- 1Department of Radiation Oncology, Zhejiang Key Laboratory of Radiation Oncology, Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital), Institute of Cancer and Basic Medicine (IBMC), Chinese Academy of Sciences, Hangzhou City, China
- 2The Second Clinical Medical College, Zhejiang Chinese Medical University, Hangzhou, China
- 3Department of Medical, Nanjing Geneseeq Technology Inc, Nanjing, China
- 4Department of Oncology, Traditional Chinese Medical Hospital of Huzhou, Huzhou, China
- 5Department of Oncology, Haining People’s Hospital, The First Affiliated Hospital of Zhejiang University, Haining, China
Purpose: Large cell neuroendocrine carcinoma (LCNEC) and classic large cell carcinoma (LCC) are two distinct entities with different histological and biological characteristics. However, the mutational profiles and the clinical behavior of the two subtypes of lung cancer remain to be explored.
Patients and Methods: Pathological diagnoses of all screened patients were finally confirmed by three or four experienced pathologists. Patients with uncertain pathological diagnoses were excluded. Finally, we genetically profiled ten patients with LCNEC and seven with LCC. ALL patients were subjected to next-generation sequencing (NGS) test, which included nine patients sequenced with a 139-gene panel and eight patients with a 425-gene panel. Including only intersected mutations from these two panels, survival analysis was further conducted.
Results: Both LCNEC and LCC showed high prevalence in male patients, with no clear association with smoking history. Potential targetable mutations in KRAS and RET were detected in the study cohort. However, LCNEC and LCC showed distinct mutational profiles with an enrichment of RB1/TP53 co-mutations in a subset of LCNEC patients. SMARCA4 and KEAP1 mutations were exclusively found in LCC patients, and RICTOR, BRAF, ROS1 and TET2 mutations were only detected in LCNEC. LCC patients in the cohort had shorter survival compared to LCNEC patients (p=0.006). Survival analysis revealed an association between SMARCA4 mutations and poor outcome in the study cohort and in the LCC subset. Mutations in BRAF were associated with a trend of increased survival in the study cohort, as well as in the LCNEC subset. Finally, TET2 mutations were associated with poor outcome in the LCNEC cohort.
Conclusion: LCC and LCNEC were both heterogeneous diseases with limited treatment options. Our study identified potential targetable mutations and prognostic biomarkers that might provide more therapeutic options and improve individualized patient care.
Introduction
Large-cell carcinoma (LCC) of the lung is an undifferentiated tumor, which lacks of the immunohistochemical and cytological characteristics of adenocarcinoma, squamous cell carcinoma or small-cell carcinoma (1). It is a relatively uncommon lung cancer that is characterized by poor prognosis and limited treatment options. Large cell neuroendocrine carcinoma (LCNEC) was initially classified as a histological variant of LCC. However, LCNEC and LCC differ in their cytological and histological features, as well as in clinical behavior to treatment. It has been reported that LCNEC was associated with worse survival outcome compared to LCC (2). As a consequence, the 2015 World Health Organization (WHO) classification significantly revised the classification of LCC and removed large cell neuroendocrine carcinoma (LCNEC) from the LCC category, establishing LCNEC as a distinct entity (3).
With the advances in next-generation sequencing (NGS), numerous actionable driver genes have emerged from the genetic landscape of different lung cancer subtypes. At the same time, many efforts have been undertaken to clarify the biological relationship between LCNEC, classic LCC and other types of lung cancer (1, 4–6). LCNEC has been segregated into small-cell lung carcinoma (SCLC)-like, characterized by the co-occurrence of TP53 and RB1 mutations, and non-small-cell lung carcinoma (NSCLC)-like, characterized by the lack of TP53/RB1 co-mutations and STK11/KEAP1/KRAS mutations (4, 6). On the other hand, classic LCCs are also genetically heterogeneous, displaying profiles characteristics of either adenocarcinoma or squamous cell carcinoma (7). In this study, we aimed to further investigate these two subtypes of lung cancer and identify potential biomarkers for treatment and prognostication of these patients.
Material and Methods
Patients and Samples
Pathological diagnoses of all screened patients were finally confirmed by three or four experienced pathologists, and patients with uncertain pathological diagnoses were excluded. Finally, a total of 17 patients, ten patients with LCNEC and seven with LCC, who were treated at Zhejiang Cancer Hospital from March 2017 to April 2020 with sufficient clinical information and follow-up, were included in the study. Samples were profiled using targeted NGS for routine diagnostic or treatment purposes at Nanjing Geneseeq Technology Inc.
