genotype was calculated. Hardy-Weinberg equilibrium was verified for all examined SNPs. Age, sex, Sokal, Hasford and Eutos risk scores, were recorded. Demographic, as well as clinical data are presented as median and range for continuous variables or frequency (percentages) for categorical variables. Hematological, cytogenetic and molecular responses were assessed and classified according to ELN guidelines13. Furthermore, molecular monitoring was carried out as established by the European guidelines and applied by all Italian LABNET network (13, 14). Time to MR was calculated from nilotinib start to the achievement of a molecular response. The time to treatment failure was defined as the interval between the initiation of nilotinib therapy and the occurrence of event that indicated that patients had failed (loss of molecular response).
Demographic and clinical prognostic features were compared across response and SNPs, using Person’s χ2 test for categorical variables and Mann-Whitney test and Kruskal-Wallis tests for continuous variables, where appropriate.
The association between the cumulative probability to obtain or lost MR3 and MR4 among genotypes or haplotypes was calculated in univariate and multivariate analysis. Univariate analysis was performed with Kaplan-Meier method to establish the relationship between SNPs and molecular response. Then, multivariate analysis was performed by COX regression model, describing as the hazard ratio and 95% confidence interval. Statistical significance was set at P<0.05.
In our cohort, a large majority of patients (74%) received nilotinib at 600 mg/day and the remaining 26% at 800 mg/die. The characteristics of patients are listed in Table 2.
Overall, 83 patients (92%) achieved MR3 during treatment, after a median time of 6 months (range, 2–82). Fourteen patients (15%) lost MR3 during treatment, with a median time of 14 months (mean 21 months, range 4–78). Sixty-seven (74%) patients receiving nilotinib achieved a DMR (deep molecular response defined as MR≥4) after a median time of 22 months (range 2–76).
Molecular response rates were not significantly affected by age or sex. The median observation time was 78 months.
We did not find any relationship between age, sex, type of transcript and the achievement or loss of molecular response.
The genotype and allele frequencies for all SNPs and their association with response to nilotinib are summarized in Tables 3 and 4.
Treatment outcomes were compared according to each candidate genotype for the seven statistical end points. To assess the association between polymorphisms and molecular response, we initially performed a survival analysis using each single variable (genotype or SNPs) and the time to reach MR3, and MR4 (data not shown). We further investigated the prognostic significance of SNPs and genotype using multiple Cox regression analysis, considering age, sex, and prognostic score as well as all molecular responses (MR3 and MR4).
When considering the effect of the genotypes on treatment outcome, some SNPs were significantly associated with higher response rate to nilotinib therapy. As reported in Table 3, ABCC2 rs374066, ABCG2 rs2231137, and ABCB1 rs1045642 SNPs affected the probability of achieving MR3. In details, the ABCC2 rs3740066 CC and CT genotypes as well as the ABCB1 rs1045642 TT genotype correlated with a high probability to achieve MR3 in a shorter time (p=0.02, p=0.004, and p=0.01) (Figures 1A–C). On the contrary, ABCG2 rs2231137 GG genotype correlated with a lower probability of achieving a molecular response (p=0.005) (Figure 1D). The same analysis was done for MR4. ABCC2 rs3740066 CC, ABCB1 rs1045642 CC and TT genotypes showed a significant positive correlation with MR4 achievement (p=0.02, p=0.007, and p=0.003) (Table 4, Figures 2A–C).
