
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 04 June 2021
Sec. Molecular and Cellular Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.666826
This article is part of the Research TopicEmerging Molecular Mechanisms of Cell Cycle Regulation in Cancer: Functions and Potential ApplicationsView all 23 articles
Background: Lung adenocarcinoma (LUAD) is a leading malignancy and has a poor prognosis over the decades. LUAD is characterized by dysregulation of cell cycle. Immunotherapy has emerged as an ideal option for treating LUAD. Nevertheless, optimal biomarkers to predict outcomes of immunotherapy is still ill-defined and little is known about the interaction of cell cycle-related genes (CCRGs) and immunity-related genes (IRGs).
Methods: We downloaded gene expression and clinical data from TCGA and GEO database. LASSO regression and Cox regression were used to construct a differentially expressed CCRGs and IRGs signature. We used Kaplan-Meier analysis to compare survival of LUAD patients. We constructed a nomogram to predict the survival and calibration curves were used to evaluate the accuracy.
Results: A total of 61 differentially expressed CCRGs and IRGs were screened out. We constructed a new risk model based on 8 genes, including ACVR1B, BIRC5, NR2E1, INSR, TGFA, BMP7, CD28, NUDT6. Subgroup analysis revealed the risk model accurately predicted the overall survival in LUAD patients with different clinical features and was correlated with immune cells infiltration. A nomogram based on the risk model exhibited excellent performance in survival prediction of LUAD.
Conclusions: The 8 gene survival signature and nomogram in our study are effective and have potential clinical application to predict prognosis of LUAD.
Lung cancer is a leading cause of cancer-related death over the world (1). It is reported that 2,206,771 lung cancer new cases and 1,796,144 deaths occurred in 2020 worldwide (2). Non-small cell lung cancer (NSCLC) is the most common histological type of lung malignancies, accounting for over 80% of all cases, and near half of NSCLC are lung adenocarcinoma (LUAD) (3, 4). The prognosis of LUAD has been disappointing over the past two decades, with a five-year survival rate below 20% (5). Mortality from LUAD has decreased substantially in recent years, coinciding with the advances in immunotherapy. Immune checkpoint inhibitors (ICIs), especially inhibitors of the programmed cell death 1 (PD-1) axis have altered the therapeutic landscape of LUAD (6, 7). The pacific clinical trial reported that PD-1 inhibitor pembrolizumab improves three-year overall survival of NSCLC from 43.5% to 57.0% (8). However, only a subset of patients achieves an impressive and durable response to immunotherapy (9).
A major molecular characteristic of human cancer is that key cell cycle proteins are frequently dysregulated. Dysfunction of the cell cycle regulators forces tumor cells enter uncontrolled cell division (10, 11). The mitotic stress and chromosomal instability result in replication errors and increased mutation load (12). Tumor mutation load and tumor-infiltrating lymphocytes (TILs) have been widely investigated as prognostic and predictive biomarkers in multiple tumor types, including lung cancer (13, 14). In addition, cell cycle-targeted therapy such as cyclin-dependent kinase (CDK) 4 or CDK6 inhibitors induce tumor cell novel-antigen and recruit TILs, indicating a possible option for immunotherapy combination. Previous studies have suggested that the cell cycle regulator RB might also be required for tumor cells’ expression of MHC Class II molecules, which can be recognized by TILs and mediate cytotoxic killing of tumor cells. Additionally, the increased tumor neoantigen observed after CDK4/6 blockade increases the efficacy of ICB. One report has reported that CDK4/6 inhibition potentiates the expression of PD-L1 in tumor cell together with a decrease of T cell function in the tumor microenvironment (15–18). Given the critical association of cell cycle regulation and tumor immunotherapy, it is highly desirable and urgent to exploit cell cycle regulators and TILs in LUAD. However, the comprehensive analysis of cell cycle-related genes (CCRGs) and immune-related genes (IRGs) or the interactions between them are still not known. In the current research, we comprehensively analyzed the CCRGs and IRGs in LUAD. In addition, we established a predictive signature based on them and further depicted the potential regulatory network between IRGs and specific CCRGs in LUAD. This work provided novel insights into predicting the prognosis and efficacy of immunotherapy in LUAD patients, which brings up a new prospect for enhancing the personalized medication for the treatment of LUAD.
The RNA-seq profiles and related clinical characteristics of LUAD patients were extracted from TCGA and GSE68465. The IRGs for further immunological analysis were based on the ImmPort database (https://www.immport.org/home) (19). The reference CCRGs set were retrieved from MSigDB2. “GO_CELL_CYCLE” was also picked from “all GO gene sets as Gene Symbols” in “c5: Ontology gene sets. The | log2 fold change | > 0.5, p value < 0.05 and false discovery rate (FDR) <0.05 were defined as the cut-off vales to screen out the differentially expressed genes (DEGs) of LUAD. Differentially expressed CCRGs and IRGs were defined as the intersection of DEGs, CCRGs and IRGs. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were used to identify the enriched function of these genes by using Enrichr database (http://amp.pharm.mssm.edu/Enrichr/) (20–22).
The STRING database (https://string-db.org/) was obtained to investigate protein-protein interactions (PPIs) based on the intersection of DEGs, CCRGs and IRGs (23). In the present study, an PPI-score greater than 0.9 was set as the threshold. The Cytoscape software was used to distinguish the hub genes and visualize the PPI results (24).
The data of LUAD cases were downloaded from GEO (GSE68465, n=439) and TCGA (n=504). The GEO database was used as a train set and the TCGA database were used as a test set. LASSO Cox regression and multivariate Cox analysis were used to identify hub genes to construct the predictive signature. First, we used univariate Cox proportional hazard regression to identify prognosis-related genes with the cut-off value of p < 0.05. To eliminate the overfit gene of the model, these prognosis-related genes from univariate Cox analysis were further included in LASSO penalized Cox proportional hazards regression via R package “glmnet”. Next, the optimal model based on the remained genes from LASSO analysis was constructed by multivariate Cox regression analysis backward stepwise regression. The formula for risk score is: represents optimal gene numbers, expression levels of gene, and regression coefficients, respectively. Meanwhile, patients were separated into low- and high-risk groups according to the average value. We employed Kaplan-Meier analysis to weigh the differences between survival of each group, and ROC curve of 1-year, 3-year and 5-year survival was performed to assess the efficacy. In addition, we did univariate and multivariate Cox regression to analyze the risk score of prognostic models and several clinicopathological characteristics for LUAD. The risk model based on train group was validated in the test group and entire group. We used the R packages “rms” to construct a nomogram to predict prognosis of LUAD patients. A calibration map was generated by comparing the nomogram predictive efficacy for the 1-year, 3-year and 5-year OS rates (25).
The data of infiltrating immune cells in the tumor microenvironment of LUAD patients were downloaded from the TIMER database (https://cistrome.shinyapps.io/timer) (26). We employed Spearman’s test to evaluate correlations between genes and the infiltrating immune cells. A two-tailed p value lower than 0.05 was set as the cut-off value for statistical significance.
The tumor mutation profile of LUAD patients were obtained from the TCGA database. The somatic mutations of LUAD were analyzed from mutation annotation format. We used the formula: tumor mutation burden to determine tumor mutation TMB for each LUAD case.
The gene expression levels of LUAD (439 samples) and normal lung tissues (19 samples) in GEO database (GSE68465) were analyzed. The results demonstrated 3,910 DEGs, 1,889 of which are upregulated and 2,021 are downregulated (Figure 1A). Then, the intersection with two sets of CCRGs and IRGs revealed 33 upregulated and 28 downregulated DEGs that are involved in regulation of cell cycle and immunity (Figure 1B). GO analysis demonstrated that the differentially expressed CCRGs and IRGs are mainly enriched in Figure 1C. The significant KEGG pathways were enriched in Figure 1D. To further explore the interactions among the 61 differentially expressed CCRGs and IRGs, we established the PPI network (Figure 2A). Ten hub genes (EGF, EGFR, STAT3, IGF1, TNF, IL10, SRC, JUN, MAPK1, MAP2K1) were identified using the Cytoscape software (Figure 2B).