DNA Extraction, Library Preparation and Sequencing
DNA extraction, sequencing library preparation, and targeted capture enrichment were carried out following the methods as previously described with modifications (8). Briefly, Genomic DNA from the white blood cells were extracted using the DNeasy Blood & Tissue Kit (Qiagen) and used as the normal control to remove germline variations. FFPE samples were de-paraffinized with xylene, and genomic DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen). DNA was quantified by Qubit 3.0 using the dsDNA HS Assay Kit (Life Technologies), and the quality was evaluated by a Nanodrop 2000 (Thermo Fisher).
Libraries were prepared by KAPA Hyper Prep kit (KAPA Biosystems), as previously described (9). Briefly, 1-2 μg of genomic DNA was sheared into ~350 bp fragments using a Covaris M220 instrument. End repair, A-tailing, and adaptor ligation of fragmented DNA were performed using the KAPA Hyper DNA Library Prep kit (Roche Diagnostics), followed by size selection with AgencourtAMPure XP beads (Beckman Coulter). DNA Libraries were then amplified by polymerase chain reaction (PCR) and purified using AgencourtAMPure XP beads.
Customized xGen lockdown probes panel (Integrated DNA Technologies) were used to selectively enrich for 139 or 425 predefined cancer-related genes (Geneseeq panel). Human cot-1 DNA (Life Technologies) and xGen Universal Blocking Oligos (Integrated DNA Technologies) were added as blocking reagents. The capture reaction was performed with Dynabeads M-270 (Life Technologies) and the xGen Lockdown Hybridization and Wash kit (Integrated DNA Technologies). Captured libraries were subjected to PCR amplification with KAPA HiFi HotStartReadyMix (KAPA Biosystems). The purified library was quantified using the KAPA Library Quantification kit (KAPA Biosystems), and its fragment size distribution was analyzed using a Bioanalyzer 2100. Target enriched libraries were sequenced on the HiSeq4000 platform (Illumina).
Mutation Calling