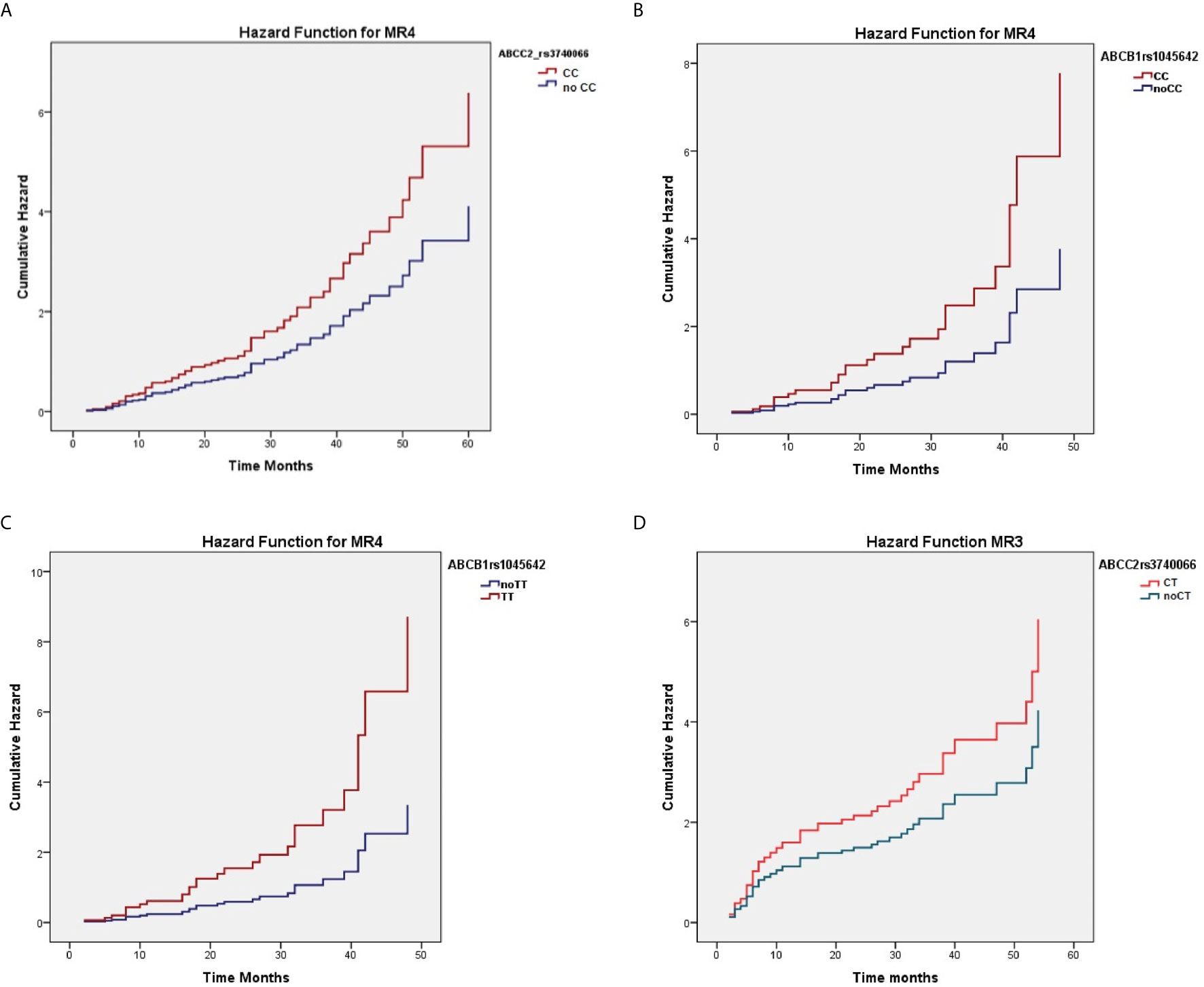
Figure 1 Correlation between ABCC2 rs3740066 CC (A), ABCC2 rs3740066 CT (B), ABCB1 rs1045642 TT (C) ABCG2 rs2231137 GG (D) genotypes and MR3 achievement.
