- 1Department of Head and Neck Surgery, Perking University Cancer Hospital and Institute, Beijing, China
- 2Department of Pathology, Peking University First Hospital, Beijing, China
- 3Department of Radiology, Massachusetts General Hospital, Boston, MA, United States
- 4Center for Cancer Research, Massachusetts General Hospital and Harvard Medical School, Boston, MA, United States
- 5Department of Otorhinolaryngology-Head and Neck Surgery, Peking University First Hospital, Beijing, China
Background/Aim: Previous literature has implicated the sustained expression of FOXM1 in numerous human cancers, including head and neck squamous cell carcinoma (HNSCC). The current study aimed to elucidate the function and regulatory mechanism of FOXM1 in HNSCC.
Methods: Western blot and RT-qPCR methods were performed to evaluate the expression of Linc-ROR, FOXM1, and LMO4 in HNSCC tissue samples and cells. The binding between FOXM1 and Linc-ROR was analyzed using a ChIP assay. Various cellular processes including proliferation and invasion abilities were assessed following alteration of FOXM1, Linc-ROR and LMO4 expression in HNSCC cells. Xenograft mouse models were established to validate the in vitro findings.
Results: Linc-ROR and FOXM1 were highly expressed in HNSCC tissues and cells. FOXM1 operated as a potential transcription factor to bind to the promoter region of Linc-ROR. Linc-ROR and FOXM1 exhibited high expression levels in both the clinical tissue samples as well as the HNSCC cells, which could facilitate the proliferation and invasion of HNSCC cells. Linc-ROR upregulated the expression of LMO4 and promoted activation of the AKT/PI3K signaling pathway, thus stimulating the proliferation and invasion of HNSCC cells. Silencing of Linc-ROR brought about a contrasting effect relative to that seen when FOXM1 was overexpressed in HNSCC in vivo.
Conclusions: Overall, FOXM1 promoted the expression of Linc-ROR and induced the activation of the LMO4-dependent AKT/PI3K signaling pathway, thus facilitating the occurrence and development of HNSCC.
Introduction
Head and neck squamous cell carcinoma (HNSCC) represents a malignancy encompassing a heterogeneous population of cancer cells that generally arise from the squamous epithelium of the oral cavity, oropharynx, larynx, as well as hypopharynx (1). HNSCC can be divided into two distinct subgroups, human papillomavirus (HPV)-positive HNSCC and HPV-negative HNSCC, based on patient HPV infection history (1). HPV-positive HNSCC is typically seen in young adults and non-smokers (2). HNSCC has been strongly associated with tobacco smoking (3). Studies have indicated that the annual incidence of HNSCC is approximately 800,000 new cases globally (4). In addition to surgical management (5), chemotherapy remains the first-line approach for advanced cases of HNSCC, with agents such as platinum combined with cetuximab, nivolumab or pembrolizumab often employed (6, 7). Despite clinical success of the aforementioned modalities, the five-year survival rate remains unsatisfactory (8).
As a nuclear transcription factor regulating cell cycle genes, the sustained expression of forkhead box M1 (FOXM1) has been highlighted as a hallmark of nearly all human cancers (9). The epigenetic signature induced by FOXM1 has been speculated to possess the clinical translational potential to function as a biomarker for HNSCC, including early cancer screening, diagnosis, and therapy, as it can mimic the cancer epigenome (10). Moreover, the knockdown of FOXM1 has been reported to encumber the proliferation and invasion of HNSCC cells, along with a more differentiated-like phenotype in three-dimensional culture and Xenograft tumor models (11). In hepatocellular carcinoma cell lines, FOXM1 was previously observed to transcriptionally activate long intergenic non-protein coding RNA regulator of reprogramming (Linc-ROR) (12). Located on 18q21.31, Linc-ROR has been reported to contribute to tumor growth by means of regulating the reprogramming of plu ripotent stem cells (13). Interestingly, the over-expression of Linc-ROR has also been emphasized in undifferentiated tumors, with studies suggesting the existence of a strong correlation with tumor recurrence and poor response to treatment (14). Certain lncRNAs have been reported to facilitate cancer progression by positively regulating LIM-only protein 4 (LMO4) (15). LMO4 is a nuclear adapter that is a pivotal factor for the assembly of multi-protein transcriptional complexes and its dysregulated expression is characteristic of a number of epithelial malignancies, including that of HNSCC (16). LMO4 possesses the ability to promote tumor cell migration and invasion in non-small-cell lung cancer via the involvement of the protein kinase B/phosphatidylinositol 3-kinase (AKT/PI3K) signaling pathway (17). The AKT/PI3K signaling pathway often exhibits hyper activation in numerous types of cancers (18). Moreover, the enhanced activity of AKT/PI3K signaling pathway, which is often altered in HPV-positive and negative HNSCC, has been linked with poor patient prognosis (19). Herein, we hypothesized that FOXM1 might participate with Linc-ROR, LMO4 and the AKT/PI3K signaling pathway in the occurrence and progression of HNSCC. We subsequently performed a series of experiments to explore and determine this hypothesis.
Methods
Ethics Statement
The current study was performed with the approval of the Ethics Committee of Peking University First Hospital, and conducted following the Declaration of Helsinki. All participants signed written informed consent documentation prior to enrollment. All animal experiments were approved by the Institutional Animal Care and Use Committee of Peking University First Hospital. Extensive efforts were made to ensure minimal suffering of the included animals.
Bioinformatics Analysis
RNA-seq expression profiles of HNSCC were obtained from the TCGA database (https://xenabrowser.net/datapages/), including 502 HNSCC samples and 44 normal samples. R package limma was applied to analyze the expression of Linc-ROR and FOXM1 in both HNSCC samples as well as the normal samples. ChIP-seq data from ENCOD TFBS were explored and analyzed in the UCSC database (https://genome-asia.ucsc.edu/index.html) to predict transcription factors for Linc-ROR.
Clinical Sample Collection
Thirty-four patients with HNSCC admitted to Peking University First Hospital from January 2016 to March 2019 were recruited for the purposes of the current study. The HNSCC biopsy specimens and adjacent normal tissues were collected. Briefly, the tissues were surgically resected, promptly flash-frozen in liquid nitrogen and subsequently stored at -80°C. The age of the patients ranged from 29 to 73 years, with all patients confirmed to have not undergone any chemotherapy or radiotherapy treatment prior to surgery. Pathologic characteristics examined included age, sex, etiology, AFP, tumor size, tumor differentiation, and stage. Tumors were classified and graded based on the pathological tumor-node-metastasis (pTNM) staging advocated by the International League (20).
Cell Culture and Lentivirus Transfection
Human tongue squamous carcinoma cell line (TSCCA, from the website of microorganism for search, http://www.biobw.org/), normal oral mucosa epithelial cell line (HNOEC/HL-047, from the American Type Culture Collection Repository [ATCC], Manassas, VA, USA), and human embryonic kidney cell line (293T, from ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA) in an incubator at 37°C with 5% CO2. Transfection of Linc-ROR, FOXM1, LMO4, negative control shRNAs, and overexpression lentivirus (pSIH1-H1-copGFP) were purchased from GenePharma (Shanghai, China). The HEK293T cells were subsequently co-transfected with packaging virus and target vector, with the supernatant collected following 48 hours of cell culture. Viruses in the logarithmic growth phase were collected and divided into 1) control group, 2) short hairpin RNA-negative control (sh-NC) group, 3) sh-Linc-ROR-1 group, 4) sh-Linc-ROR-2 group, 5) sh-FOXM1-1 group, 6) sh-FOXM1-2 group, 7) sh-LMO4-1 group, 8) sh-LMO4-2 group, 9) overexpression (oe)-NC group, 10) oe-FOXM1 group, 11) oe-Linc-ROR group, 12) oe-LMO4 group, 13) sh-Linc-ROR + oe-LMO4 group, and 14) oe-FOXM1 + sh-Linc-ROR group. Cells were cultured in 6-well plates for transfection. After 48 h of infection, the expression of the related genes in each group of cells was detected using reverse transcription quantitative polymerase chain reaction (RT-qPCR). Each experiment was performed in triplicate.
RT-qPCR
Total RNA was isolated from cultured cells by TRIzol reagents (15596-018, Solarbio, Beijing, China), while first-strand complementary DNA synthesis was performed using a cDNA Reverse Transcript kit (Reanta, Beijing, China). The cDNA levels were then amplified using the ViiA 7 Real-Time PCR detection system (DaanGene, Guangzhou, China). The primers for the qPCR reactions were synthesized by Takara (Kusatsu, Japan) and listed in Table S1. Gene expression was determined in triplicate and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The relative expression of each target gene was calculated using the 2-ΔΔCT method. Each experiment was performed in triplicate.