Sequencing data were demultiplexed by bcl2fastq (v2.19), analyzed by Trimmomatic (10) to remove low-quality (quality<15) or N bases. Then the data were aligned to the hg19 reference human genome with the Burrows-Wheeler Aligner (bwa-mem) (11) and further processed using the Picard suite (available at: https://broadinstitute.github.io/picard/) and the Genome Analysis Toolkit (GATK) (12). SNPs and indels were called by VarScan2 (13) and HaplotypeCaller/UnifiedGenotyper in GATK, with the mutant allele frequency (MAF) cutoff as 0.5%. Common variants were removed using dbSNP and the 1000 Genome project. Germline mutations were filtered out by comparing to patient’s whole blood controls.
Statistical Analysis
Comparisons of proportion between groups were performed using the Fisher’s exact test. Survival analysis was performed using Kaplan-Meier curves, and the p value was determined with the log-rank test, and hazard ratios (HRs) were calculated by Cox proportional hazards model. A two-sided p value of less than 0.05 was considered significant for all tests unless indicated otherwise. Univariable was used to study the association between different variables and PFS, and the results are presented as HRs and their 95% confidence intervals (CIs). All analyses were performed with R 3.4.0.
Results
Patient Characteristics
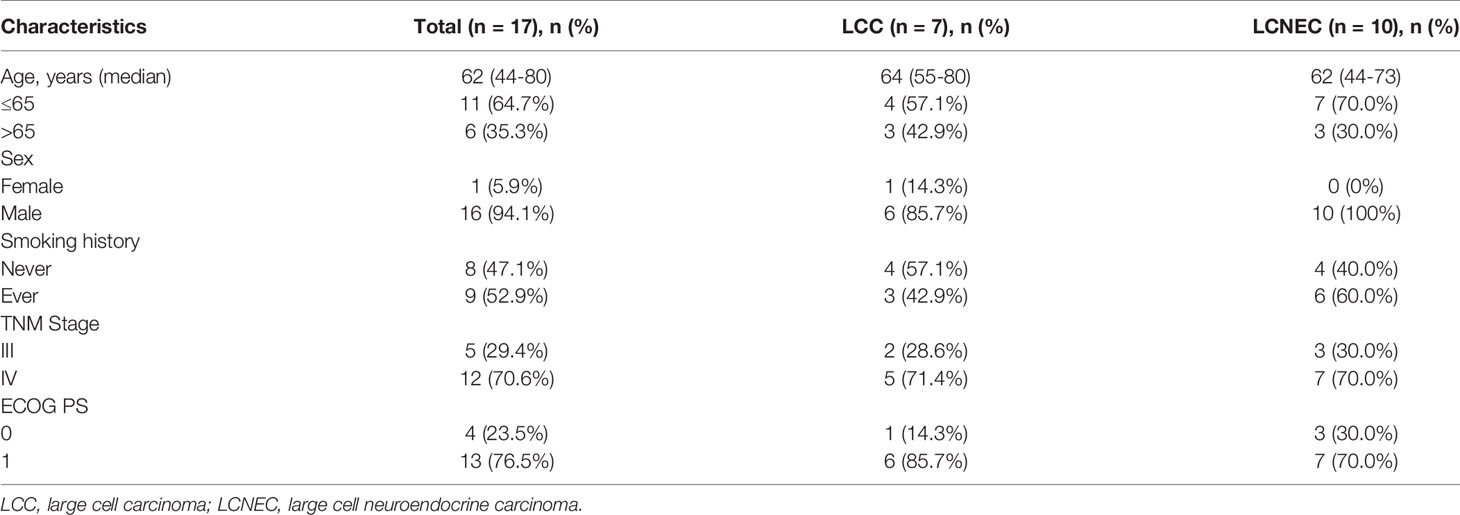
In this study, 17 patients were finally enrolled through strict screening, the clinical characteristics of these patients were summarized in Table 1. This study cohort includes ten patients with LCNEC and seven with LCC, which topical pathological images were shown in Supplementary Figure S1. The LCC and LCNEC cohorts were similar in their baseline characteristics. The median age of diagnosis of the study cohort was 62 years (LCC, 64 years; LCNEC, 62 years). There was an overall enrichment of male patients, with only one female patient in the LCC cohort. No clear association with smoking history was seen in either of the histological subtype. Stage IV patients accounted for about 70.6% of the patients included in the analysis, and stage III patients accounted for 29.4%. The correlation was analyzed between TNM stage and survival separately, and no correlation were found (p=0.420, HR 0.56, 95% CI, 0.13~2.34) (Supplementary Figure S2).