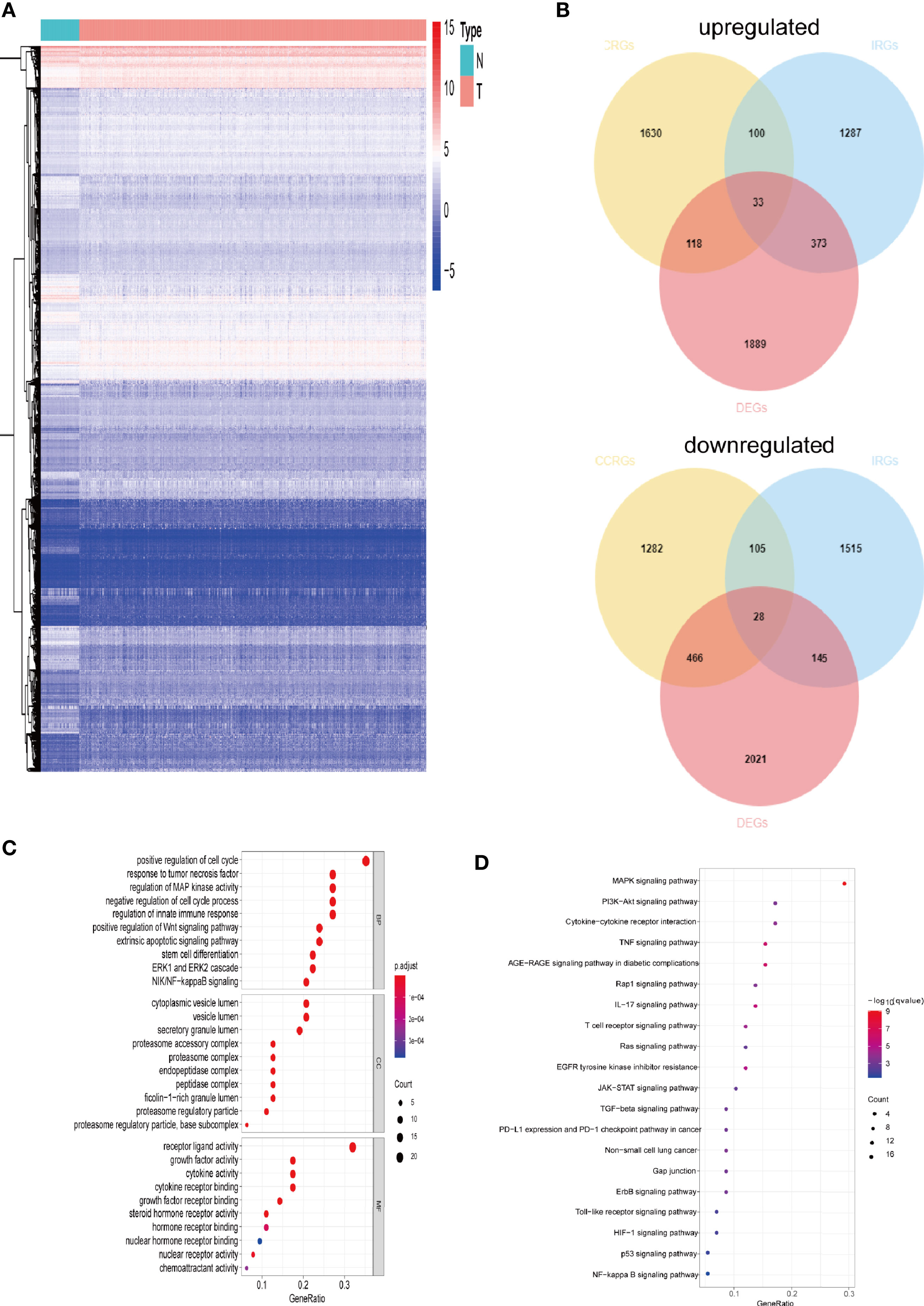
Figure 1 Comparison of gene expression profile with CCRGs and IRGs of LUAD. (A) Heatmap of differentially expressed genes in LUAD. (B) Venn diagram of up- and down-regulated differentially expressed genes based on CCRGs and IRGs. (C) GO analysis. (D) KEGG analysis.
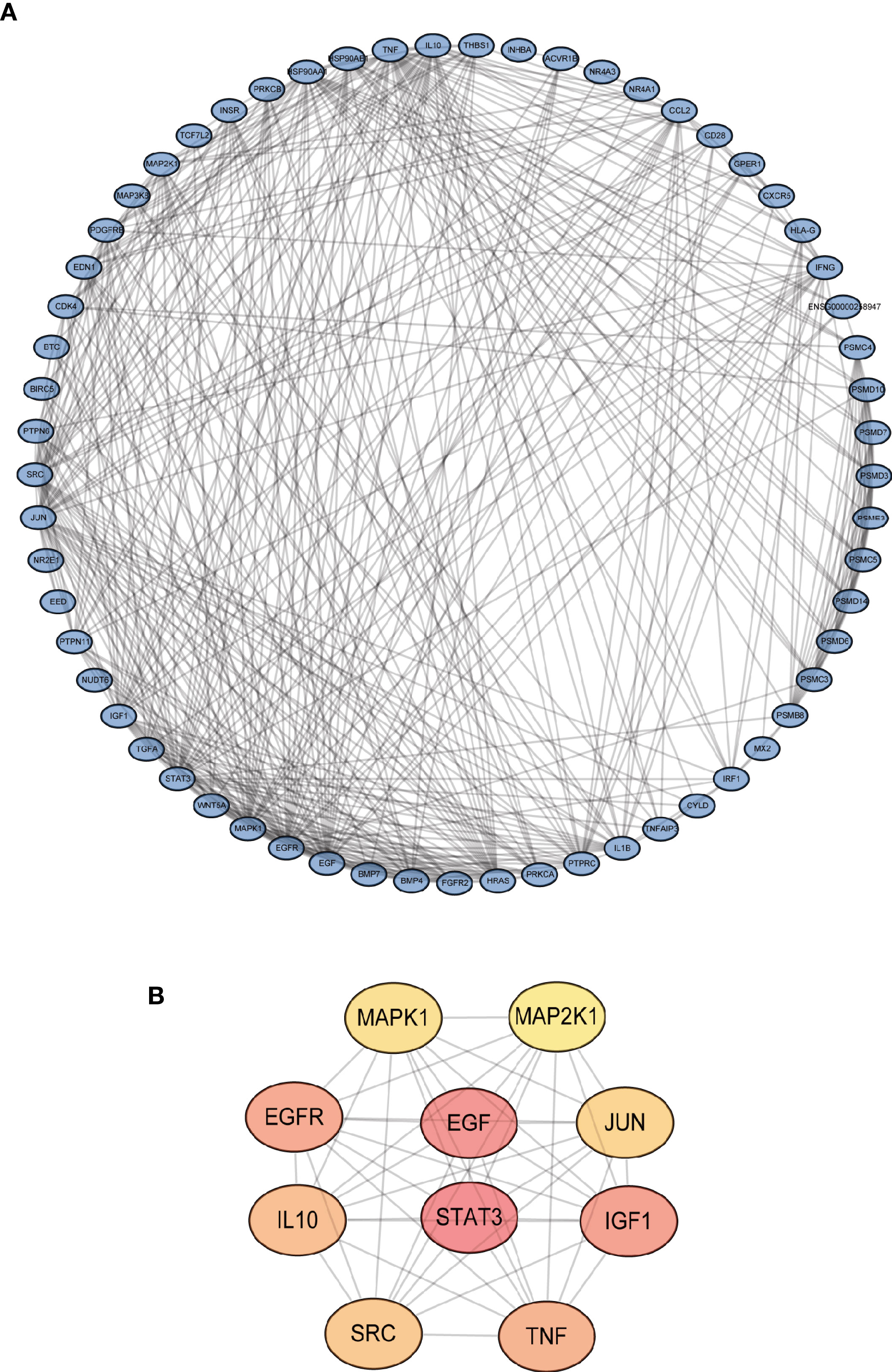
Figure 2 Identification of PPI network of the DEGs. (A) Visualization of the PPI network using STRING database and Cytoscape software. (B) Interactions of the top ten hub genes.
