
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 10 June 2021
Sec. Cancer Molecular Targets and Therapeutics
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.666597
 Chunxian Huang1,2†
Chunxian Huang1,2† Jinxiao Liang1,2†
Jinxiao Liang1,2† Shaodan Lin1,2
Shaodan Lin1,2 Dongyan Wang1,2
Dongyan Wang1,2 Qingsheng Xie1,2
Qingsheng Xie1,2 Zhongqiu Lin1,2*
Zhongqiu Lin1,2* Tingting Yao1,2*
Tingting Yao1,2*Objective: Cervical cancer is a frequently encountered gynecological malignancy as a major contributor to cancer-related deaths in women. This study focuses on how miR-193b promotes cervical cancer aggressiveness as well as the role of m6A in miR-193b silencing.
Methods: Cervical cancer samples and the matching adjacent normal cervical tissues were used to determine the significance of miR-193b in cervical cancer. The CCK-8 assay, cell cycle analysis, qRT-PCR, Western blot assay, IHC, RIP, and xenograft models were utilized to explore the impact of miR-193b in cervical cancer and how m6A regulates miR-193b expression. Luciferase reporter assays, qRT-PCR, and Western blotting were enlisted to study the interaction between miR-193b and CCND1.
Results: Our study suggested that lower miR-193b expressions were strongly linked to more advanced cervical cancer stages and the presence of deeper stromal invasion. miR-193b functions as a tumor suppressor that is regulated by m6A methylation in cervical tumors. METTL3 modulates miR-193b mature process in an m6A-dependent manner. Reintroduction of miR-193b profoundly inhibits tumorigenesis of cervical cancer cells both in vivo and in vitro through CCND1 targeting.
Conclusions: m6A associated downregulation of miR-193b promotes cervical cancer aggressiveness by targeting CCND1.
Cervical cancer is a frequently encountered gynecological malignancy as a primary cause of cancer-associated mortality in women (1). Nearly 90% of deaths occurred in developing countries, and more than half of these were recorded in Asia (2). Most patients respond to standard treatments which include surgery and radiotherapy. However, there remains a substantial risk of tumor relapse in patients who develop locally advanced cervical cancer with high risks such as positive lymph nodes or positive surgical margins. Among these groups of patients, relapse rates exceeding 50% after standard treatments have been documented. 5-year survival rates upon relapse remain dismal at only 30–60% (3, 4). Therefore, the identification of novel therapeutic methods is critical, and inevitably requires a deeper grasp of cervical cancer biology.
Gene expression has been noted to be strongly influenced by RNA modification, thereby leading to the development of the new field of RNA epigenetics (5). One ubiquitously occurring epigenetic modification is mediated by N6-methyladenosine (m6A), which methylates the adenosine N6-position in mammalian mRNA (6). m6A is necessary for a myriad of physiological processes, such as cell status regulation, sex determination, and development (7–9). Accumulating evidence indicates that m6A regulates RNA metabolism, including mRNA translation, degradation, splicing, export, and folding. Recent studies have uncovered an important post-transcriptional effect of m6A, which is the triggering of microRNA (miRNA) biogenesis (10). Alteration of m6A level was found across several cancers such as glioblastoma (9), lung cancer (11), gastric cancer (12), and hepatocellular carcinoma (13). Dysregulated m6A modification affects cancer pathogenesis and development by modulating the expression of tumor-related genes such as EGFR, BRD4, SOCS2, and MYC (14).
miRNAs are small non-coding RNAs (18-24 nucleotides) that usually negatively modulate genetic expression at the post-transcriptional stage by forming bonds to their target mRNA at the 3’-untranslated region (UTR) (15). Numerous physiological as well as pathophysiological processes, including carcinogenesis, are influenced by miRNA activity (16, 17). In this study, we found that miR-193b acts as a tumor suppressor in cervical cancer. A combination of bioinformatics algorithms, functional analyses, and xenograft models revealed the pivotal roles of METTL3/miR-193b/CCND1 signaling in cervical cancer aggressiveness.
41 pairs of cervical cancer tissues and their surrounding control tissue specimens were obtained from Sun Yat-Sen Memorial Hospital (Guangzhou, China) after surgical resection. Clinicopathological characteristics included FIGO stage, tumor grade, histologic subtype, invasion depth, lymph node metastasis, lymphatic vascular space invasion, and age were collected. Approval from the Research Ethics Committee of Sun Yat-Sen Memorial Hospital was obtained before the study. The research was performed following concepts established by the Declaration of Helsinki.
Siha and Hela cell lines (human cervical cancer cell lines) were procured from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and tested by STR Authentication. Cells were cultured in DMEM (Gibco, USA) supplemented with 10% fetal bovine serum (Biological Industries, Israel). Cultures were maintained in an incubator at 37°C with a humidified 5% CO2 environment.
The Trizol reagent (Takara, Japan) was utilized for RNA extraction followed by the use of the PrimeScript RT reagent kit (Takara, Japan) for synthesizing cDNA. Experiments were in strict compliance with protocols published by the manufacturer. miRNA and mRNA relative expression levels were confirmed using qRT-PCR as described before (18). U6 and GAPDH were used for normalization.
The sequences of miR-193b mimic (miR-193b), miRNA negative control, inhibitor targeting miR-193b (anti-miR-193b), corresponding negative control, siRNA METTL3, and siRNA negative control were listed in Supplementary Material. LipofectamineTM2000 (Invitrogen, USA) was used in cell transfection following instructions stipulated by the manufacturer.
The Cell Counting Kit 8 (CCK8) (Dojindo, Japan) was used to assess cell proliferation. Cells were incubated with CCK8 at a final concentration of 100 μl/ml for 1.5 hours at 37°C. Absorbance was recorded at 450 nm, with the absorbance at each time point used to construct cell proliferation curves.
Cells were fixed in 70% cold ethyl alcohol for 30 min at 4°C. 100 μg/ml RNase and 0.1 mg/ml propidium iodide were subsequently added. The distribution of cell cycle phases was determined by the Cell Quest software (BD Biosciences, USA).
Female nude mice of ages 4 weeks were purchased from the Sun Yat-Sen University Laboratory Animal Center (Guangzhou, China) and treated according to the National Institutes of Health guide for the care and use of laboratory animals. All animal experimental protocols were approved by the Animal Research Committee of Sun Yat-Sen University. Mice were divided into two groups (n = 4/group) randomly. 3×106 cells suspended in 200 μl PBS were administered via subcutaneous injection over the right flank region of nude mice. After the development of palpable tumors (average volume, 50 mm3), intratumoral injection of synthetic miR-193b, or negative control complexed with siPORT Amine transfection reagent (Ambion, USA) was given 6 times at a 4-day interval. Animals were sacrificed and tumors were harvested 24 days after the first injection. Tumor volumes derived as follows: V = (L × W2)/2 (L: length, W: width).