Considering only the overlapping genes in the two types of panels used in the study (Methods), the top 20 altered genes in the study cohort were shown in Figure 1, with LCC and LCNEC showed distinct mutation profiles. TP53 was the most highly altered gene in the two histologic subgroups. We observed an enrichment of TP53/RB1 co-mutations in a subset of LCNEC patients as previously reported (4, 6). In contrast, the co-occurrence of TP53 and RB1 mutations were rare in patients with classic LCC. We also noted that mutations in SMARCA4 and KEAP1 were exclusively detected in patients with classic LCC, whereas RICTOR, BRAF, ROS1 and TET2 mutations were only detected in those with LCNEC. BRAF alterations include gene copy number variation (CNV) (patient #2) and p.G460R (patient #13). ROS1 alterations include p.F1828L (patient #5) and p.Y1696C (patient #10).
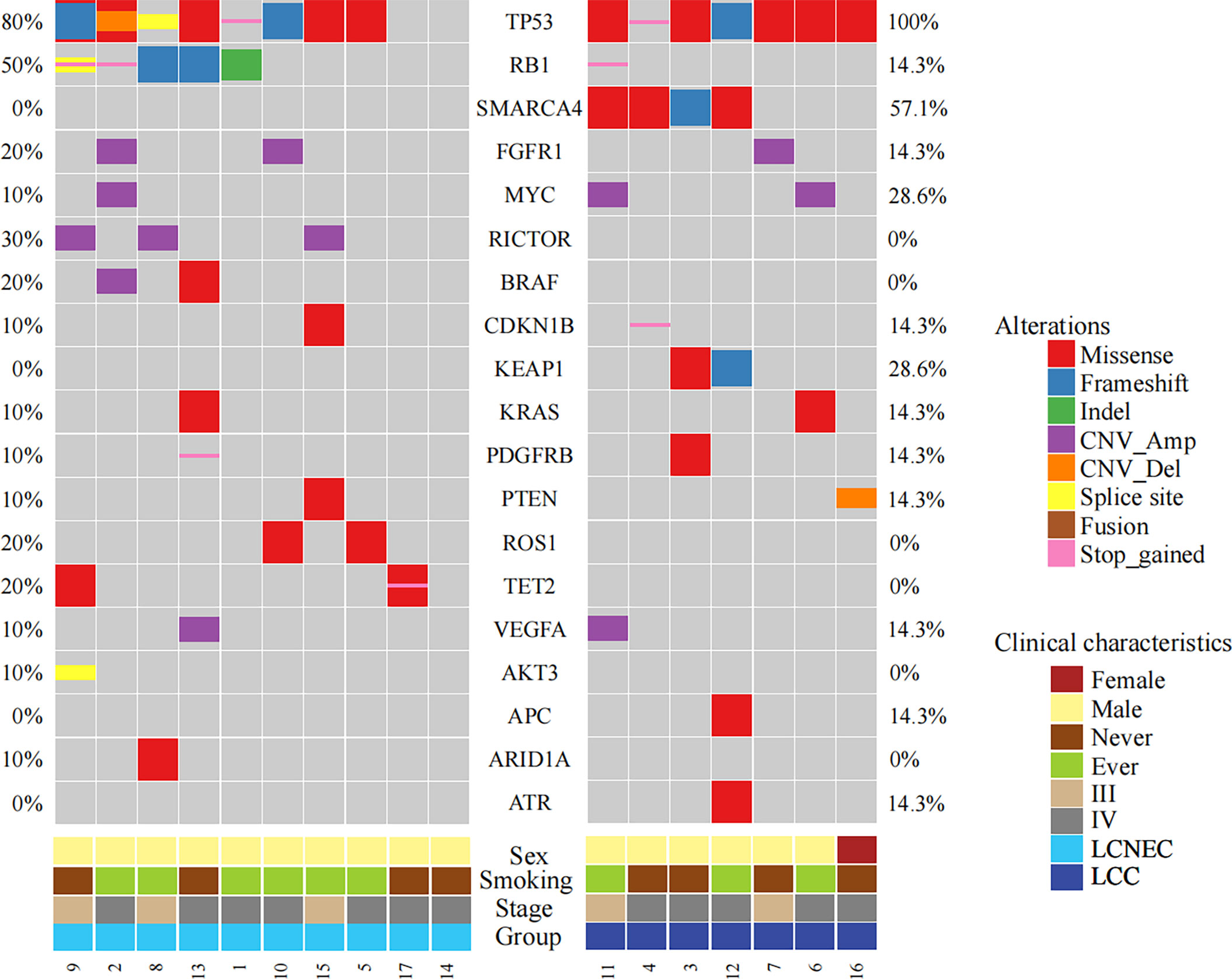
Figure 1 Mutational profiles comparing LCC and LCNEC. The top frequently mutated genes in the study cohort were shown with mutation frequencies in each subgroup indicated.
Several potentially targetable mutations were also identified, including mutations in KRAS and RET. KRAS mutations were detected in one LCC patient (p.G12V) and one LCNEC patient (p.G12C). We also identified a KIF5B-RET fusion gene in one case of LCNEC patient (patient #14).
Association Analysis of Survival
Next, we explored for biomarkers that might be associated with survival in our study cohort. Comparing the two subgroups of patients, those with LCNEC showed longer survival than those with LCC (p=0.006, Figure 2A). Mutational analysis revealed several genes might be associated with differential survival outcome between the two groups. Mutations in KEAP1 (p=0.035, Figure 2B) and SMARCA4 (p<0.001, Figure 2C), which were detected only in the LCC group, were associated with poor survival. On the other hand, mutations in RB1 (p=0.29, Figure 2D) and BRAF (p=0.17, Figure 2E), which were enriched in the LCNEC group, showed trends of increased survival.
