
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Oncol. , 09 June 2021
Sec. Molecular and Cellular Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.657965
This article is part of the Research Topic lncRNAs in Cancer Metastasis and Therapy Resistance View all 41 articles
 Ammad Ahmad Farooqi1*
Ammad Ahmad Farooqi1* Sawera Nayyab2
Sawera Nayyab2 Chiara Martinelli3
Chiara Martinelli3 Rossana Berardi4
Rossana Berardi4 Hector Katifelis5
Hector Katifelis5 Maria Gazouli5
Maria Gazouli5 William C. Cho6*
William C. Cho6*Rapidly evolving and ever-increasing knowledge of the molecular pathophysiology of pancreatic cancer has leveraged our understanding altogether to a next level. Compared to the exciting ground-breaking discoveries related to underlying mechanisms of pancreatic cancer onset and progression, however, there had been relatively few advances in the therapeutic options available for the treatment. Since the discovery of the DNA structure as a helix which replicates semi-conservatively to pass the genetic material to the progeny, there has been conceptual refinement and continuous addition of missing pieces to complete the landscape of central dogma. Starting from transcription to translation, modern era has witnessed non-coding RNA discovery and central role of these versatile regulators in onset and progression of pancreatic cancer. Long non-coding RNAs (lncRNAs) have been shown to act as competitive endogenous RNAs through sequestration and competitive binding to myriad of microRNAs in different cancers. In this article, we set spotlight on emerging evidence of regulation of different signaling pathways (Hippo, TGFβ/SMAD, Wnt/β-Catenin, JAK/STAT and NOTCH) by lncRNAs. Conceptual refinements have enabled us to understand how lncRNAs play central role in post-translational modifications of various proteins and how lncRNAs work with epigenetic-associated machinery to transcriptionally regulate gene network in pancreatic cancer.
It has been reported that only 1–2% of RNAs encode for proteins and that the great majority of them falls into the non-coding category, comprehending a large number of different structural (ribosomal RNAs, rRNAs and transfer RNAs, tRNAs) and regulatory RNAs (small conditional RNAs, scRNAs; microRNAs, miRNAs; small nucleolar RNAs, snoRNAs; long non-coding RNAs, lncRNAs) (1). Regulatory RNAs can be divided into small, medium, and long non-coding RNAs (2). LncRNAs sequences are poorly conserved and thus their genomic identification results often difficult (3). They are defined as long RNA transcripts (> 200 nucleotides) not translated in proteins (4), which are involved in the regulation of transcriptional processes by modulation of other non-coding RNAs (ncRNAs) (5). These nucleic acids are responsible also for regulating gene expression at transcriptional and post-transcriptional levels (6, 7). Even though largely debated, recent studies performed exploiting ribosome sequencing (Ribo-seq) and mass spectrometry have revealed their possible translation into proteins (8). Many researches have focused on unraveling the functions of ncRNAs and a careful classification based on their characterization has been established. In recent years, thanks to RNA sequencing and innovative methods, a great number of categories have been identified (9).
LncRNAs can originate from their own/shared promoters, from enhancers and intergenic regions and in specific cell-types upon stimuli. For example, human pancreatic β-cells contain thousands of lncRNAs that can be controlled during differentiation, maturation and upon glucose dynamic changes and are responsible for regulating gene expression in diabetes. Although their function is still under investigation, many techniques have been used for unraveling lncRNAs intracellular localization, structure and functions (10–12).
Different mechanisms that are involved in their biogenesis and peculiar subnuclear structures, called paraspeckles, have been identified. They localize in proximity of nuclear enriched abundant transcript 1 (NEAT1) lncRNAs, specific transcripts lacking introns. Recent studies identified four paraspeckle proteins required for their formation during NEAT1 synthesis and processing. Importantly, paraspeckles have been also involved in modulation of gene expression mediated by lncRNAs (13).
Interestingly, lncRNAs can be transcribed by many regions in the genome, including promoters, enhancers or long primary transcripts. Processing of lncRNAs can be carried out by ribonuclease P for generating mature 3′ ends capped by small nucleolar RNA-proteins or by formation of circular structures (14). LncRNAs can be transcribed as promoter upstream transcripts (PROMPTs) (15), enhancer RNAs (eRNAs) (16), long intervening/intergenic ncRNAs (lincRNAs) (17). When they are transcribed by RNA polymerase II (Poly II), they are produced as medium length lncRNAs with short half-life and targeted by a special nuclear degradation complex called RNA exosome. This process put their gene regulatory activity at high risk.
The most studied category of lncRNAs is lincRNAs, transcribed by Pol II from intergenic regions. These transcripts contain multiple exons and similarity to mRNAs (18), even though they present very different features: they do not possess encoding sequences, they present tissue specific expression, localize at the nucleus and have specific functions (19, 20). Other lncRNAs transcribed from the natural antisense transcripts called NATs (21).
Recently, it has been shown that lincRNAs present few histone marks and transcription factors attached to their promoters and are less spliced respect to mRNAs (22). Interestingly, some lncRNAs can be processed from long transcripts to obtain structures without 5′cap or 3′ adenosine tails (23).
LncRNAs are involved in the transcription modulation mediated by control of gene expression by attaching to DNA binding proteins and Pol II. They can interfere with many cellular mechanisms, such as cell proliferation, differentiation and development (5). Interestingly, they have been demonstrated to be associated to tumorigenesis, when mutated or dysregulated. They are involved in the onset of many types of tumors, such as colorectal, lung, liver, pancreatic, and ovarian cancer (24, 25). Some studies have shown their involvement in several diseases (26).
To provide a comprehensive overview of the interplay between lncRNAs and signaling pathways in pancreatic cancer, we have partitioned this article into three sections. In the first section, we focus on the elucidation of the molecular mechanisms used by lncRNAs to modulate signaling pathways in pancreatic cancer. This section exclusively deals with regulation of Hippo, TGFβ/SMAD, Wnt/β-catenin, Notch, JAK/STAT and TRAIL-driven pathways by lncRNAs in pancreatic cancer. In the second section, we provided a list of individual lncRNAs reportedly involved in the regulation of protein networks in pancreatic cancer. In the last section, we outlined the list of lncRNAs which served as sponges for miRNAs and sequestered target mRNAs away from inhibition by miRNAs.
LncRNAs mediated regulation of pancreatic cancer is highly intricate. Different proteins have been shown to regulate expression of lncRNAs. Likewise, lncRNAs have been reported to work with methylation specific machinery to activate or repress myriad of genes. Moreover, lncRNAs have also been noted to regulate post-translational modifications of different proteins. In this section, we exclusively focus on regulation of Hippo, TGFβ/SMAD, Wnt/β-catenin, Notch, JAK/STAT and TRAIL-driven pathways by lncRNAs in pancreatic cancer.
When the Hippo pathway is activated, LATS1/2 kinases (Large tumor suppressor-1/2) phosphorylate and inactivate YAP (Yes-associated protein) and transcriptional coactivator having PDZ-binding motifs (TAZ), the two characteristically unique downstream transducers that mediate transcriptional output of the Hippo-driven transduction cascade. Phosphorylation of YAP at 127th serine residue by LATS1 is a classical post-translational modification which induces cytoplasmic retention of YAP to inhibit its activity (27). THAP9-AS1 knockdown resulted in an increase in the phosphorylation of YAP at 127th serine residue. THAP9-AS1 increased expression of YAP by sponging miRNA-484 away from YAP. Series of experiments revealed that WW1/2 domain of YAP was essential for direct interaction with LATS1 to induce phosphorylation and subsequent retention of YAP within cytoplasm (27) (Figure 1). THAP9-AS1 interacted with YAP and blocked LATS1-YAP association. Co-immunoprecipitation assays revealed that knockdown of THAP9-AS1 potentiated the interactions between LATS1 and YAP. Although YAP/TAZ are transcriptional co-activators, they do not have DNA-binding domains. Therefore, YAP/TAZ need binding partners and usually bind with transcriptional factors such as TEAD1-4 for modulation of expression of target genes. YAP/TEAD1 complex transcriptionally upregulated the expression of THAP9-AS1 (Figure 1) (27).
