- 1Department of Medical Oncology, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China
- 2Department of Pulmonary Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China
Background: Several oncogenic drivers in non-small cell lung cancer (NSCLC) are considered actionable with available or promising targeted therapies. Although targetable drivers rarely overlap with each other, there were a minority of patients harboring co-occurring actionable oncogenic targets, whose clinical characteristics and prognosis are not yet clear.
Methods: A total of 3,077 patients with NSCLC who underwent molecular analysis by NGS were included, and their demographic and clinical data were retrospectively collected.
Results: Our study found that the frequency of NSCLC patients harboring co-occurring potentially actionable alterations was approximately 1.5% (46/3077); after excluding patients with EGFR-undetermined mutations, the incidence was 1.3% (40/3077); 80% (37/46) harbored both EGFR mutations and other potentially actionable drivers such as MET amplification (21.6%; 8/37) and alterations in ERBB2 including mutations (27%; 10/37) and amplification (21.6%; 8/37); other combinations of potentially actionable drivers including alterations in ERBB2, KRAS, MET, ALK, and RET were also identified. Additionally, de novo MET/ERBB2 amplification in patients harboring EGFR-mutant NSCLC treated with first-generation EGFR tyrosine kinase inhibitors (TKIs) was associated with shorter PFS (p < 0.05). The efficacy of TKIs in NSCLC patients harboring other co-occurring potentially actionable drivers varied across different molecular subtypes.
Conclusions: Approximately 1.5% of NSCLCs harbored co-occurring potentially actionable oncogenic drivers, commonly involving EGFR mutations. Co-occurring actionable targets may impact the efficacy of TKIs; therefore, future clinical trials in these patients should be anticipated to tailor the combination or sequential treatment strategies.
Introduction
The heterogeneity of non-small cell lung cancer (NSCLC) is mainly determined by different oncogenic drivers (1). Although dozens of oncogenic drivers are considered to be involved in the development of lung cancer, there are only several actionable targets with widely available targeted therapies, such as EGFR mutations, ALK rearrangements, ROS1 rearrangements, BRAF V600E mutation, NTRK rearrangements, and RET rearrangements (2–4). The targeted therapies for MET alterations (exon 14 splicing site mutations also known as skipping mutations or amplification), ERBB2 alterations (mutations or amplification), and KRAS G12C mutation also demonstrated promising efficacies in clinical trials, paving a way for precision medicine of NSCLC (4–8).
More and more targeted drugs were put into the first-line setting, greatly influencing the treatment strategies; however, even with the same type of actionable drivers, the efficacy of targeted therapies varies from patient to patient (9). Several studies have proved that both progression-free survival (PFS) and overall survival (OS) of EGFR mutant or ALK rearranged NSCLCs with TP53 mutations receiving EGFR or ALK TKIs, respectively, were significantly lower than those of patients without TP53 mutations (10–12). Later, increasing evidence has demonstrated that other concomitant alterations such as RB1 mutations or PIK3CA amplification also accelerated the resistance to EGFR TKIs (13, 14). In addition to these common co-existing mutations without available targeted drugs, co-occurring targetable oncogenic drivers can also be found in a small number of NSCLCs (15–18); however, there is still little evidence to make precision treatment plans for these patients, whose demographic and clinical characteristics remained largely unknown.
Based on a large population who underwent next-generation sequencing (NGS) in Shanghai Chest Hospital, our study revealed the characteristics and prognosis of NSCLC patients with co-occurring potentially actionable oncogenic drivers, trying to optimize the treatment strategies.
Patients and Methods
Patients
Between March 2018 and June 2019, patients with NSCLC analyzed for possible actionable targets by NGS in Shanghai Chest Hospital were enrolled. All patients were diagnosed as adenocarcinoma, squamous cell carcinoma, and other NSCLCs according to World Health Organization criteria assessed by experienced pathologists. The baseline clinical and demographic characteristics including age, gender, pathology, and stage were retrospectively collected. Our study has been approved by the institutional review board of Shanghai Chest Hospital. Written consent forms were obtained from patients before all invasive procedures and initiation of tyrosine kinase inhibitors (TKIs).
Next-Generation Sequencing
NGS is routinely carried out for patients with advanced NSCLCs, especially adenocarcinomas, in our center unless they refuse to do so. Patients with early stage NSCLCs can also choose to receive NGS in case of recurrence. A total of 3,077 formalin-fixed, paraffin-embedded (FFPE) tumor samples acquired from resected lung or small biopsies from NSCLCs were prepared according to standard procedure. Samples with more than 5% tumor content were sent for NGS. Tissue DNA was extracted by QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) and then evaluated with the Qubit 3.0 dsDNA assay (Life Technologies, CA, USA). DNA was fragmented by the Covaris M220 Focused-Ultrasonicator (Covaris, Woburn, MA), followed by end repair, phosphorylation, and adaptor ligation. Fragments of 200–400 bp in length were selected using Agencourt AMPure beads (Beckman Coulter, Fullerton, CA, USA), followed by hybridization with capture probes baits, hybrid selection with magnetic beads, and PCR amplification. After evaluating the quality and size of the fragments by a high-sensitivity DNA assay, the samples were sequenced on a Nextseq500 sequencer (Illumina, Inc., San Diego, CA, USA) with paired-end reads. A panel of 68 cancer-related genes described previously (19, 20) were used to detect the genetic alterations of our patients, and the details of our panel are also listed in Supplemental Table 1. The mean depth of was >1,000×. The sequencing data in the FASTQ format were mapped to the human genome (hg19) using BWA aligner 0.7.10. Local alignment optimization, variant calling, and annotation were assessed using GATK 3.2, MuTect, and VarScan, respectively. DNA translocation analysis was performed using both Tophat2 and Factera 1.4.3. Gene-level copy number variation was assessed using a t test statistic after normalizing read depths at each region by total read number and region size, and correcting GC bias using a locally estimated scatterplot smoothing (LOESS) model.