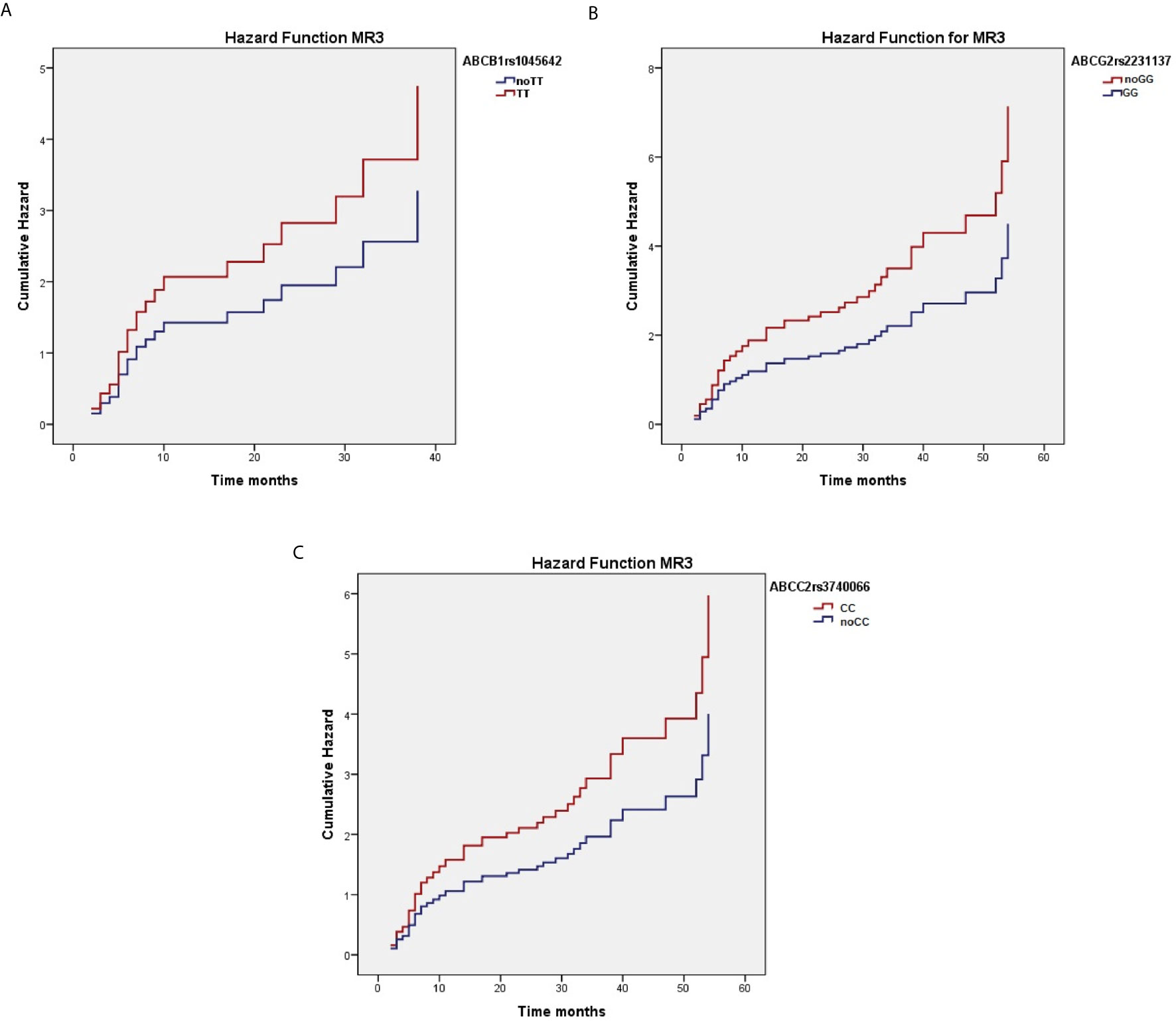
Figure 2 Correlation between ABCC2 rs3740066 CC (A), ABCB1 rs1045642 CC (B) and TT (C) genotypes and MR4 achievement.