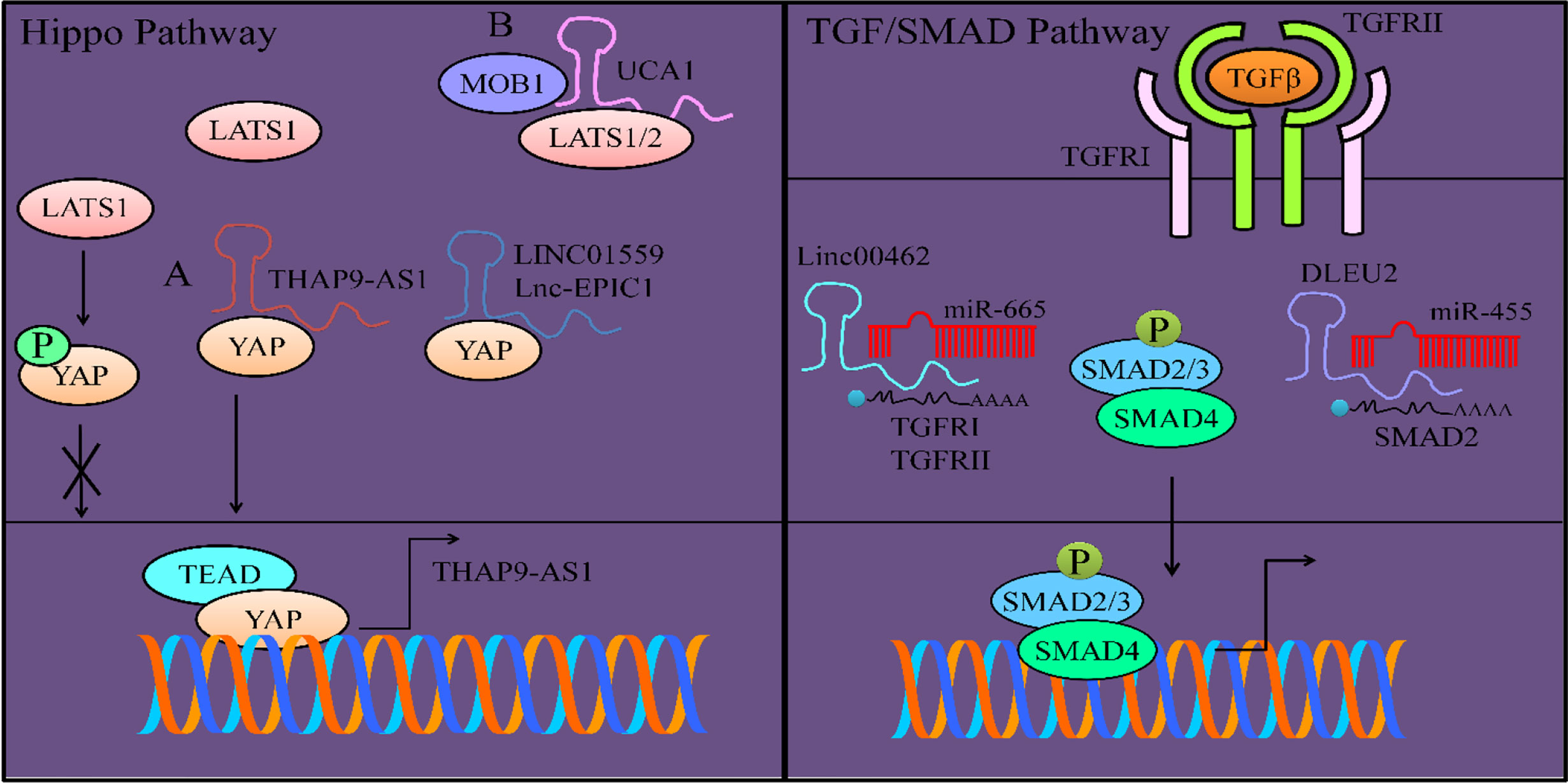
Figure 1 Regulation of Hippo pathway. LATS1 mediated phosphorylation of YAP prevented its nuclear accumulation. However, (A) THAP9-AS1 inhibited LATS1 mediated phosphorylation of YAP and promoted its nuclear accumulation. YAP interacted with TEAD and transcriptionally upregulated THAP9-AS1. LINC01559 and UCA1 also inhibit phosphorylation of YAP and promote its nuclear accumulation. (B) UCA1 formed a complex with MOB1 and LATS1/2 and not only inhibited MOB1-mediated activation of LATS1/2 but also blocked phosphorylation of YAP. TGFβ/SMAD pathway is regulated by lncRNAs. Linc00462 enhanced the expression of TGFRI and TGFRII by interfering with miR-665-mediated targeting activity. DLEU2 also served as an oncogenic lncRNA and inhibited miR-455-mediated targeting of SMAD2.
It is exciting to note that most of the lncRNAs (LINC01559 and UCA1) inhibit phosphorylation of YAP and promote its nuclear accumulation.
LINC01559 interacted with YAP, inhibited YAP phosphorylation, and enhanced YAP/induced transcriptional activities in pancreatic cancer cells (28).
YAP1 maintained the expression of MYC, whereas knockout of YAP1 caused considerable downregulation of MYC that resulted in growth arrest of pancreatic cancer cells and apoptosis (29). Lnc-EPIC1 interacted with YAP1 in pancreatic cancer cells. Lnc-EPIC1 lost its ability to promote proliferation and growth of YAP1-silenced pancreatic cancer cells (29).
Likewise, UCA1 significantly enhanced the invasive ability of PANC-1 cells (30). LATS1/2 requires a co-activator protein to achieve full activation. MOB1 (Mps One binder 1) is essential for the activation of LATS1/2. UCA1, formed a complex with LATS1, MOB1 and YAP (Figure 1). UCA1 promoted nuclear translocation of YAP in PANC-1 cells. Additionally, YAP stimulated the expression of UCA1 in pancreatic cancer cells (30). Overall, UCA1 and YAP promoted invasion of pancreatic cancer cells.
These interesting findings provided substantial evidence that different lncRNAs inhibited YAP phosphorylation and potentiated YAP-driven signaling to promote pancreatic cancer.
MST1 and MST2 promoted the phosphorylation of LATS1 and LATS2 (31). MST1 was found to be directly targeted by miR-181c-5p. miR-181c-5p promoted chemoresistance of pancreatic cancer cells through inactivation of the Hippo signaling. GAS5 interfered with miR-181c-5p-mediated inhibition of MST1 in pancreatic cancer cells. Tumor growth was significantly reduced in mice inoculated with GAS5-overexpressing PANC-1 cells (31).
TGFβ/SMAD signaling has been shown to play significant role in the onset and progression of pancreatic cancer. Here we discuss how different lncRNAs regulate TGFβ/SMAD to promote pancreatic cancer.
Linc00462 considerably enhanced invasive potential of pancreatic cancer cells via stimulating the expression of TGFβR1 and TGFβR2 (32). TGFβR1 and TGFβR2 were found to be directly targeted by miR-665. However, Linc00462 sponged away miR-665 and relived inhibitory effects of miR-665 on TGFβR1 and TGFβR2 (Figure 1). linc00462 overexpression significantly increased p-SMAD2 and p-SMAD3 whereas, overexpression of miR-665 significantly decreased p-SMAD2 and p-SMAD3 (32).
PVT1 stimulated TGFβ/SMAD signaling that sequentially induced epithelial-to-mesenchymal transition (EMT) (32). PVT1 silencing resulted in inactivation of TGFβ/SMAD signaling via reduction of p-SMAD2/3 and TGFβ1 but there was a notable increase in the levels of SMAD4 (32).
DLEU2 blocked miR-455-mediated targeting of SMAD2 in pancreatic cancer cells (Figure 1) (33). SMAD2 promoted invasive potential of pancreatic cancer cells. miR-455 overexpression or DLEU2 knockdown significantly suppressed proliferation and invasion of MIA PaCa-2 cells. Similarly, miR-455 inhibition or DLEU2 overexpression significantly induced proliferation and invasion in AsPC-1 cells, whereas SMAD2 inhibition markedly reversed the effects of miR-455 inhibition or DLEU2 overexpression (33).
Collectively, these research reports highlighted oncogenic interplay between lncRNAs and TGFβ/SMAD pathway to promote pancreatic cancer.
FZD4 and FZD6 played central role in activation of Wnt/β-catenin pathway. miR-497-5p targeted FZD4 and FZD6 and inhibited Wnt/β-catenin transduction cascade (34). DLX6-AS1 interfered with miR-497-5p-mediated targeting of FZD4 and FZD6 (Figure 2). DLX6-AS1 knockdown inhibited the metastatic capacity of pancreatic cancer cells by reducing the number of metastatic foci in the liver and lungs. However, DLX6-AS1 overexpression considerably enhanced metastatic foci in the liver and lungs of xenografted mice (34). Overall, these results indicated that DLX6-AS1 acted an oncogenic lncRNA and potentiated Wnt/β-catenin signaling.
