
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 09 August 2021
Sec. Gastrointestinal Cancers
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.668707
This article is part of the Research TopicMolecular Biomarkers for Gastric CancerView all 36 articles
 Yawei Zhang1,2†
Yawei Zhang1,2† Han Wang1,2†
Han Wang1,2† Xiaoyu Xu3
Xiaoyu Xu3 Huifang Liu1
Huifang Liu1 Tengfei Hao1
Tengfei Hao1 Songcheng Yin1*
Songcheng Yin1* Changhua Zhang1*
Changhua Zhang1* Yulong He1,2*
Yulong He1,2*Immunosuppressive molecules are valuable prognostic biomarkers across different cancer types. Leukocyte immunoglobulin like receptor subfamily B1 (LILRB1) is considered to be an immunosuppressive molecule, which is an important receptor of human leukocyte antigen G. However, the clinical significance of LILRB1 expression in gastric cancer remains unexplored. We analyzed the immunohistochemistry data of 166 gastric cancer patients to determine the clinicopathologic and survival significance of LILRB1. Immunofluorescence was conducted to detect the co-localization of LILRB1 with infiltrating immune cells. Additionally, we also assessed the immune contexture, immune cell functions and tumor microenvironment state related to LILRB1. We found that LILRB1 was mainly present in tumor stroma which was higher in tumor tissues compared with matched adjacent tissues. High-LILRB1 expression was associated with more advanced tumor stage, higher recurrence risk and worse survival. Immunohistochemistry and bioinformatic analysis showed that LILRB1 had a significant positive correlation with M2 tumor-associated macrophages (TAMs) infiltration. Immunofluorescence confirmed that M2 TAMs were the primary immune cells expressing LILRB1. Dense infiltration of LILRB1+ M2 TAMs yielded an immunosuppressive microenvironment manifested as enriched exhausted CD8+ T cells and increased immunosuppressive cytokines. Moreover, patients with high infiltration of both LILRB1+ cells and M2 TAMs indicated poor prognosis and inferior therapeutic responsiveness to adjuvant chemotherapy. In conclusion, LILRB1+ M2 TAMs were associated with a pro-tumor immune contexture and determine poor prognosis in gastric cancer. Further studies are essential to explore therapeutic targeting LILRB1+ M2 TAMs.
Gastric cancer (GC) represents the fifth most prevalent malignancy and the third leading cause of cancer mortality worldwide (1). Radical surgery combined with postoperative adjuvant chemotherapy is the main treatment for advanced GC; however, the prognosis of patients is yet poor (2). Due to the uncertainty of chemosensitivity and the consequent chemoresistance, several patients have a high recurrence rate after chemotherapy. The change in the tumor microenvironment can predict the prognosis and chemosensitivity of patients. Therefore, it is crucial to stratify prognosis and predict the treatment response based on tumor microenvironment.
In the tumor microenvironment, tumor-associated macrophages (TAMs) are the primary infiltration components with different forms of expression between inflammation and cancer. Macrophages infiltrating into tumor tissue polarize into an antitumor M1 or a pro-tumor M2 subset. Typically, M2 TAMs exert a specific role in promoting tumor growth, promoting angiogenesis, and inhibiting adaptive immunity. Our previous study also confirmed that M2 TAMs indicate poor prognosis in GC patients (3). Thus, TAMs are an attractive target for tumor therapy. However, increasing evidence showed diversity in macrophages, but distinguishing them only by the current two polarization forms is challenging. Therefore, clarifying the distribution and function of macrophage subsets in tumor tissues is essential for accurate clinical treatment targets.
Leukocyte immunoglobulin-like receptor subfamily B1 (LILRB1) is a transmembrane glycoprotein, a major receptor of human leukocyte antigen G (HLA-G) (4). It is considered to be an immunosuppressive receptor. LILRB1 combines classic and non-classic human major histocompatibility complex (MHC) molecules to exert an immunosuppressive effect. It is mainly involved in the regulation of maternal-fetal immune tolerance and induction of transplantation immune tolerance (5). The rapid growth of tumor cells is largely due to the escape of immune surveillance (6). As an immunosuppressive receptor, LILRB1 may play a major role in the process of tumor cells escaping immune surveillance. Recently, it has been found that macrophages expressing inhibitory receptor LILRB1 interact with MHC class I components on the surface of tumor cells to protect tumor cells from phagocytosis. Moreover, some studies suggested that TAMs affect the efficacy of chemotherapy in tumor patients (7, 8). However, the effect of LILRB1 on TAMs and its role in the tumor microenvironment has not yet been analyzed systematically in GC.
In this study, we detected the frequency of LILRB1 and macrophages in GC patients. The correlation between LILRB1 and clinicopathological factors, macrophage infiltration, and tumor microenvironment immune status would be analyzed. Furthermore, we evaluated the prognostic potential of LILRB1 and macrophages, and assessed the predictive value of postoperative adjuvant chemotherapy in this subpopulation.
The study recruited 166 GC patients who underwent radical surgery during 2009–2013 in the First Affiliated Hospital of Sun Yat-sen University. None of the patients received preoperative treatment, including chemotherapy or radiotherapy. Comprehensive information about the clinicopathological data and survival outcomes of all patients was obtained. The median follow-up period was 42 (range: 2–99) months in this cohort. All tumor tissues and 46 adjacent peritumoral tissues were formalin-fixed and embedded in paraffin. The tumor stages were categorized according to the 7th edition of American Joint Committee on Cancer (AJCC) TNM staging system. Adjuvant chemotherapy (ACT) was given to TNM stage II and III patients after surgery, according to the National Comprehensive Cancer Network (NCCN) guidelines and patient preference. All chemotherapy regimens were fluorouracil-based combination chemotherapy. The human studies were sanctioned by the local ethics committee at the First Affiliated Hospital of Sun Yat-sen University.
The formalin-fixed and paraffin-embedded sections were deparaffinized with xylene and then rehydrated. Antigen retrieval was performed with Tris/EDTA buffer pH 9.0 for 20 min at 95 °C in paraffin-embedded tissue sections. The slides were incubated with antibodies against CD163 (1:400; Cell Signaling Technology, #93498) and LILRB1 (1:400; Abcam, ab238145) overnight at 4°C. The reactivity was detected using Dako EnVision-HRP (Dako).
The infiltration density of LILRB1+ and CD163+ cells per field was evaluated by two independent pathologists who were blinded to the patients’ clinical data using Image-Pro Plus 6.0 (Media Cybernetics Inc.) for assistance. For each tissue core or normal section, three randomized fields of positive-stained cells were counted under a high-power field (HPF) of 400X. The density of LILRB1+ and CD163+ cells was calculated as the mean number of fields from cores or normal sections. The cut-off values of LILRB1+ and CD163+ cells density were the median values. For LILRB1+ cells,≥85 in average field was defined as high and <85 was defined as low. For CD163+ cells, ≥28 in average field was defined as high and <28 was defined as low.
We performed immunofluorescence, as described previously. Primary antibodies were used as follows: anti-human CD163 (1:100; Biolegend; 326507), anti-LILRB1 (1:200; Abcam, ab238145), anti-human-Cytokeratin 7 (1:100; Biolegend, 601601). After washing, cells were incubated with Alexa Fluor 488- or 546- or 647-labeled secondary antibodies for 1 h. Nuclei were counterstained using DAPI. The stained cells were visualized using an inverted confocal microscope, and the images were processed using ZEN2.3.
Level 4 gene expression data (RSEM normalized) of The Cancer Genome Atlas (TCGA) were downloaded from the UCSC Xena browser (https://gdc.xenahubs.net). We used TCGA database to analyze the difference of LILRB1 expression between GC and normal tissues. We calculated the scores of LILRB1+ M2 TAMs signature genes by the geomean of TCGA RSEM expression to confirm relative abundance. The correlation between LILRB1+ M2 TAMs and exhausted CD8+ T cells was analyzed by gene set enrichment analysis (GSEA, v3.0), as previously reported (9). The LILRB1+ M2 TAMs signature genes and exhausted CD8+ T cell gene set were showed in Supplementary Table 1, which were identified based on previous studies (9, 10).