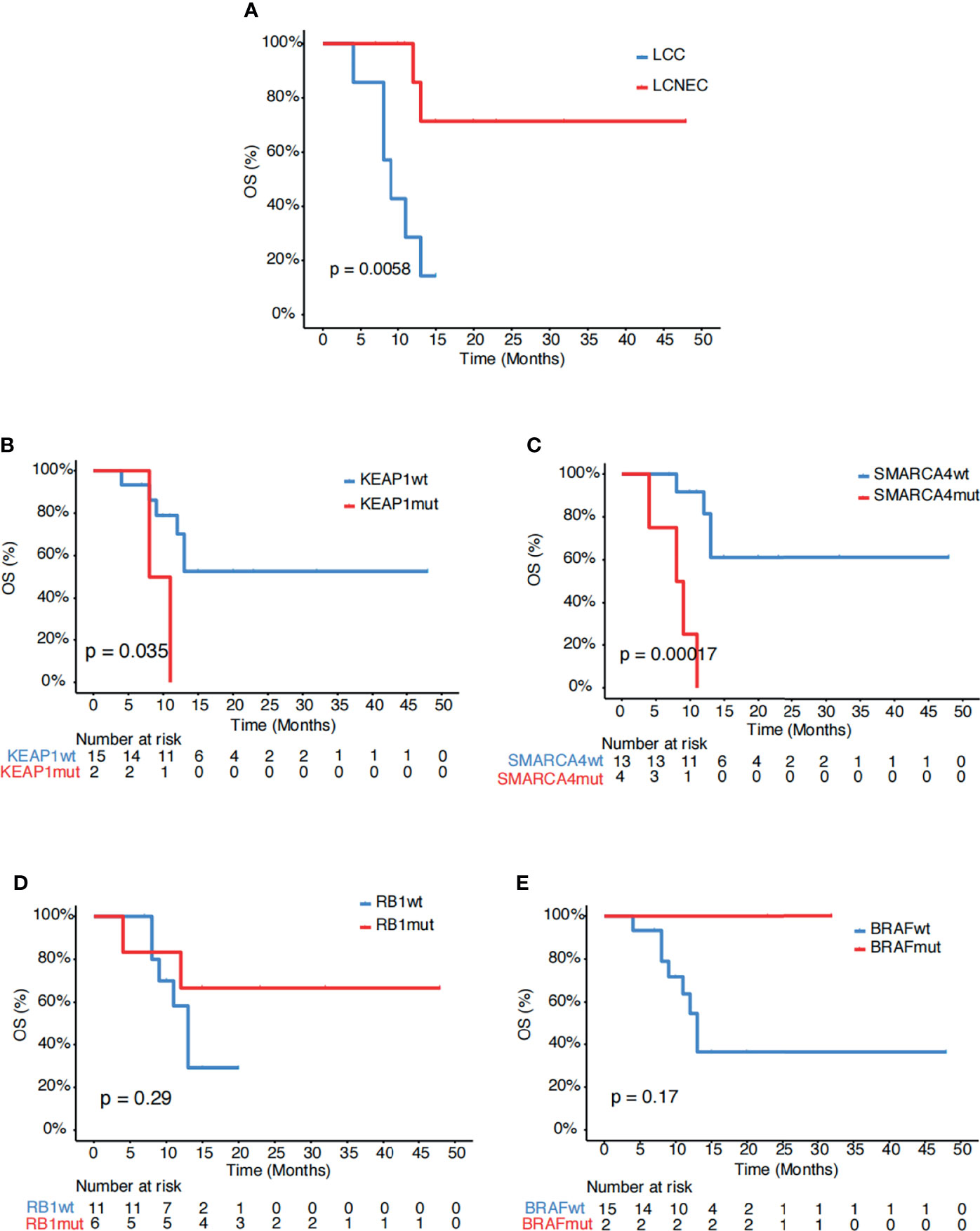
Figure 2 Associations of survival with genetic mutations. (A) Kaplan-Meier estimates of OS comparing LCC and LCNEC. (B–E) Kaplan-Meier estimates of OS comparing the subgroups with and without mutations in (B) KEAP1, (C) SMARCA4, (D) RB1 and (E) BRAF.
Further analysis showed that SMARCA4 mutations (p=0.13, Figure 3A), but not KEAP1 mutations (p=0.64, Figure 3B) remained associated with poor survival within the LCC subgroup. Within the LCNEC subgroup, BRAF mutations (p=0.34, Figure 3C), but not RB1 mutations (p=0.56, Figure 3D), still showed a trend of increased survival. In addition, mutations in TET2 were associated with poor outcome in the LCNEC subgroup (p=0.014, Figure 3E). No other genetic alterations showed associations with survival in this study cohort. Systemic treatment received in our study is depicted in Supplementary Table S1.
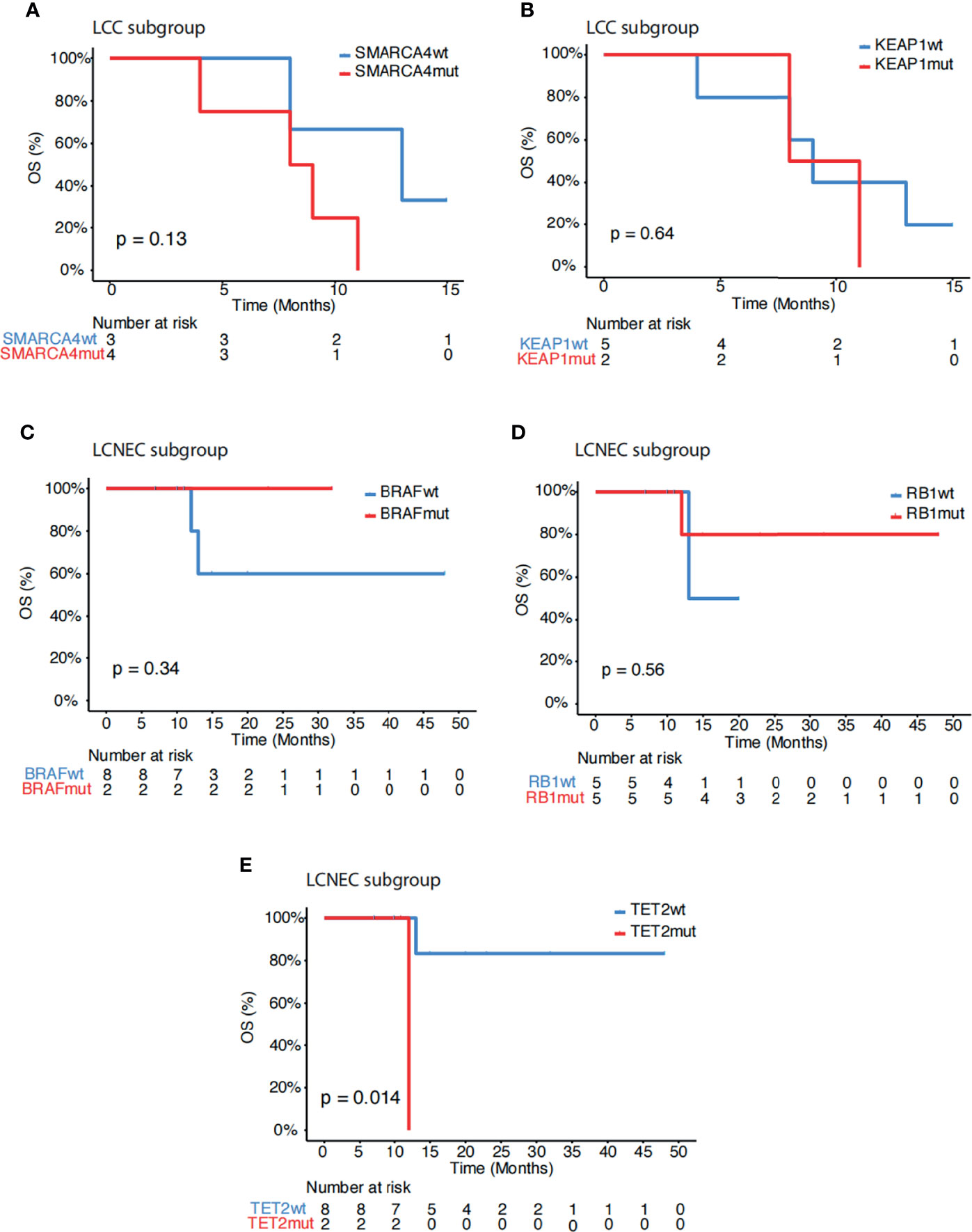
Figure 3 Subgroup association analysis of survival. (A, B) Kaplan-Meier estimates of OS comparing LCC patients with and without mutations in (A) SMARCA4 and (B) KEAP1. (C–E) Kaplan-Meier estimates of OS comparing LCNEC patients with and without mutations in (C) BRAF, (D) RB1 and (E) TET2.
Discussion
In this study, we examined the mutational profiles of LCC and LCNEC patients and conducted association analysis of survival for the identification of potential biomarkers of diagnostic and prognostic value. LCC and LCNEC are two subtypes of lung cancer of heterogenous nature and limited treatment options. Due to relatively difficult diagnosis, detailed pathological diagnosis was finally confirmed by experienced pathologists in this study. The number of cases included in the study was relatively small but ensured the authenticity of subsequent genetic testing results. In this study, we