5 μm cervical cancer paraffin-embedded tissue sections were subjected to xylene paraffin removal followed by ethanol rehydration. Freshly prepared 3% hydrogen peroxide was used to inhibit the activity of endogenous peroxidases. Sections were then incubated overnight with anti-ki-67 (Cell Signaling Technology, USA) at 4°C, rinsed thrice with PBS, and incubated with anti-rabbit & mouse IgG (DAKO, Denmark) at room temperature for 1 hour. Diaminobenzidine (DAKO, Denmark) was used to visualize the bound antibody.
Protein was harvested using lysis buffer (CWBIO, China) containing a protease inhibitor cocktail (Roche, Switzerland). 25 ug protein were separated by 10% SDS-PAGE electrophoresis and immunoblotted onto a PVDF membrane (Bio-Rad, USA). Specific primary antibodies were then incubated with the samples at 4°C for 12-16 hours after blocking endogenous reactions. Following a one-hour incubation with secondary antibodies at room temperature the next day, bound proteins were imaged with an enhanced chemiluminescence detection kit (Millipore, USA). Primary antibody for METTL3, CyclinD1, internal reference antibody for β-actin, HRP Goat-anti-Rabbit, and HRP Goat-anti-Mouse were obtained from Cell Signaling Technology (USA).
Mutant (MUT) and wild-type (WT) CCND1 3’-UTR which contained predicted miR-193b target sites were amplified by the PCR method. Primers for reporter vector construction were given in Supplementary Material. WT or MUT reporter plasmid and miR-193b or miRNA negative control were transfected into HEK-293T cells. Firefly luciferase activities were assessed 24 hours post-transfection with the Dual Luciferase Assay (Promega, USA). Renilla luciferase was used to normalize results.
After extracting total RNA from each group using the Trizol reagent (Takara, Japan), the m6A content was measured by using a commercial kit (Abcam, USA). Briefly, capture antibody solution and detection antibody solution were then added in assay dishes, where 200 ng RNAs were plated on. Absorbance was recorded at 450 nm. The m6A levels were quantified by the absorbance of each sample.
RIP experiments were carried out using the Magna RIP Kit (Millipore, USA) according to the instructions stipulated by the manufacturer. Briefly, cells were isolated and treated with deoxyribonuclease I, and then fragmented by sonication. IPs were performed with an anti-m6A antibody. RNAs were extracted and subjected to qRT-PCR using primer of pri-miR-193b and finally normalized to input.
The SPSS 20.0 software was enlisted to carry out all statistical analyses. Both the two-tailed Chi-square test and Student’s t-test were used. Statistical significance was determined by the finding of p < 0.05. All experiments were repeated independently in triplicate.