Microarray datasets GSE15459 and GSE 29272 were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) and used as a training set for the LILRB1 expressed prediction. We also estimated the proportion of immune cells used GSE62254 cohort. The RMA algorithm was applied to normalize and transform all the raw data from GEO to expression values in the R environment (v3.5.3).
The CIBERSORT algorithm was conducted to evaluate the proportion of immune cells in GC patients, as reported previously (11). This method allows sensitive and specific discrimination of 22 human immune cell phenotypes, including B cells, T cells, natural killer (NK) cells, macrophages, dendritic cells, and myeloid subsets. Briefly, the gene expression profiles were prepared using standard annotation files. Then, the data were uploaded to the CIBERSORT web portal (http://cibersort.stanford.edu/), and the algorithm was run using the default signature matrix at 1000 permutations.
The correlation between LILRB1 expression and clinicopathological characteristics of GC was evaluated by Student’s t-test. The Pearson’s correlation test was used to determine the extent of correlation between the expression of LILRB1 and that of other genes. The survival outcomes, including overall survival (OS) and disease-free survival (DFS) were analyzed using Kaplan–Meier curves, log-rank test, and univariate/multivariate Cox regression analysis. The data of all groups in the figure were expressed as mean ± SDs. Two-sided P < 0.05 was considered statistically significant. All analyses were performed using GraphPad Prism (version 6.00), R (version 3.6.1) or SPSS statistics (version 21) software.
Initially, we detected the expression of LILRB1 in GC tissues and adjacent tissues from First Affiliated Hospital of Sun Yat-sen University (FHSYSU) cohort by immunohistochemistry (Figure 1A). The current results showed that the expression of LILRB1 was higher in tumor tissues compared to the matched adjacent tissues (P < 0.01, Figure 1B). Similarly, we also detected the abnormal expression of LILRB1 in GC tissues in two other independent cohorts (TCGA and GSE29272, Figures 1C, D). Interestingly, we found that LILRB1 was mainly present in tumor stroma rather than tumor cells (Figure 1A). In order to further determine the expression distribution of LILRB1, immunofluorescence was detected, and GC cells were labeled with cytokeratin 7 (CK7). The data further confirmed that LILRB1 was mainly expressed in the tumor stromal microenvironment (Figure 1E).
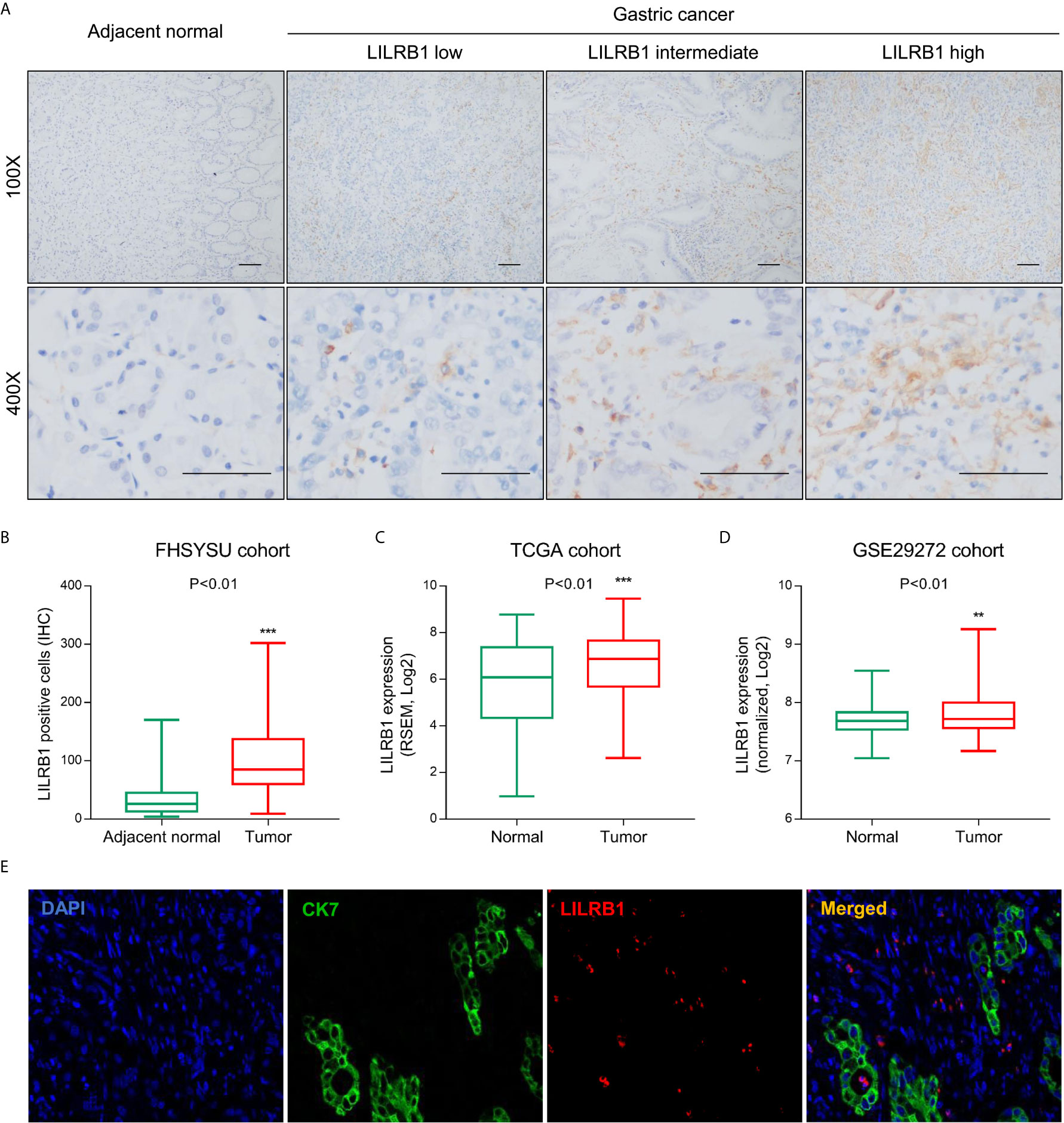
Figure 1 LILRB1 was highly expressed in stroma of GC. (A) The expression of LILRB1 in GC tissues and adjacent tissues from First Affiliated Hospital of Sun Yat-sen University (FHSYSU) cohort by IHC. (B) Box diagram showed that the count of LILRB1+ cells in GC was higher than that in gastric cancer tissues in FHSYSU cohort. (C, D) TCGA cohort and GSE29272 cohort showed higher LILRB1 expression in GC tissues than in normal gastric tissues. (E) LILRB1 was mainly distributed in the stroma of gastric cancer by tricolor immunofluorescence microscopy. **P < 0.01 and ***P < 0.001.
Additionally, we analyzed the correlation between LILRB1 and the clinicopathological characteristics of GC patients. We also observed that males with GC expressed more LILRB1 than female patients (Figure 2A). However, no significant correlation was established between LILRB1 and patients’ age or tumor grade (Figures 2B, C). The LILRB1 expression was significantly associated with large tumor size (>5 cm), deep tumor invasion, and lymph node metastasis (Figures 2D–F). Correspondingly, a higher proportion of LILRB1 was detected in stage III tumors than stage I-II tumors (Figure 2G).