Cox regression and LASSO regression were performed in the train set to establish a CCRGs and IRGs predictive signature based on the 61 DEGs to evaluate the survival of LUAD patients. First, we used univariate Cox proportional hazard regression to identify prognosis-related genes from 61 DEGs. With the cut-off value of p < 0.05, the 12 prognosis-related genes were identified (ACVR1B, BIRC5, NR2E1, PTPN11, STAT3, INSR, MAP2K1, PRKCB, TGFA, BMP7, CD28 and NUDT6). To eliminate the overfit gene of the model, these 12 prognosis-related genes from univariate Cox analysis were further included in LASSO penalized Cox proportional hazards regression via R package “glmnet” (Figures 3A, B). Next, the optimal model based on the remained genes from LASSO analysis (ACVR1B, BIRC5, NR2E1, PTPN11, STAT3, INSR, MAP2K1, PRKCB, TGFA, BMP7, CD28 and NUDT6) was constructed by multivariate Cox regression analysis backward stepwise regression. Consequently, eight genes (ACVR1B, BIRC5, NR2E1, INSR, TGFA, BMP7, CD28, NUDT6) were identified for the risk model (Figure 3C). The risk score is = (-0.1831 × ACVR1B expression) + (0.1300 × BIRC5 expression) + (0.1798 × NR2E1 expression) + (-0.2200 × INSR expression) + (0.1697 × TGFA expression) + (0.1577 × BMP7 expression) + (-0.2649 × CD28 expression) + (0.2473 × NUDT6 expression). Patients were separated into high- and low-risk groups based on the average risk score. The survival of each group was shown in Figures 4A, B. The Kaplan-Meier survival analysis and the log-rank test demonstrated that patients in the low-risk group had significantly longer survival time (p<0.05, Figure 4C). In addition, the 1-, 3-, and 5-year ROC curves were plotted as Figure 4D. The test set proved the risk model based on the train set.
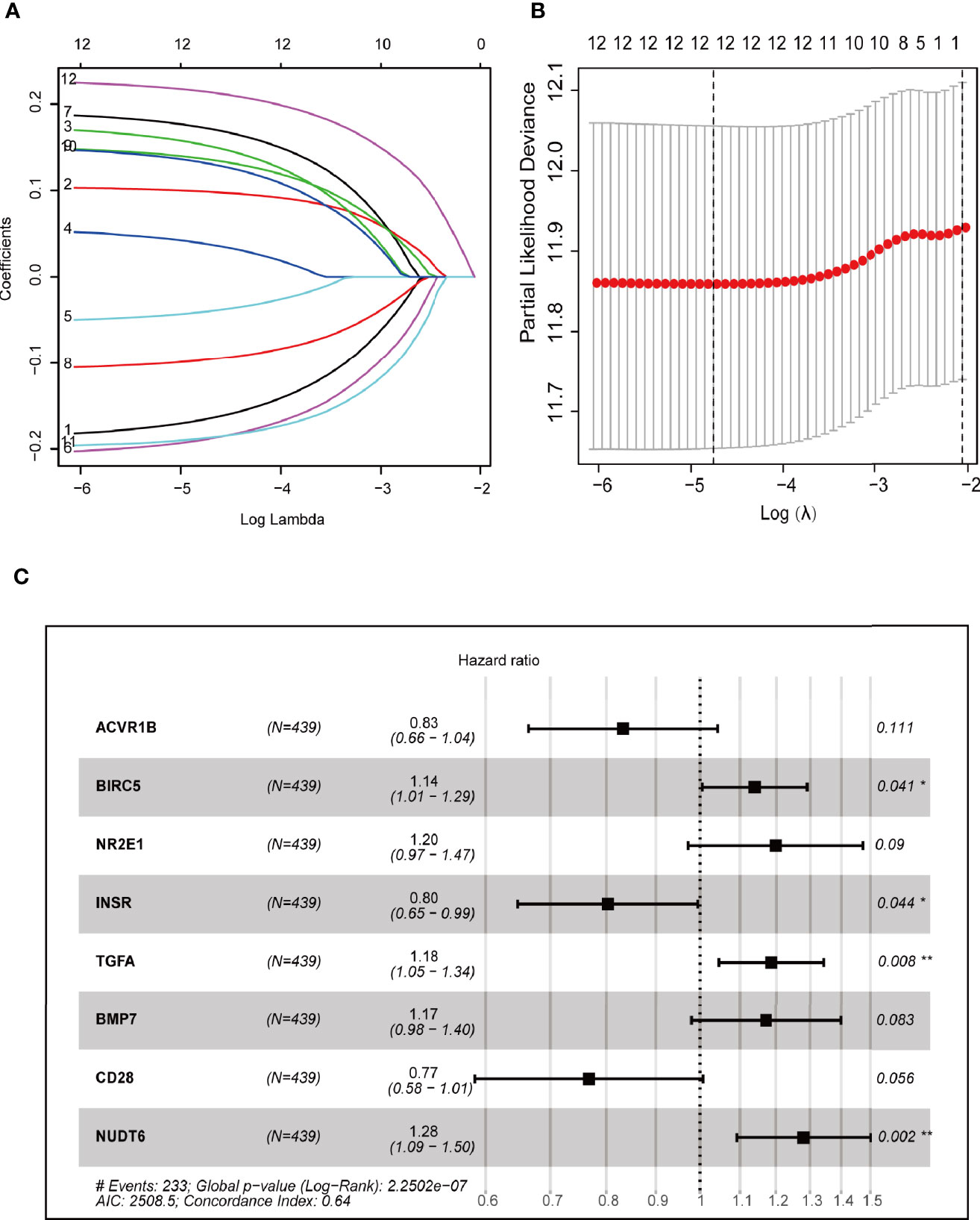
Figure 3 Construction of prognostic models based on CCRGs and IRGs. (A, B) LASSO Cox regression analysis based on OS. (C) Forest plots presenting the multivariate Cox proportional hazards regression analysis of prognostic CCRGs and IRGs in OS.