qRT-PCR was used to determine miR-193b levels in clinical cervical cancer specimens and the matching surrounding normal cervical tissues (N = 41). 26 of these tumor samples (63%) contained low miR-193b levels. These findings indicate that miR-193b suppression may be a key feature leading to the progression of human cervical cancer. Notably, lower miR-193b levels were linked to a more advanced FIGO stage (p = 0.031) and deeper stromal invasion (p = 0.006) (Table 1). However, there was no significant correlation between the low level of miR-193b and tumor grade, histologic subtype, lymphatic vascular space invasion, or lymph node metastasis (Table 1). These results show that miR-193b silencing played a crucial part during cervical cancer development.
We investigated miR-193b function by introducing miRNA or anti-miRNA into Hela and Siha cells. The efficiency of transfection was verified by qRT-PCR (Supplementary Figure 1). Reintroduction of miR-193b could markedly inhibit cell proliferation in cervical cancer (Figure 1A), while the addition of a miR-193b inhibitor (anti-miRNA) nullified this phenomenon (Figure 1A). Cell cycle assays showed that miR-193b mimics induced increases in the G2 population (Figures 1B, C), implying G2 arrest. As expected, miR-193b inhibitor significantly promoted cell cycle progression from G0–G1 into S and G2-M phase (Figures 1B, C). These results further strengthen our hypothesis of miR-193b as a tumor-suppressing molecule in cervical cancer.
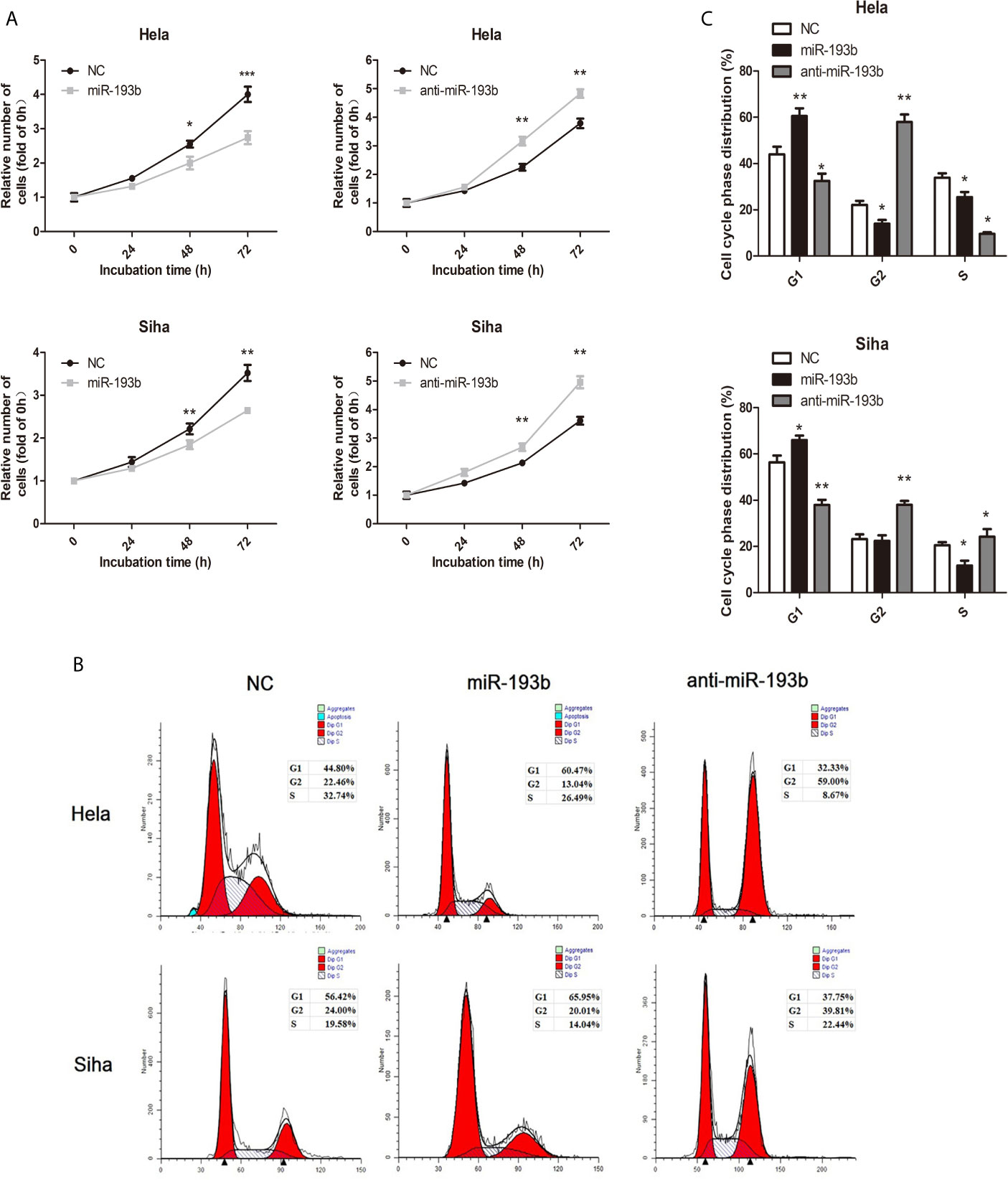
Figure 1 miR-193b is a tumor suppressor in cervical cancer. Either miR-193b or anti-miR-193b was used to transfect Hela and Siha cells as described. (A) CCK-8 assays were carried out to determine cellular proliferation. (B) Flow cytometry was used to assess distributions of cells across the cell cycle. (C) The qualifications of the cell cycle were presented. *p < 0.05, **p < 0.01, ***p < 0.001.
To determine the in vivo antitumor miR-193b impact, we first induced the formation of Siha-bearing tumors in mice. These xenograft tumors were then injected directly with the relevant miRNAs. miR-193b levels increased by 5.7 fold after this treatment (Figure 2B). After 3 weeks, miR-193b treated tumors grew more slowly than those receiving negative control miRNA injections (Figures 2A, C). We also noted a significant difference in tumor weights at the end of the treatment protocol (Figure 2D). Ki-67 staining was significantly reduced after miR-193b treatment (Figure 2E). These results suggest that miR-193b suppresses cervical cancer growth in the xenograft models.
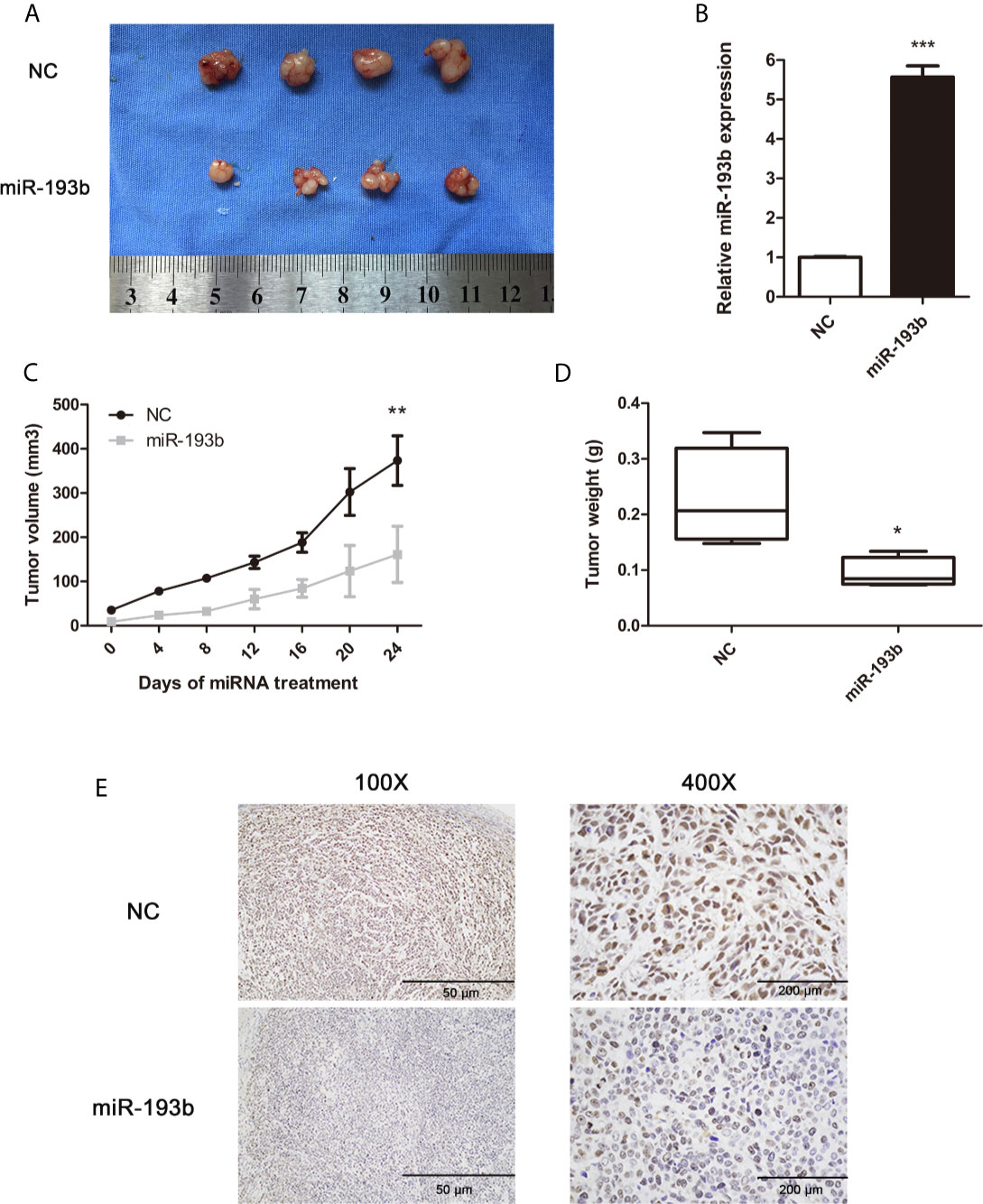
Figure 2 miR-193b suppresses cervical cancer growth in vivo. Subcutaneous Siha tumors located on mice xenograft models were directly injected with miR-193b for a total of 6 times at 4-day intervals. Tumors were harvested on the 24th day from the injection. (A) miR-193b inhibited cervical cancer growth in vivo. (B) miR-193b expression levels of each group after intratumoral miR-193b injections were measured by qRT-PCR. (C) Effect of miR-193b on tumor growth and tumor volumes. (D) The tumors were weighed after harvested. (E) The protein levels of Ki-67 were detected using IHC. *p < 0.05, **p < 0.01, ***p< 0.001.