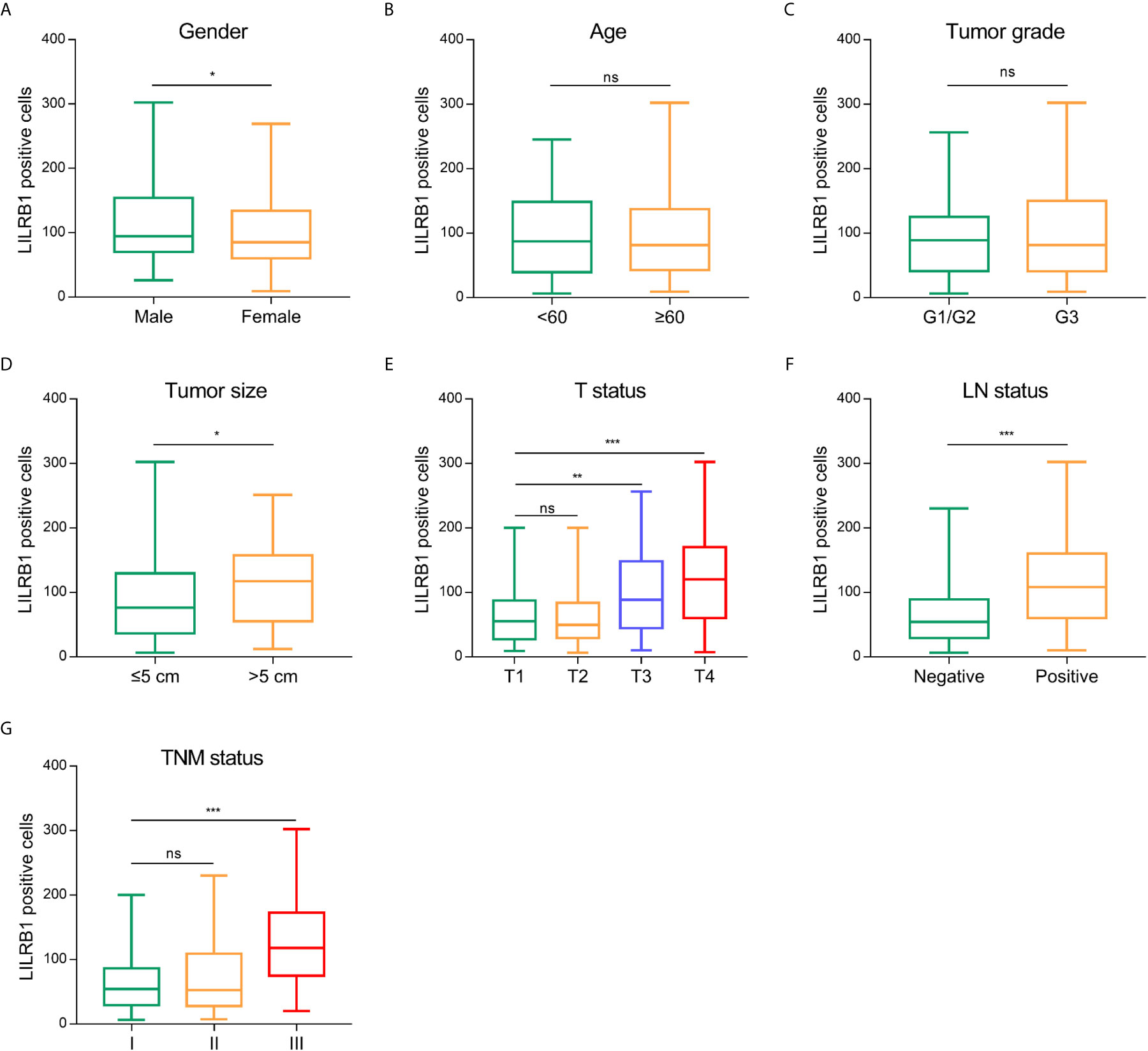
Figure 2 The correlation between LILRB1 and clinicopathological characteristics of GC patients. (A) LILRB1 expression in male patients was higher compared with female. (B, C) No significant correlation was established between LILRB1 and patients’ age or tumor grade. (D) Patients with large tumor size (>5 cm) had high-LILRB1 expression. (E) Patients with deep tumor invasion had high-LILRB1 expression. (F) Patients with lymph node metastasis had high-LILRB1 expression. (G) Higher proportion of LILRB1 was detected in stage III tumors than stage I-II tumors. *P < 0.05, **P < 0.01 and ***P < 0.001. ns, no statistical significance.
The tumor microenvironment showed an abundance of immune cell infiltration, which exerted an impact on tumor progression and prognosis of patients (12, 13). Herein, we sought to discover the influence of LILRB1 on the immune contexture in GC. Thus, CIBERSORT was employed to assess the relative proportion of 22 human hematopoietic cell phenotypes (LM22) within the GSE15459 database. The data showed that high LILRB1 expression was associated with a low level of memory B cells and memory resting CD4 T cells but a high proportion of M2 TAMs, neutrophils, and memory-activated CD4 T cells (Figure 3A). To further analyze the correlation between LILRB1 and the immune microenvironment of GC, we conducted a correlation analysis between LILRB1 and 22 types of immune cells. Next, we found an interesting phenomenon that LILRB1 had a significant positive correlation with M2 TAMs infiltration (Figure 3B), considered immunosuppressive cells that promote tumor progression (14). However, no significant correlation was established between LILRB1 and M1 macrophage infiltration, which was previously identified to possess the antitumor effect in GC (3).
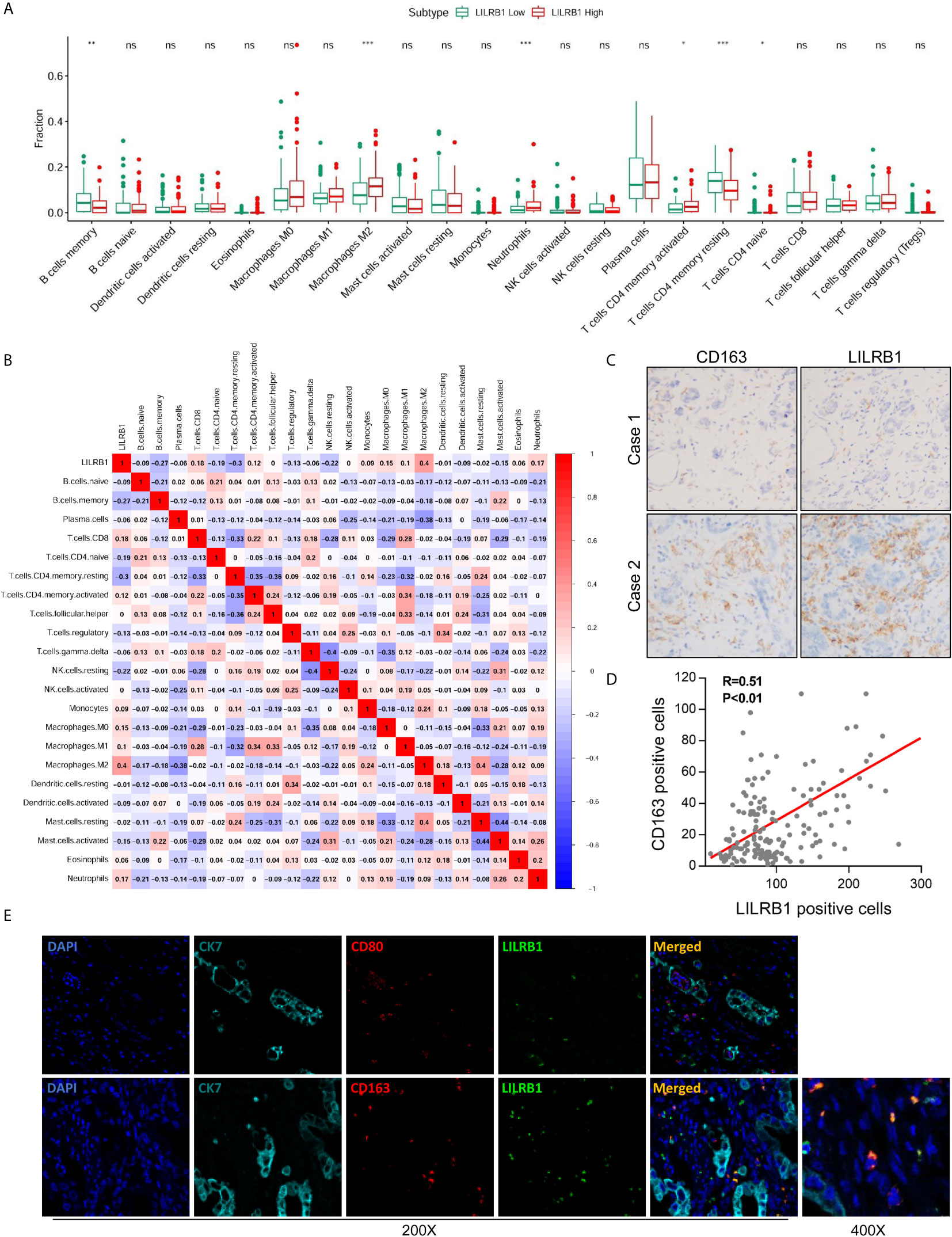
Figure 3 M2 TAMs were the primary immune cells expressing LILRB1. (A) CIBERSORT was employed to assess the difference of 22 kinds of immune cells between LILRB1 high tumors and LILRB1 low tumors. (B) The correlation analysis between LILRB1 and 22 types of immune cells. (C, D) LILRB1 expression showed a positive correlation with M2 macrophages by IHC. (E) Immunofluorescence assay showed that LILRB1 was mainly expressed in M2 TAMs. *P < 0.05, **P < 0.01 and ***P < 0.001. ns, no statistical significance.