Western Blot
Total protein was extracted from cells or tissues using high-performance RIPA lysis buffer (R0010, Solarbio), with the concentration determined using a bicinchoninic acid (BCA) assay kit (Yeasen, Shanghai, China). The proteins from each sample were then separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA) via the wet-transfer method. The membrane was blocked with 5% bovine serum albumin for 1 h at room temperature and incubated with primary antibodies against FOXM1 (ab180710, 1:1000, Abcam, Cambridge, UK), LMO4 (ab131030, 1:1000, Abcam), AKT (ab8805, 1:500, Abcam), PI3K (ab40776, 1:1000, Abcam), or phosphorylated (p)-PI3K (ab182651, 1:1000, Abcam), followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody (ab205718, 1:10000, Abcam) at room temperature for 1 h. Development was performed using VILBER FUSION FX5 (Vilber Lourmat, France). The protein bands were quantified using ImageJ 1.48 (National Institute of Health, Bethesda, MD, USA), and normalized to GAPDH. Each experiment was performed in triplicate.
Immunohistochemical (IHC) Staining
The HNSCC tissues were fixed in 4% paraformaldehyde for 12 h, cleared with xylene, and rehydrated using 100% ethanol, 95% ethanol and 75% ethanol. The tissues were then boiled in 0.01 M citrate buffer for 15-20 min, washed with phosphate-buffered saline (PBS), blocked with goat serum solution for 20 min followed by incubation with anti-FOXM1 antibody (ab180710, 1:200, Abcam) or anti-LMO4 antibody (ab229226, 1:100, Abcam) for 1 hour at room temperature. After rinsing in PBS, the tissue sections were incubated with SP-conjugated sheep anti-rabbit IgG for 1 h at room temperature followed by PBS washing. The reaction was terminated by washing with tap water for 10 min followed by 5-10 min of diaminobenzidine (DAB) development. The tissue sections were subsequently stained with hematoxylin for 2 min and hydrochloric acid alcohol, followed by washing with tap water for 10 min. After dehydration, transparency and mounting were performed, with the tissue sections analyzed under a bright-field Olympus BX-60 microscope.
In Situ Hybridization (ISH)
In situ hybridization was performed based on (21). Paraffin-embedded sections of tissues from HNSCC patients were deparaffinized and rehydrated. The sections were quenched for endogenous peroxidase activity purposes using 3% H2O2 for 30 min. After digestion with proteinase K for 5 min, the sections were fixed in 4% paraformaldehyde, rinsed in PBS, incubated in a hybridization buffer for 2 h at 60°C, then with Linc-ROR or scrambled control probe (50 nM digitoxin-labeled LNA probe, Exiqon, Vedbaek, Denmark) overnight at 60°C, and rinsed with washing buffer (50% formamide in 2× SSC and phosphate-buffered saline-Tween-20 [PBST]). Nitrotetrazolium was used for alkaline phosphate reactions. The sections were then sealed in a water-based mounting agent. Three fields of view were randomly selected and captured from each sample using a bright-field Olympus BX-60 microscope.
Xenograft Tumor in Nude Mice
BALA/c nude mice (4 weeks, 18-25 g), purchased from the SJA laboratory Animal (Hunan, China), were housed and maintained in a SPF environment. All mice were randomly separated into 5 groups: 1) oe-NC group (n = 15), 2) oe-FOXM1 group (n = 15), 3) sh-NC group (n = 15), 4) sh-Linc-ROR group (n = 15), and 5) oe-FOXM1 + sh-LincROR group (n = 15), respectively injected with 5 × 105 preconditioned TSCCA cells corresponding to the group. Volume measurement of the tumor was performed at the start of the 3rd day after injection, based on the following formula: V = (A × B2)/2 (A is the long diameter, B is the short diameter). At the end of the experiment, the mice were euthanized via carbon dioxide inhalation. A curve depicting the average volume at each time point was subsequently plotted.
Terminal Deoxynucleotidyl Transferase-Mediated dUTP-Biotin Nick End-Labeling (TUNEL) Staining
TUNEL was performed to detect the apoptosis of the nude mice. TUNEL staining was performed after the fixed-tissues (4% paraformaldehyde, overnight) had been embedded in paraffin and sectioned at a thickness of 5 mm. Five sections of the nude mouse tissues were dewaxed, followed by the addition of 50 μL of 1% proteinase K diluent, and incubated at 37°C for 30 min. The endogenous POD activity was then eliminated using 0.3% H2O2 methanol solution at 37°C for 30 min. The TUNEL reaction solution was added and incubated under dark conditions at 37°C for 1 h. Converter-POD was used at 37°C for 30 min. Finally, 2% DAB chromogenic solution was employed at room temperature for 15 min and observed under a OLYMPUS-CX43 microscope. In the event that brownish-yellow nuclei were observed, the reaction was terminated by distilled water. After hematoxylin counterstaining, and dehydration by gradient ethanol (50, 70, 90 and 100%), the sections were analyzed using a microscope. Ten fields were randomly taken from each section. The nuclei of the apoptotic cells were dyed brownish-yellow, while the nuclei of normal cells were dyed blue. The numbers of apoptosis cells in each field of view were calculated based on average findings.
Plate Colony Formation Assay
A monoclonal formation assay was employed to detect the proliferation of cells as well as different groups of TSCCA cells. The control cells at the logarithmic phase were digested with trypsin and counted. Using a 6-well plate, 1 × 103 TSCCA cells were seeded into each well with 2 mL DMEM with 10% FBS. After 7-10 days, the resulting colonies were fixed with methanol at room temperature for 15 min and subsequently stained with crystal violet. The colonies were then counted using a microscope (CX43, OLYMPUS, Japan). Each experiment was performed in triplicate.
3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium Bromide (MTT) Assay
MTT kit (C0009, Beyotime, Shanghai, China) was used to explore the proliferation of the TSCCA cells. After 24 h of transfection, 5 × 103 cells were seeded into 96-well plates in triplicate and cultured in DMEM containing 10% FBS. MTT reagent (5 mg/mL) was then added (20 μL to each well), after which the plates were incubated for 4 h. The absorbance of the supernatant was measured using a microplate reader (US6111636, Thermo Fisher Scientific) at 490 nm. The results were plotted as the average absorbance of each group/fold of control value.
Transwell Assay
ECM Matrigel (E1270-1ML, Sigma-Aldrich, St. Louis, MO, USA) was placed at 4°C overnight and diluted at a ratio of 1:9 with serum-free medium to a final concentration of 1 mg/mL. The diluted ECM (40 μL) was added to the polycarbonate membrane in the upper chamber of 24-well Transwell plate and incubated in 5% CO2, 37°C for 5 h. After ECM polymerized, the excess liquid was removed followed by the addition of 70 μL DMEM for incubation purposes in 5% CO2, 37°C for 0.5 h to rehydrate the Matrigel. Culturing in serum-free medium for 24 h, the TSCCA cells were resuspended in DMEM without FBS and seeded to the upper chamber of Transwell inserts. DMEM containing 10% FBS was added to the lower chamber. Following incubation for 24 h at 37°C, 5% CO2, the cells on the inserts and membranes were wiped off, and the remaining cells were fixed with methanol for 30 min and stained with 0.1% crystal violet staining solution for 20 min. Five fields were randomly selected under an inverted microscope (CKX41, Olympus, Tokyo, Japan) and the number of cells passing through the membrane was counted and averaged.
Cytoplasmic/Nuclear Fractionation Assay
Nuclear and cytoplasmic RNA or proteins were isolated with a nucleocytoplasmic isolation kit (K266-25, Biovision, San Francisco, CA, USA). The cells were washed twice with PBS (on ice) and then centrifuged at 500 ×g for 5 min. The cells were subsequently resuspended in cell fractionation buffer, incubated for 10 min (on ice) and centrifuged at 500 ×g for 5 min. The supernatant was then applied for purification of cytoplasmic RNA or protein. Nuclear fraction was washed with cell fractionation buffer and homogenized with cell disruption buffer (12).
Chromatin Immunoprecipitation (ChIP) Assay
The TSCCA cells were fixed with formaldehyde for 10 min to yield DNA-protein cross-linking, while an ultrasound disruptor (UP-250, Scientzbio, Ningbo, China) was used to break the cells and interrupt the chromatin into fragments (10 s ultrasound with 10 s interval, and cycled 15 times). The supernatant was then collected via centrifugation at 12000 rpm for 10 min, and divided into two tubes, which were incubated with NC rabbit IgG (ab1098489, 1:300, Abcam) or FOXM1 antibody (ab180710, 1:100, Abcam) overnight at 4°C. The DNA-protein complexes were purified with Agarose/Sepharose overnight at 65°C, and centrifuged at 12,000 g for 5 min in order to remove the supernatants. The non-specific complex was then washed, and the cross-linking was decrosslinked overnight at 65°C. The DNA fragments were extracted and purified with phenol/chloroform. The FOXM1 binding to Linc-ROR was detected by RT-qPCR using Linc-ROR-specific primers.