identified several targetable mutations in both LCC and LCNEC, including KRAS G12 mutations and a RET fusion gene. This finding suggests that genetic profiling might be necessary in such patients as it might provide more therapeutic options which is available using, tumor DNA or cell-free DNA (cfDNA) to monitor these mutations (14).
While the number of patients in our study was not large, all patients received platinum-based first-line therapy in both LCNEC and LCC. In additional, our results are similar to the observations in reported study (15), 52.9% of patients received second-line chemotherapy, which was also platinum-based. In contrast to another previous report (2), we found that LCNEC patients had better survival compared to patients with classic LCC, which might be associated with the enrichment of RB1 and BRAF mutations in the LCNEC group and the enrichment of SMARCA4 and KEAP1 mutations in the LCC group. Previous studies have shown that KEAP1 mutations were known to be a typical characteristic of NSCLC-like LCNEC, and more likely to be resistant to chemotherapy and EGFR-TKI drugs (6, 16). LCNEC samples suggest that LCNEC can be subdivided into different subtypes, partly clustering with SCLC. By comparative analysis of 69 LCNECs and 110 SCLCs, George et al. have found that despite their mutational patterns, LCNECs with KEAP1 mutations exhibit a neuroendocrine profile with closest similarity to SCLC tumors (17). Rekhtman et al. also showed that significantly higher rate of KEAP1 alterations (33%) was found in SCLC-like LCNEC, differed from SCLC with 5% of KEAP1 alterations (4). The incidence of KEAP1 mutation is not high, especially in the Chinese lung cancer patients, with the reported incidence of about 20%. KEAP1 mutation was only found in LCC patients, but not in LCNEC in this study. Possible reasons are the small sample, and the neuroendocrine profile with closest similarity to SCLC in the LCNEC subtype. This is consistent with the poor survival of LCC patients with KEAP1 mutations shown in our data.