To substantiate these findings, we performed IHC staining of LILRB1 and CD163 (classic biomarker of M2 TAMs). Consistent with the results from CIBERSORT, LILRB1 expression showed a positive correlation with M2 TAMs (Figures 3C, D, Pearson’s correlation R = 0.51, P < 0.01). Subsequently, we conducted immunofluorescence staining to evaluate the correlation between LILRB1 expression and macrophage localization. As illustrated in Figure 3E, M2 TAMs were identified as components of the LILRB1 infiltrate, expressing the CD163 M2 marker. In contrast, no co-expression of LILRB1 and CD80 (biomarker of M1 macrophages) was detected.
Further, we analyzed the macrophage markers in the TCGA database to verify the CIBERSORT results. These findings showed that LILRB1 expression was positively correlated with the expression of CD163 and CD204, M2 macrophage markers, involved in promoting tumor growth and metastasis (Figures 4A, B). On the other hand, no significant correlation was observed between LILRB1 and iNOS expression, which was widely recognized as a marker of M1 macrophages (Figure 4C). Moreover, transcription factor IRF4 induced M2-type polarization of macrophages, which was upregulated in patients with high LILRB1 expression (Figure 4D) (15).
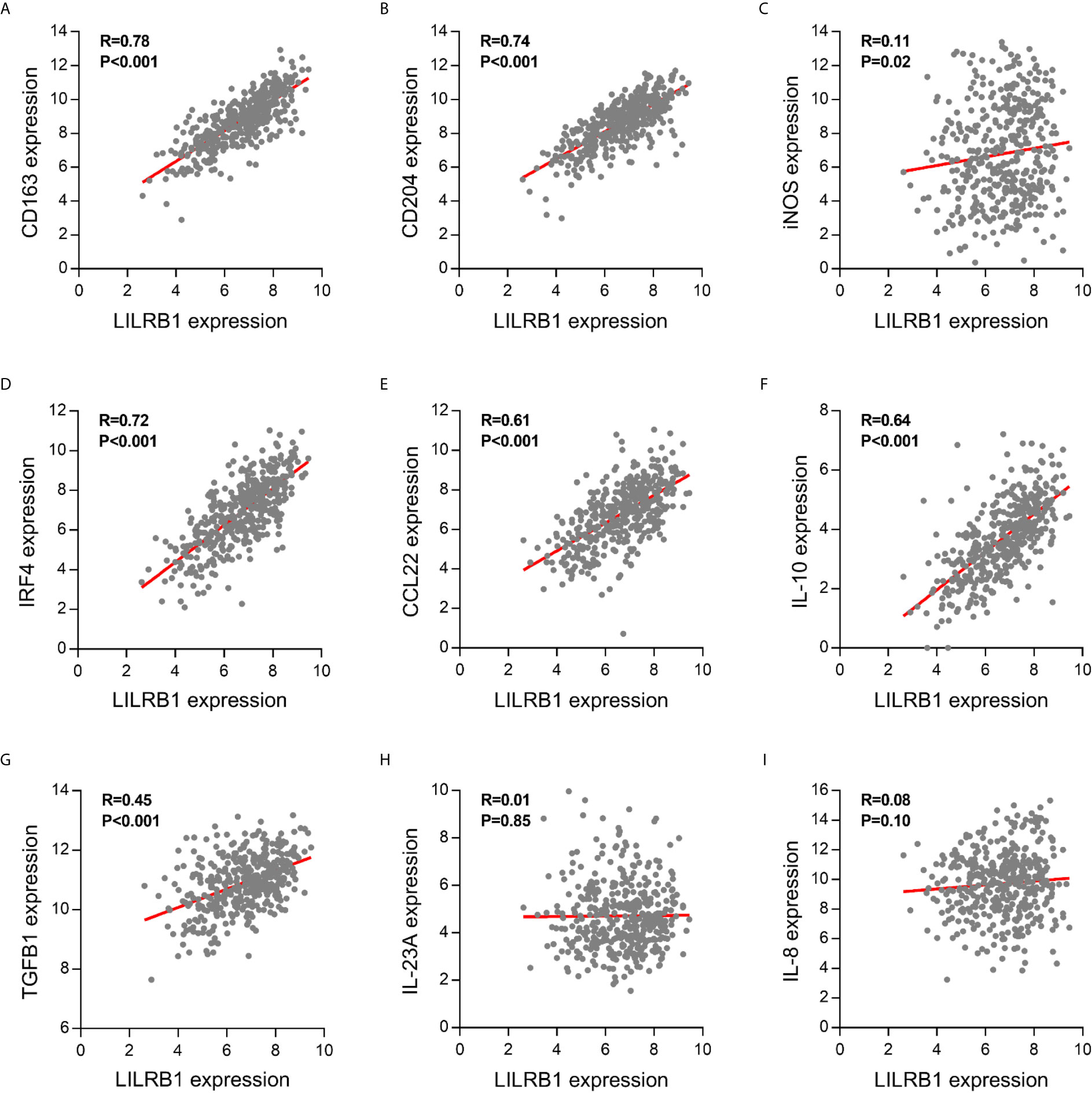
Figure 4 LILRB1 expression was correlated with multiple M2 macrophage-related molecules. Correlation between LILRB1 and M1/M2 macrophage-related molecules including (A) CD163, (B) CD204, (C) iNOS, (D) IRF4, (E) CCL22, (F) IL-10, (G) TGF-β1, (H) IL-23A, and (I) IL-8.
The imbalance between proinflammatory and anti-inflammatory cytokines secreted by M1/M2 macrophages in the tumor microenvironment promote the development of GC (16). Therefore, we hypothesized that cytokines are dysregulated in high-LILRB1 expression tumors. M2 macrophage-derived cytokines promote an immunosuppressive tumor microenvironment, including CCL22, IL-10, and TGF-β1, that were significantly correlated with LILRB1 expression GC patients (Figures 4E–G). Conversely, M1 macrophage-derived proinflammatory cytokines, such as IL-23A and IL-8, were not correlated with LILRB1 expression (Figures 4H, I). Thus, these results suggested that LILRB1 may be involved in the M2 polarization of macrophages to promote GC progression.
We next aimed to investigate the potential impact of LILRB1-expressing M2 TAMs on immune microenvironment in GC. We conducted GSEA to analyze the relationship between LILRB1+ M2 TAMs and functional status of CD8+ T cells. The result showed that exhausted CD8+ T cell gene set was significantly enriched in high LILRB1+ M2 TAMs signature (Figure 5A, FDR q = 0.008). There was no significant difference in the expression of effector molecules including CD107a and IL-17A in classification of LILRB1+ M2 TAMs signature (Figure 5B). Moreover, we evaluated the relationship between LILRB1+ M2 TAMs and immune checkpoint molecules. High LILRB1+ M2 TAMs signature exhibited abundant programmed cell death protein 1 (PD-1), cytotoxic T-lymphocyte associated protein 4 (CTLA-4), lymphocyte activation gene-3 (LAG-3), and hepatitis A virus cellular receptor 2 (HAVCR2) expression (Figure 5C). Consequently, these data indicated that LILRB1+ M2 TAMs may be involved in promoting immune escape of GC cells.