Defining MET/ERBB2 Amplification According to NGS
Although no consensus exists on the cut-off gene copy number (GCN) for MET amplification detected by NGS; however, GCN ≥4 as a cut-off for MET amplification by FISH was frequently used in several clinical trials (21, 22). Moreover, a recent study from our hospital using the same sequencing technique showed that MET-amplified patients with GCN >4 after crizotinib treatment tended to have longer PFS compared with GCN ≤4 (23). Therefore, the GCN for MET amplification in our study was greater than 4.
Similarly, there is still no recommended cut-off for ERBB2 amplification in NSCLC. However, ERBB2 amplification using in situ hybridization (ISH) in breast cancer according to 2018 ASCO/CAP clinical practice guideline was defined as ERBB2 GCN ≥6 (24). As a result, an ERBB2 GCN ≥6 was considered amplified in this study.
Treatment and Follow-Up
EGFR-sensitizing mutations included L858R, L861Q, G719X, and S768I mutations as well as exon 19 deletions; EGFR-undetermined mutations refer to other mutations without well-documented clinical significance. Some of NSCLC patients harboring EGFR-sensitizing mutations in our study were treated with first-generation EGFR TKIs including gefitinib (Iressa, AstraZeneca Pharmaceuticals) (25), erlotinib (Tarceva, Roche) (26), and icotinib (Conmana, Betta) (27) at doses of 250 mg once daily, 150 mg once daily, and 125 mg three times daily, respectively, afatinib (28) (Gilotrif, Boehringer-Ingelheim), and osimertinib (29) (Tagrisso, AstraZeneca Pharmaceuticals) at doses of 40 mg once daily and 80 mg once daily, respectively. ALK-rearranged patients were treated with crizotinib (Xalkori, Pfizer) (30) at 250 mg twice daily or alectinib (Alecensa, Roche) (31) at 600 mg twice daily. For some patients with high-level MET amplification, crizotinib at 250 mg twice daily was tried. Savolitinib (Hutchison Whampoa) at 600 or 400 mg once daily was given in a clinical trial (NCT02897479) to patients with MET exon 14 skipping mutations. Clinical evaluation was performed every 4–6 weeks according to the Response Evaluation Criteria in Solid Tumors (RECIST1.1). Progression-free survival (PFS) was defined from the initiation of TKIs to radiographic or clinical progression or the last follow-up time (January 31, 2020).
Statistical Analysis
The baseline characteristics between EGFR-mutant patients and patients with co-occurring actionable drivers receiving EGFR TKIs were compared with Chi-square test or two-sample t test as appropriate. Survival curves were generated for comparing PFS and OS by Kaplan–Meier methods and further compared by the log-rank test. A P value of <0.05 was considered statistically significant. All the analyses were performed using the Statistical Package for Social Science (SPSS, Chicago, IL, USA) version 22.0 for Windows.
Results
Baseline Characteristics of All Patients
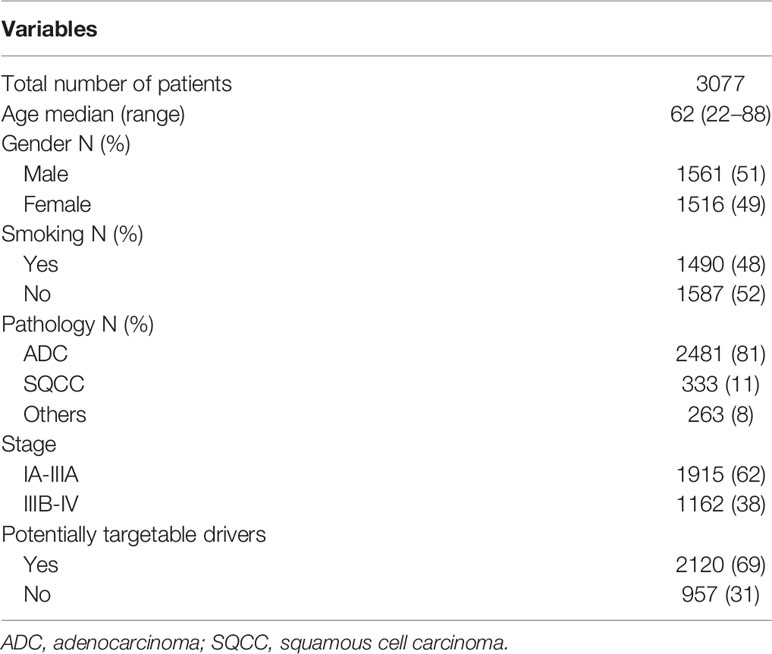
A total of 3,077 NSCLC patients were analyzed for oncogenic alterations by NGS, among whom, 81% (2481/3077), 11% (333/3077), and 8% (263/3077) were diagnosed as adenocarcinoma, squamous cell carcinoma, and other subtypes, respectively. The detailed characteristics were shown in Table 1. Of the patients, 69% (2120/3077) harbored at least one potentially actionable oncogenic drivers, namely EGFR mutations, ALK rearrangements, ROS1 rearrangements, BRAF V600E mutation, MET amplification, MET exon 14 skipping mutations, RET rearrangements, NTRK rearrangements, ERBB2 alterations (mutations and amplification), and KRAS G12C mutation. Among patients with at least one potentially actionable target, 75% (1587/2120) harbored EGFR mutations including exon 21 L858R mutation (47%; 750/1587), exon 19 deletions (40%; 634/1587), and other uncommon mutations (13%; 203/1587). The details of all potentially actionable oncogenic targets were listed in Figure 1A.
Figure 1 (A) The frequency of different oncogenic drivers in all patients. (B) Composition of EGFR-mutant NSCLC patients harboring other potentially actionable oncogenic drivers. (C) Composition of NSCLC patients harboring co-occurring potentially actionable drivers without EGFR mutations. Composition of patients with lung adenocarcinoma from MKSCC harboring co-occurring potentially actionable oncogenic drivers. double-positive, with two potentially actionable oncogenic drivers; triple-positive, with three potentially actionable oncogenic drivers; del, deletions; EGFR sensitizing, sensitizing EGFR mutations, EGFR undetermined, undetermined EGFR mutations; amp, amplification; ERBB2m, ERBB2 mutation; MET 14 skipping, MET exon 14 skipping mutation; ALKr, ALK rearrangement; NTRKr, NTRK rearrangement; RETr, RET rearrangement.