Within the LCC subgroup, the presence of SMARCA4 mutations remained associated with poor survival outcome. SMARCA4, encoding the BRG1 protein, participates in the chromatin remodeling process and DNA repair and is frequently mutated in lung cancer (18, 19). Low expression of SMARCA4 has been associated with worse prognosis in patients with non-small-cell lung cancer (18). Co-mutations occurred more frequently with SMARCA4 mutations than with SMARCA4 wild-type tumors. In our data, four cases were co-mutated with TP53 and two cases were co-mutated with KEAP1, and also the co-mutated patients showed shorter survival time. Recent studies have found that treatment with immune checkpoint inhibitors is associated with improved outcomes in patients with SMARCA4 mutated, suggesting that SMARCA4 mutated lung cancer may be more sensitive to immunotherapy. SMARCA4 mutation detection may be required in our future studies to further explore its correlation with immunotherapy (20). Within the LCNEC group, BRAF mutations showed a trend of increased survival. In addition, TET2 mutations might serve as a negative prognostic marker in the LCNEC subgroup.
Conclusion
In summary, we reported potentially targetable mutations in both LCC and LCNEC, and identified several novel genetic alterations that might serve for diagnostic and prognostic purposes. Given the relative uncommon nature of LCC and LCNEC and therefore the limited sample size in our cohort, the diagnostic and prognostic role of the reported genes would require further investigations in larger-sample cohorts.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Genome Variation Map, National Genomics Data Center, GVM000288.
Ethics Statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
XD contributed to study design and implementation. YC was the principal investigator and wrote the report. Sample collection was done by XC, DW, and YC. GX advised on the trial. Analysis and interpretation of data were done by YC, LC, MX, and LS. Writing, review, and/or revision of the manuscript were done by YC. All authors contributed to the interpretation of data. All authors contributed to the article and approved the submitted version.
Conflict of Interest
Author DW was employed by Nanjing Geneseeq Technology Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.664397/full#supplementary-material
Supplementary Figure S1 | Topical pathological images for both LCNEC and LCC of the cases in this study. Pathological examination of primary lesion biopsy. The LCC H&E staining images of patient #12 (A) and LCNEC H&E staining images of patient #11 (B) were shown. The magnificence of the images were 100X (Left) and 400X (Right) for both two patients, respectively
Supplementary Figure S2 | Kaplan-Meier curve of OS in strata of different disease stages.
References
1. Pelosi G, Fabbri A, Papotti M, Rossi G, Cavazza A, Righi L, et al. Dissecting Pulmonary Large-Cell Carcinoma by Targeted Next Generation Sequencing of Several Cancer Genes Pushes Genotypic-Phenotypic Correlations to Emerge. J Thorac Oncol (2015) 10:1560–9. doi: 10.1097/JTO.0000000000000658
2. Sun YH, Lin SW, Hsieh CC, Yeh YC, Tu CC, Chen KJ. Treatment Outcomes of Patients With Different Subtypes of Large Cell Carcinoma of the Lung. Ann Thorac Surg (2014) 98:1013–9. doi: 10.1016/j.athoracsur.2014.05.012
3. Travis WD, Brambila E, Burke AP, Marx A, Nicholson AG. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. 4th ed. Lyon, France: IARC Press (2015).
4. Rekhtman N, Pietanza MC, Hellmann MD, Naidoo J, Arora A, Won H, et al. Next-Generation Sequencing of Pulmonary Large Cell Neuroendocrine Carcinoma Reveals Small Cell Carcinoma–Like and Non–Small Cell Carcinoma–Like Subsets. Clin Cancer Res (2016) 22:3618–29. doi: 10.1158/1078-0432.CCR-15-2946
5. Zhou Z, Zhu L, Niu X, Shen S, Zhao Y, Zhang J, et al. Comparison of Genomic Landscapes of Large Cell Neuroendocrine Carcinoma, Small Cell Lung Carcinoma, and Large Cell Carcinoma. Thorac Cancer (2019) 10:839–47. doi: 10.1111/1759-7714.13011