Further experiments were carried out to discern the effect of miR-193b silencing on its initially unknown target gene. miRNA target prediction algorithms TargetScan, miRDB, and miRTarBase were used to identify the target genes of miR-193b. Venn diagrams showed 64 significantly target genes that appeared simultaneously in three prediction algorithms (Figure 3A). Gene ontology (GO) analysis was performed on 64 targeted genes and the enriched pathways were mainly associated with responses to apoptosis, protein complex binding, prolactin signaling pathway, PI3K-Akt signaling pathway, mutagenesis site, kinase activity, and cytoplasm (Figure 3B). CyclinD1 (CCND1) was a vital gene among the 64 predicted target genes given previous studies alluding to its importance in cervical cancer development (19, 20). As one of the cell cycle regulators, CCND1 is upregulated in cervical cancer and has a crucial role in enhancing tumorigenesis (21–24). CCND1 was highly expressed in low miR-193b level patients (Table 2). CCND1 expression was inhibited in Hela and Siha cells which overexpressed miR-193b, however, the converse was seen in cells with suppressed miR-193b levels (Figure 3C). A similar result was noted in CyclinD1 protein expression (Figures 3D, E), confirming that miR-193b regulated CCND1 mRNA expression in cervical cancer cells. We further sought to determine the direct miRNA binding site on CCND1 3′-UTR through luciferase reporter assays using WT and MUT miR-193b seed matched human CCND1 3′-UTR constructs. Co-transfection of miR-193b and the WT construct diminished luciferase activity. This effect was not seen with the use of the MUT construct, demonstrating that CCND1 was a direct target of miR-193b (Figure 3F). All the findings above suggest that downregulating miR-193b resulted in CCND1 modulation.
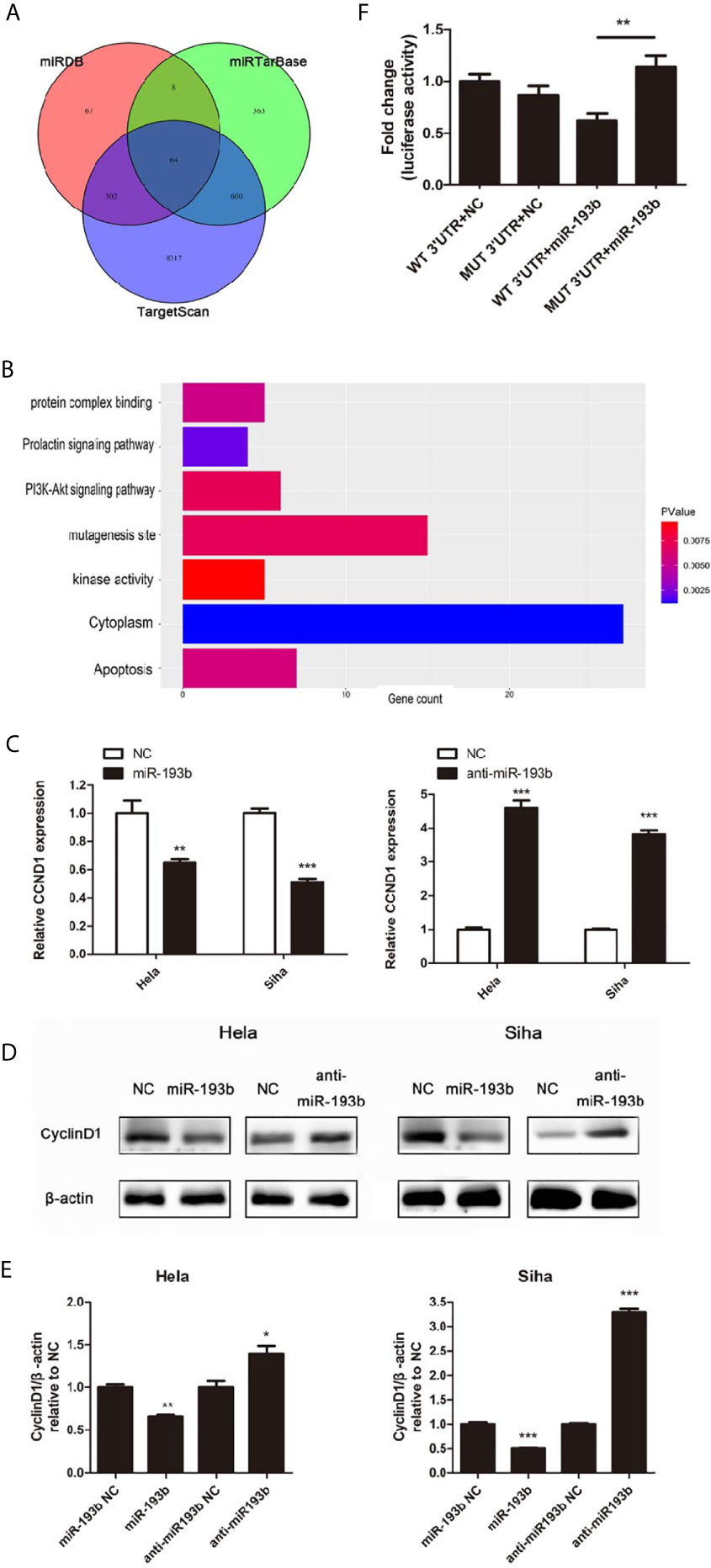
Figure 3 miR-193b regulates the expression of CCND1. Either miR-193b or anti-miR-193b was used to transfect Hela and Siha cells as described. (A) Venn diagrams showed 64 significantly target genes that appeared simultaneously in miRNA target prediction algorithms TargetScan, miRDB, and miRTarBase. (B) GO analysis was performed on 64 targeted genes. (C) CCND1 expressions were measured by qRT-PCR. (D) CyclinD1 protein expression was detected using Western blot. (E) Band density of Western blot was examined using ImageJ, and protein levels were normalized to β-actin. (F) 3′-UTR luciferase reporter assay was utilized to determine the binding of miR-193b to the 3′-UTR of CCND1. Luciferase activities of WT or MUT CCND1 3′-UTR luciferase construct co-transfected with miR-193b or NC in HEK-293T cells were measured. *p < 0.05, **p < 0.01, ***p < 0.001.