Baseline Characteristics of Patients Harboring Co-Occurring Potentially Actionable Targets
Of the patients, 1.5% (46/3077) had co-occurring potentially targetable oncogenic drivers. The characteristics of these patients were shown in Table 2. These patients are commonly seen in females (70 vs 30%), non-smokers (76 vs 24%), and adenocarcinomas (89 vs 11%). 80% (37/46) had EGFR mutations and other concomitant potentially actionable drivers, while 20% (9/46) did not harbor EGFR mutations. After excluding patients with EGFR-undetermined mutations, the remaining patients harboring co-occurring potentially actionable oncogenic drivers accounted for 1.3% (40/3077) of all NSCLCs (Figure 1B).

Table 2 The demographic and clinico-pathological characteristics of patients harboring co-occurring potentially actionable targets.
A total of 37 (80%; 37/46) patients harbored both EGFR-sensitizing (84%; 31/37) or -undetermined (16%; 6/37) mutations and other oncogenic drivers. The concomitant potentially targetable drivers included de novo MET amplification (21.6%; 8/37), de novo ERBB2 amplification (21.6%; 8/37), ERBB2 mutations (27.0%; 10/37), KRAS G12C mutation (8.1%; 3/37), ALK rearrangements (5.4%; 2/37), MET exon 14 skipping mutations (8.1%; 3/37), NTRK rearrangements (5.4%; 2/37), and RET rearrangements (2.7%; 1/37). All the molecular subtypes of these patients were shown in Figure 1B.
Among the patients harboring co-occurring potentially actionable targets without EGFR mutations, 44.4% (4/9) had both ERBB2 amplification and ERBB2 mutations; 11.1% (1/9) had KRAS G12C mutation and concurrent MET amplification; 11.1% (1/9) had MET amplification and a concurrent MET exon 14 splicing mutation; 22.2% (2/9) harbored both ALK rearrangements and ERBB2 mutations, and one patient (11.1%; 1/9) had RET rearrangement and an ERBB2 mutation (Figure 1C).
The Impact of Co-Occurring Potentially Actionable Oncogenic Drivers on Targeted Therapies
A total of 23 patients with co-occurring patterns were treated with different kinds of TKIs, whose treatment types, response, and progression events were shown in a swimmer plot (Supplemental Figure 1). A total of 17 EGFR-mutant patients with other potentially actionable oncogenic drivers were treated with first-generation EGFR TKIs. The demographic and clinical characteristics of these patients were listed in Table 3 and Supplemental Table 2. Of patients, 29% (5/17) and 29% (5/17) harbored concurrent de novo MET amplifications and ERBB2 amplifications, respectively. From March 2018 to June 2019, a total of 205 patients with EGFR mutations alone who underwent NGS analysis were treated with first-generation EGFR TKIs and had full medical records to evaluate the efficacy, whose baseline characteristics including TP53 mutation status were not significantly different from those of EGFR-mutant patients harboring other concurrent oncogenic drivers except that there were more females in the double-positive cohort (Supplemental Table 3).
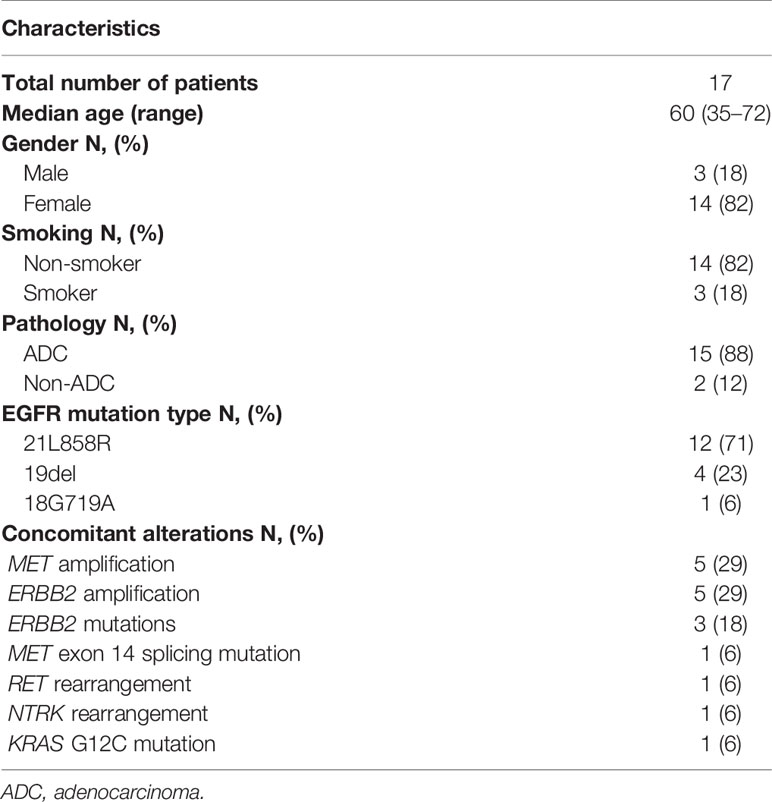
Table 3 The characteristics of EGFR mutant patients harboring other potentially actionable drivers treated with first-generation EGFR TKIs.
As shown in Figure 2A, EGFR-mutant patients with other concurrent potentially actionable drivers demonstrated significantly lower PFS compared to patients with EGFR mutations alone (5.4 vs 10.5 months; HR = 1.94, 95% CI: 1.16–3.25, p = 0.0042). Further analysis of molecular subtypes found that both de novo MET amplification (2.8 vs 10.5 months; HR = 6.03, 95% CI: 2.43–15.00; p < 0.0001) and de novo ERBB2 amplification (4.2 vs 10.5 months; HR = 2.5, 95% CI, 1.03–6.09; p = 0.0005) significantly reduced PFS of EGFR TKIs. However, EGFR-mutant patients harboring other potentially targetable drivers except MET/ERBB2 amplification showed no significant difference in PFS compared with patients harboring EGFR mutations alone (11.0 vs 10.5 months; HR = 1.13, 95% CI: 0.48–2.69; p = 0.76) (Figure 2B), although the PFS of these patients still fluctuated greatly across different molecular subtypes (Supplemental Table 2).