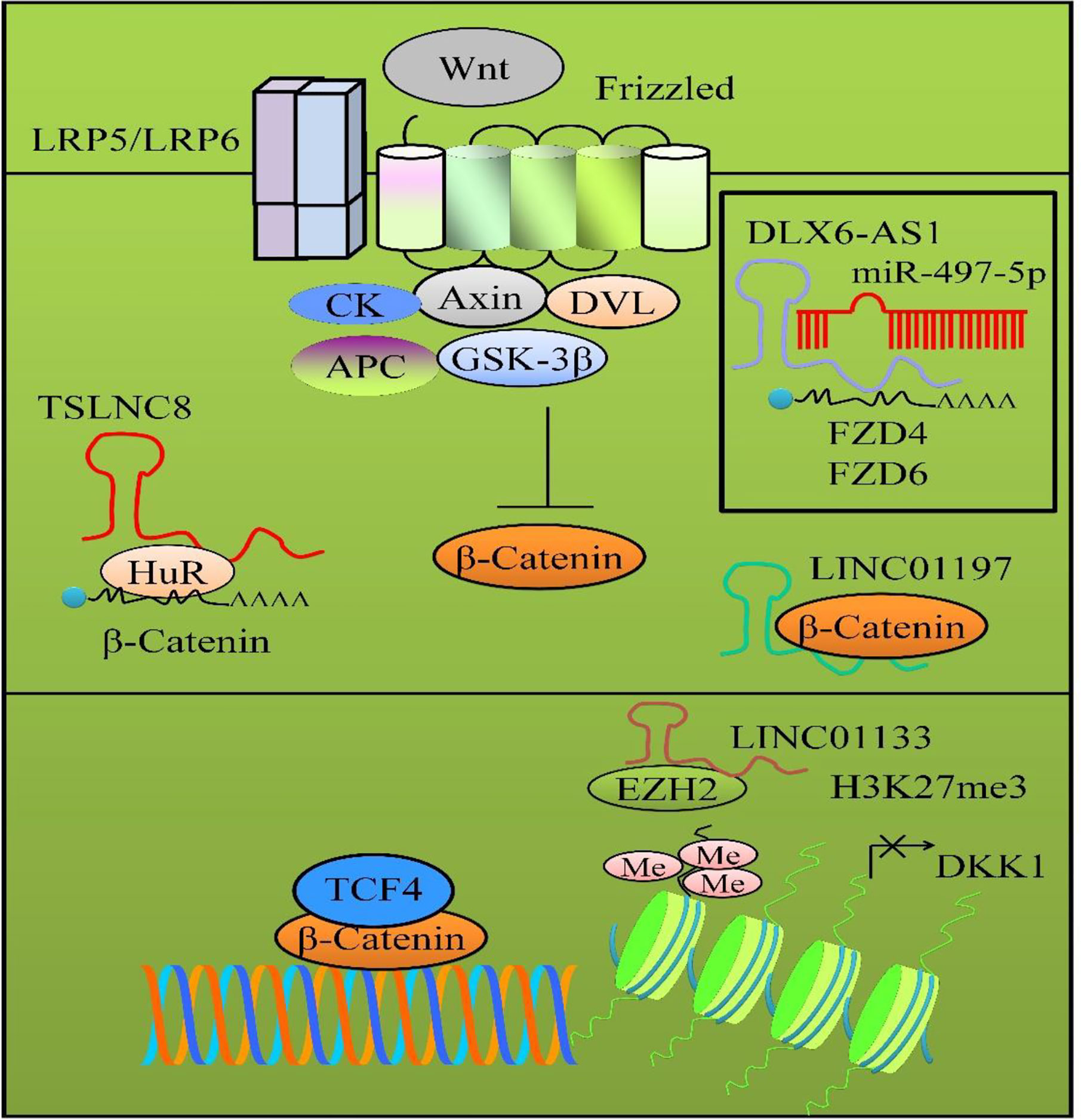
Figure 2 Wnt/β-catenin signaling in pancreatic cancer. β-catenin moves into the nucleus to transcriptionally modulate wide-ranging target gene networks. LncRNAs regulate different proteins in Wnt/β-catenin pathway. LINC01197 physically associated with β-catenin and inhibited Wnt/β-catenin signaling cascade. DLX6-AS1 interfered with miR-497-5p-mediated targeting of FZD4 and FZD6. TSLNC8 promoted the binding of HuR to β-catenin mRNA to stabilize β-catenin. LINC01133 promoted the loading of EZH2 to transcriptionally downregulate DKK1.
Linc00261 inhibited the activation of the β-catenin/TCF4 pathway and cell metastasis by blocking miR-552-5p-induced targeting of FOXO3 in pancreatic cancer cells (35). β-catenin and TCF4 were noted to be reduced in Linc00261-overexpressing pancreatic cancer cells. There was a negative relationship of FOXO3 and β-catenin/TCF4 in pancreatic cancer cells. The number of metastatic foci was reduced in the mice injected with the Linc00261-expressing PANC-1 cells (35).
TSLNC8 worked jointly with HuR and promoted the binding of HuR to β-catenin mRNA to stabilize β-catenin, thus activating WNT/β-catenin transduction cascade (Figure 2) (36).
HOTAIR inhibition increased the expression of WIF-1 (Wnt inhibitory factor 1) and enhanced radiosensitivity of pancreatic cancer cells (37).
LINC01197 physically associated with β-catenin and inhibited Wnt/β-catenin signaling cascade in PANC1 and BxPC3 cancer cells (Figure 2) (38). LINC01197 disassembled β-catenin and TCF4 in BxPC3 and PANC1 cells. LINC01197 overexpression inhibited the binding of β-catenin and TCF4 both in BxPC3 and PANC1 cells. LINC01197 overexpression induced significant inhibition of the growth of the tumors derived from BxPC3- and PANC1 cancer cells (38).
DKK1 is a soluble inhibitor of Wnt/β-catenin cascade that can bind to LRP5/6 and induce internalization of LRP proteins (39). LINC01133 promoted the loading of EZH2 to transcriptionally downregulate DKK1 in pancreatic cancer cells (Figure 2). Importantly, metastatic spread to the liver and lungs was reduced in mice inoculated with LINC01133-silenced pancreatic cancer cells (39).
SNHG1 has been reported to be significantly upregulated in pancreatic cancer cells (40). NOTCH-induced oncogenic pathway was also noted to be active in pancreatic cancer cells. SNHG1 knockdown exerted inhibitory effects on the activation of the NOTCH-driven signaling pathway and inhibited the expression of NOTCH-1, HES1, vimentin, and N-cadherin (40).
Levels of Jagged-1, HES1, HES5 were noted to be markedly reduced in RP11-567G11.1-depleted PANC-1 and BXPC-3 cells (41). Overall, these findings provided evidence that different long non-coding RNAs effectively potentiated NOTCH-driven pathway in the pancreatic cancer cells.
NOTCH3 is negatively regulated by miR-613 in pancreatic cancer cells (42). HOTAIR sequestered away miR-613 and potentiated NOTCH3 expression. miR-613 overexpression or knockdown of HOTAIR suppressed tumor growth and also reduced the expression of NOTCH3 (42).
STAT1-mediated transduction cascade played critical role in the progression of pancreatic cancer (43). miR-382-3p exerted tumor suppressive effects and directly targeted STAT1. However, PSMB8-AS1 interfered with miR-382-3p-mediated inhibition of STAT1. There was a significant increase in the growth of tumors in experimental mice xenografted with PSMB8-AS1-overexpressing PANC-1 cells. STAT1 and PD-L1 were found to be upregulated in mice xenografted with PSMB8-AS1-overexpressing PANC-1 cells (43).
H19, an oncogenic lncRNA effectively promoted STAT3-mediated signaling in pancreatic cancer cells (44). miR-675 is transcribed from the first exon of H19 and negatively regulates SOCS5 (Suppressor of cytokine signaling 5). SOCS5 is involved in the inhibition of STAT3-driven signaling. It was found that miR-675 negatively modulated SOCS5 and potentiated the expression of STAT3. H19 upregulation reduced gemcitabine chemosensitivity and lowered apoptosis in CAPAN-1 cells. However, gemcitabine chemosensitivity and the apoptosis rate were significantly increased in H19-silenced PANC-1 cells (44).
Cell surface expression of death receptors (DR4 and DR5) is of critical importance to achieve therapeutic effects of TRAIL-based therapeutics.
HOTAIR worked synchronously with EZH2 and transcriptionally downregulated DR5 in pancreatic cancer cells (45). HOTAIR inhibited DR5 transcription by enhancing EZH1-induced histone H3 trimethylation on DR5 gene. HOTAIR knockdown in the TRAIL-resistant PANC-1 cancer cells restored apoptotic cell death (45).
Here we provided a list of lncRNAs reportedly involved in the regulation of myriad of proteins in pancreatic cancer.
PLACT1 an oncogenic lncRNA has been described to repress IκBα expression mainly through increased loading of hnRNPA1 to promoter region of IκBα (46). Additionally, there was an evident increase in the trimethylated levels of lysine 27 of histone-3 that also played role in epigenetic inactivation of IκBα (Figure 3). E2F1-mediated stimulation of PLACT1 fueled progression of PDAC by sustained activity of NF-κB cascade. Expectedly, use of NF-κB signaling inhibitors caused significant suppression of PLACT1-induced sustained NF-κB activity that consequentially induced regression of tumors in xenografted mice (46).
HOTTIP (HOXA transcript at the distal tip) formed a complex with adaptor protein WDR5 and MLL1 (H3K4 methyltransferase) to trans-activate oncogenic proteins CYB5R2, KIF26A, SULT1A1, TSC22D1, and SLC1A4 by increasing the levels of trimethylated lysine-4 at histone-3 (H3K4) at their promoters (Figure 3) (47). Collectively, these findings provided concrete evidence of fundamental role of HOTTIP in promotion of PDAC progression through the HOTTIP–WDR5–MLL1 axis.