We then performed a haplotype analysis for the studied genes, by analyzing the combined effect of several SNPs, and generated a predictive model incorporating four genotypes, risk score and MR3 (Table 3). The combination of SNPs (haplotypes) of the model affected the probability and the time to molecular response, with high prognostic significance (p=0.005). The same was done for MR4 (Table 4), and the high prognostic significance of the model was confirmed (p=0.008). We did not observe any significant association between ABCC1 rs212090 (5463T>A) and MR achievement.
Subsequently, we analyzed the possible association between SNPs in ABCC1, ABCC2, ABCG2, and ABCB1 genes and the loss of molecular response. We investigated whether the different distribution of genotypes or haplotypes were associated with MR loss during nilotinib treatment. ABCG2 (rs2231142) GG genotype was associated with a decreased risk of MR3 loss, in a statistically significant fashion (Table 5).
CML patients treated with nilotinib show an improved clinical response, and most of them could reach treatment-free remission (TFR), once in deep and sustained molecular response. Several recent studies showed that TKIs can safely be discontinued, thus achieving a TFR (15–17). Moreover, TFR could be extended to patients who are in stable major molecular response (MMR), not necessarily MR4 (15). Nevertheless, a significant proportion of patients still fail to obtain or maintain a major MR (defined as ≥3 log reduction from standardized baseline) over time. For these patients, TFR will never become an achievable endpoint, and short and long term toxicities of nilotinib might thus represent a major flaw in everyday clinical practice. Although BCR/ABL mutations are the major contributory factor for the loss of response to TKIs (5), a reduced bio-availability of TKIs in leukemic stem cells may play a relevant role. In this regard, the presence of polymorphisms in drug transporters may contribute to mechanisms of drug resistance, disease progression or loss of MR. Therefore, the study of biomarkers predicting response to TKIs treatment may be of great help in treatment-decision making.
Pharmacokinetics of TKIs, has been proven to be a potential biomarker, and may partially explain the inter-individual variability in response to nilotinib (18, 19). The MDR-ABC family proteins are widely recognized for their ability to modulate the adsorption, distribution, metabolism, excretion of a broad range of compounds including TKIs. Polymorphisms in ABC transporters responsible for active drug efflux may have a role in predicting response to TKIs, even if the exact mechanism of MDR-ABC transporter-mediated resistance in CML is unclear (20).
In the current study, we examined whether genetic polymorphisms in ABCB1, ABCC1, ABCC2, and ABCG2 genes may have an impact on the outcome of CML patients treated with nilotinib. ABC transporters, in particular ABCB1 and especially ABCG2, are abundantly expressed in hematopoietic progenitor cells and are overexpressed in cancer and CML stem cells (8). We observed that the SNPs of the major multi-drug resistance genes, ABCB1, ABCC2, and ABCG2, encoding for important export pumps expressed at tissue barriers, influenced the achievement of MR3 and MR4. In our study, patients with the variant alleles of either ABCB1 rs1045642 or ABCC2 rs3740066 achieved significantly higher rates of MR3 and MR4, in a shorter period of time. Conversely, ABCG2 rs2231137 GG genotype predicted a lower MR3 rate, whereas ABCG2 rs2231137 CC genotype did not. These findings suggest that SNPs in ABCB1, ABCG2, and ABCC2 MDR genes seems to modulate the molecular response to nilotinib.
Many studies reported that the plasma levels of TKIs are associated with response to therapy in CML patients (21, 22). Clinical studies have established that ABCB1 expression levels are high in advanced stages of CML, and that high ABCB1 expression is associated with lower MMR rate and resistance to TKIs (23, 24).
Recently, Rinaldetti et al. analyzed the data of the Euro-SKY trial, in order to evaluate the expression of ABCG2, OCT1, and ABCB1, and their influence on TFR (25). Interestingly, after TKI discontinuation, ABCB1 expression was significantly downregulated in 40 patients, at the day of relapse. Furthermore, CML patients in deep molecular response showed a different expression in influx and efflux transporters after long-term imatinib treatment, compared with healthy controls. ABCB1 responded more sensitively to TKI exposure, because discontinuation of imatinib resulted in a significant downregulation or normalization of its transcription levels (25).