Recent studies have shown that the m6A RNA modification regulated the progress of diverse cancers and thousands of transcripts, including miRNAs (10). The m6A methylation level in cervical cancer is significantly higher than the adjacent normal tissues (Figure 4A). Three important subunits of the m6A methyltransferase complex, methyltransferase-like 3 (METTL3), METTL14, and WT1 associated protein (WTAP), were found different expression in tumor tissues (Figure 4B). Among them, METTL3 showed the most different mRNA levels between the normal and cancer tissues (Figure 4B). Low miR-193b and high CCND1 were found in low METTL3 patients (Table 3). So we performed siRNA-mediated downregulation of METTL3 in Hela and Siha cells. The transfection efficiency was verified by qRT-PCR (Figure 4C) and Western blot (Figures 4D, E). Increased tumor proliferation (Figure 4F) and cell cycle progression (Figures 4G, H) occurred as a result of METTL3 inhibition.
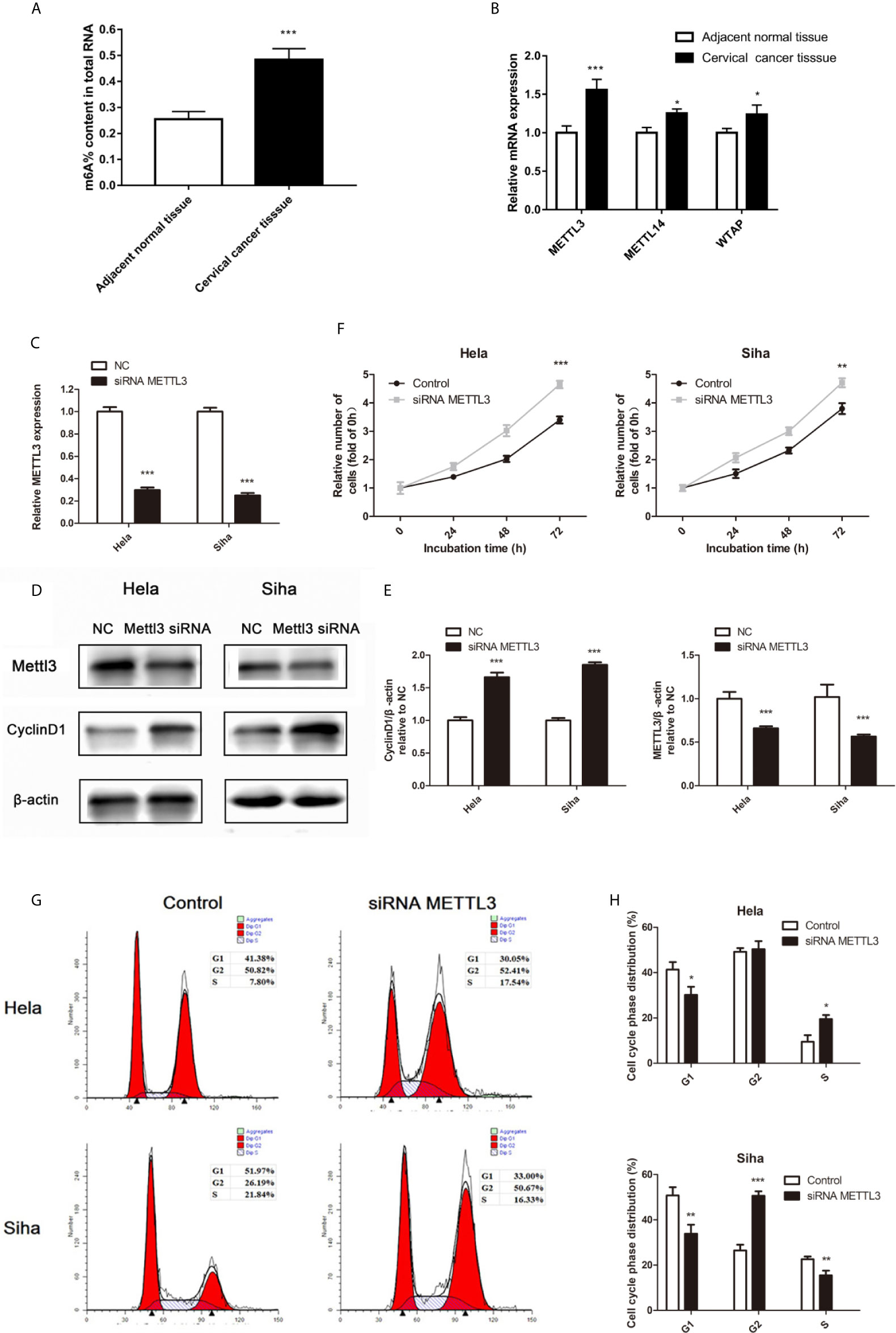
Figure 4 m6A methylation in cervical cancer. (A) The total m6A methylation level in cervical cancer tissues and adjacent normal tissues. (B) mRNA expression of METTL3, METTL14, and WTAP were measured by qRT-PCR. (C–E) Hela and Siha cells were transfected with siRNA METTL3 or negative control. METTL3 mRNA level (C), METTL3, and CCND1 protein level (D) were detected after transfected. (E) Band density of Western blot was quantified using ImageJ, and protein levels were normalized to β-actin. (F) Cellular proliferation was assessed using CCK-8 assays. (G) Flow cytometry was used to assess distributions of cells across the cell cycle. (H) The qualifications of the cell cycle were presented. *p < 0.05, **p < 0.01, ***p < 0.001.
Silencing METTL3 in cervical cancer cells inhibited miR-193b levels and increased the expression of pri-miR-193b (Figures 5A, B), suggesting that METTL3 modulated miR-193b mature process. RIP assays were used to further discover the role of METTL3 in cervical cancer. We found that METTL3 promoted pri-miR-193b m6A level (Figure 5C). METTL3 siRNA upregulated the expression of CCND1 on both protein (Figures 4D, E) and mRNA levels (Figure 5D). The results suggest that METTL3 modulates miR-193b mature process by promoted pri-miR-193b m6A methylation level.