Dual Luciferase Reporter Assay
The 3’ untranslated regions (UTR) dual-luciferase reporter plasmid of FOXM1 and mutant plasmids mutated with Linc-ROR binding site (PmirGLO-linc-ROR-WT and PmirGLO-linc-ROR-MUT) were constructed, respectively. The oe-ROXM1 plasmid or negative control plasmid was subsequently co-transfected into HEK293T cells. After transfection for 24 h, the cells were lysed and centrifuged. The supernatant was then collected after which the luciferase activity was measured using a Dual-Luciferase Reporter Assay System (E1910, Promega, Madison, WI, USA). Each experiment was performed in triplicate (12).
Statistical Analysis
All data was analyzed using SPSS 21.0 software (IBM Corp. Armonk, NY, USA). Measurement data were expressed as the mean ± standard deviation. Paired t-test methods were applied for data comparison between cancer and adjacent normal tissues. Unpaired t-test methods were used for data comparison between two groups. Tukey’s test-corrected one-way analysis of variance (ANOVA) was applied during data comparison between multiple groups. Variables were analyzed at different time points using Bonferroni-corrected repeated measure ANOVA. p < 0.05 was considered to be indicative of statistical significance.
Results
FOXM1 and Linc-ROR Are Highly Expressed in Clinical Tissues of HNSCC Patients
Previous literature has suggested that Linc-ROR induces histone methylation to promote tumorigenesis (22, 23). A previous study presented evidence indicating that FOXM1 could regulate drug resistance in head and neck cancer cells through Linc-ROR (23). The differential analysis of the 546 HNSCC data in TCGA revealed a high expression of Linc-ROR and FOXM1 in HNSCC samples (Figures 1A, B). Thus, we asserted the hypothesis that Linc-ROR and FOXM1 could promote the progression of HNSCC. We initially detected the expression of FOXM1 and Linc-ROR in HNSCC and adjacent normal tissues from 34 HNSCC patients via RT-qPCR, with the results revealing that Linc-ROR and FOXM1 were highly expressed in HNSCC tissues when compared to that of the adjacent normal tissues (Figures 1C). The ISH and IHC results revealed markedly higher expression levels of Linc-ROR and FOXM1 in the HNSCC tissues when compared with that in the adjacent normal tissues (Figures 1D, E). The aforementioned findings demonstrated that Linc-ROR and FOXM1 were highly expressed in HNSCC.
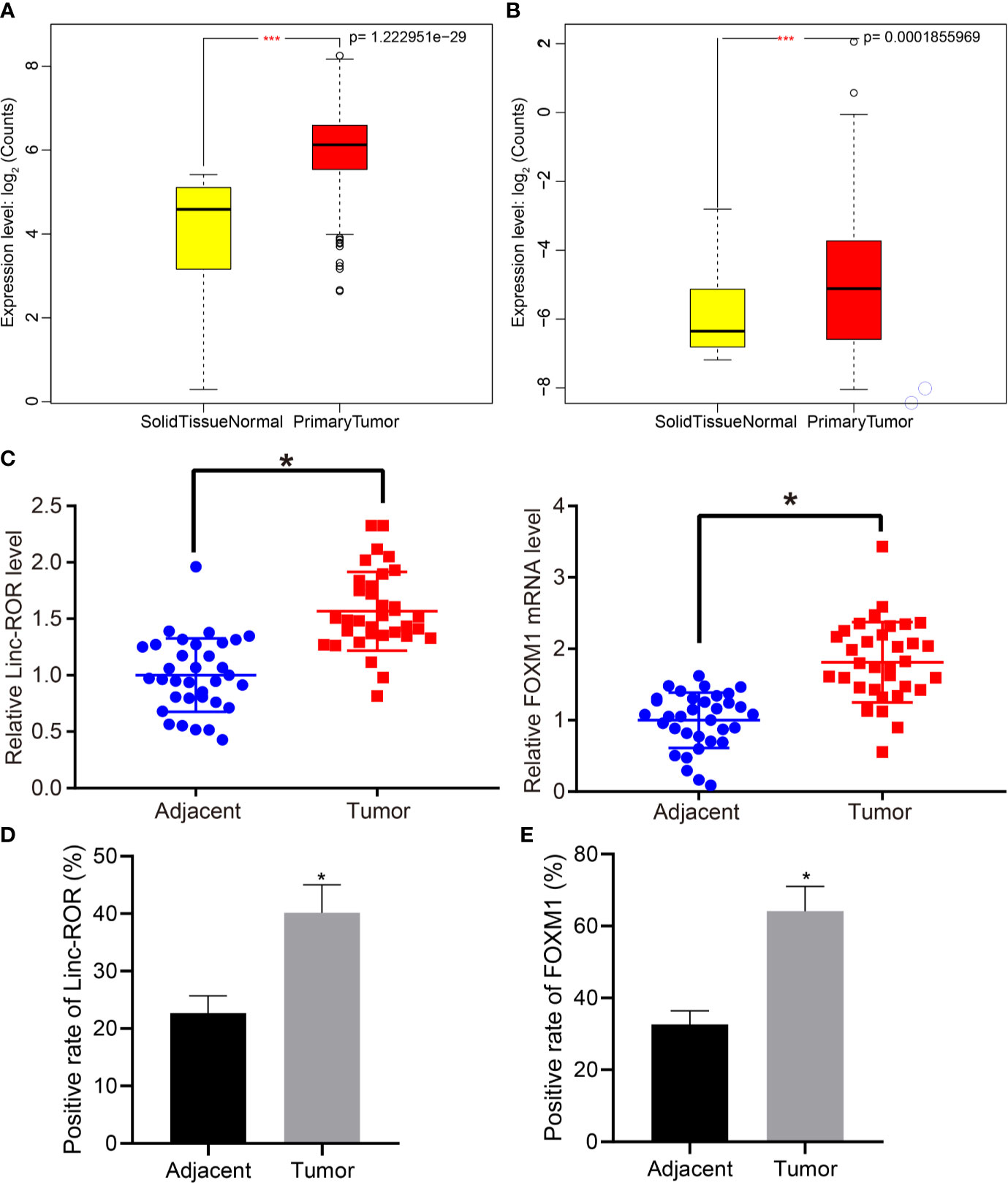
Figure 1 FOXM1 and Linc-ROR are highly expressed in clinical samples of HNSCC patients. (A) Expression of Linc-ROR of HNSCC samples in TCGA database; (B) Expression of FOXM1 in HNSCC samples in TCGA database; in (A, B), the abscissa axis indicates sample grouping, ordinate axis indicates gene expression count after log2 value; N = 546, *** indicates p < 0.001. (C) RT-qPCR detection of Linc-ROR and FOXM1 expression in HNSCC and adjacent normal tissues of 34 HNSCC patients; (D) Expression of Linc-ROR in cancer and adjacent normal tissues of 34 HNSCC patients determined by in situ hybridization; (E) Expression of FOXM1 in cancer and adjacent normal tissues of 34 HNSCC patients determined by IHC. *p < 0.05, compared with adjacent normal tissues.
Linc-ROR and FOXM1 Are Highly Expressed in HNSCC Cells and Promote HNSCC Cell Proliferation and Invasion
Next, to further determine whether Linc-ROR and FOXM1 influence the proliferation and invasion of HNSCC cells, we detected the expression of Linc-ROR and FOXM1 in HNSCC cell line TSCCA and the normal oral mucosal epithelial cell line (HNOEC/HL-047) using RT-qPCR and Western blot methods. The results revealed that the expression of Linc-ROR and FOXM1 were notably higher in TSCCA relative to that in HNOEC/HL-047 (Figures 2A, B). Additionally, the RT-qPCR and Western blot results indicated that expression of both Linc-ROR and FOXM1 were reduced in the TSCCA cells transfected with sh-Linc-ROR or sh-FOXM1, respectively (Figures 2C, D). The MTT and monoclonal formation and Transwell assay results revealed that the TSCCA cell proliferation, number of monoclonal formations as well as the invasive abilities were all significantly decreased following silencing of Linc-ROR or FOXM1, respectively (Figures 2E–J). Altogether, the results demonstrated that Linc-ROR and FOXM1 were highly expressed in HNSCC cells and promoted HNSCC cell proliferation and invasion.

Figure 2 Linc-ROR and FOXM1 are highly expressed in HNSCC cells and promote cell proliferation and invasion. (A) The expression of Linc-ROR and FOXM1 in HNOEC/HL-047 and TSCCA cells detected by RT-qPCR; (B) The protein expression of FOXM1 in HNOEC/HL-047 and TSCCA cells detected by Western blot; (C) The knockdown effects of Linc-ROR and FOXM1 detected by RT-qPCR; (D) The knockdown effects of FOXM1 detected by Western blot; (E) Proliferation ability of TSCCA cells after silencing Linc-ROR detected by monoclonal formation assay; (F) Proliferation ability of TSCCA cells after silencing FOXM1 by monoclonal formation assay; (G) Proliferation ability of TSCCA cells after silencing Linc-ROR detected by MTT assay; (H) Proliferation ability of TSCCA cells after silencing FOXM1 detected by MTT assay; (I) Invasion capacity of TSCCA cells after silencing Linc-ROR detected by Transwell assay; (J) Invasion capacity of TSCCA cells after silencing FOXM1 detected by Transwell assay. In panel (A, B), *p < 0.05, compared with the HNOEC/HL-047 cells, in panel (C–J), TSCCA cells transfected with sh-NC. All experiments were repeated three times.