In another study evaluating the effect of ABCB1 variants on response to imatinib, Dulucq et al. showed that patients with rs1128503 TT or rs2032582TT/TA genotypes achieved significantly higher MMR rate (26). However, in a subsequent, larger cohort of patients, these results were not confirmed, questioning the real impact of ABCB1 variants on response to imatinib (27). Notwithstanding, Au et al. (19) described a significant association of ABCB1 rs1045642 CC genotype with both major and complete response in a Caucasian cohort of CML patients receiving imatinib. In an attempt to explain these discrepancies, it is important to underline that the site of polymorphism may be responsible for different conformation of the influx and efflux pumps, resulting or not in a clinically measurable effect on response to imatinib (19).
A different affinity toward ABCB1 has been reported for second generation TKIs,. Nilotinib has been implied to interact with ABCB1, but this interaction is controversial. Two studies demonstrated that nilotinib is a substrate of ABCB1 (28, 29); furthermore, 1) the increased expression of ABCB1 is associated with a reduced sensitivity of the K562 cell line to nilotinib; 2) ABCB1 and ABCC6 work in concert during the development of nilotinib resistance (30). Nilotinib is also a high-affinity substrate for ABCG2 and in higher concentrations, reduces both ABCB1 and ABCG2 ATPase activity (8). ABCG2 is expressed not only in HSCs but also in hepatocytes or intestinal epithelial cells, affecting the bioavailability of TKIs. Patients with a high ABGG2 expression were shown to have a two-fold increased risk of relapse (31). ABCB1 and ABCG2 were shown to have a significant impact on the cytotoxic potential of nilotinib, dasatinib and bosutinib in vitro (8). In vitro, K562 cells were efficiently protected from nilotinib cytotoxicity by ABCG2 activity, whereas dasatinib resistance was conferred by ABCB1 and ABCG2 activity. Notably, ABCB1 and ABCG2 only slightly modified the cytotoxic effect of bosutinib, suggesting a potential different mechanism of intracellular transportation for this latter. Finally, Desilly et al. (32) evaluated the impact of three common ABCB1 variants (1236C>T-2677G<T-3435>T) on the anti-proliferative effect and intracellular influx of nilotinib, dasatinib, imatinib and ponatinib in K562 and HEK 293 cell lines. Interestingly, ABCB1 polymorphisms were associated with response to imatinib, whereas they had a weak impact on the effects of nilotinib, dasatinib and ponatinib. However, these results were generated with cell lines, namely K562 and HEK293. Accordingly, major differences in outcome are expected, since the mechanisms concurring in drug-resistance in patients are different from cell lines.
Of note, our population did not comprise any patient with multi-TKI failure, a subpopulation of CML patients with the most challenging clinical risk factors in the TKI era (33). It would be interesting, in future studies, to see if there is any correlation between SNPS in ABCB1, ABCC1, ABCC2, and ABCG2, and multi-TKI resistant CML.
In conclusion, pharmacogenetic variability in individuals might predict treatment outcome in CML patients receiving nilotinib. Our results showed the effect of the SNPs studied on the molecular response to nilotinib, significantly predicting response to therapy for MR3 and MR4 achievement and loss. These models were highly predictive. The combined effect of ABCB1 and ABCG2 polymorphisms could be synergistic and promote the achievement and/or loss of the molecular response. Further studies in larger cohorts of patients are warranted to confirm our preliminary observation.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by CERM, Comitato Etico della Regione Marche. The patients/participants provided their written informed consent to participate in this study.
FL, GV, and AI designed the study and wrote the manuscript. AR, IB, SG, AP, and MM were responsible for biological studies. FS, PP, MA, AG, SB, and EG treated the patients, collected the data, and commented on the manuscript. FF performed statistical analysis. All authors contributed to the article and approved the submitted version.