Figure 2 (A) Comparison of PFS of first-generation EGFR TKIs for patients with EGFR mutations alone and patients harboring both EGFR mutations and other potentially actionable oncogenic drivers. (B) PFS of first-generation EGFR TKIs for EGFR-mutant patients with concurrent MET amplification, ERBB2 amplification, and other oncogenic drivers.
A total of six patients with co-occurring potentially targetable oncogenic drivers were treated with other TKIs. One EGFR-mutant patient carrying concurrent de novo ERBB2 amplification was treated with afatinib, reaching a PFS of 4.6 months, while PFS of one EGFR-mutant patient with concurrent de novo MET amplification receiving osimertinib was 14.2 months. One EGFR-mutant patient with concomitant high-level MET amplification (copy number = 8.8) received crizotinib, yet with a PFS of only 2.5 months. One ALK-rearranged patient with a concurrent ERBB2 mutation was treated with alectinib, achieving partial response. One MET-amplified patient with a concomitant MET exon 14 skipping mutation was treated with crizotinib, having satisfying response with PFS of 14.4 (Table 4).
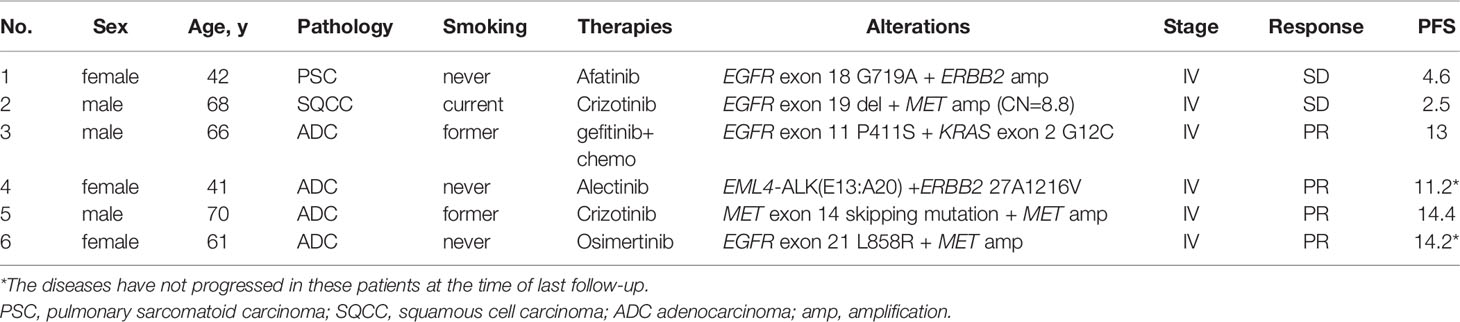
Table 4 Characteristics of patients harboring other co-occurring actionable drivers receiving targeted therapies.
Discussion
Our study found that the incidence of NSCLC patients harboring co-occurring potentially actionable alterations was approximately 1.5% (46/3077), likely to be found in females, non-smokers, and adenocarcinomas. Among these patients, 80% (37/46) harbored EGFR-sensitizing (84%; 31/37) or -undetermined (16%; 6/37) mutations and other concurrent potentially targetable oncogenic drivers such as de novo MET amplification (21.6%; 8/37) and alterations in ERBB2 including mutations (27.0%; 10/37) and amplification (21.6%; 8/37). Other concurrent potentially actionable targets in EGFR-mutant patients treated with first-generation EGFR TKIs were associated with shorter PFS (5.4 vs 10.5 months; HR = 1.94, 95% CI: 1.16–3.25, p = 0.0042), suggesting that co-occurring potentially targetable oncogenic drivers may impact the efficacy of EGFR TKIs. Further analysis showed that it was de novo MET/ERBB2 amplification that played major roles in the primary resistance to first-generation EGFR TKIs, while the third-generation EGFR TKI, osimertinib, may bring better benefits to these patients. In addition, the efficacy of TKIs in NSCLC patients harboring other co-occurring potentially actionable targets varied across different molecular subtypes with overall encouraging responses.
With the advent and rapid development of NGS, the genomic alterations of lung cancer have been fully investigated, helping to move forward to the era of precision medicine. There are three generations of TKIs for EGFR mutations, one of the most common oncogenic drivers in NSCLC. The first-generation EGFR TKIs including gefitinib, erlotinib, and icotinib were usually used as the first-line treatments in China due to their wide availability and affordable prices. Our study found that the PFS of the first-generation TKIs in EGFR-mutant patients was 10.5 months, similar to the results from previous clinical trials (25–27). Owing to the greatly heterogeneous responses to EGFR TKIs across individuals harboring EGFR mutations, many research studies were carried out to study the factors that may affect the efficacy, among which, concurrent genomic alterations were mostly investigated. For example, a retrospective study from Korea included patients who underwent molecular analysis by NGS before treatment with first-/second-generation EGFR TKIs (cohort 1) or third-generation EGFR TKIs after failure in the previous TKIs (cohort 2). Their results showed that TP53 mutations were independently associated with worse outcomes in cohort 1, while in cohort 2, TP53, RB1, and PTEN mutations as well as MDM2 amplifications all resulted in shorter PFS (11), suggesting that concurrent genomic alterations accelerated the resistance to EGFR TKIs. Additionally, co-occurring TP53 mutations were reported to be associated with worse outcomes of EGFR TKIs in several studies (10, 11, 13, 32). However, these concomitant genetic alterations in previous studies were mostly considered untargetable without available drugs. There are only a few oncogenic drivers in NSCLC having commercially available or promising targeted therapies, including alterations in EGFR, ALK,ROS1, BRAF, MET, RET, ERBB2, NTRK, and KRAS. Although these drivers were considered mutually exclusive previously (33), increasing evidence demonstrated that a minority of NSCLCs harbored co-occurring potentially actionable alterations (17, 34–36), whose overall characteristics remained largely unknown due to the small sample size of previous reports. Our findings showed that the incidence of NSCLC patients harboring co-occurring potentially actionable alterations was approximately 1.5% (46/3077), and they were commonly found in females, non-smokers, and adenocarcinomas.