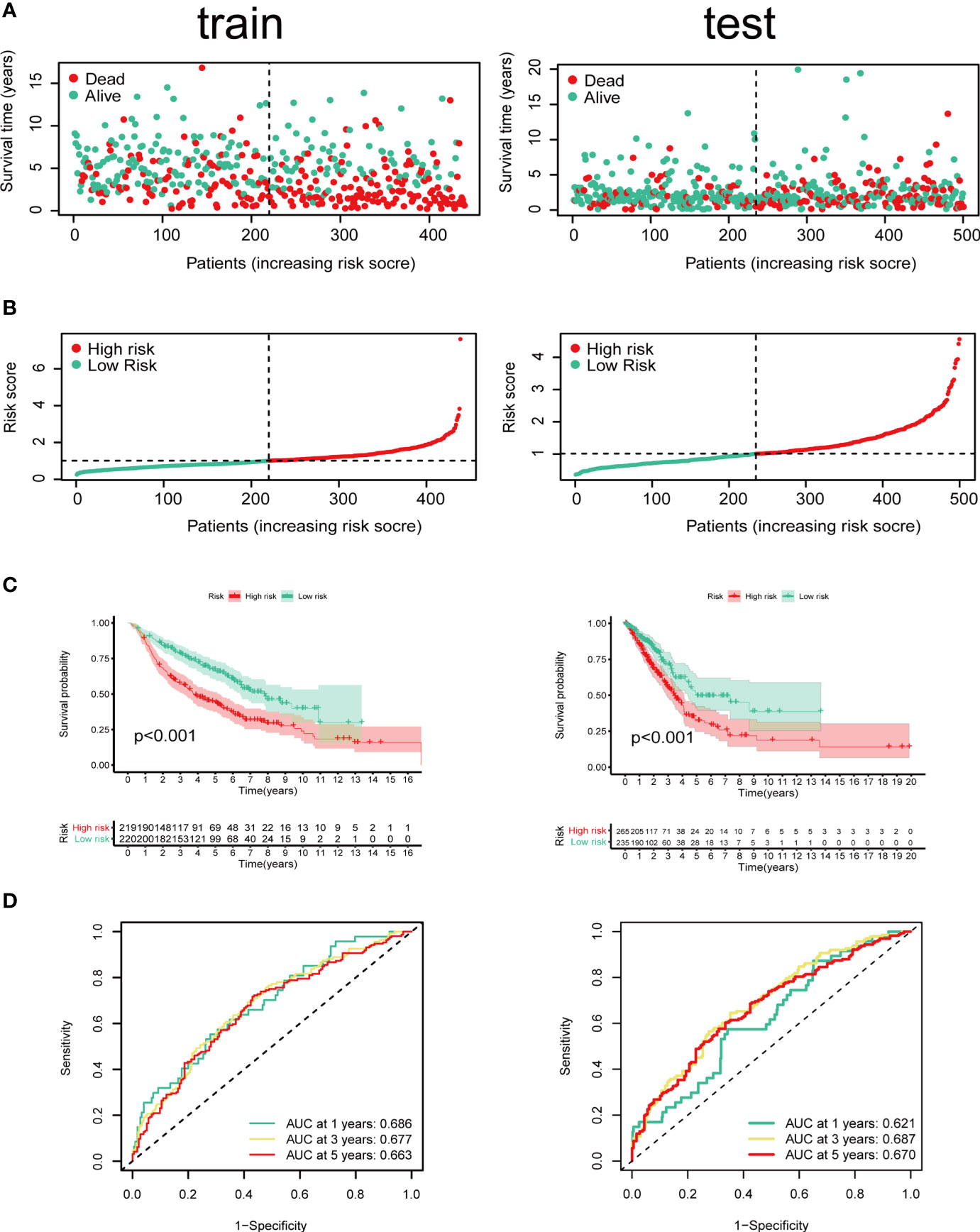
Figure 4 Identification of prognostic model in train set and test set based on OS of LUAD in GEO and TCGA cohorts. (A) The survival status and survival time of patients with LUAD ranked by risk score. (B) Rank of risk score and distribution of groups. Patients with LUAD were divided into high- and low-risk groups based on the median value of the risk score calculated. (C) Kaplan-Meier analysis and (D) time-dependent ROC curve of risk score.
We next did subgroup survival analysis based on different clinical features using the signature. These subgroups included age (≤65 or >65), gender, T stage (T1-2 or T3-4), and N stage (N0 or N1-2). As shown in Figure 5, the 5-year survival of the high-risk group stratified by the subgroups mentioned above were significantly shorter than that of the low-risk group. However, due to the small number of samples in the T3-4 group (n=36), the survival between two groups did not show significant differences.
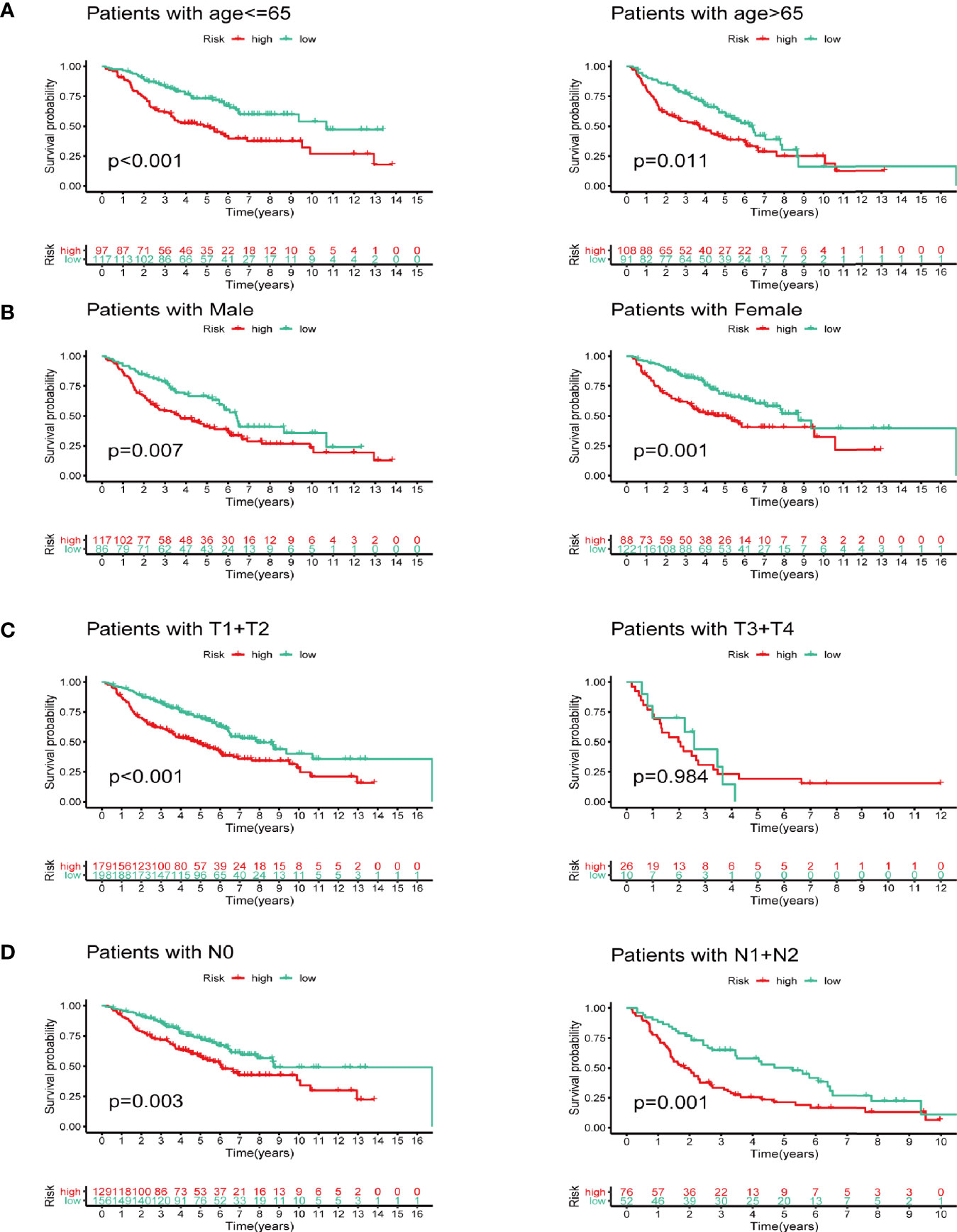
Figure 5 Subgroup survival analysis based on different clinicopathological features of LUAD. (A) Age. (B) Gender. (C) T stage. (D) N stage.
We further performed univariate and multivariate Cox analysis to investigate whether signature is an independent risk factor for the survival in LUAD. Univariate Cox analysis indicated that age, T stage, N stage and the signature were meaningful for predicting OS (p<0.001). Multivariate Cox analysis identified that the signature was an independent risk factor for predicting survival of LUAD (Figure 6A). As shown in Figure 6B, we used the clinicopathological features and risk score to generate a nomogram to predict the survival of LUAD patients. The worse prognosis is associated with higher sample scores. A calibration curve was used to evaluate the efficacy of the predictive model. It demonstrated that the signature in our study is better in predicting the 1-year, 3-year, and 5-year survival (Figure 6C).