The study was supported in part by AIL Pesaro Onlus.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Westerweel PE, Te Boekhorst PAW, Levin MD, Cornelissen JJ. New Approaches and Treatment Combinations for the Management of Chronic Myeloid Leukemia. Front Oncol (2019) 9:665. doi: 10.3389/fonc.2019.00665
2. García-Gutiérrez V, Hernández-Boluda JC. Tyrosine Kinase Inhibitors Available for Chronic Myeloid Leukemia: Efficacy and Safety. Front Oncol (2019) 9:603. doi: 10.3389/fonc.2019.00603
3. Brendel C, Scharenberg C, Dohse M, Robey RW, Bates SE, Shukla S, et al. Imatinib Mesylate and Nilotinib (Amn107) Exhibit High-Affinity Interaction With ABCG2 on Primitive Hematopoietic Stem Cells. Leukemia (2007) 21:1267–75. doi: 10.1038/sj.leu.2404638
4. Quintás-Cardama A, Kantarjian HM, Cortes JM. Mechanisms of Primary and Secondary Resistance to Imatinib in Chronic Myeloid Leukemia. Cancer Control (2009) 16(2):122–31. doi: 10.1177/107327480901600204
5. Loscocco F, Visani G, Galimberti S, Curti, Isidori A. Bcr-Abl Independent Mechanisms of Resistance in Chronic Myeloid Leukemia. Front Oncol (2019) 9:939. doi: 10.3389/fonc.2019.00939
6. Milojkovic D, Apperley J. Mechanisms of Resistance to Imatinib and Second-Generation Tyrosine Inhibitors in Chronic Myeloid Leukemia. Clin Cancer Res (2009) 15(24):7519–27. doi: 10.1158/1078-0432.CCR-09-1068
7. Ankathil R, Azlan H, Dzarr AA, Baba AA. Pharmacogenetics and the Treatment of Chronic Myeloid Leukemia: How Relevant Clinically? An Update. Pharmacogen (2018) 19:5. doi: 10.2217/pgs-2017-0193
8. Hegedűs C, Özvegy-Laczka C, Apáti Á, Magócsi M, Német K, Őrfi L, et al. Interaction of Nilotinib, Dasatinib and Bosutinib With ABCB1 and ABCG2: Implications for Altered Anti-Cancer Effects and Pharmacological Properties. Br J Pharmacol (2009) 158(4):1153–64. doi: 10.1111/j.1476-5381.2009.00383.x
9. Lopez-Fernandez LA. Atp-Binding Cassette Transporters in the Clinical Implementation of Pharmacogenetics. J Pers Med (2018) 8:40. doi: 10.3390/jpm8040040
10. Eadie LN, Saunders VA, Branford S, White DL, Hughes TP. The New Allosteric Inhibitor Asciminib is Susceptible to Resistance Mediated by ABCB1 and ABCG2 Overexpression In Vitro. Oncotarget (2018) 9(17):13423–37. doi: 10.18632/oncotarget.24393
11. Qiang W, Antelope O, Zabriskie MS, Pomicter AD, Vellore NA, Szankasi P, et al. Mechanism of Resistance to the BCR-ABL1 Allosteric Inhibitor Asciminib. Leukemia (2017) 31:2844–7. doi: 10.1038/leu.2017.264