Similar results were also shown in western populations, although the frequency of oncogenic drivers, especially EGFR mutations, in western patients with NSCLC was reported to be lower than that of eastern population (1, 37, 38). The patients with co-occurring potentially actionable targets accounted for 3.6% (31/860) of all lung adenocarcinoma according to the data presented by MSKCC (38). Among these patients, 84% (26/31) harbored EGFR-sensitizing (81%; 21/26) or -undetermined (19%; 5/26) mutations with other concurrent potentially actionable drivers including ERBB2 amplification (50%; 13/26), MET amplification (27%; 7/26), KRAS G12C mutation (11.5%; 3/26), and ERBB2 mutations (11.5%; 3/26), similar to the eastern population. There are five (16%; 5/31) patients harboring other types of co-occurring potentially actionable targets. However, the GCNs for MET/ERBB2 amplification were not known in this database.
Among NSCLC patients with co-occurring potentially oncogenic drivers, 80% (37/46) harbored EGFR mutations and other concurrent potentially targetable drivers, commonly involving de novo MET amplifications (21.6%; 8/37) and alterations in ERBB2 including mutations (27.0%; 10/37) and amplification (21.6%; 8/37). It was found that concurrent actionable drivers, especially MET/ERBB2 amplification, contributed to the primary resistance to EGFR TKIs. MET amplification often occurred in EGFR-mutant NSCLC after failure of previous TKIs as an acquired resistance mechanism by activating ERBB3 signaling (39, 40). From a cytological prospective, a small proportion of EGFR-mutant cells already harbored MET amplifications before initiation of TKIs, finally resulting in drug resistance (41). A previous study showed that approximately 3.2% (5/154) of EGFR-mutant patients harbored concurrent MET amplifications before treatment with EGFR TKIs, and the PFS for these patients was significantly shorter than that of the patients with EGFR mutations alone (17), but the sample size was too small to reach a concrete conclusion. In our study, five EGFR-mutant patients with concurrent de novo MET amplifications treated with first-generation EGFR TKIs demonstrated worse PFS compared to the patients with EGFR mutations alone, further supporting the conclusion that de novo MET amplification contributed to the primary resistance of first-generation EGFR TKIs. Additionally, one EGFR-mutant patient with high-level MET amplification received crizotinib with stable disease lasting for only 2.5 months, suggesting that these patients may respond poorly to only one targeted drug. A phase Ib/II clinical trial demonstrated promising efficacy of capmatinib plus gefitinib after failure of EGFR TKIs in patients having EGFR-mutant and MET-amplified NSCLC with acceptable toxicities (21). Therefore, the combination regimen can also be tried in EGFR-mutant patients with de novo MET amplifications in clinical practice to overcome primary resistance. Additionally, one EGFR-mutant patient with both de novo MET amplification was treated with osimertinib, achieving prolonged partial response, suggesting the third-generation EGFR TKI, osimertinib, might overcome the primary resistance from de novo MET amplification.
Similar to concurrent de novo MET amplification, de novo ERBB2 amplification was also associated with shorter PFS. Approximately 13% of patients with EGFR-mutant NSCLC will acquire ERBB2 amplification after failure of first-generation EGFR TKIs. However, the role of de novo ERBB2 amplification in EGFR-mutant NSCLC has not been fully revealed. A previous study showed that 4% (8/200) of all EGFR-mutant NSCLC harbored concurrent ERBB2 amplifications before treatment with TKIs, leading to a shorter PFS compared to patients with EGFR mutations alone (14). In our study, it was found patients having both EGFR-mutant and ERBB2 amplified NSCLC treated with first-generation EGFR TKIs reached a median PFS of 4.2 months, which was significantly shorter than that of patients with EGFR mutations alone (4.2 vs 10.5 months; HR = 2.5, 95% CI, 1.03–6.09; p = 0.0005). A recent phase II basket trial demonstrated a 51% response rate in 49 patients with ERBB2-amplified or mutant NSCLC treated with ado-trastuzumab emtansine (T-DM1), an anti-ERBB2/HER2 antibody-drug conjugate (ADC), suggesting that ADCs are effective in patients with ERBB2-aberrant NSCLC. Therefore, the future clinical trials of EGFR TKIs plus ADCs can be launched in EGFR-mutant patients with concurrent ERBB2 amplification.
Additionally, there were EGFR-mutant patients harboring other concurrent potentially actionable drivers such as KRAS mutations. Sotorasib, a KRAS inhibitor, in a recent clinical trial showed encouraging efficacy in patients with heavily pretreated advanced solid tumors harboring KRAS G12C mutation, paving a way for targeted therapy in KRAS-mutant patients. There are several gene fusions in NSCLC with corresponding targeted drugs including ALK, ROS1, RET, and NTRK (42). Although these fusions rarely overlap with other oncogenic drivers (43, 44), our study found that they can co-exist with other actionable targets such as EGFR mutations and ERBB2 mutations. Moreover, there were four patients harboring both ERBB2 mutations and ERBB2 amplifications, although a previous study showed that ERBB2 mutations were not associated with ERBB2 amplification (45).
There were some drawbacks of our study. Firstly, this was a retrospective study with a relatively small sample size, which cannot avoid selection bias and reflect the entire population with co-occurring potentially actionable oncogenic drivers. Secondly, MET and ERBB2 amplifications were not further confirmed by fluorescence in situ hybridization (FISH), although NGS was frequently applied in both clinical trials and routine practice to detect amplifications (6, 8, 21),. Furthermore, several studies showed high concordance between NGS and FISH or immunohistochemistry in detecting amplification (46–48). Additionally, the efficacy of ADCs in ERBB2-amplified NSCLC varied across different clinical trials (5, 6, 49); however, a recent study has revealed that the overall response rate by RECIST of T-DM1 in ERBB2-amplified NSCLC was 50% (5/10) (6), suggesting ERBB2 amplification was also a promising target. Moreover, the efficacy of EGFR TKIs in some of the EGFR-undetermined mutations considered potentially actionable in our study still needed to be explored. Finally, there is a potential risk of confounders by patient clinical characteristics in the survival analysis, especially in the subgroup analysis.