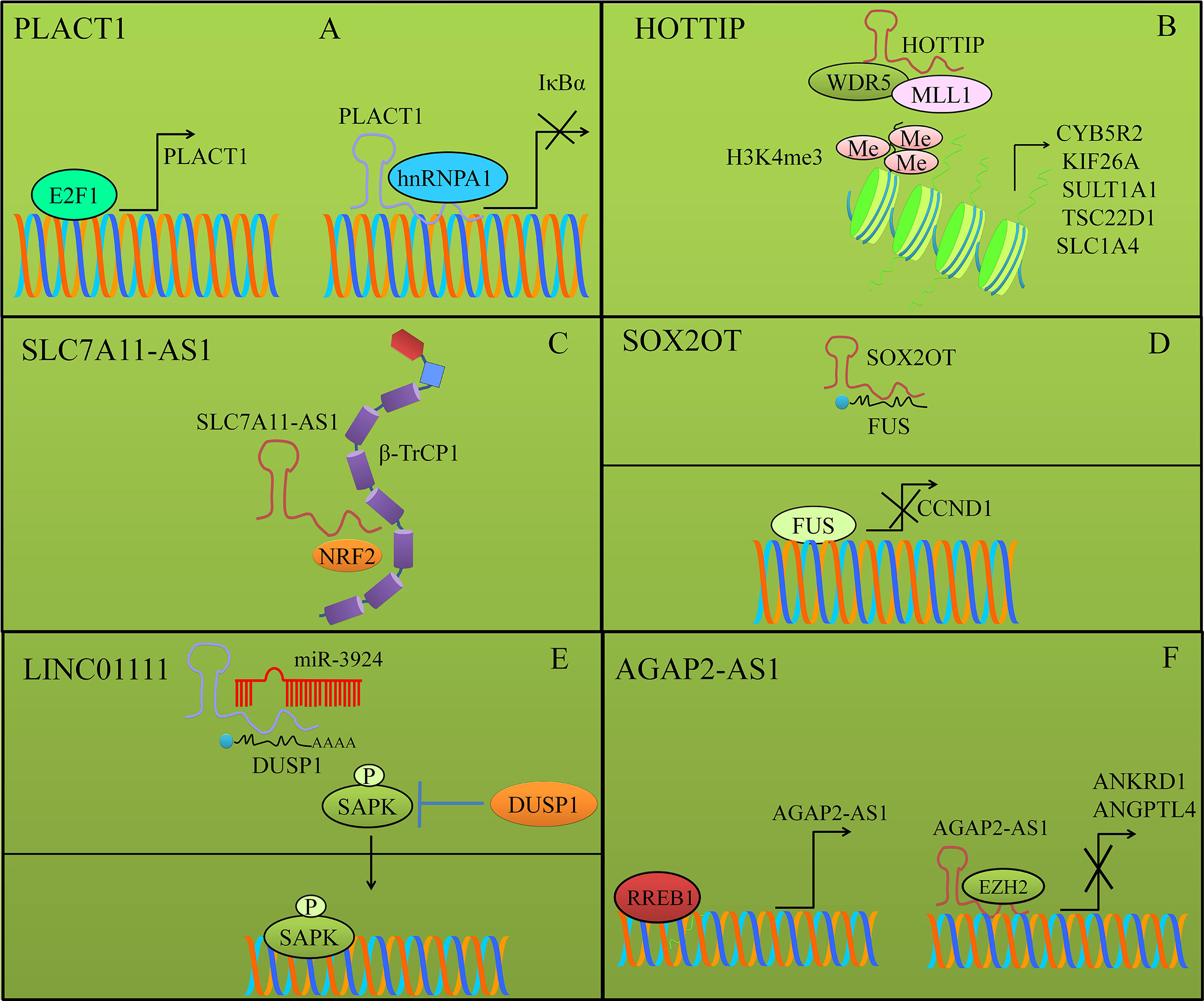
Figure 3 (A) PLACT1 mediated inhibition of IκBα by epigenetic inactivation. PLACT1 also enhanced loading of hnRNPA1 to the promoter region of IκBα. E2F1 induced activation of PLACT1. (B) HOTTIP formed a complex with adaptor protein WDR5 and MLL1 (H3K4 methyltransferase) to trans-activate oncogenic proteins by increasing the levels of trimethylated lysine-4 at histone-3 (H3K4) at their promoters. (C) SLC7A11-AS1 blocked β-TRCP-induced ubiquitination and degradation of NRF2. (D) SOX2OT destabilized FUS protein by binding directly to FUS. FUS transcriptionally repressed CCND1. (E) DUSP1 (Dual-specificity phosphatase-1) mediated dephosphorylation of SAPK resulted in inhibition of the pathway. LINC01111 sponged away DUSP1 from miR-3924 and promoted expression of DUSP1. DUSP1 dephosphorylated SAPK and prevented its nuclear accumulation. (F) RREB1-stimulated expression of AGAP2-AS1. AGAP2-AS1 interacted with EZH2 and repressed expression of ANGPTL4 and ANKRD1.
SLC7A11-AS1 promoted chemoresistance through reduction of intracellular ROS by stabilizing NRF2 (nuclear factor erythroid-2-related factor 2) (48). Proteomic studies revealed that SLC7A11-AS1 co-localized with β-TRCP1 in the nucleus. β-TrCP, an F-box protein served as substrate-recognition subunit for the SCFβ–TrCP E3 ubiquitin ligase, which mediated ubiquitylation of a broad range of substrates and post-translationally marked proteins for degradation. A series of experiments revealed that exon 3 of SLC7A11-AS1 interacted with the F-box motif of β-TRCP1. The F-box motif of β-TRCP1 acted as a critical domain that recruited β-TRCP1 to the SCFβ–TRCP E3 complex. Resultantly, this interaction prevented ubiquitination and degradation of NRF2 in the nucleus. SLC7A11-AS1 overexpression blocked SCFβ–TRCP-induced ubiquitination and degradation of NRF2 and effectively reduced intracellular levels of ROS (Figure 3) (48).
SOX2OT fueled proliferation capacity of PDAC cells by binding directly to FUS and destabilizing the FUS protein (Figure 3). SOX2OT upregulated the proliferation of the BxPC-3 and PANC-1 cells. FUS transcriptionally repressed CCND1 in pancreatic cancer cells (49).
N6-methyladenosine (m6A) has been shown to tag wide-ranging messenger RNAs in mammalian cells. Molecular studies had shown that m6A modification machinery consisted of “writers”, “erasers”, and “readers” (50). The YTH domain family (YTHDF) and IGF2BPs belonged to large families of RNA-binding proteins (RBPs) and served as readers. IGF2BPs served as a specialized family of m6A readers that targeted various mRNA transcripts. IGF2BPs stabilized and stored mRNAs marked by m6A during stress and normal situations (50). IGF2BP2 promoted pancreatic cancer cell proliferation. DANCR expression was upregulated in IGF2BP2-overexpressing cancer cells. Moreover, knockdown of IGF2BP2 suppressed DANCR expression. DANCR knockdown suppressed cell proliferation and colony formation. IGF2BP2 interacted with DANCR and stabilized it effectively. RNA methylation (m6A) occurred at 664th nucleotide of DANCR. Methylated DANCR was recognized by IGF2BP2 and resultantly, IGF2BP2 served as a reader for the methylated version of DANCR and increased its stability (50).
LINC01111 acted as a tumor suppressor lncRNA in pancreatic cancer. LINC01111 knockdown enhanced cell proliferation, invasion, and migration in vitro (51). Tumor growth was found to be significantly enhanced in mice xenografted with LINC01111-silenced pancreatic cancer cells. Higher expression levels of LINC01111 relieved repressive effects of miR-3924 on DUSP1 and effectively blocked SAPK phosphorylation and thus inactivated SAPK/JNK signaling pathway in pancreatic cancer cells. Shown in Figure 3. DUSP1 (Dual-specificity phosphatase-1) is reportedly involved in dephosphorylation of different proteins. Therefore, DUSP1-mediated dephosphorylation of SAPK resulted in inhibition of the pathway (Figure 3). Functional studies had shown that phosphorylation activated function of SAPK/JNKs. SAPK/JNKs translocated from the cytoplasm to the nucleus where they phosphorylated series of genes including c-Jun, ATF2, etc. and dramatically enhanced their transcriptional activities (51).
RREB1-binding sites have been identified in the promoter region of AGAP2-AS1 and consequently could binding of RREB1 to the promoter region of AGAP2-AS1 stimulated its expression (52). AGAP2-AS1 worked synchronously with EZH2 (enhancer of zeste homolog-2) and epigenetically inhibited ANGPTL4 and ANKRD1 and fueled proliferation and metastasis of pancreatic cancer cells (52). Shown in Figure 3.
Overexpression of XLOC_006390 promoted the protein stability of c-Myc by blocking its ubiquitination. c-Myc transcriptionally upregulated glutamate dehydrogenase-1 in pancreatic cancer cells (53, 54). Collectively, XLOC_006390 promoted pancreatic carcinogenesis and glutamate metabolism by stabilization of c-Myc (53, 54).
LINC01638 overexpression promoted, while LINC01638 silencing inhibited migratory and invasive potential of PDAC cell line (55, 56). LINC01638 overexpression increased TGFβ1, while silencing of LINC01638 markedly reduced TGFβ1 expression in pancreatic ductal adenocarcinoma cell line (55, 56).
MACC1-AS1 acted an oncogenic lncRNA and potentiated the expression of SMAD2 by sequestering it away from miR-145 (57, 58). In turn, SMAD2 stimulated the expression of MACC1-AS1 by directly binding to the promoter. Overall, these results clearly suggested that MACC1-AS1 promoted cancer by potentiating SMAD-driven signaling.