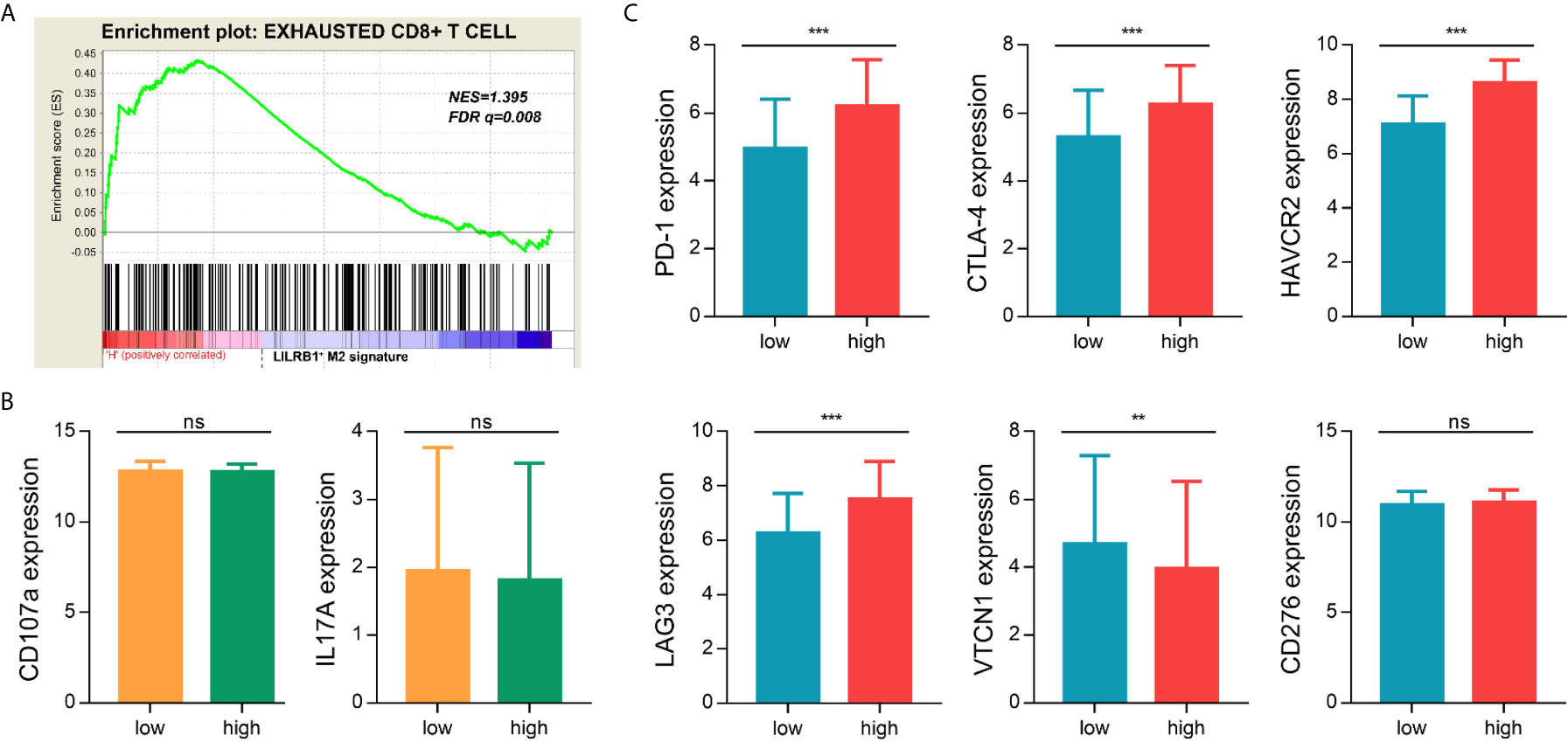
Figure 5 LILRB1+ M2 TAMs exhibited an immunosuppressive phenotype. (A) GSEA suggested that the exhausted CD8+ T cells genes enriched in LILRB1+ M2 TAMs signature high GC patients. (B) Expression difference of effector molecules (CD107a, IL-17A) between LILRB1+ M2 TAMs high signature and low signature. (C) Expression of immune checkpoint molecules (PD-1, CTLA-4, HAVCR2, LAG-3, VTCN1, and CD276) in LILRB1+ M2 TAMs high and low expression subgroup. **P < 0.01 and ***P < 0.001, ns, no statistical significance.
To further discover the clinical significance of LILRB1 and M2 TAMs infiltration in GC, we evaluated the prognosis of LILRB1+ and CD163+ cells by Kaplan–Meier analysis and log-rank test. The findings revealed that GC patients with high expression of LILRB1 had a poor OS (Figure 6A). High density of CD163+ macrophage infiltration also predicted unfavorable prognosis in OS (Figure 6B). Moreover, high levels of LILRB1 and CD163 were associated with a high risk of recurrence in GC patients (Figures 6E, F). Multivariate Cox regression analysis demonstrated that differentiation grade (OS: hazard ratio (HR): 2.025, 95% CI: 1.148-3.571, P = 0.015; DFS: HR: 3.083, 95% CI: 1.569-6.059, P = 0.001), high CD163 (OS: HR 1.866, 95% CI: 1.177-2.958, P = 0.008; DFS: HR: 1.771, 95% CI: 1.073-2.925, P = 0.025), high LILRB1 expression (OS: HR: 2.008, 95% CI: 1.262-3.195, P = 0.003; DFS: HR: 1.947, 95% CI: 1.187-3.195, P = 0.008), and TNM stage (OS: HR: 3.095, 95% CI: 1.465-6.540, P = 0.003; DFS: HR: 3.702, 95% CI: 1.619-8.464, P = 0.002) was an independent poor prognostic factor for OS and DFS (Tables 1, 2). We also evaluated the correlation between the expressions of LILRB1 and CD163 and OS by every stage. There was a significant positive correlation between LILRB1 and CD163 expression in different TNM stages. Patients with high expression of LILRB1 had poorer OS in TNM stage II and III. However, the relationship between LILRB1 and OS in TNM stage I patients was not statistically significant, possibly due to the small number of stage 1 patients included in the study. These findings were showed in Supplementary Figure 1.
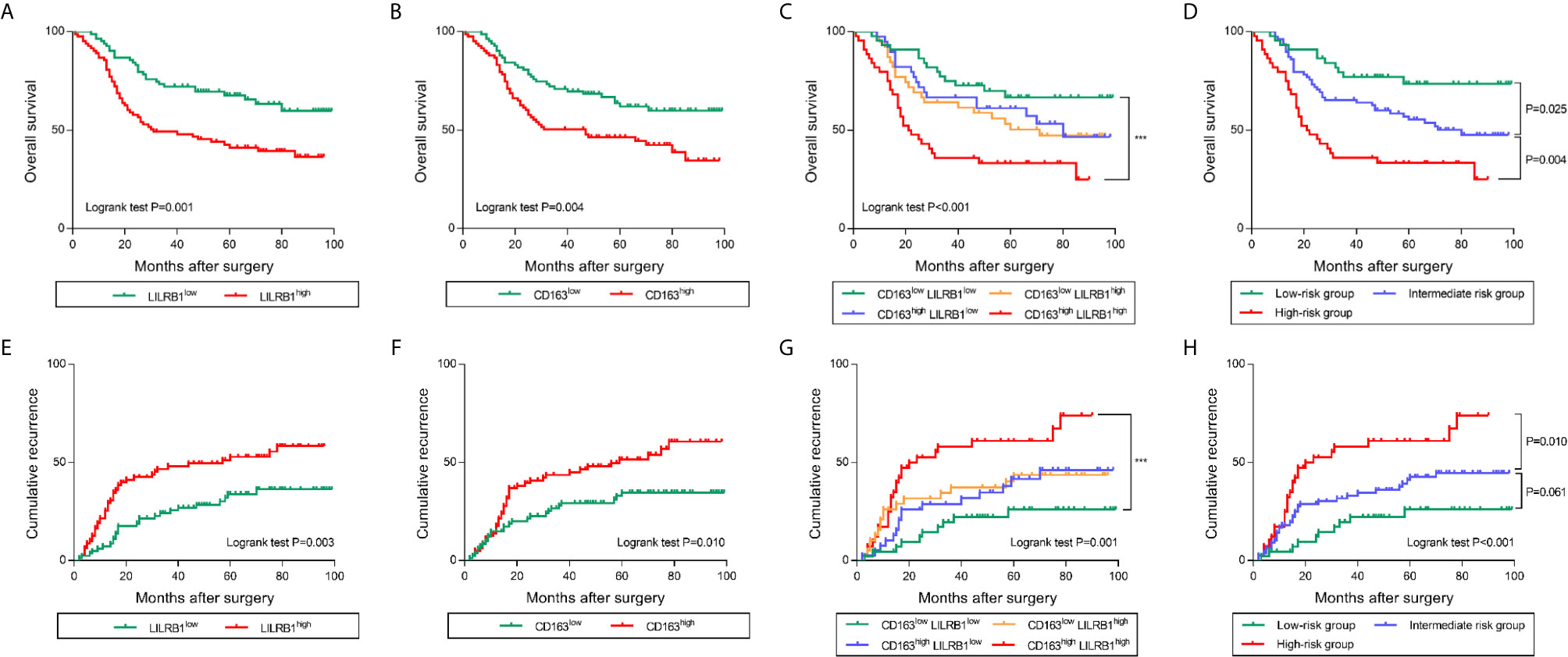
Figure 6 LILRB1 and M2 TAMs determined poor prognosis in GC patients. (A, B) Overall survival curves according to the levels of LILRB1 and CD163 distribution in GC patients. (C, D) Overall survival of GC patients stratified on the basis of LILRB1 and CD163. (E, F) Recurrence curve according to the levels of LILRB1 and CD163 distribution. (G, H) Recurrence risk stratified on the basis of LILRB1 and CD163. ***P < 0.001.