12. Ahmed S, Zhou Z, Zhou J, Chen S-Q. Pharmacogenomics of Drug Metabolizing Enzymes and Transporters: Relevance to Precision Medicine. Genom Proteom Bioinform (2016) 14(5):298–313. doi: 10.1016/j.gpb.2016.03.008
13. Hochhaus A, Baccarani M, Silver RT, Schiffer C, Apperley JF, Cervantes F, et al. European LeukemiaNet 2020 Recommendations for Treating Chronic Myeloid Leukemia. Leukemia (2020) 34(4):966–84. doi: 10.1038/s41375-020-0776-2
14. Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, Kaeda J, et al. Monitoring CML Patients Responding to Treatment With Tyrosine Kinase Inhibitors: Review and Recommendations for Harmonizing Current Methodology for Detecting Bcr-Abl Transcripts and Kinase Domain Mutations and for Expressing Results. Blood (2006) 108:28–37. doi: 10.1182/blood-2006-01-0092
15. Clark RE. Tyrosine Kinase Inhibitor Therapy Discontinuation for Patients With Chronic Myeloid Leukaemia in Clinical Practice. Curr Hematol Malig Rep (2019) 14(6):507–14. doi: 10.1007/s11899-019-00548-2
16. Cortes J, Huynh L, Mendelson E, Brandt P, Dalal D, DerSarkissian M, et al. Treatment Patterns and Deep Molecular Response in Chronic Phase - Chronic Myeloid Leukemia Patients Treated With Second-Line Nilotinib or Dasatinib: A Multi-Country Retrospective Chart Review Study. Leuk Lymphoma (2019) 13:1–10. doi: 10.1080/10428194.2019.1644332
17. Richard EC, Polydoros F, Apperley FJ, Milojkovic D, Rothwell K, Christopher Pocock C, et al. De-Escalation of Tyrosine Kinase Inhibitor Therapy Before Complete Treatment Discontinuation in Patients With Chronic Myeloid Leukaemia (Destiny): A non-Randomised, Phase 2 Trial. Lancet Haematol (2019) 6(7):e375–83. doi: 10.1016/S2352-3026(19)30094-8
18. Hassine IB, Gharbi H, Soltani I, Hadj Othman HB, Farrah A, Amouri H, et al. Molecular Study of ABCB1 Gene and it Correlation With Imatinib Response in Chronic Myeloid Leukemia. Cancer Chemother Pharmacol (2017) 80(4):829–39. doi: 10.1007/s00280-017-3424-4
19. Au A, Baba AA, Azlan H, Norsa’adah B, Ankathil R. Clinical Impact of ABCC1 and ABCC2 Genotypes and Haplotypes in Mediating Imatinib Resistance Among Chronic Myeloid Leukaemia Patients. J Clin Pharm Ther (2014) 39(6):685–90. doi: 10.1111/jcpt.12197
20. Nath A, Wang J, Huang RS. Pharmacogenetics and Pharmacogenomics of Target Therapeutics in Chronic Myeloid Leukemia. Mol Diagn Ther (2017) 21(6):621–31. doi: 10.1007/s40291-017-0292-x
21. Gambacorti-Passerini C, Zucchetti M, Russo D, Frapolli R, Verga M, Bungaro S, et al. Alpha1 Acid Glycoprotein Binds to Imatinib (STI571) and Substantially Alters its Pharmacokinetics in Chronic Myeloid Leukemia Patientsi. Clin Cancer Res (2003) 9(2):625–32.