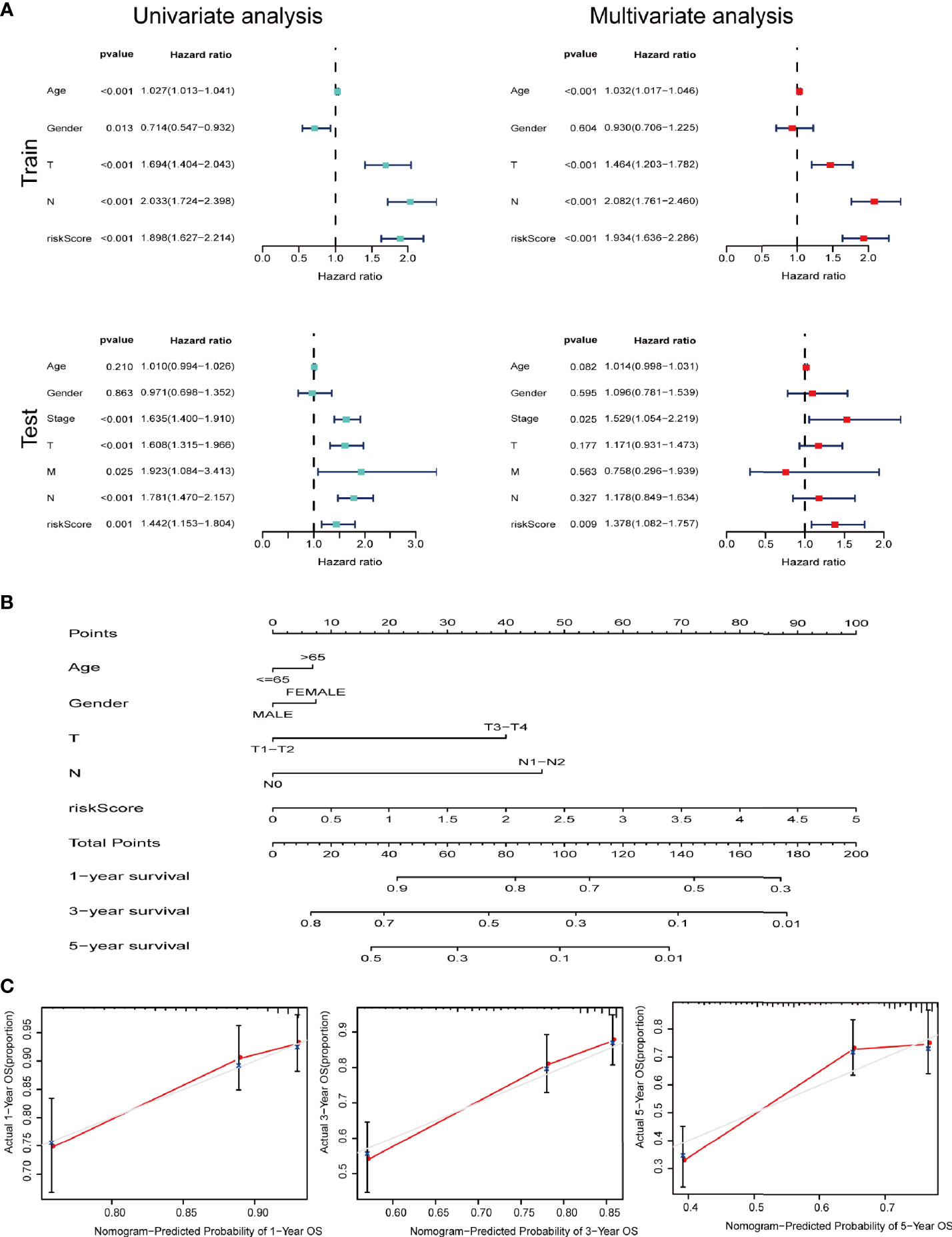
Figure 6 IRGs and CCRGs combined with other clinical factors to predict prognosis of LUAD patients based on OS model. (A) Univariate and multivariate Cox regression revealed significant survival-related clinicopathological parameters in forest plots diagram. (B) Nomogram and (C) calibration curve was constructed to verify the accuracy of predicting 1-, 3-, and 5-year survival rates.
We assessed the correlation between risk score and infiltrating immune cells in the tumor microenvironment using TIMER database. We found that patients with high-risk score was negatively correlated with infiltrating neutrophils, macrophages, dendritic cells, B lymphocytes, CD4+ T lymphocytes and CD8+ T lymphocytes in the tumor microenvironment (p<0.05), indicating a universal decrease of infiltrating immune cells (Figures 7A–F). Next, we further investigated the TMB of differentially expressed CCRGs and IRGs based on the risk signature. Immune checkpoint inhibitors have demonstrated significant overall survival benefit in LUAD. Nevertheless, a remarkable interpatient heterogeneity characterizes immunotherapy efficacy. TMB is an essential factor related to outcomes of immunotherapy of lung cancer. Since LUAD mostly occurs in non-smokers, the TMB of LUAD is relatively lower than that of lung squamous cell carcinoma, highlighting the necessity to explore factors related to TMB. We demonstrated that TMB and somatic mutation count was remarkably higher in the high-risk group (Figure 8A). TMB of genes in both groups are listed in Figure 8B. TP53, MUC16, TTN, RYR2, and CSMD3 ranked the top five TMB genes in both groups, and the TMB of them was drastically higher in high-risk group. Then, the correlations between TMB of genes were assessed. As shown in Figure 8C, TP53 and KRAS showed the most positive correlation, and RYR2 exhibited negative correlation with MUC16.
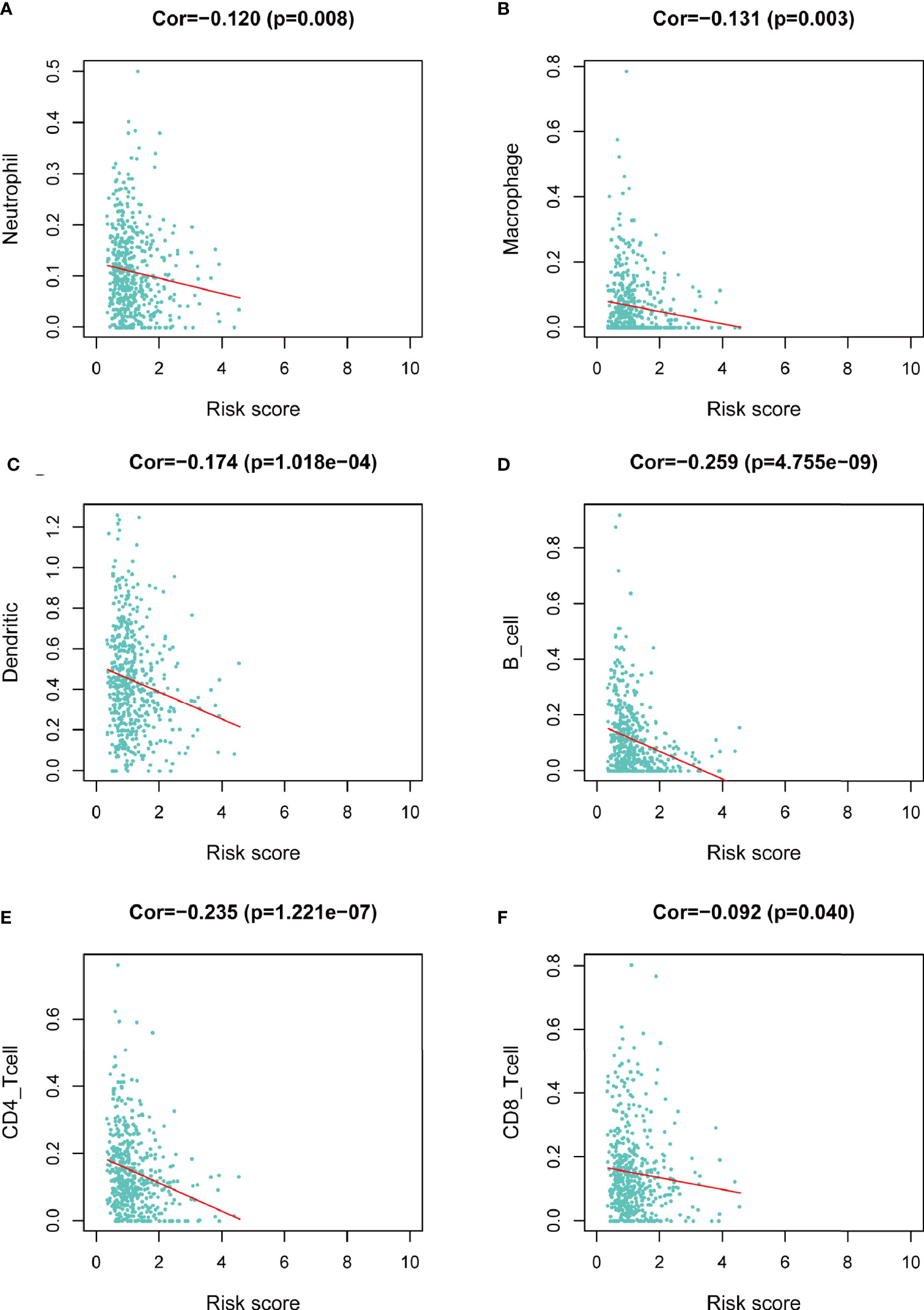
Figure 7 Correlation between risk score and infiltrating immune cells in the LUAD tumor microenvironment. (A) Neutrophil. (B) Macrophage. (C) Dendritic cell. (D) B lymphocyte. (E) CD4+ T lymphocyte. (F) CD8+ T lymphocyte.