FOXM1 Upregulates the Expression of Linc-ROR in TSCCA Cells
Previous literature has revealed that FOXM1 can promote the expression of Linc-ROR (12). In addition, retrieval on the USCS database suggests that FOXM1 acts as a transcription factor for Linc-ROR and binds to the promoter region of Linc-ROR (Supplementary Figure 1). Hence, we set out to further evaluate whether FOXM1 promoted the transcription of Linc-ROR in TSCCA cells in HNSCC. We isolated the nuclear and cytoplasmic and explored the localization of FOXM1 in TSCCA cells. FOXM1 was detected in both the nucleus and cytoplasm (Figure 3A). The RT-qPCR results revealed that upregulation of FOXM1 significantly promoted the expression of Linc-ROR, while the silencing of FOXM1 led to a significant inhibition of the expression in TSCCA cells (Figure 3B). Meanwhile, dual-luciferase reporter assay results demonstrated that upregulation of FOXM1 promoted the promoter activity of Linc-ROR in TSCCA cells (Figure 3C). Moreover, the results of ChIP assay demonstrated that FOXM1 bound to the Linc-ROR promoter directly in TSCCA and HNOEC/HL-047 cells (Figure 3D). The results suggested that FOXM1 upregulated Linc-ROR in the TSCCA cells.
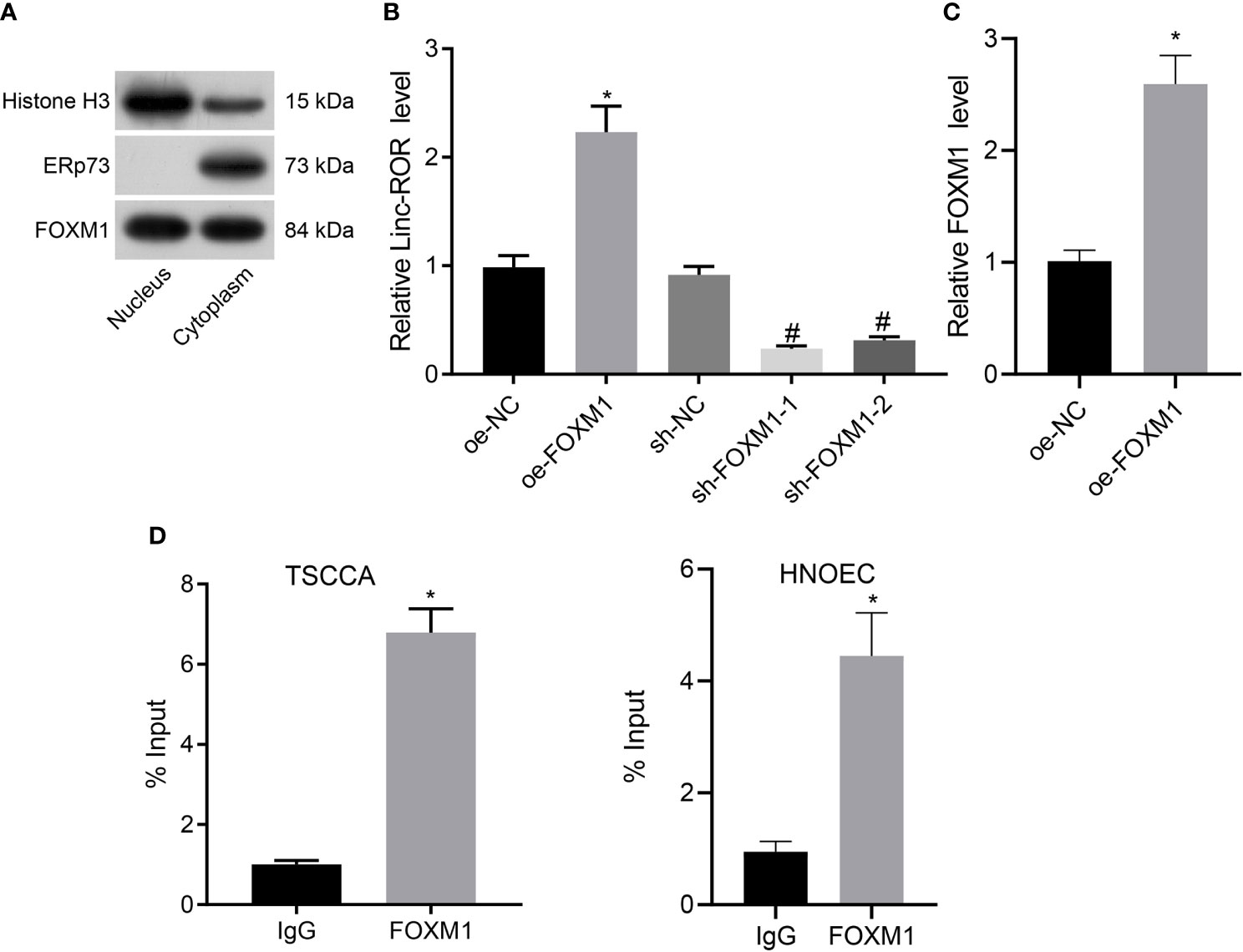
Figure 3 FOXM1 upregulates the expression of Linc-ROR in TSCCA cells. (A) Measurement of the enrichment degree of FOXM1 protein in the purified nucleus and cytoplasm. Histone H3 was considered as a nuclear marker, ERp72 was used as a cytoplasmic marker; (B) The expression of Linc-ROR in TSCCA cells by RT-qPCR, *p < 0.05, compared with the oe-NC group, #p < 0.05, compared with the sh-NC group; (C) Binding of FOXM1 to the promoter region of Linc-ROR in TSCCA cells determined by dual-luciferase reporter assay, *p < 0.05, compared with the oe-NC group; (D) The affinity of FOXM1 for the promoter of Linc-ROR in TSCCA and HNOEC/HL-047 cells determined by ChIP assay, and IgG as a negative control, *p < 0.05, compared with the IgG group. All experiments were repeated three times.
Linc-ROR Stimulates HNSCC Cell Proliferation and Invasion by Upregulating LMO4 Expression and Activating the AKT/PI3K Pathway
LMO4 has been reported to perform as a facilitator of tumor development by means activating the AKT/PI3K pathway, and moreover, LMO4 was reported as a target gene of Linc-ROR (17, 23). We hypothesized that Linc-ROR regulated HNSCC cell proliferation and invasion via the LMO4/AKT/PI3K pathway. For verification, we treated TSCCA cells with sequences of sh-Linc-ROR-1, sh-Linc-ROR-2, sh-LMO4-1 and sh-LMO4-2 and then detected their silencing efficiency by RT-qPCR. The results illustrated a decline in the expression of Linc-ROR in response to sh-Linc-ROR-1 or sh-Linc-ROR-2. The expression of LMO4 was also found to be decreased in the presence of sh-LMO4-1 or sh-LMO4-2. sh-Linc-ROR-1 and sh-LMO4-1 exhibited superior silencing efficiency (Supplementary Figure 2) and were thus selected for subsequent experiments. In addition, Linc-ROR and LMO4 exhibited high levels of expression in TSCCA cells overexpressing Linc-ROR while in response to oe-LMO4, the expression of LMO4 was increased while that of Linc-ROR was not significantly altered. The expression of Linc-ROR and LMO4 was found to be decreased in TSCCA cells treated with sh-Linc-ROR and moreover, LMO4 expression was reduced while Linc-ROR was not significantly changed in the presence of LMO4 silencing. Further, Linc-ROR and LMO4 were suppressed in TSCCA cells co-treated with sh-Linc-ROR and oe-LMO4 relative to oe-LMO4 treatment alone (Figure 4A). After overexpressing/silencing Linc-ROR and LMO4, PI3K inhibitor LY294002 was added to the TSCCA cells, and the expression of LMO4, AKT, PI3K, and the extent of AKT and PI3K phosphorylation were detected by Western blot. The results demonstrated that the expression of LMO4 and the extent of AKT and PI3K phosphorylation were increased in the presence of overexpressed LMO4 or Linc-ROR, while an opposite trend was detected in the absence of Linc-ROR or LMO4. In addition, combined treatment with sh-Linc-ROR-1 and oe-LMO4 led to a decline of LMO4 expression and the extent of AKT and PI3K phosphorylation, which was reversed in the presence of both oe-LMO4 and LY294002 (Figure 4B). The MTT and monoclonal formation assay results revealed that the cell proliferation ability was enhanced and the number of monoclonal formations was significantly increased following overexpression of Linc-ROR or LMO4, which was reversed in the absence of Linc-ROR or LMO4. Linc-ROR silencing and LMO4 overexpression simultaneously reduced the proliferation and the number of monoclonal formation. Combined treatment with oe-LMO4 and LY294002 induced higher proliferation and number of monoclonal formation than LY294002 treatment alone (Figures 4C, D). Transwell assay findings indicated that the cell invasion was increased following overexpression of Linc-ROR or LMO4 while an opposite result was obtained in the absence of Linc-ROR or LMO4. The simultaneous silencing of Linc-ROR and overexpression of LMO4 also reduced the invasion of cells while dual treatment with oe-LMO4 and LY294002 increased cell invasion (Figure 4E). These findings indicated that Linc-ROR facilitated the LMO4-mediated AKT/PI3K pathway activation thus promoting HNSCC cell proliferation and invasion.