Furthermore, we combined LILRB1+ cells with CD163+ cells for survival analysis. Notably, patients with high infiltration of both LILRB1+ and CD163+ cells indicated poor OS and high risk of recurrence (Figures 6C, G). Low levels of LILRB1+ and CD163+ cells infiltration predicted favorable survival. Patients with LILRB1highCD163low or LILRB1lowCD163high levels infiltration showed an intermediate prognosis. In order to highlight the prognostic merit and enhance clinical practicality, we trichotomized patients into three risk subgroups: low-risk group (LILRB1lowCD163low), the intermediate-risk group (LILRB1highCD163low/LILRB1lowCD163high), and high-risk group (LILRB1highCD163high). Consistent with our identification, the low-risk group showed a favorable prognosis, while high-risk group showed poor OS and maximal risk of recurrence (Figures 6D, H).
Previous studies demonstrated that the alternations within tumor infiltrating immune contexture predominantly affected the response to postoperative adjuvant chemotherapy (ACT) (17). Thus, we assessed the predictive value of different risk groups for the efficacy of fluorouracil-based ACT in GC patients. Typically, ACT produced survival benefits in the GC patients (Figure 7A). The results of stratified analysis suggested that patients in the low and intermediate risk group benefit significantly from ACT (Figures 7B, C). However, in patients with high risk (LILRB1highCD163high), OS was not improved even after ACT was applied (Figure 7D). Taken together, these findings revealed that the levels of LILRB1+ cells combined with CD163+ cells could stratify patients into various risk subgroups and predict the sensitivity of patients to ACT.
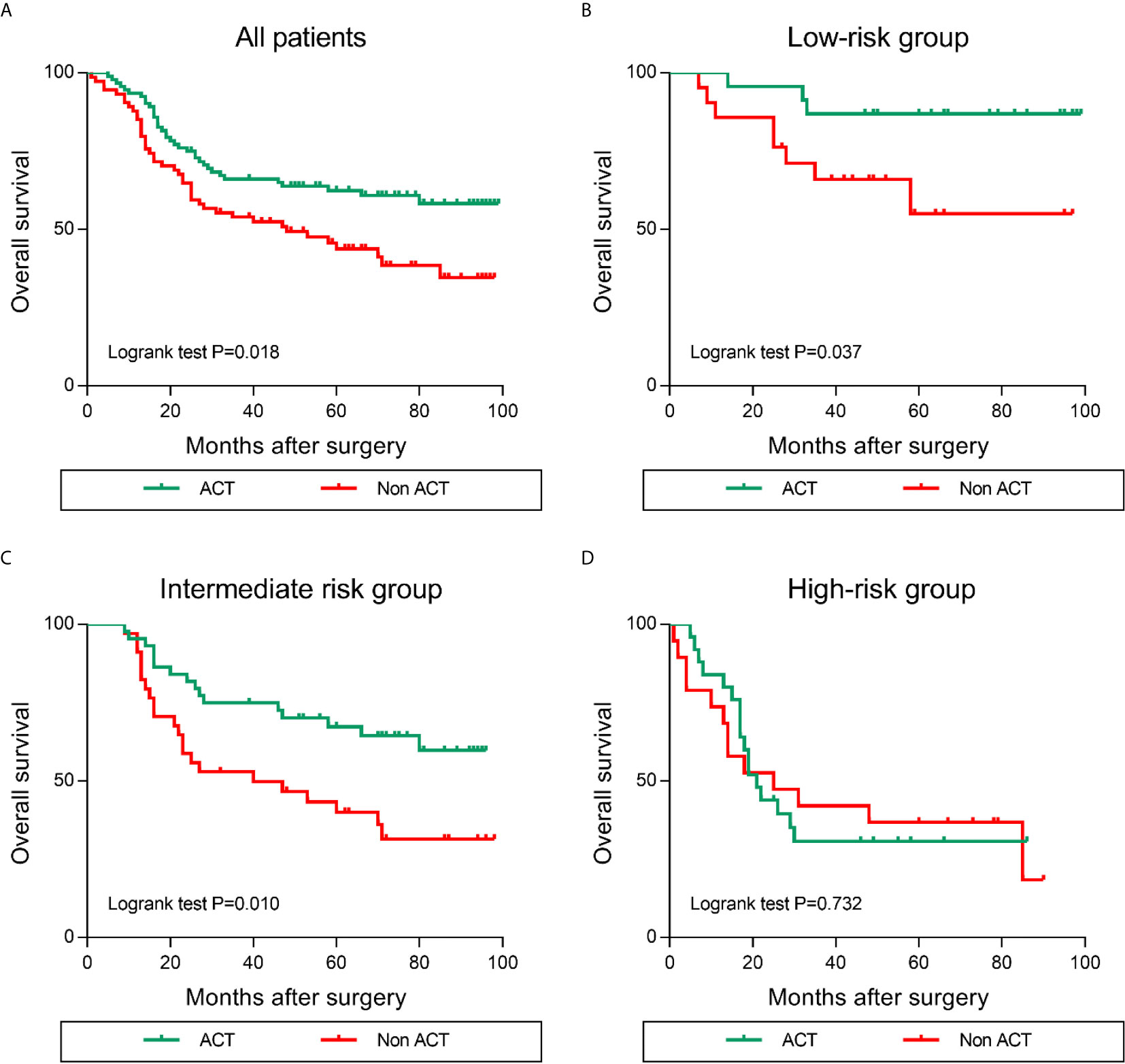
Figure 7 Increased LILRB1 and M2 TAMs infiltration predict poor efficacy of ACT (adjuvant chemotherapy) in GC patients. Overall survival for (A) all enrolled, (B) low-risk, (C) intermediate-risk, (D) high-risk GC patients with or without ACT.
Leukocyte immunoglobulin-like receptor subfamily B (LILRB) is a transmembrane glycoprotein and is a critical receptor of human leukocyte antigen G (HLA-G) molecules (18). It is widely distributed and expressed in NK cells, monocytes/macrophages, dendritic cells (DCs), and tumor cells (19). Previous studies have found that LILRB1 promotes tumor development, such as lung cancer, breast cancer, and pancreatic cancer, which can significantly enhance the movement and migration ability of cancer cells and promote tumor metastasis. Furthermore, LILRB1 is considered as an immunosuppressive receptor that can not only bind with the classical human major histocompatibility complex (MHC) molecules but also with non-classical MHC molecules (such as HLA-G and HLA-E) to exert immunosuppressive effect (20). It is mainly involved in the regulation of maternal fetal immune tolerance and induction of transplantation immune tolerance. The rapid growth of tumor cells could be attributed to the escape of immune surveillance. As a major immunosuppressive receptor, LILRB1 plays a critical role in the escaping of tumor cells from immune surveillance. Thus, it is of great significance to understand the function of LILRB1 and break the “tumor immune tolerance microenvironment.”