Conclusions
In conclusion, our study showed approximately 1.5% (46/3077) of all NSCLCs harbored co-occurring potentially actionable oncogenic drivers, commonly involving EGFR mutations; after excluding patients with EGFR-undetermined mutations, the incidence was 1.3% (40/3077). These patients are likely to be found in females, non-smokers, and adenocarcinomas. In EGFR-mutant patients, de novo MET/ERBB2 amplification was associated with shorter PFS, and the combination of EGFR and MET/ERBB2 inhibitions or third-generation EGFR TKIs can be tried in future to achieve better response. The efficacy of TKIs in NSCLC patients harboring other co-occurring potentially actionable drivers varied across different molecular subtypes. Many molecular subtypes of co-occurring actionable oncogenic drivers were found in our study, suggesting the complexity of oncogene-addicted NSCLC. In order to tailor the combination or sequential treatment strategies, future clinical trials for these patients should be anticipated.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
Ethical approval was not provided for this study on human participants because this was a retrospective study. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) and minor(s)’ legal guardian/next of kin for the publication of any potentially identifiable images or data included in this article.
Author Contributions
YZ, SW, and ZY have substantial contributions to the conception or design of the work, the collection and analysis of data, the writing and editing of the article. BH and HH contributed to interpretation of data and revision of the manuscript. The rest of the authors have given substantial contributions to the work by providing editing and writing assistance. All authors contributed to the article and approved the submitted version.
Funding
This study was supported in part by grants from the State Key Program of National Natural Science of China (8173077) and Shanghai Rising-Star Program (2YF1428100).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.665484/full#supplementary-material
Supplementary Figure 1 | A swimmer plot for patients with co-occurring patterns treated with different kinds of TKIs. EGFRm, EGFR mutations alone; double-positive, with two potentially actionable oncogenic drivers; amp, amplification.
References
1. Zhang XC, Wang J, Shao GG, Wang Q, Qu X, Wang B, et al. Comprehensive Genomic and Immunological Characterization of Chinese Non-Small Cell Lung Cancer Patients. Nat Commun (2019) 10(1):1772. doi: 10.1038/s41467-019-09762-1
2. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, et al. Lung Cancer: Current Therapies and New Targeted Treatments. Lancet (2017) 389(10066):299–311. doi: 10.1016/s0140-6736(16)30958-8
3. Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med (2018) 378(8):731–9. doi: 10.1056/NEJMoa1714448
4. Hu Z, Li M, Chen Z, Zhan C, Lin Z, Wang Q. Advances in Clinical Trials of Targeted Therapy and Immunotherapy of Lung Cancer in 2018. Transl Lung Cancer Res (2019) 8(6):1091–106. doi: 10.21037/tlcr.2019.10.17
5. Peters S, Stahel R, Bubendorf L, Bonomi P, Villegas A, Kowalski DM, et al. Trastuzumab Emtansine (T-DM1) in Patients With Previously Treated HER2-Overexpressing Metastatic Non-Small Cell Lung Cancer: Efficacy, Safety, and Biomarkers. Clin Cancer Res (2019) 25(1):64–72. doi: 10.1158/1078-0432.Ccr-18-1590
6. Li BT, Michelini F, Misale S, Cocco E, Baldino L, Cai Y, et al. Her2-Mediated Internalization of Cytotoxic Agents in ERBB2 Amplified or Mutant Lung Cancers. Cancer Discov (2020) 10(5):674–87. doi: 10.1158/2159-8290.Cd-20-0215
7. Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, et al. Kras(G12c) Inhibition With Sotorasib in Advanced Solid Tumors. N Engl J Med (2020) 383(13):1207–17. doi: 10.1056/NEJMoa1917239
8. Guo R, Luo J, Chang J, Rekhtman N, Arcila M, Drilon A. MET-Dependent Solid Tumours - Molecular Diagnosis and Targeted Therapy. Nat Rev Clin Oncol (2020) 17(9):569–87. doi: 10.1038/s41571-020-0377-z
9. Hong S, Gao F, Fu S, Wang Y, Fang W, Huang Y, et al. Concomitant Genetic Alterations With Response to Treatment and Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Patients With EGFR-Mutant Advanced Non-Small Cell Lung Cancer. JAMA Oncol (2018) 4(5):739–42. doi: 10.1001/jamaoncol.2018.0049
10. Hou H, Qin K, Liang Y, Zhang C, Liu D, Jiang H, et al. Concurrent TP53 Mutations Predict Poor Outcomes of EGFR-TKI Treatments in Chinese Patients With Advanced NSCLC. Cancer Manag Res (2019) 11:5665–75. doi: 10.2147/cmar.S201513
11. Kim Y, Lee B, Shim JH, Lee SH, Park WY, Choi YL, et al. Concurrent Genetic Alterations Predict the Progression to Target Therapy in EGFR-Mutated Advanced NSCLC. J Thorac Oncol (2019) 14(2):193–202. doi: 10.1016/j.jtho.2018.10.150
12. Kron A, Alidousty C, Scheffler M, Merkelbach-Bruse S, Seidel D, Riedel R, et al. Impact of TP53 Mutation Status on Systemic Treatment Outcome in ALK-Rearranged Non-Small-Cell Lung Cancer. Ann Oncol (2018) 29(10):2068–75. doi: 10.1093/annonc/mdy333