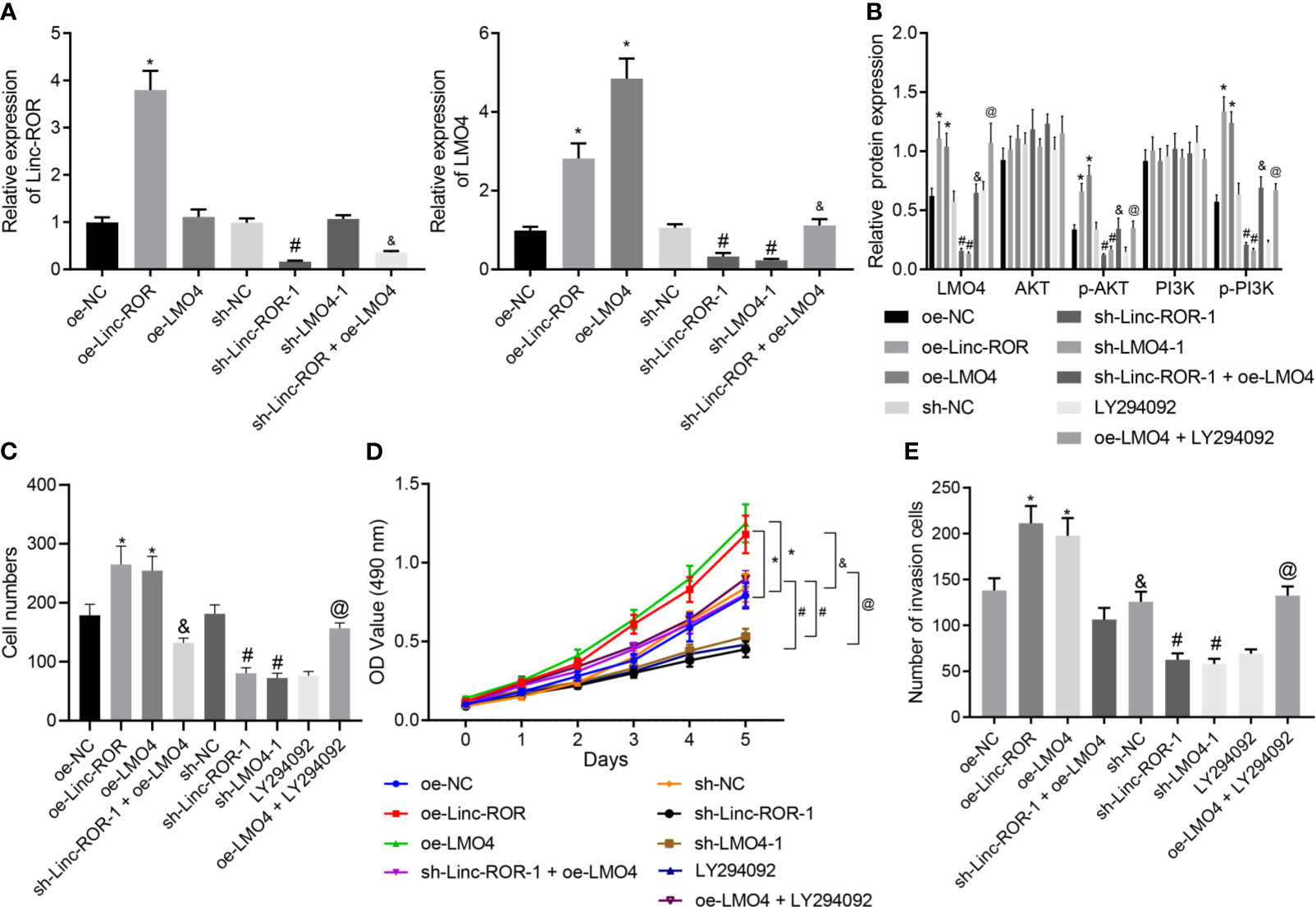
Figure 4 Linc-ROR promotes HNSCC cell proliferation and invasion via LMO4-mediated AKT/PI3K pathway activation. (A) The detection of Linc-ROR and LMO4 expression by RT-qPCR in TSCCA cells upon overexpression or silencing of LMO4 or Linc-ROR, sh-Linc-ROR + oe-LMO4, LY294002 or oe-LMO4 + LY294002. *p < 0.05, compared with the oe-NC group, #p < 0.05, compared with the sh-NC group; &p < 0.05, compared with the oe-LMO4 group; (B) Protein expression of LMO4, AKT, and PI3K along with p-AKT and p-PI3K in TSC7CA cells upon overexpression or silencing of LMO4 or Linc-ROR, sh-Linc-ROR + oe-LMO4, LY294002 or oe-LMO4 + LY294002. *p < 0.05, compared with the oe-NC group, #p < 0.05, compared with the sh-NC group; &p < 0.05, compared with the oe-LMO4 group; @p < 0.05, compared with the LY294002 group; (C) Proliferation of TSCCA cells measured by monoclonal formation assay upon overexpression or silencing of LMO4 or Linc-ROR, sh-Linc-ROR + oe-LMO4, LY294002 or oe-LMO4 + LY294002; *p < 0.05, compared with the oe-NC group, #p < 0.05, compared with the sh-NC group; &p < 0.05, compared with the oe-LMO4 group; @p < 0.05, compared with the LY294002 group; (D) Proliferation of TSCCA cells measured by MTT assay upon overexpression or silencing of LMO4 or Linc-ROR, sh-Linc-ROR + oe-LMO4, LY294002 or oe-LMO4 + LY294002. *p < 0.05, compared with the oe-NC group, #p < 0.05, compared with the sh-NC group; &p < 0.05, compared with the oe-LMO4 group; @p < 0.05, compared with the LY294002 group; (E), Invasion of TSCCA cells measured by Transwell assay upon overexpression or silencing of LMO4 or Linc-ROR, sh-Linc-ROR + oe-LMO4, LY294002 or oe-LMO4 + LY294002. *p < 0.05, compared with the oe-NC group, #p < 0.05, compared with the sh-NC group. &p < 0.05, compared with the oe-LMO4 group; @p < 0.05, compared with the LY294002 group. All experiments were repeated three times.
Silencing of Linc-ROR Reverses the Promoting Effect of Over-Expression of FOXM1 on Tumor Growth In Vivo
To further elucidate the mechanism of Linc-ROR regulating cell growth in HNSCC in vivo, we downregulated Linc-ROR and upregulated FOXM1 in TSCCA cells, and examined tumor growth by subcutaneous tumor formation assay. The results indicated that both tumor volume and weight were markedly elevated in the oe-FOXM1 group when compared to the oe-NC group, but decreased in the sh-Linc-ROR group compared with the sh-NC group; besides, tumor volume and weight were decreased in the oe-FOXM1 + sh-Linc-ROR group relative to the oe-FOXM1 group (Figures 5A, B). The RT-qPCR, Western blot and IHC results indicated that overexpression of FOXM1 upregulated the expression of Linc-ROR, LMO4, and Ki-67, while silencing of Linc-ROR downregulated the expression of LMO4 and Ki-67 as well as unchanged FOXM1 expression in the tumor tissues. The expression of FOXM1 was increased while that of Linc-ROR, LMO4, and Ki-67 was decreased in the tumor tissue of the mice in the oe-FOXM1 + sh-Linc-ROR group when compared with the oe-FOXM1 group (Figures 5C–F). The aforementioned results demonstrated that Linc-ROR knockdown could reverse the effects associated with the overexpression of FOXM1 which promoted tumor growth in vivo.