In recent years, some studies have shown that LILRB1 is expressed in a variety of tumor cells, and the expression level is significantly related to tumor growth and prognosis of patients (21). For example, LILRB1 promotes tumor progression in maintaining the stem cells and hematopoietic stem cells (22). Interestingly, LILRB1 is mainly expressed around tumor cells rather than on tumor cells in patients with GC. The analysis of GSE15459 cohort and immunohistochemical detection of 166 patients with GC in our center revealed that the expression of LILRB1 was positively correlated with M2 TAMs infiltration. Immunofluorescence further confirmed that M2 macrophages are the main immune cells expressing LILRB1, rather than M1 macrophages. M2 TAMs are the most abundant immune cells in tumor tissues. M2 TAMs stimulate natural T cells to produce Th2 type response. These cells secrete vascular endothelial growth factor (VEGF), transforming growth factor β (TGF-β), and other cytokines (23) that promote tissue repair, angiogenesis, immunosuppression, and tumor progression (24). It has been found that LILRB1 high-expressing macrophages interact with MHC class I components on tumor cell surfaces to protect tumor cells from phagocytosis (25). In addition, LILRB1 promotes M2 polarization of macrophages. These results suggested that LILRB1 mediates M2 TAMs to promote tumor immune escape in GC, which is a potential target for antitumor immunotherapy.
We further investigated the impact of LILRB1 on immune microenvironment in GC patients. LILRB1 expression was significantly correlated with several M2 macrophage-related cytokines including CCL22, IL-10, and TGF-β1. These molecules have been widely reported to promote tumor metastasis, immune escape, and angiogenesis (26–29). We next explored the difference of immune status between LILRB1+ M2 TAMs high and low subgroups. The exhausted CD8+ T cell gene set was found significantly enriched in LILRB1+ M2 TAMs signature high GC patients. Exhaustive T cells are a group of T cells with reduced effector function and continued expression of inhibitory receptors (30). It is involved in the negative regulation of tumor immunity (31). Additionally, high LILRB1+ M2 TAMs signature exhibited abundant immune checkpoint molecules expression including PD-1, CTLA-4, LAG-3, and HAVCR2. These results suggest that the application of corresponding monoclonal antibodies targeting immune regulatory points to reverse depleted T cells and restore anti-tumor immune response may benefit GC patients with high LILRB1 expression.
Prognosis evaluation is the key to selecting the appropriate treatment for cancer patients. In recent years, the prognostic significance of tumor-infiltrating immune cells has gained increasing attention because of their role in the occurrence and development of tumors (32). In the current study, we confirmed that patients with LILRB1 and CD163+ cell infiltration had poor OS rate and high recurrence rate. Moreover, we divided three risk subgroups according to the expression levels of LILRB1 and CD163. Patients with increased expression of LILRB1 and CD163 show poor prognosis. Adjuvant chemotherapy has been recommended as the standard therapy to improve the prognosis of patients with stage II/III GC. However, all patients do not benefit from adjuvant chemotherapy, and the selection criteria of candidate regimens are yet unclear. Recent studies suggested that tumor-associated macrophages affect the efficacy of chemotherapy in tumor patients (8). Therefore, we further studied the correlation between the infiltration of LILRB1 and CD163+cells and the efficacy of chemotherapy in patients with stage II/III GC. Notably, when a large number of LILRB1 and CD163+ cells infiltrate into GC tissues, patients may not benefit from chemotherapy. These results would facilitate appropriate selection of adjuvant chemotherapy for the management of GC patients.
Nevertheless, the present study has serval limitations. First, our study is based on a retrospective design. An external cohort is required to verify the prognostic significance of LILRB1 in GC patients and ACT efficacy. Moreover, there is no international unification cutoff value to identify the levels of LILRB1 and CD163 expression. Different cutoff values may affect the repeatability of the results. Moreover, we have not identified the mechanism underlying the formed and differentiated LILRB1+ macrophages, which need to be explored in future research. Previous studies have reported that the numbers of CD163+ macrophages were higher in tumor microenvironment of cases with a cytotoxic/Th1 signature (33, 34). Thus, using CD163 alone as a marker of M2 macrophages is not rigorous enough. To supplement this deficiency, we also found a correlation between LILRB1 and M2 macrophage expression using CIBERSORT algorithm. In different cohort, our analysis also found that LILRB1 was associated with the expression of other M2 macrophage marker (CD204).
This study revealed that LILRB1 is highly expressed in GC tissues and mainly expressed in M2 macrophages. Dense infiltration of LILRB1+ M2 TAMs yielded an immunosuppressive microenvironment. Patients with high infiltration of both LILRB1+ cells and M2 TAMs indicated poor prognosis and inferior therapeutic responsiveness to adjuvant chemotherapy. Further studies are essential to explore therapeutic targeting LILRB1+ M2 TAMs.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the institutional review board and ethics committee of the Seventh Affiliated Hospital of Sun Yat-sen University, Shenzhen, Guangdong, China. The patients/participants provided their written informed consent to participate in this study.
SY, YH, and CZ conceived and designed the study. SY, YZ, and HW performed experiments and analyzed the data. YZ, HW, XX, and HL wrote the paper. SY, TH, YH, and CZ edited the manuscript and provided critical comments. All authors contributed to the article and approved the submitted version.
This study was funded by grants from National Natural Science Foundation of China (82003104, 82073148, and 81772579), Guangdong Provincial Key Laboratory of Digestive Cancer Research (2021B1212040006), Guangdong Basic and Applied Basic Research Foundation (2019A1515110632), Sanming Project of Medicine in Shenzhen (SZSM201612022), Shenzhen Sustainable Project (KCXFZ202002011010593), China Postdoctoral Science Foundation (2021T140768) and Postdoctoral Foundation of the Seventh Affiliated Hospital of Sun Yat-sen University (ZSQYRSFPD0003). All these study sponsors have no roles in the study design, in the collection, analysis and interpretation of data.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.668707/full#supplementary-material
Supplementary Figure 1 | Correlation analysis of LILRB1 with CD163 expression and OS was stratified according to TNM stage. (A) Correlation between LILRB1 and CD163 expression in TNM stage I, II and III. (B) Correlation between LILRB1 and OS in TNM stage I, II and III.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric Cancer. Lancet (2020) 396(10251):635–48. doi: 10.1016/S0140-6736(20)31288-5
3. Yin S, Huang J, Li Z, Zhang J, Luo J, Lu C, et al. The Prognostic and Clinicopathological Significance of Tumor-Associated Macrophages in Patients With Gastric Cancer: A Meta-Analysis. PLoS One (2017) 12(1):e0170042. doi: 10.1371/journal.pone.0170042