6. George J, Walter V, Peifer M, Alexandrov LB, Seidel D, Leenders F, et al. Integrative Genomic Profiling of Large-Cell Neuroendocrine Carcinomas Reveals Distinct Subtypes of High-Grade Neuroendocrine Lung Tumors. Nat Commun (2018) 9(1):1048. doi: 10.1038/s41467-018-03099-x
7. Chan AW, Chau SL, Tong JH, Chow C, Kwan JSH, Chung LY, et al. The Landscape of Actionable Molecular Alterations in Immunomarker-Defined Large-Cell Carcinoma of the Lung. J Thorac Oncol (2019) 14:1213–22. doi: 10.1016/j.jtho.2019.03.021
8. Tong L, Ding N, Tong X, Li J, Zhang Y, Wang X, et al. Tumor-Derived DNA From Pleural Effusion Supernatant as a Promising Alternative to Tumor Tissue in Genomic Profiling of Advanced Lung Cancer. Theranostics (2019) 9:5532–41. doi: 10.7150/thno.34070
9. Yang Z, Yang N, Ou Q, Xiang Y, Jiang T, Wu X, et al. Investigating Novel Resistance Mechanisms to Third-Generation EGFR Tyrosine Kinase Inhibitor Osimertinib in Non-Small Cell Lung Cancer Patients. Clin Cancer Res (2018) 24:3097–107. doi: 10.1158/1078-0432.CCR-17-2310
10. Bolger AM, Lohse M, Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
11. Li H, Durbin R. Fast and Accurate Short Read Alignment With Burrows-Wheeler Transform. Bioinformatics (2009) 25:1754–60. doi: 10.1093/bioinformatics/btp324
12. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A Framework for Variation Discovery and Genotyping Using Next-Generation DNA Sequencing Data. Nat Genet (2011) 43:491–8. doi: 10.1038/ng.806
13. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: Somatic Mutation and Copy Number Alteration Discovery in Cancer by Exome Sequencing. Genome Res (2012) 22:568–76. doi: 10.1101/gr.129684.111
14. Zhuo M, Guan Y, Yang X, Hong L, Wang Y, Li Z, et al. The Prognostic and Therapeutic Role of Genomic Subtyping by Sequencing Tumor or Cell-Free DNA in Pulmonary Large-Cell Neuroendocrine Carcinoma. Clin Cancer Res (2020) 26(4):892–901. doi: 10.1158/1078-0432.CCR-19-0556
15. Fisch D, Bozorgmehr F, Kazdal D, Kuon J, Klotz LV, Shah R, et al. Comprehensive Dissection of Treatment Patterns and Outcome for Patients With Metastatic Large-Cell Neuroendocrine Lung Carcinoma. Front Oncol (2021) 11:673901. doi: 10.3389/fonc.2021.673901
16. Frank R, Scheffler M, Merkelbach-Bruse S, Ihle MA, Kron A, Rauer M, et al. Clinical and Pathological Characteristics of KEAP1- and NFE2L2-Mutated Non-Small Cell Lung Carcinoma (NSCLC). Clin Cancer Res (2018) 24:3087–96. doi: 10.1158/1078-0432.CCR-17-3416
17. Derks JL, Leblay N, Lantuejoul S, Dingemans AC, Speel EM, Fernandez-Cuesta L. New Insights Into the Molecular Characteristics of Pulmonary Carcinoids and Large Cell Neuroendocrine Carcinomas, and the Impact on Their Clinical Management. J Thorac Oncol (2018) 13:752–66. doi: 10.1016/j.jtho.2018.02.002
18. Bell EH, Chakraborty AR, Mo X, Liu Z, Shilo K, Kirste S, et al. SMARCA4/BRG1 Is a Novel Prognostic Biomarker Predictive of Cisplatin-Based Chemotherapy Outcomes in Resected Non-Small Cell Lung Cancer. Clin Cancer Res (2016) 22:2396–404. doi: 10.1158/1078-0432.CCR-15-1468
19. Medina PP, Romero OA, Kohno T, Montuenga LM, Pio R, Yokota J, et al. Frequent BRG1/SMARCA4-Inactivating Mutations in Human Lung Cancer Cell Lines. Hum Mutat (2008) 29:617–22. doi: 10.1002/humu.20730
Keywords: large cell carcinoma, SMARCA4, KEAP1, BRAF, RB1
Citation: Chen Y, Cui X, Wang D, Xia G, Xing M, Cheng L, Sheng L and Du X (2022) Molecular Characterization and Prognostication of Large Cell Neuroendocrine Carcinoma and Large Cell Carcinoma. Front. Oncol. 11:664397. doi: 10.3389/fonc.2021.664397
Received: 05 February 2021; Accepted: 27 December 2021;
Published: 14 January 2022.
Edited by:
Shinkichi Takamori, National Hospital Organization Kyushu Cancer Center, JapanReviewed by:
Petros Christopoulos, Heidelberg University Hospital, GermanyShigeki Umemura, National Cancer Center Hospital East, Japan
Copyright © 2022 Chen, Cui, Wang, Xia, Xing, Cheng, Sheng and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianghui Du, ZHVfeGlhbmdodWlAMTYzLmNvbQ==
 Ying Chen1
Ying Chen1 Xiaoying Cui
Xiaoying Cui Di Wang
Di Wang Minyan Xing
Minyan Xing