MACC1-AS1 knockdown inhibited the proliferation as well as metastasizing capacity of pancreatic cancer cells. MACC1-AS1 overexpressing pancreatic cancer cells demonstrated significantly increased mobility of cancer cells (53, 54). MACC1-AS1 stabilized protein levels of pyruvate kinase M2. NOTCH1 phosphorylation was increased in MACC1-AS1-overexpressing pancreatic cancer cells. However, phosphorylation was blocked in pyruvate kinase M2-knockdown cancer cells (53, 54). Overall, these findings suggested that MACC1-AS1 potentiated pyruvate kinase M2-driven phosphorylation of NOTCH1 (Figure 4). Oncogenic NOTCH1 pathway was activated by MACC1-AS1 through pyruvate kinase M2 in the pancreatic cancer cells.
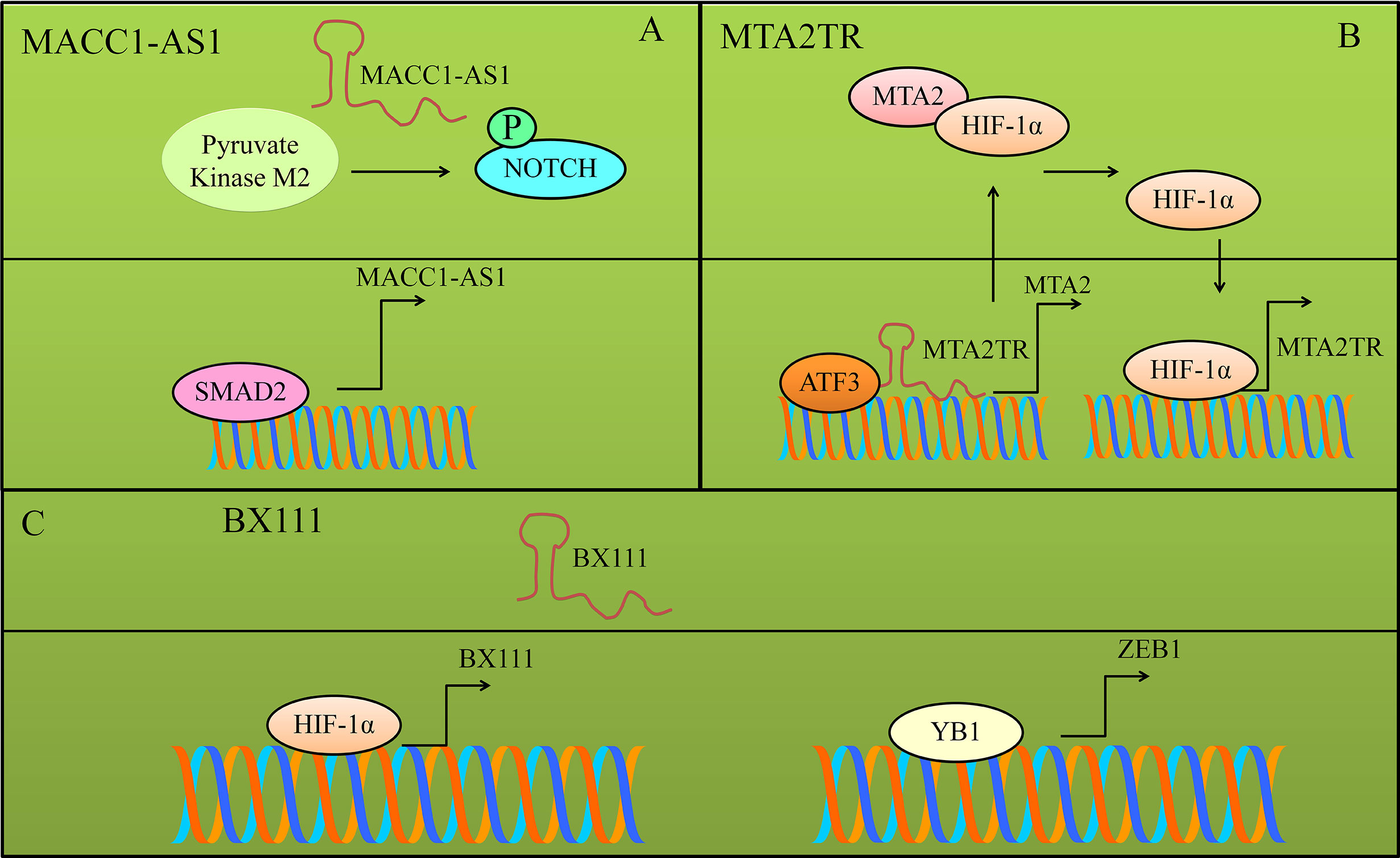
Figure 4 Different proteins transcriptionally upregulated the expression of various lncRNAs. (A) SMAD2 transcriptionally upregulated MACC1-AS1. MACC1-AS1 stabilized and promoted pyruvate kinase M2-driven phosphorylation of NOTCH1. Oncogenic NOTCH1 pathway was activated by MACC1-AS1 through pyruvate kinase M2. (B) MTA2TR and ATF3 assembled in the nucleus and stimulated the expression of MTA2. MTA2 stabilized HIF protein and consequently HIF transcriptionally activated MTA2TR. (C) BX111 was transcriptionally stimulated by HIF-1α. Furthermore, BX111 facilitated the binding of YB1 to promoter region of ZEB1.
Metastasis associated protein 2 (MTA2) transcriptional regulator lncRNA (MTA2TR) worked synchronously with ATF3 (activating transcription factor-3) and transcriptionally upregulated the expression of MTA2 (55, 56). MTA2 stabilized HIF-1α protein via deacetylation and promoted HIF-1α-induced transcriptional upregulation of MTA2TR (56). Shown in Figure 4. Overall, these results highlighted intricate role of MTA2TR in stabilization of HIF-1α via MTA2 in pancreatic cancer cells.
lnc-PCTST, a tumor suppressor lncRNA increased E-cadherin and simultaneously reduced vimentin levels (57, 58). Additionally, TACC-3 knockdown also induced an increase in the levels of E-cadherin. Functional studies revealed that lnc-PCTST was closely associated with its genomic neighboring gene TACC-3 and considerably reduced its promoter activity (57, 58).
Detailed mechanistic insights revealed that HOXA-AS2 interacted directly with EZH2 (enhancer of zeste homolog-2) and lysine specific demethylase 1 (LSD1) and synchronously promoted growth ability of pancreatic cancer cells (59).
KCNK15-AS1 markedly reduced migratory and invasive potential of BxPC-3 and MIA PaCa-2 cells (60). KCNK15-AS1 m6A enrichment was noted to be significantly higher in BxPC-3 and MIA PaCa-2 cells. ALKBH5, a versatile RNA m6A demethylase efficiently demethylated KCNK15-AS1 in pancreatic cancer cells (60). Overall, these results indicated that ALKBH5-driven demethylation of KCNK15-AS1 dramatically reduced migratory and invasive potential of pancreatic cancer cells.
BX111 was transcriptionally stimulated by HIF-1α (hypoxia-inducible factor) in response to hypoxia (61). Furthermore, BX111 participated in hypoxia-driven EMT of pancreatic cancer cells by promoting the binding of Y-box protein (YB1) to promoter region of ZEB1 (Figure 4) (61).
HOTAIR is an oncogenic lncRNA and increased expression of HOTAIR is indicative of a poor prognosis in cancer patients (62). Although its exact role is not fully understood, several studies have revealed some of its functions in pancreatic cancer. A recent study (63) showed that HOTAIR serves as up-stream regulator of HK2, an enzyme that catalyzes the first step of glycolysis (64) and thus boosting cancer cell proliferation in pancreatic cancer cells. Its overexpression also increases both ATP and lactate production as well as glucose uptake. Additionally, increased HOTAIR levels downregulated death receptor 5 (DR5) and prevented TRAIL-mediated apoptosis. EZH2 effectively catalyzed histone H3 lysine 27 trimethylation (H3K27me3), an essential epigenetic modification on histone that controlled structure of the chromatin and epigenetically silenced target genes. HOTAIR knockdown markedly decreased H3K27me3 loading to the DR5 promoter, while HOTAIR overexpression greatly enhanced H3K27me3 loading to the DR5 promoter. HOTAIR worked synchronously with EZH2 and marked promoter of DR5 with H3K27me3 to epigenetically silence DR5 (45). Moreover, polymorphisms such as rs4759314 and rs200349340 increase HOTAIR expression and promote pancreatic cancer susceptibility (65, 66).
MALAT1 and EZH2 epigenetically inactivated E-cadherin and potently enhanced invasion and migration of pancreatic cancer cells (67). Importantly, histone methylation and DNA methylation are highly dynamic mechanisms centrally involved in the reprogramming of gene networks in wide variety of cellular processes. Different chemicals are currently being tested to inhibit polycomb repressive complexes to re-program gene networks (68, 69). Furthermore, knockdown of MALAT1 inhibited proliferation, migration, invasion as well as the expression of genes involved in EMT. Knockdown of MALAT1 also results in downregulation of Snail and Slug, two transcription factors that are related to EMT (70).
Wealth of information has portrayed competing endogenous RNA (ceRNA) activity as a large-scale regulatory network across the transcriptome which has greatly expanded the functional genetic information in the human genome and played role in cancer onset and progression.