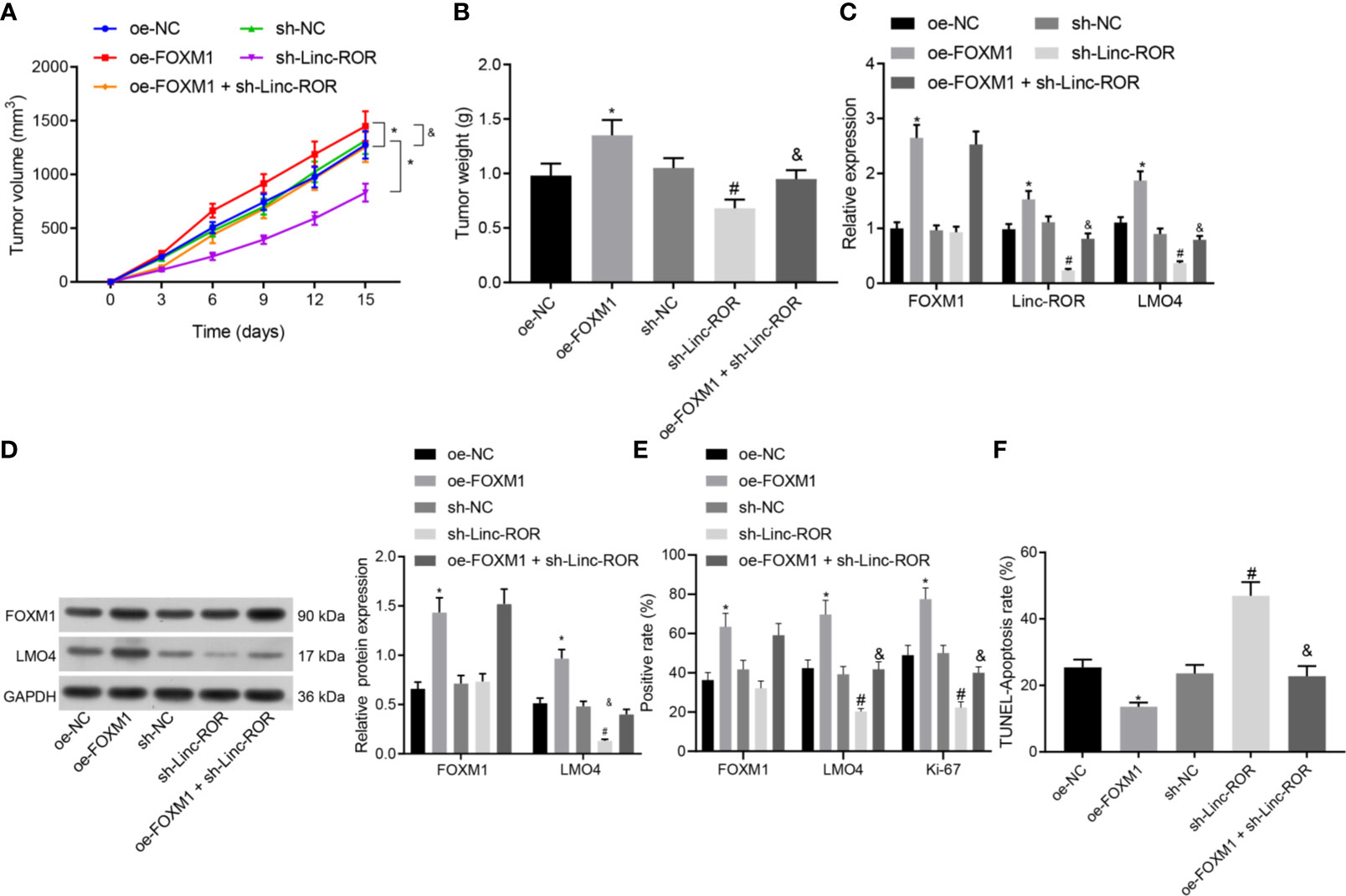
Figure 5 Silencing of Linc-ROR reverses the promoting effect of overexpressing FOXM1 on tumor growth in vivo. (A) Tumor growth curve; (B) Tumor size and weight; *p < 0.05, compared with the oe-NC group, #p < 0.05, compared with the sh-NC group; (C) Expression of FOXM1, Linc-ROR, and LMO4 in mouse tumor tissues determined by RT-qPCR; *p < 0.05, compared with the oe-NC group, #p < 0.05, compared with the sh-NC group; (D) Protein expression of FOXM1 and LMO4 in mouse tumor tissues detected by Western blot; *p < 0.05, compared with the oe-NC group, #p < 0.05, compared with the sh-NC group; &p < 0.05, compared with the oe-FOXM1 group; (E) The positive expression of FOXM1, LMO4, and Ki-67 proteins in mouse tumor tissues detected by IHC; (F) The apoptosis of cells in mouse tumor tissues detected by TUNEL assay. n = 15 for mice per group.
Discussion
HNSCC remains one of the most commonly occurring and lethal malignancies known to man, characterized by a high rate of tumor recurrence as well as a low rate of survival (24). This highlights the urgent need for more effective treatment strategies for HNSCC patients. Both our in vitro and in vivo experimental results highlighted the potential of FOXM1 to stimulate the occurrence and progression of HNSCC via Linc-ROR promotion and subsequent activation of the LMO4-dependent AKT/PI3K signaling pathway.
FOXM1 and Linc-ROR were found to be abundant in both the clinical samples of HNSCC patients as well as the cell lines. Additionally, evidence was obtained implicating these two factors in the acceleration of HNSCC cell invasion and migration. Similarly, FOXM1 has been reported to exhibit high levels of expression in tongue squamous cell carcinoma, illustrated by its contribution to the stimulation of the migration and invasion capacities of SCC9 and SCC25 cells (25). The overexpression of FOXM1 has been shown to promote cell proliferation and migration while acting to inhibit the apoptosis of hypopharyngeal squamous cell carcinoma, ultimately contributing to poor clinical prognosis (26). Linc-ROR has been highlighted to promote tumorigenesis by decoying gene-specific histone methylation (23). High levels of Linc-ROR were identified in oral squamous cell carcinomas tissues, often leading to poor treatment response and tumor recurrence (14).
Linc-ROR could be transcriptionally activated by FOXM1. The upregulation of Linc-ROR and FOXM1 has been reported to impair the sensitivity of hepatocellular carcinoma cells to sorafenib (12). The aforementioned report suggests a positive correlation between the expression of FOXM1 and the expression of Linc-ROR. Consistently, the results of our study revealed that FOXM1 could upregulate the expression of Linc-ROR in HNSCC cells, while the knockdown of Linc-ROR was shown to normalize the stimulatory effect associated with FOXM1 on tumor growth in the xenograft model of HNSCC cells. Since little progress has been made in the FOXM1/Linc-ROR signaling in the process of HNSCC, additional studies are still warranted to validate these findings.
Another key observation of the current study indicated that Linc-ROR could augment the expression of LMO4, leading to an increase in the activity of the AKT/PI3K signaling pathway. A recent study demonstrated that forced LMO4 expression could counteract breast cancer cell cycle redistribution and inhibit cell migration in vitro alongside tumor growth inhibition in vivo induced by siRNA-mediated lncRNA SNHG1 knockdown (15). Consistent with the findings of the current study, the expression of LMO4 has been reported to decrease in cells following the knockdown of Linc-ROR (23). Abundant levels of LMO4 have been highlighted in HNSCC cell lines with studies suggesting its contributory role in the metastasis, proliferation, and angiogenesis of HNSCC (16). In addition, LMO4 possesses the capacity to potentiate the proliferation and invasion of gastric cancer cells by means of activating the AKT/PI3K signaling pathway (27). Targeting the AKT/PI3K signaling pathway has been proposed as an effective therapeutic approach for patients with HNSCC since its suppression has been shown to trigger cell cycle arrest and apoptosis (28). Additionally, the crucial roles played by lncRNAs in connection with the AKT/PI3K signaling pathway have been widely documented in a numerous human cancers (29, 30). Linc-ROR has been identified as an upstream modulator or a downstream effector of major signaling pathways influencing endometrial cancer metastasis, including the AKT/PI3K signaling pathway (31). Linc-ROR has been demonstrated to trigger arsenite-transformed keratinocyte proliferation by inhibiting p53 activity and activating the AKT/PI3K signaling pathway (32). Mutations to the tumor-suppressor gene p53 represent one of the most common genomic alterations that take place in HNSCC with the mutational profile of p53 has been highlighted as an independent prognostic factor for HNSCC (33). Jin et al. proposed that p53 can directly bind to the lincRNA-p21 promoter and inhibit its expression, thus down-regulating the JAK2/STAT3 signaling pathway and subsequently inhibiting the progression of HNSCC (34). The activation of FOXM1 as well as the AKT/PI3K signaling pathway has been reported to aid in driving hepatic stellate cell activation as well as liver fibrosis (35). Based on the aforementioned information, we are further convinced that the up-regulation of FOXM1 functions as a promoter of HNSCC by means of regulating the Linc-ROR/LMO4/AKT/PI3K axis.