4. van der Touw W, Chen HM, Pan PY, Chen SH. LILRB Receptor-Mediated Regulation of Myeloid Cell Maturation and Function. Cancer Immunol Immunother (2017) 66(8):1079–87. doi: 10.1007/s00262-017-2023-x
5. Kang X, Kim J, Deng M, John S, Chen H, Wu G, et al. Inhibitory Leukocyte Immunoglobulin-Like Receptors: Immune Checkpoint Proteins and Tumor Sustaining Factors. Cell Cycle (2016) 15(1):25–40. doi: 10.1080/15384101.2015.1121324
6. Mohme M, Riethdorf S, Pantel K. Circulating and Disseminated Tumour Cells - Mechanisms of Immune Surveillance and Escape. Nat Rev Clin Oncol (2017) 14(3):155–67. doi: 10.1038/nrclinonc.2016.144
7. Halbrook CJ, Pontious C, Kovalenko I, Lapienyte L, Dreyer S, Lee HJ, et al. Macrophage-Released Pyrimidines Inhibit Gemcitabine Therapy in Pancreatic Cancer. Cell Metab (2019) 29(6):1390–99.e6. doi: 10.1016/j.cmet.2019.02.001
8. Feng Q, Chang W, Mao Y, He G, Zheng P, Tang W, et al. Tumor-Associated Macrophages as Prognostic and Predictive Biomarkers for Postoperative Adjuvant Chemotherapy in Patients With Stage II Colon Cancer. Clin Cancer Res (2019) 25(13):3896–907. doi: 10.1158/1078-0432.CCR-18-2076
9. Zhang H, Li R, Cao Y, Gu Y, Lin C, Liu X, et al. Poor Clinical Outcomes and Immunoevasive Contexture in Intratumoral IL-10-Producing Macrophages Enriched Gastric Cancer Patients. Ann Surg (2020). doi: 10.1097/SLA.0000000000004037
10. Xiao Y, Ma D, Zhao S, Suo C, Shi J, Xue MZ, et al. Multi-Omics Profiling Reveals Distinct Microenvironment Characterization and Suggests Immune Escape Mechanisms of Triple-Negative Breast Cancer. Clin Cancer Res (2019) 25(16):5002–14. doi: 10.1158/1078-0432.CCR-18-3524
11. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust Enumeration of Cell Subsets From Tissue Expression Profiles. Nat Methods (2015) 12(5):453–7. doi: 10.1038/nmeth.3337
12. Kim JY, Kim WG, Kwon CH, Park DY. Differences in Immune Contextures Among Different Molecular Subtypes of Gastric Cancer and Their Prognostic Impact. Gastric Cancer (2019) 22(6):1164–75. doi: 10.1007/s10120-019-00974-4
13. Lanzi A, Pages F, Lagorce-Pages C, Galon J. The Consensus Immunoscore: Toward a New Classification of Colorectal Cancer. Oncoimmunology (2020) 9(1):1789032. doi: 10.1080/2162402X.2020.1789032
14. Ruffell B, Coussens LM. Macrophages and Therapeutic Resistance in Cancer. Cancer Cell (2015) 27(4):462–72. doi: 10.1016/j.ccell.2015.02.015
15. Mantovani A, Allavena P. The Interaction of Anticancer Therapies With Tumor-Associated Macrophages. J Exp Med (2015) 212(4):435–45. doi: 10.1084/jem.20150295
16. Gambardella V, Castillo J, Tarazona N, Gimeno-Valiente F, Martinez-Ciarpaglini C, Cabeza-Segura M, et al. The Role of Tumor-Associated Macrophages in Gastric Cancer Development and Their Potential as a Therapeutic Target. Cancer Treat Rev (2020) 86:102015. doi: 10.1016/j.ctrv.2020.102015
17. Bruni D, Angell HK, Galon J. The Immune Contexture and Immunoscore in Cancer Prognosis and Therapeutic Efficacy. Nat Rev Cancer (2020) 20(11):662–80. doi: 10.1038/s41568-020-0285-7
18. Baia D, Pou J, Jones D, Mandelboim O, Trowsdale J, Muntasell A, et al. Interaction of the LILRB1 Inhibitory Receptor With HLA Class Ia Dimers. Eur J Immunol (2016) 46(7):1681–90. doi: 10.1002/eji.201546149
19. Dumont C, Jacquier A, Verine J, Noel F, Goujon A, Wu CL, et al. CD8(+)PD-1(-)ILT2(+) T Cells Are an Intratumoral Cytotoxic Population Selectively Inhibited by the Immune-Checkpoint HLA-G. Cancer Immunol Res (2019) 7(10):1619–32. doi: 10.1158/2326-6066.CIR-18-0764
20. Chen H, Chen Y, Deng M, John S, Gui X, Kansagra A, et al. Antagonistic Anti-LILRB1 Monoclonal Antibody Regulates Antitumor Functions of Natural Killer Cells. J Immunother Cancer (2020) 8(2). doi: 10.1136/jitc-2019-000515
21. Cheng J, Luan J, Chen P, Kuang X, Jiang P, Zhang R, et al. Immunosuppressive Receptor LILRB1 Acts as a Potential Regulator in Hepatocellular Carcinoma by Integrating With SHP1. Cancer Biomark (2020) 28(3):309–19. doi: 10.3233/CBM-190940
22. Sharma P, Allison JP. Immune Checkpoint Targeting in Cancer Therapy: Toward Combination Strategies With Curative Potential. Cell (2015) 161(2):205–14. doi: 10.1016/j.cell.2015.03.030
23. Mantovani A, Sica A. Macrophages, Innate Immunity and Cancer: Balance, Tolerance, and Diversity. Curr Opin Immunol (2010) 22(2):231–7. doi: 10.1016/j.coi.2010.01.009
24. Noy R, Pollard JW. Tumor-Associated Macrophages: From Mechanisms to Therapy. Immunity (2014) 41(1):49–61. doi: 10.1016/j.immuni.2014.06.010
25. Barkal AA, Weiskopf K, Kao KS, Gordon SR, Rosental B, Yiu YY, et al. Engagement of MHC Class I by the Inhibitory Receptor LILRB1 Suppresses Macrophages and is a Target of Cancer Immunotherapy. Nat Immunol (2018) 19(1):76–84. doi: 10.1038/s41590-017-0004-z
26. Wu S, He H, Liu H, Cao Y, Li R, Zhang H, et al. C-C Motif Chemokine 22 Predicts Postoperative Prognosis and Adjuvant Chemotherapeutic Benefits in Patients With Stage II/III Gastric Cancer. Oncoimmunology (2018) 7(6):e1433517. doi: 10.1080/2162402X.2018.1433517
27. Yang L, Dong Y, Li Y, Wang D, Liu S, Wang D, et al. IL-10 Derived From M2 Macrophage Promotes Cancer Stemness via JAK1/STAT1/NF-Kappab/Notch1 Pathway in Non-Small Cell Lung Cancer. Int J Cancer (2019) 145(4):1099–110. doi: 10.1002/ijc.32151
28. Perez LG, Kempski J, McGee HM, Pelzcar P, Agalioti T, Giannou A, et al. TGF-Beta Signaling in Th17 Cells Promotes IL-22 Production and Colitis-Associated Colon Cancer. Nat Commun (2020) 11(1):2608. doi: 10.1038/s41467-020-16363-w
29. Kindlund B, Sjoling A, Yakkala C, Adamsson J, Janzon A, Hansson LE, et al. CD4(+) Regulatory T Cells in Gastric Cancer Mucosa Are Proliferating and Express High Levels of IL-10 But Little TGF-Beta. Gastric Cancer (2017) 20(1):116–25. doi: 10.1007/s10120-015-0591-z
30. Liu F, Liu W, Sanin DE, Jia G, Tian M, Wang H, et al. Heterogeneity of Exhausted T Cells in the Tumor Microenvironment is Linked to Patient Survival Following Resection in Hepatocellular Carcinoma. Oncoimmunology (2020) 9(1):1746573. doi: 10.1080/2162402X.2020.1746573
31. Hossain MA, Liu G, Dai B, Si Y, Yang Q, Wazir J, et al. Reinvigorating Exhausted CD8(+) Cytotoxic T Lymphocytes in the Tumor Microenvironment and Current Strategies in Cancer Immunotherapy. Med Res Rev (2021) 41(1):156–201. doi: 10.1002/med.21727
32. Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The Immune Contexture in Cancer Prognosis and Treatment. Nat Rev Clin Oncol (2017) 14(12):717–34. doi: 10.1038/nrclinonc.2017.101
33. Barros MH, Hassan R, Niedobitek G. Tumor-Associated Macrophages in Pediatric Classical Hodgkin Lymphoma: Association With Epstein-Barr Virus, Lymphocyte Subsets, and Prognostic Impact. Clin Cancer Res (2012) 18(14):3762–71. doi: 10.1158/1078-0432.CCR-12-0129
Keywords: gastric cancer, LILRB1, tumor-associated macrophages, prognosis, tumor immune microenvironment
Citation: Zhang Y, Wang H, Xu X, Liu H, Hao T, Yin S, Zhang C and He Y (2021) Poor Prognosis and Therapeutic Responses in LILRB1-Expressing M2 Macrophages-Enriched Gastric Cancer Patients. Front. Oncol. 11:668707. doi: 10.3389/fonc.2021.668707
Received: 17 February 2021; Accepted: 26 July 2021;
Published: 09 August 2021.
Edited by:
Kecheng Zhang, People’s Liberation Army General Hospital, ChinaReviewed by:
Satoru Kikuchi, Okayama University, JapanCopyright © 2021 Zhang, Wang, Xu, Liu, Hao, Yin, Zhang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Songcheng Yin, eXNjMjAxNzg5QDEyNi5jb20=; Changhua Zhang, emNoMzIxOEAxMjYuY29t; Yulong He, aHlsODUxOEAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.