22. Gambacorti-Passerini C, le Coutre P, D’Incalci M. Binding of Imatinib by Alpha(1)-Acid Glycoprotein. Blood (2002) 100:367–9. doi: 10.1182/BLOOD-2002-02-0518CorpusID:12649869
23. Eadie LN, Dang P, Saunders VA, Yeung DT, Osborn MP, Grigg AP, et al. The Clinical Significance of ABCB1 Overexpression in Predicting Outcome of CML Patients Undergoing First-Line Imatinib Treatment. Leukemia (2016) 31(1):75–82. doi: 10.1038/leu.2016.179
24. da Cunha Vasconcelos F, Scheiner MAM, Moellman-Coelho A, Luiz Mencalha A, Zalcberg R, Rumjanek VM, et al. Low ABCB1 and High Oct1 Levels Play Favorable Role in the Molecular Response to Imatinib in CML Patients in the Community Clinical Practice. Leu Res (2016) 51:3–10. doi: 10.1016/j.leukres.2016.10.005
25. Rinaldetti S, Pfirrmann M, Manz K, Guilhot J, Dietz C, Panagiotidis P, et al. Effect of ABCG2, OCT1, and ABCB1 (Mdr1) Gene Expression on Treatment-Free Remission in a EURO-SKI Subtrial. Clin Lymphoma Myeloma Leuk (2018) 18(4):266–71. doi: 10.1016/j.clml.2018.02.004
26. Dulucq S, Bouchet S, Turcq B, Lippert E, Etienne G, Reiffers J. Multidrug Resistance Gene (Mdr1) Polymorphisms are Associated With Major Molecular Responses to Standard-Dose Imatinib in Chronic Myeloid Leukemia. Blood (2008) 112(5):2024–7. doi: 10.1182/blood-2008-03-147744
27. Dulucq S, Preudhomme C, Guilhot F, Mahon F-X. Response: Is There Really a Relationship Between Multidrug Resistance Gene (Mdr1) Polymorphisms and Major Molecular Response to Imatinib in Chronic Myeloid Leukemia? Blood (2010) 26):6145–6. doi: 10.1182/blood-2010-08-298794
28. Eadie LN, Saunders VA, Hughes TP, White DL. Degree of Kinase Inhibition Achieved in Vitro by Imatinib and Nilotinib is Decreased by High Levels of ABCB1 But Not ABCG2. Leukemia Lymphoma (2012) 54(3):569–78. doi: 10.3109/10428194.2012.715345
29. Kosztyu P, Dolezel P, Mlejnek P. Can P-glycoprotein Mediate Resistance to Nilotinib in Human Leukaemia Cells? Pharmacol Res (2013) 67(1):79–83. doi: 10.1016/j.phrs.2012.10.012
30. Eadie LN, Dang P, Goyne JM, Hughes TP, White DL. Abcc6 Plays a Significant Role in the Transport of Nilotinib and Dasatinib, and Contributes to TKI Resistance In Vitro, in Both Cell Lines and Primary Patient Mononuclear Cells. PloS One (2018) 13(1):e0192180. doi: 10.1371/journal.pone.0192180
31. Wang F, Wang X-K, Shi C-J, Zhang H, Hu Y-P, Chen Y-F, et al. Nilotinib Enhances the Efficacy of Conventional Chemotherapeutic Drugs in CD34+CD38– Stem Cells and ABC Transporter Overexpressing Leukemia Cells. Molecules (2014) 19(3):3356–75. doi: 10.3390/molecules19033356
32. Dessilly G, Panin N, Elens L, Haufroid V, Demoulin JB. Impact of ABCB1 1236c>T-2677>T-3435C>T Polymorphisms on the Anti-Proliferative Activity of Imatinib, Nilotinib, Dasatinib and Ponatinib. Sci Rep (2016) 6:29559. doi: 10.1038/srep29559
Keywords: chronic myeloid leukemia, nilotinib, drug resistance, MDR-ABC transporters, polymorphisms, molecular response
Citation: Loscocco F, Visani G, Ruzzo A, Bagaloni I, Fuligni F, Galimberti S, Di Paolo A, Stagno F, Pregno P, Annunziata M, Gozzini A, Barulli S, Gabucci E, Magnani M and Isidori A (2021) Clinical Relevance of ABCB1, ABCG2, and ABCC2 Gene Polymorphisms in Chronic Myeloid Leukemia Patients Treated With Nilotinib. Front. Oncol. 11:672287. doi: 10.3389/fonc.2021.672287
Received: 25 February 2021; Accepted: 21 April 2021;
Published: 13 May 2021.
Edited by:
Massimo Breccia, Sapienza University of Rome, ItalyReviewed by:
Mario Tiribelli, University of Udine, ItalyCopyright © 2021 Loscocco, Visani, Ruzzo, Bagaloni, Fuligni, Galimberti, Di Paolo, Stagno, Pregno, Annunziata, Gozzini, Barulli, Gabucci, Magnani and Isidori. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Federica Loscocco, ZmVkZXJpY2EubG9zY29jY29AZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.