Conclusion
In conclusion, the key observations of our study highlight a novel mechanism by which FOXM1 serves as an oncogenic factor in the tumorigenesis of HNSCC via regulation of the Linc-ROR/LMO4/AKT/PI3K signaling cascade. The functional mechanism of FOXM1 unveiled in the present study may provide valuable insight for the development of new targets for HNSCC (Figure 6). Further investigations into the interaction between FOXM1, Linc-ROR, LMO4 and the AKT/PI3K signaling pathway are still required to further understand and elucidate the specific mechanisms associated with FOXM1 in HNSCC, so as to validate its applicable value from a clinical practice perspective.
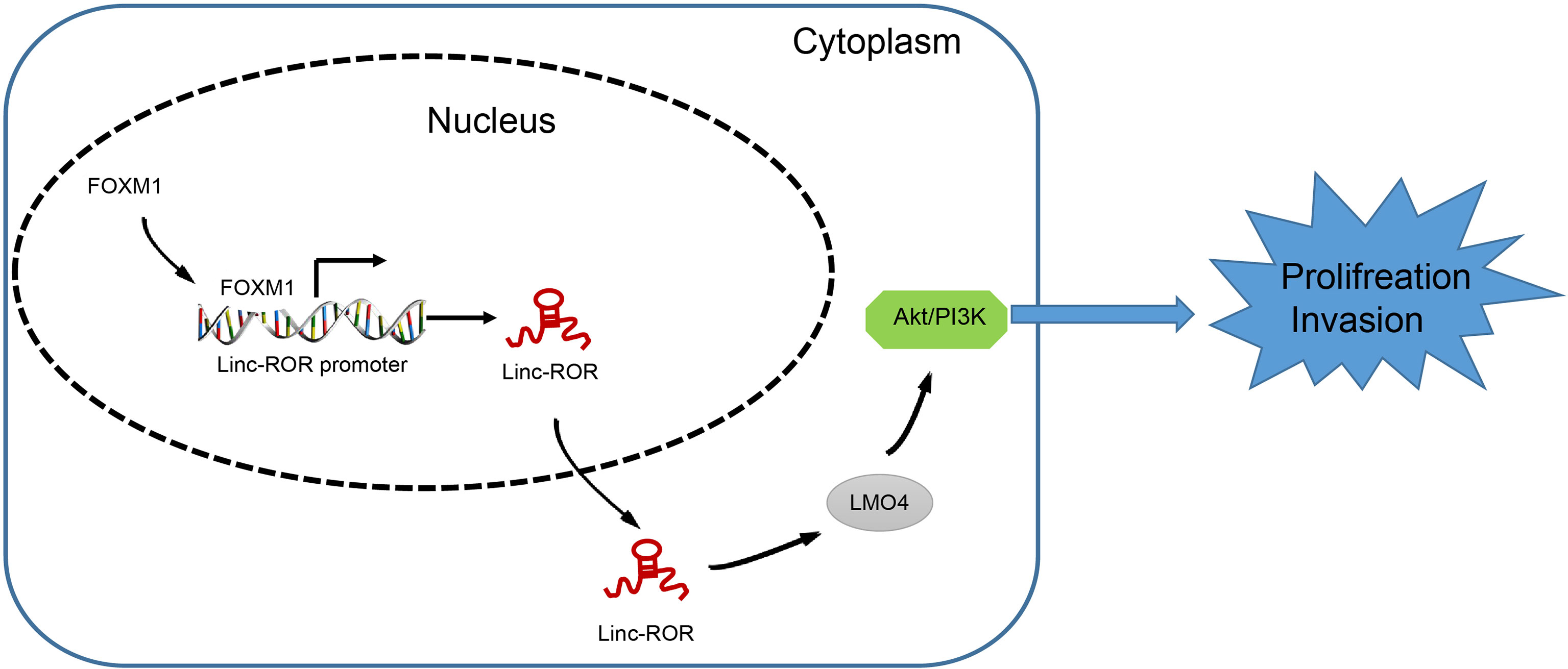
Figure 6 The mechanism of FOXM1 unveiled in the present study may provide an insight for the development of new targets for HNSCC.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Peking University First Hospital. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Ethics Committee of Peking University First Hospital.
Author Contributions
XM and HZ wrote the paper. QL and ES conceived the experiments. YQ and SX analyzed the data. TL collected and provided the sample for this study. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would also like to thank all participants enrolled in the present study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.658712/full#supplementary-material
Supplementary Figure 1 | FOXM1 bound to the promoter of Linc-ROR according to the UCSC genome database (gene assembly version hg19, DNA range: chr18: 54,729,703-54,747,249).
Supplementary Figure 2 | Silencing efficiency of sh-Linc-ROR-1, sh-Linc-ROR-2, sh-LMO4-1 and sh-LMO4-2 determined by RT-qPCR. *p < 0.05, compared with the sh-NC group.
References
1. Solomon B, Young RJ, Rischin D. Head and Neck Squamous Cell Carcinoma: Genomics and Emerging Biomarkers for Immunomodulatory Cancer Treatments. Semin Cancer Biol (2018) 52(Pt 2):228–40. doi: 10.1016/j.semcancer.2018.01.008
2. Alsbeih G, Al-Harbi N, Bin Judia S, Al-Qahtani W, Khoja H, El-Sebaie M, et al. Prevalence of Human Papillomavirus (HPV) Infection and the Association With Survival in Saudi Patients With Head and Neck Squamous Cell Carcinoma. Cancers (Basel) (2019) 11(6):820. doi: 10.3390/cancers11060820
3. El-Bayoumy K, Christensen ND, Hu J, Viscidi R, Stairs DB, Walter V, et al. An Integrated Approach for Preventing Oral Cavity and Oropharyngeal Cancers: Two Etiologies With Distinct and Shared Mechanisms of Carcinogenesis. Cancer Prev Res (Phila) (2020) 13(8):649–60. doi: 10.1158/1940-6207.CAPR-20-0096
4. Gazdzicka J, Golabek K, Strzelczyk JK, Kiczmer P, Miskiewicz-Orczyk K, Krakowczyk L, et al. Selected CDKN2A and MDM2 Polymorphisms in Oral Cavity Cancer. Acta Biochim Pol (2020) 67(2):213–8. doi: 10.18388/abp.2020_5195
5. Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, et al. Estimating the Global Cancer Incidence and Mortality in 2018: GLOBOCAN Sources and Methods. Int J Cancer (2019) 144(8):1941–53. doi: 10.1002/ijc.31937
6. Guigay J, Tahara M, Licitra L, Keilholz U, Friesland S, Witzler P, et al. The Evolving Role of Taxanes in Combination With Cetuximab for the Treatment of Recurrent and/or Metastatic Squamous Cell Carcinoma of the Head and Neck: Evidence, Advantages, and Future Directions. Front Oncol (2019) 9:668. doi: 10.3389/fonc.2019.00668
7. Galot R, Le Tourneau C, Guigay J, Licitra L, Tinhofer I, Kong A, et al. Personalized Biomarker-Based Treatment Strategy for Patients With Squamous Cell Carcinoma of the Head and Neck: EORTC Position and Approach. Ann Oncol (2018) 29(12):2313–27. doi: 10.1093/annonc/mdy452
8. Dioguardi M, Caloro GA, Laino L, Alovisi M, Sovereto D, Crincoli V, et al. Circulating miR-21 as a Potential Biomarker for the Diagnosis of Oral Cancer: A Systematic Review With Meta-Analysis. Cancers (Basel) (2020) 12(4):936. doi: 10.3390/cancers12040936
9. Black M, Arumugam P, Shukla S, Pradhan A, Ustiyan V, Milewski D, et al. FOXM1 Nuclear Transcription Factor Translocates Into Mitochondria and Inhibits Oxidative Phosphorylation. Mol Biol Cell (2020) 31(13):1411–24. doi: 10.1091/mbc.E19-07-0413
10. Teh MT, Gemenetzidis E, Patel D, Tariq R, Nadir A, Bahta AW, et al. FOXM1 Induces a Global Methylation Signature That Mimics the Cancer Epigenome in Head and Neck Squamous Cell Carcinoma. PloS One (2012) 7(3):e34329. doi: 10.1371/journal.pone.0034329
11. Roh V, Hiou-Feige A, Misetic V, Rivals JP, Sponarova J, Teh MT, et al. The Transcription Factor FOXM1 Regulates the Balance Between Proliferation and Aberrant Differentiation in Head and Neck Squamous Cell Carcinoma. J Pathol (2020) 250(1):107–19. doi: 10.1002/path.5342
12. Zhi Y, Abudoureyimu M, Zhou H, Wang T, Feng B, Wang R, et al. FOXM1-Mediated LINC-ROR Regulates the Proliferation and Sensitivity to Sorafenib in Hepatocellular Carcinoma. Mol Ther Nucleic Acids (2019) 16:576–88. doi: 10.1016/j.omtn.2019.04.008
13. Pentenero M, Bowers L, Jayasinghe R, Cheong SC, Farah CS, Kerr AR, et al. World Workshop on Oral Medicine VII: Functional Pathways Involving Differentially Expressed lncRNAs in Oral Squamous Cell Carcinoma. Oral Dis (2019) 25 (Suppl 1):79–87. doi: 10.1111/odi.13051
14. Arunkumar G, Deva Magendhra Rao AK, Manikandan M, Arun K, Vinothkumar V, Revathidevi S, et al. Expression Profiling of Long Non-Coding RNA Identifies linc-RoR as a Prognostic Biomarker in Oral Cancer. Tumour Biol (2017) 39(4):1010428317698366. doi: 10.1177/1010428317698366
15. Xiong X, Feng Y, Li L, Yao J, Zhou M, Zhao P, et al. Long Noncoding RNA SNHG1 Promotes Breast Cancer Progression by Regulation of LMO4. Oncol Rep (2020) 43(5):1503–15. doi: 10.3892/or.2020.7530