Linc00976 promoted proliferation, migration, and invasion by sequestering OTUD7B away from miR-137 in pancreatic cancer cells (71). OTUD7B, a deubiquitination enzyme efficiently deubiquitinated EGFR and activated downstream pathway. EGFR was considerably more stable in OTUD7B-overexpressing pancreatic cancer cells (71).
LOXL1-AS1 promoted pancreatic cancer by promoting the expression of Semaphorin 7A (SEMA7A) and sequestering it away from miR-28-5p (72).
Cancer susceptibility candidate 2 (CASC2) exerted tumor-suppressive effects through regulation of miR-24/MUC6 axis in pancreatic cancer cells (73). miR-24 knockdown or CASC2 overexpression suppressed pancreatic cancer cell proliferation, migration, invasion and promoted apoptosis. Mechanistically, CASC2 sponged miR-24 and relieved the repressive effects of miR-24 on MUC6 to suppress pancreatic cancer growth and progression (73).
LINC00657, an oncogenic lncRNA promoted the expression of PAK4 (p21 activated kinase-4) by protecting it from miR-433 (74). Tumor growth was significantly reduced in mice xenografted with LINC00657-silenced PDAC cells (74).
Long intergenic non-coding RNA for kinase activation (LINK-A) acted as an oncogenic lncRNA and promoted migratory and invasive potential of BxPC-3 via stimulation of ROCK1 (Rho associated coiled-coil containing protein kinase-1) (75).
HCP5, an lncRNA, stimulated the expression of hepatoma-derived growth factor (HDGF) by protecting it from targeting by miR-214-3p. HCP and HDGF promoted gemcitabine resistance in pancreatic cancer cells (76).
SBF2-AS1 knockdown inhibited proliferation, EMT and induction of apoptotic cell death in gemcitabine-resistant pancreatic cancer cells. SBF2-AS1 potentiated the expression of TWF1 by sponging away miR-142-3p (77).
The recent resurgence of public interest in herbal remedies, it was report that ginsenoside Rg3 effectively induced apoptosis in gemcitabine-resistant pancreatic cancer cells. Essentially, levels of CASC2 and PTEN were found to be considerably elevated in ginsenoside-treated pancreatic cancer cells (78).
Because of better detection and high specificity in the liquid biopsy and tissue, there is increasing interest in exploring the potential of lncRNAs in cancer patients (79, 80). Profiling of the lncRNAs derived from extracellular vesicles has helped in the identification of a diagnostic signature for the detection of pancreatic ductal adenocarcinoma (81).
Non-coding RNA biology has exploded in the recent era, and we have witnessed overwhelmingly increasing list of miRNAs and lncRNAs which regulated cancer onset and progression. Additionally, the concept of ceRNA has leveraged our understanding of the layered regulatory network of lncRNAs to another level. Cellular and molecular biologists are now focusing on identification of specialized lncRNAs which play crucial role in pancreatic cancer. Identification of most relevant lncRNAs will enable the development of mimics and antisense oligonucleotides for efficient treatment of pancreatic cancer.
SN, CM, and HK gathered and wrote the initial draft. AF, RB, MG, and WC input and edited the draft. AF designed the diagrams. AF, RB, MG, and WC checked and polished the diagrams. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Miss Maria Mariam for her assistance in English language editing.
1. Hemberg M, Gray JM, Cloonan N, Kuersten S, Grimmond S, Greenberg ME, et al. Integrated Genome Analysis Suggests That Most Conserved Non-Coding Sequences Are Regulatory Factor Binding Sites. Nucleic Acids Res (2012) 40(16):7858–69. doi: 10.1093/nar/gks477
2. Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell (2009) 136(4):629–41. doi: 10.1016/j.cell.2009.02.006
3. Al-Tobasei R, Paneru B, Salem M. Genome-Wide Discovery of Long Non-Coding RNAs in Rainbow Trout. PloS One (2016) 11(2):e0148940. doi: 10.1371/journal.pone.0148940
4. Guo X, Gao L, Wang Y, Chiu DKY, Wang T, Deng Y. Advances in Long Noncoding RNAs: Identification, Structure Prediction and Function Annotation. Brief Funct Genomics (2016) 15(1):38–46. doi: 10.1093/bfgp/elv022
5. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs Regulate Transcription? Sci Adv (2017) 3(9):eaao2110. doi: 10.1126/sciadv.aao2110
6. Mattick JS, Makunin IV. Non-Coding RNA. Hum Mol Genet (2006) 15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046
7. Esteller M. Non-Coding RNAs in Human Disease. Nat Rev Genet (2011) 12(12):861–74. doi: 10.1038/nrg3074
8. Bazin J, Baerenfaller K, Gosai SJ, Gregory BD, Crespi M, Bailey-Serres J. Global Analysis of Ribosome-Associated Noncoding RNAs Unveils New Modes of Translational Regulation. Proc Natl Acad Sci U S A (2017) 114(46):E10018–27. doi: 10.1073/pnas.1708433114
9. Kung JTY, Colognori D, Lee JT. Long Noncoding RNAs: Past, Present, and Future. Genetics (2013) 193(3):651–69. doi: 10.1534/genetics.112.146704
10. Morán I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, et al. Human β Cell Transcriptome Analysis Uncovers lncRNAs That Are Tissue-Specific, Dynamically Regulated, and Abnormally Expressed in Type 2 Diabetes. Cell Metab (2012) 16(4):435–48. doi: 10.1016/j.cmet.2012.08.010
11. Akerman I, Tu Z, Beucher A, Rolando DMY, Sauty-Colace C, Benazra M, et al. Human Pancreatic β Cell lncRNAs Control Cell-Specific Regulatory Networks. Cell Metab (2017) 25(2):400–11. doi: 10.1016/j.cmet.2016.11.016
12. Salehi S, Taheri MN, Azarpira N, Zare A, Behzad-Behbahani A. State of the Art Technologies to Explore Long Non-Coding RNAs in Cancer. J Cell Mol Med (2017) 21(12):3120–40. doi: 10.1111/jcmm.13238
13. Naganuma T, Hirose T. Paraspeckle Formation During the Biogenesis of Long Non-Coding RNAs. RNA Biol (2013) 10(3):456–61. doi: 10.4161/rna.23547
14. Wu H, Yang L, Chen LL. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet (2017) 33(8):540–52. doi: 10.1016/j.tig.2017.05.004
15. Preker P, Almvig K, Christensen MS, Valen E, Mapendano CK, Sandelin A, et al. PROMoter uPstream Transcripts Share Characteristics With mRNAs and are Produced Upstream of All Three Major Types of Mammalian Promoters. Nucleic Acids Res (2011) 39(16):7179–93. doi: 10.1093/nar/gkr370
16. Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread Transcription at Neuronal Activity-Regulated Enhancers. Nature (2010) 465(7295):182–7. doi: 10.1038/nature09033
17. Cabili M, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative Annotation of Human Large Intergenic Noncoding RNAs Reveals Global Properties and Specific Subclasses. Genes Dev (2011) 25(18):1915–27. doi: 10.1101/gad.17446611
18. Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved Function of lincRNAs in Vertebrate Embryonic Development Despite Rapid Sequence Evolution. Cell (2011) 147(7):1537–50. doi: 10.1016/j.cell.2011.11.055
19. Ulitsky I, Bartel DP. LincRNAs: Genomics, Evolution, and Mechanisms. Cell (2013) 154(1):26–46. doi: 10.1016/j.cell.2013.06.020
20. Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci (2016) 41(9):761–72. doi: 10.1016/j.tibs.2016.07.003
21. Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense Transcription in the Mammalian Transcriptome. Science (2005) 309(5740):1564–6. doi: 10.1126/science.1112009
22. Melé M, Mattioli K, Mallard W, Shechner DM, Gerhardinger C, Rinn JL. Chromatin Environment, Transcriptional Regulation, and Splicing Distinguish lincRNAs and mRNAs. Genome Res (2017) 27(1):27–37. doi: 10.1101/gr.214205.116
23. Zhang Y, Yang L, Chen LL. Life Without A Tail: New Formats of Long Noncoding RNAs. Int J Biochem Cell Biol (2014) 54:338–49. doi: 10.1016/j.biocel.2013.10.009
24. Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res (2017) 77(15):3965–81. doi: 10.1158/0008-5472.CAN-16-2634
25. Delás MJ, Hannon GJ. lncRNAs in Development And Disease: From Functions to Mechanisms. Open Biol (2017) 7(7):170121. doi: 10.1098/rsob.170121
26. Cipolla GA, de Oliveira JC, Salviano-Silva A, Lobo-Alves SC, Lemos DS, Oliveira LC, et al. Long Non-Coding RNAs in Multifactorial Diseases: Another Layer of Complexity. Noncoding RNA (2018) 4(2):13. doi: 10.3390/ncrna4020013