13. VanderLaan PA, Rangachari D, Mockus SM, Spotlow V, Reddi HV, Malcolm J, et al. Mutations in TP53, Pik3ca, PTEN and Other Genes in EGFR Mutated Lung Cancers: Correlation With Clinical Outcomes. Lung Cancer (2017) 106:17–21. doi: 10.1016/j.lungcan.2017.01.011
14. Yu HA, Suzawa K, Jordan E, Zehir A, Ni A, Kim R, et al. Concurrent Alterations in EGFR-Mutant Lung Cancers Associated With Resistance to EGFR Kinase Inhibitors and Characterization of MTOR as a Mediator of Resistance. Clin Cancer Res (2018) 24(13):3108–18. doi: 10.1158/1078-0432.Ccr-17-2961
15. Baldi L, Mengoli MC, Bisagni A, Banzi MC, Boni C, Rossi G. Concomitant EGFR Mutation and ALK Rearrangement in Lung Adenocarcinoma Is More Frequent Than Expected: Report of a Case and Review of the Literature With Demonstration of Genes Alteration Into the Same Tumor Cells. Lung Cancer (2014) 86(2):291–5. doi: 10.1016/j.lungcan.2014.09.011
16. Chen RL, Zhao J, Zhang XC, Lou NN, Chen HJ, Yang X, et al. Crizotinib in Advanced Non-Small-Cell Lung Cancer With Concomitant ALK Rearrangement and c-Met Overexpression. BMC Cancer (2018) 18(1):1171. doi: 10.1186/s12885-018-5078-y
17. Lai GGY, Lim TH, Lim J, Liew PJR, Kwang XL, Nahar R, et al. Clonal MET Amplification as a Determinant of Tyrosine Kinase Inhibitor Resistance in Epidermal Growth Factor Receptor-Mutant Non-Small-Cell Lung Cancer. J Clin Oncol (2019) 37(11):876–84. doi: 10.1200/jco.18.00177
18. Zhu YC, Liao XH, Wang WX, Xu CW, Zhuang W, Wei JG, et al. Dual Drive Coexistence of EML4-ALK and TPM3-ROS1 Fusion in Advanced Lung Adenocarcinoma. Thorac Cancer (2018) 9(2):324–7. doi: 10.1111/1759-7714.12578
19. Xiong L, Li R, Sun J, Lou Y, Zhang W, Bai H, et al. Erlotinib as Neoadjuvant Therapy in Stage IIIa (N2) EGFR Mutation-Positive Non-Small Cell Lung Cancer: A Prospective, Single-Arm, Phase II Study. Oncol (2019) 24(2):157–e64. doi: 10.1634/theoncologist.2018-0120
20. Zhao Y, Dong Y, Zhao R, Zhang B, Wang S, Zhang L, et al. Expression Profiling of Driver Genes in Female Never-Smokers With Non-Adenocarcinoma Non-Small-Cell Lung Cancer in China. Clin Lung Cancer (2020) 21(5):e355–62. doi: 10.1016/j.cllc.2020.02.005
21. Wu YL, Zhang L, Kim DW, Liu X, Lee DH, Yang JC, et al. Phase Ib/II Study of Capmatinib (Inc280) Plus Gefitinib After Failure of Epidermal Growth Factor Receptor (EGFR) Inhibitor Therapy in Patients With EGFR-Mutated, Met Factor-Dysregulated Non-Small-Cell Lung Cancer. J Clin Oncol (2018) 36(31):3101–9. doi: 10.1200/jco.2018.77.7326
22. Angevin E, Spitaleri G, Rodon J, Dotti K, Isambert N, Salvagni S, et al. A First-in-Human Phase I Study of SAR125844, a Selective MET Tyrosine Kinase Inhibitor, in Patients With Advanced Solid Tumours With MET Amplification. Eur J Cancer (2017) 87:131–9. doi: 10.1016/j.ejca.2017.10.016
23. Li J, Wang Y, Zhang B, Xu J, Cao S, Zhong H. Characteristics and Response to Crizotinib in Lung Cancer Patients With MET Amplification Detected by Next-Generation Sequencing. Lung Cancer (2020) 149:17–22. doi: 10.1016/j.lungcan.2020.08.021
24. Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS, et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J Clin Oncol (2018) 36(20):2105–22. doi: 10.1200/jco.2018.77.8738
25. Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or Carboplatin-Paclitaxel in Pulmonary Adenocarcinoma. N Engl J Med (2009) 361(10):947–57. doi: 10.1056/NEJMoa0810699
26. Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib Versus Chemotherapy as First-Line Treatment for Patients With Advanced EGFR Mutation-Positive Nn-Small-Cell Lung Cancer (OPTIMAL, CTONG-0802): A Multicentre, Open-Label, Randomised, Phase 3 Study. Lancet Oncol (2011) 12(8):735–42. doi: 10.1016/s1470-2045(11)70184-x
27. Shi YK, Wang L, Han BH, Li W, Yu P, Liu YP, et al. First-Line Icotinib Versus Cisplatin/Pemetrexed Plus Pemetrexed Maintenance Therapy for Patients With Advanced EGFR Mutation-Positive Lung Adenocarcinoma (CONVINCE): A Phase 3, Open-Label, Randomized Study. Ann Oncol (2017) 28(10):2443–50. doi: 10.1093/annonc/mdx359
28. Park K, Tan EH, O’Byrne K, Zhang L, Boyer M, Mok T, et al. Afatinib Versus Gefitinib as First-Line Treatment of Patients With EGFR Mutation-Positive Non-Small-Cell Lung Cancer (LUX-Lung 7): A Phase 2B, Open-Label, Randomised Controlled Trial. Lancet Oncol (2016) 17(5):577–89. doi: 10.1016/s1470-2045(16)30033-x
29. Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med (2018) 378(2):113–25. doi: 10.1056/NEJMoa1713137
30. Shaw AT, Kim DW, Nakagawa K, Seto T, Crinó L, Ahn MJ, et al. Crizotinib Versus Chemotherapy in Advanced ALK-Positive Lung Cancer. N Engl J Med (2013) 368(25):2385–94. doi: 10.1056/NEJMoa1214886
31. Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim DW, et al. Alectinib Versus Crizotinib in Untreated Alk-Positive Non-Small-Cell Lung Cancer. N Engl J Med (2017) 377(9):829–38. doi: 10.1056/NEJMoa1704795
32. Chen H, Liu M, Dai Z, Li S, Luo Y, Wang Y, et al. Concomitant Genetic Alterations Are Associated With Response to EGFR Targeted Therapy in Patients With Lung Adenocarcinoma. Transl Lung Cancer Res (2020) 9(4):1225–34. doi: 10.21037/tlcr-20-679