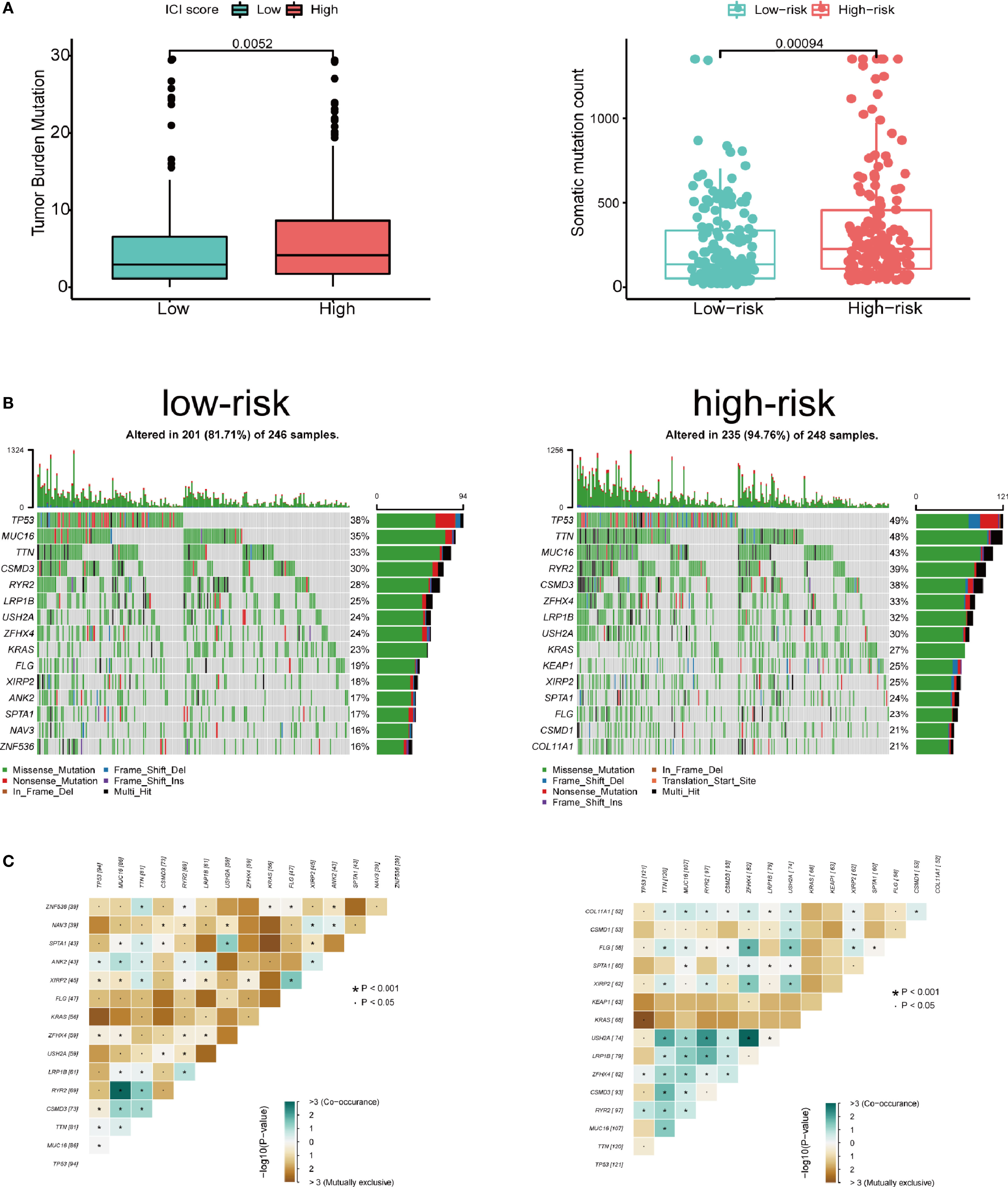
Figure 8 Tumor mutation in the high- and low-risk groups. (A) Patients in high-risk group showed significantly higher tumor mutation burden and somatic mutation count. (B) TP53, MUC16, TTN, RYR2, and CSMD3 ranked the top five TMB genes in both groups, and the TMB of them was drastically higher in high-risk group. (C) Correlations between mutated genes in both groups.
Lung cancer is characterized as one cancer type which has high morbidity and mortality. Almost 85% of lung cancer subtypes are NSCLC, of which LUAD and lung squamous cell carcinoma are the most common subtypes (3). LUAD is the most common histological subtype in never-smokers. With the progress of anti-smoking campaign, the incidence of LUAD is rapidly climbing. Despite advances in cancer treatment, the prognosis of NSCLC has been disappointing over the past decades: the 5-year survival rate for patients with metastatic LUAD was less than 5% (27). Immunotherapy has changed the landscape for treating advanced NSCLC. The application of ICBs such as antibodies against PD-1 or PD-L1 have pointed a new direction for LUAD care. Nevertheless, the response to ICBs varies among LUAD patients. Novel biomarkers to predict the outcomes of immunotherapy is urgently needed. Cell cycle dysfunction is tightly associated with tumorigenesis in lung cancer. Loss of RB protein is found in approximately 15%-20% of all NSCLC (28). The dysregulation of cell cycle drives cancer cells into uncontrolled proliferation. The rapid division and genome instability of tumor cells result in mutation of DNA mutation. TMB is reported to be a promising immunotherapy biomarker to predict survival across multiple cancer types. In addition, cell cycle-targeting drugs CDK4/6 inhibitor may enhance the expression of PD-L1 on tumor cells, indicating the significance of combining ICBs. However, the interactions between cell cycle and immune regulators are still ill-defined.
In this study, we analyzed the differentially expressed CCRGs and IRGs in LUAD of GEO and TCGA cohorts. We identified 28 upregulated and 33 downregulated differentially expressed CCRGs and IRGs. GO and KEGG functional enrichment analyses revealed potential mechanism of them in LUAD. MAPK signaling pathway is mostly involved in these DEGs. Stutvoet et al. reported that MAPK pathway plays a significant role in PD-L1 expression of LUAD and may become a target to improve the outcomes of immunotherapy (29). We subsequently did a PPI network analysis among the 61 differentially expressed CCRGs and IRGs and 10 hub genes stand out (EGF, EGFR, STAT3, IGF1, TNF, IL10, SRC, JUN, MAPK1, MAP2K1). EGF and EGFR are the most important mutations in NSCLC, especially LUAD. Target therapy against EGFR mutation have greatly improved LUAD survival (30). STAT3 can induce epithelial mesenchymal transition and participate in PD-1 signaling pathway (31).