27. Li N, Yang G, Luo L, Ling L, Wang X, Shi L, et al. LncRNA THAP9-AS1 Promotes Pancreatic Ductal Adenocarcinoma Growth and Leads to a Poor Clinical Outcome via Sponging miR-484 and Interacting with YAP. Clin Cancer Res (2019). doi: 10.1158/1078-0432.CCR-19-0674
28. Lou C, Zhao J, Gu Y, Li Q, Tang S, Wu Y, et al. LINC01559 Accelerates Pancreatic Cancer Cell Proliferation and Migration Through YAP-Mediated Pathway. J Cell Physiol (2020) 235(4):3928–38. doi: 10.1002/jcp.29288
29. Xia P, Liu P, Fu Q, Liu C, Luo Q, Zhang X, et al. Long Noncoding RNA EPIC1 Interacts with YAP1 to Regulate The Cell Cycle and Promote The Growth of Pancreatic Cancer Cells. Biochem Biophys Res Commun (2019). doi: 10.1016/j.bbrc.2019.11.167
30. Zhang M, Zhao Y, Zhang Y, Wang D, Gu S, Feng W, et al. LncRNA UCA1 Promotes Migration and Invasion in Pancreatic Cancer Cells via the Hippo Pathway. Biochim Biophys Acta Mol Basis Dis (2018) 1864(5 Pt A):1770–82. doi: 10.1016/j.bbadis.2018.03.005
31. Gao ZQ, Wang JF, Chen DH, Ma XS, Yang W, Zhe T, et al. Long Non-Coding RNA GAS5 Antagonizes the Chemoresistance of Pancreatic Cancer Cells Through Down-Regulation of miR-181c-5p. BioMed Pharmacother (2018) 97:809–17. doi: 10.1016/j.biopha.2017.10.157
32. Zhou B, Guo W, Sun C, Zhang B, Zheng F. Linc00462 Promotes Pancreatic Cancer Invasiveness Through the miR-665/TGFBR1-TGFBR2/SMAD2/3 Pathway. Cell Death Dis (2018) 9(6):706. doi: 10.1038/s41419-018-0724-5
33. Xu B, Gong X, Zi L, Li G, Dong S, Chen X, et al. Silencing of DLEU2 Suppresses Pancreatic Cancer Cell Proliferation and Invasion by Upregulating microRNA-455. Cancer Sci (2019) 110(5):1676–85. doi: 10.1111/cas.13987
34. Yang J, Ye Z, Mei D, Gu H, Zhang J. Long Noncoding RNA DLX6-AS1 Promotes Tumorigenesis by Modulating miR-497-5p/FZD4/FZD6/Wnt/β-catenin pathway in pancreatic cancer. Cancer Manag Res (2019) 11:4209–21. doi: 10.2147/CMAR.S194453
35. Chen T, Lei S, Zeng Z, Zhang J, Xue Y, Sun Y, et al. Linc00261 Inhibits Metastasis and the WNT Signaling Pathway of Pancreatic Cancer by Regulating a miR−552−5p/FOXO3 Axis. Oncol Rep (2020) 43(3):930–42. doi: 10.3892/or.2020.7480
36. Chai W, Liu R, Li F, Zhang Z, Lei B. Long Noncoding RNA TSLNC8 Enhances Pancreatic Cancer Aggressiveness By Regulating CTNNB1 Expression via Association With HuR. Hum Cell (2020). doi: 10.1007/s13577-020-00429-4
37. Jiang Y, Li Z, Zheng S, Chen H, Zhao X, Gao W, et al. The Long Non-Coding RNA HOTAIR Affects the Radiosensitivity of Pancreatic Ductal Adenocarcinoma by Regulating the Expression of Wnt Inhibitory Factor 1. Tumour Biol (2016) 37(3):3957–67. doi: 10.1007/s13277-015-4234-0
38. Ling J, Wang F, Liu C, Dong X, Xue Y, Jia X, et al. FOXO1-Regulated lncRNA LINC01197 Inhibits Pancreatic Adenocarcinoma Cell Proliferation by Restraining Wnt/β-Catenin Signaling. J Exp Clin Cancer Res (2019) 38(1):179. doi: 10.1186/s13046-019-1174-3
39. Weng YC, Ma J, Zhang J, Wang JC. Long Non-Coding RNA LINC01133 Silencing Exerts Antioncogenic Effect in Pancreatic Cancer Through The Methylation of DKK1 Promoter and the Activation of Wnt Signaling Pathway. Cancer Biol Ther (2019) 20(3):368–80. doi: 10.1080/15384047.2018.1529110
40. Cui L, Dong Y, Wang X, Zhao X, Kong C, Liu Y, et al. Downregulation of Long Noncoding RNA SNHG1 Inhibits Cell Proliferation, Metastasis, And Invasion BY Suppressing The Notch-1 Signaling Pathway in Pancreatic Cancer. J Cell Biochem (2019) 120(4):6106–12. doi: 10.1002/jcb.27897
41. Huang R, Nie W, Yao K, Chou J. Depletion of the lncRNA RP11-567G11.1 Inhibits Pancreatic Cancer Progression. BioMed Pharmacother (2019) 112:108685. doi: 10.1016/j.biopha.2019.108685
42. Cai H, Yao J, An Y, Chen X, Chen W, Wu D, et al. LncRNA HOTAIR Acts a Competing Endogenous RNA to Control the Expression of Notch3 via Sponging miR-613 in Pancreatic Cancer. Oncotarget (2017) 8(20):32905–17. doi: 10.18632/oncotarget.16462
43. Zhang H, Zhu C, He Z, Chen S, Li L, Sun C. LncRNA PSMB8-AS1 Contributes to Pancreatic Cancer Progression via Modulating miR-382-3p/STAT1/PD-L1 axis. J Exp Clin Cancer Res (2020) 39(1):179. doi: 10.1186/s13046-020-01687-8
44. Wang F, Rong L, Zhang Z, Li M, Ma L, Ma Y, et al. LncRNA H19-Derived miR-675-3p Promotes Epithelial-Mesenchymal Transition and Stemness in Human Pancreatic Cancer Cells by Targeting the STAT3 Pathway. J Cancer (2020) 11(16):4771–82. doi: 10.7150/jca.44833
45. Yang SZ, Xu F, Zhou T, Zhao X, McDonald JM, Chen Y. The Long Non-Coding RNA HOTAIR Enhances Pancreatic Cancer Resistance to TNF-Related Apoptosis-Inducing Ligand. J Biol Chem (2017) 292(25):10390–7. doi: 10.1074/jbc.M117.786830
46. Ren X, Chen C, Luo Y, Liu M, Li Y, Zheng S, et al. lncRNA-PLACT1 Sustains Activation of NF-κB Pathway Through a Positive Feedback Loop With IκBα/E2F1 Axis in Pancreatic Cancer. Mol Cancer (2020) 19(1):35. doi: 10.1186/s12943-020-01153-1
47. Wong CH, Li CH, He Q, Chan SL, Hung-Man Tong J, To KF, et al. Ectopic HOTTIP Expression Induces Noncanonical Transactivation Pathways to Promote Growth and Invasiveness in Pancreatic Ductal Adenocarcinoma. Cancer Lett (2020). doi: 10.1016/j.canlet.2020.02.038
48. Yang Q, Li K, Huang X, Zhao C, Mei Y, Li X, et al. lncRNA SLC7A11-AS1 Promotes Chemoresistance by Blocking SCFβ-TRCP-Mediated Degradation of NRF2 in Pancreatic Cancer. Mol Ther Nucleic Acids (2020) 19:974–85. doi: 10.1016/j.omtn.2019.11.035
49. Chen L, Zhang J, Chen Q, Ge W, Meng L, Huang X, et al. Long Noncoding RNA SOX2OT Promotes the Proliferation of Pancreatic Cancer by Binding to FUS. Int J Cancer (2019). doi: 10.1002/ijc.32827
50. Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, et al. IGF2BP2 regulates DANCR by Serving as an N6-Methyladenosine Reader. Cell Death Differ (2019). doi: 10.1038/s41418-019-0461-z
51. Pan S, Shen M, Zhou M, Shi X, He R, Yin T, et al. Long Noncoding RNA LINC01111 Suppresses Pancreatic Cancer Aggressiveness by Regulating DUSP1 Expression via microRNA-3924. Cell Death Dis (2019) 10(12):883. doi: 10.1038/s41419-019-2123-y
52. Hui B, Ji H, Xu Y, Wang J, Ma Z, Zhang C, et al. RREB1-Induced Upregulation of the lncRNA AGAP2-AS1 Regulates the Proliferation and Migration of Pancreatic Cancer Partly Through Suppressing ANKRD1 and ANGPTL4. Cell Death Dis (2019) 10(3):207. doi: 10.1038/s41419-019-1384-9
53. He J, Li F, Zhou Y, Hou X, Liu S, Li X, et al. LncRNA XLOC_006390 Promotes Pancreatic Carcinogenesis and Glutamate Metabolism by Stabilizing c-Myc. Cancer Lett (2020) 469:419–28. doi: 10.1016/j.canlet.2019.11.021
54. Qi C, Xiaofeng C, Dongen L, Liang Y, Liping X, Yue H, et al. Long Non-Coding RNA MACC1-AS1 Promoted Pancreatic Carcinoma Progression Through Activation of PAX8/NOTCH1 Signaling Pathway. J Exp Clin Cancer Res (2019) 38(1):344. doi: 10.1186/s13046-019-1332-7
55. Lu H, Ye J, Zhang L, Li M, Lu S, Yang D, et al. Downregulation of LINC01638 lncRNA Inhibits Migration and Invasion of Pancreatic Ductal Adenocarcinoma Cells by Reducing TGF-β Signaling. Mol Med Rep (2019) 20(5):4533–9. doi: 10.3892/mmr.2019.10699
56. Zeng Z, Xu FY, Zheng H, Cheng P, Chen QY, Ye Z, et al. LncRNA-MTA2TR Functions as a Promoter in Pancreatic Cancer via Driving Deacetylation-Dependent Accumulation of HIF-1α. Theranostics (2019) 9(18):5298–314. doi: 10.7150/thno.34559
57. Chen S, Luo X, Wu W, Li Y, Yu H, Wang Y, et al. The Long Non-Coding RNA MACC1-AS1 Promotes Nasopharyngeal Carcinoma Cell Stemness via Suppressing miR-145-Mediated Inhibition on SMAD2/MACC1-AS1 Axis. BioMed Pharmacother (2020) 125:109986. doi: 10.1016/j.biopha.2020.109986
58. Wang Y, Ding X, Hu H, He Y, Lu Z, Wu P, et al. Long Non-Coding RNA lnc-PCTST Predicts Prognosis Through Inhibiting Progression of Pancreatic Cancer by Downregulation of TACC-3. Int J Cancer (2018) 143(12):3143–54. doi: 10.1002/ijc.31657
59. Lian Y, Li Z, Fan Y, Huang Q, Chen J, Liu W, et al. The lncRNA-HOXA-AS2/EZH2/LSD1 Oncogene Complex Promotes Cell Proliferation in Pancreatic Cancer. Am J Transl Res (2017) 9(12):5496–506.