16. Simonik EA, Cai Y, Kimmelshue KN, Brantley-Sieders DM, Loomans HA, Andl CD, et al. LIM-Only Protein 4 (LMO4) and LIM Domain Binding Protein 1 (LDB1) Promote Growth and Metastasis of Human Head and Neck Cancer (LMO4 and LDB1 in Head and Neck Cancer). PloS One (2016) 11(10):e0164804. doi: 10.1371/journal.pone.0164804
17. Wang W, Wu S, Guo M, He J. LMO4 is a Prognostic Marker Involved in Cell Migration and Invasion in Non-Small-Cell Lung Cancer. J Thorac Dis (2016) 8(12):3682–90. doi: 10.21037/jtd.2016.12.22
18. Hemmat N, Mokhtarzadeh A, Aghazadeh M, Jadidi-Niaragh F, Baradaran B, Bannazadeh Baghi H. Role of microRNAs in Epidermal Growth Factor Receptor Signaling Pathway in Cervical Cancer. Mol Biol Rep (2020) 47(6):4553–68. doi: 10.1007/s11033-020-05494-4
19. Schotz U, Balzer V, Brandt FW, Ziemann F, Subtil FSB, Rieckmann T, et al. Dual PI3K/mTOR Inhibitor NVP-BEZ235 Enhances Radiosensitivity of Head and Neck Squamous Cell Carcinoma (HNSCC) Cell Lines Due to Suppressed Double-Strand Break (DSB) Repair by Non-Homologous End Joining. Cancers (Basel) (2020) 12(2):467. doi: 10.3390/cancers12020467
20. Huang J, Ji EH, Zhao X, Cui L, Misuno K, Guo M, et al. Sox11 Promotes Head and Neck Cancer Progression via the Regulation of SDCCAG8. J Exp Clin Cancer Res (2019) 38(1):138. doi: 10.1186/s13046-019-1146-7
21. Liu H, Liu Y, Liu W, Zhang W, Xu J. EZH2-Mediated Loss of miR-622 Determines CXCR4 Activation in Hepatocellular Carcinoma. Nat Commun (2015) 25(6):8494. doi: 10.1038/ncomms9494
22. Hou L, Tu J, Cheng F, Yang H, Yu F, Wang M, et al. Long Noncoding RNA ROR Promotes Breast Cancer by Regulating the TGF-Beta Pathway. Cancer Cell Int (2018) 18(18):142. doi: 10.1186/s12935-018-0638-4
23. Fan J, Xing Y, Wen X, Jia R, Ni H, He J, et al. Long Non-Coding RNA ROR Decoys Gene-Specific Histone Methylation to Promote Tumorigenesis. Genome Biol (2015) 16(1):139. doi: 10.1186/s13059-015-0705-2
24. Begg AC. Predicting Recurrence After Radiotherapy in Head and Neck Cancer. Semin Radiat Oncol (2012) 22(2):108–18. doi: 10.1016/j.semradonc.2011.12.002
25. Yang H, Wen L, Wen M, Liu T, Zhao L, Wu B, et al. FoxM1 Promotes Epithelial-Mesenchymal Transition, Invasion, and Migration of Tongue Squamous Cell Carcinoma Cells Through a C-Met/AKT-Dependent Positive Feedback Loop. Anticancer Drugs (2018) 29(3):216–26. doi: 10.1097/CAD.0000000000000585
26. Chen Y, Liu Y, Ni H, Ding C, Zhang X, Zhang Z. FoxM1 Overexpression Promotes Cell Proliferation and Migration and Inhibits Apoptosis in Hypopharyngeal Squamous Cell Carcinoma Resulting in Poor Clinical Prognosis. Int J Oncol (2017) 51(4):1045–54. doi: 10.3892/ijo.2017.4094
27. Wang N, Dong Q, Zhou XN. LMO4 Promotes the Invasion and Proliferation of Gastric Cancer by Activating PI3K-Akt-mTOR Signaling. Am J Transl Res (2019) 11(10):6534–43.
28. Yang J, Zhu XJ, Jin MZ, Cao ZW, Ren YY, Gu ZW. Osthole Induces Cell Cycle Arrest and Apoptosis in Head and Neck Squamous Cell Carcinoma by Suppressing the PI3K/AKT Signaling Pathway. Chem Biol Interact (2020) 25(316):108934. doi: 10.1016/j.cbi.2019.108934
29. Xie Y, Cheng Y. LINC01198 Facilitates Gliomagenesis Through Activating PI3K/AKT Pathway. RNA Biol (2020) 17(7):1040–52. doi: 10.1080/15476286.2020.1755112
30. Liu HT, Ma RR, Lv BB, Zhang H, Shi DB, Guo XY, et al. LncRNA-HNF1A-AS1 Functions as a Competing Endogenous RNA to Activate PI3K/AKT Signalling Pathway by Sponging miR-30b-3p in Gastric Cancer. Br J Cancer (2020) 122(12):1825–36. doi: 10.1038/s41416-020-0836-4
31. Dong P, Xiong Y, Yue J, JBH S, Kobayashi N, Todo Y, et al. Exploring lncRNA-Mediated Regulatory Networks in Endometrial Cancer Cells and the Tumor Microenvironment: Advances and Challenges. Cancers (Basel) (2019) 11(2):234. doi: 10.3390/cancers11020234
32. Li X, Zuo C, Wu M, Zhang Z. Linc-ROR Promotes Arsenite-Transformed Keratinocyte Proliferation by Inhibiting P53 Activity. Metallomics (2020) 12(6):963–73. doi: 10.1039/d0mt00076k
33. Caponio VCA, Troiano G, Adipietro I, Zhurakivska K, Arena C, Mangieri D, et al. Computational Analysis of TP53 Mutational Landscape Unveils Key Prognostic Signatures and Distinct Pathobiological Pathways in Head and Neck Squamous Cell Cancer. Br J Cancer (2020) 123(8):1302–14. doi: 10.1038/s41416-020-0984-6
34. Jin S, Yang X, Li J, Yang W, Ma H, Zhang Z. P53-Targeted lincRNA-P21 Acts as a Tumor Suppressor by Inhibiting JAK2/STAT3 Signaling Pathways in Head and Neck Squamous Cell Carcinoma. Mol Cancer (2019) 18(1):38. doi: 10.1186/s12943-019-0993-3
35. Du Z, Lin Z, Wang Z, Liu D, Tian D, Xia L. SPOCK1 Overexpression Induced by Platelet-Derived Growth Factor-BB Promotes Hepatic Stellate Cell Activation and Liver Fibrosis Through the Integrin Alpha5beta1/PI3K/Akt Signaling Pathway. Lab Invest (2020) 100(8):1042–56. doi: 10.1038/s41374-020-0425-4
Keywords: head and neck squamous cell carcinoma, FoxM1, linc-ROR, LMO4, Akt/PI3K, proliferation, invasion
Citation: Ma X, Zhang H, Li Q, Schiferle E, Qin Y, Xiao S and Li T (2021) FOXM1 Promotes Head and Neck Squamous Cell Carcinoma via Activation of the Linc-ROR/LMO4/AKT/PI3K Axis. Front. Oncol. 11:658712. doi: 10.3389/fonc.2021.658712
Received: 26 January 2021; Accepted: 25 June 2021;
Published: 10 August 2021.
Edited by:
Jozsef Dudas, Innsbruck Medical University, AustriaReviewed by:
Marco Mascitti, Marche Polytechnic University, ItalyAhmed Diab, Fred Hutchinson Cancer Research Center, United States
Jose Angelito Ugalde Hardillo, Erasmus Medical Center, Netherlands
Copyright © 2021 Ma, Zhang, Li, Schiferle, Qin, Xiao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tiancheng Li, bGl0aWFuY2hlbmdAYmptdS5lZHUuY24=
 Xiao Ma1
Xiao Ma1 Tiancheng Li
Tiancheng Li