We then generated a risk model based on 61 differentially expressed CCRGs and IRGs based on GEO cohorts, and TCGA cohorts were obtained to verify the survival predictive value of the risk model. The train set revealed eight key genes (ACVR1B, BIRC5, NR2E1, INSR, TGFA, BMP7, CD28, NUDT6). ACVR1B, also known as ALK4, encodes an activin receptor related in TGF-β superfamily or structural related signaling proteins. Mutations in this gene are associated in with progressive pancreatic cancer in mutant KRAS-induced patients (32). Similarly, a SNP variant was found in never-smoking lung cancer patients, indicating the potential driving role of ACVR1B in NSCLC (33). BIRC5, also known as survivin, is a member of the inhibitor of apoptosis gene family that prevents apoptosis and has functions in both cell survival and mitosis. BIRC5 is normally absent in mature cells but is distinctly overexpressed in tumor cells. A small-molecule inhibitor of BIRC5, YM155, was shown to have favorable safety/tolerability in NSCLC patients (34). In addition, YM-155 had radio-sensitizing effect in NSCLC cell lines (35). NR2E1 and INSR are both proliferation regulators involved in aggressive behaviors (36, 37). TGFA is a ligand for EGFR. Dopeso reported that EMT-induced upregulation of TGFA can stimulate EGFR, activate SMAD pathway and induce EMT, which forces a positive feedback loop to enhance EMT and metastasis of lung cancer (38). BMP7 encodes a ligand of TGF-beta superfamily and activate SMAD signaling pathway. CD28 is a famous protein required for T cell proliferation and mature, cytokine secretion and Th2 cell differentiation. Kamphorst showed that CD28 pathway is significant for effective PD-1 therapy. They reported that CD28 is required for the proliferation of CD8+ T cells and the increased T cells after PD-1 blockade therapy are mainly CD28 positive (39). NUDT6 is an antisense gene of fibroblast growth factor 2 (FGF2). It is reported that NUDT6 is abundantly expressed in esophageal adenocarcinoma and is associated with poor disease-free survival (40). Taken together, the eight key genes are mainly involved in TGF-β pathway. The TME is characteristically enriched by TGF-β that are secreted by cancer cells, fibroblasts, macrophages and platelets. TGF-β can inhibit the differentiation of Th1 cells and cytotoxic CD8+ T cells and block T cell proliferation. Thus, it functions as a critical suppressor in the immune system and promotes immune evasion during cancer development. However, TGF-β has a contrary role as a tumor suppressor by inducing cell cycle arrest in early cancer cells (41, 42). The complexed role of TGF-β makes it significant to clarify its function in the future.
We next did a Kaplan-Meier analysis and it indicated that the survival of patients in low-risk group is remarkably longer than that in high-risk group. Additionally, the ROC curve and AUC verified accuracy of the risk model in survival prediction. We next constructed subgroup analysis based on the eight-gene signature. The high-risk group was also related to disappointing prognosis of LUAD patients with different age, gender, T stage and N stage. In addition, the model was proved to be an independent factor for OS. However, due to the small number of enrolled patients among T3-4 stage subgroup in the dataset (n=36), we did not find any significant difference, but the signature still worked for T1-2 subgroup LUAD patients. We further established a nomogram to determine a score for predicting survival of LUAD. The calibration curve implied the signature based on the risk model achieved a promising fit and greater effectiveness in clinical applications. In addition, we obtained TIMER database to study the correlation between the signature and infiltrating immune cells. We found that the infiltrating immune cells in the tumor microenvironment of LUAD is negatively associated with the risk score. To further explore the efficacy of our signature in immunotherapy, we performed a tumor mutation analysis. We found that LUAD patients in the high-risk group exhibited significantly higher TMB and more somatic mutation count. Since the infiltrating immune cells are main effectors in cancer immunotherapy and TMB is currently considered as an effective outcome index for immunotherapy, the signature in the current research might be useful in predicting the outcomes of LUAD patients receiving ICB medication.
Although the signature seemed to be stable and practical in our study, there are some limitations. The samples in our study were collected from TCGA databases and were internally verified. Further external verifications are needed to evaluate the accuracy in other databases. And the mechanisms and interactions of the eight key genes for constructing the risk models are needed to be elucidated by future experiments.
To conclude, we constructed a signature based on eight differentially expressed CCRGs and IRGs to predict the survival of LUAD. The risk models in our study show better clinical practicability for predicting the outcomes of LUAD patients. This signature might bring about changes in personalized LUAD treatment.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
FC, JS, HC, SZ, and XS contributed to the design of the study. JS and ZY extracted the data profile. FC, JS, and BX performed the statistical analysis. FC, JS, and XS contributed to the writing and revision of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (82073344).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Jiang ZB, Huang JM, Xie YJ, Zhang YZ, Chang C, Lai HL, et al. Evodiamine Suppresses Non-Small Cell Lung Cancer by Elevating CD8(+) T Cells and Downregulating the MUC1-C/PD-L1 Axis. J Exp Clin Cancer Res (2020) 39(1):249. doi: 10.1186/s13046-020-01741-5
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
3. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-Small Cell Lung Cancer: Epidemiology, Risk Factors, Treatment, and Survivorship. Mayo Clin Proc (2008) 83(5):584–94. doi: 10.4065/83.5.584
4. Wei C, Dong X, Lu H, Tong F, Chen L, Zhang R, et al. LPCAT1 Promotes Brain Metastasis of Lung Adenocarcinoma by Up-Regulating PI3K/AKT/MYC Pathway. J Exp Clin Cancer Res (2019) 38(1):95. doi: 10.1186/s13046-019-1092-4
5. Hao CC, Xu CY, Zhao XY, Luo JN, Wang G, Zhao LH, et al. Up-Regulation of VANGL1 by IGF2BPs and miR-29b-3p Attenuates the Detrimental Effect of Irradiation on Lung Adenocarcinoma. J Exp Clin Cancer Res (2020) 39(1):256. doi: 10.1186/s13046-020-01772-y
6. Wang X, Guo G, Guan H, Yu Y, Lu J, Yu J. Challenges and Potential of PD-1/PD-L1 Checkpoint Blockade Immunotherapy for Glioblastoma. J Exp Clin Cancer Res (2019) 38(1):87. doi: 10.1186/s13046-019-1085-3
7. Rolfo C, Caglevic C, Santarpia M, Araujo A, Giovannetti E, Gallardo CD, et al. Immunotherapy in NSCLC: A Promising and Revolutionary Weapon. Adv Exp Med Biol (2017) 995:97–125. doi: 10.1007/978-3-319-53156-4_5
8. Gray JE, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, et al. Three-Year Overall Survival With Durvalumab After Chemoradiotherapy in Stage III NSCLC-Update From PACIFIC. J Thorac Oncol (2020) 15(2):288–93. doi: 10.1016/j.jtho.2019.10.002
9. Proto C, Ferrara R, Signorelli D, Lo Russo G, Galli G, Imbimbo M, et al. Choosing Wisely First Line Immunotherapy in Non-Small Cell Lung Cancer (NSCLC): What to Add and What to Leave Out. Cancer Treat Rev (2019) 75:39–51. doi: 10.1016/j.ctrv.2019.03.004
10. Ingham M, Schwartz GK. Cell-Cycle Therapeutics Come of Age. J Clin Oncol (2017) 35(25):2949–59. doi: 10.1200/JCO.2016.69.0032
11. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
12. Negrini S, Gorgoulis VG, Halazonetis TD. Genomic Instability–An Evolving Hallmark of Cancer. Nat Rev Mol Cell Biol (2010) 11(3):220–8. doi: 10.1038/nrm2858
13. Teo MY, Seier K, Ostrovnaya I, Regazzi AM, Kania BE, Moran MM, et al. Alterations in DNA Damage Response and Repair Genes as Potential Marker of Clinical Benefit From Pd-1/Pd-L1 Blockade in Advanced Urothelial Cancers. J Clin Oncol (2018) 36(17):1685–94. doi: 10.1200/JCO.2017.75.7740
14. Chen Y, Liu Q, Chen Z, Wang Y, Yang W, Hu Y, et al. Pd-L1 Expression and Tumor Mutational Burden Status for Prediction of Response to Chemotherapy and Targeted Therapy in Non-Small Cell Lung Cancer. J Exp Clin Cancer Res (2019) 38(1):193. doi: 10.1186/s13046-019-1192-1
15. Schaer DA, Beckmann RP, Dempsey JA, Huber L, Forest A, Amaladas N, et al. The CDK4/6 Inhibitor Abemaciclib Induces a T Cell Inflamed Tumor Microenvironment and Enhances the Efficacy of PD-L1 Checkpoint Blockade. Cell Rep (2018) 22(11):2978–94. doi: 10.1016/j.celrep.2018.02.053
16. Goel S, DeCristo MJ, McAllister SS, Zhao JJ. Cdk4/6 Inhibition in Cancer: Beyond Cell Cycle Arrest. Trends Cell Biol (2018) 28(11):911–25. doi: 10.1016/j.tcb.2018.07.002
17. Lu Y, Ussery GD, Muncaster MM, Gallie BL, Blanck G. Evidence for Retinoblastoma Protein (RB) Dependent and Independent IFN-Gamma Responses: RB Coordinately Rescues IFN-Gamma Induction of MHC Class II Gene Transcription in Noninducible Breast Carcinoma Cells. Oncogene (1994) 9(4):1015–9.
18. Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, et al. CDK4/6 Inhibition Triggers Anti-Tumour Immunity. Nature (2017) 548(7668):471–5. doi: 10.1038/nature23465
19. Bhattacharya S, Dunn P, Thomas CG, Smith B, Schaefer H, Chen J, et al. ImmPort, Toward Repurposing of Open Access Immunological Assay Data for Translational and Clinical Research. Sci Data (2018) 5:180015. doi: 10.1038/sdata.2018.15
20. The Gene Ontology C. Expansion of the Gene Ontology Knowledgebase and Resources. Nucleic Acids Res (2017) 45(D1):D331–8. doi: 10.1093/nar/gkw1108
21. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res (2017) 45(D1):D353–61. doi: 10.1093/nar/gkw1092
22. Wang N, Wang Y, Yang Y, Shen Y, Li A. Mirna Target Prediction Based on Gene Ontology. In: 2013 Sixth International Symposium on Computational Intelligence and Design. IEEE (2013). p. 430–3.
23. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING Database in 2017: Quality-Controlled Protein-Protein Association Networks, Made Broadly Accessible. Nucleic Acids Res (2017) 45(D1):D362–8. doi: 10.1093/nar/gkw937
24. Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J Proteome Res (2019) 18(2):623–32. doi: 10.1021/acs.jproteome.8b00702
25. Hui D. Prognostication of Survival in Patients With Advanced Cancer: Predicting the Unpredictable? Cancer Control (2015) 22(4):489–97. doi: 10.1177/107327481502200415
26. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for Analysis of Tumor-Infiltrating Immune Cells. Nucleic Acids Res (2020) 48(W1):W509–14. doi: 10.1093/nar/gkaa407
27. Arbour KC, Riely GJ. Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review. JAMA (2019) 322(8):764–74. doi: 10.1001/jama.2019.11058
28. Wistuba II, Gazdar AF, Minna JD. Molecular Genetics of Small Cell Lung Carcinoma. Semin Oncol (2001) 28(2 Suppl 4):3–13. doi: 10.1053/sonc.2001.25738
29. Stutvoet TS, Kol A, de Vries EG, de Bruyn M, Fehrmann RS, Terwisscha van Scheltinga AG, et al. MAPK Pathway Activity Plays a Key Role in PD-L1 Expression of Lung Adenocarcinoma Cells. J Pathol (2019) 249(1):52–64. doi: 10.1002/path.5280
30. da Cunha Santos G, Shepherd FA, Tsao MS. EGFR Mutations and Lung Cancer. Annu Rev Pathol (2011) 6:49–69. doi: 10.1146/annurev-pathol-011110-130206
31. Zhang X, Sai B, Wang F, Wang L, Wang Y, Zheng L, et al. Hypoxic BMSC-Derived Exosomal miRNAs Promote Metastasis of Lung Cancer Cells Via STAT3-Induced Emt. Mol Cancer (2019) 18(1):40. doi: 10.1186/s12943-019-0959-5
32. Qiu W, Tang SM, Lee S, Turk AT, Sireci AN, Qiu A, et al. Loss of Activin Receptor Type 1b Accelerates Development of Intraductal Papillary Mucinous Neoplasms in Mice With Activated Kras. Gastroenterology (2016) 150(1):218–28.e212. doi: 10.1053/j.gastro.2015.09.013
33. Spitz MR, Gorlov IP, Amos CI, Dong Q, Chen W, Etzel CJ, et al. Variants in Inflammation Genes Are Implicated in Risk of Lung Cancer in Never Smokers Exposed to Second-Hand Smoke. Cancer Discovery (2011) 1(5):420–9. doi: 10.1158/2159-8290.CD-11-0080
34. Giaccone G, Zatloukal P, Roubec J, Floor K, Musil J, Kuta M, et al. Multicenter Phase II Trial of YM155, A Small-Molecule Suppressor of Survivin, in Patients With Advanced, Refractory, Non-Small-Cell Lung Cancer. J Clin Oncol (2009) 27(27):4481–6. doi: 10.1200/JCO.2008.21.1862
35. Iwasa T, Okamoto I, Suzuki M, Nakahara T, Yamanaka K, Hatashita E, et al. Radiosensitizing Effect of YM155, A Novel Small-Molecule Survivin Suppressant, in Non-Small Cell Lung Cancer Cell Lines. Clin Cancer Res (2008) 14(20):6496–504. doi: 10.1158/1078-0432.CCR-08-0468
36. Johansson E, Zhai Q, Zeng ZJ, Yoshida T, Funa K. Nuclear Receptor TLX Inhibits TGF-Beta Signaling in Glioblastoma. Exp Cell Res (2016) 343(2):118–25. doi: 10.1016/j.yexcr.2016.03.028
37. Heckl SM, Wiesener V, Behrens HM, Ulase D, Kruger S, Rocken C. The Expression of the Insulin Receptor in Gastric Cancer Correlates With the HER2 Status and May Have Putative Therapeutic Implications. Gastric Cancer (2019) 22(6):1130–42. doi: 10.1007/s10120-019-00964-6
38. Dopeso H, Jiao HK, Cuesta AM, Henze AT, Jurida L, Kracht M, et al. Phd3 Controls Lung Cancer Metastasis and Resistance to EGFR Inhibitors Through Tgfalpha. Cancer Res (2018) 78(7):1805–19. doi: 10.1158/0008-5472.CAN-17-1346
39. Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, et al. Rescue of Exhausted CD8 T Cells by PD-1-Targeted Therapies Is CD28-Dependent. Science (2017) 355(6332):1423–7. doi: 10.1126/science.aaf0683
40. Zhang SC, Barclay C, Alexander LA, Geldenhuys L, Porter GA, Casson AG, et al. Alternative Splicing of the FGF Antisense Gene: Differential Subcellular Localization in Human Tissues and Esophageal Adenocarcinoma. J Mol Med (Berl) (2007) 85(11):1215–28. doi: 10.1007/s00109-007-0219-9
41. Colak S, Ten Dijke P. Targeting TGF-Beta Signaling in Cancer. Trends Cancer (2017) 3(1):56–71. doi: 10.1016/j.trecan.2016.11.008
Keywords: lung adenocarcinoma, cell cycle, immune infiltration, prognostic signature, bioinformatics
Citation: Chen F, Song J, Ye Z, Xu B, Cheng H, Zhang S and Sun X (2021) Integrated Analysis of Cell Cycle–Related and Immunity-Related Biomarker Signatures to Improve the Prognosis Prediction of Lung Adenocarcinoma. Front. Oncol. 11:666826. doi: 10.3389/fonc.2021.666826
Received: 11 February 2021; Accepted: 05 May 2021;
Published: 04 June 2021.
Edited by:
Mingyan Zhu, Affiliated Hospital of Nantong University, ChinaCopyright © 2021 Chen, Song, Ye, Xu, Cheng, Zhang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinchen Sun, c3VueGluY2hlbkBuam11LmVkdS5jbg==; Shu Zhang, emhhbmdzaHVAbmptdS5lZHUuY24=; Hongyan Cheng, Y2hlbmdob25neWFuNjhAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.