60. He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, et al. ALKBH5 Inhibits Pancreatic Cancer Motility by Decreasing Long Non-Coding RNA KCNK15-AS1 Methylation. Cell Physiol Biochem (2018) 48(2):838–46. doi: 10.1159/000491915
61. Deng SJ, Chen HY, Ye Z, Deng SC, Zhu S, Zeng Z, et al. Hypoxia-Induced LncRNA-BX111 Promotes Metastasis and Progression of Pancreatic Cancer Through Regulating ZEB1 Transcription. Oncogene (2018) 37(44):5811–28. doi: 10.1038/s41388-018-0382-1
62. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long Non-Coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature (2010) 464(7291):1071–6. doi: 10.1038/nature08975
63. Ma Y, Hu M, Zhou L, Ling S, Li Y, Kong B, et al. Long Non−Coding RNA HOTAIR Promotes Cancer Cell Energy Metabolism in Pancreatic Adenocarcinoma by Upregulating Hexokinase−2. Oncol Lett (2019) 18(3):2212–9. doi: 10.3892/ol.2019.10551
64. Roberts DJ, Miyamoto S. Hexokinase II Integrates Energy Metabolism and Cellular Protection: Akting on Mitochondria and TORCing to Autophagy. Cell Death Differ (2015) 22(2):248–57. doi: 10.1038/cdd.2014.173
65. Jiang D, Xu L, Ni J, Zhang J, Cai M, Shen L. Functional Polymorphisms in LncRNA HOTAIR Contribute to Susceptibility of Pancreatic Cancer. Cancer Cell Int (2019) 47. doi: 10.1186/s12935-019-0761-x
66. Moschovis D, Vasilaki E, Tzouvala M, Karamanolis G, Katifelis H, Legaki E, et al. Association Between Genetic Polymorphisms in Long Non-Coding RNAs and Pancreatic Cancer Risk. Cancer Biomark (2019) 24(1):117–23. doi: 10.3233/CBM-181959
67. Han T, Jiao F, Hu H, Yuan C, Wang L, Jin ZL, et al. EZH2 Promotes Cell Migration and Invasion But Not Alters Cell Proliferation by Suppressing E-cadherin, Partly Through Association with MALAT-1 in Pancreatic Cancer. Oncotarget (2016) 7(10):11194–207. doi: 10.18632/oncotarget.7156
68. Passaro F, De Martino I, Zambelli F, Di Benedetto G, Barbato M, D’Erchia AM, et al. YAP Contributes to DNA Methylation Remodeling Upon Mouse Embryonic Stem Cell Differentiation. J Biol Chem (2020) 2. doi: 10.1074/jbc.RA120.015896
69. Testa G, Russo M, Di Benedetto G, Barbato M, Parisi S, Pirozzi F, et al. Bmi1 Inhibitor PTC-209 Promotes Chemically-Induced Direct Cardiac Reprogramming of Cardiac Fibroblasts Into Cardiomyocytes. Sci Rep (2020) 10(1):7129. doi: 10.1038/s41598-020-63992-8
70. Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, et al. Elevated Expression Level of Long Noncoding RNA MALAT-1 Facilitates Cell Growth, Migration and Invasion in Pancreatic Cancer. Oncol Rep (2014) 32:2485–92. doi: 10.3892/or.2014.3518
71. Lei S, He Z, Chen T, Guo X, Zeng Z, Shen Y, et al. Long Noncoding RNA 00976 Promotes Pancreatic Cancer Progression Through OTUD7B by Sponging miR-137 Involving EGFR/MAPK Pathway. J Exp Clin Cancer Res (2019) 38(1):470. doi: 10.1186/s13046-019-1388-1394
72. Liu Y, Guo C, Li F, Wu L. LncRNA LOXL1-AS1/miR-28-5p/SEMA7A Axis Facilitates Pancreatic Cancer Progression. Cell Biochem Funct (2019). doi: 10.1002/cbf.3449
73. Xu DF, Wang LS, Zhou JH. Long Non−Coding RNA CASC2 Suppresses Pancreatic Cancer Cell Growth and Progression by Regulating the miR−24/MUC6 Axis. Int J Oncol (2019). doi: 10.3892/ijo.2019.4937
74. Bi S, Wang Y, Feng H, Li Q. Long Noncoding RNA LINC00657 Enhances the Malignancy of Pancreatic Ductal Adenocarcinoma by Acting as a Competing Endogenous RNA on microRNA-433 to Increase PAK4 Expression. Cell Cycle (2020) 1–16. doi: 10.1080/15384101.2020.1731645
75. Zhang M, Wang R, Zhao X, Lu L, Wang T. LncRNA LINK-A Regulates ROCK1 Expression in Early-Stage Pancreatic Adenocarcinoma. Exp Ther Med (2020) 19(3):1933–9. doi: 10.3892/etm.2019.8400
76. Liu Y, Wang J, Dong L, Xia L, Zhu H, Li Z, et al. Long Noncoding RNA HCP5 Regulates Pancreatic Cancer Gemcitabine (GEM) Resistance By Sponging Hsa-miR-214-3p To Target HDGF. Onco Targets Ther (2019) 12:8207–16. doi: 10.2147/OTT.S222703
77. Hua YQ, Zhu YD, Xie GQ, Zhang K, Sheng J, Zhu ZF, et al. Long Non-Coding SBF2-AS1 Acting as a Competing Endogenous RNA to Sponge microRNA-142-3p to Participate in Gemcitabine Resistance in Pancreatic Cancer via Upregulating TWF1. Aging (Albany NY) (2019) 11(20):8860–78. doi: 10.18632/aging.102307
78. Zou J, Su H, Zou C, Liang X, Fei Z. Ginsenoside Rg3 Suppresses the Growth of Gemcitabine-Resistant Pancreatic Cancer Cells by Upregulating lncRNA-CASC2 and Activating PTEN Signaling. J Biochem Mol Toxicol (2020), e22480. doi: 10.1002/jbt.22480
79. Liu Y, Feng W, Liu W, Kong X, Li L, He J, et al. Circulating lncRNA ABHD11-AS1 Serves as a Biomarker for Early Pancreatic Cancer Diagnosis. J Cancer (2019) 10(16):3746–56. doi: 10.7150/jca.32052
80. Wang Y, Zhou L, Lu J, Jiang B, Liu C, Guo J, et al. Research Progress on Long Non-Coding RNAs and Their Roles as Potential Biomarkers for Diagnosis and Prognosis in Pancreatic Cancer. Cancer Cell Int (2020) 20:457. doi: 10.1186/s12935-020-01550-y
Keywords: lncRNA, apoptosis, signaling pathways, microRNA, pancreatic cancer
Citation: Farooqi AA, Nayyab S, Martinelli C, Berardi R, Katifelis H, Gazouli M and Cho WC (2021) Regulation of Hippo, TGFβ/SMAD, Wnt/β-Catenin, JAK/STAT, and NOTCH by Long Non-Coding RNAs in Pancreatic Cancer. Front. Oncol. 11:657965. doi: 10.3389/fonc.2021.657965
Received: 24 January 2021; Accepted: 23 March 2021;
Published: 09 June 2021.
Edited by:
Palmiro Poltronieri, Italian National Research Council, ItalyReviewed by:
Massimo Mallardo, University of Naples Federico II, ItalyCopyright © 2021 Farooqi, Nayyab, Martinelli, Berardi, Katifelis, Gazouli and Cho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ammad Ahmad Farooqi, ZmFyb29xaWFtbWFkYWhtYWRAZ21haWwuY29t; William C. Cho, Y2hvY3NAaGEub3JnLmhr; d2lsbGlhbWNzY2hvQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.