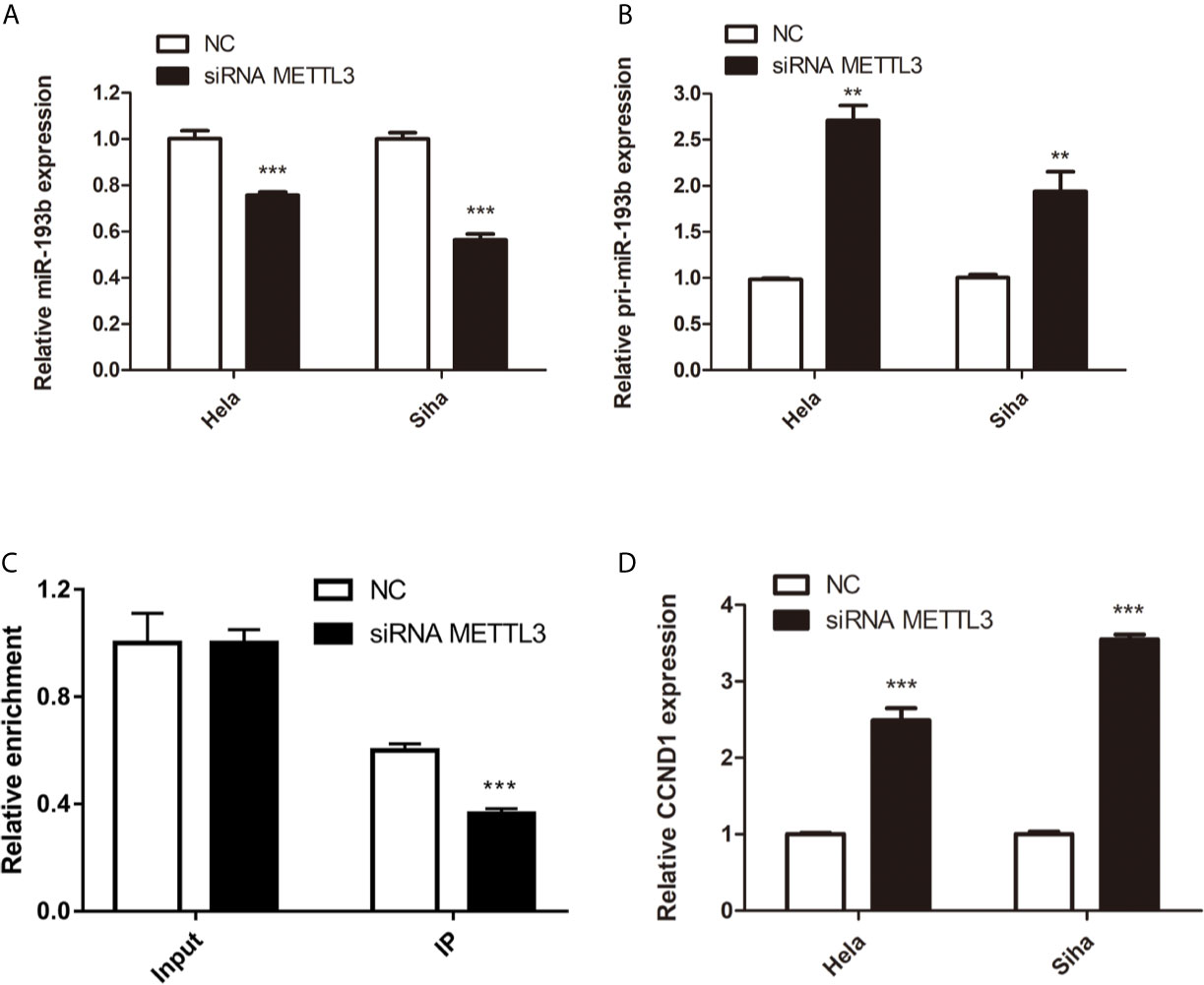
Figure 5 miR-193b is regulated by m6A methylation. Hela and Siha cells were transfected with siRNA METTL3 or negative control. Expression levels of miR-193b (A), pri-miR-193b (B), and CCND1 (D) were measured by qRT-PCR. (C) Immunoprecipitation of m6A modified RNA in control or METTL3 downregulated cells followed by qRT-PCR to assess the pri-miR-193b m6A modification levels. **p < 0.01, ***p < 0.001.
Tumor biology research has revealed miRNAs to be able to exert potent oncogenic or tumor-suppressing properties (25, 26). This investigation found that downregulated miR-193b was significantly associated with advanced FIGO stage and deep stromal invasion in cervical cancer. As expected, miR-193b served as a tumor suppressor in cervical cancer through induction of cell cycle G1 phase arrest and inhibition of cellular proliferation. miRNAs are predicted to regulate more than 60% of all human mRNAs and are involved in almost all biological processes in mammal systems (27). Functional characterization of miRNA target networks as well as identification of dysregulated miRNA expression has both highlighted the critical role of miRNAs in malignancy (28). Through binding to target sites in mRNA 3’-UTR regions, miRNAs can modulate protein-coding gene expressions (15, 29). Using the miRNA target prediction algorithm, we found CCND1 as one of the miR-193b target genes and verified it in the following experiments.
As a cell cycle regulator, CCND1 is associated with cyclin-dependent kinases 4 or 6 (CDK4/6). The CCND1/CDK4/6 complex phosphorylated the retinoblastoma protein to release the E2F transcription factor that is responsible for triggering G1-S phase cell transition (30, 31). Accumulating evidence demonstrated that cervical cancer specimens harboring raised CCND1 levels were linked to lower overall patient survival (21–24). Interestingly, genetic amplification has been mostly ruled out as a cause of overexpression of CCND1, suggesting the presence of a post-transcriptional process (32–34). Numbers of miRNAs involved in cell cycle control were identified in recent studies. In gastric cancer, miR-218 was noted to inhibit cell cycle progression via the CDK6/CyclinD1/E2F1 axis (35). Up-regulation of miR-15a and miR-16-1 induced leukemic cell death by resulting in the downregulation of the target gene CyclinD1 (36). In our study, we indicated that miR-193b not only led to a G1 arrest in Hela and Siha cells through CCND1 targeting but also acted to suppress malignant features of cervical cancer in xenograft models.