33. Gainor JF, Varghese AM, Ou SH, Kabraji S, Awad MM, Katayama R, et al. ALK Rearrangements Are Mutually Exclusive With Mutations in EGFR or KRAS: An Analysis of 1,683 Patients With Non-Small Cell Lung Cancer. Clin Cancer Res (2013) 19(15):4273–81. doi: 10.1158/1078-0432.Ccr-13-0318
34. Chen M, Xu Y, Zhao J, Zhong W, Zhang L, Bi Y, et al. Concurrent Driver Gene Mutations as Negative Predictive Factors in Epidermal Growth Factor Receptor-Positive Non-Small Cell Lung Cancer. EBioMedicine (2019) 42:304–10. doi: 10.1016/j.ebiom.2019.03.023
35. Tiseo M, Gelsomino F, Boggiani D, Bortesi B, Bartolotti M, Bozzetti C, et al. EGFR and EML4-ALK Gene Mutations in NSCLC: A Case Report of Erlotinib-Resistant Patient With Both Concomitant Mutations. Lung Cancer (2011) 71(2):241–3. doi: 10.1016/j.lungcan.2010.11.014
36. Cai W, Lin D, Wu C, Li X, Zhao C, Zheng L, et al. Intratumoral Heterogeneity of ALK-Rearranged and ALK/EGFR Coaltered Lung Adenocarcinoma. J Clin Oncol (2015) 33(32):3701–9. doi: 10.1200/jco.2014.58.8293
37. Wen S, Dai L, Wang L, Wang W, Wu D, Wang K, et al. Genomic Signature of Driver Genes Identified by Target Next-Generation Sequencing in Chinese Non-Small Cell Lung Cancer. Oncol (2019) 24(11):e1070–81. doi: 10.1634/theoncologist.2018-0572
38. Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov (2017) 7(6):596–609. doi: 10.1158/2159-8290.Cd-16-1337
39. Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of Tumor Specimens At the Time of Acquired Resistance to EGFR-TKI Therapy in 155 Patients With EGFR-Mutant Lung Cancers. Clin Cancer Res (2013) 19(8):2240–7. doi: 10.1158/1078-0432.Ccr-12-2246
40. Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET Amplification Leads to Gefitinib Resistance in Lung Cancer by Activating ERBB3 Signaling. Science (New York NY) (2007) 316(5827):1039–43. doi: 10.1126/science.1141478
41. Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and Clonal Selection of MET Amplification in EGFR Mutant NSCLC. Cancer Cell (2010) 17(1):77–88. doi: 10.1016/j.ccr.2009.11.022
42. Gainor JF, Shaw AT. Novel Targets in Non-Small Cell Lung Cancer: ROS1 and RET Fusions. Oncol (2013) 18(7):865–75. doi: 10.1634/theoncologist.2013-0095
43. Lin JJ, Ritterhouse LL, Ali SM, Bailey M, Schrock AB, Gainor JF, et al. Ros1 Fusions Rarely Overlap With Other Oncogenic Drivers in Non-Small Cell Lung Cancer. J Thorac Oncol (2017) 12(5):872–7. doi: 10.1016/j.jtho.2017.01.004
44. Entrectinib OK’d for Cancers With NTRK Fusions, NSCLC. Cancer Discov (2019) 9(10):Of2. doi: 10.1158/2159-8290.Cd-nb2019-101
45. Li BT, Ross DS, Aisner DL, Chaft JE, Hsu M, Kako SL, et al. Her2 Amplification and HER2 Mutation Are Distinct Molecular Targets in Lung Cancers. J Thorac Oncol (2016) 11(3):414–9. doi: 10.1016/j.jtho.2015.10.025
46. Cenaj O, Ligon AH, Hornick JL, Sholl LM. Detection of ERBB2 Amplification by Next-Generation Sequencing Predicts Her2 Expression in Colorectal Carcinoma. Am J Clin Pathol (2019) 152(1):97–108. doi: 10.1093/ajcp/aqz031
47. Hoda RS, Bowman AS, Zehir A, Razavi P, Brogi E, Ladanyi M, et al. Next-Generation Assessment of Human Epidermal Growth Factor Receptor 2 Gene (ERBB2) Amplification Status in Invasive Breast Carcinoma: A Focus on Group 4 by Use of the 2018 American Society of Clinical Oncology/College of American Pathologists HER2 Testing Guideline. Histopathology (2020) 78(4):498–507. doi: 10.1111/his.14241
48. Robinson CL, Harrison BT, Ligon AH, Dong F, Maffeis V, Matulonis U, et al. Detection of ERBB2 Amplification in Uterine Serous Carcinoma by Next-Generation Sequencing: An Approach Highly Concordant With Standard Assays. Mod Pathol (2020) 34(3):603–12. doi: 10.1038/s41379-020-00695-5
49. Kris MG, Camidge DR, Giaccone G, Hida T, Li BT, O’Connell J, et al. Targeting HER2 Aberrations as Actionable Drivers in Lung Cancers: Phase II Trial of the Pan-HER Tyrosine Kinase Inhibitor Dacomitinib in Patients With HER2-Mutant or Amplified Tumors. Ann Oncol (2015) 26(7):1421–7. doi: 10.1093/annonc/mdv186
Keywords: non-small cell lung cancer, actionable oncogenic drivers, EGFR, tyrosine kinase inhibitors, next-generation sequencing
Citation: Zhao Y, Wang S, Yang Z, Dong Y, Wang Y, Zhang L, Hu H and Han B (2021) Co-Occurring Potentially Actionable Oncogenic Drivers in Non-Small Cell Lung Cancer. Front. Oncol. 11:665484. doi: 10.3389/fonc.2021.665484
Received: 08 February 2021; Accepted: 16 April 2021;
Published: 16 June 2021.
Edited by:
Hideharu Kimura, Kanazawa University, JapanReviewed by:
Jiajia Zhang, Johns Hopkins University, United StatesAlessandro Morabito, Istituto Nazionale Tumori Fondazione G. Pascale (IRCCS), Italy
Copyright © 2021 Zhao, Wang, Yang, Dong, Wang, Zhang, Hu and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baohui Han, MTg5MzA4NTgyMTZAMTYzLmNvbQ==; Hai Hu, aHVoYWlAbWFpbC5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
 Yiming Zhao1,2†
Yiming Zhao1,2† Shuyuan Wang
Shuyuan Wang Lele Zhang
Lele Zhang Hai Hu
Hai Hu Baohui Han
Baohui Han