The N6-position of adenosine methylation is one of the main epigenetic aberrations in the inactivation of tumor suppressors (37). In our study, low miR-193b expression was found in cervical cancer. The miR-193b precursor is located on chromosome 16p13.12, where the loss of heterozygosity (LOH) is rare in cervical cancer (38, 39). Therefore, we hypothesized that the expression of miR-193b was epigenetically silenced by m6A. m6A mark triggers the formation of miRNAs in a post-transcriptional manner (10). Biogenesis of miRNAs begins when pri-miRNAs are processed by the microprocessor complex, comprising of type III RNase DROSHA and the RNA-binding protein DGCR8 (40, 41). DGCR8 is first required to detect stem junctions in flanking single-stranded RNAs of the pri-miRNA hairpin (42). Subsequently, DROSHA recruiting results in RNA duplex cleaving to yield pre-miRNA (42). During pri-miRNA processing, METTL3, an important component of the m6A methylase complex, methylates pri-miRNAs and labels them failure for DGCR8 identification and processing (10). The efficiency of m6A mark in regulating pri-miRNA processing has been confirmed in in vitro experiments (10). However, relatively little is known about how m6A modulates miRNA expression in cervical cancer. As expected, methylation of pri-miR-193b could be detected by using METTL3 siRNA to mediate the loss of miR-193b and the accumulation of pri-miR-193b. Correspondingly, up-regulated CCND1 and suppressed cancer aggressiveness were observed after METTL3 RNAi. The evidence supports the notion that m6A modification associated epigenetic silencing of miR-193b occurs during the development and progression of cervical cancer.
In conclusion, our results suggest that cervical cancer may be critically modified by tumor-suppressing miR-193b which is in turn regulated by m6A modification. Reintroduction of miR-193b markedly suppressed in vitro and in vivo tumorigenicity of cervical cancer cells through CCND1 targeting. These findings show that miR-193b may be used as a miRNA-based tumor therapy target in the future.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the Research Ethics Committee of Sun Yat-Sen Memorial Hospital. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Animal Research Committee of Sun Yat-Sen University.
CH, JL, ZL, and TY designed the experiments. CH and JL finished the first draft of the manuscript. ZL and TY completed the final version. CH focused on the CCK-8 assay, cell cycle analysis, qRT-PCR, Western blot assay, IHC, and Luciferase reporter assay. JL and SL collected tissue samples as well as clinic data, and completed statistical analysis. DW and QX performed most of the animal studies. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (81972433), and the Natural Science Foundation of Guangdong Province (Doctoral Research Start-up Fund Program) (2016A030310178).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.666597/full#supplementary-material
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global Cancer Statistics, 2012. CA Cancer J Clin (2015) 65:87–108. doi: 10.3322/caac.21262
2. Koh WJ, Abu-Rustum NR, Bean S, Bradley K, Campos SM, Cho KR, et al. Cervical Cancer, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw (2019) 17:64–84. doi: 10.6004/jnccn.2019.0001
3. Monk BJ, Cha DS, Walker JL, Burger RA, Ramsinghani NS, Manetta A, et al. Extent of Disease as an Indication for Pelvic Radiation Following Radical Hysterectomy and Bilateral Pelvic Lymph Node Dissection in the Treatment of Stage IB and IIA Cervical Carcinoma. Gynecol Oncol (1994) 54:4–9. doi: 10.1006/gyno.1994.1157
4. Cheng X, Cai S, Li Z, Tang M, Xue M, Zang R. The Prognosis of Women With Stage IB1-IIB Node-Positive Cervical Carcinoma After Radical Surgery. World J Surg Oncol (2004) 2:47. doi: 10.1007/BF02739674
5. He C. Grand Challenge Commentary: RNA Epigenetics? Nat Chem Biol (2010) 6:863–5. doi: 10.1038/nchembio.482
6. Meyer KD, Jaffrey SR. The Dynamic Epitranscriptome: N6-methyladenosine and Gene Expression Control. Nat Rev Mol Cell Biol (2014) 15:313–26. doi: 10.1038/nrm3785
7. Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, et al. M(6)a Modulates Neuronal Functions and Sex Determination in Drosophila. Nature (2016) 540:242–7. doi: 10.1038/nature20568
8. Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, et al. Temporal Control of Mammalian Cortical Neurogenesis by M(6)a Methylation. Cell (2017) 171:877–89. doi: 10.1016/j.cell.2017.09.003
9. Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. M(6)a RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep (2017) 18:2622–34. doi: 10.1016/j.celrep.2017.02.059
10. Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-Methyladenosine Marks Primary microRNAs for Processing. Nature (2015) 519:482–5. doi: 10.1038/nature14281
11. Du Y, Hou G, Zhang H, Dou J, He J, Guo Y, et al. Sumoylation of the M6a-RNA Methyltransferase METTL3 Modulates its Function. Nucleic Acids Res (2018) 46:5195–208. doi: 10.1093/nar/gky156
12. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-Mediated M(6)a Modification of HDGF mRNA Promotes Gastric Cancer Progression and has Prognostic Significance. GUT (2020) 69:1193–205. doi: 10.1136/gutjnl-2019-319639
13. Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, et al. Microrna-145 Modulates N(6)-Methyladenosine Levels by Targeting the 3’-Untranslated Mrna Region of the N(6)-Methyladenosine Binding YTH Domain Family 2 Protein. J Biol Chem (2017) 292:3614–23. doi: 10.1074/jbc.M116.749689
14. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its Role in Cancer. Mol Cancer (2019) 18:176. doi: 10.1186/s12943-019-1109-9
15. Rigoutsos I. New Tricks for Animal microRNAS: Targeting of Amino Acid Coding Regions at Conserved and Nonconserved Sites. Cancer Res (2009) 69:3245–8. doi: 10.1158/0008-5472.CAN-09-0352
16. O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and Pathological Roles for microRNAs in the Immune System. Nat Rev Immunol (2010) 10:111–22. doi: 10.1038/nri2708
17. Tornesello ML, Faraonio R, Buonaguro L, Annunziata C, Starita N, Cerasuolo A, et al. The Role of Micrornas, Long non-Coding RNAs, and Circular Rnas in Cervical Cancer. Front Oncol (2020) 10:150. doi: 10.3389/fonc.2020.00150
18. Chen K, Liu MX, Mak CS, Yung MM, Leung TH, Xu D, et al. Methylation-Associated Silencing of miR-193a-3p Promotes Ovarian Cancer Aggressiveness by Targeting GRB7 and MAPK/ERK Pathways. Theranostics (2018) 8:423–36. doi: 10.7150/thno.22377
19. Nichols GE, Williams ME, Gaffey MJ, Stoler MH. Cyclin D1 Gene Expression in Human Cervical Neoplasia. Mod Pathol (1996) 9:418–25.
20. Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a Therapeutic Target in Cancer. Nat Rev Cancer (2011) 11:558–72. doi: 10.1038/nrc3090
21. Cheung TH, Yu MM, Lo KW, Yim SF, Chung TK, Wong YF. Alteration of Cyclin D1 and CDK4 Gene in Carcinoma of Uterine Cervix. Cancer Lett (2001) 166:199–206. doi: 10.1016/S0304-3835(01)00457-8
22. Carreras R, Alameda F, Mancebo G, Garcia-Moreno P, Marinoso ML, Costa C, et al. A Study of Ki-67, c-erbB2 and Cyclin D-1 Expression in CIN-I, Cin-III and Squamous Cell Carcinoma of the Cervix. Histol Histopathol (2007) 22:587–92. doi: 10.14670/HH-22.587
23. Conesa-Zamora P, Domenech-Peris A, Orantes-Casado FJ, Ortiz-Reina S, Sahuquillo-Frias L, Acosta-Ortega J, et al. Effect of Human Papillomavirus on Cell Cycle-Related Proteins P16, Ki-67, Cyclin D1, p53, and Proex C in Precursor Lesions of Cervical Carcinoma: A Tissue Microarray Study. Am J Clin Pathol (2009) 132:378–90. doi: 10.1309/AJCPO0WY1VIFCYDC
24. Sommer G, Dittmann J, Kuehnert J, Reumann K, Schwartz PE, Will H, et al. The RNA-binding Protein La Contributes to Cell Proliferation and CCND1 Expression. Oncogene (2011) 30:434–44. doi: 10.1038/onc.2010.425
25. Rupaimoole R, Slack FJ. MicroRNA Therapeutics: Towards a New Era for the Management of Cancer and Other Diseases. Nat Rev Drug Discovery (2017) 16:203–22. doi: 10.1038/nrd.2016.246
26. Duica F, Condrat CE, Danila CA, Boboc AE, Radu MR, Xiao J, et al. Mirnas: A Powerful Tool in Deciphering Gynecological Malignancies. Front Oncol (2020) 10:591181. doi: 10.3389/fonc.2020.591181
27. Krol J, Loedige I, Filipowicz W. The Widespread Regulation of microRNA Biogenesis, Function and Decay. Nat Rev Genet (2010) 11:597–610. doi: 10.1038/nrg2843
28. Calin GA, Croce CM. MicroRNA Signatures in Human Cancers. Nat Rev Cancer (2006) 6:857–66. doi: 10.1038/nrc1997
29. Ambros V. The Evolution of Our Thinking About Micrornas. Nat Med (2008) 14:1036–40. doi: 10.1038/nm1008-1036
30. Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: Polymorphism, Aberrant Splicing and Cancer Risk. Oncogene (2006) 25:1620–8. doi: 10.1038/sj.onc.1209371
31. Musgrove EA. Cyclins: Roles in Mitogenic Signaling and Oncogenic Transformation. Growth Factors (2006) 24:13–9. doi: 10.1080/08977190500361812
32. Inge LJ, Rajasekaran SA, Wolle D, Barwe SP, Ryazantsev S, Ewing CM, et al. alpha-Catenin Overrides Src-dependent Activation of Beta-Catenin Oncogenic Signaling. Mol Cancer Ther (2008) 7:1386–97. doi: 10.1158/1535-7163.MCT-07-2029
33. Frost P, Shi Y, Hoang B, Gera J, Lichtenstein A. Regulation of D-cyclin Translation Inhibition in Myeloma Cells Treated With Mammalian Target of Rapamycin Inhibitors: Rationale for Combined Treatment With Extracellular Signal-Regulated Kinase Inhibitors and Rapamycin. Mol Cancer Ther (2009) 8:83–93. doi: 10.1158/1535-7163.MCT-08-0254
34. Cowling VH. Enhanced mRNA Cap Methylation Increases Cyclin D1 Expression and Promotes Cell Transformation. Oncogene (2010) 29:930–6. doi: 10.1038/onc.2009.368
35. Deng M, Zeng C, Lu X, He X, Zhang R, Qiu Q, et al. miR-218 Suppresses Gastric Cancer Cell Cycle Progression Through the CDK6/Cyclin D1/E2F1 Axis in a Feedback Loop. Cancer Lett (2017) 403:175–85. doi: 10.1016/j.canlet.2017.06.006
36. Abraham M, Klein S, Bulvik B, Wald H, Weiss ID, Olam D, et al. The CXCR4 Inhibitor BL-8040 Induces the Apoptosis of AML Blasts by Downregulating ERK, Bcl-2, MCL-1 and cyclin-D1 Via Altered miR-15a/16-1 Expression. Leukemia (2017) 31:2336–46. doi: 10.1038/leu.2017.82
37. Yu S, Li X, Liu S, Yang R, Liu X, Wu S. N(6)-Methyladenosine: A Novel RNA Imprint in Human Cancer. Front Oncol (2019) 9:1407. doi: 10.3389/fonc.2019.01407
38. Kloth JN, Oosting J, van Wezel T, Szuhai K, Knijnenburg J, Gorter A, et al. Combined Array-Comparative Genomic Hybridization and Single-Nucleotide Polymorphism-Loss of Heterozygosity Analysis Reveals Complex Genetic Alterations in Cervical Cancer. BMC Genomics (2007) 8:53. doi: 10.1186/1471-2164-8-53
39. Kersemaekers AM, Hermans J, Fleuren GJ, van de Vijver MJ. Loss of Heterozygosity for Defined Regions on Chromosomes 3, 11 and 17 in Carcinomas of the Uterine Cervix. Br J Cancer (1998) 77:192–200. doi: 10.1038/bjc.1998.33
40. Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA Biogenesis: Isolation and Characterization of the Microprocessor Complex. Methods Mol Biol (2006) 342:33–47. doi: 10.1385/1-59745-123-1:33
41. Huang JT, Wang J, Srivastava V, Sen S, Liu SM. Microrna Machinery Genes as Novel Biomarkers for Cancer. Front Oncol (2014) 4:113. doi: 10.3389/fonc.2014.00113
Keywords: m6A, METTL3, miR-193b, CCND1, cervical cancer
Citation: Huang C, Liang J, Lin S, Wang D, Xie Q, Lin Z and Yao T (2021) N6-Methyladenosine Associated Silencing of miR-193b Promotes Cervical Cancer Aggressiveness by Targeting CCND1. Front. Oncol. 11:666597. doi: 10.3389/fonc.2021.666597
Received: 10 February 2021; Accepted: 17 May 2021;
Published: 10 June 2021.
Edited by:
Edward Wenge Wang, City of Hope National Medical Center, United StatesReviewed by:
Dawn Sijin Nin, National University of Singapore, SingaporeCopyright © 2021 Huang, Liang, Lin, Wang, Xie, Lin and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tingting Yao, eWFvdHRpbmdAbWFpbC5zeXN1LmVkdS5jbg==; Zhongqiu Lin, bGluLXpob25ncWl1QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.